SUMMARY
The plant pathogenic fungus Magnaporthe grisea is able to enter its host via appressorium‐mediated penetration. Earlier investigations have shown that these infection structures are rich in the cell wall polysaccharide chitin. Previously, we have described how the transcription of a class VII chitin synthase‐encoding gene CHS7 is completely dependent on the putative transcription factor Con7p during the germination of conidia, and how con7 − mutants are unable to form appressoria under any conditions tested. Because of the pleiotropic effects of the con7 − mutation, we examined the consequences of the targeted deletion of CHS7. The chs7 − mutants generated were unable to form appressoria on artificial surfaces, except following the application of the exogenous inducers 1,16‐hexadecanediol and cyclic adenosine monophosphate. The appressoria formed had a reduced chitin content and were often found to be smaller and misshapen compared with the wild‐type. chs7 − mutants were significantly reduced in their ability to enter rice plants, but growth in planta was not affected. Reverse transcriptase‐polymerase chain reaction analysis demonstrated that CHS7 transcription was strongly induced on germination of spores, and a green fluorescent protein‐tagged Chs7p protein was found to be produced abundantly during infection‐related morphogenesis. Together, these data suggest that the class VII chitin synthase Chs7p of M. grisea is required for normal appressorium formation and function.
INTRODUCTION
Chitin, a polysaccharide formed of β‐1,4‐linked N‐acetyl‐ glucosamine (GlcNAc) units, is a constituent of a diverse range of organisms, including fungi. Within α‐chitin, the antiparallel arrangement of chitin chains is found in most fungi studied; hydrogen bonding also occurs along the chains and between adjacent chains to form structures known as microfibrils. Chitin is an integral part of the fungal cell wall, with microfibrils typically found in the innermost wall layer and embedded within an amorphous matrix. This ‘two‐phase’ network is predicted to have immense strength and is expected to contribute significantly to the structural attributes of the cell wall. Within ascomycete fungi, chitin typically comprises 2%–20% of the cell wall dry weight, although much higher levels have been recorded within cell walls of a few species (Bartnicki‐Garcia, 1968; Klis, 1994).
The synthesis of chitin depends on the activity of chitin synthase (CS) enzymes (EC 2.4.1.16), which catalyse the formation of chitin from uridine diphosphate (UDP)‐GlcNAc. All fungal CSs studied to date are known or predicted to be plasma membrane associated and, where studied, their activity is generally associated with subcellular membrane‐bound particles, which have been termed chitosomes (for a review, see Bartnicki‐Garcia, 2006). Based on the structure of the predicted CSs from a number of different fungal species, these enzymes have been grouped into seven classes (classes I–VII) by Choquer et al. (2004), expanding on the classification originally proposed by Bowen et al. (1992). Some CSs have been found to exist as zymogens which must be proteolytically activated. Based on this characteristic, CSs can be more broadly divided into zymogen and non‐zymogen types (Orlean, 1987).
In the filamentous ascomycete fungi, CSs have been studied in most detail in Aspergillus nidulans and A. fumigatus where, in both cases, eight predicted enzymes with representatives of all seven different classes of CS are recognized (Choquer et al., 2004; and references cited therein). In A. nidulans, mutants have been created which lack single or multiple members of five of these CSs. Although most of the genes studied are individually dispensable, a mutant lacking the class III CS‐encoding gene, chsB, has a severely restricted growth rate (Borgia et al., 1996; Yanai et al., 1994). Further studies in A. nidulans have revealed that several of the other CSs have partially overlapping functions. For example, the class I and II CSs (chsC and chsA gene products, respectively) play a partially redundant role in septum formation (Ichinomiya et al., 2005). The class IV CS (chsD gene product), meanwhile, has functions that can partly compensate for the effects of the loss of chsB on growth and sporulation (Ichinomiya et al., 2002). Double csmA −/csmB − mutants, which carry mutations within the class V and class VI CS‐encoding genes, are not viable (Takeshita et al., 2006). These genes may be expressed co‐ordinately from a common promoter, and the products, both of which have a myosin motor‐like domain (a defining characteristic of the class V and VI CSs), may act together to supply the chitin required for polarized growth.
Within phytopathogenic fungi, a number of CSs that influence the outcome of the interaction with the host plant have been identified. Disruption of the class I CS‐encoding gene Bcchs1 leads to a loss of virulence in Botrytis cinerea (Souliéet al., 2003). The bcchs1 − mutant exhibits a normal growth rate in culture, and it is possible that, in this case, the reduced virulence is caused by a weakened cell wall, which may compromise the ability to survive within hosts. Deletion of a class III CS‐encoding gene, Bcchs3a, from B. cinerea gave rise to a mutant with markedly reduced CS activity and virulence (Souliéet al., 2006). It is probable that, in this case, the reduced virulence reflects the reduced growth rate of the mutant. CSs have also been studied in the vascular wilt pathogen Fusarium oxysporum, where a reduced pathogenicity in chs2− (class II) and a loss of pathogenicity in chsV − (class V) and chsVb − (class VI; however, referred to as a class VII enzyme by these authors according to the nomenclature of Niño‐Vega et al., 2004) mutants have been reported (Madrid et al., 2003; Martín‐Urdíroz et al., 2004, 2008). In Colletotrichum graminicola, a mutant lacking the class V CS is also non‐pathogenic (Werner et al., 2007). A further F. oxysporum gene, chs7, was proposed by its homology to the genes CHS7 of Candida albicans and Saccharomyces cerevisiae to regulate CS trafficking, and a strain lacking this gene also has a reduced ability to cause disease (Martín‐Urdíroz et al., 2004). Extensive analysis of CSs has also been performed in the basidiomycete Ustilago maydis, a maize pathogen. Mutants of all the predicted CSs from this species were generated and, although all showed some loss of virulence, the most severely affected mutants were chs6 and msc1, the two class V CSs, and chs7, one of the two class IV CSs (Weber et al., 2006). Taking all of these studies together, it seems apparent that the myosin motor‐like domain CSs (class V and VI) play a critical role in determining the outcome of the interaction between pathogenic fungi and their hosts, and this may be a result of effects on cell wall integrity, which seem to be common to all such mutants generated to date.
The focus of this study is CHS7, a class VII CS‐encoding gene of the phytopathogenic fungus Magnaporthe grisea. We have found previously that an M. grisea con7− mutant, lacking the Con7p transcription factor, does not transcribe the CHS7 gene during germination, and that germinated spores of this strain have a reduced chitin content (Odenbach et al., 2007). In order to clarify the role of CHS7 in chitin biosynthesis and infection‐related morphogenesis, we created a strain lacking this gene. Our results suggest that Chs7p is required for normal appressorial chitin content and for the normal formation and function of these infection structures.
RESULTS
CHS7 is predicted to encode the sole class VII CS of M. grisea
Given previous observations made using the con7 − mutant (Odenbach et al., 2007), we reasoned that the predicted Chs7p CS might contribute significantly to the chitin produced during disease‐related development. We firstly set out to confirm the sequence and intron–exon structure of CHS7 by obtaining a full‐length cDNA sequence from germinated conidia of the wild‐type strain 70‐15. By comparing this 3388‐bp CHS7 cDNA sequence with the M. grisea genome sequence (Broad Institute, Cambridge, MA, USA; http://www.broad.mit.edu/annotation/genome/magnaporthe_grisea/MultiHome.html), we were able to confirm the presence of two introns, one of 132 bp and one of 128 bp, located 37 bp and 204 bp 3′ of the start codon, respectively. An open reading frame within the cDNA is capable of coding for an 827‐amino‐acid, 93.5‐kDa protein with striking similarity to known or predicted CSs. The highest levels of similarity were found to an uncharacterized hypothetical protein from Neurospora crassa (accession number EAA32522; 66% of amino acids identical), and the best match to a characterized protein was to chsD a (class VII CS‐encoding gene) of A. fumigatus (P78746; 64% of amino acids identical). The predicted Chs7p protein differs from the protein sequence predicted by automated annotation of the M. grisea genome sequence (ID: MGG_06064.6; Broad Institute) in that the first intron is shorter than the intron predicted by automated annotation, resulting in a predicted protein that is longer by 14 amino acids. In addition, a different amino acid, valine rather than phenylalanine, is predicted at position 196.
In order to understand the genetic potential of M. grisea in terms of the numbers and types of predicted CSs, we conducted similarity searches using known CS sequences. This analysis revealed that M. grisea has seven predicted CSs, with one representative of each of the seven classes of CS (following the classification of Choquer et al., 2004). CHS1–CHS4 are predicted to encode enzymes from classes I–IV, respectively, and were identified previously using a degenerate polymerase chain reaction (PCR)‐based approach (Vidal‐Cros and Boccara, 1998). A further previously identified gene, CSM1, is predicted to encode a class V CS (Park et al., 1999). The CHS7 gene has been proposed previously to be a CS based on the results of homology‐based searches (Choquer et al., 2004). Our in silico analysis further revealed that M. grisea has a gene predicted to encode a class VI CS, linked in a head‐to‐head arrangement with CSM1 (the class V gene), an arrangement that seems to be conserved among all the euascomycetes in which a whole genome sequence is available. Extending our in silico analysis to other filamentous ascomycete fungi, it is apparent that the presence of at least one member of each of classes I–VII is typical (Fig. 1). Attempts to identify further non‐annotated CSs within the genome sequence of M. grisea using tblastn interrogations or degenerate PCRs yielded no further CS‐encoding genes (data not shown). Real‐time reverse transcriptase (RT)‐PCR experiments confirmed that each of these predicted M. grisea CS‐encoding genes is transcribed during growth in complete medium (CM) agar and/or during infection‐related development, and that, of the corresponding transcripts, the CHS7 message is the most abundant within spores 2 h post‐germination (data not shown).
Figure 1.
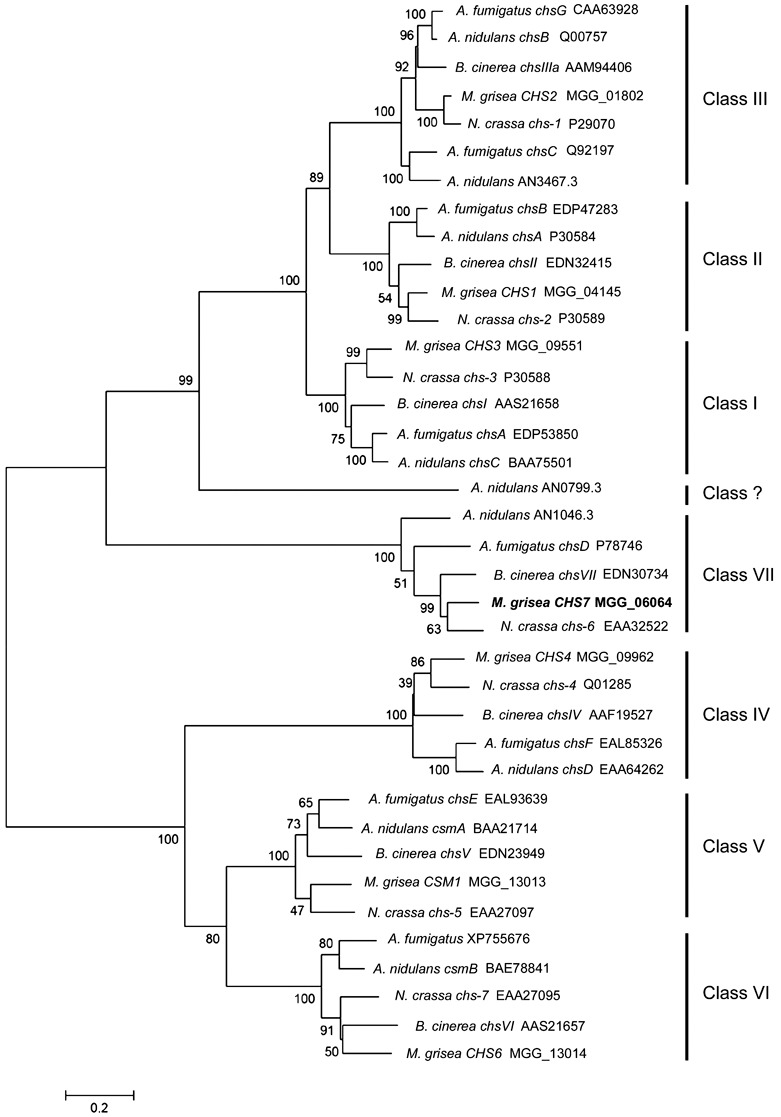
Phylogenetic tree based on alignment of the predicted Magnaporthe grisea CHS7 gene product with the predicted gene products of known and predicted chitin synthase (CS)‐encoding genes. Tree reliability was tested using bootstrap resampling with 1000 replications. Together with gene names, where assigned, the GenBank or Swissprot accession numbers are given, or, alternatively, and for all the Magnaporthe predicted proteins used, the Broad Institute database ID number is given. Note that AN0799.3 (marked Class ?) seems to be a divergent CS which, although it has characteristics of the class I, II and III CSs [PFAM domains PF1644 (chitin synthase 1) and PF08407 (chitin synthase 1 N‐terminal domain)], lacks the close similarity that the other members of these classes exhibit. This protein shares only 28% of its amino acid sequence with Aspergillus nidulans ChsC protein (class I). For comparison, the A. nidulans ChsA (class II) and ChsC proteins have 48% of amino acids in common, a value more typical between the class I and II CSs, whereas, within the various CS classes, typically 60%–70% of amino acids are conserved between species. Numbers on the branches indicate the bootstrap value for each branch and the bar indicates 0.2 distance units.
The length and domain structure of Chs7p are typical of a CS of this class (Choquer et al., 2004). The predicted Chs7p protein lacks the myosin head‐like region present in class V and VI enzymes and the cytochrome b5‐like domain present in class IV–VI CSs. Chs7p has six predicted transmembrane regions at positions 14–34, 55–77, 99–121, 447–467, 473–491 and 501–521. The conserved fungal CS domain (PFAM ID: PF03142), including the catalytic core of the enzyme, is located within the central region between residues 141 and 439, and matches the consensus sequences of processive β‐glycosyl transferases, as reported previously (Choquer et al., 2004). A prediction of membrane topology using the program mtop (Hartmann et al., 1989) suggests that the approximately 340‐amino‐acid‐long C‐terminal end of the protein will be located inside the membrane.
Construction and morphological studies of a chs7 − mutant strain
In order to analyse the effects of the CHS7 gene deletion, we created a construct in which the region coding for the C‐terminal end of Chs7p from amino acid 178 to beyond the predicted STOP codon was replaced with the hygromycin resistance gene (HPT) cassette flanked by the sequences 3′ and 5′ to the deleted region (Fig. 2A). The sequences removed included most of the conserved catalytic domain. This construct was introduced into wild‐type strain 70‐15 (hereafter referred to simply as WT) using Agrobacterium tumefaciens‐mediated transformation (ATMT). Homologous recombination of the gene deletion construct was confirmed by Southern blotting for 14 of the 22 transformants analysed. A shift in the size of the hybridizing band as a result of the deletion of two XhoI sites in the regions excised is indicative of the deletion of CHS7 by homologous recombination (Fig. 2B). Three chs7− mutants were selected for further phenotypic analysis. A complemented chs7− mutant was also constructed in which the CHS7 gene, with its own 1.2‐kb‐long promoter and approximately 0.5‐kb terminator sequence, was reintroduced ectopically (Fig. 2B).
Figure 2.
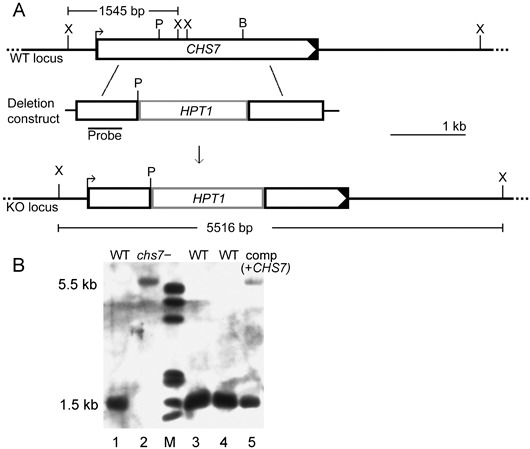
Magnaporthe grisea CHS7 gene deletion strategy and Southern analysis of chs7− transformants and complemented mutant strains. (A) A gene deletion construct was created in vitro by replacing a 1.2‐kb PmlI (P)–BglII (B) fragment within the CHS7 gene with a 1.5‐kb PmlI‐BamHI‐ended hygromycin phosphotransferase cassette (HPT1). The BglII restriction site is not re‐formed by ligation to a BamHI fragment, and this was confirmed by restriction digests. This construct was introduced into the wild‐type strain 70‐15 (WT). (B) The integration of the gene deletion construct by homologous recombination was predicted to lead to the loss of two XhoI sites present in the WT locus. This was expected to result in a ‘band‐shift’ when a probe derived from the 3′ end of CHS7 was hybridized to XhoI‐digested DNA from a CHS7‐deleted strain. Genomic DNA was isolated from strains transformed with the deletion construct, digested with XhoI, fractionated and transferred to a nylon membrane, which was then hybridized to a CHS7 probe (see Fig. 2A). Transformant strain 2 (lane 2) had a hybridization pattern indicative of the deletion of CHS7. Several other transformants with a hybridization pattern indicative of CHS7 deletion were identified (not shown). WT DNA digested with XhoI was loaded in lanes 1, 3 and 4. Lane 5 (labelled comp + CHS7) shows the hybridization pattern of a complemented strain derived from a chs7 − mutant, where the WT CHS7 gene, promoter and associated transcriptional signals were reintroduced ectopically.
We explored the consequences of gene deletion on the morphogenesis of the chs7 − strains obtained at the macroscopic and microscopic level. The chs7 − strains grew at rates indistinguishable from WT on CM agar plates and under hypo‐ and hyperosmotic stress (CM plates plus sorbitol in the range 0–1.8 m). Sporulation of the mutant strain also occurred at levels comparable with that of WT, and conidia had a WT size and morphology. We found no altered sensitivity of mycelium or of germinating conidia to the CS inhibitor Nikkomycin Z. The mutant strain also exhibited no altered sensitivity to sodium dodecylsulphate (SDS) or H2O2, as has been reported for CS‐deleted mutants of other ascomycete fungi (Fujiwara et al., 2000; Madrid et al., 2003). Dramatic effects of CHS7 gene deletion were apparent on investigation of disease‐related morphogenesis. In stark contrast with WT, the chs7 − mutant formed no appressoria in response to an artificial hydrophobic surface (Fig. 3A,D). We were able to restore appressorium formation on these surfaces by application of the cutin monomer 1,16‐hexadecanediol (1,16‐HDD; Fig. 3B,E), the diacylglycerol 1,2‐dioctanoyl‐rac‐glycerol (DOG; not shown) or 8‐(4‐chlorophenylthio)‐cyclic adenosine monophosphate (8‐CPT‐cAMP; Fig. 3C,F), or by incubation on the surface of untreated onion epidermis or rice leaves, but not on chloroform‐treated onion epidermis (Table 1; Fig. 3G–J). Appressoria formed in response to these inducers, or on both untreated onion epidermis and rice leaves, were generally smaller (mean appressorial diameter: WT, 9.6 ± 2.1 µm; chs7 − mutant, 5.0 ± 2.6 µm) and often elongated and/or misshapen compared with WT appressoria (Fig. 3E,F,J). They also formed at a later time point than those of WT, as chs7 − appressoria were only apparent after a 16‐h incubation and were not found after an 8‐h incubation when more than 95% of WT spores had germinated and formed appressoria. Appressoria of the mutant strain typically appeared after an extended period of germ tube elongation (mean germ tube length: WT, 23.0 ± 7.2 µm; chs7 − mutant, 80.6 ± 19.5 µm).
Figure 3.
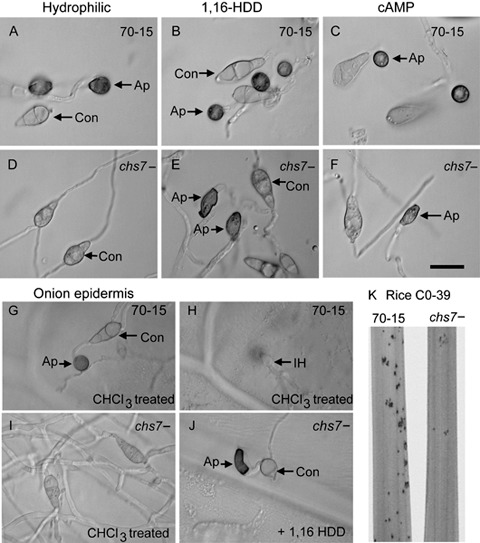
Morphogenetic defects of the Magnaporthe grisea chs7 − strain. Freshly harvested conidia (Con) were incubated on hydrophobic microscope slides for 16 h. Wild‐type (WT) spores germinated (A) and formed appressoria (Ap), but the chs7 − germinated spores failed to undergo swelling or appressorium formation (D). When the spore suspensions were supplemented with 200 ng/mL 1,16‐hexadecanediol (1,16‐HDD; B, E) or 40 µg/mL 8‐(4‐chlorophenylthio)‐cyclic adenosine monophosphate (8‐CPT‐cAMP; C, F), the mutant formed appressoria that were either misshapen or much smaller than WT appressoria. The ability to form appressoria on plant surfaces and to penetrate these surfaces was assessed by inoculation of conidial suspensions onto rice leaves (not shown) or onto onion epidermis (G–J). These experiments indicated that WT formed appressoria and penetrated as expected [G and H show the same field of view; H is focused within the onion cell and shows the primary infection hyphae (IH)]. Meanwhile, the mutant formed no appressoria on chloroform‐treated onion (I) and was only able to form misshapen or small appressoria on addition of 1,16‐HDD (J) or 8‐CPT‐cAMP (not shown), or by inoculation onto untreated onion (note: both WT and mutant cannot penetrate untreated onion). Bar, 20 µm. The virulence of chs7 − mutants was assessed using seedlings of rice cultivar CO‐39 inoculated with conidial suspensions of WT and the mutant. Seedlings were incubated for 5 days and the lesion numbers per plant were recorded. Representative leaf sections are shown (K). These infection assays were repeated three times and, each time, 40 plants per strain were analysed.
Table 1.
Appressorium formation of the chs7 −mutant and wild‐type (WT) in the presence and absence of different surfaces and inducers.
| Strain | Hydrophobic No inducer | Hydrophobic 1,16‐HDD | Hydrophobic 8‐CPT‐cAMP | Hydrophobic DOG | Onion epidermis (untreated) | Onion epidermis (treated) |
|---|---|---|---|---|---|---|
| 70‐15 | 99.7 ± 0.58 | 99.3 ± 1.4 | 99.7 ± 0.58 | 98.33 ± 0.6 | 88.4 ± 3.5 | 83.9 ± 5.8 |
| chs7 | 0 | 80.3 ± 2.1 | 71.0 ± 3.61 | 77.67 ± 3.1 | 70.7 ± 1.7 | 0 |
Spores were incubated at 105 conidia/mL for 24 h on the surface indicated. The percentages of spores that formed one or more appressoria are shown. No appressoria were formed by either strain on a hydrophilic surface (glass microscope slides). The data shown are the mean values obtained by counting 300 spores for each strain, and the standard deviation is given. The experiment was repeated three times with very similar results. The inducers used were the cutin monomer 1,16‐hexadecanediol (1,16‐HDD; 200 ng/mL), the diacylglycerol 1,2‐dioctanoyl‐rac‐glycerol (DOG; 20 µg/mL) and the cyclic adenosine monophosphate (cAMP) analogue 8‐(4‐chlorophenylthio)‐cAMP (8‐CPT‐cAMP; 40 µg/mL).
To test whether chs7 − mutants were affected in their ability to cause disease in rice plants, we spray‐inoculated spores of the mutant and WT strain onto the susceptible rice cultivar CO‐39. A consistent reduction of approximately 80% in disease lesion development was apparent (t‐test, P < 0.001; Fig. 3K). Inoculation of abraded leaf segments indicated that there was no obvious difference in the ability of the mutant strain to cause disease if the need to penetrate the plant leaf cuticle was circumvented (data not shown).
All defects described above were common to all three of the chs7 − strains analysed, and were fully complemented within two independently isolated chs7 −‐derived strains by reintroduction of the CHS7 gene and transcriptional signals.
CHS7 gene is expressed during disease‐related morphogenesis
In order to further investigate the transcriptional regulation of CHS7 during disease‐related morphogenesis, real‐time RT‐PCR was used to compare CHS7 transcript abundance at various time points following germination on an artificial hydrophobic surface. By comparison with the MgATG3/AUT1 gene, whose expression levels have been found to be reproducibly constant during disease‐related morphogenesis (D. Odenbach et al., unpublished data), there was a strong induction in CHS7 transcription on germination (Fig. 4). CHS7 transcription abundance peaked at an early time point, and we were able to detect the transcript within spores that had germinated and formed appressoria 24 h post‐germination (Fig. 4).
Figure 4.
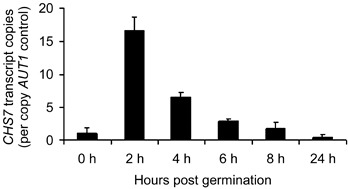
Transcription of the Magnaporthe grisea CHS7 gene during germination within the parental strain 70‐15, assessed by real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis. Transcription of CHS7 during germination within the wild‐type (WT) strain was analysed by real‐time RT‐PCR using RNA extracted from spores applied to hydrophobic plastic slides. Transcript abundance was determined using the mathematical model of Pfaffl (2001). The transcript abundance relative to that of the control gene AUT1 is indicated. Values were obtained from three independent biological replicates, and standard errors are indicated by the error bars.
A green fluorescent protein (GFP)‐tagged Chs7p protein is expressed within growing cells and maturing appressoria
In order to investigate the localization of the CHS7 gene product, a construct containing a 1.1‐kb‐long CHS7 promoter sequence and the entire CHS7 ORF fused in frame with the GFP‐encoding gene was introduced into the wild‐type strain 70‐15 by transformation. This construct fuses the predicted C‐terminus of Chs7p to GFP. An ectopic single integration of the construct was proven by PCR and Southern blotting (data not shown). Of three transformants selected for further study, none showed any defects in growth, germination or pathogenic development; however, they all exhibited GFP fluorescence, which was apparent within discrete subcellular bodies within the apical cells of ungerminated spores (Fig. 5A,B), nascent germ tubes (Fig. 5C–F) and germ tubes prior to the hooking that precedes appressorium formation after 2–3 h of incubation (Fig. 5E–H). Chs7p‐GFP fluorescence was also pronounced within the conidial cell from which the germ tube emerged for some time following germination (Fig. 5E–H). Chs7p‐GFP fluorescence was also particularly marked within both nascent and mature appressoria (Fig. 5I–L), where, in the latter, a punctate distribution towards the periphery of the cell was apparent. A much lower level of Chs7p‐GFP fluorescence was observed within the mycelium, where again it had a punctate distribution (Fig. 6A–C). No particular association of this faint Chs7p‐GFP fluorescence with hyphal tips at the colony margin (in contrast with the association with germ tube tips) or with septa was detected (data not shown). Within forming conidiophores, a faint and diffuse Chs7p‐GFP fluorescence was observed (Fig. 6D,E).
Figure 5.
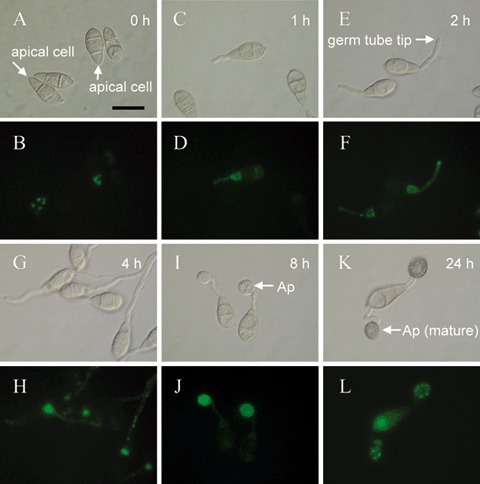
Localization of a Magnaporthe grisea Chs7p‐GFP fusion protein during infection‐related morphogenesis. A fusion of the M. grisea CHS7 gene with the green fluorescent protein gene (GFP), driven by the CHS7 promoter, was constructed. Localization of Chs7p‐GFP fluorescence within ungerminated conidia was apparent predominantly as large ‘spots’ within the apical cell (A, B), 1 h following germination. Chs7p‐GFP fluorescence was associated with spots within the germ tube and with the conidial cell from which the germ tube arose (C, D). Two hours (E, F) and 4 h (G, H) following germination, Chs7p‐GFP fluorescence was primarily associated with the germ tube apex and/or an apparently vacuolated region of the conidial cell that had given rise to the germ tube (E–H). Nascent appressoria (8 h post‐germination; I, J) showed a strong diffuse Chs7p‐GFP fluorescence, whereas mature appressoria (24 h post‐germination; K, L) showed a punctate distribution of Chs7p‐GFP fluorescence located towards the periphery of the appressorium. Scale bar, 20 µm.
Figure 6.
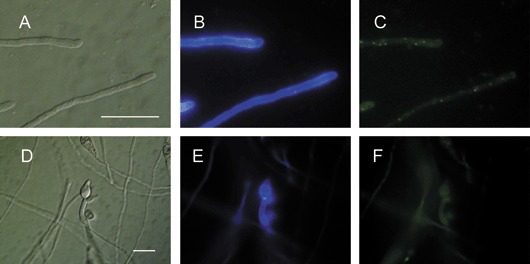
Localization of a Magnaporthe grisea Chs7p‐GFP fusion protein within growing hyphae and during asexual reproduction. Localization of Chs7p‐GFP fluorescence within hyphae after 3 days of growth in liquid corn meal (CM) on a glass surface had a punctate distribution (A–C). Nascent conidiophores were formed following the inoculation of conidia onto hydrophobic coverslips in water after 3 days. In these cells, a faint diffuse Chs7p‐GFP fluorescence was apparent (D, E). (B) and (E) show the Calcofluor staining pattern and (C) and (F) show Chs7p‐GFP fluorescence. Note that this Chs7p‐GFP fluorescence was much fainter than that seen during infection‐related morphogenesis (Fig. 5), although this is not readily apparent from this figure because the exposure time was dramatically increased to document the Chs7p‐GFP fluorescence within mycelium and conidiophores. Scale bar, 20 µm.
Chitin content is reduced in appressoria of the chs7 − mutant
The consequences of CHS7 deletion on chitin accumulation were explored using both Calcofluor staining and direct chitin quantification. These analyses both indicated that the appressorial chitin content was reduced in chs7 − strains when compared with WT levels. On a hydrophobic surface in the presence of 1,16‐HDD, the bright staining of WT appressoria using Calcofluor (Fig. 7A–D) and tetramethylrhodamine isothiocyanate‐labelled wheat germ agglutinin (TRITC‐WGA) (not shown) was much less obvious in the appressorium‐like swellings produced by the chs7 − strains (Fig. 7E–H). Direct biochemical quantification confirmed the reduced chitin content within spores that had been allowed to germinate and form appressoria. The chitin present in this material, which is a mixture of spores, germ tubes and appressoria, was reduced in the mutant to about 37% of that of the WT strain (Fig. 7). This decrease seems to be specific to appressoria, because no reduction was seen in the chitin content of the mutant when spores alone (both ungerminated or germinated) and mycelium were examined (Fig. 7).
Figure 7.
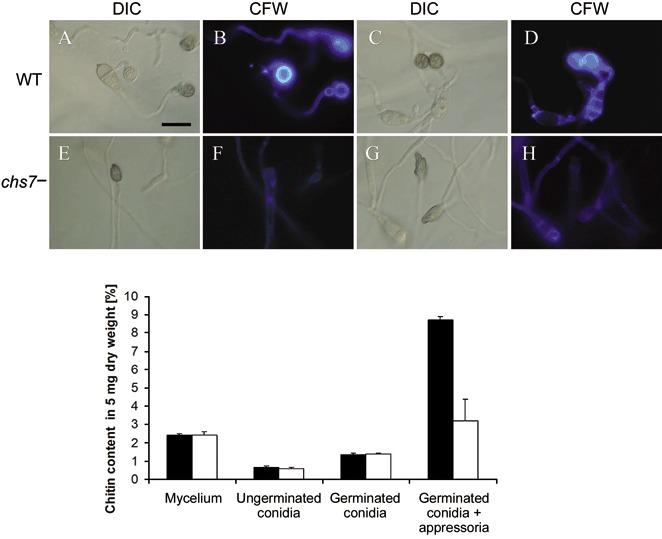
Chitin distribution in the Magnaporthe grisea chs7 − mutant. Top: chitin distribution in the M. grisea wild‐type 70‐15 (WT) (A–D) and chs7 − (E–H) strains visualized by staining with Calcofluor white (CFW). Conidia were inoculated onto the surface of untreated onion and incubated for 48 h. Bright staining in appressoria of the parental strain was apparent (A–D). Appressoria formed by the chs7− conidia (E–H) showed reduced CFW staining. Scale bar, 20 µm. Bottom: altered chitin content within the M. grisea chs7 − mutant. Chitin was quantified colorimetrically according to Bulik et al. (2003). This analysis indicated that, relative to WT (black bars), the chs7 − mutant (white bars) showed reduced chitin content within appressoria. The chitin content of mycelium, ungerminated spores and germinated spores was unaffected.
Pathogenicity of chs7− mutants is reduced because of a reduced ability to penetrate
The ability to penetrate the plant cuticle was assayed with spores incubated on the surface of chloroform‐treated onion epidermis for 48 h, with the induction of appressorium formation using 1,16‐HDD. After this time point, most of the WT appressoria had ruptured the cuticle and showed intracellular infection hyphae growing away from the penetration site (Fig. 3G,H). In contrast, penetration by chs7 − appressoria (Fig. 3J) was only approximately 25% of that of WT (t‐test, P < 0.05). Even after an extended incubation time (72 h), penetration by the majority of mutant appressoria was not seen. The behaviour of the mutant in terms of the number and morphology of appressoria formed and the ability of these appressoria to penetrate the rice leaf was very similar to that on treated onion epidermis in the presence of 1,16‐HDD, indicating that these responses were not an artefact of the non‐host‐based assay. There was no difference in the penetration frequency when misshapen chs7 − appressoria were compared with the small wild‐type‐shaped chs7 − appressoria. These data suggest that appressorium formation is delayed in the absence of the CHS7 gene product, and that the majority of mutant appressoria are non‐functional.
DISCUSSION
We have shown previously that the transcription of the CHS7 gene of M. grisea is dependent on the predicted transcription factor Con7p using microarray‐based expression analysis. We show here that the class VII CS is required for normal appressorium formation and function. Of the seven CS‐encoding genes of M. grisea, this is the first whose function has been investigated by targeted gene deletion.
The predicted product of the CHS7 gene shows striking similarities to several predicted class VII CSs. To date, the single class VII CS‐encoding gene analysed via targeted gene deletion is chsD of the opportunistic human pathogen A. fumigatus (Mellado et al., 1996), where no morphogenetic or pathogenic abnormalities were found to be associated with mutation despite an approximately 20% reduction in mycelial chitin (Mellado et al., 1996). Here, we present evidence that the class VII CS of M. grisea contributes to the chitin present in appressoria, but not significantly to that present within the mycelium. It is also possible that Chs7p contributes to the chitin present within mycelium, but that, in its absence, an increased activity of other CSs may compensate; however, the low levels of both the CHS7 transcript and the Chs7p‐GFP protein within mycelium suggest that the contribution of Chs7p is probably minor within this material. This must be taken as an indication that the class VII CSs have acquired different, stage‐specific, functions within different filamentous ascomycete species. In contrast with the situation reported for the chsD − mutant of A. fumigatus, we show here that the chs7 − strain of M. grisea has a number of obvious morphogenetic abnormalities. Firstly, it fails to respond to the hydrophobicity trigger that efficiently stimulates appressorium formation in WT. Secondly, the appressoria formed by the mutant in the presence of a plant surface or inducers of appressorium formation do not have a WT morphology, and give rise to successful penetration events with a much lower frequency than do WT appressoria. Thirdly, both Calcofluor staining and chitin quantification suggest that appressoria formed by the chs7 − strain do not contain the comparatively high levels of chitin obvious within WT appressoria. In many ways, the phenotype of the chs7 − strain is reminiscent of that of a strain lacking the predicted chitin binding protein Cbp1p (Kamakura et al., 2002). In both cases, the mutant strains created are unable to form infection structures efficiently on artificial surfaces, but do so on the plant or on the application of known inducers. Interestingly, the expression of CHS7 may also mirror that of CBP1 as, in both cases, the corresponding mRNA was detected in germinated spores, but not within mycelium (Kamakura et al., 2002; D. Odenbach et al., unpublished data). Furthermore, both genes are not transcribed at a detectable level in germinated spores of the con7 − mutant, suggesting a co‐regulation by the predicted transcription factor Con7p (Odenbach et al., 2007). However, the cbp1 − and chs7 − phenotypes are not identical as, in the former, appressoria eventually do form, albeit at a lower frequency than in the parental strain, unlike the situation described here for the chs7 − strain. In addition, in the presence of 3‐isobutyl‐1‐methylxanthine (IBMX), a non‐specific inhibitor of phosphodiesterases, the appressoria of the cbp1 − strain resemble morphologically those of the parental strain, whereas the chs7 − mutant does not form appressoria with WT morphology under any conditions tested. Furthermore, we have shown that chs7 − appressoria form efficiently on onion epidermis, but not on this substrate following chloroform treatment. This suggests that chloroform‐soluble components of the plant can induce appressorium formation of the mutant. As known inducers of appressorium formation, cutin monomers are obvious candidates (Gilbert et al., 1996). Furthermore, the cutin monomer 1,16‐HDD was the most effective inducer of chs7 − appressorium formation tested. A common feature of both the cbp1 − and chs7 − strains seems to be a loss of response to hydrophobicity, which is overridden by these chemical cues.
Given the similarities between the chs7 − and cbp1 − phenotypes, the co‐regulation of these genes and the link in both cases to chitin, we could speculate that there is an interaction between the Cbp1p protein and the chitin formed by Chs7p. Cbp1p may mediate the sensing of physical cues for appressorium formation and, in the absence of normal appressorial chitin content in the chs7 − strain, it may not associate effectively with the cell surface. The greater severity of the chs7 − phenotype may reflect functional redundancy amongst the several predicted chitin‐binding proteins of M. grisea (Dean et al., 2005). Previous microarray‐based transcription analyses have suggested that several of the chitin‐binding protein‐encoding genes are expressed following germination (D. Odenbach et al., unpublished data). It is, of course, highly likely that the severity of the chs7 − phenotype in comparison with that of the cbp1 − strain is a result of the fact that chitin within the appressorium has a function other than to act as an anchor point for these chitin‐binding proteins. As well as the possibility that other cell wall‐associated proteins might associate with chitin, the structural significance of chitin itself should not be overlooked. Indeed, the observations that the appressoria formed by the chs7 − strain are often deformed would suggest that chitin plays an important role in determining the structure of these cells.
The analyses of gene transcription and the localization of the Chs7p‐GFP fusion protein suggest an involvement of the gene product in infection‐related morphogenesis. The fusion protein was prominent within germ tubes and appressoria, but present only at low levels within mycelium. Similarly, the transcript was abundant in germinated spores, but present at very low levels in mycelium. The fluorescence signal within germ tubes was especially concentrated at the tip, but also occurred as spots along the whole germ tube. Interestingly, very little Chs7p‐GFP fluorescence was associated with the cell periphery (plasma membrane). The subcelluar localization of Chs7p‐GFP contrasts with that of a GFP‐tagged class VI enzyme from C. graminicola, which associates with the cell periphery and the tips of hyphal branches, but not with the tips of germ tubes (Amnuaykanjanasin and Epstein, 2006). An association with vegetative hyphal tips was reported for the A. nidulans CsmA class V and VI proteins (Takeshita et al., 2005, 2006), whereas the class I and II CSs of this species localize to the forming septum and, in the case of the class I protein only, also to hyphal tips. Although a comprehensive analysis of the localization of all CSs from a single filamentous ascomycete species is lacking, it is apparent from studies to date that the temporal and spatial distribution of CSs of different classes varies. The exception to this may be the CSs of classes V and VI, which may act cooperatively. The observations made here further support the view that different classes of CS are involved in the synthesis of chitin within different stages and cell types.
The occurrence of the Chs7p‐GFP protein within discrete subcellular particles running along the length of the growing germ tube and the subsequent bright fluorescence of the nascent appressorium suggest that transportation may be occurring, perhaps towards sites of new cell wall deposition or cell wall reinforcement. The accumulation of the particles within the mature appressorium may suggest that they act as a reservoir of chitin biosynthetic machinery that could be rapidly mobilized following penetration. Similar types of subcellular CS‐containing particles have been reported in other filamentous fungi, including a class VI enzyme from C. graminicola (Amnuaykanjanasin and Epstein, 2006) and the class I, II, V and VI proteins of A. nidulans (Ichinomiya et al., 2005; Takeshita et al., 2005, 2006). Although these CSs are not produced in the same cell types as M. grisea Chs7p and are of different classes, it is possible that similar CS‐containing particles exist in different stages, but that the specific CSs present differ.
Unlike the cbp1 − strain discussed above, the chs7 − strain has a significantly reduced ability to cause disease using rice as a host. This may be a result of a reduced penetration frequency by appressoria of the chs7 − mutant. Consistent with this view, the lesions formed by the mutant on wounded plant leaves were indistinguishable from those of WT, suggesting that the deletion of CHS7 had no impact on growth in planta. Delayed appressorium formation could contribute to the reduced virulence, as this would allow more time for the plant to raise an effective defence response. A defect specifically affecting the ability to gain entry to the plant is a novel phenotype amongst the CS mutants generated within phytopathogenic fungi to date. So far, when virulence has been found to be affected by CS gene deletion, this has been considered to be caused by either a reduced growth rate within the plant and/or an increased sensitivity to antifungal agents (Madrid et al., 2003; Martín‐Urdíroz et al., 2004; Souliéet al., 2006; Weber et al., 2006).
The reduced penetration of the chs7 − strain raises the possibility that there is an impact on turgor generation. Appressorial melanin is a critical determinant of the ability of these cells to maintain internal turgor (Howard and Ferrari, 1989). A link between chitin biosynthesis and melanin deposition in the cell wall of the human pathogen Exophiala (Wangiella) dermatitidis has been reported (Wang et al., 1999). However, we found no deleterious effects on melanization of the chs7 − appressoria using bright field microscopy (data not shown). A more direct approach to quantify melanin production might offer further insight as to whether the reduced penetration by the mutant is a result of disturbed melanin deposition. Unfortunately, it has proven difficult to assess turgor generation by chs7 − appressoria using a simple cytorrhysis assay because of the altered morphology of many of these cells.
There is considerable interest in chitin biosynthesis as a target for drug intervention in the control of fungal diseases of plants and animals. In order to test the suitability of CSs as drug targets, it is necessary to establish a direct link between chitin synthesis and pathogenic development. In a number of cases, as discussed earlier, such a direct link has been established by the analysis of mutants deleted for specific CSs. Analyses of CSs from various pathogenic fungi have implicated these enzymes in maintaining normal growth rates and in the ability to withstand antifungal agents. In this study, we have shown that a class VII CS of M. grisea is specifically produced during pathogenic development, and is required for normal appressorial formation and function and consequent virulence.
EXPERIMENTAL PROCEDURES
Strains, culture conditions, chemicals and DNA manipulations
All the strains described in the present study were derived from the M. grisea wild‐type strain 70‐15 (Chao and Ellingboe, 1991). Fungal strains were routinely grown on CM (Talbot et al., 1993) agar plates. The fungal strains were grown at 28 °C unless otherwise indicated. Standard procedures for DNA manipulations were followed (Sambrook et al., 1989). Escherichia coli strain XL1‐Blue (Stratagene, La Jolla, CA, USA) was used for routine bacterial transformations and maintenance of plasmids. Oligonucleotides were designed with the aid of the program PrimerSelect (DNA Star Inc., Madison, WI, USA), and were obtained from MWG‐Biotech (Ebersberg, Germany). Unless otherwise stated, chemicals were sourced from Sigma‐Aldrich (Hamburg, Germany). Where sensitivity to SDS was tested, 1 µm to 1 mm was used. H2O2 was tested in the range 0.001%–0.2%, and Nikkomycin Z was tested from 10 ng/mL to 100 µg/mL. 1,16‐HDD was added to spores at 200 ng/mL, 8‐CPT‐cAMP at 40 µg/mL and DOG at 20 µg/mL.
Transformations and pathogenicity assays
Transformation of M. grisea was conducted using ATMT (de Groot et al., 1998; Rho et al., 2001). Vectors based on pCAMBIA‐0380 (CAMBIA, Canberra, Australia) were introduced into A. tumefaciens strain AGL1 using the ‘freeze–thaw’ method (Holsters et al., 1978). Transformants were checked for the presence of the expected plasmid and were then co‐cultivated with conidia of M. grisea. Selection of blastacidin‐resistant transformants of M. grisea was carried out using the method of Kimura et al. (1995) as described previously (Odenbach et al., 2007). For pathogenicity tests, spores (conidia) were harvested from 12‐day‐old CM agar cultures and sprayed onto 20‐day‐old seedlings of the rice cultivar CO‐39 or 14‐day‐old seedlings of the barley cultivar Golden Promise at 1 × 105 conidia/mL in 0.2% gelatin. The incubation and examination of intact and wounded plants were carried out as described previously (Foster et al., 2003). Assays of penetration using onion epidermis were conducted according to Chida and Sisler (1987).
Collection of spores and growth of fungal material
Twelve‐day‐old CM agar cultures were used as a source of asexual spores (conidia), which were harvested in water by rubbing the surface of the plate with a glass spreader, followed by filtration through Miracloth (Calbiochem, Darmstadt, Germany). When infection‐related morphogenesis was studied or to prepare extracts from such material (for chitin quantification or for the preparation of RNA extracts), the spores were inoculated at 5 × 105 conidia/mL onto the surface of hydrophobic plastic slides (Cellstar, Greiner Bio One, Kremsmünster, Austria) and incubated for the time indicated. When germinated spores without appressoria were required, the spores were inoculated into liquid CM and incubated for the time stated. Following the incubation period, the biomass was collected by centrifugation (10 min, 4000 g). Mycelium for DNA extraction was obtained from cultures grown for 3 days in liquid CM with shaking at 120 r.p.m. For analysis of mycelial chitin content, cultures used spores as an inoculum and were grown for 16 h in liquid CM with shaking on a rocking platform.
Construction of plasmids and analysis of transformants
The CHS7 deletion construct was constructed as described previously (Odenbach et al., 2007). For Southern analysis, genomic DNA was prepared using a DNAeasy Plant Miniprep Kit (Qiagen, Hilden, Germany). A DIG‐labelled probe was generated using the primers CHS7probFOR (TTGGCCGAGGTTGAAGAAGTGAAA) and CHS7probREV (GCCGAATCGTCCAATGCTC). For the production of a vector for complementation of the chs7 − mutant, a 4.4‐kb CHS7 promoter–gene–terminator fragment was amplified from 70‐15 genomic DNA using primers CHS7comp_FOR (CGGAGGCGCGATATGAGATG) and CHS7compREV (CTGCGACTTGGCGAGATGCT). The PCR product generated was ligated into pGEM‐Teasy, and the insert from the resultant vector, pCHS7‐Comp, was subcloned into the EcoRI site of pCAMB‐BSD (Odenbach et al., 2007) to yield the final vector pCAMB‐CHS7‐COMP. The construct used to generate strains expressing a GFP‐tagged Chs7p protein was produced by PCR amplification of the CHS7 ORF plus 5′ untransformed region and promoter regions, using plasmid pCAMB‐CHS7‐COMP as a template and the primers CHS7gfpFOR (GCCCCAATAACATTGTCTGTCTGCC) and CHS7gfpREVhind (AAGCTTCCGTTGCTGATGGCCGCCG). The primer CHS7gfpREVhind removes the CHS7 stop codon and includes a HindIII site necessary for later fusion of the eGFP ORF to the C‐terminus of Chs7p. The PCR product was ligated to pGEMT‐easy and the resultant plasmid, pGEM‐CHS7‐GFP, was linearized using HindIII and SpeI, and ligated to a HindIII‐SpeI‐ended eGFP ORF and terminator fragment, generated by PCR amplification using pAJF‐GFP (Odenbach et al., 2007) as a template and primers GFP_FOR_hind (AAGCTTATGGTGAGCAAGGGCGAGGAGC) and GFP_REV (GCGGCCGCTCTAGAACTAGTGGATCC). The ApaI‐SpeI insert from the resultant plasmid was then subcloned into pCAMBIA‐HPT (Odenbach et al., 2007) by digestion with ApaI and SpeI to generate the completed vector pCAMB‐Chs7p‐gfp.
RNA isolation
Conidia were harvested as described above and applied to hydrophobic plastic slides (Cellstar, Greiner Bio One) at 5 × 105 conidia/mL, and incubated for the time stated, after which the biomass was collected by centrifugation (3000 g). Cells were broken using glass beads and rapid agitation, generated by a Fast Prep device (Bio101) set to maximum shaking using 5 × 20‐s pulses with cooling on ice in between each shaking pulse. The RNA was then immediately isolated using an RNAeasy Kit according to the manufacturer's instructions (Qiagen). RNA quality was assessed using a nanochip and an Agilent Bioanalyzer 2100 (Agilent, Walbronn, Germany). Only high‐quality RNAs, as determined by rRNA profiles, were used for cDNA synthesis.
cDNA amplification and analysis
Amplification of the 5′ and 3′ cDNA fragments was performed using the SMART cDNA Amplification Kit (BD Biosciences, Clontech, Oxford, UK), according to the manufacturer's instructions. The RNA template used for cDNA synthesis was extracted from germinated conidia (2 h post‐germination). The amplicons obtained were cloned into pGEMTeasy vector (Promega, Mannheim, Germany). The inserts present within the resultant plasmids were sequenced completely on both strands using two independent clones containing independently generated PCR products. The sequences were assembled using the program SeqManII v5.06 (DNA Star Inc., Madison, WI, USA).
RT‐PCR analysis
RT‐PCR was conducted using the Quanti Tect™ SYBR® Green RT‐PCR method (Qiagen) and a Lightcycler (Roche Diagnostics, Mannheim, Germany), according to the manufacturers’ recommendations. One hundred nanograms of RNA were used as a template for the synthesis of cDNA at 50 °C (20 min), and all amplification steps used the following cycling regime: denaturation (15 s, 95 °C), annealing (25 s, temperature varied according to primer set), elongation (30 s, 72 °C). A melting curve analysis was also performed for each reaction, and products were gel fractionated to check the product size and for contamination with genomic DNA (an intron‐spanning primer was always used as a control). Reactions were performed using three independent biological replicates. Comparison of transcript abundance was calculated using the mathematical model of Pfaffl (2001). The following primers were employed: for CHS7, CHS7RTFOR (CTGCCGTCCTGGAGTGGTTC) and CHS7RTREV (CGGCCTAGCTGGTTGGTGAC); for the control gene AUT1/ATG3 (MGG_02959.5; D. Odenbach et al., unpublished data), Aut1exp2for (CGCCGGCGATGCAGGTCTTG) and Aut1exp2rev (GCTTCAGTGCGGCATCGGCTCGGTCTA).
Measurement of the chitin content
The chitin content was determined using the method of Bulik et al. (2003), and biomass was generated and processed as described previously (Odenbach et al., 2007), except that, in order to generate the material for the analysis of appressorial chitin content, appressorium formation was induced by supplementation with 1,16‐HDD and material was collected at 16 h post‐germination. Analysis was repeated three times on separate occasions, in each case using independently grown biomass.
Microscopy
For microscopic analysis, conidia were applied to either glass (non‐inductive of appressorium formation) or plastic (inductive) coverslips. Visualization of chitin was achieved with TRITC‐WGA (Sigma, Hamburg, Germany) or Calcofluor white (CFW; Sigma) at 10 µg/mL. Samples were observed and photographed with a fluorescence microscope (Zeiss Axioskop 2, HBO mercury lamp burner, Jena, Germany) and Zeiss filter sets 02 (excitation at 365 nm and emission at 420 nm for GFP and TRITC‐WGA) and 09 (excitation at 450–490 nm and emission at 515 nm for Calcofluor).
Sequence analysis and comparisons
Sequence comparisons were made using blastp and tblastn searches (Altschul and Lipman, 1990) via the National Center for Biotechnology Information (NCBI) non‐redundant database. Comparisons with other fungal genomes were conducted via http://www.broad.mit.edu (Broad Institute) for M. grisea (Dean et al., 2005), N. crassa (Galagan et al., 2003) and B. cinerea (http://www.broad.mit.edu/annotation/genome/botrytis_cinerea/Home.html), and via http://www.sanger.ac.uk/cgi‐bin/blast/blast_server for A. fumigatus (Nierman et al., 2005). Phylogenetic analyses were conducted with the aid of the program mega v.3.1 (Kumar et al., 2004). The program ClustalW (Thompson et al., 1994), using the Gonnet series protein weight matrix, was used to generate a multiple sequence alignment. This alignment was then used as the basis for construction of a neighbour‐joining tree, and the reliability of the resultant tree was tested using bootstrap resampling with 1000 replications.
Nucleotide sequence accession number
The M. grisea CHS7 nucleotide sequence (cDNA) for strain 70‐15 has been deposited in the GenBank database under the accession number EU935590.
ACKNOWLEDGMENTS
Nikkomycin Z was a kind gift from Professor H. P. Fiedler (University of Tübingen, Germany).
REFERENCES
- Altschul, S.F. and Lipman, D.J. (1990) Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. USA, 87, 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amnuaykanjanasin, A. and Epstein, L. (2006) A class Vb chitin synthase in Colletotrichum graminicola is localized in the growing tips of multiple cell types, in nascent septa, and during septum conversion to an end wall after hyphal breakage. Protoplasma, 227, 155–164. [DOI] [PubMed] [Google Scholar]
- Bartnicki‐Garcia, S. (1968) Cell wall chemistry, morphogenesis and taxonomy of filamentous fungi. Annu. Rev. Microbiol. 22, 97–108. [DOI] [PubMed] [Google Scholar]
- Bartnicki‐Garcia, S. (2006) Chitosomes: past, present and future. FEMS Yeast Res. 6, 957–965. [DOI] [PubMed] [Google Scholar]
- Borgia, P.T. , Iartchouk, N. , Riggle, P.J. , Winter, K.R. , Koltin, Y. and Bulawa, C.E. (1996) The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 20, 193–203. [DOI] [PubMed] [Google Scholar]
- Bowen, A.R. , Chen‐Wu, J.L. , Momany, M. , Young, R. , Szaniszlo, P.J. and Robbins, P.W. (1992) Classification of fungal chitin synthases. Proc. Natl. Acad. Sci. USA, 89, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik, D.A. , Olczak, M. , Lucero, H.A. , Osmond, B.C. , Robbins, P.W. and Specht, C.A. (2003) Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell, 2, 886–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, C.C.T. and Ellingboe, A.H. (1991) Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 69, 2130–2134. [Google Scholar]
- Chida, T. and Sisler, H.D. (1987) Restoration of appressorial penetration ability by melanin precursors in Pyricularia oryzae treated with antipenetrants and in melanin‐deficient mutants. J. Pestic. Sci. 12, 49–55. [Google Scholar]
- Choquer, M. , Boccara, M. , Goncalves, I.R. , Soulié, M.C. and Vidal‐Cros, A. (2004) Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 271, 2153–2164. [DOI] [PubMed] [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. , Thon, M. , Kulkarni, R. , Xu, J.R. , Pan, H. , Read, N.D. , Lee, Y.H. , Carbone, I. , Brown, D. , Oh, Y.Y. , Donofrio, N. , Jeong, J.S. , Soanes, D.M. , Djonovic, S. , Kolomiets, E. , Rehmeyer, C. , Li, W. , Harding, M. , Kim, S. , Lebrun, M.H. , Bohnert, H. , Coughlan, S. , Butler, J. , Calvo, S. , Ma, L.J. , Nicol, R. , Purcell, S. , Nusbaum, C. , Galagan, J.E. and Birren, B.W. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- Foster, A.J. , Jenkinson, J.M. and Talbot, N.J. (2003) Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea . EMBO J. 22, 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, M. , Ichinomiya, M. , Motoyama, T. , Horiuchi, H. , Ohta, A. and Takaqi, M. (2000) Evidence that the Aspergillus nidulans class I and II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J. Biochem. (Tokyo), 127, 359–366. [DOI] [PubMed] [Google Scholar]
- Galagan, J.E. , Calvo, S.E. , Borkovich, K.A. Selker, E.U. , Read, N.D. , Jaffe, D. , FitzHugh, W. , Ma, L.J. , Smirnov, S. , Purcell, S. , Rehman, B. , Elkins, T. , Engels, R. , Wang, S. , Nielsen, C.B. , Butler, J. , Endrizzi, M. , Qui, D. , Ianakiev, P. , Bell‐Pedersen, D. , Nelson, M.A. , Werner‐Washburne, M. , Selitrennikoff, C.P. , Kinsey, J.A. , Braun, E.L. , Zelter, A. , Schulte, U. , Kothe, G.O. , Jedd, G. , Mewes, W. , Staben, C. , Marcotte, E. , Greenberg, D. , Roy, A. , Foley, K. , Naylor, J. , Stange‐Thomann, N. , Barrett, R. , Gnerre, S. , Kamal, M. , Kamvysselis, M. , Mauceli, E. , Bielke, C. , Rudd, S. , Frishman, D. , Krystofova, S. , Rasmussen, C. , Metzenberg, R.L. , Perkins, D.D. , Kroken, S. , Cogoni, C. , Macino, G. , Catcheside, D. , Li, W. , Pratt, R.J. , Osmani, S.A. , DeSouza, C.P. , Glass, L. , Orbach, M.J. , Berglund, J.A. , Voelker, R. , Yarden, O. , Plamann, M. , Seiler, S. , Dunlap, J. , Radford, A. , Aramayo, R. , Natvig, D.O. , Alex, L.A. , Mannhaupt, G. , Ebbole, D.J. , Freitag, M. , Paulsen, I. , Sachs, M.S. , Lander, E.S. , Nusbaum, C. and Birren, B. (2003) The genome sequence of the filamentous fungus Neurospora crassa . Nature, 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Gilbert, R.D. , Johnson, A.M. and Dean, R.A. (1996) Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea . Phys. Mol. Plant Pathol. 48, 335–346. [Google Scholar]
- De Groot, M.J. , Bundock, P. , Hooykaas, P.J. , and Beijersbergen, A.G. (1998) Agrobacterium tumefaciens‐mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842. [DOI] [PubMed] [Google Scholar]
- Hartmann, E. , Rapoport, T.A. and Lodish, H.F. (1989) Predicting the orientation of eukaryotic membrane‐spanning proteins. Proc. Natl. Acad. Sci. USA, 86, 5786–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters, M. , DeWaele, D. , Depicker, A. , Messens, E. , Van Montagu, M. and Schell, J. (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187. [DOI] [PubMed] [Google Scholar]
- Howard, R.J. and Ferrari, M.A. (1989) Role of melanin in appressorium function. Exp. Mycol. 13, 403–418. [Google Scholar]
- Ichinomiya, M. , Motoyama, T. , Fujiwara, M. , Takagi, M. , Horiuchi, H. and Ohta, A. (2002) Repression of chsB expression reveals the functional importance of class IV chitin synthase gene chsD in hyphal growth and conidiation of Aspergillus nidulans. Microbiology, 148, 1335–1347. [DOI] [PubMed] [Google Scholar]
- Ichinomiya, M. , Ohta, A. and Horiuchi, H. (2005) Expression of asexual developmental regulator gene abaA is affected in the double mutants of classes I and II chitin synthase genes, chsC and chsA, of Aspergillus nidulans . Curr. Genet. 48, 171–183 [DOI] [PubMed] [Google Scholar]
- Kamakura, T. , Yamaguchi, S. , Saitoh, K. , Teraoka, T. and Yamaguchi, I. (2002) A novel gene, CBP1, encoding a putative extracellular chitin‐binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant–Microbe Interact. 15, 437–444. [DOI] [PubMed] [Google Scholar]
- Kimura, M. , Izawa, K. , Yoneyama, K. , Arie, T. , Kamakura, T. and Yamaguchi, I. (1995) A novel transformation system for Pyricularia oryzae: adhesion of regenerating fungal protoplasts to collagen‐coated dishes. Biosci. Biotechnol. Biochem. 59, 1177–1180. [Google Scholar]
- Klis, F.M. (1994) Review: cell wall assembly in yeast. Yeast, 10, 851–869. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Tamura, K. and Nei, M. (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163. [DOI] [PubMed] [Google Scholar]
- Madrid, M.P. , Di Pietro, A. and Roncero, M.I. (2003) Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47, 257–266. [DOI] [PubMed] [Google Scholar]
- Martín‐Urdíroz, M. , Madrid, M.P. and Roncero, M.I. (2004). Role of chitin synthase genes in Fusarium oxysporum. Microbiology, 150, 3175–3187. [DOI] [PubMed] [Google Scholar]
- Martín‐Urdíroz, M. , Roncero, M.I. , González‐Reyes, J.A. and Ruiz‐Roldán, C. (2008) ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum . Eukaryot. Cell, 7, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado, E. , Specht, C.A. , Robbins, P.W. and Holden, D.W. (1996) Cloning and characterization of chsD, a chitin synthase‐like gene of Aspergillus fumigatus. FEMS Microbiol. Lett. 143, 69–76. [DOI] [PubMed] [Google Scholar]
- Nierman, W.C. , Pain, A. , Anderson, M.J. , Wortman, J.R. , Kim, H.S. , Arroyo, J. , Berriman, M. , Abe, K. , Archer, D.B. , Bermejo, C. , Bennett, J. , Bowyer, P. , Chen, D. , Collins, M. , Coulsen, R. , Davies, R. , Dyer, P.S. , Farman, M. , Fedorova, N. , Fedorova, N. , Feldblyum, T.V. , Fischer, R. , Fosker, N. , Fraser, A. , Garcia, J.L. , Garcia, M.J. , Goble, A. , Goldman, G.H. , Gomi, K. , Griffith‐Jones, S. , Gwilliam, R. , Haas, B. , Haas, H. , Harris, D. , Horiuchi, H. , Huang, J. , Humphray, S. , Jimenez, J. , Keller, N. , Khouri, H. , Kitamoto, K. , Kobayashi, T. , Konzack, S. , Kulkarni, R. , Kumagai, T. , Lafton, A. , Latge, J.P. , Li, W.X. , Lord, A. , Majoros, W.H. , May, G.S. , Miller, B.L. , Mohamoud, Y. , Molina, M. , Monod, M. , Mouyna, I. , Mulligan, S. , Murphy, L. , O’Neil, S. , Paulsen, I. , Penalva, M.A. , Pertea, M. , Price, C. , Pritchard, B.L. , Quail, M.A. , Rabbinowitsch, E. , Rawlins, N. , Rajandream, M.A. , Reichard, U. , Renauld, H. , Robson, G.D. , De Cordoba, S.R. , Rodriguez‐Pena, J.M. , Ronning, C.M. , Rutter, S. , Salzberg, S.L. , Sanchez, M. , Sanchez‐Ferrero, J.C. , Saunders, D. , Seeger, K. , Squares, R. , Squares, S. , Takeuchi, M. , Tekaia, F. , Turner, G. , De Aldana, C.R.V. , Weidman, J. , White, O. , Woodward, J. , Yu, J.H. , Fraser, C. , Galagan, J.E. , Asai, K. , Machida, M. , Hall, N. , Barrell, B. and Denning, D.W. (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus . Nature, 438, 1151–1156. [DOI] [PubMed] [Google Scholar]
- Niño‐Vega, G.A. , Carrero, L. and San‐Blas, G. (2004) Isolation of the CHS4 gene of Paracoccidioides brasiliensis and its accommodation in a new class of chitin synthases. Med. Mycol. 42, 51–57. [DOI] [PubMed] [Google Scholar]
- Odenbach, D. , Breth, B. , Thines, E. , Weber, R.W.S. , Anke, H. and Foster A.J. (2007) The transcription factor Con7p is a central regulator of infection‐related morphogenesis in the rice blast fungus Magnaporthe grisea . Mol. Microbiol. 64, 293–307. [DOI] [PubMed] [Google Scholar]
- Orlean, P. (1987) Two chitin synthases in Saccharomyces cerevisiae . J. Biol. Chem. 262, 5732–5739. [PubMed] [Google Scholar]
- Park, C. , Horiuchi, H. , Hwang, C.W. , Yeh, W.H. , Ohta, A. , Ryu, J.C. and Takagi, M. (1999) Isolation of csm1 encoding a class V chitin synthase with a myosin motor‐like domain from the rice blast fungus, Pyricularia grisea . FEMS Microbiol. Lett. 170, 131–139. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho, H.S. , Kang, S. and Lee, Y.H. (2001) Agrobacterium tumefaciens‐mediated transformation of the plant pathogenic fungus, Magnaporthe grisea . Mol. Cells, 12, 407–411. [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989). Molecular Cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Soulié, M.‐C. , Piffeteau, A. , Choquer, M. , Boccara, M. and Vidal‐Cros, A. (2003) Disruption of Botrytis cinerea class I chitin synthase gene Bcchs1 results in cell wall weakening and reduced virulence. Fungal Genet. Biol. 40, 38–46. [DOI] [PubMed] [Google Scholar]
- Soulié, M.‐C. , Perino, C. , Piffeteau, A. , Choquer, M. , Malfatti, P. , Cimerman, A. , Kunz, C. , Boccara, M. and Vidal‐Cros, A. (2006) Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a ). Cell Microbiol. 8, 1310–1321. [DOI] [PubMed] [Google Scholar]
- Takeshita, N. , Ohta, A. and Horiuchi, H. (2005) CsmA, a class V chitin synthase with a myosin motor‐like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans . Mol. Biol. Cell, 16, 1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, N. , Yamashita, S. , Ohta, A. and Horiuchi, H. (2006) Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor‐like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 59, 1380–1394. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J. , Ebbole, D.J. , and Hamer, J.E. (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell, 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal‐Cros, A. and Boccara, M. (1998) Identification of four chitin synthase genes in the rice blast disease agent Magnaporthe grisea . FEMS Microbiol. Lett. 165, 1, 103–109. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Zheng, L. , Hauser, M. , Becker, J.M. and Szaniszlo, P.J. (1999) WdChs4p, a homolog of chitin synthase 3 in Saccharomyces cerevisiae, alone cannot support growth of Wangiella (Exophiala) dermatitidis at the temperature of infection. Infect. Immun. 67 (12), 6619–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, I. , Assmann, D. , Thines, E. and Steinberg, G. (2006) Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis . Plant Cell, 18, 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, S. , Sugui, J.A. , Steinberg, G. and Deising, H.B. (2007) A chitin synthase with a myosin‐like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola . Mol. Plant–Microbe Interact. 20, 1555–1567. [DOI] [PubMed] [Google Scholar]
- Yanai, K. , Kojima, N. , Takaya, N. , Horiuchi, H. , Ohta, A. and Takagi, M. (1994) Isolation and characterization of two chitin synthase genes from Aspergillus nidulans . Biosci. Biotechnol. Biochem. 58, 1828–1835. [DOI] [PubMed] [Google Scholar]


