SUMMARY
Citrus exocortis viroid (CEVd) is the causal agent of exocortis disease of citrus. CEVd has a wide host range that includes woody and herbaceous species. A new CEVd strain (CEVdCOL), phylogenetically clustering with CEVd variants of Class A inducing severe symptoms in tomato, was identified in Colombia and shown to induce only extremely mild symptoms in Etrog citron indicator plants. Using site‐directed mutagenesis, two nucleotide substitutions (314A → G and 315U → A) in the lower strand of the P domain of the predicted CEVdCOL secondary structure resulted in a severe artificial CEVdMCOL variant. Conversely, two nucleotide exchanges (314G → A and 315A → U) in the same region of the severe variant CEVdE‐117 resulted in a symptomless artificial CEVdME‐117 variant. Infectivity assays conducted with the natural and mutated variants showed that all induced severe symptoms in Gynura aurantiaca, tomato and chrysanthemum. This is the first report of the identification of pathogenic determinants of CEVd in citrus, and shows that these pathogenicity determinants are host dependent.
Citrus exocortis viroid (CEVd) is the causal agent of exocortis disease of citrus, a bark scaling disorder that affects, among others, trifoliate orange [Poncirus trifoliata (L.) Raf.] and its hybrids (Troyer and Carrizo citranges) and Rangpur lime (Citrus limonia Osb.), which are all widely used as rootstocks in commercial orchards. CEVd belongs to the genus Pospiviroid, family Pospiviroidae, and is a covalently closed, circular RNA with about 370 nucleotides and a highly base‐paired, rod‐like secondary structure (revised by Duran‐Vila and Semancik, 2003). Like the other members of this genus, the predicted secondary structure of CEVd conforms to the model of the five structural domains (Terminal left, TL; Pathogenicity, P; Central, C; Variable, V; and Terminal right, TR) defined by Keese and Symons (1985), with a central conserved region (CCR) and a terminal conserved region (TCR) within the C and TL domains, respectively (Fig. 1) (Flores et al., 2005). CEVd has a wide experimental host range, including woody and herbaceous species, with sensitive hosts displaying symptoms of stunting, leaf epinasty and distortion. Etrog citron (Citrus medica L.) has been widely used for biological indexing purposes and, following infection, displays a severe syndrome characterized by pronounced stunting, strong leaf epinasty, and midvein, petiole and stem necrosis. Based on the symptoms induced in tomato (Solanum lycopersicum L.) as an experimental host, CEVd variants have been classified into severe ‘Class A’ and mild ‘Class B’ (Visvader and Symons, 1985). Such variants have been found to differ in a minimum of 26 nucleotides, mainly affecting two regions (PL and PR) located, respectively, in the P and V domains of the viroid secondary structure (Fig. 1). Infectivity assays conducted with chimeric cDNA clones have shown that the changes in the PL region are responsible for symptom modulation (Visvader and Symons, 1986). This result was further confirmed using Gynura aurantiaca as an experimental host, in which a set of five nucleotides located in the P domain discriminated between variants inducing severe symptoms and those inducing mild symptoms (Chaffai et al., 2007; Skoric et al., 2001). The limited information available regarding the modulation of symptom expression of CEVd in citrus indicates that ‘Class A’ and Class B’ variants induce similar symptoms in trifoliate orange (Vernière et al., 2004), suggesting that the pathogenicity determinants identified using tomato and G. aurantiaca do not necessarily affect symptom expression in citrus hosts.
Figure 1.
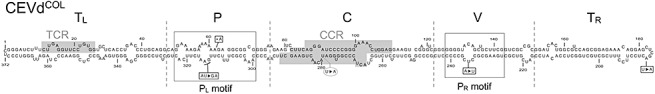
Proposed secondary structure of minimum free energy of CEVdCOL. Discontinuous lines divide the secondary structure into the five structural domains (Terminal left, TL; Pathogenicity, P; Central, C; Variable, V; and Terminal right, TR). Conserved regions [central conserved region (CCR) and terminal conserved region (TCR)] are shaded in the TL and C domains. PL and PR are boxed in the P and V domains. Boxed nucleotides (square boxes) show nucleotide changes found in CEVdCOL when compared with the severe variant CEVdE‐117. Boxed nucleotide (round box) shows the nucleotide change found in the CEVd variants recovered from inoculated tomatoes.
Recently, an unusual CEVd isolate (CEVdCOL) has been identified in a symptomless Etrog citron tree from Colombia. Unexpectedly, the consensus sequence of CEVdCOL (EU512994), constructed with the sequences of full‐length clones showing some variability, was highly similar (96.5%) to the reference sequence (M30868) of ‘Class A’ variants (Murcia et al., 2007). As viroids infect their hosts as populations of closely related variants, the dominant CEVd variant present in CEVdCOL was identified as follows. Briefly, nucleic acid preparations from the symptomless Etrog citron infected with CEVdCOL were subjected to reverse transcriptase‐polymerase chain reaction (RT‐PCR) using primers CEVdF1 and CEVdR1, as described by Bernad and Duran‐Vila (2006), and the DNA amplicons were cloned in Bluescript II KS+ plasmid (Promega®, Madison, WI, USA). The dominant CEVd variant, identified by sequencing six clones, was found to be identical to the consensus sequence reported by Murcia et al. (2007) .
To compare the biological properties of this new variant (CEVdCOL) with that of CEVdE‐117, a variant which induces severe symptoms in Etrog citron as well as in other citrus hosts, and is characterized as ‘class A’ with a nucleotide identity of 98.2% to the reference sequence (M30868) (nucleotide changes are boxed in Fig. 1) (Duran‐Vila and Semancik, 1990; Gandía et al., 2005; Vernière et al., 2004), an assay was performed to compare symptom expression on Etrog citron. The inocula consisted of in vitro‐synthesized dimeric transcripts of each variant. Briefly, monomeric DNAs of each clone were recovered as blunt‐end PCR products using the phosphorylated primers CEVd‐R1 and CEVd‐F1 and Pfu DNA polymerase. The amplification products were ligated with T4 DNA ligase and the dimeric molecules were cloned into pBluescript II KS (+) digested with EcoRV. The recombinant plasmids from transformed cells were sequenced to verify the head‐to‐tail organization of the dimeric inserts and, according to their orientation, these plasmids were linearized with EcoRI and used as a template in transcription reactions with 1 mm deoxynucleoside triphosphates (NTPs), 1 mm dithiothreitol (DTT) and 50 U of RNA polymerase T7 to produce dimeric transcripts. Three Etrog citrons were slash‐inoculated using 50 ng of each transcript per plant, and kept for 6 months in the glasshouse at 28–32°C; three additional noninoculated plants were maintained under the same conditions as controls. Infection was confirmed by Northern blot hybridization, as described by Murcia et al. (2009b) (data not shown), and the stability of the progeny was assessed by sequencing RT‐PCR amplicons from nucleic acid extracts of each plant. In order to monitor symptom expression, all the plants were cut at the level of the second internode and the symptoms in the second flush of growth were evaluated. Three months later, plants infected with CEVdE‐117 disclosed the characteristic CEVd syndrome (Fig. 2). In contrast, plants infected by CEVdCOL disclosed an almost imperceptible leaf distortion (Fig. 2). Northern blot hybridization analysis (Murcia et al., 2009b) of these plants using a CEVd‐specific probe showed that infected plants accumulated similar viroid titres, and that the drastic difference between the severe symptom expression in CEVdE‐117‐infected plants and the virtually symptomless condition in CEVdCOL‐infected plants was unrelated to viroid titres (Fig. 2). Therefore, subtle differences in the nucleotide composition of CEVdE‐117 and CEVdCOL must be responsible for their distinct biological properties.
Figure 2.

Top: symptom expression in Etrog citron plants infected with natural variants (CEVdCOL, CEVdE‐117) and the corresponding mutated variants (CEVdMCOL, CEVdME‐117) obtained by site‐directed mutagenesis. Bottom: viroid titres in plants infected with CEVdCOL, CEVdE‐117, CEVdMCOL and CEVdME‐117, as determined by Northern blot hybridization with CEVd‐specific DNA probes. Ethidium bromide staining of a nondenaturing polyacrylamide gel shows that RNA levels (6S and 5S RNAs) in all preparations are comparable.
In order to verify which nucleotide changes were responsible for the differences in biological properties observed, an approach using site‐directed mutagenesis was used. As the PL region located in the P domain has been demonstrated to be responsible for symptom modulation in herbaceous hosts (Visvader and Symons, 1986), the changes identified in positions 314 and 315 of the lower strand of this region were chosen to synthesize two mutants. Mutant CEVdMCOL was designed by introducing, into the sequence of CEVdCOL, the substitutions 314G → A and 315A → U, which are characteristic of CEVdE‐117. Similarly, the mutant CEVdME‐117 was designed by introducing, into the sequence of CEVdE‐117, the substitutions 314A → G and 315U → A, which are characteristic of CEVdCOL. These mutants were generated following a PCR‐based protocol (Byrappa et al., 1995) with minor modifications (Gago et al., 2005). Briefly, 5 ng of plasmid pBluescript II KS (+) containing the full‐length sequences of either CEVdCOL or CEVdE‐117 were amplified using each pair of adjacent phosphorylated primers in which the appropriate changes (shown in bold) had been introduced in the forward primers F‐MCO (5′ATATCTTCACTGCTCTCCGGGCG3′) and F‐ME117 (5′GAATCTTCACTGCTCTCCGGGCG3′). In both instances, the reverse primer was CEVd‐R (5′AAGAAAAGCGGTTTGGGGTTGAAGC3′). The PCR cycling profile to amplify the complete plasmid with Pfu Turbo DNA polymerase consisted of 30 cycles of 30 s at 94°C, 30 s at 60°C and 3.5 min at 72°C, with an initial denaturation at 94°C for 2 min and a final extension at 72°C for 10 min. After electrophoresis in 1% agarose gels, PCR‐amplified products of plasmid length were purified with the QIAquick kit (Qiagen®, Valencia, CA, USA), circularized with T4 ligase and used for transformation. Sequencing confirmed that the plasmids contained inserts with only the desired mutations. Dimeric transcripts of each mutant were generated with the strategy described above, using, as template, the plasmid containing the insert of the mutants (CEVdMCOL and CEVdME‐117). Three Etrog citron plants were slash‐inoculated with 50 ng of each transcript per plant and kept for 6 months in the glasshouse at 28–32°C; three plants inoculated with CEVdCOL, three plants inoculated with CEVdE‐117 and three noninoculated plants were maintained under the same conditions as positive and negative controls. Infection was confirmed by Northern blot hybridization (data not shown), and the stability of the progeny was assessed by sequencing RT‐PCR amplicons obtained with nucleic acid preparations from each plant. In order to monitor viroid‐induced symptoms, all the plants were cut at the level of the second internode, and the second flush of tissue was evaluated for symptom expression. Three months later, positive control plants displayed the characteristic symptoms of CEVdE‐117 and CEVdCOL, as described above. Plants infected with CEVdMCOL disclosed the characteristic syndrome induced by CEVdE‐117 (Fig. 2), whereas plants infected with CEVdME‐117 were symptomless (Fig. 2) and indistinguishable from the noninoculated controls. Northern blot hybridization showed that the differences observed in symptom expression were unrelated to viroid titres (Fig. 2) and were therefore a result of differences in nucleotides 314 and 315, acting as pathogenicity determinants. However, it should be mentioned that, although plants infected with CEVdME‐117 were indistinguishable from negative controls, plants infected with CEVdCOL displayed an extremely subtle leaf distortion. This observation suggests that other positions in the viroid molecule may also play a role in the modulation of symptom expression.
Although the molecular bases involved in symptom expression are still unknown, it is generally accepted that viroid‐induced symptoms are caused by specific interference with host gene expression. This hypothesis is supported by the results obtained from macroarray‐based and differential display approaches, which showed that the regulation of specific host genes was altered in viroid‐infected plants (Itaya et al., 2002; Tessitori et al., 2007). Studies conducted with different strains of Potato spindle tuber (PSTVd), the type member of the genus Pospiviroid, allowed the identification of the virulence‐modulating (VM) region located in the P domain, in which as few as one or two nucleotides appear to be responsible for symptom modulation (Góra et al., 1996; Góra‐Sochacka et al., 1997; Lakshman and Tavantzis, 1993; Owens et al., 1991). This supports the hypothesis that specific viroid sequences/structures (Owens et al., 1996) probably interact with yet‐to‐be‐identified host factors. The results reported here on the modulation of symptom expression in CEVd‐infected Etrog citron plants show the same trend as those described for PSTVd. It was also observed that CEVdCOL‐infected plants displayed extremely subtle leaf distortion, suggesting that other positions of the viroid molecule may also play a role in the modulation of symptom expression, as already reported for the TL, TR and C domains of PSTVd (Qi and Ding, 2003; Sano et al., 1992) and for the TL domain of Citrus viroid V (CVd‐V) and Citrus dwarfing viroid (CDVd), two members of the genus Apscaviroid (Murcia et al., 2009a; Serra et al., 2009).
As most of the information available regarding the identification of pathogenicity determinants of viroids has been obtained using experimental herbaceous hosts, additional assays were performed to determine the effect of inoculation of CEVdCOL, CEVdE‐117, CEVdMCOL or CEVdME‐117 into G. aurantiaca, chrysanthemum (Chrysanthemum morifolium) and tomato plants. Nucleic acid preparations from infected Etrog citron plants were used to inoculate these three herbaceous hosts. The inoculated plants (five plants per inoculum source and host species) and five noninoculated controls were maintained for 3 months in the glasshouse at 28–32°C. Three months later, infection was assessed by Northern blot hybridization, which showed that most infected plants accumulated similar viroid titres (Fig. 3). However, unlike that observed with Etrog citron, all CEVd‐infected plants disclosed symptoms regardless of the inoculum source (Fig. 3). Within each host, the symptoms induced by natural variants and by artificial mutants were indistinguishable from each other. It should be noted that, although tomato plants infected with CEVdME‐117 presented slightly lower viroid titres, the symptoms were indistinguishable from those induced by the other CEVd variants (CEVdCOL, CEVdE‐117 and CEVdMCOL). The stability of the inoculated variants in each host was assessed by RT‐PCR and amplicon sequencing, showing that the symptoms observed were not associated with reversion events. It should be noted, however, that all the CEVd variants recovered from tomato contained a 279U → A transition in the lower strand of the C domain (boxed in Fig. 1) that did not affect symptom expression. As already reported by Semancik et al. (1993), this change may be the result of differences in host selection pressures.
Figure 3.

Top: symptom expression in Gynura aurantiaca (A), Chrysanthemum morifolium (B) and Solanum lycopersicum (C) infected with natural variants (CEVdCOL, CEVdE_117) and the corresponding mutated variants (CEVdMCOL, CEVdME‐117) obtained by site‐directed mutagenesis. Bottom: viroid titres in plants infected with CEVdCOL, CEVdE‐117, CEVdMCOL and CEVdME‐117, as determined by Northern blot hybridization with a CEVd‐specific DNA probe. Ethidium bromide staining of a nondenaturing polyacrylamide gel shows that RNA levels (6S and 5S RNAs) in all preparations are comparable.
The overall results reported here, in addition to identifying the first pathogenic determinants of CEVd in citrus, illustrate that the modulation of symptom expression is host dependent. Although the natural variants, CEVdCOL and CEVdE‐117, and their respective mutants, CEVdMCOL and CEVdME‐117, induced different responses in Etrog citron, their effect on the experimental herbaceous hosts tested was always severe. Recently, a new CEVd variant recovered from citrus (Bernad et al., 2005) has been shown to act as a very mild strain in herbaceous hosts, whereas it induced severe symptoms in Etrog citron (L. Bernad et al., in preparation). The lack of correlation between symptom expression in Etrog citron and other experimental hosts has also been found in a field assay in which, over a 12‐year period, the response of clementine trees on trifoliate orange inoculated with a severe (Class A) CEVd variant was compared with that of trees inoculated with a mild (Class B) CEVd variant (Vernière et al., 2004). The two CEVd variants induced comparable reductions in tree size and harvest, and no differences in agronomic parameters were observed.
The mechanisms underlying viroid pathogenesis are still unclear, and different hypotheses have been advanced on how viroid infection elicits the cascade of events leading to symptom expression in sensitive hosts. As viroid RNAs must interact with host proteins to produce a systemic infection, it is plausible that interactions of this kind could be the primary signal of pathogenesis. X‐Ray crystal and nuclear magnetic resonance studies have shown that most RNA loops and bulges are highly structured motifs stabilized by non‐Watson–Crick base pairing, base stacking and other noncanonical interactions; these motifs most probably serve as the major sites for RNA–RNA, RNA–protein and RNA–small ligand interactions (Leontis et al., 2006). Although the nucleotide changes associated with pathogenesis in Etrog citron do not alter the predicted secondary structure of CEVd, they modify the primary structure of a specific loop and may affect its internal structure. A second mechanism in viroid pathogenesis involves RNA silencing and proposes that small viroid‐derived RNAs (vd‐sRNAs) guide the RNA‐induced silencing complex to inactivate certain messenger RNAs of the host (Wang et al., 2004). Our results are also consistent with this sequence‐specific mechanism, because it is possible that vd‐sRNAs from CEVdE‐117 and CEVdME‐117 might target different host RNAs.
From a practical point of view, one should be cautious when using biological indexing for viroid detection. As shown in this work, certain viroid variants may infect and replicate latently in Etrog citron, the most widely used indicator species for viroid detection in citrus certification programmes. We recommend the concomitant use of bioassays and additional molecular methods.
ACKNOWLEDGEMENTS
This research was supported by grant AGL2008‐01491 from the Ministerio de Educación y Ciencia. P. Serra and L. Bernad are recipients of fellowships from the Consellería de Agricultura—IVIA. The authors would like to acknowledge J.M. Bové for critical reading of the manuscript and R. Carbó for technical assistance.
REFERENCES
- Bernad, L. and Duran‐Vila, N. (2006) A novel RT‐PCR approach for detection and characterization of citrus viroids. Mol. Cell. Probes, 20, 105–113. [DOI] [PubMed] [Google Scholar]
- Bernad, L. , Moreno, P. , Bové, J.M. and Duran‐Vila, N. (2005) Viroids in Gummy bark sources from the Sultanate of Oman In: Proceedings of the 16th Conference of the International Organization of Citrus Virologists (IOCV) (Hilf M.E., Duran‐Vila N. and Rocha‐Peña M.A., eds), pp. 272–279. Riverside, CA: IOCV. [Google Scholar]
- Byrappa, S. , Gavin, D.K. and Gupta, K.C. (1995) A highly efficient procedure for site‐specific mutagenesis of full‐length plasmids using Vent DNA polymerase. PCR Methods Appl. 5, 404–407. [DOI] [PubMed] [Google Scholar]
- Chaffai, M. , Serra, P. , Gandía, M. , Hernández, C. and Duran‐Vila, N. (2007) Molecular characterization of CEVd strains that induce different phenotypes in Gynura aurantiaca: structure–pathogenicity relationships. Arch. Virol. 152, 1283–1294. [DOI] [PubMed] [Google Scholar]
- Duran‐Vila, N. and Semancik, J.S. (1990) Variations on the ‘cross protection’ effect between two strains of citrus exocortis viroid. Ann. Appl. Biol. 17, 367–377. [Google Scholar]
- Duran‐Vila, N. and Semancik, J.S. (2003) Citrus viroids In: Viroids (Hadidi A., Flores R., Randles J.W. and Semancik J.S., eds), pp. 178–194. Collingwood, Australia: CSIRO. [Google Scholar]
- Flores, R. , Hernández, C. , Martínez de Alba, A.E. , Darós, J.A. and Di Serio, F. (2005) Viroids and viroid–host interactions. Annu. Rev. Phytopathol. 43, 117–139. [DOI] [PubMed] [Google Scholar]
- Gago, S. , De La Peña, M. and Flores, R. (2005) A kissing‐loop interaction in hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA, 11, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandía, M. , Rubio, L. , Palacio, A. and Duran‐Vila, N. (2005) Genetic variation and population structure of an isolate of citrus exocortis viroid (CEVd) and of the progenies of two infectious sequences variants. Arch. Virol. 150, 1945–1957. [DOI] [PubMed] [Google Scholar]
- Góra, A. , Candresse, T. and Zagórski, W. (1996) Use of intramolecular chimeras to map molecular determinants of symptom severity of potato spindle tuber viroid (PSTVd). Arch. Virol. 141, 2045–2055. [DOI] [PubMed] [Google Scholar]
- Góra‐Sochacka, A. , Kierzez, A. , Candresse, T. and Zagórski, W. (1997) The genetic stability of potato spindle tuber viroid (PSTVd) molecular variants. RNA, 3, 68–74. [PMC free article] [PubMed] [Google Scholar]
- Itaya, A. , Matsuda, Y. , Gonzales, R.A. , Nelson, R.S. and Ding, B. (2002) Potato spindle tuber viroid strains of different pathogenicity induce and suppress expression of common and unique genes in infected tomato. Mol. Plant–Microbe Interact. 15, 990–999. [DOI] [PubMed] [Google Scholar]
- Keese, P. and Symons, R.H. (1985) Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA, 82, 4582–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshman, D.K. and Tavantzis, S.M. (1993) Primary and secondary structure of a 360‐nucleotide isolate of potato spindle tuber viroid. Arch. Virol. 128, 319–331. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B. , Lescoute, A. and Westhof, E. (2006) The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 16, 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia, N. , Bernad, L. , Caicedo, A. and Duran‐Vila, N. (2007) Citrus viroids in Colombia In: Proceedings 17th Conference of the IOCV. IOCV, Riverside (in press). [Google Scholar]
- Murcia, N. , Bernad, L. , Serra, P. , Bani Hashemian, S.M. and Duran‐Vila, N. (2009a) Molecular and biological characterization of natural variants of Citrus dwarfing viroid. Arch. Virol. 154, 1329–1334. [DOI] [PubMed] [Google Scholar]
- Murcia, N. , Serra, P. , Olmos, A. and Duran‐Vila, N. (2009b) A novel hybridization approach for detection of citrus viroids. Mol. Cell. Probes, 23, 95–102. [DOI] [PubMed] [Google Scholar]
- Owens, R.A. , Thomsom, S.M. and Steger, G. (1991) Effects of random mutagenesis upon potato spindle tuber viroid replication and symptom expression. Virology, 185, 18–31. [DOI] [PubMed] [Google Scholar]
- Owens, R.A. , Steger, G. , Hu, Y. , Fels, A. , Hammond, R.W. and Riesner, D. (1996) RNA structural features responsible for potato spindle tuber viroid pathogenicity. Virology, 22, 144–158. [DOI] [PubMed] [Google Scholar]
- Qi, Y. and Ding, B. (2003) Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell, 15, 1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, T. , Candresse, T. , Hammond, R.W. , Diener, T.O. and Owens, R.A. (1992) Identification of multiple structural domains regulating viroid pathogenicity. Proc. Natl. Acad. Sci. USA, 89, 1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semancik, J.S. , Szychowski, J.A. , Rakowski, A.G. and Symons, R.H. (1993) Isolates of citrus exocortis viroid recovered by host and tissue selection. J. Gen. Virol. 74, 2427–2436. [DOI] [PubMed] [Google Scholar]
- Serra, P. , Bani Hashemian, S.M. , Pensabene‐Bellavia, G. , Gago, S. and Duran‐Vila, N. (2009) An artificial chimeric derivative of citrus viroid V involves the terminal left domain in pathogenicity. Mol. Plant Pathol. 10, 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoric, D. , Conerly, M. , Szychowski, J.A. and Semancik, J.S. (2001) CEVd induced symptom modification as a response to a host‐specific temperature‐sensitive reaction. Virology, 280, 115–123. [DOI] [PubMed] [Google Scholar]
- Tessitori, M. , Maria, G. , Capasso, C. , Catara, G. , Rizza, S. , De Luca, V. , Catara, A. , Capasso, A. and Carginale, V. (2007) Differential display analysis of gene expression in Etrog citron leaves infected by Citrus viroid III. Biochim. Biophys. Acta, 1769, 228–235. [DOI] [PubMed] [Google Scholar]
- Vernière, C. , Perrier, X. , Dubois, C. , Dubois, A. , Botella, L. , Chabrier, C. , Bové, J.M. and Duran‐Vila, N. (2004) Citrus viroids: symptom expression and effect on vegetative growth and yield on clementine trees grafted on trifoliate orange. Plant Dis. 88, 709–713. [DOI] [PubMed] [Google Scholar]
- Visvader, J.E. and Symons, R.H. (1985) Eleven new sequence variants of citrus exocortis viroid and the correlation of sequence with pathogenicity. Nucleic Acids Res. 13, 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader, J.E. and Symons, R.H. (1986) Replication of in vitro constructed viroid mutants: location of the pathogenicity‐modulating domain of citrus exocortis viroid. EMBO J. 5, 2051–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M.B. , Bian, X.Y. , Wu, L.M. , Liu, L.X. , Smith, N.A. , Isenegger, D. , Wu, R.M. , Masuta, C. , Vance, V.B. , Watson, J.M. , Rezaian, A. , Dennis, E.S. and Waterhouse, P.M. (2004) On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA, 101, 3275–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]


