SUMMARY
Rhizoctonia solani Kühn is a soil‐borne fungal pathogen that causes disease in a wide range of plants worldwide. Strains of the fungus are traditionally grouped into genetically isolated anastomosis groups (AGs) based on hyphal anastomosis reactions. This article summarizes aspects related to the infection process, colonization of the host and molecular mechanisms employed by tobacco plants in resistance against R. solani diseases.
Taxonomy: Teleomorph: Thanatephorus cucumeris (Frank) Donk; anamorph: Rhizoctonia solani Kühn; Kingdom Fungi; Phylum Basidiomycota; Class Agaricomycetes; Order Cantharellales; Family Ceratobasidiaceae; genus Thanatephorus.
Identification: Somatic hyphae in culture and hyphae colonizing a substrate or host are first hyaline, then buff to dark brown in colour when aging. Hyphae tend to form at right angles at branching points that are usually constricted. Cells lack clamp connections, but possess a complex dolipore septum with continuous parenthesomes and are multinucleate. Hyphae are variable in size, ranging from 3 to 17 µm in diameter. Although the fungus does not produce any conidial structure, ellipsoid to globose, barrel‐shaped cells, named monilioid cells, 10–20 µm wide, can be produced in chains and can give rise to sclerotia. Sclerotia are irregularly shaped, up to 8–10 mm in diameter and light to dark brown in colour.
Disease symptoms: Symptoms in tobacco depend on AG as well as on the tissue being colonized. Rhizoctonia solani AG‐2‐2 and AG‐3 infect tobacco seedlings and cause damping off and stem rot. Rhizoctonia solani AG‐3 causes ‘sore shin’ and ‘target spot’ in mature tobacco plants. In general, water‐soaked lesions start on leaves and extend up the stem. Stem lesions vary in colour from brown to black. During late stages, diseased leaves are easily separated from the plant because of severe wilting. In seed beds, disease areas are typically in the form of circular to irregular patches of poorly growing, yellowish and/or stunted seedlings.
Resistance: Knowledge is scarce regarding the mechanisms associated with resistance to R. solani in tobacco. However, recent evidence suggests a complex response that involves several constitutive factors, as well as induced barriers controlled by multiple defence pathways.
Management: This fungus can survive for many years in soil as mycelium, and also by producing sclerotia, which makes the management of the disease using conventional means very difficult. Integrated pest management has been most successful; it includes timely fungicide applications, crop rotation and attention to soil moisture levels. Recent developments in biocontrol may provide other tools to control R. solani in tobacco.
INTRODUCTION
The soil‐borne basidiomycete fungus Rhizoctonia solani Kühn (teleomorph: Thanatephorus cucumeris) causes disease on many economically important crop plants worldwide. Isolates of this species‐complex are traditionally classified into genetically isolated anastomosis groups (AGs) based primarily on hyphal anastomosis reactions (Sneh et al., 1991). Over the last 30 years, a comprehensive literature has been produced dealing with the taxonomy, molecular systematics, genetics, pathology and ecology of Thanatephorus cucumeris that reflects its broad host range and prevalence in different agroecosystems. The establishment of R. solani hyphal AGs has greatly assisted in identification and epidemiological studies (Carling, 1996; Ogoshi, 1987). Further support for these genetic groupings has come from more recent internal transcribed spacer (ITS) rRNA gene sequence polymorphism analyses (Carling et al., 2002; González et al., 2001; Kuninaga et al., 2000; Salazar et al., 2000). Rhizoctonia solani AG‐2‐2 and AG‐3 are the main causal agents of leaf spot and root rot in tobacco (Nicotiana tabacum L.), where they cause damping off and stem rot in young transplants, sore shin in older field plants and a foliar disease named ‘target spot’ (Lucas, 1975; Sneh et al., 1996).
The leaf spot and root rot in tobacco were described for the first time in US tobacco crops in 1904 (Lucas, 1975). Although this disease is not considered to be critically important for tobacco cultivation, it nevertheless occurs every year in many fields of this crop (Lucas, 1975). As a result of the broad host range reported for both R. solani AG‐2‐2 and AG‐3, it is likely that this fungus may be dispersed in cultivated soils worldwide. However, little is known about the molecular components responsible for the susceptibility or resistance of N. tabacum to R. solani. This knowledge would be very desirable in order to obtain resistant genotypes that could be included in different integrated management strategies. This article summarizes aspects related to the infection process, colonization of the host and molecular mechanisms employed by tobacco plants in resistance against Rhizoctonia diseases.
TAXONOMIC HISTORY
In 1858, Julius Kühn described a fungus on diseased potatoes and placed it in the genus Rhizoctonia, which had been described earlier by the Swiss mycologist A. P. De Candolle (1815), naming it Rhizoctonia solani. As Kühn's original description of R. solani was brief and purportedly contained descriptions of a secondary organism (Parmerter and Whitney, 1970), and as the mycelia of some ascomycetes may closely resemble those of R. solani (Moreau and Moreau, 1956; Whitney and Parmeter, 1964), the description of R. solani was later revised (Duggar, 1915; Parmerter et al., 1967) to help minimize misidentification. The current species concept for this taxon suggests that the diagnostic features for the species are as follows: (i) hyphal pigmentation of different brownish tones; (ii) branching near the distal septum of cells in young vegetative hyphae; (iii) constriction of hyphae and the formation of septa near the point of origin of hyphal branches; (iv) dolipore septa; (v) the number of nuclei close to the tips of young vegetative hyphae is greater than two (Parmerter and Whitney, 1970; Sneh et al., 1996; Fig. 1C,D). Most, but not all, isolates have characteristics such as monilioid cells, sclerotia, rapid growth rate and pathogenicity (Sneh et al., 1996). In addition to the above morphological characteristics, the following features are never present and may be helpful to rule out other species with similar morphology: clamp connections, conidia, sclerotia differentiated into rind and medulla, and rhizomorphs (Parmerter and Whitney, 1970; Parmerter et al., 1967). Concerning its systematic relationships, Thanatephorus is actually included (together with the genera Ceratobasidium and Waitea) in the family Ceratobasidiaceae, considered by some authors (Roberts, 1999; Rogers, 1935; Weiss and Oberwinkler, 2001) as one of the most primitive group of Holobasidiomycetes, because of their characteristic basidial morphology, with large and sometimes septate sterigmata, close to some phragmobasidiomycetous groups. The evolutionary relationship between the genera Thanatephorus and Ceratobasidium remains controversial. Employing classical taxonomic approaches, some authors (Roberts, 1999; Stalpers and Andersen, 1996) have considered both genera as part of a generic complex, where delimitation between the two genera presents some difficulties, and differences in morphometric features and ecological behaviour are gradual along the several taxa within the two genera.
Figure 1.
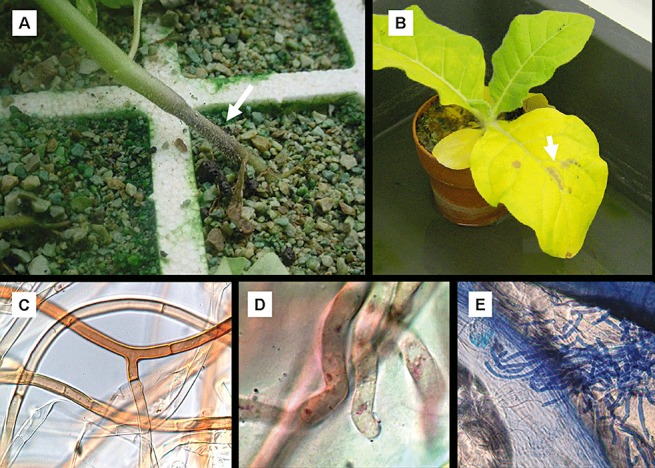
Symptoms and morphological features of plant pathogenic Rhizoctonia solani (Thanatephorus cucumeris). (A) Damping off of a tobacco seedling in a floating tray. A characteristic necrosis symptom is visible on the infected stem (arrow). (B) Target spot in tobacco leaf (arrow). (C) Typical morphology of R. solani hyphae. (D) Hyphae of R. solani showing the number of nuclei stained with Giemsa. (E) Hyphal branching of R. solani colonizing a leaf of Nicotiana tabacum. The plant and fungal tissues were stained with trypan blue.
Isolates of R. solani can vary greatly in phenotypic and genotypic characteristics, but have been traditionally arranged in genetically related groups based on hyphal anastomosis criteria. To date, 14 different AGs have been recognized (González‐García et al., 2006) for R. solani, although some authors (Roberts, 1999) have suggested the convenience of splitting the complex into several biological species. Thus, they have proposed four taxonomic epithets to cover all the ‘classical’R. solani AGs. In this sense, T. microsclerotium (G.F. Weber) Boidin, Mugnier & Canales is proposed for AG‐1B, T. sasakii (Shirai) C.C. Tu & Kimbr. is preferred to name AG‐1A isolates, T. praticola (Kotila) Flentje for AG‐4, and T. cucumeris is preferred to name most of the rest of the described AGs.
In general terms, teleomorphic stages are usually difficult to obtain in vitro for this group of fungi (Adams and Butler, 1983), and sexual fruitbodies are also scarce in natural substrates. Thus, the identification of R. solani isolates is typically based on the comparison of the anamorphic features mentioned previously. However, the species T. cucumeris has been designated as the teleomorphic counterpart for R. solani (Donk, 1956). Sexual fruitbodies of this taxon are typically characterized by the presence of a hypochnoid, thin basidiomata possessing a hymenium made up of successive layers of basidia rising from vertically branching, cymose hyphae just above the basal hyphae (subiculum), and typically with four sterigmata, sometimes septate, and about the same length as the metabasidia or shorter (González‐García et al., 2006; Roberts, 1999; Sneh et al., 1996). Hymenia produce basidia bearing four ellipsoid to oblong, hyaline basidiospores [4–5.5(6.5) µm × 7–10 µm] (Roberts, 1999). Moore (1987) proposed the grouping of anamorphs with perfect stages into the genera Thanatephorus and Waitea in the genus Moliniopsis Ruhland, a higher priority generic name erected to accommodate M. aderholdii, an ancient synonym of R. solani. Subsequently, Moore (1996) restricted Moliniopsis to species with Thanatephorus teleomorphs. However, the name Rhizoctonia has been proposed to be conserved against Moliniopsis (Stalpers et al., 1998), making Rhizoctonia solani the current name for the most well‐known and studied species in the complex.
SYMPTOMS IN TOBACCO
Rhizoctonia solani can survive for many years in soils by way of sclerotia or as a saprophyte, colonizing soil organic matter. Sclerotia and/or mycelium present in soil and/or plant tissue can eventually activate to produce vegetative hyphae that can attack a wide range of crops (Keijer, 1996). In certain situations, R. solani can produce basidiospores that will cause disease and also serve as a source for rapid and long‐distance dispersal of the fungus. The basidiospores germinate to produce hyphae that infect leaves during periods of high relative humidity (Fig. 2). Although most Rhizoctonia diseases are initiated by mycelium and/or sclerotia, several important diseases of tobacco and other crops, such as beans and sugar beet, are a result of basidiospore infection (González‐García et al., 2006; Harveson et al., 2009).
Figure 2.
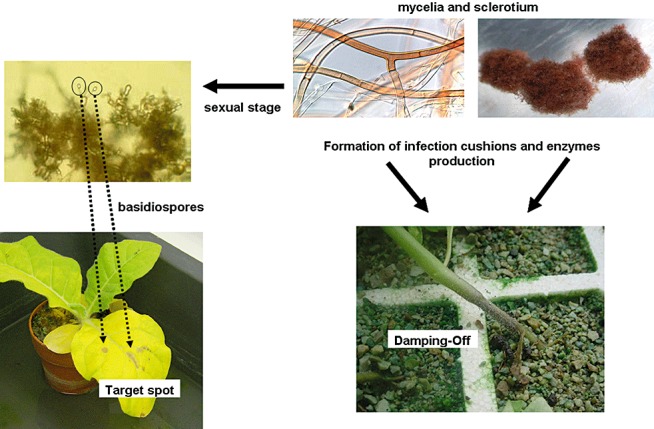
Disease cycle of Rhizoctonia solani and (Thanatephorus cucumeris) on tobacco.
Rhizoctonia solani causes damping off and stem rot in young transplants and a disease of the lower stem and root, called ‘sore shin’, in older field plants (Shew, 1991; Sneh et al., 1996). Damping off (Fig. 1A) is the most widespread symptom caused by R. solani observed in tobacco. In general, seedlings are susceptible to parasitization during the first few weeks of their development, and become progressively less susceptible as they mature through the development of biochemical and physical defence mechanisms and/or barriers. In damping‐off events, severely infected seeds usually do not germinate, and infected seedlings can be killed either before or after emergence. Seedlings killed after emergence often appear to have fallen over the soil surface as a result of excessive rainfall (Sneh et al., 1991). The transplantation of infected seedlings is a significant factor in the spread of sore shin in field plants; however, the latter infections can also be caused by R. solani that is already present in the field (Elliott et al., 2008). Isolates of R. solani causing stem and root rot symptoms have typically been associated with AG‐1, AG‐2‐2 and AG‐4 (Stevens et al., 1993).
Target spot is a foliar disease that first appeared in the USA in the 1980s and causes economically important losses in tobacco production (Elliott et al., 2008; Shew, 1991). This disease is caused by infection with basidiospores of T. cucumeris that are produced from hymenia on the soil surface or infected plant tissue (Elliott et al., 2008; Shew and Main, 1990). Symptoms begin as small water‐soaked lesions on leaves which can expand to large circular spots with concentric rings (Fig. 1B). Target spot can occur on tobacco seedlings in high‐humidity environments, particularly when leaves have grown close together to form a canopy in glasshouse environments. In some severe cases, the pathogen may grow from leaf tissue into the stem, resulting in plant death. Isolates of R. solani causing stem and root rot symptoms have been characterized by anastomosis as groups AG‐1, AG‐2‐2 and AG‐4, whereas target spot is typically associated in the glasshouse and the field as group AG‐3 (Stevens et al., 1993).
INFECTION PROCESS
Regardless of the type of host plant, in most of the disease events caused by R. solani isolates, the generalized infection process includes some consecutive steps identified as adhesion, penetration and colonization (Keijer, 1996). However, experimental data available on infection processes have shown that there are few differences among AGs with regard to the infection process in R. solani (González‐García et al., 2006). The initial step in the infection process is characterized by hyphal growth over the surface of the host plant. Such growth is easily washed off the host. In contrast, the attachment of hyphae in subsequent infections steps is characterized by flattened hyphae that are closely associated with the host. After attachment, hyphae grow following contiguous epidermal cells (Armentrout and Downer, 1987; Marshall and Rush, 1980), a probable response of R. solani to the topography of the host surface, leakage of host nutrients or other stimulatory components, or a combination of the above (Armentrout et al., 1987). T‐shaped hyphal branching is typically one of the morphological precursors prior to the formation of infection structures, such as short swollen hyphae, appressoria, infection cushions or repetitive T‐shaped branches, in part as a specific response to the host (Fig. 1E). Finally, infection pegs are produced that allow the fungus to penetrate and enter intact plant tissue through the cuticle and epidermal cell walls or, more rarely, through the stomata or wounds (Matsuura, 1986; Weinhold and Sinclair, 1996).
During the course of infection, the fungus produces extracellular hydrolytic enzymes capable of degrading the cell wall in advance of the invading hyphae. It has been shown that, during the earlier stages of the infection process, R. solani AG‐4 secretes pectinolytic and cellulolytic enzymes, such as endopectin lyase, which have been reported to be associated with tissue degradation in later stages of infection (Marcus et al., 1986). Altogether, at least 10 different extracellular enzymes have been identified to be produced by R. solani (Bertagnolli et al., 1996; Lister et al., 1975). Together with cell wall damage, changes in the cytoplasm of cortical cells can be detected before colonization events occur. For example, cytological changes in infected plant cells include the formation of reaction zones, plasmolysis and the collapse of the cytoplasm (González‐García et al., 2006). As the pathogenic process of R. solani is characterized by the death of plant cells, both before and after penetration and colonization events, it is interesting to speculate that the fungus relies on both a necrotrophic and hemibiotrophic lifestyle for pathogenicity, as has been shown previously with other fungal pathogen models (Bolton et al., 2006).
Infection by basidiospores occurs in target spot of tobacco. Although the infection process has not been studied in detail in tobacco, Naito and Sugimoto (1978) followed the course of basidiospore infection of R. solani AG‐2‐2 IV in sugar beet, which may be similar to infection in tobacco. In sugar beet, basidiospores originate from basidia on the soil surface or on host plants, and land on the surface of the host plant. After germination, penetration occurs via appressoria. From such primary infection sites, invading hyphae grow on the leaf surface and enter stomata to create secondary lesion sites. Spores are formed, thereby completing the life cycle and generating inoculum to infect other susceptible hosts (Naito and Sugimoto, 1978).
NICOTIANA SPP. DEFENCE AGAINST R. SOLANI INFECTION
Several constitutive factors (formed prior to infection), including cuticle and epicuticular wax thickness (Reddy, 1980; Yang et al., 1992), cell wall calcium content (Bateman and Lumsden, 1965) and tolerance of cuticle and epicuticular wax to pathogen enzymes and toxins, have been reported to function in resistance against R. solani (Kenning and Hanchey, 1980). In addition, induced mechanisms of resistance (formed after infection), such as hypersensitive responses (HRs) (Marshall and Rush, 1980) and an increased production of pathogenesis‐related proteins (PRPs), may also be involved (Anuratha et al., 1996).
So far, the interaction of R. solani with rice is the best studied pathosystem (Chang‐Jiang et al., 2008; Lee et al., 2006). Unfortunately, there is little information regarding the mechanisms associated with resistance to R. solani in plants of the Solanaceae family to which Nicotiana spp. belong. Genetic resistance to R. solani in tobacco is scarce. Recently, 97 genotypes of tobacco and related Nicotiana spp. were evaluated for seedling resistance to stem rot and target spot caused by R. solani (Elliott et al., 2008). Significant differences in disease incidence were initially found among the genotypes for both stem rot and target spot. However, resistance to target spot was not observed when disease pressure was high. Partial resistance to stem rot was observed in several genotypes in repeated tests (Elliott et al., 2008). A cDNA library, using suppression subtractive hybridization, was generated from transcripts that were differentially expressed during a compatible and incompatible interaction between N. tabacum cv. ‘Sumatra’ and R. solani (Chacón et al., 2010). This allowed the isolation of a protein kinase cDNA that was downregulated during a compatible and upregulated during an incompatible interaction. A functional study showed that this cDNA was directly related to resistance to R. solani (Chacón et al., 2010).
Nicotiana plumbaginifolia Viv. plants silenced for the ATP‐binding cassette transporter gene NpPDR1 showed an increased sensitivity to R. solani infection compared with wild‐type plants. The infiltration of pathogen suspension through the leaf stomata of mature wild‐type plants of N. plumbaginifolia induced NpPDR1 expression after 4 days, and no symptoms were observed in infiltrated zones. Similarly, the expression of an NpPDR1 orthologue in leaves of mature wild‐type plants of N. tabacum was activated by R. solani (Bultreys et al., 2009).
Resistance to R. solani in genetically modified plants under glasshouse conditions has also been reported. The first success in the production of a Rhizoctonia‐resistant plant was obtained with tobacco expressing an endochitinase from bean (Broglie et al., 1991). This was soon followed by a study in which a cauliflower mosaic virus (CaMV) 35S‐driven tobacco chitinase A was overexpressed in N. sylvestris Speg. & Comes. Transgenic plants showed less colonization and a smaller loss of fresh weight than controls (Vierheilig et al., 1993). Recently, a chitinase 1 gene from the entomopathogenic fungus Metarhizium anisopliae (Metschn.) Sorokïn was expressed in tobacco under the control of the CaMV 35S promoter. Transgenic plants showed enhanced resistance to R. solani, providing the first example of transgenic plants inducing resistance through the expression of a chitinase from Metarhizium anisopliae, an entomopathogenic and acaricide fungus (Kern et al., 2010). Overexpression of individual or combined cDNAs encoding the barley (Hordeum vulgare L.) class II chitinase, class II β‐1,3‐glucanse and type I ribosome‐inactivating protein, driven by the CaMV 35S promoter, in tobacco resulted in enhanced protection against R. solani (Jach et al., 1995; O'Brien et al., 2001). Overexpression of sarcotoxin IA, a bactericidal peptide from Sarcophaga peregrina, enhanced the resistance of transgenic tobacco plants to bacterial (Erwinia carotovora ssp. carotovora and Pseudomonas syringae pv. tabaci) and fungal pathogens (Mitsuhara et al., 2000). Interestingly, the heterologous overexpression of the Gastrodia antifungal protein (GAFP; gastrodianin) provides a broad‐spectrum resistance to various pathogens, such as R. solani, Phytophthora nicotianae Breda de Haan and Meloidogyne incognita Kofoid & White, but no resistance was observed against Ralstonia solanacearum (Cox et al., 2006). A cDNA clone from a novel CCCH‐type zinc finger protein (GhZFP1) of Gossypium hirsutum L. was isolated from salt‐induced cotton using differential hybridization screening. Overexpression of GhZFP1 in transgenic N. tabacum cv. NC89 enhanced tolerance to salt stress and resistance to R. solani (Guo et al., 2009). Transgenic plants expressing the thaumatin gene from Thaumatoccus daniellii (Benn.) Benth, under the control of the CaMV 35S promoter, displayed enhanced resistance and delayed disease symptoms against fungal diseases caused by Pythium aphanidermatum (Edson) Fitzp. and R. solani (Rajam et al., 2007). Finally, a decrease in resistance to a compatible strain of R. solani was observed in type III knockdown tobacco lines targeting the calmodulin (CaM) gene NtCaM13. The expression of jasmonic acid (JA)‐ and/or ethylene (ET)‐inducible basic PR genes was not affected in this line, suggesting that type III CaM isoforms are probably involved in basal defence against necrotrophic pathogens, independent of JA and ET signalling (Takabatake et al., 2007). These results indicate a complex response to challenge by R. solani that involves the simultaneous induction of proteins from multiple defence pathways (Table 1).
Table 1.
Genes from Nicotiana spp. involved in Rhizoctonia solani defence.
| Gene | Effects on disease development | Reference |
|---|---|---|
| Class II chitinase, class II β‐1,3‐glucanase and type I ribosome‐inactivating protein | Overexpression in transgenic plants increases protection against diseases | Jach et al. (1995) |
| Type III calmodulin | Knockdown of this gene compromises disease resistance | Takabatake et al. (2007) |
| NpPDR1 | Silencing of this gene increases sensitivity to the fungus in Nicotiana plumbaginifolia and N. tabacum | Bultreys et al. (2009) |
| GhZFP1 (CCCH‐type zinc finger protein) | Overexpression in transgenic plants enhances resistance | Guo et al. (2009) |
| NtPK protein kinase | Overexpression enhances resistance to damping off produced by an aggressive R. solani strain, and silencing compromises the resistance to a nonaggressive Rhizoctonia solani strain | Chacón et al. (2010) |
MANAGEMENT MEASURES
The control of this soil‐borne fungus has been difficult to achieve using traditional means, such as breeding plants for resistance, crop rotation and fungicides. Generally, the most successful practices include integrated pest management and, in this case, it is very important to know the main sources of inoculum in order to control them. The growth of healthy transplants is an essential step in tobacco production. The main system for the production of tobacco seedlings worldwide is through the use of floating trays. Such Styrofoam trays are filled with a soil‐less medium, seeded with pelletized seed, and floated on a shallow water reservoir. These trays are perforated on the bottom to allow for water and nutrient uptake. This soil‐less medium procedure has been identified as the principal source of inoculum because it can harbour R. solani sclerotia and hyphae (Gutierrez et al., 1997). This makes it important to quantify the pathogen potentially present in soil‐less medium before its use and to implement proper disinfection before re‐use (Gutierrez et al., 1997).
Integrated pest management also relies on timely chemical application. For example, fungicides must be applied prior to infection to control diseases caused by R. solani AG‐2‐2 IIIB and IV in sugar beet. Recently, Bolton et al. (2010) identified soil temperature and moisture thresholds as necessary for infection. Because R. solani is affected by these environmental parameters, growers may be able to evaluate soil conditions for the optimal application of fungicide. The fungicide mancozeb was used to control damping off and target spot in glasshouse production. However, the fungicide iprodione demonstrated, in a comparison of the activity of several fungicides against R. solani, excellent control of these diseases (Csinos and Stephenson, 1999).
Finally, biological control is another important aspect of R. solani management in tobacco. The efficacy of this method has been demonstrated in R. solani using Trichoderma sp. (Cole and Zvenyika, 1988; Elad et al., 1980; Hadar et al., 1979). Isolates from T. harzianum Rifai reduced the growth of R. solani and enhanced the disease control in tobacco plants (Cole and Zvenyika, 1988). Seed quality and levels of cultivar resistance are other aspects to be taken into account. All of the above management measures will not totally eliminate the pathogen, but will cause a reduction in inoculum levels. However, once the plant is infected during a cropping season, management options become limited.
CONCLUDING REMARKS
A deeper understanding of the molecular interaction between Nicotiana spp. and R. solani will further our knowledge of the defence mechanisms that occur during fungal infection. The genome sequence should open up new possibilities, and important improvement for the future would be to determine the main biochemical and molecular mechanisms involved in the pathogenicity and virulence of R. solani. A knowledge of the possible mechanisms and genes suppressed in the host by the virulent pathogen may further our understanding of potential defences of the host. The identification of crucial genes may lead to useful tools for molecular breeding or for the development of transgenic varieties. It may also aid in the elimination of susceptible individuals during initial stages in the management of diseases.
ACKNOWLEDGEMENTS
The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information, and does not imply recommendation or endorsement by the USDA. The authors would like to thank Dr Ryohei Terauchi for comments and critical reading of the manuscript. The authors thank the anonymous reviewers and editor for useful suggestions.
REFERENCES
- Adams, G.C. and Butler, E.E. (1983) Environmental factors influencing the formation of basidia and basidiospores in Thanatephorus cucumeris . Phytopathology, 73, 152–155. [Google Scholar]
- Anuratha, C.S. , Zen, K.C. , Cole, K.C. , Mew, T. and Muthukrishnan, S. (1996) Induction of chitinases and β‐1,3‐glucanases in Rhizoctonia solani‐infected rice plants: isolation of an infection related chitinase cDNA clone. Physiol. Plant. 97, 39–46. [Google Scholar]
- Armentrout, V.N. and Downer, A.J. (1987) Infection cushions development by Rhizoctonia solani on cotton. Phytopathology, 77, 619–623. [Google Scholar]
- Armentrout, V.N. , Downer, A.J. , Grasmick, D.L. and Weinhold, A.R. (1987) Factors affecting infection cushion development by Rhizoctonia solani on cotton. Phytopathology, 77, 623–630. [Google Scholar]
- Bateman, D.F. and Lumsden, R.D. (1965) Relation of calcium content and nature of the pectic substances in bean hypocotyls of different ages to susceptibility to an isolate of Rhizoctonia solani . Phytopathology, 55, 734–738. [Google Scholar]
- Bertagnolli, B.L. , Dal Soglio, F.K. and Sinclair, J.B. (1996) Extracellular enzyme profiles of the fungal pathogen Rhizoctonia solani isolate 2B‐12 and of two antagonists, Bacillus megaterium strain B153‐2‐2 and Trichoderma harzianum isolate Th008.I. Possible correlations with inhibition of growth and biocontrol. Physiol. Mol. Plant Pathol. 48, 145–160. [Google Scholar]
- Bolton, M.D. , Thomma, B.P.H. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Bolton, M.D. , Panella, L.W. , Campbell, L.G. and Khan, M.F. (2010) Temperature, moisture, and fungicide effects in managing Rhizoctonia root and crown rot of sugar beet. Phytopathology, 100, 689–697. [DOI] [PubMed] [Google Scholar]
- Broglie, K. , Chet, I. , Holliday, M. , Cressman, R. , Biddle, P. , Knowlton, S. , Mauvais, C.J. and Broglie, R. (1991) Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani . Science, 254, 1194–1197. [DOI] [PubMed] [Google Scholar]
- Bultreys, A. , Trombik, T. , Drozak, A. and Boutry, M. (2009) Nicotiana plumbaginifolia plants silenced for the ATP‐binding cassette transporter gene NpPDR1 show increased susceptibility to a group of fungal and oomycete pathogens. Mol. Plant Pathol. 10, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling, D.E. (1996) Grouping in Rhizoctonia solani by hyphal anastomosis reaction In: Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control (Sneh B., Jabaji‐Hare S., Neate S. and Dijst G., eds), pp. 35–47. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Carling, D.E. , Kuninaga, S. and Brainard, K.A. (2002) Hyphal anastomosis reactions, rDNA‐internal transcribed spacer sequences, and virulence levels among subsets of Rhizoctonia solani anastomosis group‐2 (AG‐2) and AG‐BI. Phytopathology, 92, 43–50. [DOI] [PubMed] [Google Scholar]
- Chacón, O. , González, M. , López, Y. , Portieles, R. , Pujol, M. , González, E. , Schoonbeek, H.J. , Metraux, J.P. and Borrás‐Hidalgo, O. (2010) Overexpression of a protein kinase gene enhances the defense of tobacco against Rhizoctonia solani . Gene, 452, 54–62. [DOI] [PubMed] [Google Scholar]
- Chang‐Jiang, Z. , Ai‐Rong, W. , Yu‐Jun, S. , Liu‐Qing, W. , Wen‐De, L. , Zong‐Hua, W. and Guo‐Dong, L. (2008) Identification of defense‐related genes in rice responding to challenge by Rhizoctonia solani . Theor. Appl. Genet. 116, 501–516. [DOI] [PubMed] [Google Scholar]
- Cole, J.S. and Zvenyika, Z. (1988) Integrated control of Rhizoctonia solani and Fusarium solani in tobacco transplants with Trichoderma harzianum and triadimenol. Plant Pathol. 37, 271–277. [Google Scholar]
- Cox, K.D. , Layne, D.R. , Scorza, R. and Schnabel, G. (2006) Gastrodia anti‐fungal protein from the orchid Gastrodia elata confers disease resistance to root pathogens in transgenic tobacco. Planta, 224, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Csinos, A.S. and Stephenson, M.G. (1999) Evaluation of fungicides and tobacco cultivar resistance to Rhizoctonia solani incited target spot, damping off and sore shin. Crop Prot. 18, 373–377. [Google Scholar]
- De Candolle, A.P. (1815) Mémoire sur les rhizoctones, noveau genre de champignons qui attaque les racines, des plantes et en particulier celle de la luzerne cultivée. Mem. Mus. d'Hist. Nat. 2, 209–216. [Google Scholar]
- Donk, M.A. (1956) Notes on resupinate fungi II. The tullasneloid fungi. Reinwardtia, 3, 363–379. [Google Scholar]
- Duggar, B.M. (1915) Rhizoctonia crocorum (Pres.) DC and R. solani Kühn (Corticium vagum B. & C.) with notes on other species. Ann. Mo. Bot. Gard. 2, 403–458. [Google Scholar]
- Elad, Y. , Chet, I. and Katan, J. (1980) Trichoderma harzianum: a biocontrol agent effective against Sclerotium rolfsii and Rhizoctonia solani . Phytopathology, 7, 119–121. [Google Scholar]
- Elliott, P.E. , Lewis, R.S. , Shew, H.D. , Gutierrez, W.A. and Nicholson, J.S. (2008) Evaluation of tobacco germplasm for seedling resistance to stem rot and target spot caused by Thanatephorus cucumeris . Plant Dis. 92, 425–430. [DOI] [PubMed] [Google Scholar]
- González, D. , Carling, D.E. , Kuninaga, S. , Vilgalys, R. and Cubeta, M.A. (2001) Ribosomal DNA systematics of Ceratobasidium and Thanatephorus with Rhizoctonia anamorphs. Mycologia, 93, 1138–1150. [Google Scholar]
- González‐García, V. , Portal‐Onco, M.A. and Rubio, V. (2006) Biology and systematics of the form genus Rhizoctonia . Span. J. Agric. Res. 4, 55–79. [Google Scholar]
- Guo, Y.H. , Yu, Y.P. , Wang, D. , Wu, C.A. , Yang, G.D. , Huang, J.G. and Zheng, C.C. (2009) GhZFP1, a novel CCCH‐type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 183, 62–75. [DOI] [PubMed] [Google Scholar]
- Gutierrez, W.A. , Shew, H.D. and Melton, T.A. (1997) Sources of inoculum and management for Rhizoctonia solani damping‐off on tobacco transplants under greenhouse conditions. Plant Dis. 81, 604–606. [DOI] [PubMed] [Google Scholar]
- Hadar, Y. , Chet, I. and Henis, Y. (1979) Biological control of Rhizoctonia solani damping‐off with wheat bran culture of Trichoderma harzianum . Phytopathology, 69, 64–68. [Google Scholar]
- Harveson, R.M. , Hanson, L.E. and Hein, G.L. (2009) Compendium of Beet Diseases and Pests. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Jach, G. , Görnhardt, B. , Mundy, J. , Logemann, J. , Pinsdorf, E. , Leah, R. and Maas, C. (1995) Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8, 97–109. [DOI] [PubMed] [Google Scholar]
- Keijer, J. (1996) The initial steps of the infection process in Rhizoctonia solani In: Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control (Sneh B., Jabaji‐Hare S., Neate S. and Dijst G., eds), pp. 149–162. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Kenning, L.A. and Hanchey, P. (1980) Ultrastructure of lesion formation in Rhizoctonia infected bean hypocotyls. Phytopathology, 70, 998–1004. [Google Scholar]
- Kern, M.F. , Maraschin, S.D. , Vom Endt, D. , Schrank, A. , Vainstein, M.H. and Pasquali, G. (2010) Expression of a chitinase gene from Metarhizium anisopliae in tobacco plants confers resistance against Rhizoctonia solani . Appl. Biochem. Biotechnol. 160, 1933–1946. [DOI] [PubMed] [Google Scholar]
- Kuninaga, S. , Carling, D.E. , Takeuchi, T. and Yokosawa, R. (2000) Comparison of rDNA‐ITS sequences between potato and tobacco strains in Rhizoctonia solani AG‐3. J. Gen. Plant Pathol. 66, 2–11. [Google Scholar]
- Lee, J. , Bricker, T.M. , Lefevre, M. , Pinson, S.R.M. and Oard, J.H. (2006) Proteomic and genetic approaches to identifying defense‐related proteins in rice challenged with the fungal pathogen Rhizoctonia solani . Mol. Plant Pathol. 7, 405–416. [DOI] [PubMed] [Google Scholar]
- Lister, N. , Katan, J. and Henis, Y. (1975) Sequential production of polygalacturonase, cellulase and pectinlyase by Rhizoctonia solani . Can. J. Microbiol. 21, 298–304. [DOI] [PubMed] [Google Scholar]
- Lucas, G.B. (1975) Diseases of Tobacco, 3rd edn. Raleigh, NC: Biological Consulting Associates. [Google Scholar]
- Marcus, L. , Barash, I. , Sneh, B. , Koltin, Y. and Finkler, A. (1986) Purification and characterization of pectinolytic enzymes produced by virulent and hypovirulent isolates of Rhizoctonia solani Kuhn. Physiol. Mol. Plant Pathol. 29, 325–336. [Google Scholar]
- Marshall, D.S. and Rush, M.C. (1980) Relation between infection by Rhizoctonia solani and R. oryzae and disease severity in rice. Phytopathology, 70, 941–946. [Google Scholar]
- Matsuura, K. (1986) Scanning electron microscopy of the infection process of Rhizoctonia solani in leaf sheaths of rice plants. Phytopathology, 76, 811–814. [Google Scholar]
- Mitsuhara, I. , Matsufuru, H. , Ohshima, M. , Kaku, H. , Nakajima, Y. , Murai, N. , Natori, S. and Ohashi, Y. (2000) Induced expression of sarcotoxin IA enhanced host resistance against both bacterial and fungal pathogens in transgenic tobacco. Mol. Plant–Microbe Interact. 13, 860–868. [DOI] [PubMed] [Google Scholar]
- Moore, R.T. (1987) The genera of Rhizoctonia‐like fungi: Ascorhizoctonia, Ceratorhiza gen. nov., Epulorhiza gen. nov., Moliniopsis, and Rhizoctonia . Mycotaxon, 29, 91–99. [Google Scholar]
- Moore, R.T. (1996) The dolipore/parenthesome septum in modern taxonomy In: Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control (Sneh B., Jabaji‐Hare S., Neate S. and Dijst G., eds), pp. 13–35. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Moreau, C. and Moreau, M. (1956) Examen comparatif du mycelium et des sclerotes chez diverses souches du Rhizoctonia solani Kiihn et du Morchella hortensis Boud. Bull. Soc. Bot. Fr. 103, 117–120. [Google Scholar]
- Naito, S. and Sugimoto, T. (1978) Basidiospore infection and lesion development on sugar beet leaves by Thanatephorus cucumeris (Frank) Donk. Ann. Phytopathol. Soc. Jpn. 44, 426–431. [Google Scholar]
- O'Brien, P.A. , MacNish, G.C. and Milton, N.M.K. (2001) Transgenic tobacco plants show different resistance to Rhizoctonia solani AG 4 and AG 8. Australas. Plant Pathol. 30, 221–225. [Google Scholar]
- Ogoshi, A. (1987) Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kühn. Annu. Rev. Phytopathol. 25, 125–143. [Google Scholar]
- Parmerter, J.R. and Whitney, H.S. (1970) Taxonomy and nomenclature of the imperfect state In: Rhizoctonia Solani: Biology and Pathology (Parmerter J.R., ed.), p. 225 Berkeley, CA: University of California Press. [Google Scholar]
- Parmerter, J.R. , Whitney, H.S. and Platt, W.D. (1967) Affinities of some Rhizoctonia species that resemble mycelium of Thanatephorus cucumeris . Phytopathology, 57, 218–223. [Google Scholar]
- Rajam, M.V. , Chandola, N. , Goud, P.S. , Singh, D. , Kashyap, V. , Choudhary, M.L. and Sihachakr, D. (2007) Thaumatin gene confers resistance to fungal pathogens as well as tolerance to abiotic stresses in transgenic tobacco plants. Biol. Plant. 51, 135–141. [Google Scholar]
- Reddy, M.N. (1980) Studies on groundnut hypocotyl exudates and the behavior of Rhizoctonia solani influencing the diseases. Plant Soil, 55, 445–454. [Google Scholar]
- Roberts, P. (1999) Rhizoctonia‐Forming Fungi: A Taxonomic Guide. Kew: Royal Botanical Gardens; p. 239. [Google Scholar]
- Rogers, D.P. (1935) Notes on the lower Basidiomycetes. Studies Natural History University Iowa 17, 1–43. [Google Scholar]
- Salazar, O. , Hyakumachi, M. and Rubio, V. (2000) Phylogenetic grouping of cultural types of Rhizoctonia solani AG 2‐2 based on ribosomal ITS sequences. Micologia, 92, 505–509. [Google Scholar]
- Shew, H.D. (1991) Target spot In: Compendium of Tobacco Diseases (Shew H.D. and Lucas G.B., eds), pp. 90–92. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Shew, H.D. and Main, C.E. (1990) Infection and development of target spot of flue‐cured tobacco caused by Thanatephorus cucumeris . Plant Dis. 74, 1009–1013. [Google Scholar]
- Sneh, B. , Burpee, L. and Ogoshi, A. (1991) Identification of Rhizoctonia Species. St. Paul: The American Phytopathological Society, APS Press, p. 134. [Google Scholar]
- Sneh, B. , Jajabi‐Hare, S. , Neate, S. and Dijst, G. (1996) Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control. Dordrecht: Kluwer Academic Publishers, p. 578. [Google Scholar]
- Stalpers, J.A. and Andersen, T.F. (1996) A synopsis of the taxonomy of teleomorphs connected with Rhizoctonia s.l In: Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control (Sneh B., Jabaji‐Hare S., Neate S. and Dijst G., eds), pp. 49–63. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Stalpers, J.A. , Andersen, T.F. and Gams, W. (1998) Two proposals to conserve the names Rhizoctonia and R. solani . Taxon, 47, 725–726. [Google Scholar]
- Stevens, J.S. , Jones, R.K. , Shew, H.D. and Carling, D.E. (1993) Characterization of populations of Rhizoctonia solani AG‐3 from potato and tobacco. Phytopathology, 83, 854–858. [Google Scholar]
- Takabatake, R. , Karita, E. , Seo, S. , Mitsuhara, I. , Kuchitsu, K. and Ohashi, Y. (2007) Pathogen induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 48, 414–423. [DOI] [PubMed] [Google Scholar]
- Vierheilig, H. , Alt, M. , Neuhaus, J.M. , Boller, T. and Wiemken, A. (1993) Colonization of transgenic Nicotiana sylvestris plants, expressing different forms of Nicotiana tabacum chitinase, by the root pathogen Rhizoctonia solani and by the mycorrhizal symbiont Glomus mosseae . Mol. Plant–Microbe Interact. 6, 261–264. [Google Scholar]
- Weinhold, A.R. and Sinclair, J.B. (1996) Rhizoctonia solani: penetration, colonization and host response In: Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Diseases Control (Sneh B., Jajabi‐Hare S., Neate S. and Dijst G., eds), p. 578 Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Weiss, M. and Oberwinkler, F. (2001) Phylogenetic relationships in Auriculariales and related groups—hypotheses derived from nuclear ribosomal DNA sequences. Mycol. Res. 105, 403–415. [Google Scholar]
- Whitney, H.S. and Parmeter, J.R. (1964) The perfect stage of Rhizoctonia hiemalis . Mycologia, 56, 114–118. [Google Scholar]
- Yang, J. , Verma, P.R. and Lees, G.L. (1992) The role of cuticle and epidermal cell wall in resistance of rapeseed and mustard to Rhizoctonia solani . Plant Soil, 142, 315–321. [Google Scholar]


