SUMMARY
In commercial maize, there are at least two different alleles of the chiA gene that encode alloforms of ChitA chitinase, a protein that is abundant in developing seed. Both known alloforms are modified by Bz‐cmp, a chitinase‐modifying protein (cmp) secreted by the fungal pathogen Bipolaris zeicola. One alloform (ChitA‐B73) is also modified by Stm‐cmp, a protein secreted by the fungal pathogen Stenocarpella maydis, whereas the other (ChitA‐LH82) is resistant. The two ChitA alloforms possess six differences or polymorphisms (P1–P6). To determine whether the P2 polymorphism in the chitin‐binding domain is responsible for resistance or susceptibility to modification by Stm‐cmp, and to determine whether Stm‐cmp and Bz‐cmp are proteases, heterologous expression strains of the yeast Pichia pastoris that produce recombinant maize ChitA (rChitA) alloforms and mutant rChitAs were created. rChitA alloforms and mutant rChitAs were purified from yeast cultures and used as substrates in assays with Stm‐cmp and Bz‐cmp. As with native protein, Bz‐cmp modified both rChitA‐LH82 and rChitA‐B73, whereas Stm‐cmp modified rChitA‐B73 only. Mutant rChitAs, in which the P2 amino acids were changed to those of the other alloform, resulted in a significant exchange in Stm‐cmp susceptibility. Amino‐terminal sequencing of unmodified and modified rChitA‐B73 demonstrated that Stm‐cmp cleaves the peptide bond on the amino‐terminal side of the P2 alanine, whereas Bz‐cmp cleaves in the poly‐glycine hinge region, the site of P3. The results demonstrate that Stm‐cmp and Bz‐cmp are proteases that truncate ChitA chitinase at the amino terminus, but at different sites. Both sites correspond to polymorphisms in the two alloforms, suggesting that the sequence diversity at P2 and P3 is the result of selective pressure to prevent truncation by fungal proteases.
INTRODUCTION
Maize (Zea mays L.) kernels develop on ears of mature plants and are susceptible to ear rot infection caused by specialized fungi (Koehler, 1959). Commercial maize resistance to ear rot is mainly the result of traditional breeding. Although breeding programmes now use molecular biology and bioinformatics to link performance in the field with DNA‐based molecular markers (Eathington et al., 2007), little is known about the molecular mechanisms of fungal–maize interactions or how maize genetic diversity contributes to the observed phenotypes. Uncovering the molecular mechanisms used by fungal proteins to manipulate maize biology will help breeders understand the relationship between molecular markers and plant resistance.
The maize ChitA protein—encoded by the chiA gene (GRMZM2G051943; Schnable et al., 2009)—is a class IV plant chitinase that is abundant in developing maize kernels. Plants contain many chitinases. They are divided into classes on the basis of the amino acid sequence and predicted structure (Neuhaus et al., 1996). Like all class IV chitinases, maize ChitA has an amino‐terminal sequence that directs the protein to be secreted. This sequence is removed during secretion, resulting in a mature, secreted protein with two functional domains that are separated by a flexible hinge. The smaller, amino‐terminal domain functions to bind insoluble chitin. The larger, carboxy‐terminal domain hydrolyses chitin.
The biological role of maize ChitA chitinases has not been established. As fungal cell walls contain chitin, ChitA may contribute to plant defence against fungi by directly inhibiting fungal growth (Huynh et al., 1992; Schlumbaum et al., 1986) or by cleaving chitin‐containing molecules from the surface of hyphae to produce signalling molecules that bind plant chitin receptors (Kaku et al., 2006; Shibuya and Minami, 2001). However, class IV chitinases have also been implicated in embryogenesis (Jong et al., 1992; Wiweger et al., 2003), defence against bacterial pathogens (Gerhardt et al., 1997) and abiotic stress (Gerhardt et al., 2004). Thus, the biological role of ChitA is probably complex.
In previous studies, it has been shown that ChitA is modified by fungal proteins secreted by two distinct members of the class Dothideomycetes: Bipolaris zeicola (G.L. Stout) Shoemaker (Holomorph: Cochliobolus carbonum R.R. Nelson) and Stenocarpella maydis (Berk.) B. Sutton [syn. Diplodia maydis (Berk.) Sacc.] (Naumann and Wicklow, 2010; Naumann et al., 2009). In healthy ears of a commercial maize hybrid (FS 6873RR), two different forms of ChitA were present. When inoculated with B. zeicola, the causative agent of northern corn leaf spot, which causes both ear rot and leaf spot of maize, both forms of ChitA were modified by the secreted fungal protein Bz‐cmp (B. zeicola chitinase‐modifying protein) into new forms that do not bind insoluble chitin, but can hydrolyse chitin. When inoculated with S. maydis, the most prevalent ear rot pathogen in the world (Rossouw et al., 2009), one form of ChitA was modified by the secreted fungal protein Stm‐cmp, whereas the other was not. It was determined that the two ChitAs are alloforms, proteins encoded by different alleles of the chiA gene. Maize inbred B73 contains an allele that encodes the Stm‐cmp‐susceptible ChitA‐B73 alloform, whereas inbred LH82 contains an allele that encodes the resistant ChitA‐LH82.
ChitA‐LH82 and ChitA‐B73 possess six differences or polymorphisms (P1–P6). As cmp modification has been shown to affect the chitin‐binding ability of ChitA (Naumann et al., 2009) and the P2 polymorphism is within the core of the chitin‐binding domain (Aboitiz et al., 2004), P2 may be in the amino acid region targeted by cmps and may be responsible for ChitA‐LH82 resistance to Stm‐cmp modification—but this has not been tested. And while the most obvious explanation of cmp modification of ChitA is that cmp proteins are proteases that remove the small chitin‐binding domain from the protein, this mechanism has not been proven. This article describes the creation of strains of the methylotrophic yeast Pichia pastoris that produce and secrete recombinant ChitA (rChitA) alloforms and mutants, and their purification from expression cultures. Purified rChitAs were used to test two hypotheses. The first hypothesis is that the P2 aspartic acid residue in the chitin‐binding domain of ChitA‐LH82 is responsible for its resistance to Stm‐cmp modification. The second hypothesis is that Stm‐cmp and Bz‐cmp are proteases that remove the chitin‐binding domain of ChitA.
RESULTS
Expression of rChitA alloforms in P. pastoris and purification from expression cultures
Heterologous expression strains of the yeast P. pastoris were constructed to produce rChitA alloforms from chiA cDNAs. Introns were deleted from existing genomic clones (Fig. 1A) (Naumann and Wicklow, 2010) and chiA cDNAs were inserted into expression vectors (Fig. 1B). In the expression vectors, the maize secretion signal sequence (SSS) is replaced by one from yeast which is fused to Q27 of ChitA with a restriction site at the junction. When produced in yeast, the protein is secreted and the yeast SSS is removed, resulting in secreted rChitA that is identical to secreted maize ChitA, but with two additional amino acids (E and F) at the amino terminus that are encoded by the six bases of the restriction site. rChitA does not contain the P1 polymorphism as it is in the maize SSS.
Figure 1.
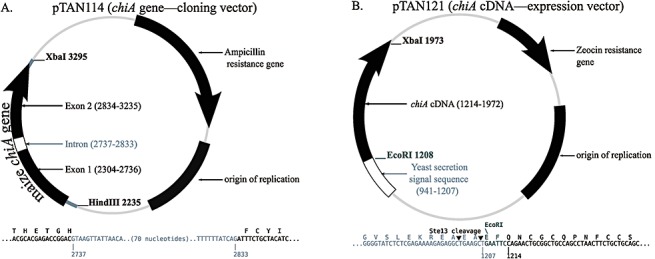
Plasmids used in this study. (A) Cloning plasmids containing the chiA gene cloned from either maize inbred LH82 (pTAN114) or B73 (pTAN112; not shown). The exon–intron borders are shown below; the amino acids encoded by the exons are displayed above the DNA sequence with the amino acid letter above the first letter of the codon. (B) Expression plasmids. The two chiA cDNAs were cloned into pPICZαA, creating pTAN121 (LH82 allele) and pTAN122 (B73 allele; not shown). The fusion of the yeast secretion signal sequence to the chiA cDNA is shown below; an EcoRI restriction endonuclease site connects the secretion signal to the ChitA‐encoding chiA sequence; the Ste13 protease of Pichia pastoris removes the secretion signal during secretion from the yeast.
When grown in liquid medium containing methanol, strains with integrated chiA cDNAs secreted rChitA into the medium (Fig. 2). rChitAs were purified from the medium in four steps: production of cell‐free medium (CFM) by centrifugation and filtration; precipitation from CFM by salting out, followed by dialysis to form a dialysed, resuspended pellet (D‐RP); cation exchange chromatography; and re‐precipitation by salting out (re‐ppt). The procedure typically yielded more than 8 mg of protein in a 1‐mL solution from a 1‐L culture.
Figure 2.
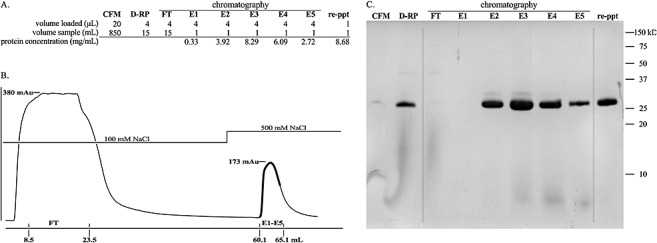
Purification of rChitA. (A) The total volume of solution at each stage of purification, the volume of sample loaded in Tricine‐sodium dodecylsulphate‐polyacrylamide gel electrophoresis (Tricine‐SDS‐PAGE) and the protein concentrations of elution fractions and reprecipitated (re‐ppt) protein are listed. (B) Chromatogram of cation exchange chromatography. The y‐axis is the absorbance at 280 nm; the absorbance maxima of the flow‐through (FT) and elution (E1–E5) peaks are shown. The x‐axis is the volume of buffer (mL); the beginning and end of FT and fractions E1–E5 are shown; the dark line on the chromatogram traces the absorbance of E1–E5. (C) Tricine‐SDS‐PAGE analysis of purification. The first lane is cell‐free medium (CFM). The second lane is the dialysed, resuspended ammonium sulphate pellet (D‐RP). The third lane is the flow through from the cation exchange column (FT). The fourth to eighth lanes are the fractions that eluted after increasing the concentration of NaCl from 100 to 500 mm (E1–E5). The last lane is the purified protein, the result of combining fractions E2, E3, E4, E5 and reprecipitation of rChitA with ammonium sulphate (re‐ppt).
Modification of rChitA‐LH82 and rChitA‐B73 by Stm‐cmp and Bz‐cmp
Each of the purified rChitAs was incubated with either Stm‐cmp‐containing protein extracts or purified Bz‐cmp (Fig. 3A). Samples were taken at three time points. Like maize‐produced ChitAs, the rChitA‐LH82 alloform was resistant to Stm‐cmp modification, whereas rChitA‐B73 was susceptible (Fig. 3A, top gel). When rChitA‐LH82 was incubated with Stm‐cmp, products were not detected at the earliest time point; in the later two time points, only a minor product band was visible. When rChitA‐B73 was incubated with Stm‐cmp, a minor product band was detected at the earliest time point, and increasing amounts of product were visible in the later two time points. Bz‐cmp reactions with rChitAs resulted in modification of both rChitAs, with increasing amounts of modified product appearing with increasing time (Fig. 3A, bottom gel). The strong qualitative substrate preference that is seen with Stm‐cmp was not present in these reactions. Therefore, one or more of the five polymorphisms that distinguish between the two proteins is highly likely to be responsible for the difference in alloform susceptibility to Stm‐cmp modification without affecting Bz‐cmp modification (Fig. 3B).
Figure 3.
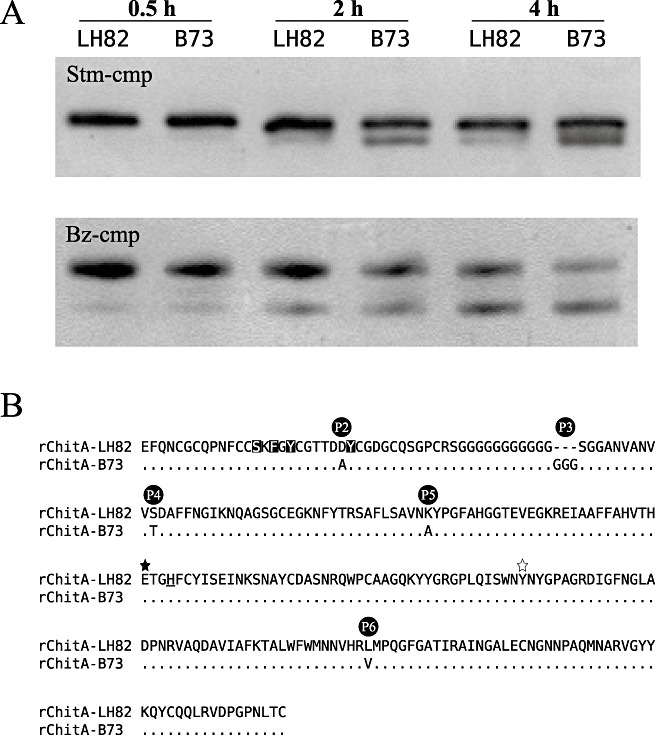
Modification of rChitA alloforms by fungal chitinase‐modifying proteins (cmps). (A) rChitA‐LH82 and rChitA‐B73 were incubated with either Stm‐cmp‐containing protein extracts from Stenocarpella maydis (top gel) or purified Bz‐cmp (cmp from Bipolaris zeicola) (bottom gel). Aliquots from each of the four reactions were taken at 0.5, 2 and 4 h. (B) Sequence alignment of the two rChitA alloforms. The five differences between the proteins are labelled P2 to P6. The rChitA‐LH82 sequence is shown in the top row; the rChitA‐B73 sequence is aligned below. Dots indicate homology; dashes indicate gaps; letters indicate amino acid differences. The P1 polymorphism is in the secretion signal sequence of ChitA and is not present in rChitA. The serine, phenylalanine and two tyrosine amino acids that are inverted are predicted to be involved in chitin binding (Aboitiz et al., 2004). The filled star marks a glutamic acid that is predicted to be essential for catalysis; the open star marks a tyrosine that probably forms part of the active site of the enzyme (Verburg et al., 1992). The underlined histidine marks the end of exon 1 and the start of exon 2 (Fig. 1A).
Modification of rChitA P2 mutants by Stm‐cmp
Strains were constructed to express two mutant ChitAs in which the P2 amino acid of each alloform was mutated to that of the other. The rChitA‐LH82(D48A) mutant has alanine at P2, whereas rChitA‐B73(A47D) has aspartic acid. The engineered strains were grown under inducing conditions and the mutant rChitAs were purified. Mutant rChitAs were incubated with Stm‐cmp and their modification was compared with the rChitA‐LH82 and rChitA‐B73 alloforms (Fig. 4). Samples were taken at 2‐h time points and the products were analysed by Tricine‐sodium dodecylsulphate‐polyacrylamide gel electrophoresis (Tricine‐SDS‐PAGE). Mutation of the P2 aspartic acid to alanine in the rChitA‐LH82 background led to an increase in Stm‐cmp modification (Fig. 4; second lane). Mutation of the P2 alanine to aspartic acid in the rChitA‐B73 background led to a decrease in Stm‐cmp modification (Fig. 4; last lane). Both mutant proteins changed the susceptibility to degradation substantially, but not completely. This indicates that the remaining four polymorphisms make some contribution to the difference in Stm‐cmp modification susceptibility. However, in both backgrounds, ChitAs with aspartic acid at P2 were more resistant to Stm‐cmp degradation than were ChitAs with alanine.
Figure 4.
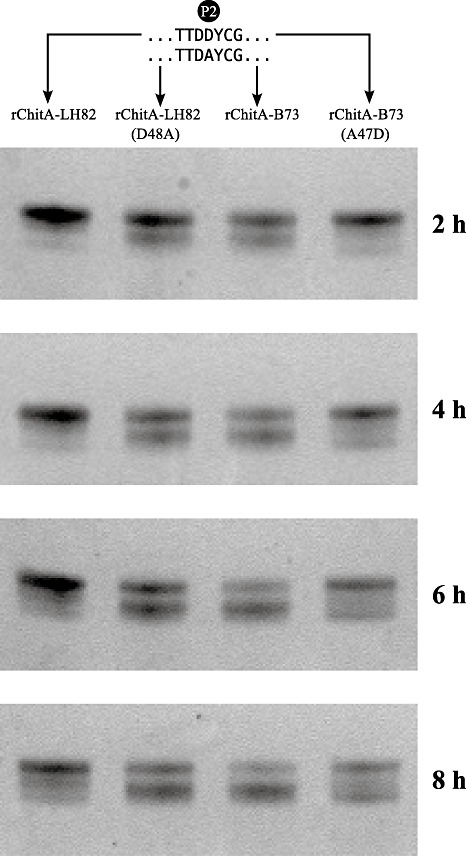
P2 amino acid mutations change Stm‐cmp (Stenocarpella maydis chitinase‐modifying protein) susceptibility. The two rChitA alloforms and proteins with mutations at P2 were incubated with secreted protein extracts from S. maydis. Samples were taken every 2 h and analysed by Tricine‐sodium dodecylsulphate‐polyacrylamide gel electrophoresis (Tricine‐SDS‐PAGE). The gels were stained, and digital images were obtained.
Amino‐terminal sequencing of rChitA‐B73 and modified forms
rChitA‐B73 was either incubated alone in buffer or in the presence of Bz‐cmp or Stm‐cmp. Amino‐terminal sequencing of unmodified, Bz‐cmp‐modified and Stm‐cmp‐modified forms of rChitA‐B73 was performed by Edman degradation (Edman and Begg, 1967) (Fig. 4).
Analysis of the first five amino acids from unmodified rChitA‐B73 confirmed that the purified protein had the expected amino terminus (Fig. 1B). The first two amino acids were the EcoRI site‐encoded glutamic acid (E) and phenylalanine (F). The third and fourth were the glutamine (Q) and asparagine (N) of ChitA. Analysis of the fifth amino acid resulted in the absence of a significant signal. A lack of signal is consistent with the fifth residue being cysteine (C) as this amino acid is destroyed by Edman degradation and is not detected. Analysis of Stm‐cmp‐modified rChitA‐B73 revealed that it was truncated and began in the chitin‐binding domain. The first amino acid was the P2 alanine residue (A), followed by tyrosine (Y); the third amino acid did not have significant signal [consistent with cysteine (C)]; the fourth and fifth amino acids were glycine (G) and aspartic acid (D). Analysis of Bz‐cmp‐modified rChitA‐B73 showed that it was also truncated, but differently than when modified by Stm‐cmp. The amino terminus began with five glycine (G) residues, corresponding to cleavage in the hinge region. The hinge region separated the chitin‐binding domain from the chitinase domain and was the site of P3 (Fig. 3B). This demonstrates that Stm‐cmp and Bz‐cmp are proteases that truncate ChitA by cleaving at P2 and P3, respectively.
DISCUSSION
This study shows that the aspartic acid residue in the chitin‐binding domain of maize ChitA‐LH82 is largely responsible for its resistance to Stm‐cmp modification. When the aspartic acid of rChitA‐LH82 was mutated to alanine, it greatly increased the amount of Stm‐cmp product. When the alanine of rChitA‐B73 was mutated to aspartic acid, it greatly decreased the amount of product (Fig. 4). This study also shows that Stm‐cmp and Bz‐cmp are proteases; furthermore, it demonstrates that the fungal proteases cleave ChitA in different locations. Stm‐cmp destroys the chitin‐binding domain by cleaving the peptide bond on the amino‐terminal side of the P2 amino acid, whereas Bz‐cmp separates the chitin‐binding and catalytic domains by cleaving in the poly‐glycine hinge region, the site of P3 (Fig. 5). This mechanism explains why ChitA‐LH82 is resistant to Stm‐cmp modification, but not Bz‐cmp modification: the two proteases are not homologous proteins secreted by different fungi (orthologues), but rather are two different types of protease that cleave ChitA in different locations. Although the P3 polymorphism does not have a significant effect on susceptibility to Bz‐cmp truncation, it does link Bz‐cmp activity to this site. In addition to the difference in the sequences of ChitA‐LH82 and ChitA‐B73 at P3, molecular evolutionary studies of chiA sequences from maize's ancestor, the annual teosinte Zea mays ssp. parviglumis, have shown that the hinge‐encoding region is highly diverse (Moeller and Tiffin, 2008; Tiffin, 2004). The demonstration that Stm‐cmp and Bz‐cmp are proteases and that both cleave ChitA at sites of sequence diversity suggests that this diversity resulted from selective pressure to prevent truncation by fungal proteases. It further suggests that the regions of chiA that encode P2 and P3 may be useful as molecular markers to guide the breeding of maize hybrids with increased ear rot resistance.
Figure 5.
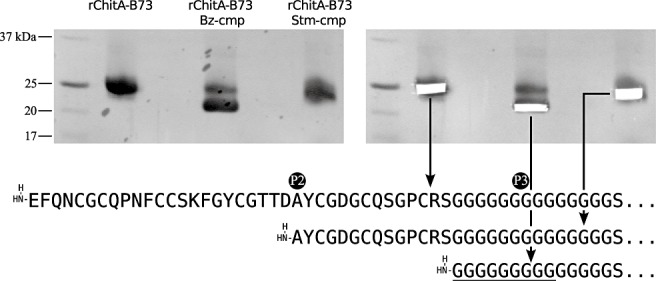
Amino‐terminal sequencing of rChitA‐B73 and its fungal‐modified forms. rChitA‐B73 and its Stm‐cmp and Bz‐cmp (Stenocarpella maydis and Bipolaris zeicola chitinase‐modifying protein)‐modified forms were separated by Tricine‐sodium dodecylsulphate‐polyacrylamide gel electrophoresis (Tricine‐SDS‐PAGE), transferred to a poly(vinylidene difluoride) (PVDF) membrane and stained with Coomassie (top left). Unmodified protein and each modified form were removed with a scalpel (top right) and analysed by amino‐terminal sequencing (bottom). The underlined glycines may or may not be present in Bz‐cmp‐modified ChitA.
To my knowledge, this is the third report of a plant class IV chitinase that is proteolytically degraded at the amino terminus, and the first to demonstrate that it is the result of fungal proteases. The first report was of the PvChi4 protein of bean (Phaseolus vulgaris L. cv. Saxa) roots that had been infected by the fungal pathogen Fusarium solani f. sp. phaseoli (Lange et al., 1996). In that study, the authors found that fungal infestation resulted in the conversion of PvChi4 into three different, catalytically active forms as a result of proteolysis. Amino‐terminal sequencing showed that the smallest active form was missing both the chitin‐binding domain and hinge, whereas the intermediate active form began in the hinge region (like Bz‐cmp‐modified ChitA). The amino terminus of the largest active form could not be determined, but probably resulted from cleavage in the chitin‐binding domain (like Stm‐cmp‐modified ChitA). However, the study did not establish whether PvChi4 was truncated by fungal proteases or by induced plant proteases. Although it is tempting to speculate that fungal proteases were the cause, another group studying a class IV chitinase, NtChitIV, demonstrated that it was cleaved in the hinge region by endogenous tobacco proteases (Shinya et al., 2007). This suggests that maize and other plants may produce their own proteases that cleave ChitA and other class IV chitinases, and that truncated forms without chitin‐binding domains are not disabled proteins but post‐translationally modified proteins with an important biological role. It further suggests that Stm‐cmp and Bz‐cmp are not simply proteases that destroy ChitA, but—like many bacterial virulence proteins (Abramovitch et al., 2006; Mudgett, 2005)—are mimics of plant proteins that function to suppress the immune response in a more elegant way.
EXPERIMENTAL PROCEDURES
Engineering of rChitA expression strains
Engineering of rChitA expression strains began with two pBluescript KS cloning plasmids (GenBank accession X52331) (Agilent Technologies, Santa Clara, CA, USA) that contain the chiA genes from either maize inbred LH82 (pTAN114) or B73 (pTAN112), as described previously (Naumann and Wicklow, 2010). The intron was removed from the chiA genes of both plasmids. The intron was deleted from pTAN114 by a PCR‐based method called splicing overlap extension (Horton et al., 1990); the oligonucleotide primers used are listed in Table 1. The resulting plasmid, pTAN114ΔI, contained the chiA cDNA from maize inbred LH82. To remove the intron from pTAN112, both pTAN112 and pTAN114ΔI were digested with restriction enzymes SmaI and BglII. The small fragment from pTAN114ΔI was ligated into the large vector fragment of pTAN112 to create pTAN112ΔI, which contains the chiA cDNA from maize inbred B73.
Table 1.
Oligonucleotide sequences.
| Name | Sequence 5′–3′ | Description |
|---|---|---|
| chiA intron deletion | ||
| Exon2Fwd | ACGCACGAGACCGGACATTTCTGCTACATCAGC | Exon2 forward primer |
| Exon1Rev | GATGTAGCAGAAATGTCCGGTCTCGTGCGTGAC | Exon1 reverse primer |
| Exon1Fwd | TCAACGGCATCAAGAACCAG | Exon1 forward primer |
| Exon2Rev | GTCTTGAACGCGATCACGGC | Exon2 reverse primer |
| ChitA amino acid mutagenesis | ||
| rChitA‐B73(A47D)Fwd | GCGGCACGACCGACGACTACTGCGGCGACGGGT | pTAN112ΔI(A47D) |
| rChitA‐B73(A47D)Rev | CGTCGCCGCAGTAGTCGTCGGTCGTGCCGCAGT | pTAN112ΔI(A47D) |
| rChitA‐LH82(D48A)Fwd | GCGGCACGACCGACGCCTACTGCGGCGACGGGT | pTAN114ΔI(D48A) |
| rChitA‐LH82(D48A)Rev | CGTCGCCGCAGTAGGCGTCGGTCGTGCCGCAGT | pTAN114ΔI(D48A) |
| FwdMut | ATGGCAAACGCGCCGAGGATCCTG | Both mutants |
| RevMut | CGATCTCGCGCTTGCCCTCCACCT | Both mutants |
| Expression plasmid cloning | ||
| LH82ExpFwd | GGATGAATTCCAGAACTGCGGCTGCCAGCCTAAC | pTAN121 and pTAN121(D48A) cloning |
| B73ExpFwd | GGATGAATTCCAGAACTGCGGCTGCCAGCCAAAC | pTAN122 and pTAN122(A47D) cloning |
| ExpRev | CCATTCTAGACTAGCAAGTGAGGTTGGGCCCTGG | All four expression clones |
Single nucleotides from each of pTAN114ΔI and pTAN112ΔI were changed by site‐directed mutagenesis to create pTAN114ΔI(D48A) and pTAN112ΔI(A47D). As in the intron deletion step, the splicing overlap extension technique was used. Oligonucleotide primers are listed in Table 1.
The four expression plasmids were created by PCR amplification of chiA cDNAs from pTAN114ΔI, pTAN112ΔI, pTAN114ΔI(D48A) and pTAN112ΔI(A47D), ligating the amplified DNA into the pPICZαA expression vector (Invitrogen, Carlsbad, CA, USA) to create pTAN121, pTAN122, pTAN121(D48A) and pTAN122(A47D). The PCR products were ligated into pPICZαA between the EcoRI and XbaI restriction sites. In the expression plasmids, the maize SSS is replaced by one that is present in pPICZαA. The identity of the maize SSS was determined by the SignalP 3.0 server (Bendtsen et al., 2004), which predicts it to be the first 26 amino acids [D‐score, 0.919; hidden Markov model (HMM), 1.000]. For each of the four plasmids, DNA was prepared from Escherichia coli cultures, linearized and transformed into competent P. pastoris strain X‐33 by electroporation. Isolates with integrated chiA genes were selected by plating transformed cells on yeast peptone dextrose (YPD) agar supplemented with sorbitol and zeocin (1% yeast extract, 2% peptone, 2% glucose, 1 m sorbitol, 2% agar, 100 µg/mL zeocin). Table 2 describes all the plasmids used in this study.
Table 2.
Plasmids used in this study.
| Plasmid name | Description |
|---|---|
| pBluescript KS(+) cloning plasmids | |
| pTAN112 | chiA gene from maize inbred B73 |
| pTAN114* | chiA gene from maize inbred LH82 |
| pTAN112ΔI | chiA cDNA from maize inbred B73 |
| pTAN114ΔI | chiA cDNA from maize inbred LH82 |
| pTAN112ΔI(A47D) | chiA from B73 with A47D mutation |
| pTAN114ΔI(D48A) | chiA from LH82 with D48A mutation |
| pPICZαA expression plasmids | |
| pTAN121* | Expression of rChitA‐LH82 |
| pTAN122 | Expression of rChitA‐B73 |
| pTAN121(D48A) | Expression of rChitA‐LH82 with D48A mutation |
| pTAN122(A47D) | Expression of rChitA‐B73 with A47D mutation |
Plasmids shown in Fig. 1.
Expression and purification of rChitAs from P. pastoris
Secreted expression of maize ChitA proteins was guided by the easyselect Pichia expression kit manual (Invitrogen). Small starter cultures with 25 mL of buffered glycerol complex medium (BMGY) [1% yeast extract, 2% peptone, 100 mm potassium phosphate (pH 6.0), 1.34% yeast nitrogen base, 4 × 10% biotin, 1% glycerol] were inoculated with a single colony from a zeocin‐containing YPD agar plate. These cultures were grown at 28 °C for 16 h. Cultures were then used to inoculate a 1 L BMGY culture at an optical density (OD) of 0.1. The larger BMGY cultures were grown at 25 °C to OD = 5. Cells were pelleted by centrifugation (3150 g; 5 min). Cell pellets were resuspended in 1 L of buffered methanol complex medium (BMMY) [1% yeast extract, 2% peptone, 100 mm potassium phosphate (pH 6.0), 1.34% yeast nitrogen base, 4 × 10–5% biotin, 0.5% methanol] to induce secreted expression of ChitA. Expression cultures were grown at 25 °C for 48 h.
The first step in the purification of rChitAs was the production of CFM. Cultures were centrifuged (3150 g; 5 min) and the supernatants were filtered through a 0.45‐µm, HV durapore membrane (Millipore, Bedford, MA, USA). CFMs were chilled to 4 °C and proteins were salted out by the gradual addition of ammonium sulphate to 60% saturation (390 g/L); after salt had been added, the solutions were incubated with stirring for 1 h; after incubation, the proteins were precipitated by centrifugation (4900 g; 20 min). Precipitated proteins were resuspended in 10 mL of sodium acetate buffer [10 mm sodium acetate (pH 5.2) + 100 mm sodium chloride] and dialysed against sodium acetate buffer for 20 h. ChitA proteins were bound to a cation exchange column [HiTrap SP XL (5 mL), GE Healthcare, Waukesha, WI, USA] and the column was washed with an additional 20 mL of sodium acetate buffer. Bound proteins were eluted from the column by increasing the sodium chloride concentration from 100 to 500 mm. Elution fractions of 1 mL were collected. Fractions that contained ChitA were combined and ammonium sulphate was added to 40% saturation. ChitA was pelleted by centrifugation (14 000 g; 2 min) and resuspended in sodium acetate buffer, followed by dialysis. The protein concentrations of elution fractions and final purified ChitA were determined using the DC Protein Assay (Bio‐Rad, Hercules, CA, USA) with bovine serum albumin (BSA) as a standard.
Assays of cmp
Assays of cmp contained 20 µg rChitA (74 µm) and either 5 µg of secreted protein extract from S. maydis NRRL 43670 or 0.1 µg of purified Bz‐cmp from B. zeicola NRRL 47238 in 10 µL of sodium acetate buffer. The growth of S. maydis cultures and the preparation of secreted protein extracts were performed as described previously (Naumann and Wicklow, 2010). The purification of Bz‐cmp from B. zeicola has been reported previously (Naumann et al., 2009). Stm‐cmp was not purified because secreted protein extracts already exhibit alloform‐specific modification of ChitA and a method for its purification has not been developed. Reactions were incubated at 30 °C for the indicated amount of time. Reactions were stopped by combining aliquots from the reaction with SDS‐PAGE loading dye, and heating in boiling water for 1 min. Reactions were analysed by Tricine‐SDS‐PAGE (12% T; 3% C) (Schagger, 2006).
Amino‐terminal sequencing of proteins
rChitA‐B73 was incubated (30 °C; 16 h) either alone or with the addition of Stm‐cmp or Bz‐cmp. The reactions were loaded in a Tricine‐SDS‐PAGE gel, and the proteins were separated at 90 V for 2.5 h. The proteins were transferred from the gel to a poly(vinylidene difluoride) (PVDF) membrane (Immun‐Blot; Bio‐Rad) for 1 h at 50 V in a semi‐dry transfer unit with N‐cyclohexyl‐3‐aminopropanesulphonic acid (CAPS) buffer [10 mm CAPS (pH 11.0), 10% methanol]. The membrane was stained in a Coomassie solution (10 mg Coomassie Brilliant Blue R‐250, 1 mL acetic acid, 40 mL methanol, 60 mL water) for 30 s, followed by destaining for 10 min (50% methanol). Protein bands were removed from the membrane with a scalpel, placed in microcentrifuge tubes and submitted for analysis by Edman degradation (Protein Sciences Facility, University of Illinois, Urbana, IL, USA).
Mention of a trade name or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
REFERENCES
- Aboitiz, N. , Vila‐Perello, M. , Groves, P. , Asensio, J.L. , Andreu, D. , Canada, F.J. and Jimenez‐Barbero, J. (2004) NMR and modeling studies of protein–carbohydrate interactions: synthesis, three‐dimensional structure, and recognition properties of a minimum hevein domain with binding affinity for chitooligosaccharides. Chembiochem, 5, 1245–1255. [DOI] [PubMed] [Google Scholar]
- Abramovitch, R.B. , Anderson, J.C. and Martin, G.B. (2006) Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Nielsen, H. , von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Eathington, S.R. , Crosbie, T.M. , Edwards, M.D. , Reiter, R.S. and Bull, J.K. (2007) Molecular markers in a commercial breeding program. Crop Sci. 47, S‐154–S‐163. [Google Scholar]
- Edman, P. and Begg, G. (1967) A protein sequenator. Eur. J. Biochem. 1, 80–91. [DOI] [PubMed] [Google Scholar]
- Gerhardt, L.B. , Sachetto‐Martins, G. , Contarini, M.G. , Sandroni, M. , de Lima, V.M. , Cordeiro, M.C. , de Oliveira, D.E. and Margis‐Pinheiro, M. (1997) Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris . FEBS Lett. 419, 69–75. [DOI] [PubMed] [Google Scholar]
- Gerhardt, L.B. , Magioli, C. , Perez, A.B. , Margis, R. , Sachetto‐Martins, G. and Margis‐Pinheiro, M. (2004) AtchitIV gene expression is stimulated under abiotic stresses and is spatially and temporally regulated during embryo development. Genet. Mol. Biol. 27, 118–123. [Google Scholar]
- Horton, R.M. , Cai, Z.L. , Ho, S.N. and Pease, L.R. (1990) Gene splicing by overlap extension: tailor‐made genes using the polymerase chain reaction. Biotechniques, 8, 528. [PubMed] [Google Scholar]
- Huynh, Q.K. , Hironaka, C.M. , Levine, E.B. , Smith, C.E. , Borgmeyer, J.R. and Shah, D.M. (1992) Antifungal proteins from plants: purification, molecular cloning, and antifungal properties of chitinases from maize seed. J. Biol. Chem. 267, 6635–6640. [PubMed] [Google Scholar]
- Jong, A.D. , Cordewener, J. , Schiavo, F.L. , Terzi, M. , Vandekerckhove, J. , Kammen, A.V. and Vries, S.D. (1992) A carrot somatic embryo mutant is rescued by chitinase. Plant Cell, 4, 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA, 103, 11 086–11 091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, B. (1959) Corn ear rots in Illinois. Ill. Agric. Exp. Stn. Bull. 639, 87 pp. [Google Scholar]
- Lange, J. , Mohr, U. , Wiemken, A. , Boller, T. and Vogeli‐Lange, R. (1996) Proteolytic processing of class IV chitinase in the compatible interaction of bean roots with Fusarium solani . Plant Physiol. 111, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller, D.A. and Tiffin, P. (2008) Geographic variation in adaptation at the molecular level: a case study of plant immunity genes. Evolution, 62, 3069–3081. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531. [DOI] [PubMed] [Google Scholar]
- Naumann, T.A. and Wicklow, D.T. (2010) Allozyme specific modification of a maize seed chitinase by a protein secreted by the fungal pathogen Stenocarpella maydis . Phytopathology, 100, 645–654. [DOI] [PubMed] [Google Scholar]
- Naumann, T. , Wicklow, D. and Kendra, D. (2009) Maize seed chitinase is modified by a protein secreted by Bipolaris zeicola . Phys. Mol. Plant Pathol. 74, 134–141. [Google Scholar]
- Neuhaus, J. , Fritig, B. , Linthorst, H. , Meins, F. , Mikkelsen, J. and Ryals, J. (1996) A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 14, 102–104. [Google Scholar]
- Rossouw, J.D. , Pretorius, Z.A. , Silva, H.D. and Lamkey, K.R. (2009) Breeding for resistance to Stenocarpella ear rot in maize. Plant Breed. Rev. 31, 223–246. [Google Scholar]
- Schagger, H. (2006) Tricine‐SDS‐PAGE. Nat. Protoc. 1, 16–22. [DOI] [PubMed] [Google Scholar]
- Schlumbaum, A. , Mauch, F. , Vogeli, U. and Boller, T. (1986) Plant chitinases are potent inhibitors of fungal growth. Nature, 324, 365–367. [Google Scholar]
- Schnable, P.S. , Ware, D. , Fulton, R.S. , Stein, J.C. and Wei, F. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science, 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- Shibuya, N. and Minami, E. (2001) Oligosaccharide signalling for defence responses in plant. Phys. Mol. Plant Pathol. 59, 223–233. [Google Scholar]
- Shinya, T. , Hanai, K. , Galis, I. , Suzuki, K. , Matsuoka, K. , Matsuoka, H. and Saito, M. (2007) Characterization of NtChitIV, a class IV chitinase induced by β‐1,3‐,1,6‐glucan elicitor from Alternaria alternata 102: antagonistic effect of salicylic acid and methyl jasmonate on the induction of NtChitIV. Biochem. Biophys. Res. Commun. 353, 311–317. [DOI] [PubMed] [Google Scholar]
- Tiffin, P. (2004) Comparative evolutionary histories of chitinase genes in the genus Zea and family Poaceae. Genetics, 167, 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verburg, J. , Smith, C. , Lisek, C. and Huynh, Q. (1992) Identification of an essential tyrosine residue in the catalytic site of a chitinase isolated from Zea mays that is selectively modified during inactivation with 1‐ethyl‐3‐(3‐dimethylaminopropyl)‐carbodiimide. J. Biol. Chem. 267, 3886–3893. [PubMed] [Google Scholar]
- Wiweger, M. , Farbos, I. , Ingouff, M. , Lagercrantz, U. and Arnold, S.V. (2003) Expression of Chia4‐Pa chitinase genes during somatic and zygotic embryo development in Norway spruce Picea abies: similarities and differences between gymnosperm and angiosperm class IV chitinases. J. Exp. Bot. 54, 2691–2699. [DOI] [PubMed] [Google Scholar]


