SUMMARY
Taxonomy: Pepino mosaic virus (PepMV) belongs to the Potexvirus genus of the Flexiviridae family.
Physical properties: PepMV virions are nonenveloped flexuous rods that contain a monopartite, positive‐sense, single‐stranded RNA genome of 6.4 kb with a 3′ poly‐A tail. The genome contains five major open reading frames (ORFs) encoding a 164‐kDa RNA‐dependent RNA polymerase (RdRp), three triple gene block proteins of 26, 14 and 9 kDa, and a 25‐kDa coat protein.
Genome diversity: Four PepMV genotypes, with an intergenotype RNA sequence identity ranging from 78% to 95%, can be distinguished: the original Peruvian genotype (LP); the European (tomato) genotype (EU); the American genotype US1; and the Chilean genotype CH2.
Transmission: PepMV is very efficiently transmitted mechanically, and a low seed transmission rate has been demonstrated. In addition, bumblebees have been associated with viral transmission.
Host range: Similar to other Potexviruses, PepMV has a rather narrow host range that is thought to be largely restricted to species of the Solanaceae family. After originally being isolated from pepino (Solanum muricatum), PepMV has been identified in natural infections of the wild tomato species S. chilense, S. chmielewskii, S. parviflorum and S. peruvianum. PepMV is causing significant problems in the cultivation of the glasshouse tomato Solanum lycopersicum, and has been identified in weeds belonging to various plant families in the vicinity of tomato glasshouses.
Symptomatology: PepMV symptoms can be very diverse. Fruit marbling is the most typical and economically devastating symptom. In addition, fruit discoloration, open fruit, nettle‐heads, leaf blistering or bubbling, leaf chlorosis and yellow angular leaf spots, leaf mosaic and leaf or stem necrosis have been associated with PepMV. The severity of PepMV symptoms is thought to be dependent on environmental conditions, as well as on the properties of the viral isolate. Minor nucleotide sequence differences between isolates from the same genotype have been shown to lead to enhanced aggressiveness and symptomatology.
Control: Prevention of infection through strict hygiene measures is currently the major strategy for the control of PepMV in tomato production. Cross‐protection can be effective, but only under well‐defined and well‐controlled conditions, and the effectiveness depends strongly on the PepMV genotype.
INTRODUCTION
Pepino mosaic virus (PepMV) was observed for the first time in tomato (Solanum lycopersicum) crops in the Netherlands only a decade ago [European Plant Protection Organization (EPPO), 2000], and is currently a major disease of glasshouse tomato crops worldwide (Cotillon et al., 2002; French et al., 2001; Hanssen et al., 2008; Hasiów et al., 2008; Ling, 2006; Ling et al., 2008; Maroon‐Lango et al., 2005; Mumford and Metcalfe, 2001; Pagán et al., 2006; van der Vlugt et al., 2000). The economic impact of PepMV on the tomato industry has been strongly debated, as the impact largely depends on the structure of the tomato market, more specifically on the marketability and economic value of lower quality fruits, which differs considerably between growing areas (Jones and Lammers, 2005; Spence et al., 2006). Furthermore, the large variability in the nature and severity of symptom display complicates a reliable determination of the economic impact of PepMV on the tomato industry. In a questionnaire conducted among Belgian tomato growers, yield losses caused by PepMV were estimated to be between 5% and 10% in the 2006 growth season and negligible in 2005, whereas fruit quality losses were more pronounced in 2005 (Hanssen et al., 2009a). Glasshouse trials conducted in the UK from 2001 to 2003 revealed considerable differences in damage between subsequent years, with the percentage of downgraded tomato fruit caused by PepMV‐induced quality loss ranging from 6% to 38%.
In this pathogen profile, we review the current knowledge on PepMV biology, genome diversity, population dynamics, symptomatology, transmission and control.
HOST RANGE AND SYMPTOMATOLOGY
As indicated by its name, PepMV was originally isolated in Peru from pepino (Solanum muricatum) that showed yellow leaf mosaic symptoms (Jones et al., 1980). Its host range is thought to be mainly restricted to Solanaceae species (Salomone and Roggero, 2002; Soler et al., 2002; Verhoeven et al., 2003). In a survey in central and southern Peru, the virus has been identified in natural infections of the wild tomato species S. chilense, S. chmielewskii, S. parviflorum and S. peruvianum (Soler et al., 2002). Furthermore, by performing mechanical inoculations, the host range of PepMV has been shown to contain eggplant (Solanum melongena), potato (Solanum tuberosum) and species from the genera Nicotiana (e.g. N. benthamiana), Datura (e.g. D. stramonium), Capsicum (C. annuum) and Physalis (P. floridana) (Jones et al., 1980; Martin and Mousserion, 2002; Salomone and Roggero, 2002; Verhoeven et al., 2003). So far, basil (Ocimum basilicum; Lamiaceae) is the only reported natural host that does not belong to the Solanaceae, with plants displaying interveinal chlorosis (Davino et al., 2009). Furthermore, in a survey of 42 native weed species growing in or around tomato production sites in Spain, PepMV infection was found in 18 weed species, including those belonging to the Amaranthaceae (e.g. Chenopodium murale), Convolvulaceae (e.g. Calystegia sepium), Brassicaceae (e.g. Diplotaxis erucoides), Boraginaceae (e.g. Heliotropium europaeum), Asteraceae (e.g. Sonchus tenerrimus), Plantaginaceae (Plantago afra) and Polygonaceae (Rumex sp.) (Córdoba et al., 2004). Interestingly, a recent study has revealed that co‐inoculation with EU (European genotype) and CH2 (Chilean genotype) isolates extended the host range beyond that of the single isolates (Gómez et al., 2009). More specifically, neither the EU isolate Sp13, nor the CH2 isolate PS5, could establish infection in N. glutinosa or N. tabacum, whereas both host plants appeared to be susceptible on inoculation with a mixture of the two isolates (Gómez et al., 2009).
PepMV symptomatology has been most extensively studied in cultivated tomato. A wide range of symptoms have been associated with PepMV infection. Fruit marbling is generally considered to be the most devastating symptom of PepMV infection as it diminishes the economic value of the crop, but other fruit symptoms, such as discoloration (blotchy ripening or flaming) and the occurrence of ‘open fruit’ (fruit that splits shortly after setting, such that the seeds become visible), can be similarly devastating (Fig. 1; Hanssen et al., 2008; Spence et al., 2006). Symptoms on vegetative plant parts may comprise nettle‐heads (upper young leaves distorted, serrated and upright with a reduced surface), leaf blistering or bubbling, chlorosis and yellow angular leaf spots, but also severe leaf mosaics and even leaf or stem necrosis (Fig. 1; Hanssen et al., 2009b; Hasiów et al., 2008; Hasiów‐Jaroszewska et al., 2009a; Roggero et al., 2001; Spence et al., 2006; van der Vlugt et al., 2000). In addition, it has been suggested that the so‐called ‘tomato collapse’ disease, a sudden and progressive wilt of tomato which eventually leads to plant death, is caused by necrosis of the vascular system as a result of PepMV accumulation (Soler‐Aleixandre et al., 2005).
Figure 1.
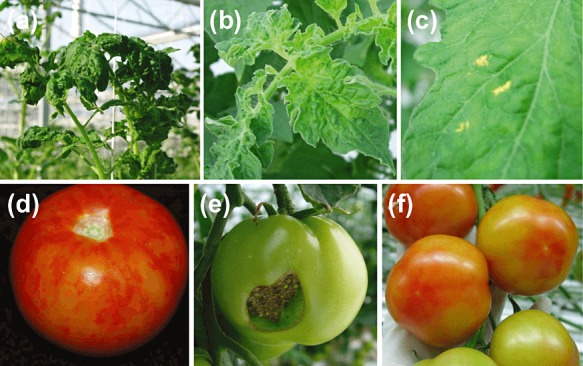
Typical Pepino mosaic virus (PepMV) symptoms on tomato: (a) nettlehead of young top leaves; (b) leaf bubbling; (c) yellow spots; (d) fruit marbling; (e) open fruit; (f) fruit discoloration (flaming).
TRANSMISSION
PepMV is efficiently transmitted mechanically (Jones et al., 1980). The virus is highly contagious in tomato, as it easily spreads by the standard crop handling procedures in a glasshouse through contaminated tools, hands and clothing and by direct plant‐to‐plant contact (Spence et al., 2006; Wright and Mumford, 1999). Therefore, once the virus enters a tomato production facility, the containment of further spread is virtually impossible and it is usual for all plants to become infected eventually. It has been shown that bumblebees, often used for pollination in commercial tomato production, contribute to the spread of the virus (Lacasa et al., 2003; Shipp et al., 2008). In infected glasshouse tomato crops, nearly all bumblebees have been shown to carry PepMV, and vectoring of the virus to noninfected plants has been demonstrated (Shipp et al., 2008). On the basis of infection levels in flowers, fruits and leaves, it has been suggested that the infection occurs first in pollinated flowers and then spreads to other parts of the plant. Whether infection occurs through direct injury to the flowers or through fertilization with infected pollen could not be determined (Shipp et al., 2008).
Recently, the root‐infecting parasitic fungus Olpidium virulentus, which has been implicated in the transmission of several plant viruses, has been shown to be able to enhance PepMV spread (Alfaro‐Fernández et al., 2009). In addition, it has been reported that PepMV can be efficiently transmitted by nutrient solution in a closed recirculation system, leading to the infection of healthy tomato plants, although the virus itself was not detectable directly in the nutrient solution (Schwarz et al., 2009).
Seed transmission of PepMV has been demonstrated in several studies, with rates up to ∼2% depending on the time of seed harvest, the tomato variety and the seed cleaning or disinfection methods applied (Córdoba‐Sellés et al. 2007; Hanssen et al., 2009d; Krinkels, 2001;Ling, 2008). In a recently performed grow‐out trial with over 87 000 seedlings, a seed transmission rate of 0.026% was found for seeds cleaned according to industry standards without disinfection (Hanssen et al., 2009d). Interestingly, the rate of transmission increased as the interval between the infection of the mother crop and seed harvest increased. Disinfection treatments have been shown to efficiently reduce the seed transmission rate (Córdoba‐Sellés et al. 2007). Although the efficiency of seed transmission is low, the highly infectious nature of PepMV implies a substantial risk associated with tomato seeds harvested from an infected crop. Therefore, strict regulations aimed at eliminating the risk of viral spread through seeds are in place in the European Union (Commission Decision 2001/536/EC and 2004/200/EC). In addition, sensitive quantitative TaqMan reverse transcriptase‐polymerase chain reaction (RT‐PCR) detection methods to screen tomato seed lots for the presence of PepMV have been developed (Gutiérrez‐Aguirre et al., 2009; Ling et al., 2007). As a consequence of these measures, long‐distance dissemination of PepMV most probably occurs by the transfer of young infected plants from the nursery to the grower through infected grafts, cuttings or fruits, and even through seed‐to‐seedling transmission (Córdoba‐Sellés et al., 2007).
GENOME ORGANIZATION AND DIVERSITY
PepMV belongs to the Potexvirus genus of the Flexiviridae family. Virions are nonenveloped flexuous rods of 508 nm (Jones et al., 1980). The positive single‐stranded RNA genome is 6.4 kb in length and consists of five open reading frames (ORFs), 5′ and 3′ untranslated regions (UTRs) and a 3′ poly‐A tail. The ORFs encode a 164‐kDa RNA‐dependent RNA polymerase (RdRp) which contains the characteristic methyltransferase, nucleoside triphosphate (NTP)‐binding and polymerase motifs, three triple gene block (TGB) proteins of 26, 14 and 9 kDa (assigned TGBp1, TGBp2 and TGBp3, respectively) and a 25‐kDa coat protein (Fig. 2; Aguilar et al., 2002; Cotillon et al., 2002). Phylogenetic analyses of replicase, TGBp1 and coat protein amino acid sequences have revealed that PepMV is closely related to Narcissus mosaic virus (NMV), Scallion virus X (SVX), Cymbidium mosaic virus (CymMV) and Potato aucuba mosaic virus (PAMV) (Cotillon et al. 2002). The highest overall nucleotide identities are with NMV and CymMV (Aguilar et al., 2002). Initially, the genetic characteristics, symptomatology and host range of different European PepMV isolates showed high similarity, suggesting a common origin of these isolates (Mumford and Metcalfe, 2001; Verhoeven et al., 2003). Nucleotide sequence comparisons of coat protein genes of 15 isolates originating from different European countries revealed 99% identity among the isolates, but these isolates shared only 96%–97% identity with the original Peruvian pepino isolate (BBA1137; Mumford and Metcalfe, 2001). Therefore, and because the Peruvian pepino isolate does not cause symptoms in tomato, European isolates were considered to be a distinct PepMV type (Mumford and Metcalfe, 2001; van der Vlugt et al., 2000). A comparative symptomatology and host range study of 15 PepMV tomato isolates and the original pepino isolate BBA1137 confirmed that the pepino isolate differed from the tomato isolates, as only the pepino isolate (occasionally) caused symptoms in N. tabacum, C. annuum and P. floridana (Verhoeven et al., 2003). The comparison of the complete nucleotide sequences of two tomato isolates with an isolate from L. peruvianum (LP‐2001), which was symptomless in tomato, demonstrated that the tomato isolates shared over 99% identity, whereas they shared approximately 96% identity with the LP‐2001 genome (Soler et al., 2002). In addition, a two‐nucleotide deletion and some polymorphisms were identified in the 5′ UTR, and TGBp3 contained two extra amino acids. These differences have been suggested to play a role in the differential biological characteristics (López et al. 2005). At that time, only part of the sequence of the original PepMV isolate from pepino, BBA1137, had been determined. As partial sequence comparison revealed a high identity between LP‐2001 and BBA1137, the sequence of LP‐2001 was considered to be a reference for the original pepino strain. Complete sequence determination of BBA1137 confirmed that both isolates share nucleotide sequence homologies of over 99%, and can thus be considered as isolates from the distinct pepino or Peruvian type of PepMV, which is further referred to as LP (Pagán et al., 2006).
Figure 2.
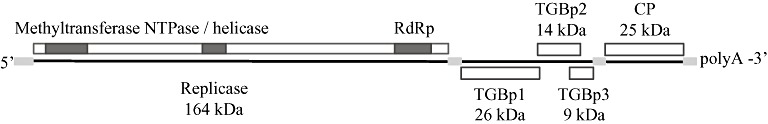
Schematic overview of the Pepino mosaic virus (PepMV) genome organization, displaying the encoded gene products (adapted from Cotillon et al., 2002). Untranslated regions are shown as light grey bars. CP, coat protein; NTPase, nucleosidetriphosphatase; RdRp, RNA‐dependent RNA polymerase; TGBp, triple gene block protein. The size and overlap of the proteins are proportional to the actual sizes.
In 2005, two distinct isolates originating from US tomato production, designated US1 and US2, were described that shared only 86% sequence identity (Table 1; Maroon‐Lango et al., 2005). Moreover, they shared only 78% and 81% sequence identities with the so‐called European tomato isolates (Table 1; Maroon‐Lango et al., 2005). As both US isolates caused disease in tomato, the designation ‘tomato strain’, which had been used for the European isolates until then, was no longer appropriate, and most authors started to refer to this group of isolates as the ‘European (tomato) genotype (EU)’ (Hanssen et al., 2008; Maroon‐Lango et al., 2005; Pagán et al., 2006; Pospieszny and Borodynko, 2006). In addition to EU, US1 and US2 genotypes, a divergent genotype was isolated from tomato seeds originating from Chile, and designated CH2 (Ling, 2006). This CH2 isolate shared 78%–80% nucleotide sequence identity with the LP and EU genotypes and 78% identity with US1 (Table 1). Phylogenetic analyses revealed two main clusters, one containing the EU and LP genotypes and the other consisting of the more recently described US and CH2 genotypes, suggesting two distinct evolutionary routes (Fig. 3). As nucleotide sequence comparisons suggested that US2 was a recombinant of US1 and CH2, it was proposed that four PepMV genotypes could be distinguished: the original Peruvian genotype (LP); the European (tomato) genotype (EU); the American genotype US1; and the Chilean genotype CH2 (Hanssen et al., 2009d).
Table 1.
Nucleotide sequence identities between type isolates of the different Pepino mosaic virus (PepMV) genotypes.
| PepMV genotype | GenBank accession | LP (LP2001) | CH2 | US1 | US2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G* | R† | T‡ | C§ | G | R | T | C | G | R | T | C | G | R | T | C | ||
| EU¶ | AJ438767 | 95 | 95 | 96 | 97 | 79 | 78 | 79 | 77 | 82 | 81 | 85 | 83 | 79 | 79 | 80 | 77 |
| LP** | AJ606361 | 79 | 78 | 80 | 77 | 82 | 81 | 85 | 84 | 79 | 78 | 80 | 77 | ||||
| CH2†† | DQ000985 | 78 | 77 | 80 | 80 | 90 | 88 | 92 | 99 | ||||||||
| US1‡‡ | AY509926 | 86 | 87 | 86 | 80 | ||||||||||||
| US2‡‡ | AY509927 | ||||||||||||||||
Genome (complete sequence).
Replicase gene.
Triple gene block.
Coat protein gene.
Figure 3.
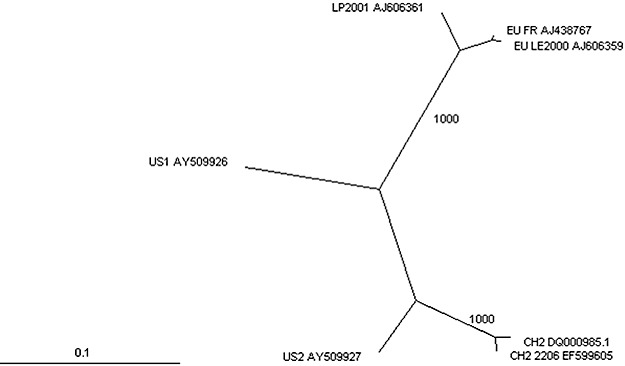
Unrooted distance tree of complete nucleotide sequences from various Pepino mosaic virus (PepMV) genotypes, including sequences from isolates FR (Cotillon et al., 2002) and LE2000 (López et al., 2005) of the EU genotype, isolates CH2 (Ling, 2006) and the Belgian ‘2206/06/A1’ (Hanssen et al., 2008) of the CH2 genotype, LP2001 (López et al., 2005) of the LP genotype, and US1 and US2 (Maroon‐Lango et al., 2005). GenBank accession numbers are indicated in the figure. The tree was generated using ClustalX with 1000 bootstrap values, and visualized using Treeview. The scale bar represents 0.1 changes per nucleotide.
Pep MV POPULATION DYNAMICS
Since first appearing in glasshouse tomato crops in the Netherlands in 1999, PepMV has rapidly established itself in tomato‐producing countries. An unprecedented, worldwide series of PepMV outbreaks within just a few years' time was reported, with disease reports from the UK, France, Italy and Spain, but also China, Canada and the USA (Aguilar et al., 2002; Cotillon et al., 2002; French et al., 2001; Jordáet al., 2001; Mumford and Metcalfe, 2001; Roggero et al., 2001; Yaoliang and Zhongjian, 2003). Initially, all reported outbreaks were caused by the EU genotype of PepMV and the reported symptoms were rather mild. However, a study on the genetic structure of the PepMV population in Spain in 2005 revealed that the population was more diverse than assumed (Pagán et al., 2006). Although the EU genotype was dominant in Spanish tomato production, the LP genotype appeared to be present on the Canary Islands already in 2000, and US2‐like isolates were present in peninsular Spain in 2004 (Pagán et al., 2006). In addition, the occurrence of mixed infections with two different genotypes (combinations of EU and US2‐like, and of LP and US2‐like), and with intergenotype recombinants, was revealed. In 2006, the genetic diversity of the PepMV population in Belgian glasshouses was studied, revealing the occurrence of isolates belonging to the EU and CH2 genotypes, often in mixed infections, and the presence of recombinants (Hanssen et al., 2008). A remarkable finding was the dominance of the CH2 genotype, which had not been reported in Europe until then, occurring in 85% of infected crops, whereas the EU genotype occurred in less than 50% of these crops (Hanssen et al., 2008). In addition, the majority of recent PepMV isolates from Dutch and French tomato crops that were genotyped belonged to the CH2 genotype (I. M. Hanssen et al., unpublished data). In 2002 and 2005, two distinct Polish PepMV isolates were obtained from tomato, and were shown to belong to the EU genotype and CH2 genotype, respectively (Pospieszny and Borodynko, 2006; Pospieszny et al., 2002). Two additional Polish PepMV strains, isolated in 2007, that differed from the previously identified PepMV isolates in host range and symptomatology, appeared to belong to the CH2 genotype (Hasiów‐Jaroszewska et al., 2009a). Altogether, these results are indicative of a shift in the PepMV population, with the EU genotype gradually being overtaken by the CH2 genotype. It has been suggested that the CH2 genotype has a biological advantage over the EU genotype, as it seems to spread more rapidly within a crop (Hanssen et al., 2008). This was confirmed by a recent study on the evolutionary dynamics of the PepMV population in Spain, in which RT‐quantitative PCR analyses in inoculated tomato plants showed that a CH2 isolate (PS5) accumulated more rapidly and to higher viral loads than an EU isolate (Sp13) (Gómez et al., 2009). This study further revealed that PepMV populations in Spain are composed of isolates belonging to the EU and CH2 genotypes, and that the CH2 type is predominant. Interestingly, EU isolates were shown to persist in mixed infections, and it was suggested that mixed infections contribute to the shaping of the population structure (Gómez et al., 2009). In addition, mutational analyses revealed very few nonsynonymous substitutions, reflecting strong purifying selection. These results are in line with the observation that the number of mutations in the RNA sequence of PepMV isolates throughout a glasshouse trial period is rather limited, and that most of the mutations that take place have no clear biological relevance (Hanssen et al., 2009b).
Recent data on the Belgian PepMV population showed a further dominance of the CH2 genotype as, in the 2008 growth season, the EU genotype was detected in only 7% of infected crops, whereas the CH2 genotype occurred in 90% of infected crops. Interestingly, the LP genotype was also detected, with an incidence of 10% (I. M. Hanssen et al., unpublished data). The sudden occurrence of the LP genotype in Belgian tomato production may perhaps be linked to the use of a mild LP isolate in the Netherlands for cross‐protection in commercial tomato glasshouses (Hanssen et al., 2009c).
In retrospect, the US2‐like sequences reported from Spain (Pagán et al., 2006) displayed high sequence identity to the later described CH2 genotype (Ling, 2006), suggesting that CH2, rather than US2, was present in Spain in 2004, indicating that the current PepMV CH2 epidemic in Europe had been initiated in or before 2004. Intriguingly, a recent population study on PepMV isolates in 31 infected North American glasshouse tomato crops revealed the occurrence of the EU, US1, US2 and CH2 genotypes, with a clear dominance of the EU genotype, which was identified in all 31 crops (Ling et al., 2008). The remaining genotypes were found only rarely and exclusively in mixed infections. The low incidence of the CH2 genotype in North America is remarkable, and might reflect different PepMV dissemination pathways linked to a different, less intensive structure of tomato growth facilities in North America relative to Europe, where PepMV is mainly prevalent in dense glasshouse tomato cultivation areas. In the American situation, mechanical transmission through workers or bumblebees may be subordinate to the long‐distance transmission through young plants and seeds.
The factors contributing to PepMV population dynamics are currently unknown. However, recently, the existence of a population bottleneck during seed transmission has been reported, with an apparent advantage of the EU genotype in transmission through seeds harvested from a mother crop co‐infected by the EU and CH2 genotypes (Hanssen et al., 2009c). As seed transmission was suggested to be a major dissemination route of PepMV in 1999 and 2000, before strict sanitary regulations were in place (Córdoba‐Sellés et al., 2007), this putative population bottleneck might be related to the original dominance of the EU genotype in European countries.
Recently, the US1 genotype has been isolated in the Canary Islands from glasshouse tomato crops displaying leaf blistering and mosaic (Alfaro‐Fernández et al., 2008); this is the first time that this genotype has been isolated in a different location from that originally reported (North America; Maroon‐Lango et al., 2005).
THE DIVERSITY OF SYMPTOM SEVERITY
The symptom intensity in PepMV‐infected tomato crops is highly variable, ranging from asymptomatic infections to very severe symptomatology (Hanssen et al., 2008; Jordáet al., 2001; Soler et al., 2000; Soler‐Aleixandre et al., 2005). Observations in commercial tomato production have led to the hypothesis that environmental conditions play an important role in symptom severity. Low environmental temperatures and low light conditions are thought to result in more severe damage (Jordáet al., 2001). Furthermore, the PepMV‐associated ‘tomato collapse’ is thought to be enhanced by temperature fluctuations throughout the growth season (Soler‐Aleixandre et al., 2005). PepMV‐induced leaf scorching was attributed to a period of high light intensity followed by a period of dull weather (Spence et al., 2006). However, a PepMV trial conducted under high‐light conditions in 2003 resulted in considerably more damage than a trial conducted under lower light conditions in 2001–02 (Spence et al., 2006). Although growers in Belgium and the Netherlands have confirmed the importance of light and temperature, the interplay of environmental factors contributing to PepMV damage appears to be complex and remains to be elucidated.
As the impact of environmental growth conditions and tomato genotype on PepMV symptom development is not yet fully understood, it is not clear whether the differences in symptom display in commercial tomato glasshouses should also be attributed to the viral isolate that invades the crop. In a population study conducted in Spanish tomato crops, no correlation between PepMV genotypes and symptomatology was found (Pagán et al., 2006). In a similar study of Belgian tomato crops, no significant differences in symptom severity were detected between EU and CH2 isolates (Hanssen et al., 2008). However, tomato crops that were simultaneously infected with isolates of both genotypes showed significantly enhanced symptom display on all plant parts when compared with crops infected with a single isolate (Hanssen et al., 2008). Nevertheless, increasing evidence is accumulating that shows a clear role of the viral isolate in PepMV symptomatology. Studies on Polish PepMV isolates revealed clear differences in host range and symptomatology of different isolates belonging to the CH2 genotype (Hasiów‐Jaroszewska et al., 2009a; Pospieszny et al., 2008). Three necrotic CH2 isolates sharing over 99% sequence identity with non‐necrotic isolates from the CH2 genotype have been identified recently (Hasiów‐Jaroszewska et al., 2009a). Evidence for a role of the viral isolate in PepMV symptomatology has also been found in recent glasshouse inoculation experiments (Hanssen et al., 2009b). A CH2 isolate selected on the basis of mild symptom expression in the crop of origin caused only mild symptoms in the trial, whereas another isolate with a sequence identity of 99.4%, which was selected on the basis of severe symptom display in the crop of origin, caused significantly more severe symptoms in the same trial, including nettle‐head and a high incidence of premature leaf senescence, open fruit and fruit flaming. These results demonstrate that minor differences at the nucleotide level can account for considerable differences in symptomatology between isolates that infect crops under the same conditions. Nevertheless, currently, it remains unclear which regions of the PepMV genome are important for symptomatology. The recent development of an infectious clone derived from a necrotic CH2 isolate is an important step forward to elucidate the role of certain regions and residues in PepMV symptomatology (Hasiów‐Jaroszewska et al., 2009b).
HOST RESPONSES TO Pep MV
Global transcriptional profiling, for instance with the use of micro‐arrays, can provide an insight into the cellular biology of the host on pathogen infection (van Baarlen et al., 2008; Quirino and Bent, 2003; Wise et al., 2007). As viruses establish infection in plants by exploiting the cellular components of the host, viruses can induce a wide range of alterations in host gene expression (Whitham et al., 2003). To date, micro‐array studies have been undertaken for a limited number of viral interactions with their hosts (Whitham et al., 2006; Wise et al., 2007). In most compatible plant–virus interactions, a general virus‐induced host gene repression occurs shortly after infection (Maule et al., 2002). However, genes related to cell death, cell rescue, defence, ageing and stress are often induced in response to viral infection (Marathe et al., 2004; Senthil et al., 2005; Whitham et al., 2003). Another important virus‐induced host response is the induction of the RNA silencing machinery of the plant, which degrades or modifies viral RNAs to block the translation of viral proteins (Baulcombe, 2004). This virus‐induced post‐transcriptional gene silencing (PTGS) mechanism involves the processing of viral double‐stranded RNA (dsRNA) by Dicer‐like enzymes (DCL) into small interfering RNAs (siRNAs), which are subsequently incorporated into protein complexes containing endonucleolytic Argonaute enzymes (Ding and Voinnet, 2007). PTGS is thought to be the mechanism behind the long‐known ‘recovery’ phenomenon, first described by Wingard (1928), who observed that the upper leaves of tobacco plants infected with tobacco ringspot virus were asymptomatic and resistant to secondary infection (Baulcombe, 2004; Ratcliff et al., 1999). Host‐adapted viruses have evolved strategies to counteract PTGS in their hosts by encoding viral suppressors of RNA silencing (Ding and Voinnet, 2007). As viruses are inducers, suppressors and targets of the RNA silencing mechanism, virus‐induced symptom development in infected plants can be influenced by siRNA pathways in many different ways (Baulcombe, 2004), for example by perturbation of the endogenous microRNA (miRNA) function (Whitham et al., 2006). Moreover, it has been shown recently that virus resistance induced by a nucleotide binding‐leucine‐rich repeat (NB‐LRR)‐type disease resistance gene is mediated by Argonaute4‐dependent inhibition of translation of virus‐encoded proteins (Bhattacharjee et al., 2009). Therefore, PTGS components appear to be key factors in both compatible and noncompatible plant–virus interactions.
A custom‐designed Affymetrix tomato GeneChip array (Syngenta Biotechnology, Inc., Research Triangle Park, NC, USA), which contains probe sets to interrogate over 22 000 tomato transcripts (van Esse et al., 2007), was used to study changes in the tomato transcriptome in response to inoculation with mild and aggressive PepMV isolates of the CH2 genotype (I. M. Hanssen et al., unpublished data). Over‐representation analysis demonstrated a severe down‐regulation of host genes involved in photosynthesis and energetic processes on PepMV infection, whereas defence and stress responses were clearly induced. This reinforces the notion that, like bacteria and fungi, compatible viruses induce basal plant defence, although the mechanism to recognize the pathogen is likely to be different (Ascencio‐Ibáñez et al., 2008; Whitham et al., 2006). Intriguingly, the induction of defence and stress responses was stronger and more persistent in plants that were inoculated with the aggressive CH2 PepMV isolate when compared with plants that were inoculated with the mild CH2 isolate, although viral loads were similar (I. M. Hanssen et al., unpublished data). Interestingly, DCL2, a key factor in antiviral PTGS, was strongly induced by the aggressive isolate and only moderately induced by the mild isolate. In addition, several Argonautes were differentially regulated, suggesting that PTGS plays an important role in the interaction between PepMV and its host tomato. Nevertheless, these defence responses did not result in PepMV containment. Moreover, these results suggest that some of the symptoms provoked by the aggressive isolate may be caused by a more elaborate host defence response or, perhaps, a more severe perturbation of the plant miRNA function through PepMV‐encoded silencing suppressors that have not yet been identified.
Another interesting observation is that PepMV infection results in the differential regulation of genes that code for several key enzymes in the flavanoid and lycopene biosynthesis pathway (I. M. Hanssen et al., unpublished data). This may possibly explain the impact of the virus on fruit symptoms, such as fruit marbling and flaming. Although the use of microarrays has made it possible to profile changes in transcriptional activity of thousands of genes simultaneously, to link expression profiles to biological pathways as they occur in the cell remains a challenge (van Baarlen et al., 2008). Therefore, functional analysis of candidate genes is needed to reveal their role in viral defence and symptomatology.
CONTROL STRATEGIES
Sources varying from moderate to full resistance have been identified in specific wild Solanum accessions, including S. pseudocapsicum, S. chilense, S. peruvianum and S. habrochaites (Ling and Scott, 2007; Soler‐Aleixandre et al., 2007). In particular, the resistance that segregates in accession LA1731 from S. habrochaites is thought to be promising, because segregants of this accession display resistance against the CH1, CH2 and EU PepMV genotypes (Ling and Scott, 2007). As the introduction of the identified resistance into cultivated tomato by breeding is a time‐consuming process, commercial resistant varieties are not yet available. Therefore, prevention through hygiene currently remains the most important strategy for the control of PepMV in commercial tomato production. However, as a result of the high infectivity of the virus, the prevention of infection through hygiene measures is a challenge, especially in dense tomato‐growing areas.
Many tomato growers, especially in the Netherlands, have chosen to inoculate their crops with a mild PepMV isolate in an attempt to protect their crops from severe damage on natural infection by an aggressive isolate based on cross‐protection (Hanssen et al., 2009a; Spence et al., 2006). In addition to the cross‐protection effect which is aimed for, many growers feel that an infection early in the growing season is less harmful than an infection that occurs later in the growing season. In support of this, glasshouse trials conducted in the UK from 2001 to 2003 showed that the time of infection had an impact on PepMV‐associated damage, as inoculations in May were more damaging than inoculations in February (Spence et al., 2006). In addition, from a questionnaire conducted among Belgian tomato growers, it seems that early infections result in less damage than late infections (Hanssen et al., 2009a). Whether this is a result of plant age, the activation of PTGS‐based plant defences, resulting in (partial) recovery, or climate conditions has not been clarified. Although disease symptoms in infected crops sometimes decrease or disappear after a certain period, the virus remains detectable even in asymptomatic plant parts. However, a recent cross‐protection study based on glasshouse trials revealed that efficient cross‐protection against an aggressive isolate belonging to the CH2 genotype could only be achieved by pre‐inoculation with a mild isolate from the same genotype. By contrast, enhanced symptom severity was observed when plants were pre‐inoculated with a mild isolate belonging to the EU or LP genotype (Hanssen et al., 2009c). These results suggest that the PepMV cross‐protection efficacy largely depends on RNA sequence identity, as shown for other plant–virus interactions (Desbiez and Lecoq, 1997; Wang et al., 1991; Yeh and Gonsalves, 1984). The role of PTGS in cross‐protection was demonstrated by the observation that two viral constructs derived from different viruses, but sharing a common sequence, could suppress each other when co‐inoculated in plants (Ratcliff et al., 1999). It has been suggested that cross‐protection is mediated by the pre‐activation of the siRNA‐induced silencing complex, thus inhibiting replication of the challenge isolate (Gal‐On and Shiboleth, 2006).
Thus, although cross‐protection can be efficient, the enhanced symptom severity in the case of limited nucleotide sequence identity between protector and challenge isolate undermines the potential of cross‐protection as a general PepMV control strategy. Cross‐protection can only be used successfully in areas in which one single PepMV genotype is dominant, provided that continuous monitoring of the PepMV population is performed and that strict hygiene measures are undertaken.
Future strategies to combat PepMV epidemics in tomato production might also include transgenic approaches. Coat protein‐mediated resistance (CPMR), by which the expression of the viral coat protein confers resistance, could be an efficient strategy. However, the protection efficiency obtained ranges from immunity to the delay or attenuation of symptoms, and the mechanisms are not fully understood (Prins et al., 2008). In addition, the expression of replicase or dysfunctional movement proteins in transgenic plants can sometimes lead to resistance or symptom attenuation (Golemboski et al., 1990; Lapidot et al., 1993). A more promising strategy could be the introduction of an inverted repeat (IR) transgene, derived from viral sequences, into the plant genome. The generation of long dsRNA precursors from these IR fragments will induce siRNAs and the PTGS machinery, thus conferring sequence‐specific antiviral resistance (Prins et al., 2008).
CONCLUSION
Since first appearing in protected tomato crops in Europe in 1999, PepMV has displayed a high potential to adapt to diverse environmental conditions. In only a few years' time, not only the original EU genotype, but also the more recently described CH2 genotype, have become established in tomato‐producing regions worldwide, and a recent shift in the PepMV population reveals a dynamic interplay between the different PepMV genotypes and their host. Symptoms can be very diverse in terms of both severity and nature. Although recent studies have shown that small differences in nucleotide sequence can account for large differences in biological properties and host responses, the host and viral factors that play a role in symptom severity remain unknown. Functional studies using host and viral mutants could identify viral factors that have an impact on the biological characteristics and increase our understanding of the host responses to PepMV infection. Unravelling the role of PTGS and viral‐encoded silencing suppressors in differential symptom severity may shed light on the interplay between different genotypes in mixed infections, and thus may contribute to the further development of a sound cross‐protection strategy. Although resistance sources have been identified in wild tomato species, commercial resistant varieties are not yet available and PepMV control is largely restricted to hygiene measures. However, currently applied prevention strategies often fail, demonstrating that our understanding of PepMV dissemination pathways is still too limited to contain the spread of the virus.
ACKNOWLEDGEMENTS
The authors thank IWT Vlaanderen (IWT 60669; Brussels, Belgium) and the European Commission (PEPEIRA; EC contract no. 044189) for financial support. B.P.H.J.T. is supported by a Vidi Grant of the Research Council for Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO).
REFERENCES
- Aguilar, J.M. , Hernandez‐Gallardo, M.D. , Cenis, J.L. , Lacasa, A. and Aranda, M.A. (2002) Complete sequence of the Pepino mosaic virus RNA genome. Arch. Virol. 147, 2009–2015. [DOI] [PubMed] [Google Scholar]
- Alfaro‐Fernández, A. , Cebrián, M.C. , Córdoba‐Sellés, C. , Herrera‐Vásquez, J.A. and Jordá, C. (2008) First report of the US1 strain of Pepino mosaic virus in tomato in the Canary Islands, Spain. Plant Dis. 92, 11. [DOI] [PubMed] [Google Scholar]
- Alfaro‐Fernández, A. , Córdoba‐Selles, M.C. , Herrera‐Vásquez, J.A. , Cebrián, M.C. and Jordá, C. (2009) Transmission of Pepino mosaic virus by the fungal vector Olpidium virulentus . J. Phytopathol. doi: 10.1111/j.1439‐0434.2009.01605.x. [Google Scholar]
- Ascencio‐Ibáñez, J.T. , Sozzani, R. , Lee, T.J. , Chu, T.M. , Wolfinger, R.D. , Cella, R. and Hanley‐Bowdoin, L. (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting the pathogen response and cell cycle controls during geminivirus infection. Plant Physiol. 148, 436–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Baarlen, P. , Van Esse, H.P. , Siezen, R.J. and Thomma, B.P.H.J. (2008) Challenges in plant cellular pathway reconstruction based on gene expression profiling. Trends Plant Sci. 13, 44–50. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, S. , Zamora, A. , Ahzar, M.T. , Sacco, M.A. , Lamber, L.H. and Moffet, P. (2009) Virus resistance induced by NB‐LRR proteins involves Argonaute4‐dependent translational control. Plant J. 58, 940–951. [DOI] [PubMed] [Google Scholar]
- Córdoba, M.C. , Martínez‐Priego, L. and Jordá, C. (2004) New natural hosts of Pepino mosaic virus in Spain. Plant Dis. 88, 906. [DOI] [PubMed] [Google Scholar]
- Córdoba‐Sellés, M.C. , García‐Rández, A. , Alfaro‐Fernández, A. and Jordá‐Gutiérrez, C. (2007) Seed transmission of Pepino mosaic virus and efficacy of tomato seed disinfection treatments. Plant Dis. 91, 1250–1254. [DOI] [PubMed] [Google Scholar]
- Cotillon, A.C. , Girard, M. and Ducouret, S. (2002) Complete nucleotide sequence of the genomic RNA of a French isolate of Pepino mosaic virus (PepMV). Arch. Virol. 147, 2231–2238. [DOI] [PubMed] [Google Scholar]
- Davino, S. , Accotto, G.P. , Masenga, V. , Torta, L. and Davino, M. (2009) Basil (Ocimum basicilum), a new host of Pepino mosaic virus. Plant Pathol. 58, 407. [Google Scholar]
- Desbiez, C. and Lecoq, H. (1997) Zucchini yellow mosaic virus. Plant Pathol. 46, 809–829. [Google Scholar]
- Ding, S.W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esse, H.P. , Bolton, M.D. , Stergiopoulos, I. , De Wit, P.J.G.M. and Thomma, B.P.H.J. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant–Microbe Interact. 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- EPPO (2000) EPPO Alert list‐viruses. Pepino mosaic potexvirus. http://www.eppo.org/QUARANTINE/Alert_List/viruses/PEPMV0.htm. [accessed on October 2009].
- French, C.J. , Bouthillier, M. , Bernardy, M. , Ferguson, G. , Sabourin, M. , Johnson, R.C. , Masters, C. , Godkin, S. and Mumford, R. (2001) First report of Pepino mosaic virus in Canada and the United States. Plant Dis. 85, 1121. [DOI] [PubMed] [Google Scholar]
- Gal‐On, A. and Shiboleth, Y.M. (2006) Cross protection In: Natural Resistance Mechanisms of Plants to Viruses (Loebenstein G. and Carr J.P., eds), pp. 261–268. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Golemboski, D.B. , Lomonossoff, G.P. and Zaitlin, M. (1990) Plants transformed with tobacco mosaic virus nonstructural gene sequence are resistant to the virus. Proc. Natl. Acad. Sci. USA, 87, 6311–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, P. , Sempere, R.N. , Elena, S.F. and Aranda, M.A. (2009) Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J. Virol. doi: 10.1128/JVM.01486‐09 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez‐Aguirre, I. , Mehle, N. , Delic, D. , Gruden, K. , Mumford, R. and Ravnikar, M. (2009) Real‐time quantitative PCR based sensitive detection and genotype discrimination of Pepino mosaic virus . J. Virol. Methods, 162, 46–55. [DOI] [PubMed] [Google Scholar]
- Hanssen, I.M. , Gutiérrez‐Aguirre, I. , Paeleman, A. , Goen, K. , Wittemans, L. , Lievens, B. , Vanachter, A.C.R.C. , Ravnikar, M. and Thomma, B.P.H.J. (2009c) Co‐infection of greenhouse tomato with different Pepino mosaic virus isolates results either in cross‐protection or enhanced symptom display. Plant Pathol. doi: 10.1111/j.1365‐3059.2009.02190.x (in press). [Google Scholar]
- Hanssen, I.M. , Mumford, R. , Blystad, D.‐R. , Cortez, I. , Hasiów‐jareszewska, B. , Hristova, D. , Pagán, I. , Pepeira, A.‐M. , Peters, J. , Pospieszny, H. , Ravnikar, M. , Stijger, I. , Tomassoli, L. , Varveri, C. , Van Der Vlugt, R. and Nielsen, S.L. (2009d) Seed transmission of Pepino mosaic virus in tomato. Eur. J. Plant Pathol. doi: 10.1007/s10658‐009‐9528‐x. [Google Scholar]
- Hanssen, I.M. , Paeleman, A. , Van Bergen, L. , Vandewoestijne, E. , Wittemans, L. , Goen, K. , Vanachter, A.C.R.C. and Thomma, B.P.H.J. (2009a) Survey of symptom expression and damage caused by Pepino mosaic virus (PepMV) in commercial tomato production in Belgium. Acta Hort. 808, 185–192. [Google Scholar]
- Hanssen, I.M. , Paeleman, A. , Vandewoestijne, E. , Van Bergen, L. , Bragard, C. , Lievens, B. , Vanachter, A.C.R.C. and Thomma, B.P.H.J. (2009b) Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol. 58, 450–460. [Google Scholar]
- Hanssen, I.M. , Paeleman, A. , Wittemans, L. , Goen, K. , Lievens, B. , Bragard, C. , Vanachter, A.C.R.C. and Thomma, B.P.H.J. (2008) Genetic characterization of Pepino mosaic virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur. J. Plant Pathol. 121, 131–146. [Google Scholar]
- Hasiów, B. , Borodynko, N. and Pospieszny, H. (2008) Complete genomic RNA sequence of the Polish Pepino Mosaic Virus isolate belonging to the US2 strain. Virus Genes, 36, 209–214. [DOI] [PubMed] [Google Scholar]
- Hasiów‐Jaroszewska, B. , Borodynko, N. and Pospieszny, H. (2009b) Infectious RNA transcripts derived from cloned cDNA of a pepino mosaic virus isolate. Arch. Virol. 154, 853–856. [DOI] [PubMed] [Google Scholar]
- Hasiów‐Jaroszewska, B. , Pospieszny, H. and Borodynko, N. (2009a) New necrotic isolates of Pepino mosaic virus representing the CH2 genotype. J. Phytopathol. 157, 494–496. [Google Scholar]
- Jones, D.R. and Lammers, W. (2005) Pest Risk Analysis for Pepino Mosaic Virus. York: Central Science Laboratory; http://www.defra.gov.uk/planth/pra.htm [Google Scholar]
- Jones, R.A.C. , Koenig, R. and Lesemann, D.E. (1980) Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum). Ann. Appl. Biol. 94, 61–68. [Google Scholar]
- Jordá, C. , Lázaro‐Perez, A. and Martinez‐Culebras, P.V. (2001) First report of Pepino mosaic virus on tomato in Spain. Plant Dis. 85, 1292. [DOI] [PubMed] [Google Scholar]
- Krinkels, M. (2001) PepMV causes sticky problem. Prophyta, May, 30–33. [Google Scholar]
- Lacasa, A. , Guerrero, M.M. , Hita, I. , Martinéz, M.A. , Jordá, C. , Bielza, P. , Contreras, J. , Alcázar, A. and Cano, A. (2003) Implicationes de los abejorros (Bombus spp.) en la dipersión del virus del mosaico del pepino dulce (Pepino mosaic virus) en cultivos de tomato. Bol. Sanid. Veg., Plagas, 29, 393–402. [Google Scholar]
- Lapidot, M. , Gafny, R. , Ding, B. , Wolf, S. , Lucas, W.J. and Beachy, R.N. (1993) A dysfunctional movement protein of tobacco mosaic virus that partially modifies the plasmodesmata and limits virus spread in transgenic plants. Plant J. 4, 959–970. [Google Scholar]
- Ling, K. (2006) Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes, 34, 1–8. [DOI] [PubMed] [Google Scholar]
- Ling, K. (2008) Pepino mosaic virus on tomato seed: virus location and mechanical transmission. Plant Dis. 9, 1701–1705. [DOI] [PubMed] [Google Scholar]
- Ling, K. and Scott, J.W. (2007) Sources of resistance to Pepino mosaic virus in tomato accessions. Plant Dis. 91, 749–753. [DOI] [PubMed] [Google Scholar]
- Ling, K. , Wechter, W.P. and Jordan, R. (2007) Development of a one‐step immunocapture real‐time TaqMan RT‐PCR assay for the broad spectrum detection of Pepino mosaic virus. J. Virol. Methods, 144, 65–72. [DOI] [PubMed] [Google Scholar]
- Ling, K. , Wintermantel, W.M. and Bledsoe, M. (2008) Genetic composition of Pepino mosaic virus population in North American greenhouse tomatoes. Plant Dis. 92, 1683–1688. [DOI] [PubMed] [Google Scholar]
- López, C. , Soler, S. and Nuez, F. (2005) Comparison of the complete sequences of three different isolates of Pepino mosaic virus: size variability of the TGBp3 protein between tomato and L. peruvianum isolates. Arch. Virol. 150, 619–627. [DOI] [PubMed] [Google Scholar]
- Marathe, R. , Guan, Z. , Anandalakshmi, R. , Zhao, H. and Dinesh‐Kumar, S.P. (2004) Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol Biol. 55, 501–520. [DOI] [PubMed] [Google Scholar]
- Maroon‐Lango, C.J. , Guaragna, M.A. , Jordan, R.L. , Hammond, J. , Bandla, M. and Marquardt, S.K. (2005) Two unique US isolates of Pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (EU like) US isolate. Arch. Virol. 150, 1187–1201. [DOI] [PubMed] [Google Scholar]
- Martin, J. and Mousserion, C. (2002) Potato varieties which are sensitive to the tomato strains of Pepino mosaic virus (PepMV). Phytoma Défense Végétaux, 552, 26–28. [Google Scholar]
- Maule, A. , Leh, V. and Lederer, C. (2002) The dialogue between viruses and host in compatible interactions. Curr. Opin. Plant Biol. 5, 1–6. [DOI] [PubMed] [Google Scholar]
- Mumford, R.A. and Metcalfe, E.J. (2001) The partial sequencing of the genomic RNA of a UK isolate of Pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch. Virol. 146, 2455–2460. [DOI] [PubMed] [Google Scholar]
- Pagán, I. , Córdoba‐Sellés, M.C. , Martinez‐Priego, L. , Fraile, A. , Malpica, J.M. , Jordá, C. and García‐Arenal, F. (2006) Genetic structure of the population of Pepino mosaic virus infecting tomato crops in Spain. Phytopathology, 96, 274–279. [DOI] [PubMed] [Google Scholar]
- Pospieszny, H. and Borodynko, N. (2006) New Polish isolates of Pepino mosaic virus highly distinct from European tomato, Peruvian and US2 strains. Plant Dis. 90, 1106. [DOI] [PubMed] [Google Scholar]
- Pospieszny, H. , Borodynko, N. and Palczewska, M. (2002) Occurrence of Pepino mosaic virus in Poland. Phytopathologia Polonica, 26, 91–94. [Google Scholar]
- Pospieszny, H. , Hasiów, B. and Borodyndo, N. (2008) Characterization of two distinct Polish isolates of Pepino mosaic virus. Eur. J. Plant Pathol. 122, 443–445. [Google Scholar]
- Prins, M. , Laimer, M. , Noris, E. , Schubert, J. , Wasseneger, M. and Tepfer, M. (2008) Strategies for antiviral resistance in transgenic plants. Mol. Plant Pathol. 9, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino, B.F. and Bent, A.F. (2003) Deciphering host resistance and pathogen virulence: the Arabidopsis/Pseudomonas interaction as a model. Mol. Plant Pathol. 4, 517–530. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F.G. , MacFarlane, S. and Baulcombe, D.C. (1999) Gene silencing without DNA: RNA‐mediated cross‐protection between viruses. Plant Cell, 11, 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggero, P. , Masenga, V. , Lenzi, R. , Coghe, F. , Ena, S. and Winter, S. (2001) First report of Pepino mosaic virus in tomato in Italy. Plant Pathol. 50, 798. [Google Scholar]
- Salomone, A. and Roggero, P. (2002) Host range, seed transmission and detection by ELISA and lateral flow of an Italian isolate of Pepino mosaic virus. J. Plant Pathol. 84, 65–68. [Google Scholar]
- Schwarz, D. , Paschek, U. , Bandte, M. , Büttner, C. and Obermeier, C. (2009) Detection, spread, and interactions of Pepino mosaic virus and Pythium aphanidermatum in the root environment of tomato in hydroponics. Acta Hort. 808, 163–170. [Google Scholar]
- Senthil, G. , Liu, H. , Puram, V.G. , Clark, A. , Stromberg, A. and Goodin, M.M. (2005) Specific and common changes in Nicotiana benthamiana gene expression in response to infection by enveloped virus. J. Gen. Virol. 86, 2615–2625. [DOI] [PubMed] [Google Scholar]
- Shipp, J.L. , Buitenhuis, R. , Stobbs, L. , Wang, K. , Kim, W.S. and Ferguson, G. (2008) Vectoring of Pepino mosaic virus by bumble‐bees in tomato greenhouses. Ann. Appl. Biol. 53, 149–155. [Google Scholar]
- Soler, S. , Cebolla‐Cornejo, J. , Prohens, J. and Nuez, F. (2000) El Pepino Mosaic Virus (PepMV), una nueva amenaza para el cultivo del tomate. II. Vida Rural, 119, 48–52. [Google Scholar]
- Soler, S. , Prohens, J. , Díez, M.J. and Nuez, F. (2002) Natural occurrence of Pepino mosaic virus in Lycopersicon species in central and southern Peru. J. Phytopathol. 150, 49–53. [Google Scholar]
- Soler‐Aleixandre, S. , López, C. , Cebolla‐Cornejo, J. and Nuez, F. (2007) Sources of resistance to Pepino mosaic virus (PepMV) in tomato. HortScience, 42, 40–45. [Google Scholar]
- Soler‐Aleixandre, S. , López, C. , Díez, M.J. , Pérez de Castro, A. and Nuez, F. (2005) Association of Pepino mosaic virus with tomato collapse. J. Phytopathol. 153, 464–469. [Google Scholar]
- Spence, N.J. , Basham, J. , Mumford, R.A. , Hayman, G. , Edmondson, R. and Jones, D.R. (2006) Effect of Pepino mosaic virus on the yield and quality of glasshouse‐grown tomatoes in the UK. Plant Pathol. 55, 595–606. [Google Scholar]
- Van Der Vlugt, R.A.A. , Stijger, C.C.M.M. and Verhoeven, J.T.J. (2000) First report of Pepino Mosaic Virus on tomato. Plant Dis. 84, 103. [DOI] [PubMed] [Google Scholar]
- Verhoeven, J.T.J. , Van Der Vlugt, R. and Roenhorst, J.W. (2003) High similarity between tomato isolates of Pepino mosaic virus suggests a common origin. Eur. J. Plant Pathol. 109, 419–425. [Google Scholar]
- Wang, H.L. , Gonsalves, D. , Provvidenti, R. and Lecoq, H.L. (1991) Effectiveness of cross protection by a mild strain of zucchini yellow mosaic virus in cucumber, melon, and squash. Plant Dis. 75, 203–207. [Google Scholar]
- Whitham, S.A. , Quan, S. , Chang, H.S. , Cooper, B. , Estes, B. , Zhu, T. , Wang, X. and Hou, Y.M. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Yang, C. and Goodin, M.M. (2006) Global impact: elucidating plant responses to viral infection. Mol. Plant–Microbe Interact. 19, 1207–1215. [DOI] [PubMed] [Google Scholar]
- Wingard, S.A. (1928) Hosts and symptoms of ring spot, a virus disease of plants. J. Agric. Res. 37, 127–153. [Google Scholar]
- Wise, R.P. , Moscou, M.J. , Bogdanove, A.J. and Whitham, S.A. (2007) Transcript profiling in host–pathogen interactions. Annu. Rev. Phytopathol. 45, 329–369. [DOI] [PubMed] [Google Scholar]
- Wright, D. and Mumford, R. (1999) Pepino mosaic Potexvirus (PepMV): first records in tomato in the United Kingdom. Plant Disease Notice No. 89. York: Central Science Laboratory.
- Yaoliang, Z. and Zhongjian, S. (2003) Preliminary characterization of Pepino mosaic virus Shanghai isolate (PepMV‐Sh) and its detection with ELISA. Acta Agric. Shanghai, 19, 90–92. [Google Scholar]
- Yeh, S.D. and Gonsalves, D. (1984) Evaluation of induced mutants of Papaya ringspot virus for control by cross protection. Phytopathology, 74, 1086–1091. [Google Scholar]


