SUMMARY
We provide the first conclusive evidence that Xanthomonas axonopodis pv. citri Asiatic strain (Xac‐A) and, in particular, Xac‐Aw, a unique citrus canker A strain isolated from Key lime in Wellington, Florida, induces a hypersensitive reaction (HR) in grapefruit leaves. Using the heterologous tomato pathogen X. perforans, as a recipient of the Xac‐Aw genomic library, we identified a 1599‐bp open reading frame responsible for HR in grapefruit, but not Key lime, and designated it avrGf1. Xac‐AwΔavrGf1 produced typical, although visibly reduced, citrus canker symptoms (i.e. raised pustules) in grapefruit and typical canker symptoms in Key lime. We also determined that the X. perforans transconjugant carrying an Xac‐Aw hrpG elicited HR in grapefruit and Key lime leaves, and that xopA in X. perforans was partly responsible for HR. Xac‐A transconjugants carrying the X. perforans xopA were reduced in ability to grow in grapefruit leaves relative to wild‐type Xac‐A. The X. perforans xopA appears to be a host‐limiting factor. An avrBs3 homologue, which contained 18.5 repeats and induced HR in tomato, was designated avrTaw. This gene, when expressed in a pustule‐minus Xac‐Aw, did not complement pustule formation; however, pthAw, a functional pthA homologue, complemented the mutant strain to produce typical pustules in Key lime, but markedly reduced pustules in grapefruit. Both avrBs3 homologues, when expressed in a typical Xac‐A strain, resulted in typical citrus canker pustules in grapefruit, indicating that neither homologue suppressed pustule size in grapefruit. Xac‐Aw contains other unidentified factors that suppress development in grapefruit.
INTRODUCTION
Until recently, citrus canker was known to be initiated by two pathovars of Xanthomonas axonopodis. These pathovars, i.e. X. axonopodis pv. citri (Xac) and X. axonopodis pv. aurantifolii (Xaa), have been distinguished by genetic differences and phenotypic traits. Recently, a change in nomenclature of these two pathovars has been proposed to X. citri ssp. citri and X. fuscans ssp. aurantifolii, respectively (Schaad et al., 2005). Groups of strains exist within each pathovar that have different pathogenicities. The most destructive strains causing Asiatic citrus canker, which are members of the pathovar citri, are termed the A form and are designated Xac‐A. Xac‐A strains have a broad host range within Rutaceae, including grapefruit and Key lime (Table 1). Other strains exist that are genetically very similar to Xac‐A, but have a limited host range (Sun et al., 2004; Verniere et al., 1998). Verniere et al. (1998) designated strains restricted to Key/Mexican lime as Xac‐A*, based on their physiological and genetic similarities and serological differences from Xac‐A. Sun et al. (2004) designated strains that were pathogenic on Key lime, but that did not cause canker symptoms on grapefruit, as Xac‐Aw (Table 1). More distantly related strains associated with citrus canker include Cancrosis B, or false canker (formerly known as B‐strain canker), discovered on lemon (Citrus limon) in Argentina in 1923 (Civerolo, 1984), and found primarily on C. limon and C. aurantiifolia, but also affecting C. aurantium. Another strain, found to be associated with Key/Mexican lime cancrosis (formerly known as C‐strain canker), was observed in Brazil on Key/Mexican lime (C. aurantiifolia) in 1963 and only infects this citrus host (Stall and Civerolo, 1991). The causal bacteria of the latter diseases that produce a very similar canker syndrome on their limited citrus hosts are genetically related to each other, but different from Xac‐A, and therefore have been referred to as strains of X. axonopodis pv. aurantifolii (Xaa) (Gabriel et al., 1989).
Table 1.
Reaction of citrus xanthomonads and Xanthomonas perforans on citrus.
| Xanthomonad groups used in this study | Disease reaction in: | ||
|---|---|---|---|
| Grapefruit | Key lime | Tomato | |
| X. axonopodis pv. citri | |||
| Xac‐A | + | + | (HR) |
| Xac‐Aw | − | + | (HR) |
| Xac‐A* | − | + | |
| Xaa‐B | ± | + | |
| Xaa‐C | − | + | (HR) |
| X. perforans | − (NR) | − (NR) | + |
+, typical disease reaction; ±, weak citrus bacterial canker disease; −, inability to cause typical disease reaction on host; HR, hypersensitive reaction; NR, null reaction.
The symptoms of citrus canker include erumpent lesions on fruit, foliage and young stems of susceptible citrus cultivars (Gottwald et al., 1997, 2002a, 2002b). The occurrence of citrus canker lesions on fruit rind decreases the commercial quality, and infected fruit is not accepted by most important markets (Canteros, 2004). Warm weather, together with rains accompanied by the strong winds that occasionally occur, creates ideal conditions for the spread of Xac‐A (Gottwald et al., 1997, 2002a, 2002b). Optimum temperatures for infection range between 20 and 30 °C (Koizumi, 1985).
The pathogen of Asiatic citrus canker was brought to North America in 1910 and became distributed throughout the Gulf States, according to the review by Loucks (1934). The eradication of the pathogen began in Florida in 1914 and was declared to be complete in North America in 1947 (Dopson, 1964). The Xac‐A pathogen was rediscovered in 1986 in Manatee County, Florida and declared to be eradicated in 1994, but was found again in the area later (Cubero and Graham, 2002; Schubert et al., 1996). A new focus of a different strain of Xac‐A appeared in Miami, Florida, in 1995 (Gottwald et al., 2002a; Schubert et al., 2001), and an eradication programme was begun again. Successful eradication of Xac‐A has occurred in the USA, South Africa, Australia and New Zealand. Xac‐A is endemic to all countries in eastern and southern Asia, Argentina, Paraguay and Uruguay. An eradication programme currently exists in Brazil (Schubert et al., 2001). As a result of the catastrophic weather conditions in 2004 and 2005, Xac‐A spread extensively in Florida and eradication efforts were suspended in 2006.
Schubert et al. (1996) described at least two separate introductions of Xac in Florida after 1986. A group of strains that is pathogenic to Key lime and alemow plants, but not to grapefruit and orange, occurred in one of the introductions. The group was characterized (Cubero and Graham, 2002; Sun et al., 2004) and found to be related genetically to Xac, rather than Xaa, and designated as Xac‐Aw to represent the unique group of strains associated with Key lime but not grapefruit trees in Wellington, Florida. Sun et al. (2004) distinguished the Xac‐Aw strains from the Xac‐A* strains by physiological differences.
The aim of this work was to investigate the presence of host‐limiting factors in the Xac‐Aw strain. The Xac‐Aw strain was thought to cause a hypersensitive reaction (HR) in grapefruit based on the observations of Sun et al. (2004), and was investigated. An HR caused by bacteria is usually the result of an avirulence gene in the bacterial genome interacting with a resistance gene (R) in the host (Bonas et al., 1993; Crute, 1985; Minsavage et al., 1990; Staskawicz et al., 1984, 1987). If avirulence of the Xac‐Aw strain in grapefruit is the result of the presence of a single gene in the bacterium, a change of virulence might occur by mutation (Dahlbeck and Stall, 1979; Gassmann et al., 2000; Kearney and Staskawicz, 1990; Swords et al., 1996). Analysis of the Xac‐Aw strain was undertaken to determine whether a change in virulence to grapefruit of the Xac‐Aw strain would occur. In addition, isolation of a postulated avirulence gene in the genome of Xac‐Aw, which interacted with grapefruit leaves in a hypersensitive manner, was undertaken. Using a heterologous bacterial host (i.e. X. perforans, a tomato pathogen, which causes a null reaction when infiltrated into citrus leaves) as a recipient of the Xac‐Aw library, factors associated with host specificity were identified and are presented.
RESULTS
HR in grapefruit leaves inoculated with Xac‐Aw
Fleck lesions occurred in grapefruit leaves and typical canker lesions occurred in Key lime leaves when they were infiltrated with bacterial suspensions of Xac‐Aw strain 12879 of approximately 103 colony‐forming units (cfu)/mL (Fig. 1). Following isolation from approximately 30 representative fleck lesions, the bacterium was recovered from less than 25% of the lesions. Representative colonies from the remaining isolations, when inoculated into grapefruit and Key lime, produced typical canker symptoms in Key lime, but a necrotic reaction in grapefruit, indicative of an incompatible interaction. After infiltration of grapefruit leaves with inoculum adjusted to 5 × 108 cfu/mL, internal bacterial populations of Xac‐A (strain A 40) and Xac‐Aw (strain 12879) were similar through the second day, but populations of Xac‐A were significantly greater than those of Xac‐Aw after 6 days (similar to the data shown in Fig. 4, bottom). Electrolyte leakage from leaf tissue samples inoculated with Xac‐Aw was similar to that of leaf tissue samples inoculated with Xac‐A after 2 days. However, electrolyte leakage values for leaves infiltrated with the Xac‐Aw bacterium were significantly greater than those of leaves infiltrated with the Xac‐A bacterium at 4 and 6 days (similar to the data presented in Fig. 4, bottom). The data are typical for HR caused by the Xac‐Aw strain, and similar to HR occurring in other plants and caused by other xanthomonads. However, HR in grapefruit leaves differs from that in other systems by being slow to occur (Astua‐Monge et al., 2000; Minsavage et al., 1990, 2004).
Figure 1.

Fleck lesions (above) and typical citrus canker lesions (below) caused by Xanthomonas citri ssp. citri strain Xac‐Aw in grapefruit and Key lime leaves, respectively, 30 days after infiltration of the top half of each leaf with inoculum (103 cfu/mL).
Figure 4.
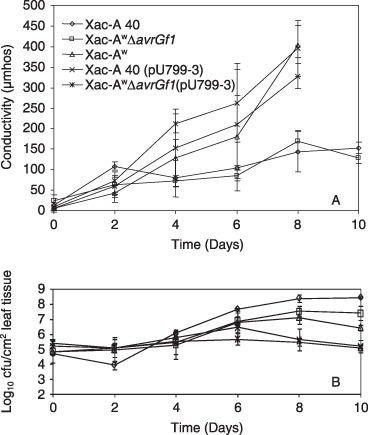
Conductivity readings (top) and bacterial populations (bottom) in grapefruit leaves at different times after inoculation with 5 × 108 cfu/mL of Xac‐A 40, Xac‐AwΔavrGf1, Xac‐Aw 12879, Xac‐A 40 (pU799‐3) or Xac‐AwΔavrGf1 (pU799‐3).
Selection of host‐limiting genes from the strain Xac‐Aw library
Three clones were selected from a genomic library of the 12879 strain of Xac‐Aw that caused rapid necrosis in grapefruit leaves, but not in tomato leaves when they were expressed in X. perforans. These three clones were designated pL22, pL689 and pL799. All three clones were successfully transferred from Escherichia coli by triparental matings into a strain (Xac‐A 40) that causes Asiatic citrus canker in grapefruit and Key lime leaves. Only one of the three clones (pL799) caused HR in grapefruit leaves and a susceptible reaction in Key lime leaves when expressed in the Xac‐A strain. The disease reaction of Xac‐A containing the pL799 clone, when infiltrated into grapefruit leaves, was typical of the reaction of the Xac‐Aw strain on these hosts. The presence of pL22 and pL689 clones in Xac‐A 40 had no effect on disease reaction. Four clones were selected that caused HR in tomato, but not in grapefruit. All of these clones were investigated further.
DNA analysis of pL799
A 1599‐bp open reading frame (ORF) was found within the nucleotide sequence of DNA from a 2.3‐kb subclone from pL799 that caused HR in grapefruit leaves. The complete sequence of the ORF, designated as avrGf1, was submitted to GenBank and was assigned accession number DQ275469. The upstream region of avrGf1 does not contain an hrp box, but does contain an imperfect PIP box, TTCGT‐N10‐TTCGC (Huguet and Bonas, 1997), 80 bp upstream of the start codon. A GenBank search identified significant homology of avrGf1 with gene XCC3600, which is a hypothetical protein (AM428701) in the genome of X. campestris pv. campestris (Xcc33913), a hypothetical protein (ABM30884.1) in Acidovorax avenae ssp. citrulli, HopG1 (AAZ34904.1) in Pseudomonas syringae pv. phaseolicola, HopG1 (NP_794468.1) in P. syringae pv. tomato, a putative type III effector protein (CAD17474.2) in Ralstonia solanacearum, and a hypothetical conserved protein (ABC92870.1) in Rhizobium etli. Alignment analysis of avrGf1 with AM428701 from X. campestris pv. campestris using Clustal (1.82) multiple sequence alignment indicated 84.99% identity at the amino acid level.
Presence of avrGf1 in xanthomonads pathogenic to citrus
Primers were selected for the amplification of a fragment containing avrGf1 by polymerase chain reaction (PCR). A fragment of the expected size was amplified from the genomic DNA of four Xac‐Aw strains, but not from the genomic DNA of three Xac‐A strains. In Southern hybridizations with the labelled DNA of clone pU799‐3 as a probe, the probe hybridized strongly with the DNA of the Xac‐Aw strain, but not with the DNA of Xac‐A strains. Reactions did not occur with the DNA of a strain of each of the B and C groups of Xaa, with the DNA of four strains of the Xac‐A* group or with the DNA of a strain of X. axonopodis pv. citrumelo, the pathogen causing bacterial spot of citrus (Fig. 2). The Xaa‐B and X. axonopodis pv. citrumelo strains did not cause rapid necrosis in grapefruit leaves. The Xaa‐C strain caused rapid necrosis in grapefruit leaves phenotypically different from that caused by Xac‐Aw. In addition, some Xac‐A* strains caused rapid necrosis in grapefruit leaves, and one of the strains, Xac‐A* 1974, was included in probing. The probe hybridized to a band in the DNA of X. campestris pv. campestris (Xcc) strain 8004, but the hybridization pattern was different from that of the DNA of Xac‐Aw (Fig. 2). Strain Xcc 8004 contained gene XCC3600 described above and caused rapid necrosis in grapefruit leaves. The avrGf1 probe did not hybridize to the plasmid DNA of Xac‐Aw strain 12879 (Fig. 2).
Figure 2.

Hybridization (right) of the subclone pU799‐3 containing avrGf1 with total genomic DNA of Xanthomonas strains digested with HindIII and plasmid DNA (left). Lane 1, Xac‐Aw 12879; lane 2, Xac‐AwΩ; lane 3, Xac‐A 40; lane 4, Xac‐A 306; lane 5, Xac‐A* 1974; lane 6, Xac‐A* 1975; lane 7, Xaa‐B; lane 8, Xaa‐C; lane 9, Xac‐E 1887; lane 10, Xcc 8004; lane 11, Xac‐Aw 12879; lane 12, plasmid Xac‐Aw; lane 13, plasmid Xac‐A 40; lane 14, λ marker digested with EcoRI and HindIII.
Effect of knockout of avrGf1 in Xac‐Aw on disease reaction in grapefruit
A strain with a mutated avrGf1 gene (Xac‐AwΔavrGf1) was compared with the wild‐type Xac‐A and Xac‐Aw strains by inoculation into grapefruit leaves by a pin‐prick method with inocula adjusted to 5 × 108 cfu/mL. Visually, the symptoms caused by the Xac‐AwΔavrGf1 strain were more similar to those produced by the wild‐type Xac‐A strain than to those produced by the wild‐type Xac‐Aw strain, in that raised pustules were observed and an expanding lesion developed (Fig. 3a). The symptoms of Xac‐AwΔavrGf1 were less severe than those produced by the Xac‐A strain. Nevertheless, mutation of the avrGf1 gene in Xac‐Aw resulted in a disease reaction in grapefruit leaves.
Figure 3.

Disease symptoms. (A) Grapefruit leaf after inoculation by needle punctures with Xac‐Aw (a), Xac‐AwΔavrGf1 (b) and Xac‐A 40 (c). (B) Key lime leaf following inoculation with Xac‐AwΔavrGf1ΔavrTawDpthA w::pUFR034 (a), ΔavrGf1ΔavrTawDpthA w::pAW5.2 (b) and ΔavrGf1ΔavrTawDpthA w::pUFR80.1 (c). (C) Grapefruit leaf following inoculation with Xac‐AwΔavrGf1ΔavrTawDpthA w::pAW5.2 (a), Xac‐AwΔavrGf1ΔavrTawDpthA w::pUFR80.10 (b) and Xac‐AwΔavrGf1ΔavrTawDpthA w:: pUFR034 (c). (D) Grapefruit leaf following inoculation with Xac‐AwΔavrGf1 (a) and Xac‐AwΔavrGf1ΔavrTawDpthA w::pAW5.2 (b). (E) Grapefruit leaf following inoculation with Xac‐A40::pAW5.2 (a), Xac‐A40::pUFR80.1 (b) and Xac‐A40::pUFR034 (c). (F) Grapefruit leaf following inoculation with Xanthomonas perforans strains 91‐118DxopA (a), 91‐118 (b), 91‐118+hrpG (c) and 91‐118 DxopA+hrpG (d).
Internal bacterial populations and electrolyte leakage from grapefruit leaves infiltrated with strains Xac‐A 40, Xac‐Aw 12879, Xac‐A 40 (pU799‐3), Xac‐AwΔavrGf1 and Xac‐AwΔavrGf1 (pU799‐3), at a concentration of 5 × 108 cfu/mL, were compared (Fig. 4). Populations of all strains were about equal for the first 4 days after inoculation. However, at day six, the population of the Xac‐Aw strain was significantly smaller than that of the Xac‐A strain. At day ten, the population of the Xac‐A strain was largest, and the population of the Xac‐Aw strain was about 1.5 log units lower. The population of the Xac‐AwΔavrGf1 strain was intermediate between those of the Xac‐A and Xac‐Aw strains. The strains containing the pU799‐3 clone (avrGf1) showed the smallest populations. Complementation of the mutated Xac‐Aw strain, Xac‐AwΔavrGf1, with pU799‐3 was successful. The strains Xac‐A 40, Xac‐Aw 12879, Xac‐AwΔavrGf1 and Xac‐AwΔavrGf1 (pU799‐3) all multiplied equally for 12 days in Key lime leaves after infiltration of a low concentration (5 × 105 cfu/mL) of inoculum into the leaves (data not included).
The electrolyte leakage from leaf tissue inoculated with the Xac‐Aw strain and strains containing avrGf1 (pU799‐3) started to increase at day four relative to the leakage from leaf tissue infiltrated with the Xac‐A and Xac‐AwΔavrGf1 strains (Fig. 4). At day eight, the strains with avrGf1 caused significantly greater electrolyte leakage than the strains that did not contain the gene. Electrolyte leakage caused by the Xac‐A and Xac‐AwΔavrGf1 strains was similar.
avrGf1 requires a functional type III secretion system (TTSS) for HR
Transconjugants of X. perforans strains 91‐118hrp − and 91‐118hrp + containing the clone pU799‐3 (avrGf1) were grown overnight, and inocula were prepared and adjusted to 5 × 108 cfu/mL and were infiltrated into grapefruit leaves. The hrp − strain did not cause HR, but the hrp + strain did. Thus, an active TTSS appears to be necessary for HR produced by avrGf1.
Identification of non‐host effector by overexpression of X. citri ssp. citri HrpG in X. perforans
Clones pL22 and pL689 were determined to be overlapping on the basis of their similar restriction enzyme digestion fragment profiles using electrophoresis. Partial digestion of pL22 with Sau3A resulted in a subclone containing 5041 bp, which produced an HR in grapefruit leaves when expressed in X. perforans. The nucleotide sequence of the subclone contained ORFs that were homologous to hrpX, hrpG and Hsp90Xo (heat shock protein molecular chaperone) genes. Subsequently, a subclone containing only hrpG (p0346) was obtained, which produced an HR in grapefruit leaves when expressed in X. perforans, but not in cells of Xac‐A. As hrpG is a key regulatory gene for the transcriptional activation of pathogenicity genes (Wengelnik et al., 1996, 1999), a factor in addition to hrpG in X. perforans was speculated to be necessary to elicit HR in grapefruit leaves. Therefore, we attempted to determine the factor(s) involved in the elicitation of HR by X. perforans in grapefruit leaves when hrpG was present in trans. The possibility that xopA might be involved was suggested from a similar study with X. axonopodis pv. glycines (Kim et al., 2003). The gene hpaG, together with an additional copy of hrpG, was necessary for HR in tobacco caused by that bacterium. Therefore, a clone (pLXOPA) containing xopA, a homologue of hpaG, was identified in a pLAFR3 cosmid library of X. perforans 91‐118 by PCR. The clones p0346 (hrpG) and pLXOPA (xopA) were conjugated into Xac‐A 306 individually or in combination. A strong HR occurred in grapefruit leaves inoculated with Xac‐A 306 containing both p3046 and pLXOPA, but not with Xac‐A 306 cells alone, or containing p0346 or pLXOPA alone. The populations of the four strains in grapefruit leaves were also determined at different times after the infiltration of leaves with inocula adjusted to 5 × 105 cfu/mL. The populations of Xac‐A 306 were 100‐fold higher than those of Xac‐A 306 containing both p0346 and pLXOPA, 15 days after inoculation of the leaves (Fig. 5). The populations of Xac‐A 306 were 10‐fold higher than those of Xac‐A 306 containing pLXOPA alone. The populations of Xac‐A 306 containing p0346 were significantly larger than those of Xac‐A 306.
Figure 5.

Populations of Xac‐A 306, Xac‐A 306 containing p0346 (plasmid containing hrpG from Xac‐Aw), p0346+pLXOPA (pLAFR3 cosmid that contains the xopA gene from Xanthomonas perforans) and pLXOPA alone in grapefruit leaves at different times after infiltration of 5 × 105 cfu/mL of each strain into the mesophyll.
In order to determine the role of the xopA gene in X. perforans, the gene was deleted by suicide‐assisted mutagenesis, and the clone containing hrpG was conjugated into the mutated X. perforans strain 91‐118::ΔxopA. Following the infiltration of grapefruit leaves with a cell suspension of the mutant strain adjusted to 5 × 108 cfu/mL, there was an observable reduction in necrosis compared with wild‐type X. perforans containing an additional hrpG clone (Fig. 3f). The reaction could not be considered as a null reaction, but the deletion of xopA had a pronounced effect on the development of HR. However, there may be another gene in the pLXOPA clone that also contributes to HR in grapefruit leaves caused by wild‐type X. perforans containing an additional hrpG.
Role of avrBs3 homologues from Xac‐Aw in tomato HR and in virulence in grapefruit and Key lime
Strains of both Xac‐A and Xac‐Aw caused HR in tomato. Four clones (pL80, pL104, pL115 and pL622) were selected from the Xac‐Aw library that caused HR in tomato (Fig. 6), but not in grapefruit (data not shown), when expressed in X. perforans. These clones all contained an avrBs3 homologue, based on hybridization to a probe containing avrBs3‐2 from X. euvesicatoria (Jones et al., 2004) in Southern hybridizations. We determined that the avrBs3 homologue, pthAw (pAW5.2), from Xac‐Aw, previously determined to be necessary for typical canker lesions including typical raised pustules (Al‐Saadi et al., 2007; Table 2), when expressed in X. perforans, resulted in HR in tomato (Fig. 6). None of the four clones, when expressed in a strain of X. axonopodis pv. citrumelo following conjugation by triparental matings, caused symptoms similar to Xac‐A when infiltrated into grapefruit leaves (data not shown). The insert from pL80 was subcloned and transferred to pUFRO34 to contain a single ORF which included the avrBs3 homologue, and was designated pUFR80.1. The ORF in pUFR80.10 was sequenced and was determined to contain an avrBs3 homologue with 18.5 tandem repeats consisting of a combination of 33 and 34 amino acids; it was designated avrTaw for HR in tomato by an avrBs3 homologue from the Wellington strain of X. citri ssp. citri group A. The sequence for the tandem repeats was assigned accession number FJ360749. This clone, pUFR80.1, when expressed in Xac‐AwΔavrGf1ΔavrTawΔPthaA w, mutated in avrGf1, and both copies of the avrBs3 homologues (Duan et al., 2004), deficient in pustule formation, did not complement pustule formation in grapefruit or Key lime (Fig. 3b,c), whereas pAW5.2 containing pthA w complemented the triple mutant strain for pustule formation in Key lime, but not in grapefruit (Fig. 3c). Pustules were produced in grapefruit with Xac‐AwΔavrGf1 (Fig. 3a), in which both Xac‐Aw avrBs3 homologues, avrTaw and pthaA w, were present. In a separate experiment in which we inoculated grapefruit leaves by pin‐prick with Xac‐AwΔavrGf1 or Xac‐AwΔavrGf1ΔavrTawΔpthaA w:pAW5.2, typical pustules were only formed when Xac‐AwΔavrGf1 was used as inoculum (Fig. 3d). It is clear that, at least one other host‐limiting factor is present in Xac‐Aw, given the pronounced reduction in pustule formation associated with Xac‐AwΔavrGf1 in grapefruit (Fig. 3a) relative to Key lime (Fig. 3c).
Figure 6.

Reaction of Bonny Best tomato leaflet following infiltration with Xanthomonas perforans transconjugants carrying pAW80.1 (subclone of pL80), pthAw and pLAFR. Note that clones carrying pAW80.1 and pAW5.2 induced a hypersensitive reaction.
Table 2.
List of bacterial strains and plasmids used in this study.
| Bacterium or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Xanthomonas. perforans | ||
| 91‐118 | Wild‐type, pathogenic to tomato, Rifr | Jones et al. (2004) |
| 91‐118hrp− | Same as 91‐118, hrp cluster mutated with NTG, complemented with hrp clone, Rifr | G. V. Minsavage |
| 91‐118ΔxopA X. axonopodis pv. citri | XopA‐RifR | Hert (2007) |
| Xac‐A 40 | Wild‐type, Asiatic strain, isolated in Argentina, Rifr | DPI |
| Xac‐A 306 | Wild‐type, Asiatic strain, isolated in Brazil, Rifr | DPI |
| Xac‐Aw 12879 | Wild‐type, Wellington strain, pathogenic to Key lime | DPI |
| Xac‐AwΔavrGf1 | Wellington strain, Ω cassette inserted into avrGf1 | This work |
| Xac‐AwΩ4 | Wellington strain, Ω cassette inserted into avrGf1, pthAw and avrTaw | This work |
| Xac‐A* 1974 | A* strain (Xc290) | (Sun et al., 2004; Vernier et al., 1998), DPI |
| Xac‐A* 1975 | A* strain (Xc280) | (Sun et al., 2004; Vernier et al., 1998), DPI |
| X. axonopodis pv.aurantifolii | ||
| Xaa‐B 1622 | B‐69, isolated in Argentina | DPI |
| Xaa‐C 5979 | Xc 70, isolated in Brazil | DPI |
| X. axonopodis pv. citrumelo | ||
| Xac‐E 1887 | Wild‐type | DPI |
| X. campestris pv. campestris | ||
| Xcc 8004 | Wild‐type, Rifr | Metz et al. (2005) |
| Escherichia coli | ||
| DH5α | F−recA hsdR17(rk‐mk+) Φ80dLacZ | Bethesda Research Laboratories, Bethesda, MD, USA |
| HB101 | F− recA | Maniatis et al. (1982) |
| Plasmids | ||
| pLAFR3 | Tra−Mob+, RK2 replicon, tetr | Staskawicz et al. (1987) |
| pLAFR6 | pLAFR1 with trp terminators | Bonas et al. (1989) |
| pUFR034 | Inc W, Kmr, Mob+, lacZα, Par+, cosmid | De Feyter et al. (1990) |
| pUFR051 | pLAFR3 derivative | De Feyter et al. (1990) |
| pBluescript II SK(+) | Phagemid, pUC derivative, Ampr | Stratagene (La Jolla, CA, USA) |
| pHOKmGus | Kmr Apr Tn3‐uidA fusion | Bonas et al. (1989) |
| pUC8Ω | Vector with Ω cassette | B. Staskawicz |
| PGEM®T Easy Vector | Multiple site cloning vector | Promega |
| pOK1 | Suicide vector, pKNG101 derivative, Smr/Sucs | Huguet et al. (1998) |
| pRK2073 | ColE1 replicon, TraRK+, Mob+, Specr | Figurski and Helinski (1979) |
| pAW5.2 | 5‐kb EcoRI‐KpnI fragment containing pthA w from X0053 cloned in pUFR047 that complements pustule formation in a pustule minus strain | Al‐Saadi et al. (2007) |
| pL799 | pLAFR3 with DNA fragment from Xac‐Aw that contains avrGf1 | This study |
| pL799‐1 | pL799 with Tn3‐Gus insert in avrGf1 | This study |
| pL799‐2 | 3.0‐kb fragment from pL799 in pLAFR6 | This study |
| pU799‐3 | 2.3‐kb fragment containing avrGf1 in pUFR034 | This study |
| pBs3.0 | pBluescript II KS with 3.0‐kb insert from pL799‐1 containing Tn3‐KmGus | This study |
| pL22 | XacAw pLAFR3 cosmid that contains hrpX, hrpG and hsp90Xo homologues | This study |
| pL22E | Xac‐Aw pLAFR3 clone containing hrpG | This study |
| p0346 | Xac‐Aw pUFR034 clone containing hrpG | This study |
| pL689 | Similar to pL22 | This study |
| pLXOPA | 91‐118 pLAFR3 cosmid that contains the xopA gene | This study |
| pL80 | XacAw pLAFR3 clone that contains avrBs3 homologue, designated avrTaw | This study |
| pL104 | Similar to pL80 | This study |
| pL115 | Similar to pL80 | This study |
| pL622 | Similar to pL80 | This study |
| pUFR80.1 | Subclone of pL80 containing only avrTaw homologue that does not complement pustule formation | This study |
| pLAT211 | pLAFR3 containing avrBs3‐2 | Bonas et al. (1991) |
DPI, Division of Plant Industry of the Florida Department of Agriculture and Consumer Services, Gainesville, FL, USA; NTG, N‐methyl‐N′‐nitro‐N‐nitrosoguanidine; B. Staskawicz, University of California, Berkeley, CA, USA.
Given that pustule formation in grapefruit by Xac‐AwΔavrGf1 was significantly reduced relative to that in Key lime, we attempted to determine what other gene(s), in addition to avrGf1, was responsible for the reduced host range. Shiotani et al. (2007) demonstrated that an avrBs3 homologue with 15.5 repeats was associated with reduced pustule formation in C. grandis. It was logical to determine whether the avrBs3 homologues present in pUFR80.1 were involved in any way with reduced disease development in grapefruit. We transferred both avrBs3 homologues present in Xac‐Aw, avrTaw and pthaAw expressed in pUFR80.1 and pAW5.2, respectively, to the Xac‐A strain by conjugation, and determined that pustule formation was not reduced in grapefruit when inoculated with the Xac‐A transconjugants carrying either avrBs3 homologue (Fig. 3e).
DISCUSSION
The multiplication patterns of the two strains of X. axonopodis pv. citri (Xac‐Aw and Xac‐A) in Duncan grapefruit and Key lime leaves that occurred in this study were very similar to those reported by Sun et al. (2004). Maximum populations of the Xac‐Aw strain were 10–100‐fold less than those of the Xac‐A strain in grapefruit leaves. The multiplication of each strain was no different after infiltration into Key lime leaves.
The relationship between confluent necrosis and the cell population, after high concentrations (5 × 108 cfu/mL) of Xac‐A and Xac‐Aw bacteria were infiltrated separately into leaves of grapefruit and Key lime, has not been reported previously (Sun et al., 2004), and is indicative of an incompatible interaction in grapefruit leaves (Minsavage et al., 1990). Necrosis, as determined by electrolyte leakage, occurred earlier and with smaller populations after inoculation of grapefruit leaves with cells of the Xac‐Aw strain relative to those of the Xac‐A strain. The time to necrosis and the cell populations were similar for each strain (data not presented) after inoculation of Key lime leaves. These data are consistent with the concept that the Xac‐Aw strain caused HR in grapefruit leaves. The more rapid necrosis in grapefruit caused by Xac‐Aw relative to Xac‐A could easily be discerned visually. It should be noted that necrosis associated with both HR and the susceptible reaction occurs relatively slowly in grapefruit leaves inoculated with the citrus canker bacterium relative to that observed in some other host–pathogen systems (Minsavage et al., 1990). This is the first report of the association of an avirulence gene with this type of interaction in citrus, although Khalaf et al. (2008) identified a general non‐host interaction between X. citri ssp. citri and Fortunella margarita.
Conclusive evidence for hypersensitivity in citrus leaves to strains of the citrus canker bacterium has not been reported previously. HR was suggested to be caused by a strain of Xaa‐C when a suspension of 108 cfu/mL was infiltrated into grapefruit leaves (Stall et al., 1982). The resulting rapid necrosis was compared with the relatively slow necrosis caused by cells of either Xac‐A or Xaa‐B strains in the same test. However, data on the relationship of populations of bacteria to necrosis were not reported. This relationship still needs to be determined for these strains.
A genomic library of a Xac‐Aw strain was successfully produced and incorporated into E. coli DH5α. For successful screening of the library for an avirulence gene, one would normally transfer each clone into a strain that was pathogenic to the plant in question, in this case, into a strain of Xac‐A. However, in previous experiments, the transfer of clones in the pLAFR3 cosmid library to strains of Xac‐A from E. coli DH5α by triparental matings did not occur at high frequency. Clones of the library were transferred from E. coli to strain 91‐118 of X. perforans, a highly promiscuous recipient, by triparental conjugations to circumvent this problem. The strain of X. perforans causes a null reaction in leaves of grapefruit, even though it contains the hrp genes necessary for the transfer of effector proteins into host cells (Bonas et al., 1991). It was thought that an avirulence gene in a clone of genomic DNA of Xac‐Aw would result in HR in grapefruit leaves when present in the strain of X. perforans. In fact, an avirulence gene in the Xac‐Aw library was found. As this procedure was successful, it could possibly be used to locate other avirulence genes when the transfer of clones to a pathogen occurs very infrequently during triparental matings with E. coli.
The clone that was expressed in Xac‐A and caused HR in grapefruit leaves did not cause HR in leaves of Key lime. This reflected the host range of the Xac‐Aw strain. The importance of the avirulence gene in the determination of the host specificity of the Xac‐Aw strain was further investigated by determining the presence of the gene in other bacterial strains pathogenic to citrus. Using PCR and Southern hybridization techniques, we could only detect the avirulence gene in Xac‐Aw strains, although we did observe weak hybridization with the X. campestris pv. campestris strain which contained gene 3600. When the gene was mutated in a Xac‐Aw strain, the symptoms caused by the mutated strain were similar to those caused by the Xac‐A strains. However, the symptoms appeared to be less pronounced.
The growth of the Xac‐Aw strain with avrGf1− (Xac‐AwΔavrGf1) in grapefruit leaves was significantly greater than that of the wild‐type Xac‐Aw strain, but lower than that of the Xac‐A strain, when both high and low levels of inoculum were used. The differences in the cell populations also reflected differences in the number of cells present at the time of necrosis in grapefruit leaves. Complementation of the mutant strain with the avrGf1 clone restored avirulence in grapefruit leaves for both the multiplication of cells and electrolyte leakage. The growth of Xac‐AwΔavrGf1 in Key lime leaves was equal to that of wild‐type Xac‐A and Xac‐Aw strains (data not presented). Therefore, avrGf1 function did not appear to affect the virulence of the Xac‐Aw strain in Key lime. The gene avrGf1 seems to be important in the delimitation of the host specificity of the Xac‐Aw strain, but not in the virulence in Key lime.
Mutation of a hypothetical avirulence gene in the Xac‐Aw strain would nullify HR and result in a susceptible reaction in grapefruit leaves, as has been observed in other systems (Dahlbeck and Stall, 1979; Gassmann et al., 2000; Swarup et al., 1992). Bacterial cells of Xac‐Aw were treated with N‐methyl‐N′‐nitro‐N‐nitrosoguanidine (NTG) (Rybak, 2005). This is a powerful mutagenic agent that increases the frequency of mutation. NTG treatment increased the frequency of streptomycin mutants and white colonies in the population used to inoculate grapefruit leaves, attesting to the effectiveness of treatment. Rybak (2005) noted that no mutants of the Xac‐Aw strain were found in these tests that were pathogenic on grapefruit leaves. This may indicate that an additional factor exists in Xac‐Aw.
An assumption could be made that other resistance genes exist in grapefruit leaves that prevent the symptoms of the mutated avrGf1 strain from being equal to those of the wild‐type Xac‐A strain, and prevent the populations of the two strains from being equal. Other host‐limiting genes in the genomic library of Xac‐Aw that interact with resistance genes in grapefruit may be present in the library, because only 300 clones were screened. Clearly, more than one gene is present in Xac‐Aw that affects its ability to cause typical citrus canker on grapefruit; however, it does not appear to be associated with HR and thus would not have been identified using the screening procedure employed to identify the other clones. We examined the possibility that avrBs3 homologues could be involved in the suppression of growth in grapefruit, given that Shiotani et al. (2007) determined that a member of the avrBs3/pthA family, designated hssB3.0, with 14.5 tandem repeats, suppressed virulence, as manifested by a decrease in lesion size. However, neither of the avrBs3 homologues (pUFR80.1 and pAW5.2) in Xac‐Aw suppressed pustule formation in grapefruit when expressed in an Xac‐A strain (Fig. 3d). Further work is necessary to identify other factors associated with the limitation of Xac‐Aw in grapefruit.
We also provided indirect evidence that the avrBs3 homologue, present in pUFR80.1, which contains 18.5 tandem repeats, is necessary for pustule formation in grapefruit. When Xac‐Xac‐AwΔavrGf1ΔavrTawΔPthaA w (deletion in avrGf1 and both avrBs3 homologues), complemented with pAW5.2, was inoculated into grapefruit, pustules were not produced; they were only formed at sites inoculated with Xac‐AwΔavrGf1, which contained both copies of the avrBs3 homologue. This second avrBs3 homologue, although not essential for pustule formation in Key lime, appears to be important for pustule formation in grapefruit.
The protein expressed from avrGf1 may be an effector, which is delivered via TTSS into plant cells and has a role in virulence (Kjemtrup et al., 2000). Proteins expressed from other avirulence genes in Xanthomonas have been determined to be transferred to host cells by TTSS (Metz et al., 2005; Mudgett et al., 2000; Szurek et al., 2002), and thus were determined to be effectors that incited HR. Strains with mutation in the hrp gene cluster were used to prevent the transfer of proteins to host cells by TTSS in the experiments. HR did not occur in grapefruit leaves after the avrGf1 clone was conjugated into a strain of X. perforans that was mutated in the hrp locus. More work is needed to confirm that avrGf1 is transferred into grapefruit cells by the Xac‐Aw strain, and thus is an effector.
The pL22 clone was unique because HR occurred in grapefruit leaves when it was expressed in X. perforans, but not when it was expressed in the Xac‐A strain. Interestingly, the presence of a hrpG sequence in pL22 caused cells of X. perforans to incite HR rather than the null reaction in grapefruit leaves. As clone pL22, when expressed in X. perforans, but not in Xac‐A, incited HR in grapefruit leaves, and did not contain an avirulence gene, it should be emphasized that any suspected clone must be transferred to a pathogen of the host to confirm that an avirulence gene exists.
Kim et al. (2003) reported that X. axonopodis pv. glycines did not produce HR in tobacco until a clone from X. oryzae pv. oryzae, which contained both hrpX and hrpG, was conjugated into X. axonopodis pv. glycines. The importance of either hrpX, hrpG or both in the production of HR in tobacco was not determined in this work. In our work, hrpX clearly was not necessary for HR in citrus caused by X. perforans. Kim et al. (2003) also gave evidence that the hrpX and hrpG clones increased the function of hpaG, a homologue of hrpN of Erwinia amylovora and a known harpin gene, in X. axonopodis pv. glycines and, subsequently, HR in tobacco. The hpaG nucleotide sequence is a homologue of xopA in X. campestris pv. vesicatoria strain 85‐10, but the XopA protein produced in X. axonopodis pv. glycines did not cause HR in tobacco (Kim et al., 2003). Although both X. campestris pv. vesicatoria and X. perforans cause bacterial spot of tomato, they are genetically different (Jones et al., 2004). However, the DNA sequences of xopA from both bacteria are almost identical (data not presented). The xopA gene in X. perforans, in conjunction with an additional hrpG copy, is at least partly involved in inducing HR in grapefruit leaves, but has no apparent effect on tomato. HrpG is reported to be the response regulator of a putative two‐component system in which the unidentified sensor presumably detects environmental variables which induce the hrc and hrp genes (Wengelnik et al., 1996). HrpG is thought to be at the top of the hrc/hrp regulatory cascade, as the mutant HrpG* constitutively activates all other hrc and hrp genes (Wengelnik et al., 1999). Multiple copies of hrpG in trans may alter hrp gene expression and may perhaps lead to constitutive expression; this may have caused the increased delivery of Hrp‐related proteins, such as XopA, which may have resulted in HR in grapefruit. Furthermore, expression of xopA and associated genes in Xac‐A, with or without additional copies of HrpG, resulted in reduced growth in grapefruit. Therefore, xopA and/or other genes in the pLXOPA clone may be host‐limiting factor(s) in citrus. A similar response was observed with X. axonopodis pv. glycines, in which the wild‐type elicited HR in pepper, whereas the hpaH mutant did not (Kim et al., 2003). It remains to be determined whether XopA has harpin‐like activity in grapefruit leaves.
Four clones that caused HR in tomato were obtained from the 300 clones of the Xac‐Aw library. All four of the clones contained an avrBs3 homologue. Two homologues of this class are plasmid borne in Xac‐Aw (Duan et al., 2004), and therefore may have been over‐represented in our library. All four of our clones were conjugated into a strain of X. axonopodis pv. citrumelo, a pathogen of citrus, and none of our clones caused the erumpent‐type lesions in grapefruit or Key lime leaves when expressed in a strain of X. axonopodis pv. citrumelo, as do the avrBs3 homologues designated as pthA (Swarup et al., 1991, 1992). Two different avrBs3 homologues have been reported to exist in the Xac‐Aw strains (Duan et al., 2004), and one was cloned and found to be related to pthA (Al‐Saadi et al., 2007). The second avrBs3 homologue, avrTaw1, which was cloned by us and caused HR in tomato, did not produce the pthA effect in citrus leaves. This finding agrees with that of Al‐Saadi et al. (2007), who determined that the pustule‐forming genes, pthA homologues, contained 17.5 repeats, unlike avrTaw1 which contained 18.5 repeats. It was not determined whether the pthAw gene cloned by Duan et al. (2004) from another strain of Xac‐Aw caused HR in tomato. It is known that the different avrBs3 homologues have different specificities in host reaction causing HR and pathogenicity in plants (Kay et al., 2005). Recently, Shiotani et al. (2007) identified a pthA homologue that induces a defence response in C. grandis. Perhaps one or more of these homologues, isolated from the Xac‐Aw strain, is associated with the induction of a defence response in grapefruit. This could help to explain why the mutation experiments did not result in the identification of one or more mutants that were fully virulent on grapefruit. Further work is needed to address the issue of whether or not these pthA homologues are host‐limiting factors.
EXPERIMENTAL PROCEDURES
Plants
Plants of grapefruit (C. paradisi), cultivar Duncan, and Key lime (C. aurantifolia) were kept in 12‐in pots in a quarantine glasshouse at the Florida Department of Agriculture and Consumer Services, Division Plant Industry, Gainesville, FL, USA, which was kept between 20 and 35 °C. The plants were kept in vigorous growth by the application of ‘Osmocote’ fertilizer (19 : 6 : 12 ratio of nitrogen : phosphorus : potassium) as needed. Before inoculations, the plants were pruned to stimulate new growth. Leaves on the newly developed shoots were inoculated 14–21 days after the shoots began growth. The leaves chosen for inoculations were fully expanded or nearly so, but soft to the touch and not as fully green as mature leaves. By using this procedure, all the inoculated leaves were in a similar developmental stage. Tomato plants (cultivar Bonny Best) inoculated with clones in X. perforans were approximately 6 weeks of age. After inoculation with the clones, the plants were incubated in a growth room at 28 ± 2 °C.
Bacterial strains
The strains used in this research are listed in Table 1. All strains were maintained at −80 °C, and subcultured, when needed, on nutrient agar (NA). Antibiotic‐resistant strains were obtained by plating 500 µL of 109 cfu/mL onto NA supplemented with appropriate antibiotic. Escherichia coli strains were cultured on Luria–Bertani (LB) medium (Maniatis et al., 1982). Conjugations were performed on nutrient‐yeast‐glycerol agar (NYGA) (Daniels et al., 1984). The antibiotics and concentrations used in the media were as follows: rifamycin, 75 µg/mL; tetracycline, 12.5 µg/mL; kanamycin, 25 µg/mL.
Preparation of inocula
Suspensions of strains were each transferred to NA to determine the purity of the cultures. Several colonies from pure cultures were transferred to nutrient broth. After shaking overnight at 28 °C, the bacterial suspensions were centrifuged and the cells were resuspended in sterile tap water, and standardized (0.3A at 600 nm wavelength) in a Spectronic 20 (UNICAM, Rochester, NY, USA) spectrophotometer. This optical density corresponded to a bacterial concentration of 5 × 108 cfu/mL. Other concentrations of inocula were obtained by dilution.
Electrolyte leakage
Leaves of grapefruit were inoculated with 5 × 108 cfu/mL by infiltrating areas of c. 5 cm2 with a syringe and 27‐gauge needle (Klement, 1963). The inoculated area was outlined by pin‐pricking the infiltrated area with a needle. Each strain was placed in each leaf. Replicates involved different leaves. After 2 h and 2, 4, 6, 8 and 10 days, electrolyte leakage was measured from each of three leaves. Electrolyte leakage was determined as an increase in electrical conductivity over a 2‐h period of a bath containing six 0.5‐cm2 leaf discs from an inoculated area that were added to 3 mL of de‐ionized water (Klement et al., 1990). The baths in 16 × 100‐mm tubes were shaken at 200 r.p.m. A zero‐time determination was subtracted from the 2‐h determination to give the final value. The mean of the three values was used as the conductivity at each time period, but each value was used to determine the experimental error.
Bacterial populations
The populations of strains in grapefruit leaves were determined in the same leaves and at the same times as the electrolyte leakage determinations. From each leaf, 0.5 cm2 of inoculated area was taken and triturated in 1 mL of sterile tap water; after appropriate 10‐fold dilutions with sterile tap water, 50 µL were plated onto NA medium. The colonies were counted 3 days later. Three replicates were included at each time period. The experiments on population and electrolyte leakage were repeated three times, but only one experiment was included in the figures.
Recombinant DNA techniques
The techniques used for enzyme digestion, ligation, Southern transfer, plasmid alkaline lysis and agarose gel electrophoresis have been described by Maniatis et al. (1982). A genomic library of Xac‐Aw (strain 12879) was constructed in the vector pLAFR3, as described by Metz et al. (2005). Individual clones were transfected into E. coli DH5α and maintained on LB medium. Each clone contained approximately a 25‐kb fragment of Xac‐Aw DNA. The helper plasmid pRK2073 in E. coli HB101 was used in conjugations involving triparental matings. Individual clones in E. coli DH5α were conjugated into strain 91‐118 of X. perforans (Jones et al., 2004). Each transconjugant was infiltrated into a 1‐cm2 area of leaf tissue with a syringe and 27‐gauge needle. Grapefruit leaves were inoculated with c. 300 transconjugants individually. Five transconjugants were infiltrated into each leaf, which also contained a control suspension of strain 91‐118. Tomato leaves were inoculated similarly as controls, because the Xac‐Aw strain also elicits HR in tomato leaves. A clone that caused HR in grapefruit or tomato leaves was detected via necrosis in the infiltrated area of the leaf, which occurred more rapidly than with the control suspension of strain 91‐118.
Subcloning of pL799
A clone (pL799) with HR activity in grapefruit, but not tomato, was subcloned to contain only the DNA necessary for HR activity. To identify the location of the avirulence gene in pL799, the gene responsible for HR activity was inactivated by the transposon pHoKmGus by the procedure described by Huguet and Bonas (1997). About 160 clones with kanamycin resistance were transferred to X. perforans and screened for the lack of HR in grapefruit. Three clones were selected for a lack of HR and one clone (pL799‐1) was selected from the three for further work. The clone pL799‐1 was restricted with each of several enzymes to find a fragment that contained the Tn3‐KmGus insert. A HindIII restriction fragment contained the Tn3‐KmGus insert and about 3.0 kb of DNA. This fragment was ligated into pBluescript II KS and labelled pBs3.0. A portion of the 3.0‐kb insert in pBs3.0 was then sequenced (see below) using forward and reverse primers from the cloning vector. Custom oligonucleotide primers were designed for completion of the sequencing of the HindIII fragment.
An ORF was identified in the sequence of the HindIII fragment. However, the ORF was at the end and was not complete. When the HindIII fragment was ligated into pLAFR6 (pL799‐2) and conjugated into X. perforans, no HR occurred in grapefruit leaves. Primers were selected for sequencing beyond the end of the HindIII fragment in the original pL799 clone to obtain the complete ORF. Primers were then selected to amplify, by PCR, a 2.3‐kb fragment that contained the complete ORF, which was then ligated into pGEM®T Easy Vector (Promega, Madison, WI, USA). The 2.3‐kb insert was removed from pGEM®T Easy Vector with EcoRI enzyme and then ligated into the vector pUFR034, and designated pU799‐3. The pUFR034 cosmid was used as a vector because DNA inserts in this vector could be conjugated into strains of Xac‐Aw, whereas pLAFR3 derivatives could not be transferred to strains of Xac‐Aw. The pU799‐3 clone in X. perforans and Xac‐A 40 caused HR in grapefruit leaves.
DNA sequence analysis
Sequencing of the DNA fragments was completed at the sequencing facility [Interdisciplinary Center for Biotechnology Research (ICBR), University of Florida, Gainesville, FL, USA] with an Applied Biosystems model 373 system (Foster City, CA, USA). Custom primers were synthesized at the ICBR facility with an Applied Biosystem model 394 DNA synthesizer to complete the sequencing of both strands of DNA. The computer program SeqAid II version 3.81 was used to analyse nucleotide sequence data and predicted protein products of the 2.3‐kb region that contained avrGf1. A search for nucleotide and amino acid sequence homology in the gene bank was conducted using the blastn and blastp 2.2.11 programs (Altschul et al., 1997).
Presence of avrGf1 in other xanthomonads
Primers were designed and custom‐made to amplify a 199‐bp fragment from within avrGf1 for its detection by PCR in the DNA of xanthomonads that cause disease in citrus plants. The primers used were as follows: forward, 5′‐CGCCGGTTTCTGTCACTTG‐3′; reverse, 5′‐GCCGCCTTTGCCATCGACCAG‐3′. PCRs were performed in a thermocycler (M.J. Research, Watertown, MA, USA).
Southern hybridization experiments were performed on Nytran (Schleicher and Scheull, Keene, NH, USA) membranes using the GENIUS non‐radioactive DNA labelling and detection kit, according to the manufacturer's instructions (Boeheringer Mannheim Biochemicals, Indianapolis, IN, USA). Genomic DNA extractions were made using the Genomic Prep TM Cells and Tissue DNA Isolation Kit (Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA). The isolated DNA preparations were restricted with HindIII endonuclease. Plasmid extraction from Xac‐A and Xac‐Aw strains was performed as described previously (Stall et al., 1986). The restricted DNA and plasmids were electrophoresed in 0.75% agarose gel.
Mutation of avrGf1 in Xac‐Aw
The avrGf1 gene in Xac‐Aw was mutated to investigate the role of the gene in the host specificity of the bacterium. Mutation in avrGf1 in pGEM®T Easy Vector occurred by first cloning an Omega cassette (from pUC8Ω) into a BamHI site in the gene. Then, the inactive gene was exchanged in the Xac‐Aw strain using the suicide vector pOK1, following the procedure described by Huguet et al. (1998). Eventually, Xac‐Aw (strain 12879) with the mutated avirulence gene was obtained and labelled Xac‐AwΔavrGf1.
avrGf1 activity in a hrp − strain
A strain of X. perforans 91‐118 that had been mutated in the hrp locus was on hand. This strain was obtained by NTG mutation and selected for not being pathogenic or hypersensitive on tomato. The strain was complemented to pathogenicity and hypersensitivity after the addition of plasmid pXV9, which contained the hrp cluster (Bonas et al., 1991). The clone pU799‐3 (avrGf1) was conjugated into X. perforans strains 91‐118hrp − and 91‐118hrp + and tested for reaction in grapefruit leaves as described above.
Hypersensitivity of pL22 in grapefruit
Clones pL22 and pL689, which caused HR in grapefruit when in cells of X. perforans, were restricted with several enzymes. Based on the restriction patterns, the clones were similar. Clone pL22 was chosen for further analysis and contained about 27 kb of inserted DNA. A partial digest of this cosmid was obtained by restricting it with Sau3A1 to obtain fragments of about 5 kb. The fragments were then ligated into pUFR051, transformed into competent cells of E. coli DH5α and conjugated into cells of X. perforans. Of the 180 conjugants, two gave HR in grapefruit leaves. These two clones did not produce HR in grapefruit when conjugated into strain 40 of Xac‐A. The insert of one of the clones contained about 5 kb, which contained two BamHI fragments. Each BamHI fragment was cloned into pBluescript and sequenced. Three ORFs were found. With appropriate enzyme digestion, a fragment containing each ORF was transferred to pLAFR3, transformed into cells of E. coli DH5α, conjugated into X. perforans and inoculated into grapefruit leaves at a concentration of 5 × 108 cfu/mL.
Role of X. perforans XopA region in host specificity
A clone in pLAFR3 containing a xopA gene from X. perforans was selected from a previously developed library of strain 91‐118 of X. perforans in pLAFR3 (Astua‐Monge et al., 2000). The clone was identified by amplifying by PCR the gene from pools of clones from the 91‐118 library. The primers used in the amplification were as follows: forward, 5′‐CATCTCGGAAAAGCARCTGGACC‐3′; reverse, 5′‐CATTCTKCGCCTGKARAATSTCTCC‐3′. The clone containing xopA was labelled pLXOPA.
The xopA gene was deleted in strain 91‐118. In the procedure, the xopA gene was amplified using the following primers: forward, 5′‐GGGAAGCTTTGCTGCAAGAGGAAAAGCG‐3′; reverse, 5′‐GGGGAATTCAATCCGCGCGTGCGA‐3′. The amplified product (1800 bp) was cloned into pGEM®T Easy Vector. The enzyme AvaI was used to delete 706 bp, which included the xopA gene (353 bp), 103 bp downstream and 250 bp upstream. The upstream deletion included a PIP promoter for hpaH running in the opposite direction. The deleted fragment was excised from the pGEM®T Easy Vector clone with enzymes ApaI and SpeI, and ligated into pOKI suicide vector that was cut with ApaI and XbaI. The suicide vector with the deletion fragment was then conjugated into strain 91‐118 for deletion of xopA in that strain. This procedure was described by Huguet et al. (1998). Deletion of xopA in strain 91‐118 was confirmed by PCR.
Clones with avirulence to tomato from Xac‐Aw
The DNAs of four clones from the Xac‐Aw library, which caused HRs in tomato leaves when conjugated in X. perforans, were digested with BamHI and KpnI restriction enzymes and subjected to electrophoresis. All four clones were similar based on the restriction fragment profiles. The DNAs of the clones were then hybridized with a probe (pLAT 211) (Bonas et al., 1993) containing avrBs3‐2 by the procedure described above. Genomic DNA from strain 82‐8 of X. euvesicatoria and from a strain of Xac‐Aw was included in the hybridization. These strains were known to contain an avrBs3 homologue (Bonas et al., 1989; Duan et al., 2004).
Role of Bs3 homologues in HR and virulence
pUFR80.1 identified in this study and pAW5.2 containing the avrBs3 homologue from Xac‐Aw (Al‐Saadi et al., 2007) were transferred to X. perforans by triparental mating, and suspensions from the resulting transconjugants were infiltrated into Bonny Best tomato at 5 × 108 cfu/mL. The reactions in tomato were compared with the reactions caused by inoculations with X. perforans at the same concentration.
Both pUFR80.1 and pAW5.2 were also transferred to strain Xac‐AwΔavrGf1ΔavrTawΔpthA w, which was also inactivated in avrGf1 and in both copies of the avrBs3 homologues present in XacAw, and which was deficient for pustule formation. The resulting transconjugants and Xac‐AwΔavrGf1 were inoculated into grapefruit and Key lime by infiltration and the pin‐prick method. The pin‐prick method involves placing a drop of bacterial suspension on the adaxial surface of Key lime and grapefruit leaves and piercing a needle through the drop and through the leaf. The inoculated plants were incubated in the glasshouse and observed for disease development.
ACKNOWLEDGMENTS
This research was supported by the USDA special grant (J. B. Jones, R. E. Stall, and X. Sun, USDA 2001‐34446‐10781‐S).
REFERENCES
- Al‐Saadi, A. , Reddy, J.D. , Duan, Y.P. , Brunings, A.M. , Yuan, Q. and Gabriel, D.W. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host‐range variation. Mol. Plant–Microbe Interact. 20, 934–943. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astua‐Monge, G. , Minsavage, G.V. , Stall, R.E. , Davis, M.J. , Bonas, U. and Jones, J.B. (2000) Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant‐inducible avirulence gene. Mol. Plant–Microbe Interact. 13, 911–921. [DOI] [PubMed] [Google Scholar]
- Bonas, U. , Stall, R.E. and Staskawicz, B. (1989) Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria . Mol. Gen. Genet. 218, 127–136. [DOI] [PubMed] [Google Scholar]
- Bonas, U. , Schulte, R. , Fenselau, D. , Minsavage, G.V. , Staskawicz, B. and Stall, R.E. (1991) Isolation of a cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant–Microbe Interact. 4, 81–85. [Google Scholar]
- Bonas, U. , Conrads‐Strauch, J. and Balbo, I. (1993) Resistance in tomato to Xanthomonas campestris pv. vesicatoria is determined by alleles of the pepper‐specific avirulence gene avrBs3 . Mol. Gen. Genet. 238, 261–269. [DOI] [PubMed] [Google Scholar]
- Canteros, B.I. (2004) Management of citrus canker in Argentina. A review In Proceedings of the Tenth International Society of Citriculture Congress, Agadir, Morocco, 15–20 February 2004 (Ait‐Oubahou A. and El‐Otmani M. eds) Vol. 1, pp. 448–451. Agadir, Morocco: International Society of Citriculture. [Google Scholar]
- Civerolo, E.L. (1984) Bacterial canker disease of citrus. J. Rio Grande Valley Hortic. Soc. 37, 127–145. [Google Scholar]
- Crute, I.R. (1985) Mechanisms of Resistance to Plant Diseases (Fraser R.S.S., ed.), pp. 80–142. Dordrecht: Nijhoff & Junk. [Google Scholar]
- Cubero, J. and Graham, J.H. (2002) Genetic relationship among world wide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl. Environ. Microbiol. 68, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlbeck, D. and Stall, R.E. (1979) Mutations for change of race in cultures of Xanthomonas vesicatoria. Phytopathology, 69, 634–636. [Google Scholar]
- Daniels, M.J. , Barber, C.E. , Turner, D.C. , Cleary, W.G. and Sawzyc, M. (1984) Isolation of mutants of Xanthomonas campestris showing altered pathogenicity. J. Gen. Microbiol. 130, 2447–2455. [Google Scholar]
- De Feyter, R. , Kado, C.I. and Gabriel, D.W. (1990) Small, stable shuttle vectors for use in Xanthomonas . Gene, 88, 65–72. [DOI] [PubMed] [Google Scholar]
- Dopson, R.N. (1964) The eradication of citrus canker. Plant Dis. 48, 30–31. [Google Scholar]
- Duan, Y.P. , Al‐Saadi, A. and Gabriel, D.W. (2004) Role of pthA homologues in Xanthomonas citri pv. citri A(W) in hyperplasia and necrosis on Key lime and grapefruit. Phytopathology, 94, S26. Publication no. P2004‐0174‐AMA. [Google Scholar]
- Figurski, D. and Helinski, D.R. (1979) Replication of an origin‐containing derivative of plasmid RK2 dependent on a plasmid function provided in trans . Proc. Natl. Acad. Sci. USA, 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, D.W. , Kingsley, M.T. , Hunter, J.E. and Gottwald, T.R. (1989) Reinstatement of Xanthomonas citri (ex Hasse) and X. phaseoli (ex Smith) to species and reclassification of all X. campestris pv. citri strains. Int. J. Syst. Bacteriol. 39, 14–22. [Google Scholar]
- Gassmann, W. , Dahlbeck, D. , Chesnokova, O. , Minsavage, G.V. , Jones, J.B. and Staskawicz, B.J. (2000) Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 182, 7053–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald, T.R. , Graham, J.H. and Schubert, T.S. (1997) An epidemiological analysis of the spread of citrus canker in urban Miami, Florida, and synergistic interaction with the Asian citrus leafminer. Fruits, 52, 371–378. [Google Scholar]
- Gottwald, T.R. , Sun, X. , Riley, T. , Graham, J.H. , Ferrandino, F. and Taylor, E.L. (2002a) Geo‐referenced spatiotemporal analysis of the urban citrus canker epidemic in Florida. Phytopathology, 92, 361–377. [DOI] [PubMed] [Google Scholar]
- Gottwald, T.R. , Graham, J.H. and Schubert, T.S. (2002b) Citrus canker: the pathogen and its impact. Plant Health Progress Online. doi: 10.1094/PHP-2002-0812-01-RV. [DOI] [Google Scholar]
- Hert, A.P. (2007) Evaluation of bacteriocins in Xanthomonas perforans for use in biological control of Xanthomonas euvesicatoria . Dissertation. Gainesville, FL: University of Florida. [Google Scholar]
- Huguet, E. and Bonas, U. (1997) hrpF of Xanthomonas campestris pv. vesicatoria encodes an 87‐kDa protein with homology to NolX of Rhizobium fredii. Mol. Plant–Microbe Interact. 10, 488–498. [DOI] [PubMed] [Google Scholar]
- Huguet, E. , Hahn, K. , Wengelnik, K. and Bonas, U. (1998) hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host‐specific hypersensitive reaction. Mol. Microbiol. 29, 1379–1390. [DOI] [PubMed] [Google Scholar]
- Jones, B.J. , Lacy, G.H. , Bouzar, H. , Stall, R.E. and Schaad, N.W. (2004) Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 27, 755–762. [DOI] [PubMed] [Google Scholar]
- Kay, S. , Bock, J. and Bonas, U. (2005) Characterization of AvrBs3‐like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol. Plant–Microbe Interact. 18, 838–848. [DOI] [PubMed] [Google Scholar]
- Kearney, B. and Staskawicz, B.J. (1990) Characterization of IS416 and its role in bacterial spot disease of pepper and tomato. J. Bacteriol. 172, 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf, A. , Moore, G.A. , Jones, J.B. and Gmitter, F.G. (2008) New insights into the resistance of Nagami kumquat to canker disease. Physiol. Mol. Plant Pathol. 71, 240–250. [Google Scholar]
- Kim, J‐G. , Park, B.K. , Yoo, C‐H. , Jeon, E. , Oh, J. and Hwang, I. (2003) Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185, 3155–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjemtrup, S. , Nimchuk, Z. and Dangl, J.L. (2000) Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3, 73–78. [DOI] [PubMed] [Google Scholar]
- Klement, S. , Stall, R.E. , Novacky, A.J. , Ersek, T. , Fett, W.F. , Huang, J.S. and Beckman, C.H. (1990) In: Methods of Phytobacteriology. Chapter III.4. Mechanism of Resistance (Klement Z., Rudolph K. and Sands D.C., eds), pp. 469–493. Budapest: Akademiai Kiado. [Google Scholar]
- Klement, Z. (1963) Rapid detection of the pathogenicity of phytopathogenic pseudomonads. Nature, 199: 299–300. [DOI] [PubMed] [Google Scholar]
- Koizumi, M. (1985) Citrus canker: the world situation In: Citrus Canker: An International Perspective (Timmer L.W. ed.), pp. 2–7. Lake Alfred, FL: Citrus Research & Education Center, University of Florida. [Google Scholar]
- Loucks, K.W. (1934) Citrus Canker and its Eradication in Florida. Unpublished manuscript. Original copy in the files of the Division of Plant Industry, Florida Department of Agriculture, Gainesville, FL. [Google Scholar]
- Maniatis, T. , Fritsch, E.F. and Sambrook, J. (1982) Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Metz, M. , Dahlbeck, D. , Morales, C. , Al Sady, B. , Clark, E. , and Staskawicz, B. (2005) The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana . Plant J. 41, 801–814. [DOI] [PubMed] [Google Scholar]
- Minsavage, G.V. , Dahlbeck, D. , Whalen, M.C. , Kearney, B. , Bonas, U. , Staskawicz, B.J. and Stall, R.E. (1990) Gene‐for‐gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria–pepper interactions. Mol. Plant–Microbe Interact. 3, 41–47. [Google Scholar]
- Minsavage, G.V. , Mudgett, M.B. , Stall, R.E. and Jones, J.B. (2004) Importance of opgHXcv of Xanthomonas campestris pv. vesicatoria in host–parasite interactions. Mol. Plant–Microbe Interact. 17, 152–161. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. , Chesnokova, O. , Dahlbeck, D. , Clark, E.T. , Rossier, O. , Bonas, U. and Staskawicz, B.J. (2000) Molecular signals are required for type III secretion and translocation of the Xanthomonas campestris AvrBs2 protein to pepper plants. Proc. Natl. Acad. Sci. USA, 97, 13 324–13 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak, M. (2005) Genetic determinants of host range specificity of the Wellington strain of Xanthomonas axonopodis pv. citri . Gainesville, FL: University of Florida. [Google Scholar]
- Schaad, N.C. , Postnikova, E. , Lacy, G.H. , Sechler, A. , Agarkova, I. , Stromberg, P.E. , Stromberg, V.K. , and Vidaver, A.K. (2005) Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29, 690–695. [DOI] [PubMed] [Google Scholar]
- Schubert, T.S. , Miller, J.W. and Gabriel, D.W. (1996) Another outbreak of bacterial canker on citrus in Florida. Plant Dis. 80, 1208. [Google Scholar]
- Schubert, T.S. , Rizvi, S.A. , Sun, X.A. , Gottwald, T.R. , Graham, J.H. and Dixon, W.N. (2001) Meeting the challenge of eradicating citrus canker in Florida—again. Plant Dis. 85, 340–356. [DOI] [PubMed] [Google Scholar]
- Shiotani, H. , Fujikawa, F. , Ishiihara, H. , Tsuyumu, S. and Ozaki, K. (2007) A pthA homolog from Xanthomonas axonopodis pv. citri responsible for host‐specific suppression of virulence. J. Bacteriol. 189, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stall, R.E. and Civerolo, E.L. (1991) Research relating to the recent outbreak of Citrus canker in Florida. Annu. Rev. Phytopathol. 29, 339–420. [DOI] [PubMed] [Google Scholar]
- Stall, R.E. , Miller, J.W. , Marco, G.M. and Canteros, B.I. (1982) Pathogenicity of three strains of the citrus canker organism on grapefruit In Fifth International Conference on Plant Pathogenic Bacteria (Lozano J.C. and Gwin P.J.), pp. 334–340. Cali, Colombia: Centro Internacional de Agricultura Tropical. [Google Scholar]
- Stall, R.E. , Loschke, D.C. and Jones, J.B. (1986) Linkage of copper resistance and avirulence loci on self‐transmissible plasmid in Xanthomonas campestris pv. vesicatoria . Phythopathology, 76, 240–243. [Google Scholar]
- Staskawicz, B. , Dahlbeck, D. , Keen, N. and Napoli, C. (1987) Molecular characterization of cloned virulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea . J. Bacteriol. 169, 5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J. , Dahlbeck, D. , and Keen, N.T. (1984) Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race‐specific incompatibility on Glycine max (L.) Merr. Proc. Natl. Acad. Sci. USA, 81, 6024–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Stall, R.E. , Jones, J.B. , Cubero, J. , Gottwald, T.R. , Graham, J.H. , Dixon, W.N. , Schubert, T.S. , Chaloux, P.H. , Stromberg, V.K. , Lacy, G.H. and Sutton, B.D. (2004) Detection and characterization of a new strain of citrus canker bacteria from Key/Mexican lime and Alemow in South Florida. Plant Dis. 88, 1179–1188. [DOI] [PubMed] [Google Scholar]
- Swarup, S. , De Feyter, R. , Brlansky, R.H. and Gabriel, D.W. (1991) A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit lesions on citrus. Phytopathology, 81, 802–809. [Google Scholar]
- Swarup, S. , Yang, Y. , Kingsley, M.T. and Gabriel, D.W. (1992) A Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant–Microbe Interact. 5, 204–213. [DOI] [PubMed] [Google Scholar]
- Swords, K.M. , Dahlbeck, D. , Kearney, B. , Roy, M. and Staskawicz, B. (1996) Spontaneous and induced mutations in a single open reading frame alter both virulence and avirulence in Xanthomonas campestris pv. vesicatoria avrBs2 . J. Bacteriol. 178, 4661–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurek, B. , Rossier, O. , Hause, G. and Bonas, U. (2002) Type III‐dependent translocation of the Xanthomonas AvrBs3 protein into plant cell. Mol. Microbiol. 46, 13–23. [DOI] [PubMed] [Google Scholar]
- Verniere, C. , Hartung, O.P. , Pruvost, E.L. , Civerolo, A.M. , Alvarez, P. , Maestri, P . and Luisetti, J. (1998) Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. Eur. J. Plant Pathol. 104: 477–487. [Google Scholar]
- Wengelnik, K. , Van den Ackerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria, is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. , Rossier, O. and Bonas, U. (1999) Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181, 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]


