SUMMARY
Rhizosphere‐competent fungi of the genus Trichoderma are widely used as biofertilizers and biopesticides in commercial formulates because of the multiple beneficial effects on plant growth and disease resistance. In this work, we demonstrate that genetic variability among wild and cultivated tomato lines affects the outcome of the interaction with two ‘elite’ biocontrol strains of T. atroviride and T. harzianum. The beneficial response, which included enhanced growth and systemic resistance against Botrytis cinerea, was clearly evident for some, but not all, the tested lines. At least in one case (line M82), treatment with the biocontrol agents had no effect or was even detrimental. Expression studies on defence‐related genes suggested that the fungus is able to trigger, in the responsive lines, a long‐lasting up‐regulation of the salicylic acid pathway in the absence of a pathogen, possibly activating a priming mechanism in the plant. Consequently, infection with B. cinerea on plants pretreated with Trichoderma is followed by enhanced activation of jasmonate‐responsive genes, eventually boosting systemic resistance to the pathogen in a plant genotype‐dependent manner. Our data indicate that, at least in tomato, the Trichoderma induced systemic resistance mechanism is much more complex than considered so far, and the ability of the plant to benefit from this symbiotic‐like interaction can be genetically improved.
INTRODUCTION
The massive and ever increasing application of synthetic chemicals for plant pest control causes serious problems to both human health and the environment (Budnik and Baur, 2009; Debenest et al., 2010). In this context, the use of biological control agents (BCAs) as an alternative to conventional practices for disease management is a declared objective of agricultural politics throughout the world. Many biopesticide and biofertilizer products are now available on the market, the majority of which are based on beneficial symbionts of the genus Trichoderma (Woo et al., 2006). These fungi are well known for their ability to kill plant pathogens (Benitez et al., 2004; Verma et al., 2007), as well as to promote plant growth and resistance against biotic and abiotic stresses (Harman et al., 2004a; Shoresh et al., 2010). It has been reported that some Trichoderma strains can activate induced systemic resistance (ISR) (Hanson and Howell, 2004; Mandal and Mitra, 2007; Segarra et al., 2007) a mechanism triggered after root colonization by nonpathogenic rhizobacteria or fungi and regulated by a specific signal transduction cascade (Pieterse et al., 1996; Segarra et al., 2009). In addition, plants whose roots are colonized by selected Trichoderma isolates are ‘sensitized’ and respond faster and/or more intensely to pathogen attack, following a mechanism known as priming (Ahn et al., 2007; Conrath et al., 2006). The molecular cross‐talk that is established between the fungal BCA and the plant provides benefits to both players, with the latter receiving protection and more available nutrients and the fungus organic compounds. Following root penetration by rhizosphere‐competent Trichoderma strains, which set the stage for the exchange of bioactive compounds, plants control the endophytic colonization of the fungus by callose deposition, increased chitinase and peroxidase activities, and the accumulation of antimicrobial phenolics, in some cases associated with the expression of phenylalanine ammonia lyase (PAL), hydroxyperoxide lyase and glucanase genes (Segarra et al., 2007; Shoresh et al., 2005; 1999, 2003). The current hypothesis is that plants initially perceive root endophytic colonization by Trichoderma as a potential pathogen attack, and react with the activation of typical local and systemic defence mechanisms that involve signal transduction pathways responding to jasmonic acid (JA) and ethylene (Et) (Shoresh et al., 2005). After successful limitation of fungus penetration to the first few layers of root cortical cells, the expression of some defence‐related genes and antimicrobial activity return to pre‐infection levels (Shoresh et al., 2005; 1999, 2003). This demonstrates the existence of a molecular cross‐talk between the plant and the fungus involving Trichoderma‐produced elicitors that are recognized by the plant and stimulate defence responses (Woo et al., 2006). However, the mechanism by which Trichoderma is then able to remain within the root system as an avirulent symbiont is still unclear, and possibly depends on fungal effectors that suppress plant defences.
Some selected strains of Trichoderma also promote plant growth and development, and thus increase yields (Harman et al., 2004a; Lorito et al., 2010). This additional beneficial effect, which is very popular among bioinoculum users, has been related to the control of deleterious soil microflora, the degradation of toxic compounds, the direct stimulation of root development by the production of hormone‐like compounds or effects on certain plant hormone synthetic pathways, and/or the promotion of water and nutrient uptake (Bae et al., 2009; Contreras‐Cornejo et al., 2009; Verma et al., 2007). An increased availability of phosphorus and several micronutrients, associated with enhanced solubility but also other mechanisms, has been demonstrated in the case of plants treated with strains of Trichoderma (Altomare et al., 1999; Gravel et al., 2007; Yedidia et al., 2001). This ability of some Trichoderma strains to improve the efficiency of fertilizer use by crops provides the opportunity to reduce chemical input in agriculture, as well as the pollution of ground and surface water.
Even in the case of strains such as T. harzianum T22, one of the most widely used biopesticides, there is still a substantial lack of knowledge on how the expected beneficial effects of Trichoderma application depend on the treated plant genotype. The importance of this concept has been proven already in maize, to the point that there are varieties for which the use of T22‐based products may be either recommended or counter‐indicated (Harman, 2006; Harman et al., 2004b). Despite the obvious significance for agriculture, there is still a lack of knowledge on how the plant response to Trichoderma spp., especially to the strains currently used as biopesticides/biofertilizers, is influenced by the plant genotype in terms of enhanced systemic resistance and growth promotion. In order to shed light on this important issue, we studied the interaction of T. harzianum T22 and T. atroviride P1, two microbes extensively used for agricultural and scientific purposes, with five diverse tomato lines: M82 and TA209, two inbred processing tomato varieties with a determinate growth habit, long used in research and breeding, and carrying the resistance genes I and Ve, with medium‐sized cylindrical or square fruits, respectively; one landrace of Corbarino, an indeterminate, stress (mainly drought)‐tolerant Italian Regional inbred variety with cherry‐like fruits, used for both processing and the fresh market; SM36, an advanced breeding line, tolerant to high and low temperatures and carrying the resistance genes I and Ve, with a determinate growth habit and cylindrical fruits, derived from a cross between Solanum pimpinellifolium and the traditional processing tomato cultivar San Marzano; and LA1777, a Peruvian accession of the allogamous wild species S. habrochaites, characterized by an indeterminate growth habit, very dense pubescence on all the above‐ground parts of the plant, small green fruits, and resistance to chilling and late blight.
In this work, we demonstrate that substantial differences in the response to the symbiotic interaction with two selected strains of Trichoderma spp. occur when different tomato varieties are tested. The effect of T. harzianum T22 and T. atroviride P1 on the growth and systemic resistance against Botrytis cinerea was found to be dependent on plant genotype. Finally, we report data on the induction of defence response pathways that may help the selection or breeding of tomato lines with enhanced ability to benefit from the interaction with Trichoderma.
RESULTS
Effects on growth and development
The response of four genetically distant cultivated S. lycopersicum lines and one wild S. habrochaites accession, differing with regard to their growth habit, to the growth‐promoting fungi T. atroviride strain P1 and T. harzianum strain T22 was assessed. Plant development was evaluated, for each experiment, on two sets of 2‐month‐old plants, one of which was not inoculated with the pathogen in order to determine the effect of the treatments on the shoot dry weight. The other set was used for the other assays (growth promotion, pathogen resistance, gene expression, etc.).
As expected, the canopy size of all tested lines was generally stimulated, although to a different extent, by at least one of the two Trichoderma strains in the assay conditions used (Fig. 1a). Similarly, shoot dry weight, as tested in parallel experiments, was found to be increased in almost all plant genotype–Trichoderma species' interactions (Table 1). In addition, in terms of stem length, analysis of variance (anova) and Duncan test revealed significant differences among genotypes and treatments (Fig. 1b). When data from the same treatment (T22, P1 or controls) of the five lines tested were combined and averaged, only T. harzianum T22 was found to stimulate significantly tomato stem growth (P≤ 0.05), whereas T. atroviride P1‐treated plants did not differ statistically from untreated controls. However, statistically significant differences (P≤ 0.05) were detected among the five genotypes in their individual responses to either one of the two Trichoderma strains. More significant stem growth stimulation was obtained for SM36 and TA209 with T. harzianum T22, and for TA209 with T. atroviride P1 (Fig. 1b). Interestingly, the stem growth of Corbarino, M82 and LA1777 was inhibited by P1, although the difference was statistically significant only for LA1777 (Fig. 1b).
Figure 1.
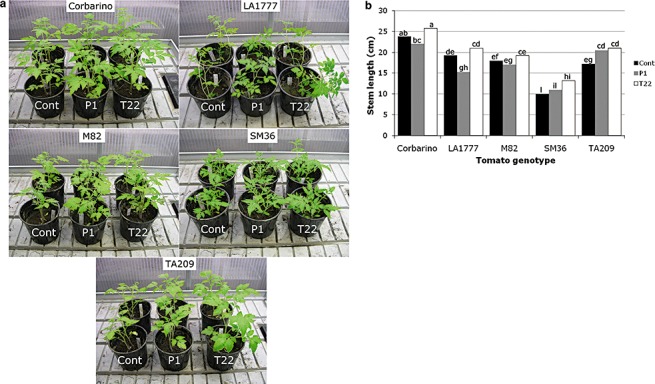
Effects of Trichoderma spp. treatments on the growth and development of tomato plants. The canopy size (a) and stem length (b) of 2‐month‐old Solanum lycopersicum (Corbarino, TA209, M82 and SM36) and S. habrochaites (LA1777) lines, developed from untreated (Cont), T. atroviride P1‐treated (P1) or T. harzianum T22‐treated (T22) seeds, are presented. Values indicated by the same letter are not statistically significantly different for P≤ 0.01 according to the Duncan test.
Table 1.
Effects of Trichoderma spp. treatments on shoot development of tomato plants.
| Tomato genotype | Shoot dry weight (% of untreated control) | |
|---|---|---|
| P1 | T22 | |
| Corbarino | 130.7 ± 15.4 | 130.1 ± 11.3 |
| LA1777 | 129.7 ± 4.6 | 83.2 ± 0.3 |
| M82 | 112.6 ± 3.9 | 156.9 ± 15.2 |
| SM36 | 139.5 ± 5.9 | 158.1 ± 11.0 |
| TA209 | 75.6 ± 3.7 | 113.8 ± 5.5 |
Changes in shoot dry weight of 2‐month‐old Solanum lycopersicum (Corbarino, TA209, M82 and SM36) and S. habrochaites (LA1777) lines, developed from T. atroviride P1‐treated (P1) or T. harzianum T22‐treated (T22) seeds, are reported as a percentage of the untreated control ± SE.
Trichoderma atroviride P1‐ and T. harzianum T22‐treated plants were also evaluated in terms of root development in comparison with the untreated controls. The fresh and dry weights of the roots were collected and subjected to statistical analysis. The root systems of the various tomato lines responded differentially to the two Trichoderma species (P≤ 0.05 for the interaction plant genotype × treatment). As both dry and fresh weights were similarly affected, only data on dry weight are reported. The lines Corbarino and TA209 showed a highly significant (P≤ 0.01) increase in root dry weight caused by T. atroviride P1 (Fig. 2a). The only significantly negative effect was detected on TA209 treated with T22, whereas all the other samples were no different from the controls (Fig. 2a).
Figure 2.
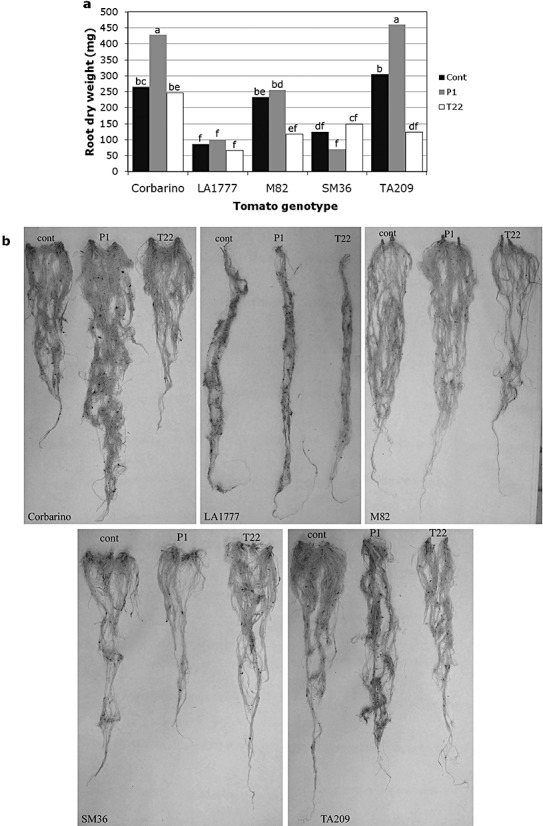
Effects of Trichoderma spp. treatments on the growth and development of tomato roots. The root dry weight (a) and development (b) of 2‐month‐old Solanum lycopersicum (Corbarino, TA209, M82 and SM36) and S. habrochaites (LA1777) lines, developed from untreated (Cont), T. atroviride P1‐treated (P1) or T. harzianum T22‐treated (T22) seeds, are presented. Values indicated by the same letter are not statistically significantly different for P≤ 0.01 according to the Duncan test.
The application of either Trichoderma species also produced remarkable differences on the root system architecture of the various tomato genotypes. Corbarino, LA1777 and M82 were found to be highly responsive to strain P1 in terms of increased root elongation, whereas the same strain significantly reduced the length of the SM36 root system. Strain T22 enhanced lateral root development in lines SM36 and Corbarino, and reduced it in TA209 and M82, but, in most cases, had no effect on root length (Fig. 2b).
Effects on tolerance to pathogens
Two‐month‐old plants of the five tomato lines, treated with either T. atroviride P1 or T. harzianum T22, were artificially inoculated with the foliar pathogen B. cinerea. The progress of infection was recorded at regular time intervals, but only results at 48 and 96 h post‐infection (hpi) are shown. In the case of accession LA1777, the presence of many trichomas caused the dispersal of the inoculated conidia, and thus only about 17% of the inoculation points developed into necrotic lesions in both control and T. harzianum T22‐treated plants, whereas no symptom development was observed for the T. atroviride P1 treatment. The average area of necrotic lesions on T22‐treated plants of this line at 96 hpi was 2 mm2 vs. 28 mm2 in the control (Fig. 3c). The values at 48 and 96 hpi, as reported in Fig. 3, indicated that the remaining four tomato genotypes had a different susceptibility to B. cinerea (P≤ 0.01) and that the genotype–treatment interaction was statistically significant at both 48 and 96 hpi (P≤ 0.05). This result suggests that the plant genotype is a key determinant of the effect of Trichoderma‐based treatments on plant resistance to pathogens, at least in tomato. As expected, the interaction at the rhizosphere level between S. lycopersicum lines and Trichoderma species overall increased resistance against B. cinerea leaf infection (Fig. 3a,b). However, remarkable differences were detected among the four tomato genotypes. In the case of T. harzianum T22, the development of B. cinerea lesions was delayed in all tested lines at 48 hpi, although not significantly in Corbarino (Fig. 3a). At 96 hpi, this positive effect was overcome by the pathogen in lines M82 and SM36, which showed average lesion areas not significantly different from the control, whereas, in T22‐treated Corbarino, the lesion reduction became highly significant (P≤ 0.01) (Fig. 3b). Line TA209 demonstrated the most effective interaction with T. harzianum T22 in terms of systemic resistance at both 48 and 96 hpi (Fig. 3a,b). As for T. atroviride P1, it was less effective than T22 against the foliar pathogen. At 48 hpi, P1 reduced the lesion size to a significant extent only in line TA209 (Fig. 3a), but this positive effect was not significant at 96 hpi (Fig. 3b). Noticeably, B. cinerea necrotic lesions on line M82 were much larger in P1‐treated plants than in the control (Fig. 3a,b). Thus, our data confirm that, in tomato, the protective effects of Trichoderma spp. against foliar pathogens are influenced by the genetic background of the plant.
Figure 3.
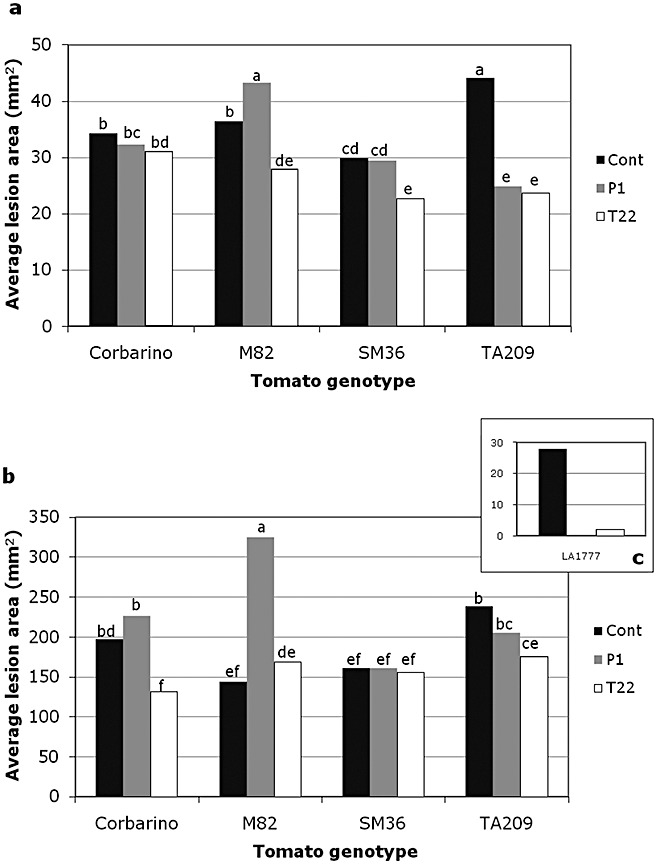
Effects of Trichoderma spp. treatments on plant resistance to the pathogen Botrytis cinerea. Two‐month‐old plants of Solanum lycopersicum (Corbarino, TA209, M82 and SM36) lines, developed from untreated (Cont), T. atroviride P1‐treated (P1) or T. harzianum T22‐treated (T22) seeds, were artificially inoculated with a suspension of B. cinerea spores, and lesion development was measured at 48 h (a) and 96 h (b) post‐inoculation. Values indicated by the same letter are not statistically significantly different for P≤ 0.01 according to the Duncan test. Average lesion areas at 96 h post‐inoculation on control and T22‐treated plants of the wild S. habrochaites LA1777 line are reported in the inset (c).
Effects on the expression of defence‐related genes
Many of the most active Trichoderma strains have shown the ability to enhance systemically plant resistance to microbial pathogens. To test whether the tomato response to Trichoderma involves a differential activation of defence‐related genes, we analysed markers of both salicylic acid (SA) (PR1b1 and PR‐P2) and JA (PINI, PINII, TomLoxA and TomLoxC) pathways by relative quantitative reverse transcription real‐time polymerase chain reaction (qRT‐PCR). Changes in gene expression were considered to be significant only for values twofold or more for up‐regulation and ≤0.5‐fold for down‐regulation relatively to the untreated control (calibrator).
Up‐regulated genes included PR1b1 in LA1777, SM36 and TA209 in response to either T22 or P1, with the wild accession LA1777 responding to both strains; PR‐P2 in both P1‐ and T22‐treated plants of Corbarino, LA1777 and TA209; PINI in Corbarino, LA1777 and SM36, but only in T22‐treated plants; PINII only in Corbarino in response to both strains; TomLoxA in all genotypes, except SM36, in response to either T22 or P1, with the wild accession LA1777 responding to both strains; and TomLoxC only in SM36 with both strains (Fig. 4). Generally, the most responsive lines to Trichoderma spp. were LA1777, Corbarino and TA209 (one wild and two cultivated lines), which also showed strong symptom reduction following inoculation with B. cinerea (Fig. 3). With regard to down‐regulation, neither the pathogenesis‐related (PR) genes nor TomLoxA was significantly affected, and the expression levels of PIN genes were frequently reduced, mainly by strain P1. This fungus also inhibited the transcription of TomLoxC, but only in line LA1777. Line M82 was confirmed to be the least responsive to the interaction with Trichoderma, as treatment with the two biocontrol strains did not induce a substantial variation in the expression of defence‐related genes before B. cinerea infection (Fig. 4) or decrease the disease symptoms after pathogen challenge (Fig. 3). Although the five tomato genotypes responded very differently to Trichoderma spp., the above results indicate that, in the absence of pathogen challenge, the SA‐mediated defence response is generally up‐regulated by Trichoderma treatment (i.e. PR genes are stimulated in most cases), whereas JA‐related genes are less responsive, although with a few exceptions (mainly for TomLoxA) (Fig. 4). This profile changes substantially after pathogen inoculation (see below). Notably, the two Trichoderma strains were able to affect defence‐related genes in tomato (a PR1 and a PR4) for at least 2 months after their application as seed coating, suggesting a long‐lasting effect on the readiness of the defence machinery.
Figure 4.
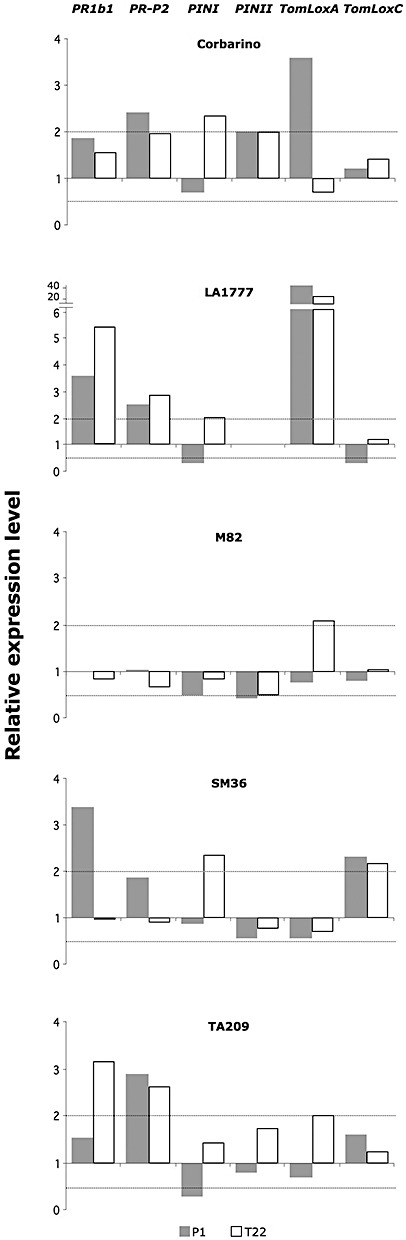
Effects of Trichoderma spp. treatments on the transcription of defence‐related genes in tomato. The relative expression of PR1b1, PR‐P2, PINI, PINII, TomLoxA and TomLoxC was measured by quantitative reverse transcription real‐time polymerase chain reaction (qRT‐PCR) in 2‐month‐old plants of Solanum lycopersicum (Corbarino, TA209, M82 and SM36) and S. habrochaites (LA1777) lines, developed from untreated (Cont) or T. atroviride P1‐treated (P1) or T. harzianum T22‐treated (T22) seeds. Expression levels for each gene are reported as the fold increase relative to those of the untreated control. Values within the dotted lines were not considered to be statistically significantly different from the control. Data on PINII expression in the wild tomato line LA1777 are missing as no amplification of this gene could be obtained with the primer pair used. The results shown are from one representative experiment out of four.
When untreated control and Trichoderma‐treated plants were challenged with B. cinerea, the transcription rate of the tested defence genes changed in a peculiar way for each tomato line, as reported in Fig. 5. Generally, the PR genes, which were pre‐activated in Trichoderma‐treated plants before pathogen inoculation (Fig. 4), showed a negative peak at 24 hpi, whereas the JA‐responsive genes were up‐regulated (Fig. 5). This indicates that, in the presence of Trichoderma, the SA pathway is repressed and the JA pathway is stimulated by B. cinerea infection at 24 hpi. At the end of the assay (48 hpi), the commercial biocontrol agent T22 caused a more consistent repression of SA‐ and stimulation of JA‐dependent genes compared with the untreated control plants across the five tomato lines (Fig. 5). By contrast, P1, a strain never developed commercially, showed a more variable effect (Fig. 5).
Figure 5.
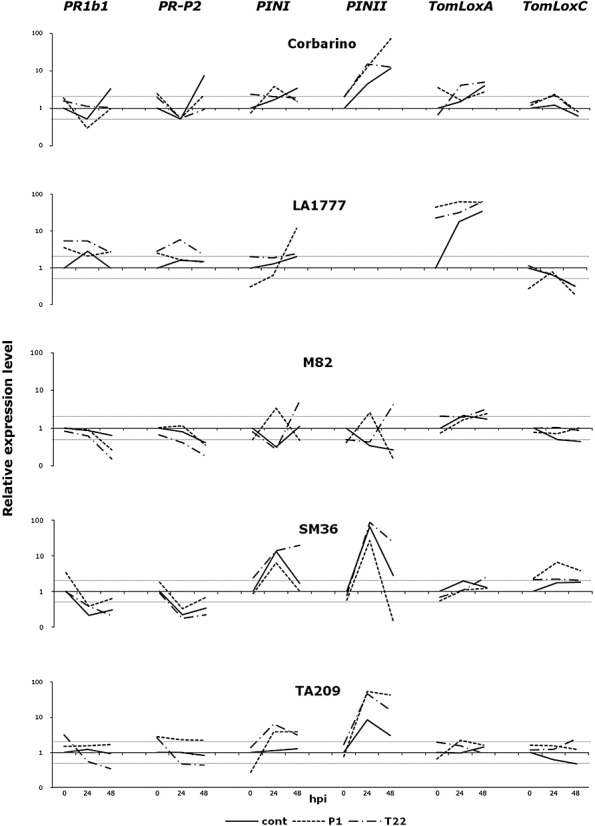
Transcriptional analysis of defence‐related genes in tomato plants inoculated with the pathogen Botrytis cinerea and pretreated or not with Trichoderma spp. The relative expression of PR1b1, PR‐P2, PINI, PINII, TomLoxA and TomLoxC was measured by quantitative reverse transcription real‐time polymerase chain reaction (qRT‐PCR) at different times after pathogen infection [0, 24 and 48 h post‐inoculation (hpi)] in 2‐month‐old plants of Solanum lycopersicum (Corbarino, TA209, M82 and SM36) and S. habrochaites (LA1777) lines, developed from untreated (cont), T. atroviride P1‐treated (P1) or T. harzianum T22‐treated (T22) seeds. The expression levels for each gene are reported as the fold increase relative to that of the control plants not treated with Trichoderma before infection with the pathogen (0 hpi), in a semi‐logarithmic scale. Values within the dotted lines were not considered to be significantly different from the control. Data on PINII expression in the wild tomato line LA1777 are missing as no amplification of this gene could be obtained with the primer pair used. The results shown are from one representative experiment out of four.
In contrast with the results at time zero, the changes in PR gene expression observed after B. cinerea inoculation did not correlate with resistance to the pathogen (3, 5). For instance, line TA209 treated with T22 showed reduced transcription of both PR1b1 and PR‐P2 (Fig. 5), but increased resistance (Fig. 3), whereas treatment with P1 increased PR expression (Fig. 5), but had no significant effect on resistance at the end of the assay (Fig. 3). However, transcriptional activation of the JA‐related genes PINI and PINII matched the increased tolerance to B. cinerea, as observed for T22‐treated SM36 plants (3, 5) and for TA209 treated with both strains (3, 5). Accordingly, P1‐treated M82 plants were more susceptible to the pathogen and showed a remarkable decrease in PINI and PINII expression at 48 hpi. Further, pretreatment of the same tomato line with T22 induced increased transcription of PIN genes at 48 hpi (Fig. 5), which probably limited the spread of B. cinerea lesions, although only in an early phase (Fig. 3a). In addition, in the case of the other two JA‐responsive genes, TomLoxA and TomLoxC, the up‐regulation often matched Trichoderma‐mediated tolerance to B. cinerea, whereas the opposite was not always true (3, 5).
These results indicate that the effect of defence gene expression in Trichoderma‐treated tomato plants during B. cinerea infection is complex and involves both systemic acquired resistance (SAR) and ISR, probably with the SA‐responsive genes contributing to maintain the readiness of the early reaction, and the JA‐responsive genes being more significant and effective at a later stage.
DISCUSSION
It is known that some Trichoderma strains can stimulate plant growth, at least in part by increasing the nutrient uptake and efficiency of nitrogen use (Altomare et al., 1999; Yedidia et al., 2001). Our data confirm the ability of the commercial biocontrol agent T. harzianum strain T22 to increase canopy and stem growth in tomato and indicate that, in this respect, T22 is more effective than T. atroviride strain P1, a fungus mainly used for laboratory work and not developed commercially to a significant extent. Moreover, our findings demonstrate that the extent of growth stimulation is largely dependent on the tomato genotype, suggesting that the response to Trichoderma spp. is under genetic control. Shoot dry weight was increased in most lines, except for TA209 with P1 and for the wild accession LA1777 with T22. Interestingly, P1 was able to reduce the stem length of LA1777 and had a similar, although not significant, effect on the other indeterminate line under study, Corbarino (Table 1 and Fig. 1b). The importance of the plant genetic background has already been reported for the interaction between maize and T. harzianum T22 (Harman et al., 2004b). Commercial trials on several T22‐treated hybrids and inbred lines have revealed the expected yield increases in most genotypes, with a few actually showing a yield reduction (Harman et al., 2004b). Genetic analysis has demonstrated that the maize response is largely conditioned by dominant genes (Harman, 2006).
The importance of the tomato genotype for the outcome of the beneficial plant–fungus interaction was also assessed for root growth promotion, with a significant increase in root dry weight obtained by treating Corbarino and TA209 with strain P1 and a significant decrease for M82 and TA209 with strain T22 (Fig. 2a). Trichoderma spp. also modified the root architecture of tomato in a differential way among the five tested lines (Fig. 2b). In three of the five lines, T. atroviride P1 increased root length and reduced lateral development (Fig. 2b). By contrast, in most cases, root length was unaffected by T. harzianum T22, whereas lateral development was stimulated by this fungus in lines Corbarino and SM36 and inhibited in the other three lines (Fig. 2b). We have not investigated the mechanisms underlying these differential responses. A recent study on Arabidopsis thaliana suggested that two strains of T. atroviride and T. virens stimulated lateral root development and reduced primary root length by producing indole‐3‐acetic acid (IAA) and auxin‐like compounds (Contreras‐Cornejo et al., 2009). Preliminary data indicate that both Trichoderma strains used in this work synthesize IAA (results not shown), and we have demonstrated previously their ability to release secondary metabolites with an auxin‐like effect on plants (2008a, 2008b). Further, Gravel et al. (2007) suggested that another strain of T. atroviride stimulates tomato root growth in a ‘controlled manner’ by balancing the synthesis and degradation of IAA and/or by limiting Et synthesis through hydrolysis of its precursor molecule 1‐aminocyclopropane‐1‐carboxylic acid (ACC). However, cytokinins could also be involved, as demonstrated for plant growth‐promoting rhizobacteria (PGPR) (Ortiz‐Castro et al., 2009), as the production of cytokinin‐like molecules, e.g. zeatin, and of GA3 or GA3‐related gibberellins has been reported as being possibly correlated with the biofertilization potential of Trichoderma (Benitez et al., 2004). An antagonistic auxin–cytokinin cross‐talk may explain the opposite effects of T. harzianum T22 on the shoot and root development of the tomato lines TA209 and, possibly, M82 (1, 2 and Table 1). Given that the ability of Trichoderma to promote a balanced growth of plants depends on a fine regulation of hormones and hormone‐like compounds in the rhizosphere, our data indicate that other factors, such as the genotype and the physiological status of the plant, or the cultural conditions (i.e. hydroponics vs. soil or high vs. low fertilizer content), are involved.
Beside the well‐characterized direct biocontrol activity of Trichoderma species on soil pathogens, it has more recently become clear that root colonization by these microorganisms induces plant resistance to foliar diseases, in the absence of direct contact between Trichoderma and the pathogen (Bigirimana et al., 1997; Harman et al., 2004a; Shoresh et al., 2010). Our results confirm that damage from B. cinerea infection on tomato leaves can be limited by rhizosphere colonization with either T. harzianum T22 or T. atroviride P1, although the former species appears to be more effective. However, major differences were also detected among the five tested tomato lines with regard to the Trichoderma‐induced tolerance to the pathogen. Apart from the wild species line LA1777, whose very low susceptibility to B. cinerea was further boosted by interaction with either Trichoderma strain, lesion expansion was initially limited by T22 in all tested lines. At later times, T22 reduced significantly the average lesion area in only TA209 and Corbarino, i.e. the two lines which are the least able to defend themselves from the pathogen in sterilized soil (Fig. 3b). Corbarino and LA1777, being a local variety selected by farmers in low‐input agricultural conditions and a wild species accession, respectively, might be more ecologically fit and therefore able to benefit the most from rhizosphere interaction with beneficial microflora. Information about the pedigree of the seed company‐bred line TA209 is too sparse to confirm or disprove this hypothesis. The susceptible line TA209 also showed alleviation of disease symptoms with T. atroviride P1, which was not effective for the other S. lycopersicum lines, and even increased the pathogen susceptibility of M82, a result that is worthy of further investigation. Notably, the very limited or negative Trichoderma‐induced responses to B. cinerea of line M82 also extended to the transcription of defence genes (4, 5). The ability of different strains of T. harzianum, including T22, to significantly restrict the development of B. cinerea symptoms on tomato has been reported previously (De Meyer et al., 1998; Dik and Elad, 1999; O'Neill et al., 1996; Seaman et al., 2003). However, only one study was performed on different tomato varieties (two), but the effect of the genotypic differences was not investigated (Dik and Elad, 1999). In addition, reports on the genotypic variation of the maize response to T. harzianum T22 did not address the variability in induced plant resistance (Harman, 2006; Harman et al., 2004b). Our results clearly highlight the existence of a genetic component of the plant response to Trichoderma spp. in terms of the induction of systemic resistance. This is in agreement with reports demonstrating that genetic background affects the response of different cucumber varieties to PGPR species (Liu et al., 1995), which actually are considered to share with Trichoderma spp. similar mechanisms of ISR (Harman et al., 2004a).
In order to further study the Trichoderma‐mediated systemic plant increase in pathogen tolerance, we analysed the transcriptional activation of several defence genes in tomato leaves: PR1b1, a PR1 gene used as a marker for SAR activation and SA‐mediated responses (Tornero et al., 1997); PR‐P2, a PR4 gene mainly induced by SA in tomato (Bertini et al., 2003; Fiocchetti et al., 2006; Linthorst et al., 1991; Van Kan et al., 1995); PINI and PINII, which are induced through the JA signal transduction pathway (Doares et al., 1995; Fidantsef et al., 1999); and TomLoxA and TomLoxC, encoding two enzymes of the lipoxygenase (LOX) family, which is involved in JA response and defence (Feussner and Wasternack, 2002; Porta and Rocha‐Sosa, 2002). Root penetration by Trichoderma is known to increase the expression of defence‐related genes and the production of antimicrobial compounds, but only transiently (Shoresh et al., 2005; 1999, 2003). However, our results demonstrate that rhizosphere colonization by Trichoderma can support the transcription of some defence‐related genes at low but significant levels for a relatively long period of time (60 days), at least in some plant genotype–Trichoderma species' interactions. This effect was particularly strong for PR1b1 and PR‐P2 expression (Fig. 4), suggesting that the long‐term response to Trichoderma in tomato may involve SA signalling. Similarly, transcriptomic analysis revealed constitutive PR5 up‐regulation in 7‐week‐old tomato plants sown in T. hamatum 382‐inoculated soil (Alfano et al., 2007).
When plants are challenged with a pathogen soon after the establishment of the interaction with Trichoderma, they are primed to react more strongly, increasing defence gene expression and the activity of protective enzymes sooner and to higher levels than in untreated plants (Shoresh et al., 2005; 1999, 2003). Our data demonstrate that this effect lasts long after the onset of the interaction between Trichoderma and the plant (60 days), as reduced lesion size (Fig. 3) and higher expression levels of defence genes in systemic leaves (Fig. 5) were detected in lines LA1777, TA209 and Corbarino. Generally, enhanced accumulation of RNA concerned PIN and, to some extent, lox genes, suggesting the involvement of an ISR response 2 months after treatment with the biocontrol agent. Higher induction of PR proteins/activities by pathogens in plants treated with Trichoderma spp. has been reported in cucumber and maize (Harman et al., 2004b; Shoresh et al., 2005). However, in this work, the expression levels of PR1b1 and PR‐P2 were higher than in controls on plants treated only with Trichoderma (Fig. 4), but, in most cases, decreased below the control value after subsequent inoculation with the pathogen (Fig. 5). These data are in agreement with those from proteomic analysis of a three‐player interaction (pathogen, plant and Trichoderma) (Marra et al., 2006), and strongly suggest that the presence of the biocontrol agent reduces the intensity of some plant responses to pathogen infection. Concerning the response of lox genes, T. asperellum was reported to up‐regulate lox1 transcription in roots, but not leaves, of cucumber (Shoresh et al., 2005) and of lox2 in plants of Arabidopsis thaliana (Segarra et al., 2009) soon after treatment. Based on these results, the authors proposed that Trichoderma‐enhanced plant defences depend on JA signalling and hence the activation of the phenylpropanoid pathway (through activation of PAL genes transcription), eventually leading to induced accumulation of phytoalexins and other antimicrobial metabolites (Shoresh et al., 2005; Yedidia et al., 2003). We also found that an increased resistance of the various tomato lines to B. cinerea corresponded to enhanced expression of lox genes, although the opposite was not always true (3, 5). In the case of the PINI and PINII genes, their expression was often induced and maintained at a level higher than that in controls in Trichoderma‐treated and B. cinerea‐inoculated tomato, and generally correlated with increased pathogen tolerance.
Several lines of evidence indicate that the plant defence reaction mediated by Trichoderma involves the JA pathway and stimulation of Et‐responsive genes (Korolev et al., 2008; Shoresh et al., 2005). However, the reported activation of Pal1 (Shoresh et al., 2005; Yedidia et al., 2003) could also increase SA biosynthesis. Indeed, Martinez et al. (2001) proposed that the reduction of powdery mildew symptoms in melon cotyledons by a cellulase from T. longibrachiatum occurs through two parallel mechanisms: active cellulase induces an oxidative burst and hence increases PAL activity and the synthesis of SA and phenolic compounds; at the same time, it induces LOX activity, JA synthesis and Et accumulation, thus additionally stimulating PAL and the phenylpropanoid metabolism (Martinez et al., 2001). It has also been suggested that ISR may be mediated by different signalling routes, depending on the BCA and challenging pathogen (Korolev et al., 2008).
In terms of the activation of different signal transduction pathways, our gene expression data suggest the following mechanism for the tomato response: (i) in the absence of pathogen infection, a preventative treatment of the plant with Trichoderma, as typically used in agriculture, induces an increase in the expression of some PR genes, such as the SAR markers PR1b1 and PR‐P2, indicative of a priming of the defence reaction; (ii) when plants are challenged by B. cinerea, pretreatment with Trichoderma might mitigate the SA‐dependent gene expression; (iii) soon after infection, expression of PINI, PINII, TomLoxA and TomLoxC is enhanced, possibly as a consequence of the priming effect, which results in an increased systemic resistance, probably caused by promotion of the JA‐mediated response.
Taken together, our data demonstrate that the plant response to ‘elite’ strains of Trichoderma spp., such as the commercial biocontrol agent T. harzianum T22 or the widely studied T. atroviride P1, is affected by plant genetic variability and thus is under genetic control in Solanum species of section Lycopersicon. Genotypic differences in any of the plant components of the complex cross‐talk with Trichoderma can be evoked to explain this effect, including the genotype ability to attract and sustain root colonization by the fungus, different sensitivities to the effectors produced by the BCA, variability in the perception and signal transduction of any of the hormones whose concentrations are controlled by Trichoderma spp., and so on. An explanation of the mechanisms that underlie plant genetic control of the interaction was beyond the scope of this work and will require a massive research effort. However, our findings hold promise that studies aimed at the identification of the major plant genetic determinants involved in the interaction with Trichoderma spp. will lead to the selection and breeding of tomato genotypes with an improved capacity to benefit from rhizosphere colonization by these microorganisms.
EXPERIMENTAL PROCEDURES
Cultivation of Trichoderma spp. and B. cinerea
Trichoderma atroviride strain P1 (ATCC 74058), isolated from wood chips and selected for its tolerance to the fungicide iprodion (Tronsmo, 1991), and T. harzianum T22 (ATCC 20847; Stasz et al., 1988) were used in this study. Pathogen B. cinerea strain 309, isolated from tobacco, was obtained from the culture collection of the Department ArBoPaVe of the University of Naples, Italy.
Fungi were grown on potato dextrose agar (PDA) and colonies were allowed to sporulate at 25 °C in the dark for 7 days. Spores were collected by washing the plates with sterile distilled water and brought to a concentration of 106 mL−1.
Culture of tomato plants
Seeds of the cultivated tomato (Solanum lycopersicum L.) lines Corbarino, M82, SM36 and TA209, and of the wild S. habrochaites accession LA1777, kindly provided by the Tomato Genetic Resource Center, University of California at Davis, CA, USA (M82, TA209 and LA1777) and by Professor L. Frusciante, University of Naples Federico II, Italy (Corbarino and SM36), were sterilized in 2% sodium hypochlorite for 20 min and thoroughly washed in sterile distilled water. Sterilized seeds were incubated in a 106 mL−1 fresh spore suspension of either T. atroviride P1 or T. harzianum T22 (coating), or in water (control seeds). Coated and control seeds were air dried for 24 h and then sown in sterilized soil in 40‐well polystyrene trays maintained in a growth chamber at 25 °C, 80% relative humidity, with a photoperiod of 16 h light. After 3 weeks, tomato seedlings were transplanted in 14‐cm‐diameter plastic pots in sterilized soil and grown for 5 weeks in the same controlled environmental conditions.
Botrytis cinerea inoculation and biometric assays
Untreated control and Trichoderma‐treated 2‐month‐old tomato plants were infected with B. cinerea by inoculating the third true leaf with 10 µL of a 106 mL−1 spore suspension of the pathogen. Three replicated plants for each treatment were used for inoculation. Immediately before infection, the fourth leaf of control and Trichoderma‐treated plants of each replicate was collected at 0 hpi as an uninfected control, immediately frozen in liquid nitrogen and stored at −80 °C until use in molecular analyses. Inoculated plants were enveloped in transparent plastic bags to achieve high relative humidity conditions and incubated in a growth chamber at 18 °C with a photoperiod of 16 h light.
At 24 and 48 hpi, the fifth or sixth leaf from each replicate plant was collected, immediately frozen in liquid nitrogen and stored at −80 °C until use in molecular analyses (24 and 48 hpi samples).
The disease spread was recorded at 48, 72 and 96 hpi by measuring necrotic lesions with an electronic calliper and calculating the area as an ellipse. The disease severity on the different treatments (control, T. atroviride P1 and T. harzianum T22) was expressed as the mean area of necrotic lesions.
Inoculation experiments were repeated four times.
At the end of each inoculation experiment, the stimulation of plant growth was measured in terms of stem height and root fresh and dry weight of untreated control and Trichoderma‐treated 2‐month‐old tomato plants. After visual evaluation of root architecture, roots were cut at the soil line and immediately weighed (fresh weight). The dry weight was recorded after drying the roots at 60 °C. As inoculation experiments with B. cinerea affect the leaf system, shoot dry weight was measured on a separate set of uninoculated plants.
Data on B. cinerea lesion size and plant growth were analysed by anova in a split‐plot design with three replicates, using plant genotype and Trichoderma treatment as experimental factors. Means were separated according to the Duncan test.
RNA isolation from tomato plants
Uninfected and B. cinerea‐infected tomato leaves, collected and stored as described above, were ground in liquid nitrogen and used for total RNA extraction with the Purelink Micro‐to‐Midi Total RNA Purification System (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA quality was checked by UV visualization after denaturing agarose gel electrophoresis in the presence of ethidium bromide. Total RNA concentration was measured with a NanoDrop 1000 Spectrophotometer (Thermo Fischer Scientific, Wilmington, DE, USA) and the purity was estimated by the A 260/A 280 absorbance ratio. After RNA purification, RNase‐free deoxyribonuclease I (DNase I) treatment was performed to eliminate residual genomic DNA in a final volume of 10 µL using 1 U of DNase I (Invitrogen), 1 µL of 10X DNase I Reaction Buffer (200 mm Tris‐HCl pH 8.4, 20 mm MgCl2, 500 mm KCl) added to 1 µg of total RNA in DEPC‐treated water. The DNA digestion was performed at room temperature for 15 min, and then DNase I was inactivated by the addition of 1.2 µL of 25 mm ethylenediaminetetraacetic acid (EDTA) solution and incubation at 65 °C for 10 min.
qRT‐PCR analysis of gene expression
The expression of selected defence genes was monitored in each of the four inoculation experiments through qRT‐PCR; thus four biological replicates were analysed.
Following total RNA isolation, RT for first‐strand cDNA synthesis was carried out in a final volume of 20 µL by mixing 1 µg of DNase‐treated total RNA and 500 ng of oligo(dT)12–18 primers (Invitrogen) at 70 °C for 10 min. After 5 min of incubation on ice, 4 µL of 5X First‐Strand RT buffer (250 mm Tris‐HCl pH 8.3, 375 mm KCl, 15 mm MgCl2), 1 µL of 10 mm of each deoxynucleoside triphosphate (dNTP), 2 µL of 100 mm dithiothreitol and 160 U SuperScript II Reverse Transcriptase (Invitrogen) were added. RT reactions were incubated at 42 °C for 1 h and then stopped at 70 °C for 10 min. For each RNA sample, a reaction without RT was performed as a control for contamination by genomic DNA.
The reference housekeeping gene coding for actin and six target genes coding for defence proteins were chosen: LOXA and LOXC (TomLoxA and TomLoxC); proteinase inhibitors I and II (PINI and PINII); pathogenesis‐related 1 and 4 (PR1b1 and PR‐P2, respectively). The corresponding tomato nucleotide sequences were obtained from the GenBank database and used to design gene‐specific primer pairs (Table 2) employing Primer3 software (Rozen and Skaletsky, 1998) and checking for self‐annealing by Primer Express 2.0 software (Applied Biosystems, Foster City, CA, USA). In the set‐up step, primers forming dimers were excluded after testing in control reactions without cDNA.
Table 2.
Gene‐specific primers used in quantitative reverse transcription real‐time polymerase chain reaction (qRT‐PCR).
| Gene name | Accession number | Forward primer | Reverse primer | Amplicon size (bp) |
|---|---|---|---|---|
| actin | BT013524 | CACCACTGCTGAACGGGAA | GGAGCTGCTCCTGGCAGTTT | 100 |
| PR1b1 | Y08804 | GCACTAAACCTAAAGAAAAATGGG | AAGTTGGCATCCCAAGACATA | 177 |
| PR‐P2 | X58548 | GGAACAGGAACACAAGAAACAGTGA | CCCAATCCATTAGTGTCCAATCG | 104 |
| PINI | K03290 | TGAAACTCTCATGGCACGAAAAG | GGCCACATTTGTTTTCCTTCG | 102 |
| PINII | K03291 | GGCCAAATGCTTGCACCTTT | CGTGGTACATCCGGTGGGATA | 101 |
| TomLoxA | U09026 | TGAACCATGGTGGGCTGAAA | CTGCCCGAAATTGACTGCTG | 106 |
| TomLoxC | U37839 | TCCGGCAACACCGTTTACTC | GTCAATGGCCGGAAAATGTG | 102 |
Amplifications were performed using the 7900HT Fast Real‐Time PCR System (Applied Biosystems). Reactions were prepared in a total volume of 20 µL with 10 µL of the 2X Power SYBR Green PCR Master Mix (Applied Biosystems), 0.2 pmol of target gene primers or 0.4 pmol of actin primers, and 4 µL of 1:4 diluted cDNA template. Three independent reactions were performed from each cDNA sample and qRT‐PCR experiments were carried out in triplicate. The thermal cycling programme started with a step of 10 min at 95 °C for Taq polymerase activation and initial template denaturation, followed by 40 cycles of two steps: 95 °C for 15 s and 60 °C for 1 min. A dissociation kinetics analysis was performed after each assay in order to check the specificity of the amplification products. The melting curve programme was from 60 to 95 °C with a 2% heating rate and a continuous fluorescence measurement. Reaction products were also resolved in agarose gel to verify amplicon size. The primer pairs used and the size of the expected amplicons are shown in Table 2. The quantification of gene expression relative to the untreated control sample at 0 hpi (calibrator) was carried out using the 2−ΔΔ Ct method (Livak and Schmittgen, 2001), where ΔΔCt= (Ct of target gene −Ct of reference gene)sample− (Ct of target gene −Ct of reference gene)calibrator and Ct is the threshold cycle of each transcript, defined as the point at which the amount of amplified target reaches a fixed threshold above the background fluorescence. The actin housekeeping gene was used as an endogenous reference gene for the normalization of the expression levels of the target genes, after verifying its constant expression throughout the time points and the treatments of our experiments. In order to check PCR efficiency, standard curves based on Ct values vs. log(cDNA dilution) were constructed using serial 10‐fold dilution of cDNA for each pair of selected primers, where a 100% PCR efficiency corresponds to a slope of −3.32. In the present study, all the PCRs displayed efficiencies close to 100%, which allowed normalization and realistic comparisons.
ACKNOWLEDGEMENTS
The work of MT, MDP and LDM was partially supported by the Italian Ministries of Education, University and Scientific Research (project GenoPOM, D.D. 14/03/2005 prot. 602) and of Agricultural, Food and Forestry Politics (project PROM, CIPE n. 17/2003). The work of MR and ML was supported by the following grants: FIRB 2002 prot. RBNE01K2E7; PRIN 2003 prot. 2003070719‐003; MIUR PON project no. DD12935 of 02/08/2002; MIUR PON project no. DD1219 of 05/10/2004; MIUR PON project no. DD1801 of 31/12/2004; EU TRICHOEST QLK3‐2002‐02032; EU 2E‐BCA2. We thank Gianluca Caruso (Dip. DISSPAPA, Università di Napoli Federico II, Italy) for help with statistical analysis and the Genomic Platform of the GenoPOM Laboratory (Consiglio Nazionale delle Ricerche, Istituto Genetica Vegetale, Portici and Dip. DISSPAPA, Università di Napoli Federico II, Italy) for technical assistance.
This is Contribution n. 354 from the CNR‐Istituto di Genetica Vegetale, Portici (NA), Italy.
REFERENCES
- Ahn, I.P. , Lee, S.W. and Suh, S.C. (2007) Rhizobacteria‐induced priming in Arabidopsis is dependent on ethylene, jasmonic acid, and NPR1. Mol. Plant–Microbe Interact. 20, 759–768. [DOI] [PubMed] [Google Scholar]
- Alfano, G. , Ivey, M.L. , Cakir, C. , Bos, J.I. , Miller, S.A. , Madden, L.V. , Kamoun, S. and Hoitink, H.A. (2007) Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology, 97, 429–437. [DOI] [PubMed] [Google Scholar]
- Altomare, C. , Norvell, W.A. , Bjorkman, T. and Harman, G.E. (1999) Solubilization of phosphates and micronutrients by the plant‐growth‐promoting and biocontrol fungus Trichoderma harzianum Rifai 1295–22. Appl. Environ. Microbiol. 65, 2926–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, H. , Sicher, R.C. , Kim, M.S. , Kim, S.H. , Strem, M.D. , Melnick, R.L. and Bailey, B.A. (2009) The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao . J. Exp. Bot. 60, 3279–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez, T. , Rincon, A.M. , Limon, M.C. and Codon, A.C. (2004) Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 7, 249–260. [PubMed] [Google Scholar]
- Bertini, L. , Leonardi, L. , Caporale, C. , Tucci, M. , Cascone, N. , Di Berardino, I. , Buonocore, V. and Caruso, C. (2003) Pathogen‐responsive wheat PR4 genes are induced by activators of systemic acquired resistance and wounding. Plant Sci. 164, 1067–1078. [Google Scholar]
- Bigirimana, J. , De Meyer, G. , Poppe, J. , Elad, Y. and Höfte, M. (1997) Induction of systemic resistance on bean (Phaseolus vulgaris) by Trichoderma harzianum . Med. Fac. Landbouww. Univ. Gent. 62, 1001–1007. [Google Scholar]
- Budnik, L.T. and Baur, X. (2009) The assessment of environmental and occupational exposure to hazardous substances by biomonitoring. Dtsch Arztebl. Int. 106, 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath, U. , Beckers, G.J.M. , Flors, V. , Garcia‐Agustin, P. , Jakab, G. , Mauch, F. , Newman, M.A. , Pieterse, C.M.J. , Poinssot, B. , Pozo, M.J. , Pugin, A. , Schaffrath, U. , Ton, J. , Wendehenne, D. , Zimmerli, L. and Mauch‐Mani, B. (2006) Priming: getting ready for battle. Mol. Plant–Microbe Interact. 19, 1062–1071. [DOI] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. , Cortes‐Penagos, C. and Lopez‐Bucio, J. (2009) Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin‐dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer, G. , Bigirimana, J. , Elad, Y. and Hofte, M. (1998) Induced systemic resistance in Trichoderma harzianum T39 biocontrol of Botrytis cinerea . Eur. J. Plant Pathol. 104, 279–286. [Google Scholar]
- Debenest, T. , Silvestre, J. , Coste, M. and Pinelli, E. (2010) Effects of pesticides on freshwater diatoms. Rev. Environ. Contam. Toxicol. 203, 87–103. [DOI] [PubMed] [Google Scholar]
- Dik, A.J. and Elad, Y. (1999) Comparison of antagonists of Botrytis cinerea in greenhouse‐grown cucumber and tomato under different climatic conditions. Eur. J. Plant Pathol. 105, 123–137. [Google Scholar]
- Doares, S.H. , Narvaez‐Vasquez, J. , Conconi, A. and Ryan, C.A. (1995) Salicylic‐acid inhibits synthesis of proteinase‐inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 108, 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner, I. and Wasternack, C. (2002) The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. [DOI] [PubMed] [Google Scholar]
- Fidantsef, A.L. , Stout, M.J. , Thaler, J.S. , Duffey, S.S. and Bostock, R.M. (1999) Signal interactions in pathogen and insect attack: expression of lipoxygenase, proteinase inhibitor II, and pathogenesis‐related protein P4 in the tomato, Lycopersicon esculentum . Physiol. Mol. Plant Pathol. 54, 97–114. [Google Scholar]
- Fiocchetti, F. , Caruso, C. , Bertini, L. , Vitti, D. , Saccardo, F. and Tucci, M. (2006) Over‐expression of a pathogenesis‐related protein gene in transgenic tomato alters the transcription patterns of other defence genes. J. Hortic. Sci. Biotechnol. 81, 27–32. [Google Scholar]
- Gravel, V. , Antoun, H. and Tweddell, R.J. (2007) Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: possible role of indole acetic acid (IAA). Soil Biol. Biochem. 39, 1968–1977. [Google Scholar]
- Hanson, L.E. and Howell, C.R. (2004) Elicitors of plant defense responses from biocontrol strains of Trichoderma virens . Phytopathology, 94, 171–176. [DOI] [PubMed] [Google Scholar]
- Harman, G.E. (2006) Overview of mechanisms and uses of Trichoderma spp. Phytopathology, 96, 190–194. [DOI] [PubMed] [Google Scholar]
- Harman, G.E. , Howell, C.R. , Viterbo, A. , Chet, I. and Lorito, M. (2004a) Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. [DOI] [PubMed] [Google Scholar]
- Harman, G.E. , Petzoldt, R. , Comis, A. and Chen, J. (2004b) Interactions between Trichoderma harzianum strain T22 and maize inbred line Mo17 and effects of these interactions on diseases caused by Pythium ultimum and Colletotrichum graminicola . Phytopathology, 94, 147–153. [DOI] [PubMed] [Google Scholar]
- Korolev, N. , David, D.R. and Elad, Y. (2008) The role of phytohormones in basal resistance and Trichoderma‐induced systemic resistance to Botrytis cinerea in Arabidopsis thaliana . Biocontrol, 53, 667–683. [Google Scholar]
- Linthorst, H.J.M. , Danhash, N. , Brederode, F.T. , Van Kan, J.A.L. , De Wit, P.J.G.M. and Bol, J.F. (1991) Tobacco and tomato Pr proteins homologous to win and Pro‐hevein lack the hevein domain. Mol. Plant–Microbe Interact. 4, 586–592. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Kloepper, J.W. and Tuzun, S. (1995) Induction of systemic resistance in cucumber by plant growth‐promoting rhizobacteria: duration of protection and effect of host resistance on protection and root colonization. Phytopathology, 85, 1064–1068. [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔC T method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorito, M. , Woo, S.L. , Harman, G.E. and Monte, E. (2010) Translational research on Trichoderma: from ‘Omics’ to the field. Annu. Rev. Phytopathol. 48, 395–417. [DOI] [PubMed] [Google Scholar]
- Mandal, S. and Mitra, A. (2007) Reinforcement of cell wall in roots of Lycopersicon esculentum through induction of phenolic compounds and lignin by elicitors. Physiol. Mol. Plant Pathol. 71, 201–209. [Google Scholar]
- Marra, R. , Ambrosino, P. , Carbone, V. , Vinale, F. , Woo, S.L. , Ruocco, M. , Ciliento, R. , Lanzuise, S. , Ferraioli, S. , Soriente, I. , Gigante, S. , Turra, D. , Fogliano, V. , Scala, F. and Lorito, M. (2006) Study of the three‐way interaction between Trichoderma atroviride, plant and fungal pathogens by using a proteomic approach. Curr. Genet. 50, 307–321. [DOI] [PubMed] [Google Scholar]
- Martinez, C. , Blanc, F. , Le Claire, E. , Besnard, O. , Nicole, M. and Baccou, J.C. (2001) Salicylic acid and ethylene pathways are differentially activated in melon cotyledons by active or heat‐denatured cellulase from Trichoderma longibrachiatum . Plant Physiol. 127, 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, T.M. , Niv, A. , Elad, Y. and Shtienberg, D. (1996) Biological control of Botrytis cinerea on tomato stem wounds with Trichoderma harzianum . Eur. J. Plant Pathol. 102, 635–643. [Google Scholar]
- Ortiz‐Castro, R. , Contreras‐Cornejo, H.A. , Macias‐Rodriguez, L. and Lopez‐Bucio, J. (2009) The role of microbial signals in plant growth and development. Plant Signal. Behav. 4, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M. , van Wees, S.C. , Hoffland, E. , van Pelt, J.A. and van Loon, L.C. (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis‐related gene expression. Plant Cell, 8, 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta, H. and Rocha‐Sosa, M. (2002) Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 130, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H.J. (1998) Primer3. Code available at http://www‐genome.wi.mit.edu/genome_software/other/primer3.html. [DOI] [PubMed]
- Seaman, A. , Stivers, L. , Shail, J. and Price, H. (2003) Efficacy of OMRI‐approved products for tomato foliar disease control. NY State Integr. Pest Manag. Program, 129, 164–167. [Google Scholar]
- Segarra, G. , Casanova, E. , Bellido, D. , Odena, M.A. , Oliveira, E. and Trillas, I. (2007) Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics, 7, 3943–3952. [DOI] [PubMed] [Google Scholar]
- Segarra, G. , Van der Ent, S. , Trillas, I. and Pieterse, C.M.J. (2009) MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 11, 90–96. [DOI] [PubMed] [Google Scholar]
- Shoresh, M. , Yedidia, I. and Chet, I. (2005) Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology, 95, 76–84. [DOI] [PubMed] [Google Scholar]
- Shoresh, M. , Harman, G.E. and Mastouri, F. (2010) Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. [DOI] [PubMed] [Google Scholar]
- Stasz, T.E. , Harman, G.E. and Weeden, N.F. (1988) Protoplast preparation and fusion in two biocontrol strains of Trichoderma harzianum . Mycologia, 80, 141–150. [Google Scholar]
- Tornero, P. , Gadea, J. , Conejero, V. and Vera, P. (1997) Two PR‐1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis‐related gene during the hypersensitive response and development. Mol. Plant–Microbe Interact. 10, 624–634. [DOI] [PubMed] [Google Scholar]
- Tronsmo, A. (1991) Biological and integrated controls of Botrytis cinerea on apple with Trichoderma harzianum . Biol. Control, 1, 59–62. [Google Scholar]
- Van Kan, J.A.L. , Cozijnsen, T. , Danhash, N. and De Wit, P.J.G.M. (1995) Induction of tomato stress protein messenger‐RNAs by ethephon, 2,6‐dichloroisonicotinic acid and salicylate. Plant Mol. Biol. 27, 1205–1213. [DOI] [PubMed] [Google Scholar]
- Verma, M. , Brar, S.K. , Tyagi, R.D. , Surampalli, R.Y. and Valero, J.R. (2007) Antagonistic fungi, Trichoderma spp.: panoply of biological control. Biochem. Eng. J. 37, 1–20. [Google Scholar]
- Vinale, F. , Sivasithamparam, K. , Ghisalberti, E.L. , Marra, R. , Barbetti, M.J. , Li, H. , Woo, S.L. and Lorito, M. (2008a) A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 72, 80–86. [Google Scholar]
- Vinale, F. , Sivasithamparam, K. , Ghisalberti, E.L. , Marra, R. , Woo, S.L. and Lorito, M. (2008b) Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 40, 1–10. [Google Scholar]
- Woo, S.L. , Scala, F. , Ruocco, M. and Lorito, M. (2006) The molecular biology of the interactions between Trichoderma spp., phytopathogenic fungi, and plants. Phytopathology, 96, 181–185. [DOI] [PubMed] [Google Scholar]
- Yedidia, I. , Benhamou, N. and Chet, I. (1999) Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum . Appl. Environ. Microbiol. 65, 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia, I. , Srivastva, A.K. , Kapulnik, Y. and Chet, I. (2001) Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant Soil, 235, 235–242. [Google Scholar]
- Yedidia, I. , Shoresh, M. , Kerem, Z. , Benhamou, N. , Kapulnik, Y. and Chet, I. (2003) Concomitant induction of systemic resistance to Pseudomonas spingae pv. lachrymans in cucumber by Trichoderma asperellum (T‐203) and accumulation of phytoalexins. Appl. Environ. Microbiol. 69, 7343–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]


