SUMMARY
Cyclic di‐GMP [(bis‐(3′–5′)‐cyclic di‐guanosine monophosphate)] is an almost ubiquitous second messenger in bacteria that is implicated in the regulation of a range of functions that include developmental transitions, aggregative behaviour, adhesion, biofilm formation and virulence. Comparatively little is known about the mechanism(s) by which cyclic di‐GMP exerts these various regulatory effects. PilZ has been identified as a cyclic di‐GMP binding protein domain; proteins with this domain are involved in regulation of specific cellular processes, including the virulence of animal pathogens. Here we have examined the role of PilZ domain proteins in virulence and the regulation of virulence factor synthesis in Xanthomonas campestris pv. campestris (Xcc), the causal agent of black rot of crucifers. The Xcc genome encodes four proteins (XC0965, XC2249, XC2317 and XC3221) that have a PilZ domain. Mutation of XC0965, XC2249 and XC3221 led to a significant reduction of virulence in Chinese radish. Mutation of XC2249 and XC3221 led to a reduction in motility whereas mutation of XC2249 and XC0965 affected extracellular enzyme production. All mutant strains were unaffected in biofilm formation in vitro. The reduction of virulence following mutation of XC3221 could not be wholly attributed to an effect on motility as mutation of pilA, which abolishes motility, has a lesser effect on virulence.
The production of virulence determinants by pathogenic bacteria is strictly regulated and probably occurs as an adaptation to particular environmental changes. There is a great deal of interest in the signals that activate virulence gene expression and in the processes of signal perception and signal transduction. Cyclic di‐GMP [bis‐(3′–5′)‐cyclic di‐guanosine monophosphate] is a novel second messenger in bacteria that was first described as an allosteric activator of cellulose synthase in Gluconacetobacter xylinus (Ross et al., 1987). It is now established that this nucleotide regulates a range of functions in diverse bacteria, including the virulence of bacterial pathogens of animals and plants (for recent reviews see Jenal and Malone, 2006; Römling et al., 2005; Römling and Amikam, 2006; Ryan et al., 2006b). Cellular levels of cyclic di‐GMP are controlled through synthesis, catalysed by the GGDEF protein domain, and degradation by EAL or HD‐GYP domains (Christen et al., 2005; Paul et al., 2004; Ryan et al., 2006a; Ryjenkov et al., 2005; Schmidt et al., 2005; Tamayo et al., 2005). Proteins with GGDEF, EAL and HD‐GYP domains are involved in the regulation of bacterial properties that include developmental transitions, aggregative behaviour, adhesion and biofilm formation in addition to synthesis of virulence factors and virulence.
Comparatively little is known regarding the mechanisms by which cyclic di‐GMP exerts its diverse actions within the bacterial cell. Bioinformatics studies carried out by Amikam and Galperin (2006) identified the PilZ domain, named after the PilZ protein from Pseudomonas aeruginosa, as a candidate player in cyclic di‐GMP signalling mechanisms. PilZ domain proteins from a number of bacteria have been shown to bind cyclic di‐GMP and in some cases have been implicated in the regulation of specific cellular processes such as motility, polysaccharide synthesis, biofilm formation and virulence of animal pathogens (Benach et al., 2007; Christen et al., 2007; Merighi et al., 2007; Pratt et al., 2007; Ramelot et al., 2007; Ryjenkov et al., 2006). It is unlikely that PilZ domain proteins mediate all of the cellular effects of cyclic di‐GMP however; PelD of Pseudomonas aeruginosa is a cyclic di‐GMP receptor that mediates regulation of polysaccharide (PEL) biosynthesis but lacks a recognizable PilZ domain (Lee et al., 2007).
The aim of the present study was to expand our understanding of cyclic di‐GMP signalling in plant pathogenesis by addressing the role of PilZ domain proteins in Xanthomonas campestris pv. campestris (Xcc), the causal agent of black rot in crucifers. We have previously examined the contribution of all proteins with GGDEF, EAL or HD‐GYP domains to virulence and virulence factor production in Xcc (Ryan et al., 2007). Genes with significant roles in virulence to plants include those encoding proteins whose probable function is in cyclic di‐GMP synthesis as well as others, including the HD‐GYP domain regulator RpfG, implicated in cyclic di‐GMP degradation. Mutation of rpfG in Xcc has pleiotropic effects, leading to significant reduction in the synthesis of the extracellular cell‐wall‐degrading enzymes protease and endoglucanase, reduction in motility and enhanced biofilm formation in rich medium (Dow et al., 2003; Ryan et al., 2007; Slater et al., 2000). Other GGDEF‐EAL domain proteins influence motility but have no effect on virulence factor synthesis (Ryan et al., 2007). Here we examine the role of the different PilZ domain proteins of Xcc in controlling these various phenotypes.
The Xcc genome encodes four proteins with a PilZ domain (Amikam and Galperin, 2006); XC0965, XC2249 and XC3221 comprise ‘stand‐alone’ domains whereas XC2317 has an additional N‐terminal YcgR domain. These proteins show no significant amino acid sequence similarity with each other on pairwise searches using BLASTP. Nevertheless, in all cases proteins with related amino acid sequence are found in other bacteria, although for XC2249 these homologues only occur in the proteomes of other xanthomonads (data not shown). Searches against a protein fold library using the FUGUE algorithm (Shi et al., 2001) showed predicted structural homology of all Xcc proteins with the PilZ domain proteins VCA0042 from Vibrio cholerae (PDB 1yln) and PP4397 from Pseudomonas putida (PDB 2gjg) at above the 99% confidence level.
The functions of XC0965, XC2249, XC2317 and XC3221 were examined using mutant strains in which the genes were disrupted using either Tn5gusA (XC2249, XC3221), or pK18mobsac carrying an internal fragment of the gene amplified by PCR (XC0965, XC2317) (Table 1). XC2317 and XC2449 are predicted to be in single transcriptional units whereas XC0965 and XC3221 are the most downstream genes in the predicted XC0960–XC0965 and XC3224–XC3221 operons (Qian et al., 2005). This indicates that mutation of the genes under investigation is unlikely to have polar effects. For complementation studies, genes were amplified using the PCR primers given in Table 1 (which also have sites for BamHI and HindIII) and cloned into the TOPO TA vector (Invitrogen). After verification by sequencing, inserts were excised with BamHI and HindIII and were ligated into pBBR1MCS (Kovach et al., 1995) cut with the same enzymes to create pXC0965, pXC2249, pXC2317 and pXC3221. This orientation allowed expression of the inserted gene in these constructs to be driven by the vector (lac) promoter. Constructs were introduced into Xcc strains by triparental mating.
Table 1.
Strains, plasmids and primers used in this study.
| Strain/plasmid | Genotype/Phenotype | Reference |
|---|---|---|
| X. campestris pv. campestris 8004 | Wild‐type RifR | Qian et al. (2005) |
 rpfG
rpfG
|
rpfG deletion mutant of 8004, RifR | Slater et al. (2000) |
 hrpG
hrpG
|
hrpG deletion mutant of 8004, RifR; KmR | Xu et al. (2008) |
 hrpX
hrpX
|
hrpX deletion mutant of 8004, RifR; KmR | Xu et al. (2008) |
| pilA | XC3823::Tn5gusA5; RifR; TcR | Ryan et al. (2007) |
| XC2317 | pK18mobkan, XC2317; RifR; KmR | This study |
| XC2249 | XC2249::Tn5gusA5; RifR; TcR | This study |
| XC3221 | XC3221::Tn5gusA5; RifR; TcR | This study |
| XC0965 | pK18mobkan, XC0965; RifR; KmR | This study |
| XC0965 (pXC0965) | Complemented mutant ClR; RifR; KmR | This study |
| XC2317 (pXC2317) | Complemented mutant ClR; RifR; KmR | This study |
| XC2249 (pXC2249) | Complemented mutant ClR; RifR; KmR | This study |
| XC3221 (pXC3221) | Complemented mutant ClR; RifR; KmR | This study |
| Primers sets used in construction of clones for complementation (5′–3′) | ||
| Target gene | Forward | Reverse |
| XC0965 | GGATCC ATGATCCAGGACACCCGCC | AAGGCTGAGCATCTGGTCTAG |
| XC2249 | GGATCCGTGCAGCGCATGGACGCCAA | AAGGCTTTCAGCGCTGGCGACGGGCG |
| XC2317 | GGATCCGTGCTTGTGCCCATGTCCGAA | AAGGCTTCAGAACACGCCGCTCTT |
| XC3221 | GGATCCATGAGTGCAATGAATGCACG | AAGCTTTTACATCGTGTGGGTCGGCTTG |
| RT‐PCR primer sets (5′–3′) | ||
| Target gene | Forward | Reverse |
| XC0965 | CTGTATCAGCTGCGGTTCG | AGATGCTCTTCCGCAATCC |
| XC2249 | CCAAACTCGACCTCATCCTG | ACAATTCAGGAAGCCAGTCC |
| XC2317 | CATCGAACAGGCCAAGTACC | GAATCGGTGATGGGTGTTTC |
| XC3221 | GCGATGAGGTGTTTCTGCTT | GCCAGCAGTGTTTCGATCTT |
| 16S rDNA | GTCAGGAGTCGGTGCTCAGT | CCTTCCAGATCGACCCAGTA |
Key: Cl, chloramphenicol; Rif, rifampicin; Km, kanamycin; Tc: tetracycline.
Restriction enzyme sites for BamHI or HindIII within the primer sequences are underlined.
The virulence of each mutant was tested by measurement of the lesion length after bacteria were introduced into the vascular system of Chinese radish by leaf clipping as previously described (Dow et al., 2003; Ryan et al., 2007). Each strain was tested on 60 leaves (20 leaves for each strain with three independent replicates). Although mutation of XC2317 had no effect, strains with mutations in XC0965, XC2249 and XC3221 had significantly reduced virulence compared with the wild‐type (Fig. 1A,B). Introduction of the cloned genes into the respective mutants restored virulence to wild‐type levels (Fig. 1).
Figure 1.
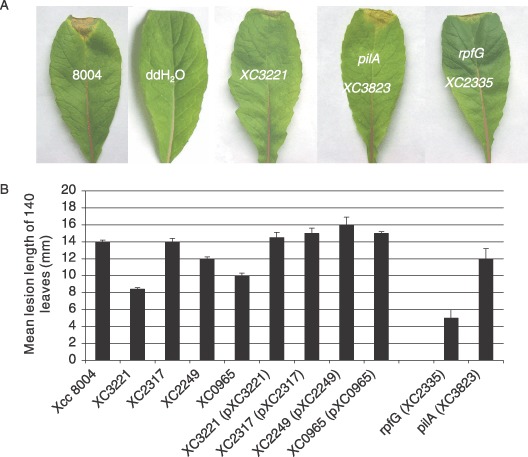
The effects of mutation of genes encoding the PilZ domain proteins XC0965, XC2249, XC2317 and XC3221 on the virulence of Xcc to Chinese radish. The virulence of each mutant was tested by measurement of the lesion length after bacteria were introduced into the vascular system of Chinese radish by leaf clipping. Values given are the means and SD of 60 measurements. Also shown are the effects of mutation of rpfG, which encodes an HD‐GYP domain regulatory protein and pilA encoding the major pilin. (A) Representative virulence assays for (from left to right) Xcc wild‐type strain 8004, negative control (H2O), XC3221::Tn5gusA5 mutant, pilA mutant (XC3823::Tn5gusA5) and rpfG deletion mutant. (B) Mutation of XC3221, XC2249 and XC0965 gave a significant reduction in virulence in repeated tests, although mutation of XC2317 had no effect. Introduction of the cloned genes restores virulence of these mutants to wild‐type levels, but had no influence on the XC2317 mutant, which retained wild‐type virulence.
These studies were extended to examine the influence of mutation on specific factors associated with Xcc virulence including extracellular enzyme production, motility and biofilm formation. Levels of β‐1,4‐endoglucanase from culture supernatants were estimated by radial diffusion assays using carboxymethylcellulose as substrate. For these experiments bacteria were grown to an OD at 600 nm of 2.0 in NYGB medium. Relative protease production was measured by growth of colonies on NYG agar plates supplemented with skimmed milk (Ryan et al., 2007; Slater et al., 2000). Mutation of XC2317 led to a small but reproducible reduction in protease production, although mutation of XC0965, XC2249 or XC3221 had no apparent effect (Fig. 2B). Mutation of XC0965 and XC2317 led to a reduction in endoglucanase levels, although mutation of XC2249 or XC3221 had no effect (Fig. 2C). These effects of XC0965 and XC2317 mutation on extracellular enzyme production were much less pronounced, however, than those seen after mutation of rpfG (Fig. 2C; Slater et al., 2000). Introduction of the cloned XC0965 or XC2317 genes into the respective mutant restored production of extracellular enzymes to wild‐type levels (Fig. 2C), whereas introduction of these constructs into the wild‐type had no effect (data not shown). Motility of different strains was assessed on 0.6% Eiken agar plates as described by Ryan et al. (2007). Mutation of XC2249 and XC3221 but not XC0965 and XC2317 negatively affected motility (Fig. 3). Introduction of the cloned XC2249 or XC3221 gene into the respective mutant restored the wild‐type phenotype (Fig. 3). However, introduction of the cloned XC2249 gene into the XC3221 mutant or the cloned XC3221 gene into the XC2249 mutant had no effect. Taken together these findings suggest that different PilZ domain proteins have independent action in the regulation of motility.
Figure 2.
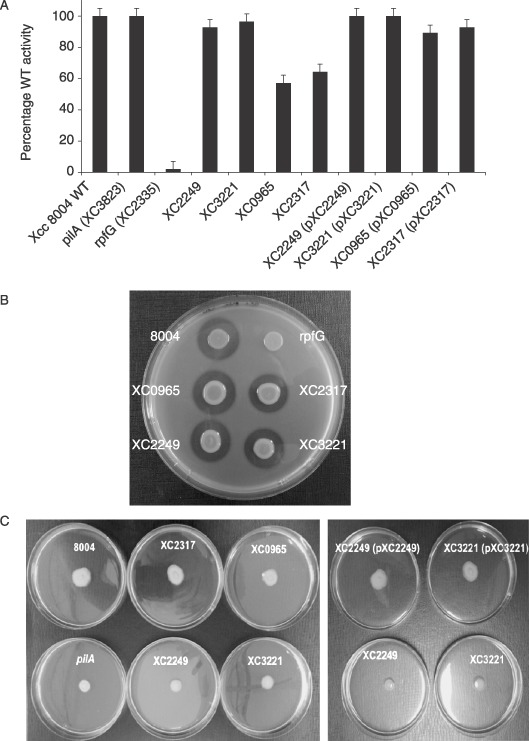
Mutation of genes encoding PilZ domain proteins has effects on motility and the synthesis of extracellular enzyme virulence factors by Xcc. (A) XC0965 and XC2317 mutants have lower levels of extracellular endoglucanase than wild‐type; the complemented strains XC0965/pXC0965 and XC2317/pXC2317 have wild‐type levels. The effects of mutation of XC0965 and XC2317 are much less than those seen after mutation of rpfG (which is XC2335), whereas mutation of pilA (which is XC3823) has no effect on endoglucanase activity. (B) Mutation of XC2317 led to a small reduction in protease production whereas mutation of XC0965, XC2249 and XC3221 had no apparent effect. (C) Mutation of XC2249 and XC3221 influence motility on 0.5% Eiken agar‐NYG plates. These effects are reversed by complementation with the cloned XC2249 and XC3221 genes respectively.
Figure 3.
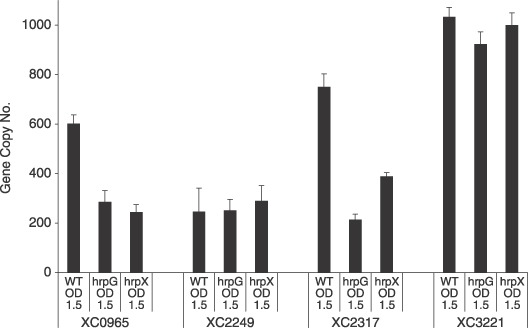
Effect of mutation of hrpG and hrpX on the expression of genes encoding PilZ domain proteins. Transcript levels of all genes were measured by RT‐PCR. Bacterial strains were grown to an OD at 600 nm of 1.5 in MME medium before RNA extraction. All RT‐PCR results were normalized using the Ct values obtained for the 16S rRNA amplifications run in the same plate. The relative amounts of selected genes and relative levels of gene transcripts were determined by a standard curve (i.e. Ct values plotted against logarithm of the DNA copy number). The results shown are averages of three repetitions.
Wild‐type Xcc 8004 does not form biofilms in liquid L medium although the rpfG mutant does (Dow et al., 2003). Ectopic expression in wild‐type Xcc of wspR19, which encodes a GGDEF domain protein from Pseudomonas fluorescens (Aldridge et al., 2003), gives a phenocopy of the rpfG mutant with increased cellular levels of cyclic di‐GMP, increased biofilm formation and aggregation in liquid culture, decreased motility and decreased production of extracellular enzymes (Ryan et al., 2006a; and data not shown). To examine any potential role of PilZ domain proteins in biofilm formation, wspR19 cloned in pVSP61 (Table 1) was introduced into the wild‐type and panel of mutants. Expression of wspR19 promoted biofilm formation equally in the wild‐type and all the mutant strains (data not shown). As expected, expression of wspR19 reduced endoglucanase levels and motility in wild‐type Xcc. Similar effects on endoglucanase production were seen in all of the mutant backgrounds, and all strains carrying wspR19 showed no motility. These findings suggest that none of the PilZ domain proteins individually has a significant role in the repression of extracellular enzyme production and motility by elevated cellular levels of cyclic di‐GMP.
In some cases genes encoding cyclic di‐GMP signalling systems with a role in disease may only be expressed within the eukaryotic host (Osorio et al., 2005). We have previously shown that HrpG, the master regulator of the hrp regulon (for hypersensitive response and pathogenicity), significantly influences expression of pdeA, encoding a GGDEF‐EAL domain protein that contributes to Xcc virulence. The hrpG gene is expressed in minimal media, shows increased expression in the plant environment but is repressed in rich media (Wengelnik et al., 1996, 1999). HrpG also affects the expression of hrpX, which encodes an AraC‐type transcriptional activator that regulates expression of a subset of genes within the hrp operon. Comparison was made of the expression levels of XC0965, XC2249, XC2317 and XC3221 in wild‐type, hrpG and hrpX deletion mutants (Xu et al., 2008) grown in MME minimal medium using RT‐PCR. Primers for RT‐PCR are given in Table 1 and procedures and medium composition are given in Ryan et al. (2007). The findings indicated that both HrpG and HrpX significantly influenced expression of XC0965 and XC2317 (Fig. 3). This raises the possibility that expression of these particular genes may be elevated in the plant environment.
Mutation of XC3221, XC2249 and XC0965 affects the virulence of Xcc, suggesting that there is no redundancy of action between these ‘stand‐alone’ PilZ domain proteins, which also do not show any significant amino acid sequence homology (see above). As mutation of XC2249 had a substantial effect on motility, we examined the relationship between motility and virulence in Xcc. The form of motility observed on Eiken agar plates is pilus‐dependent and is abolished by mutation of pilA (Ryan et al., 2007). Furthermore, wild‐type Xcc 8004 does not show swimming motility, which is flagellum‐dependent, when assessed in 0.1% (w/v) agar. Mutation of pilA led to a reduction in virulence similar to that seen after mutation of XC2249 (Fig. 1A,B). This indicates the possibility that the effects of mutation of XC2249 on virulence could be due to an effect on motility. In contrast, mutation of XC3221, which has only a small effect on motility in vitro, has a larger effect on virulence than mutation of pilA (Fig. 1B). This suggests that XC3221 influences further virulence functions that remain obscure. Mutation of XC0965 and XC2317 did not affect motility but each had a modest effect on production of extracellular enzymes in vitro. It is plausible that this effect may be more pronounced in planta, as both of these genes are under the regulation of HrpG and HrpX (Fig. 3). However, despite this co‐regulation and apparent similarities in the phenotypes of the mutants, only mutation of XC0965 had an effect on virulence.
Although a number of reports have described the role of PilZ domain proteins in regulation of factors associated with bacterial virulence, only in a few cases has the role of these proteins been directly examined in virulence models. Pratt and colleagues have shown that a subset of PilZ domain proteins has been shown to contribute to colonization of infant mouse small intestine by Vibrio cholerae (Pratt et al., 2007). Of particular relevance to the present work, Wang et al. (2007) identified through a transposon mutant screen that the homologue of XC3221 in the rice pathogen Xanthomonas oryzae pv. oryzicola contributes to virulence. Our findings thus contribute to a growing body of work that establishes a role in virulence for cyclic di‐GMP signal transduction via PilZ domain proteins. The mechanisms by which these various PilZ domain proteins exert their action, however, remains largely unknown and this topic, together with other aspects of cyclic di‐GMP signal transduction, are likely to be the focus of a considerable future research effort.
ACKNOWLEDGMENTS
This work was supported by Principal Investigator Awards from the Science Foundation of Ireland (to J.M.D.), grants from the National Natural Science Foundation of China Key Programme (30130010) and The National High Technology Research and Development Programme of China (2001AA223051 and 2004AA223060) (to J.L.T.) and by the China‐Ireland Science and Technology Research Collaboration Programme (to J.M.D. and J.L.T.).
REFERENCES
- Aldridge, P. , Paul, R. , Goymer, P. , Rainey, P. and Jenal, U. (2003) Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus . Mol. Microbiol. 47, 1695–1708. [DOI] [PubMed] [Google Scholar]
- Amikam, D. and Galperin, M.Y. (2006) PilZ domain is part of the bacterial c‐di‐GMP binding protein. Bioinformatics, 22, 3–6. [DOI] [PubMed] [Google Scholar]
- Benach, J. , Swaminathan, S.S. , Tamayo, R. , Handelman, S.K. , Folta‐Stogniew, E. , Ramos, J.E. , Forouhar, F. , Neely, H. , Seetharaman, J. , Camilli, A. and Hunt, J.F. (2007) The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26, 5153–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen, M. , Christen, B. , Folcher, M. , Schauerte, A. and Jenal, U. (2005) Identification and characterization of a cyclic di‐GMP‐specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–30837. [DOI] [PubMed] [Google Scholar]
- Christen, M. , Christen, B. , Allan, M.G. , Folcher, M. , Jeno, P. , Grzesiek, S. and Jenal, U. (2007) DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter cresentus. Proc. Natl Acad. Sci. USA, 104, 4112–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, J.M. , Crossman, L. , Findlay, K. , He, Y.Q. , Feng, J.X. , and Tang, J.L. (2003) Biofilm dispersal in Xanthomonas campestris is controlled by cell‐cell signaling and is required for full virulence to plants. Proc. Natl Acad. Sci. USA, 100, 10995–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal, U. and Malone, J. (2006) Mechanisms of cyclic‐di‐GMP signaling in bacteria. Annu. Rev. Genet. 40, 385–407. [DOI] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. 2nd and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Lee, V.T. , Matewish, J.M. , Kessler, J.L. , Hyodo, M. , Hayakawa, Y. and Lory, S. (2007) A cyclic‐di‐GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65, 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi, M. , Lee, V.T. , Hyodo, M. , Hayakawa, Y. and Lory, S. (2007) The second messenger bis‐(3–5)‐cyclic‐GMP and its PilZ domain‐containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895. [DOI] [PubMed] [Google Scholar]
- Osorio, C.G. , Crawford, J.A. , Michalski, J. , Martinez‐Wilson, H. , Kaper, J.B. and Camilli, A. (2005) Second‐generation recombination‐based in vivo expression technology for large‐scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73, 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, R. , Weiser, S. , Amiot, N.C. , Chan, C. , Schirmer, T. , Giese, B. and Jenal, U. (2004) Cell cycle‐dependent dynamic localization of a bacterial response regulator with a novel di‐guanylate cyclase output domain. Genes Dev. 18, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, J.T. , Tamayo, R. , Tischler, A.D. and Camilli, A. (2007) PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282, 12860–12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W. , Jia, Y. , Ren, S.X. , He, Y.Q. , Feng, J.X. Lu, L.F. , Sun, Q.H. , Ying, G. , Tang, D. J. , Tang, H. , Wu, W. , Hao, P. , Wang, L. , Jiang, B.L. , Zeng, S. , Gu, W.Y. , Lu, G. , Rong, L. , Tian, Y. , Yao, Z. , Fu, G. , Chen, B. , Fang, R. , Qiang, B. , Chen, Z. , Zhao, G.P. , Tang, J.L. and He, C. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris . Genome Res. 15, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot, T.A. , Yee, A. , Cort, J.R. , Semesi, A. , Arrowsmith, C.H. and Kennedy, M.A. (2007) NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c‐di‐GMP binding protein. Proteins, 66, 266–271. [DOI] [PubMed] [Google Scholar]
- Römling, U. and Amikam, D. (2006) Cyclic di‐GMP as a second messenger. Curr. Opin. Microbiol. 9, 1–11. [DOI] [PubMed] [Google Scholar]
- Römling, U. , Gomelsky, M. and Galperin, M.Y. (2005) C‐di‐GMP: the dawning of a novel bacterial signaling system. Mol. Microbiol. 57, 629–639. [DOI] [PubMed] [Google Scholar]
- Ross, P. , Weinhouse, H. , Aloni, Y. , Michaeili, D. , Weinberger‐Ohana P. and Mayer, P. (1987) Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature, 325, 279–281. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Fouhy, Y. , Lucey, J.F. , Crossman L.C., Spiro, S. , He, Y.W. , Zhang, L.H. , Heeb, S. , Camara, M. , Williams, P. and Dow, J.M. (2006a) Cell–cell signalling in Xanthomonas campestris involves an HD‐GYP domain protein that functions in cyclic di‐GMP turnover. Proc. Natl Acad. Sci. USA, 103, 6712–6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ryan, R.P. , Fouhy, Y. , Lucey, J.F. and Dow, J.M. (2006b) Cyclic di‐GMP signalling in bacteria: recent advances and new puzzles. J. Bacteriol. 188, 8327–8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. , Fouhy, Y. , Lucey, J.F. , Jiang, B.‐L. , He, Y.‐Q. , Feng, J.X. , Tang, J.‐L. and Dow, J. M. (2007) Cyclic di‐GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris . Mol. Microbiol. 63, 429–442. [DOI] [PubMed] [Google Scholar]
- Ryjenkov, D.A. , Simm, R. , Römling, U. and Gomelsky, M. (2006) The PilZ domain is a receptor for the second messenger c‐di‐GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281, 30310–30314. [DOI] [PubMed] [Google Scholar]
- Ryjenkov, D.A. , Tarutina, M. , Moskvin, O.M. and Gomelsky, M. (2005) Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A.J. , Ryjenkov, D.A. and Gomelsky, M. (2005) Ubiquitous protein domain EAL encodes cyclic diguanylate‐specific phosphodiesterase: enzymatically active andinactive EAL domains. J. Bacteriol. 187, 4774–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Blundell, T.L. and Mizuguchi, K. (2001) FUGUE: sequence‐structure homology recognition using environment‐specific substitution tables and structure‐dependent gap penalties. J. Mol. Biol. 310, 243–257. [DOI] [PubMed] [Google Scholar]
- Slater, H. , Alvarez‐Morales, A. , Barber, C.E. , Daniels, M.J. , and Dow, J.M. (2000) A two‐component system involving an HD‐GYP domain protein links cell‐cell signalling to pathogenicity gene expression in Xanthomonas campestris . Mol. Microbiol. 38, 986–1003. [DOI] [PubMed] [Google Scholar]
- Tamayo, R. , Tischler, A.D. and Camilli, A. (2005) The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280, 33324–33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Makino, S. , Subedee, A. and Bogdanove, A.J. (2007) Novel candidate virulence factors in rice pathogen Xanthomonas oryzae pv. oryzicola as revealed by mutational analysis. Appl. Environ. Microbiol. 73, 8023–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Rossier, O. and Bonas, U. (1999) Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181, 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Van den Ackerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- Xu, R.Q. , Blanvillain. S. , Feng, J.X. , Jiang, B.L. , Li, X.Z. , Wei, H.Y. , Kroj, T. , Lauber, E. , Roby, D. , Chen, B. , He, Y.Q. , Lu, G.T. , Tang, D.J. , Vasse, J. , Arlat, M. and Tang, J.L. (2008) AvrACXcc8004, a type III effector with a leucine‐rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col‐0. J. Bacteriol. 190, 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]


