SUMMARY
A 3′‐terminal, 77‐nucleotide sequence of Bamboo mosaic virus (BaMV) minus‐strand RNA (Ba‐77), comprising a 5′ stem‐loop, a spacer and a 3′‐CUUUU sequence, can be used to initiate plus‐strand RNA synthesis in vitro. To understand the mechanism of plus‐strand RNA synthesis, mutations were introduced in the 5′ untranslated region of BaMV RNA, resulting in changes at the 3′ end of minus‐strand RNA. The results showed that at least three uridylate residues in 3′‐CUUUU are required and the changes at the penultimate U are deleterious to viral accumulation in Nicotiana benthamiana protoplasts. Results from UV‐crosslinking and in vitro RNA‐dependent RNA polymerase competition assays suggested that the replicase preferentially interacts with the stem structure of Ba‐77. Finally, CMV/83 + UUUUC, a heterologus RNA, which possesses about 80 nucleotides containing the 3′‐CUUUU pentamer terminus, and which folds into a secondary structure similar to that of Ba‐77, could be used as template for RNA production by the BaMV replicase complex in vitro.
INTRODUCTION
Bamboo mosaic virus (BaMV), a single‐stranded, positive‐sense RNA virus, belongs to the genus Potexvirus of the Flexiviridae family. The 5′‐capped genome of BaMV encodes five open reading frames (ORFs) and has a 3′‐poly(A) tail. ORF1 encodes a 155‐kDa polypeptide comprising three domains, a guanosine triphosphate (GTP) methyltransferase/S‐adenosylmethionine (AdoMet)‐dependent guanylyltransferase domain (Huang et al., 2004, 2005; Li et al., 2001a), an RNA helicase‐like domain with RNA 5′‐triphosphatase activity (Gorbalenya and Koonin, 1989; Hodgman, 1988; Li et al., 2001b) and an RNA‐dependent RNA polymerase (RdRp) domain (Argos, 1988; Koonin, 1991; Li et al., 1998).
After a positive‐sense RNA virus has entered the host cell, minus‐strand RNA is generated from genomic RNA and, in turn, serves as the template for the synthesis of genomic and subgenomic RNAs. Viral polymerase and/or host factors recognize various primary, secondary and tertiary RNA structures in the promoter region to initiate viral RNA synthesis (Ahlquist et al., 2003; Dreher, 1999). Both the partially purified Cucumber necrosis virus replicase from plants and the recombinant RdRp of Turnip crinkle virus (TCV) can use the double‐stranded RNA templates to transcribe viral plus‐stranded RNA in vitro (Panavas et al., 2006). The terminal AU‐rich sequence has been proposed to be important for opening the double‐stranded RNA structure around the plus‐strand initiation promoter for positive‐strand RNA viruses (Panavas et al., 2006).
The BaMV polymerase has been shown to interact with specific sequences and structures at the 3′‐end of both plus‐ and minus‐strand RNA to initiate RNA synthesis (Huang et al., 2001; Lin et al., 2005a; Tsai et al., 1999). The promoter elements at the 3′‐end of the minus‐strand RNA have been mapped up to the last 77 nucleotides (Ba‐77), which fold into a 5′ large stem‐loop and a 3′ small unstable stem‐loop (Lin et al., 2005a, 2005b). In this study, we have investigated the promoter elements and their interactions with BaMV RdRp in further detail.
RESULTS
At least three adenylates in the 5′‐terminus of BaMV genomic RNA are required for efficient accumulation of viral products in vivo
Ba‐77 RNA (Fig. 1) has been shown previously to be a sufficient template for the initiation of positive‐strand RNA synthesis (Lin et al., 2005a). The terminal cis‐acting pentamer 3′‐CUUUU plays an important role in this process. In order to determine in detail the possible mechanisms of the terminal pentamer involved in viral accumulation in plant cells, the minimal number of U residues within the 3′‐CUUUU sequence was investigated. The infectious cDNA clone, pBaMV40A (Tsai et al., 1999), was used to construct pBaMVΔ1A, pBaMVΔ2A, pBaMVΔ3A and pBaMVΔ4A mutants, which gave rise to minus‐strand RNA with 3′‐CUUUG‐, 3′‐CUUG‐, 3′‐CUG‐ and 3′‐CG‐, respectively, during the replication cycle. pBaMV40A and its derivatives (Fig. 2A) showed similar levels of BaMV replicase protein in in vitro wheatgerm translation reactions, indicating that the mutations introduced concurrently at the 5′‐terminus of the plus‐strand RNA did not interfere with the translation efficiency of the viral ORF. These full‐length RNA transcripts were subsequently used to inoculate protoplasts. Northern blotting analysis showed that similar levels of positive‐strand RNA were detected in protoplasts inoculated with mutant BaMVΔ1A and BaMV40A RNA. However, inoculation with mutants BaMVΔ2A, BaMVΔ3A and BaMVΔ4A led to less than 2% plus‐strand RNA accumulation of that of the wild‐type (Fig. 2B and Table 1). These results suggest that at least three adenylate residues in the 5′‐GAAAA‐ stretch of genomic RNA are required for efficient viral RNA accumulation in vivo. As mutations introduced into the 5′‐terminus of genomic RNA did not interfere with their translation (Fig. 2A), the impairment of the viral accumulation of some mutants is more likely to be a result of effects on the minus‐strand promoter for plus‐strand RNA synthesis (Lin et al., 2005a).
Figure 1.
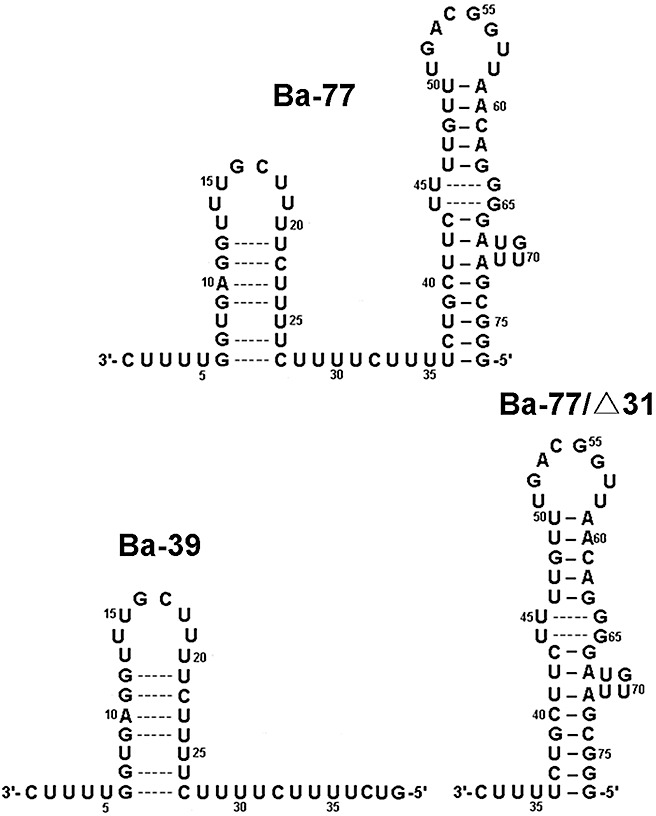
Secondary structures of Ba‐77, the 3′‐terminal 77 nucleotides of Bamboo mosaic virus (BaMV) minus‐strand RNA, and its derivatives, Ba‐39 and Ba‐77/Δ31. Nucleotides are numbered from the 3′‐end cytosine.
Figure 2.
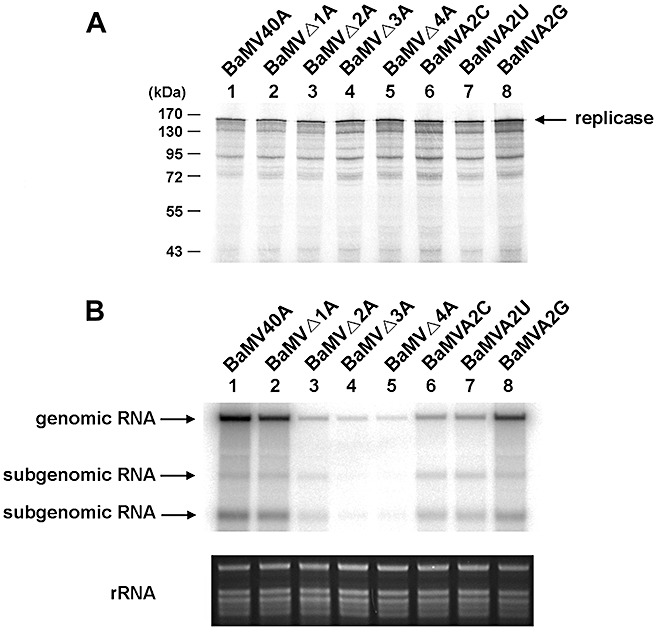
In vitro translation assay of the full‐length BaMV40A infectious RNA transcript and its derivatives, and Northern blotting analysis of their RNA accumulation in Nicotiana benthamiana protoplasts. (A) In vitro translation assay. The products of in vitro translation in wheatgerm extract were separated by 10% sodium dodecylsulphate‐polyacrylamide gel electrophoresis and analysed by a PhosphorImager (Fujifilm BAS 1500). The molecular masses of the markers are indicated on the left. The arrow indicates the position of the replicase. The smaller sized bands are nonspecific products. (B) Northern blotting analysis. Total RNA was prepared from N. benthamiana protoplasts inoculated with the RNA transcripts and probed with a 32P‐labelled RNA transcript complementary to a 0.6‐kb region of the 3′‐end of the genomic RNA. The 6.4‐kb genomic and 2.0‐ and 1.0‐kb subgenomic RNAs are indicated. The RNA transcripts used are indicated above each lane.
Table 1.
The levels of accumulation of viral products of Bamboo mosaic virus (BaMV) and its derivatives in protoplasts of Nicotiana benthamiana leaves.
| Mutant | Genomic RNA* | Genomic RNA† | Minus‐strand RNA‡ | Plus/minus§ |
|---|---|---|---|---|
| BaMV40A | 1 | 1 | 1 | 1 |
| BaMVΔ1A | 0.91 ± 0.24 | 0.95 ± 0.25 | 0.93 ± 0.25 | 1.02 |
| BaMVΔ2A | 0.02 ± 0.01 | 0.03 ± 0.02 | 0.05 ± 0.03 | 0.6 |
| BaMVΔ3A | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.01 | ND¶ |
| BaMVΔ4A | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.02 ± 0.01 | ND |
| BaMVA2C | 0.07 ± 0.05 | 0.10 ± 0.03 | 0.13 ± 0.03 | 0.77 |
| BaMVA2U | 0.07 ± 0.02 | 0.09 ± 0.02 | 0.14 ± 0.07 | 0.64 |
| BaMVA2G | 0.40 ± 0.12 | 0.45 ± 0.01 | 0.60 ± 0.01 | 0.75 |
Genomic RNA accumulation level in protoplasts, detected by Northern blotting with a probe complementary to 0.6 kb of the 3′‐end of BaMV RNA. The level of wild‐type BaMV40A genomic RNA was set at unity. All data are the average (± standard deviation) of at least three independent experiments.
Genomic RNA accumulation level in protoplasts, detected by quantitative real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR) with TaqMan probe.
Minus‐strand genomic RNA accumulation level in protoplasts, detected by qRT‐PCR with SYBR green I dye.
The plus/minus value results from the levels of genomic RNA over the minus‐strand RNA, both measured by qRT‐PCR.
ND, not determined. The level of plus‐ and minus‐strand RNA accumulation is too low to be counted effectively.
The U+2 residue of BaMV minus‐strand RNA is involved in BaMV RNA accumulation in protoplasts
Nucleotides at the 3′‐terminus of BaMV minus‐strand RNA are designated 3′‐C+1U+2U+3U+4U+5‐, as explained in Experimental procedures. The penultimate nucleotide of the minus‐strand template has been reported to play a major role in the accumulation of Brome mosaic virus (BMV) genomic and subgenomic RNA in protoplasts (Hema and Kao, 2004). Therefore, we examined whether U+2 is important for BaMV viral RNA accumulation in vivo. Mutants BaMVA2C, BaMVA2U and BaMVA2G were constructed to generate the genomic transcripts with 5′‐GCAAA‐, 5′‐GUAAA‐ and 5′‐GGAAA‐, respectively, leading to minus‐strand RNA terminating with 3′‐CGUUU‐, 3′‐CAUUU‐ and 3′‐CCUUU‐, respectively. Results of in vitro translation analysis showed that the mutation introduced at the second nucleotide in the 5′ untranslated region (UTR) of genomic RNA did not affect the efficiency of the translation (Fig. 2A). Northern blotting analysis indicated that substitution of the penultimate 3′ nucleotide U of minus‐strand RNA with the other pyrimidine C (BaMVA2G) resulted in RNA accumulation to about 40% relative to that of the wild‐type, whereas substitution with the purines A or G (BaMVA2U or BaMVA2C) resulted in only about 7% accumulation relative to that of the wild‐type in protoplasts (Table 1). As the levels of viral RNA accumulation were reduced significantly with mutations at this position when transfected into Nicotiana benthamiana protoplasts, we conclude that U+2 plays a role in the regulation of viral RNA accumulation (Fig. 2B).
Mutations introduced into the 3′‐terminus 3′‐CUUUU pentamer are defective in plus‐strand RNA synthesis
To test whether shorter Us or the penultimate mutations introduced at the terminal 3′‐CUUUU of minus‐strand RNA are specific for defects on plus‐strand RNA synthesis, we compared the accumulation ratio of plus/minus genomic RNAs. The accumulation levels of both plus‐ and minus‐strand genomic RNAs were measured by quantitative real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR; Table 1). The accumulation levels of plus‐strand genomic RNA measured by two different techniques, Northern blots with radiolabelled probe and qRT‐PCR with TaqMan probe, were almost the same (Table 1). According to the value of the plus/minus ratio (both plus and minus levels were derived from qRT‐PCR), mutants can be classified into two categories. The ratio for BaMVΔ1A was nearly identical to that for the wild‐type (Table 1). The rest of the mutants showed ratios less than that for the wild‐type. Among these, the accumulation levels of mutants BaMVΔ3A and BaMVΔ4A were too low to be counted effectively. These results indicate that the accumulation levels of minus‐strand RNA are affected by the mutations, but less so than those of plus‐strand RNAs. On the basis of these results, we conclude that mutations introduced at the 3′‐terminus of minus‐strand RNA interfere with the accumulation of plus‐strand RNA, possibly at the initiation step of RNA synthesis.
The helicase‐like domain of the BaMV replicase is responsible for binding at the promoter region
As we had previously demonstrated that the promoter sequence for minus‐strand RNA synthesis (the 3′ UTR of BaMV genomic RNA) specifically interacts with RdRp and the helicase‐like domains of replicase (Chen et al., 2003; Huang et al., 2001), we wanted to determine whether the replicase could interact with the promoter sequence for plus‐strand RNA synthesis. A UV‐crosslinking assay was undertaken to analyse these interactions. Equal amounts of the purified proteins, RdRp and the helicase domain were incubated with labelled Ba‐77, followed by UV treatment. The results indicated that both RdRp and helicase‐like domains can interact with Ba‐77 (Fig. S1A). However, the RdRp domain was shown to be degraded during purification and gave a weaker signal in the UV‐crosslinking experiment. Therefore, we focused on the interaction of the helicase‐like domain and Ba‐77 (Fig. S1).
In order to determine the specificity of the interaction between the helicase‐like domain and Ba‐77, the UV‐crosslinking condition was optimized (Fig. S1B) and the unlabelled RNAs were used as competitors in the reactions (Fig. 3). Considering the double‐stranded structures present in Ba‐77, a double‐stranded polyIC ribopolymer was included in our competition experiments, together with four single‐stranded homoribopolymers polyA, polyG, polyU and polyC. One‐fold mass excess of polyIC is sufficient to compete out 70% of the Ba‐77 probe (Fig. 3A). However, a five‐fold mass excess of polyA homopolymer is required to compete out 50% of Ba‐77. A 10‐fold mass excess of polyU and polyG and a 100‐fold mass excess of polyC are needed to compete out 50% and 30% of Ba‐77, respectively. These results suggest that the helicase‐like domain prefers binding double‐stranded rather than single‐stranded RNA.
Figure 3.
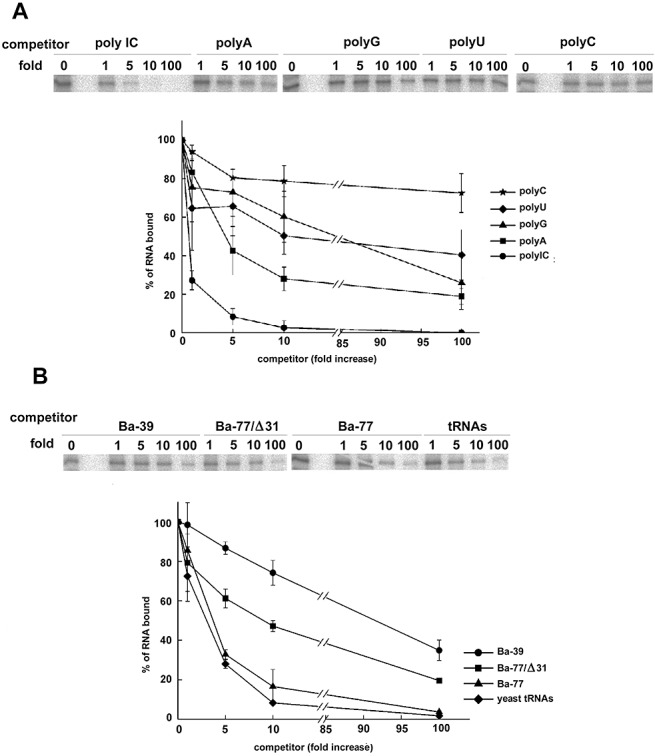
UV‐crosslinking competition analysis of the interaction between the helicase‐like domain of Bamboo mosaic virus (BaMV) replicase and Ba‐77 with unlabelled competitors. The purified helicase‐like domain (1 nmol) was pre‐incubated with unlabelled RNAs with the mass (A) or molar (B) excess amount as indicated above each lane. (A) Competitors are the unlabelled double‐stranded RNA (polyIC) and single‐stranded RNAs (polyA, polyG, polyU and polyC). (B) Competitors are the unlabelled Ba‐39 (the small stem‐loop), Ba‐77/Δ31 (the large stem‐loop), Ba‐77 and yeast tRNA. After UV‐crosslinking for 10 min, samples were digested with RNase A and then loaded onto a 12% sodium dodecylsulphate‐polyacrylamide gel. All UV‐crosslinking data presented are the average and error bars of at least three independent reactions.
To localize the region on Ba‐77 at which the helicase‐like domain binds, two competitors, Ba‐39 and Ba‐77/Δ31, derived from Ba‐77, were used in the reactions (Fig. 1). Ba‐39 is the 3′‐end 39‐nucleotide RNA of the minus‐strand containing the small stem‐loop of Ba‐77. Ba‐77/Δ31 is the other section of Ba‐77 (nucleotides 32–77) containing the large stem‐loop. Ba‐39 and Ba‐77/Δ31 require 70‐ and 10‐fold molar excess of transcripts, respectively, to compete out 50% of Ba‐77 probe, whereas only about three‐fold molar excess of unlabelled Ba‐77 is needed to compete out the same amount of probe (Fig. 3B and Table S1). These results indicate that the large stem‐loop of Ba‐77 is the major position interacting with the helicase‐like domain of BaMV replicase. The small stem‐loop, Ba‐39, could also contribute some minor binding activity. The nonspecific competitor, yeast tRNAs, showed a surprising binding capability, which was as good as that of Ba‐77 (Fig. 3B and Table S1). These results suggest that tRNAs, by virtue of their double‐stranded stems, exhibit a better interaction with the helicase‐like domain of BaMV replicase than does Ba‐77.
If the result of binding of the Escherichia coli‐expressed helicase‐like domain to the large stem is correct, it can be tested by using the entire functional replicase complex for an in vitro RdRp competition assay. Approximately 40% of the RNA products derived from Ba‐77 RNA were competed out by adding a five‐fold molar excess of Ba‐39 (Fig. 4). The template activity of Ba‐39 is barely detected even with five‐fold more template than that of Ba‐77 in the reaction. Ba‐77/Δ31, containing both the large stem‐loop and the terminal 3′‐CUUUU, reduced Ba‐77 template activity by about 60% at five‐fold molar excess. The RdRp products of Ba‐77/Δ31 can be detected in the reaction (Fig. 4). However, 70% of the Ba‐77 template activity was also competed out by yeast tRNAs, although no RNA replication was initiated from yeast tRNA in the in vitro RdRp template activity assay (Fig. 4, lanes 8 and 9). Both Ba‐77 and yeast tRNAs are rich in secondary structures, suggesting that the RdRp complex prefers to bind to stem‐loops.
Figure 4.
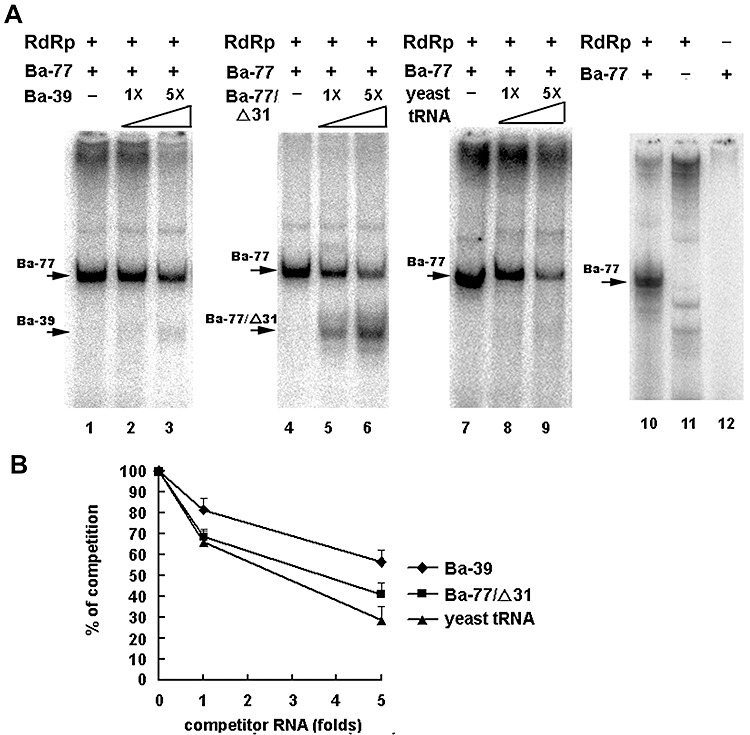
In vitro Bamboo mosaic virus (BaMV) RNA template activity assay‐based competition assay of Ba‐77 mutants. (A) Autoradiograph of BaMV RNA template activity assay with Ba‐77 alone (lanes 1, 4, 7, and 10) or in the presence of competitors Ba‐39 (lanes 2 and 3), Ba‐77/Δ31 (lanes 5 and 6) and yeast tRNA (lanes 8 and 9) at the different molar excesses indicated above each lane. Control with RNA‐dependent RNA polymerase (RdRp) alone is shown in lane 11 or Ba‐77 without RdRp in the reaction is shown in lane 12. (B) The signals in (A) were quantified and plotted as the percentage of the levels of Ba‐77 in the absence of any competitors. The data presented are the average and error bars of at least three independent reactions.
On consideration of all of these results, we propose that the replicase complex uses the helicase‐like domain to interact with the double‐stranded region and the connecting RdRp domain to recognize the terminal 3′‐CUUUU in order to initiate RNA synthesis. The nucleotides between these two cis‐acting elements, the large stem‐loop and 3′‐CUUUU, allow for appropriate steric interaction with the two domains of BaMV replicase.
The addition of the terminal 3′‐CUUUU to heterologous RNA conditionally transforms it into a template recognized by the BaMV replicase complex
Based on the model, we proposed that the BaMV RdRp complex could interact with the stem‐loop structure and the adjacent 3′‐CUUUU sequence for initiation. If a heterologous RNA fulfils this criterion, it can then be used as a template for in vitro transcription with the BaMV RdRp complex. The CMV/263 sequence with the 3′‐end 263 nucleotides containing the entire tRNA‐like structure of Cucumber mosaic virus (CMV) RNA was used to construct various mutants (Fig. 5A) to test this hypothesis. The secondary structure‐enriched CMV/263 is not a template for the BaMV replicase complex (Fig. 5C). Furthermore, no RNA accumulation was detected when the 3′‐CUUUU sequence was added to the 3′‐terminus of CMV/263 (Fig. 5C). Notably, the 3′‐end 83 nucleotides of CMV RNA were predicted to fold into a secondary structure similar to that of Ba‐77 (Fig. 5B). RNA accumulation was detected from CMV/83 + UUUUC with the 3′‐end 83 nucleotides of CMV RNA fused to the 3′‐CUUUU motif in the in vitro BaMV RdRp activity assay (Fig. 5C). Further deletion as in CMV/22 + UUUUC resulted in no RNA products (Fig. 5C). These results strengthen our hypothesis that a simple double‐stranded region can be recognized by the helicase‐like domain of replicase, and the 3′‐CUUUU motif is spaced in such a way that it corresponds collinearly with the RdRp domain of the replicase to initiate RNA synthesis.
Figure 5.
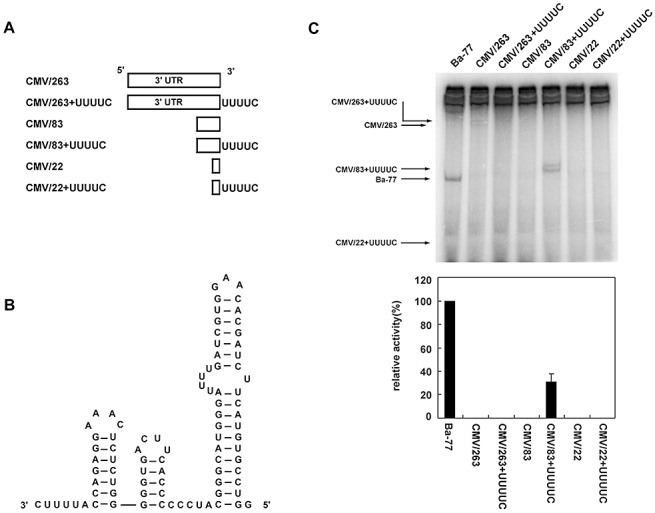
In vitro Bamboo mosaic virus (BaMV) RNA template activity assay of CMV/263‐derived mutants. (A) A diagram of the various constructs derived from the entire 3′ untransformed region of CMV RNA (CMV/263). (B) mFOLD‐predicted secondary structure of the 83‐nucleotide fragment of CMV/263 fused with an extra UUUUC sequence at the 3′‐end. (C) In vitro RNA‐dependent RNA polymerase (RdRp) template activity assay. For RNA transcripts of Ba‐77 and the CMV/263‐derived constructs, products of the BaMV RNA template activity assay were labelled with [α‐32P]UTP, separated by polyacrylamide gel electrophoresis and detected by autoradiography. Quantification of the RdRp template activity assay products. The intensities of the bands detected in (B) were quantified. The level of the Ba‐77 product was arbitrarily designated as 100 and used to normalize the other signals. Data presented are the average and error bars of at least three independent reactions.
DISCUSSION
It is believed that the 3′‐terminal region of viral RNA contains the cis‐acting elements for viral RdRp to initiate RNA synthesis (Dreher, 1999). The structural elements within the 3′‐end 77 nucleotides of BaMV minus‐strand (Ba‐77) have been shown to be important for the initiation of genomic RNA synthesis (Lin et al., 2005a). The results of the UV‐crosslinking competition assay in vitro could not clearly demonstrate whether direct interaction occurs between the E. coli‐expressed RdRp and the UUUUC pentamer (data not shown). Results from the protoplast inoculation of BaMV40A and its derivatives (BaMVA2C, BaMVA2U, BaMVA2G, BaMVΔ1A, BaMVΔ2A, BaMVΔ3A and BaMVΔ4A) suggested that U+2 and the number of Us in the 3′‐CUUUU pentamer played substantial roles in directing efficient positive‐sense RNA accumulation (most probably in RNA synthesis initiation) in plant cells (Fig. 2). The role of the terminal 3′‐CUUUU sequence in BaMV is similar to that of the CCA box in Turnip yellow mosaic virus (TYMV) RNA, which can be recognized by TYMV, TCV and Qβ RdRp to initiate RNA synthesis in vitro without any specific upstream sequence (Yoshinari et al., 2000). However, the mechanism of initiation of plus‐strand RNA synthesis in BaMV is much more similar to that in BMV, whose CCA box requires an upstream‐specific element, namely stem C, for RNA synthesis (Chapman and Kao, 1999). Furthermore, viral RNA accumulation in protoplasts was hindered by shortening the distance between stem C and the CCA box, but not by inserting a short sequence in BMV RNA (Hema and Kao, 2004), suggesting that a spacer of suitable length between these two elements is required for efficient RNA synthesis. The two similar key elements in BaMV, 3′‐CUUUU and the upstream large stem‐loop, also required a suitable spacing to initiate plus‐strand RNA synthesis efficiently (Lin et al., 2005a). This hypothesis is supported by results from this study that CMV/83 + UUUUC, predicted to form a structure similar to that of Ba‐77 (1, 5), is the only construct derived from heterologous viral RNA that gives rise to viral RNA accumulation in an in vitro template activity assay (Fig. 5C). CMV/263 + UUUUC, the tRNA‐like structure fused with UUUUC at the 3′‐terminus, may fold into an unfavourable structure for recognition by the BaMV replicase complex. However, the structural modules of the shorter CMV/22 + UUUUC RNA may not be sufficiently distinct and/or stable to interact with the BaMV replicase complex. These results suggest that the size of the RNA molecule may also be important. The three RNA molecules, Ba‐77 (77 nucleotides), CMV/83 + UUUUC (88 nucleotides) and tRNAs (∼80 nucleotides), shown to be competitive in interacting with an RdRp preparation, have a similar size. Overall, the terminal 3′‐CUUUU sequence most probably serves as the initiation template for BaMV plus‐strand RNA synthesis, whereas the upstream stem‐loop structure stabilizes the protein–RNA complex and enhances the efficiency of RNA synthesis.
The promoter in the BaMV 3′‐UTR for minus‐strand RNA synthesis requires both a specific sequence and tertiary structures for efficient RNA synthesis in vitro and in vivo (Chen et al., 2003; Cheng and Tsai, 1999; Huang et al., 2001; Tsai et al., 1999). In contrast, the promoter elements for plus‐strand RNA synthesis do not possess this complicated recognition process. These simple recognition modules, a stem‐loop and a terminal pentamer, in the promoter may account, at least in part, for the efficient RNA synthesis, and result in often‐observed 10‐ to 100‐fold accumulation of plus‐strand RNA over that of minus‐strand RNA for positive‐sense RNA viruses.
In summary, we have demonstrated that the terminal 3′‐CUUUU sequence of minus‐strand RNA can plausibly serve as the initiation template for viral plus‐strand RNA synthesis by the BaMV replicase complex. In addition, the secondary structures upstream of the 3′‐CUUUU sequence also play an important role in directing efficient RNA synthesis in vitro.
EXPERIMENTAL PROCEDURES
In vitro transcription
PCR‐generated DNA templates for the in vitro transcription of Ba‐77 RNA and its derivatives were obtained using the pBaMV40A template, which contains a full‐length cDNA copy of the BaMV‐O RNA genome. BaMV 5′(+) (5′‐GAAAACCACTCCAAACGAAA‐3′) in combination with T7‐Ba77 and T7‐Ba39 (5′‐TAATACGACTCACTATAGGGTCTTTTCTTTTCTTTTCTTTTCG‐3′; nonviral sequence is shown in bold type) were used as PCR primers for Ba‐77 and Ba‐39, respectively. Primers for Ba‐77/Δ31 were BaMV 5′(+)32 (5′‐GAAAAGACGAAGAAAACAAA‐3′) and T7‐Ba77.
The 3′‐end nucleotides of BaMV minus‐strand RNA were designated as 3′‐C+1U+2U+3U+4U+5 based on the position of their complementary nucleotides in positive‐strand RNA. Primer sets for PCR to insert an extra nucleotide at the 3′‐end of C+1 were T7‐Ba77 and BaMV5′+G, +A, +T or +C [5′‐(G/A/T/C)GAAAACCACTCCACTCCAAACGAAA‐3′]. Ba‐77 mutants with abolished or restored upper stem and lower stem (Fig. 1) were obtained by a similar procedure with primer pairs BaMV5′(+)51 (5′‐GAAAACCACTCCAAACGAAAAGAAAAGAAAAGAAAAGACGAAGAAAACAAA‐3′) and BaMVT7(−)72M (5′‐TAATACGACTCACTATAGGTGCTTTGTAGGGGACAATTGGCAGTTTGTTTTCT‐3′; the changed nucleotide is shown in bold type) for Ba‐77/LM, BaMV5′(+)41R (5′‐GAAAACCACTCCAAACGAAAAGAAAAGAAAAGAAAAGCGCTAGAAAACAAA‐3′) and BaMVT7(−)72M for Ba‐77/LR, BaMV5′(+)51 and BaMVT7(−)60M (5′‐TAATACGACTCACTATAGGGCGATTGTAGGGGTGTATTGGCAGTTTGTTTTCTTCGT‐3′) for Ba‐77/UM, and BaMV5′(+)49R (5′‐GAAAACCACTCCAAACGAAAAGAAAAGAAAAGAAAAGACGAAGAAATGTAACTGCCAA‐3′) and BaMVT7(−)60R (5′‐TAATACGACTCACTATAGGGCGATTGTAGGGGTGTATTGGCAGTTACAT‐3′) for Ba‐77/UR. All PCR‐generated DNA fragments were gel‐purified and employed as templates for in vitro transcription using T7 RNA polymerase. The transcription products were gel‐purified and quantified by UV spectrophotometry.
For infectious clones with mutations at the U+5 to U+2 positions, DNA templates were generated from pBaMV40A (Chiu et al., 2002) The primers for the U+2 substitution mutants were BaMV5′/T7A1G, BaMV5′/T7A1T or BaMV5′/T7A1C [5′‐GTCTAGATAATACGACTCACTATAG(G/T/C)AAACCACTCCAAA3′] and BaMV/(−)421 (5′‐ATCTCCTCTTCTCCGGAA‐3′), and for the U+5 to U+2 deletion mutants were BaMV5′/T7Δ1A, BaMV5′/T7Δ2A, BaMV5′/T7Δ3A or BaMV5′/T7Δ4A [5′‐GCTCTAGATAATACGACTCACTATA(GAAA/GAA/GA/G)CCACTCC‐3′] and BaMV(−)421. The amplified DNA fragments were gel‐purified and cloned into the pGEM‐T Easy vector (Promega, Madison, WI, USA). The sequence of each mutant was verified before subcloning into the BspEI‐XbaI‐cut pBaMV40A vector. The four BaMV deletion mutants were designated BaMV/Δ1U, BaMV/Δ2U, BaMV/Δ3U and BaMV/Δ4U, respectively.
In vitro BaMV RNA template activity assay
Partially purified RdRp from BaMV‐infected N. benthamiana leaves (Cheng et al., 2001; Lin et al., 2005b) was used to analyse the template activity of various exogenous RNA transcripts. The exogenous RNA template activity assay was performed in a 50‐µL reaction containing 20 µL of MNase‐treated RdRp, 10 mm dithiothreitol (DTT), 3 mm MgCl2, 2 mm of ATP, CTP and GTP, 2 µm UTP, 66 µm[α‐32P]UTP (111 TBq/mmol), 300 µg of bentonite, 120 units of RNase inhibitor and 50–200 ng of RNA template (Cheng et al., 2001; Lin et al., 2005b). Reactions were incubated at 30 °C for 1 h and stopped by phenol–chloroform extraction, followed by ethanol precipitation. The RNA products were subjected to polyacrylamide gel electrophoresis and analysed by a PhosphorImager (Fujifilm BAS‐2500, Tokyo, Japan).
In vitro translation
In vitro translation was performed using the TnT Transcription/Translation System (Promega). The 12.5‐µL reaction mixtures contained 0.5 µg of plasmid, 185 KBq of L‐[35 S]‐methionine (37 TBq/mmol; Amersham, Piscatway, NJ, USA) and nuclease‐treated wheatgerm extract (Promega) supplemented with all amino acids except methionine. Incubations were carried out at 30 °C for 1 h and terminated by adding Laemmli sample buffer. The translation products were resolved by 10% sodium dodecylsulphate‐polyacrylamide gel electrophoresis. Gels were fixed, dried and analysed by a PhosphorImager (Fujifilm BAS‐2500, Tokyo, Japan).
Protoplast inoculation and viral RNA quantification
Four grams of sliced N. bethamiana leaves were digested with 25 mL of enzyme solution [0.1% bovine serum albumin, 0.6 mg/mL pectinase and 12 mg/mL cellulase in 0.55 M mannitol–2‐(N‐morpholino)ethanesulphonic acid buffer, pH 5.7] at 25 °C overnight. The mesophyll cells were isolated and transfected with 5 µg of RNA transcripts (about 4 × 105 cells for each sample) with the help of polyethyleneglycol. Finally, the transfected protoplasts were incubated under constant light at 25 °C for 48 h. For Northern blotting analysis, total RNA was extracted from protoplasts (2.5 × 105 cells), glyoxalated, electrophoresed through a 1% agarose gel and transferred to a membrane (Zeta‐Probe; Bio‐Rad, Hercules, CA, USA), as described previously (Chen et al., 2005). The hybridization probe, a 0.6‐kb 32P‐labelled RNA transcript derived from the HindIII‐linearized pBaMV‐O/SB2.6 (Huang and Tsai, 1998), is complementary to the 3′‐end of positive‐strand BaMV RNA. The banding signals were scanned and quantified using a PhosphorImager (Fujifilm BAS‐2500, Tokyo, Japan).
The qRT‐PCR technique was used to detect both BaMV plus‐ and minus‐strand genomic RNAs. The cDNA synthesis reaction was performed according to the manufacturer's instructions for SuperScript® III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) with the primers BaMV‐2002 (5′‐ATGTATCACGGAAATAAGAGTT‐3′) and BaMV+51 (5′‐ACTGCCAATTGTCCCCTACA‐3′) for the plus‐ and minus‐strand, respectively. qPCR for BaMV genomic RNA detection, the specific primers BaMV+1 (5′‐GAAAACCACTCCAAACGAAA‐3′) and BaMV‐282 (5′‐TGTGCTGAACGGGTTATGAG‐3′), and TaqMan probe BaMV+85 (5′‐TGCGGCAACAATGGCACTCGT‐3′), with the fluorescent dyes 5‐carboxyfluorescein on the 5′‐end and Black Hole Quencher‐1 on the 3′‐end, were designed by the software Primer 3 with the sequence of BaMV40A. For minus‐strand genomic RNA detection, primers BaMV+1766 (5′‐CACATCCGGCACTTACCA‐3′) and BaMV‐2002 were used in the reaction containing a 20 000 times dilution of SYBR green I (Cambrex Bio Science Rockland Inc., ME, USA). qPCR was performed in 0.2‐mL PCR tubes with 0.6 µm (for minus‐strand) or 0.9 µm (for plus‐strand) of each primer, 0.2 mm of each deoxynucleoside triphosphate (dNTP), 10 mm Tris‐HCl (pH 8.8), 1.5 mm MgCl2, 50 mm KCl, 0.1% Triton X‐100, 4 µL of cDNA, 3 units of Taq DNA polymerase (Promega) and RNase‐free water up to a final volume of 20 µL. The reaction for plus‐strand also contained 312.5 nm of TaqMan probe. Cycling conditions began with an initial hold at 95 °C for 5 min, followed by 35 cycles consisting of 94 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s. Reactions were carried out in a RotorGene 3000 (Corbett Research, Sydney, Australia) with data acquisition at 72 °C on the channel with excitation at 470 nm and detection at 585 nm using a high‐pass filter for both plus‐ and minus‐strand. All samples were run at least twice, and the reactions without template or reverse transcriptase were performed as negative controls.
UV‐crosslinking and competition assay
The purified proteins (RdRp or helicase‐like domain) were incubated with labelled Ba‐77 for 5 min at room temperature in binding buffer (25 mm Tris‐HCl, pH 8.0, 175 mm NaCl, 5% glycerol), and then irradiated with a 254‐nm‐wavelength UV lamp (Stratagene, LaJolla, CA, USA, UV stratalinker TM 1800) on ice for 10 min. After irradiation, the samples were treated with 40 µg of boiled RNase A for 20 min at 37 °C, boiled in Laemmli buffer and electrophoresed on a 12% sodium dodecylsulphate‐polyacrylamide gel. In the competition reactions, various amounts of unlabelled competitor RNAs were pre‐incubated with the proteins for 10 min prior to the addition of 32P‐labelled RNA probe.
Supporting information
Fig. S1 UV‐crosslinking assay of the purified proteins with the Ba‐77 RNA probe. (A) The same amount of purified glutathione transferase (GST)‐fused RdRp (indicated as RdRp) and helicase‐like domain (indicated as helicase) shown above each lane was UV‐crosslinked with 32P‐labelled Ba‐77 RNA (190 fmol). After treatment with RNase A, the labelled proteins were resolved on a 12% sodium dodecylsulphate‐polyacrylamide gel and analysed using a PhosphorImager. The arrows indicate the position of crosslinked proteins. In addition to the full‐length GST‐fusion RdRp protein, the degradation product of lower molecular weight was also linked. (B) Optimization for the UV‐crosslinking reaction was examined with various salt concentrations indicated above each lane. The condition used in the rest of the UV‐crosslinking experiments was 175 mm NaCl which shows less background.
Table S1 The binding activity of the Escherichia coli‐expressed helicase‐like domain of Bamboo mosaic virus (BaMV) replicase with Ba‐77 probe and competitors.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
ACKNOWLEDGEMENTS
We are grateful to Dr Pei‐Yu Lee at the Institute of Medical Biotechnology, Central Taiwan University of Sciences and Technology for editorial help. This work was supported by grants from the National Science Council (NSC 94‐2752‐B‐005‐013‐PAE and 95‐2752‐B‐005‐012‐PAE).
REFERENCES
- Ahlquist, P. , Noueiry, A.O. , Lee, W.M. , Kushner, D.B. and Dye, B.T. (2003) Host factors in positive‐strand RNA virus genome replication. J. Virol. 77, 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos, P. (1988) A sequence motif in many polymerases. Nucleic Acids Res. 16, 9909–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, M.R. and Kao, C.C. (1999) A minimal RNA promoter for minus‐strand RNA synthesis by the Brome mosaic virus polymerase complex. J. Mol. Biol. 286, 709–720. [DOI] [PubMed] [Google Scholar]
- Chen, I.H. , Chou, W.J. , Lee, P.Y. , Hsu, Y.H. and Tsai, C.H. (2005) The AAUAAA motif of Bamboo mosaic virus RNA is involved in minus‐strand RNA synthesis and plus‐strand RNA polyadenylation. J. Virol. 79, 14555–14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, I.H. , Meng, M. , Hsu, Y.H. and Tsai, C.H. (2003) Functional analysis of the cloverleaf‐like structure in the 3′ untranslated region of bamboo mosaic potexvirus RNA revealed dual roles in viral RNA replication and long distance movement. Virology 315, 415–424. [DOI] [PubMed] [Google Scholar]
- Cheng, C.P. and Tsai, C.H. (1999) Structural and functional analysis of the 3′ untranslated region of bamboo mosaic potexvirus genomic RNA. J. Mol. Biol. 288, 555–565. [DOI] [PubMed] [Google Scholar]
- Cheng, J.H. , Ding, M.P. , Hsu, Y.H. and Tsai, C.H. (2001) The partial purified RNA‐dependent RNA polymerases from bamboo mosaic potexvirus and potato virus X infected plants containing the template‐dependent activities. Virus. Res. 80, 41–52. [DOI] [PubMed] [Google Scholar]
- Chiu, W.W. , Hsu, Y.H. and Tsai, C.H. (2002) Specificity analysis of the conserved hexanucleotides for the replication of bamboo mosaic potexvirus RNA. Virus Res. 83, 159–167. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W. (1999) Functions of the 3′‐untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37, 151–174. [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A.E. and Koonin, E.V. (1989) Viral proteins containing the purine NTP‐binding sequence pattern. Nucleic Acids. Res. 17, 8413–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hema, M. and Kao, C.C. (2004) Template sequence near the initiation nucleotide can modulate Brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 78, 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman, T.C. (1988) A new superfamily of replicative proteins. Nature 333, 22–23. [DOI] [PubMed] [Google Scholar]
- Huang, C.Y. and Tsai, C.H. (1998) Evolution of Bamboo mosaic virus in a nonsystemic host results in mutations in the helicase‐like domain that cause reduced RNA accumulation. Virus Res. 58, 127–136. [DOI] [PubMed] [Google Scholar]
- Huang, C.Y. , Huang, Y.L. , Meng, M. , Hsu, Y.H. and Tsai, C.H. (2001) Sequences at the 3′ untranslated region of bamboo mosaic potexvirus RNA interact with the viral RNA‐dependent RNA polymerase. J. Virol. 75, 2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.L. , Han, Y.T. , Chang, Y.T. , Hsu, Y.H. and Meng, M. (2004) Critical residues for GTP methylation and formation of the covalent m7GMP‐enzyme intermediate in the capping enzyme domain of Bamboo mosaic virus . J. Virol. 78, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.L. , Hsu, Y.H. , Han, Y.T. and Meng, M. (2005) mRNA guanylation catalyzed by the S‐adenosylmethionine‐dependent guanylyltransferase of Bamboo mosaic virus . J. Biol. Chem. 280, 13153–13162. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. (1991) The phylogeny of RNA‐dependent RNA polymerases of positive‐strand RNA viruses. J. Gen. Virol. 72, 2197–2206. [DOI] [PubMed] [Google Scholar]
- Li, Y.I. , Chen, Y.J. , Hsu, Y.H. and Meng, M. (2001a) Characterization of the AdoMet‐dependent guanylyltransferase activity that is associated with the N terminus of Bamboo mosaic virus replicase. J. Virol. 75, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Cheng, Y.M. , Huang, Y.L. , Tsai, C.H. , Hsu, Y.H. and Meng, M. (1998) Identification and characterization of the Escherichia coli‐expressed RNA‐dependent RNA polymerase of Bamboo mosaic virus . J. Virol. 72, 10093–10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.I. , Shih, T.W. , Hsu, Y.H. , Han, Y.T. , Huang, Y.L. and Meng, M. (2001b) The helicase‐like domain of plant potexvirus replicase participates in formation of RNA 5′ cap structure by exhibiting RNA 5′‐triphosphatase activity. J. Virol. 75, 12114–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.W. , Chiu, H.N. , Chen, I.H. , Chen, T.C. , Hsu, Y.H. and Tsai, C.H. (2005a) Structural and functional analysis of the cis‐acting elements required for plus‐strand RNA synthesis of Bamboo mosaic virus . J. Virol. 79, 9046–9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.W. , Hsu, Y.H. and Tsai, C.H. (2005b) Characterization of the infectivity of Bamboo mosaic virus with its correlation to the in vitro replicase activities in Nicotiana benthamiana . Virus Res. 112, 77–84. [DOI] [PubMed] [Google Scholar]
- Panavas, T. , Stork, J. and Nagy, P.D. (2006) Use of double‐stranded RNA templates by the tombusvirus replicase in vitro: implications for the mechanism of plus‐strand initiation. Virology 352, 110–120. [DOI] [PubMed] [Google Scholar]
- Tsai, C.H. , Cheng, C.P. , Peng, C.W. , Lin, B.Y. , Lin, N.S. and Hsu, Y.H. (1999) Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J. Virol. 73, 2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari, S. , Nagy, P.D. , Simon, A.E. and Dreher, T.W. (2000) CCA initiation boxes without unique promoter elements support in vitro transcription by three viral RNA‐dependent RNA polymerases. RNA 6, 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 UV‐crosslinking assay of the purified proteins with the Ba‐77 RNA probe. (A) The same amount of purified glutathione transferase (GST)‐fused RdRp (indicated as RdRp) and helicase‐like domain (indicated as helicase) shown above each lane was UV‐crosslinked with 32P‐labelled Ba‐77 RNA (190 fmol). After treatment with RNase A, the labelled proteins were resolved on a 12% sodium dodecylsulphate‐polyacrylamide gel and analysed using a PhosphorImager. The arrows indicate the position of crosslinked proteins. In addition to the full‐length GST‐fusion RdRp protein, the degradation product of lower molecular weight was also linked. (B) Optimization for the UV‐crosslinking reaction was examined with various salt concentrations indicated above each lane. The condition used in the rest of the UV‐crosslinking experiments was 175 mm NaCl which shows less background.
Table S1 The binding activity of the Escherichia coli‐expressed helicase‐like domain of Bamboo mosaic virus (BaMV) replicase with Ba‐77 probe and competitors.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


