SUMMARY
Plants possess two distinct types of immune receptor. The first type, pattern recognition receptors (PRRs), recognizes microbe‐associated molecular patterns (MAMPs) and initiates pattern‐triggered immunity (PTI) on recognition. FLS2 is a PRR, which recognizes a part of bacterial flagellin. The second type, resistance (R) proteins, recognizes pathogen effectors and initiates effector‐triggered immunity (ETI) on recognition. RPM1, RPS2 and RPS5 are R proteins. Here, we provide evidence that FLS2 is physically associated with all three R proteins. Our findings suggest that signalling interactions occur between PTI and ETI at very early stages and/or that FLS2 forms a PTI signalling complex, some components of which are guarded by R proteins.
INTRODUCTION
Plants can sense microbial organisms by detecting microbe/pathogen‐associated molecular patterns (MAMPs/PAMPs) with pattern recognition receptors (PRRs) (Ausubel, 2005; Medzhitov and Janeway, 1997; Tsuda and Katagiri, 2010), which are often leucine‐rich repeat receptor kinases (LRR‐RKs). One well‐studied PRR in Arabidopsis is FLS2, which is an LRR‐RK and recognizes a conserved 22‐amino‐acid fragment (flg22) of bacterial flagellin (Chinchilla et al., 2006; Gomez‐Gomez and Boller, 2000; Zipfel et al., 2004). flg22 perception by FLS2 triggers a chain of signalling events that eventually results in induced immunity called pattern‐triggered immunity (PTI) (Chinchilla et al., 2007a; Jones and Dangl, 2006; Tsuda and Katagiri, 2010; Zipfel et al., 2004).
Another mode of induced immunity in plants is called effector‐triggered immunity (ETI) (Jones and Dangl, 2006), also known as gene‐for‐gene resistance (Flor, 1971). ETI signalling is initiated on the basis of the perception of pathogen effectors by plant disease resistance (R) proteins, either directly or indirectly (Dangl and Jones, 2001; Jones and Dangl, 2006). Most R proteins are members of the nucleotide‐binding site/leucine‐rich repeat (NB‐LRR) family (Dangl and Jones, 2001). Examples of well‐studied R proteins in Arabidopsis include RPM1, RPS2 and RPS5 (Bent et al., 1994; Grant et al., 1995; Mindrinos et al., 1994; Warren et al., 1998). Indirect recognition of effectors by R proteins is explained by the ‘guard hypothesis’, in which R proteins ‘guard’ certain host proteins (‘guardees’) that are manipulated by pathogen effectors (Dangl and Jones, 2001). One well‐known ‘guardee’ is RIN4, which is targeted by at least three bacterial effector proteins (AvrRpm1, AvrB and AvrRpt2) and ‘guarded’ by two R proteins (RPM1 and RPS2) (Axtell and Staskawicz, 2003; Mackey et al., 2002, 2003). Another ‘guardee’ is PBS1, which is targeted by the bacterial effector protein AvrPphB and ‘guarded’ by RPS5 (Shao et al., 2003). Accumulated evidence suggests an arms race between pathogens and plants, in which pathogens interfere with plant PTI using effectors and plants evolve to mount strong ETI responses on recognition of effectors (Boller and He, 2009; Chisholm et al., 2006; Jones and Dangl, 2006). However, not much is known about how PTI and ETI interplay to coordinate plant immunity. Here, we show evidence that the PTI and ETI receptors can reside in the same protein complex, which raises the possibility that PTI and ETI signalling interact at the very beginning.
RESULTS
Previously, we have developed a method to purify RPS2 protein complexes using a biotin tag called the ‘HPB’ tag and identified putative complex components by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) (Qi and Katagiri, 2009). Among eight replicate assays, we identified peptides of BSK1 (At4g35230, IPI00530358; two peptide hits) and BSK8 (At5g41260, IPI00529310; one peptide hit) at a 95% probability level, once each in the samples pulled down with RPS2, but not in negative control samples even at a 50% confidence level (Fig. S1, see Supporting Information) (Qi and Katagiri, 2009). The BSK family consists of 12 members (Tang et al., 2008), including BSK1 and BSK8, and the family members belong to the receptor‐like cytoplasmic kinase (RLCK) subfamily XII (Shiu et al., 2004). Some BSKs, including BSK1, positively participate in brassinosteroid signalling through a physical association with the BRI1 receptor (Tang et al., 2008). Both FLS2 and BRI1 are LRR‐RKs and physically associate with an LRR‐RK, BAK1 (Chinchilla et al., 2007b; Li et al., 2002; Nam and Li, 2002). Furthermore, BSK8 was phorsphorylated on flg22 treatment (Benschop et al., 2007). These lines of evidence prompted us to investigate the possibility that BSKs physically associate with FLS2.
First, the subcellular localization of BSK8 was examined using Arabidopsis plants expressing the BSK8::YFP::HA transgene and confocal microscopy. Full‐length BSK8::YFP::HA protein was detected (Fig. S2, see Supporting Information) and clearly localized to the plasma membrane (PM) (Fig. 1A), which is the same as the localization of other BSK members (Tang et al., 2008). As the PM localization of BSK8 was consistent with the PM localization of FLS2 (Gohre et al., 2008; Robatzek et al., 2006), we next examined a possible physical association between BSK8 and FLS2 by a pulldown assay. FLS2::HPB (Qi and Katagiri, 2009) and BSK8::GFP were transiently expressed in Nicotiana benthamiana. HPB‐tagged proteins are biotinylated in plant cells and thus readily captured by streptavidin beads (Qi and Katagiri, 2009). Indeed, BSK8::GFP was pulled down by FLS2::HPB with streptavidin beads from the sample in which both proteins were co‐expressed, but not from the negative control sample where only BSK8::GFP was expressed (Fig. 1B). In addition, N‐terminus Myc‐tagged BSK8 (Myc::BSK8) was pulled down by FLS2::HPB (Fig. S3, see Supporting Information).
Figure 1.
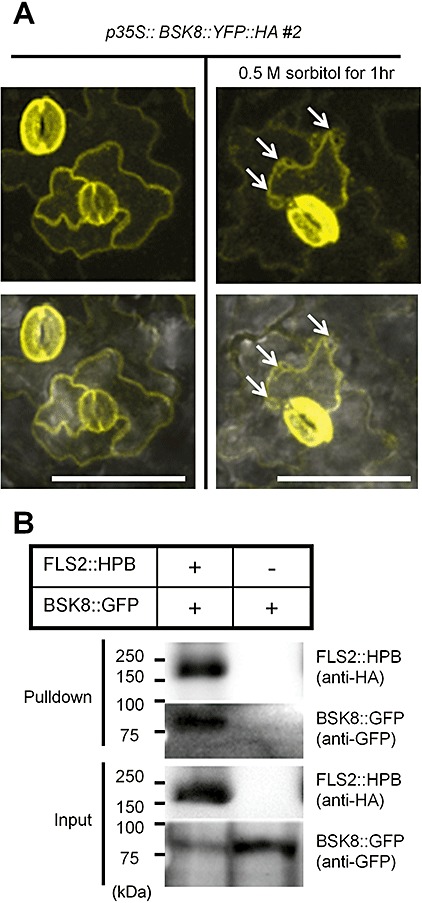
Plasma membrane (PM)‐localized BSK8 is physically associated with FLS2 in planta. (A) BSK8 is localized to PM. Leaves of 5‐week‐old T3 homozygous p35S::BSK8::YFP::HA line #2 plants were employed for the detection of yellow fluorescent protein (YFP) signal in epidermal cells using confocal microscopy. The top panel shows the YFP channel signal. The bottom panel shows a merged photograph of both the YFP channel and the bright field channel. Pretreatment with 0.5 m sorbitol for 1 h resulted in the detachment of PM from the cell wall (shown by white arrows), indicating that BSK8 is localized to PM, not to the cell wall. Bar, 100 µm. (B) BSK8::GFP forms a protein complex with FLS2::HPB in Nicotiana benthamiana. BSK8::GFP and FLS2::HPB were transiently expressed in N. benthamiana leaves, and FLS2::HPB was pulled down with streptavidin beads. Nicotiana benthamiana leaves expressing only BSK8::GFP were used as a negative control. Both pulldown samples and input samples were analysed by immunoblotting using anti‐haemagglutinin (HA) and anti‐green fluorescent protein (GFP) antibodies. Both experiments were performed twice with similar results.
The finding that BSK8 appears to physically associate with both FLS2 and RPS2 led us to hypothesize that RPS2 and FLS2 may reside in the same protein complex. To test this idea, we performed a pulldown assay using a plant line transgenically expressing RPS2::HPB from the RPS2 promoter in the rpm1 rps2 double‐mutant background (Qi and Katagiri, 2009). FLS2 was pulled down by RPS2::HPB from rpm1 rps2 RPS2::HPB plants, but not from negative control rpm1 rps2 plants (Fig. 2A). Therefore, FLS2 and RPS2 are physically associated. Next, we determined whether FLS2 forms protein complexes with RPM1 and RPS5, using a pulldown assay with transgenic plants expressing RPM1::HPB and RPS5::HPB (Qi and Katagiri, 2009). FLS2 was also pulled down by RPM1::HPB and RPS5::HPB (Fig. 2A). The anti‐FLS2 antibody recognized a band of ∼175 kDa in Col wild‐type plants, but none in fls2 mutants (Fig. 2B), indicating that the antibody is specific to FLS2, as shown previously (Chinchilla et al., 2006). RIN4, which is known to physically interact with RPS2 and RPM1 (Axtell and Staskawicz, 2003; Mackey et al., 2002, 2003), served as a positive control and was clearly pulled down by both RPS2::HPB and RPM1::HPB (Fig. 2A). All of the Arabidopsis lines used in the study showed high expression of the wild‐type 3‐methylcrotonyl CoA carboxylase (MCCA), from which the biotinylation site of the HPB tag was derived (Qi and Katagiri, 2009). As FLS2 was not pulled down from the plant lines without R::HPB transgenes (although a large amount of MCCA was present in the samples), FLS2 did not physically associate with the biotinylation site of the HPB tag. In addition, as the amount of FLS2 pulled down did not correlate with the amount of R::HPB expressed (RPM1::HPB pulled down more FLS2 than RPS2::HPB, whereas RPS2::HPB accumulated more than RPM1::HPB; Fig. 2A), it is very unlikely that FLS2 was pulled down through a physical association with the HPB tag. To prove that the FLS2–R protein complex formation does not depend on the HPB tag, we immunoprecipitated RPM1::Myc, which does not contain any part of the HPB tag, from extracts of an Arabidopsis RPM1::Myc transgenic line using an anti‐Myc antibody to determine whether FLS2 would be pulled down by RPM1::Myc. As shown in Fig. 2C, FLS2 was indeed pulled down by RPM1::Myc. We thus conclude that the FLS2–R protein complexes are not artefacts arising from the HPB tag.
Figure 2.
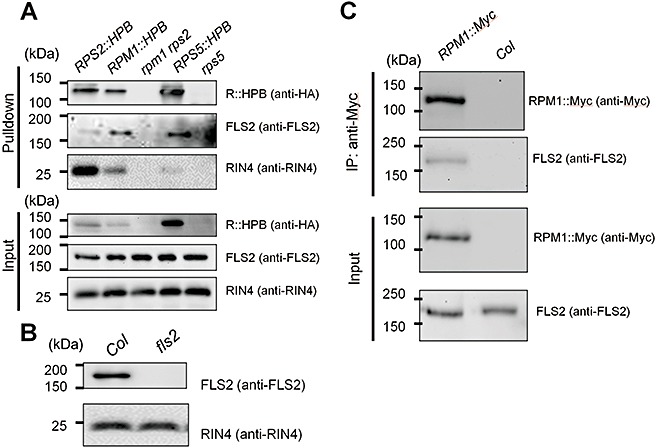
FLS2 was pulled down by three distinct R proteins in Arabidopsis. (A) The pulldown assays were performed using solubilized membrane proteins prepared from 5‐week‐old Arabidopsis plants of RPS2::HPB rpm1 rps2 (RPS2::HPB), RPM1::HPB rpm1 rps2 (RPM1::HPB), rpm1 rps2 (control), RPS5::HPB rps5 (RPS5::HPB) and rps5 (control). The HPB‐tagged proteins were pulled down with streptavidin beads. The pulldown and input samples were examined by immunoblotting with anti‐haemagglutinin (HA) and anti‐FLS2 antibodies. (B) Anti‐FLS2 antibody specifically recognizes FLS2, but not other proteins. Detection of RIN4 by an anti‐RIN4 antibody was used as a loading control. (C) The pulldown assay was performed using solubilized membrane proteins prepared from 4‐week‐old Arabidopsis plants of RPM1::Myc and Col (control). RPM1::Myc was pulled down with an anti‐Myc antibody. Both pulldown and input proteins were examined by immunoblotting with anti‐Myc and anti‐FLS2 antibodies. The experiments shown in (A) and (B) were performed three times with similar results, and that in (C) was performed twice with similar results.
To further confirm the R protein–FLS2 association in vivo, a reciprocal pulldown assay was conducted between RPM1 and FLS2. The FLS2::HPB plant line (Qi and Katagiri, 2009) was crossed to a plant line expressing RPM1::Myc (Boyes et al., 1998). Pulldown was then conducted using F1 plants which express both tagged proteins and control plants expressing only RPM1::Myc. RPM1::Myc was pulled down by FLS2::HPB from F1 plants, but not from the RPM1::Myc control plant (Fig. 3). Thus, we demonstrated RPM1–FLS2 association in a reciprocal manner.
Figure 3.
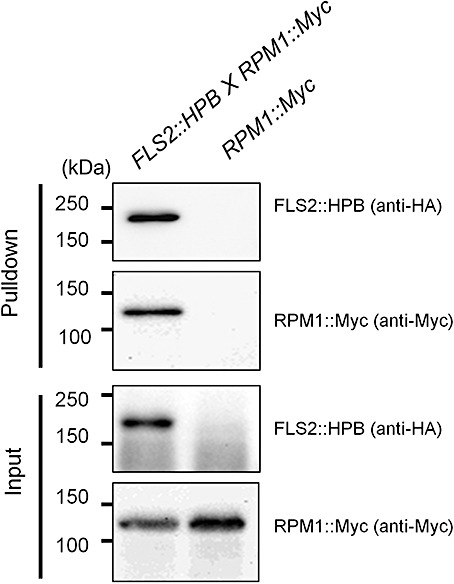
RPM1 was pulled down by FLS2 in Arabidopsis. The pulldown assays were performed using solubilized membrane proteins prepared from 5‐week‐old Arabidopsis plants of genotypes FLS2::HPB×RPM1‐Myc (F1 generation) and RPM1‐Myc (control). FLS2::HPB was pulled down with streptavidin beads. The pulldown and input samples were examined by immunoblotting with anti‐haemagglutinin (HA) and anti‐Myc antibodies. This experiment was performed three times with similar results.
It has been shown that RIN4 interacts with both RPS2 and RPM1 (Fig. 2A) (Axtell and Staskawicz, 2003; Mackey et al., 2002, 2003; Qi and Katagiri, 2009) and participates in FLS2 signalling (Kim et al., 2005). We therefore examined whether RIN4 was also residing in the FLS2–protein complex. We conducted a pulldown assay with FLS2::HPB plants and used an fls2 mutant as a control. We were able to pull down RIN4 with FLS2::HPB when the cross‐linker dithiobis(succinimidyl propionate) (DSP) was applied (Fig. 4). It should be noted that the cross‐link was cleaved before resolving the pulled down proteins by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE). As DSP cross‐linking greatly increased the pulldown of RIN4 by both RPS2 (Qi and Katagiri, 2009) and RPM1 (data not shown), the results presented here indicate that RIN4 is probably associated with FLS2, but its associations with RPS2, RPM1 or FLS2 are relatively weak and mostly disrupted by the stringent wash conditions used in the pulldown assay.
Figure 4.
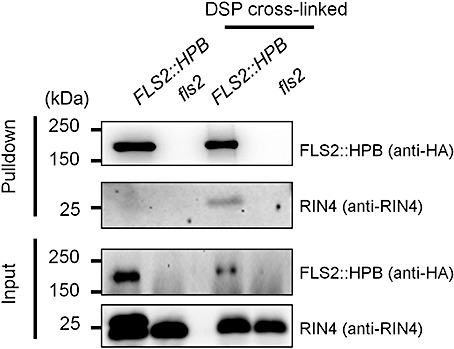
FLS2 and RIN4 may reside in the same protein complex. The pulldown assays were performed using solubilized membrane proteins [either without or with dithiobis(succinimidyl propionate) (DSP) cross‐linking] prepared from 5‐week‐old Arabidopsis plants of FLS2::HPB fls2 (FLS2::HPB) and fls2 (control). FLS2::HPB was pulled down with streptavidin beads. The pulldown and input samples were examined by immunoblotting with anti‐haemagglutinin (HA) and anti‐RIN4 antibodies. This experiment was performed three times with similar results.
DISCUSSION
We have demonstrated that the PRR FLS2 forms a complex with the R proteins RPS2, RPM1 and RPS5. For the pulldown assay results shown in 2, 3, 4, the tagged proteins were transgenically expressed from their own promoters (except for RPM1::HPB, for which the RPS2 promoter was used), which substantially reduces the possibility of artefactual associations as a result of misexpression of the proteins. We also used stringent wash conditions in the pulldown assay: 1% Nonidet P40 (NP40), which is known to solubilize the protein‐rich microdomains of membranes called lipid rafts (Rixon et al., 2004), and 0.5% sodium deoxycholate, which is an ionic detergent, in two different salt concentrations (150 and 50 mm NaCl). The fact that the membrane‐integrated protein RIN4 was barely pulled down by FLS2 without cross‐linking (Fig. 4) strongly suggests that such membrane microdomains were efficiently disrupted in our standard pulldown assay procedure, which does not include protein cross‐linking. It is likely that the protein complexes containing FLS2 and R proteins observed here were formed through strong physical associations.
RPS2, RPM1 and RPS5 are all PM‐localized R proteins of the coiled‐coil (CC)‐NB‐LRR class (Axtell and Staskawicz, 2003; Boyes et al., 1998; Dangl and Jones, 2001; Holt et al., 2005). We have discovered another characteristic common to these three R proteins: the ability to associate with the PTI receptor FLS2. Whether the physical association between two types of immune receptor is a general phenomenon is an intriguing question. Although FLS2 was not included among the RPS2 complex component candidates, one receptor‐like kinase (At4g08850) was (Qi and Katagiri, 2009). At4g08850 might be an unidentified PRR and, if so, PRRs other than FLS2 might form complexes with RPS2 and other R proteins.
What is the biological significance of PTI and ETI receptors residing in the same protein complex(es)? There are at least two hypotheses, which are not mutually exclusive. In the first hypothesis, such an association may be a result of the fact that PTI and ETI share signalling components. For example, recent work has shown that PTI and ETI share a highly overlapping signalling network (Tsuda et al., 2009). Residence in the same protein complex may simply be a fortuitous consequence, as both types of receptor physically associate with the common signalling components. Alternatively, there could be functional significance in having two types of receptor in a single protein complex: PTI and ETI signalling may interact with each other through this protein complex. For example, the activation of PTI signalling might sensitize ETI signalling. This is an attractive hypothesis, and it is likely that PTI and ETI signalling interact in some ways. However, without knowing the exact composition and functional modes of this presumptive protein complex, it is very difficult to pinpoint such signalling interactions occurring within the protein complex. Therefore, it will be important to elucidate details of the protein complex to evaluate its functional significance.
The second hypothesis is that different R proteins are physically associated with FLS2 because the R proteins ‘guard’ particular components in the FLS2–protein complex. It has been shown recently that RLCKs, including BIK1, PBS1 and other close members, are involved in FLS2 signalling by physically associating with FLS2 (Lu et al., 2010; Zhang et al., 2010). Thus, in the case of RPS5, it ‘guards’ PBS1 (Shao et al., 2003), which participates in FLS2 signalling (Zhang et al., 2010). The physical association between RPS5 and FLS2 suggests that RPS5, PBS1 and FLS2 may reside in the same protein complex, which is consistent with the ‘guard hypothesis’. In the case of RPS2 and RPM1, both ‘guard’ RIN4 (Axtell and Staskawicz, 2003; Mackey et al., 2002, 2003), which has been shown to be involved in the flg22‐induced response (Kim et al., 2005). Our results suggest the existence of ‘guard’ complexes containing FLS2, its signalling partners (PBS1 and RIN4) and R proteins ‘guarding’ the partners.
EXPERIMENTAL PROCEDURES
Constructs and plants
All the plants used in this study had the genetic background of accession Col‐0.
The BSK8 cDNA sequence without a stop codon was amplified with the primers At5g41260‐5 (ATGGGTTGTGAGGTTTCAAAGTTATCTGCA) and At5g41260‐3 (CAAAGGGTTTCTTTTGCTTTCAAGCAT) by reverse transcriptase‐polymerase chain reaction (RT‐PCR), cloned into pCR®8/GW/TOPO® (Invitrogen, Carlsbad, CA, USA) and recombined into the Gateway® destination vector pEG101 (Earley et al., 2006) to yield BSK8::YFP::HA expressed from the cauliflower mosaic virus (CaMV) 35S promoter or pMDC83 (Curtis and Grossniklaus, 2003) to yield BSK8::GFP expressed from the CaMV 35S promoter. Agrobacterium tumefaciens strain GV3101/pMP90 was transformed with both constructs. The transformed A. tumefaciens strain with pEG101‐BSK8 was used to transform a bsk8 mutant (SALK_090812) (Alonso et al., 2003) using the floral dip method (Clough and Bent, 1998). T1 transgenic plants were selected by spraying LIBERTY 200 SL herbicide (18.19% glufosinate ammonium; Bayer Cropscience, Kansas City, MO, USA) at a 1 : 2000 dilution in water. T3 homozygous plants were selected in the same manner.
The RPM1::Myc plant was prepared by Boyes et al. (1998). The following genotypes have been described previously in Qi and Katagiri (2009): RPS2::HPB line #1, RPM1::HPB line #2, FLS2‐HPB line #19, rpm1 rps2, fls2 and rps5.
Pulldown assay
Pulldown assay using Arabidopsis material
The procedure (either with or without DSP cross‐linking) was conducted as described previously, except that the amount of plant material was scaled down to 5 g per sample, except for the experiment in Fig. 2C (Qi and Katagiri, 2009).
The co‐immunoprecipitation experiment in Fig. 2C was performed similarly with the following exceptions. Ten grams per sample of plant material were used without cross‐linking. Fifty microlitres of anti‐c‐Myc monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA; clone 9E10) were added to each solubilized microsomal protein sample (2 mL) and incubated at 4 °C on a rotator for 2 h. Then, 50 µL of protein A/G agarose (Santa Cruz Biotechnology) was added to each sample, which was incubated for another 4 h. All the samples were spun down at 1000 g for 2 min, washed with RIPA buffer 1 [50 mm Tris/HCl, pH 7.4, 150 mm NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 1% NP40, 0.5% sodium deoxycholate, 1 mm phenylmethylsulphonylfluoride (PMSF)] twice and then RIPA buffer 2 [same as RIPA buffer 1, except that 50 mm (instead of 150 mm) NaCl was used] twice. The precipitated proteins were eluted by heating agarose in 50 µL of 1 × SDS sample buffer at 99 °C for 10 min.
Pulldown assay using N. benthamiana material
Equal amounts of cultured A. tumefaciens strain GV3101/pVP90 carrying different expression plasmids were suspended with 2‐(N‐morpholino)ethanesulphonic acid (MES) buffer (10 mm MES‐KOH, 10 mm MgCl2, 150 µm acetosyringone, pH 5.6) to a final optical density at 600 nm (OD600) of 0.4, so that the concentration of each strain was OD600= 0.2. For the negative control, the final OD600 was 0.2 as only one construct was used. The bacterial suspension was infiltrated into 4‐week‐old N. benthamiana leaves for transient expression. Two days later, 1 g of each infiltrated leaf sample was collected and quickly frozen in liquid nitrogen, and then ground with a mortar and pestle in 2 mL of extraction buffer [50 mm N‐2‐hydroxyethylpiperazine‐N′‐2‐ethanesulphonic acid (HEPES)‐KOH, pH 7.6, 150 mm NaCl, 1% NP40, 0.5% sodium deoxycholate, 1 mm dithiothreitol (DTT)] with protease inhibitors (1 tablet/10 mL Complete Mini® and 1 mm PMSF, 1 µg/mL leupeptin, 1 µg/mL pepstatin, 1 µg/mL E64; all from Roche, Indianapolis, IN, USA). The ground sample in extraction buffer (∼2 mL) was incubated at 4 °C on a rotator for 30 min for solubilization, followed by centrifugation at 16 000 g at 4 °C for 30 min to obtain the supernatant as a protein extract. Meanwhile, 100 µL per sample of Dynabeads® M‐280 (Invitrogen) were washed with extraction buffer three times and resuspended in 100 µL per sample of extraction buffer. Each protein extract was mixed with 100 µL prewashed Dynabeads® M‐280 and incubated on a rotator at 4 °C for 2 h. The streptavidin beads were then washed three times with RIPA buffer 1 and three times with RIPA buffer 2. The bead‐captured proteins were eluted by heating the beads in 60 µL of 1 × SDS sample buffer at 99 °C for 10 min.
Immunoblot
Protein samples of equal volume were resolved by 10% SDS‐PAGE, and then transferred to poly(vinylidene difluoride) (PVDF) membrane (Bio‐Rad, Hercules, CA, USA) by semi‐dry electrophoretic transfer using the TRANS‐BLOT® SD (Bio‐Rad) device. For detection of the proteins on the blot, the blots were cut into strips according to the expected molecular weight of the proteins, and the strips of the blots were probed with the corresponding antibodies separately. The following antibodies or reagents were used: anti‐c‐Myc monoclonal antibody (Santa Cruz Biotechnology, clone 9E10) at 1 : 200 dilution and goat anti‐mouse horseradish peroxidase (HRP) conjugate (Pierce, Rockford, IL, USA) at 1 : 5000 dilution; anti‐haemagglutinin (HA) high‐affinity monoclonal antibody (Roche, clone 3F10) at 1 : 500 dilution and goat anti‐rat immunoglobulin G (IgG) (H+I) HRP conjugate (Bethyl, Montgomery, TN, USA) at 1 : 5000 dilution; anti‐green fluorescent protein (GFP) polyclonal antibody (Invitrogen), anti‐FLS2 (Chinchilla et al., 2006) at 1 : 500 dilution, anti‐RIN4 (Mackey et al., 2002) at 1 : 2000 dilution and goat anti‐rabbit HRP conjugate (Pierce). SuperSignal® West Femto Maximum Sensitivity Substrate (Pierce) was used for detection and images were recorded using a chilled CCD‐camera.
Confocal microscopy
Rosette leaves from 5‐week‐old Arabidopsis plants were cut into approximately 5 × 5 mm2 squares and mounted between slides and cover glasses with water. The samples were then excited with a 514‐nm laser and signals were filtered through a yellow fluorescent protein (YFP) (543‐nm) filter, using an Eclipse C1si Spectral Imaging Confocal Microscope (Nikon, Melville, NY, USA). The images were collected using EZ‐C1 software (Nikon).
Supporting information
Fig. S1 Identification of BSK1 (A) and BSK8 (B) by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) in the RPS2‐HPB pulldown samples. This figure shows original database search results viewed in the Scaffold viewer ( http://www.proteomesoftware.com/Scaffold/Scaffold_viewer.htm). BSK1 protein was identified with 100% probability and BSK8 protein was identified with 94% probability (top left panel) based on the identification of two BSK1 peptides and one BSK8 peptide at a 95% probability level (top right panel).
Fig. S2 Detection of BSK8::YFP::HA protein from the p35S::BSK8::YFP::HA transgenic Arabidopsis line #2. Leaf tissues of a control bsk8 plant (SALK_090812) and a p35S::BSK8::YFP::HA #2 plant (in SALK_090812 background) were flash‐frozen in liquid nitrogen and ground to a fine powder. Then, 2 × Laemmli buffer [4% sodium dodecylsulphate (SDS), 20% glycerol, 10% 2‐mercaptoethanol, 0.004% bromophenol blue, 0.125 M Tris‐HCl, pH 6.8] was added at a ratio of 1 g tissue to 2 mL buffer. The samples were boiled for 6 min and centrifuged at 16 000 g for 10 min. The supernatant was employed for immunoblotting using anti‐green fluorescent protein (GFP) antibody. This experiment was performed twice with similar results.
Fig. S3 Myc::BSK8 forms a protein complex with FLS2::HPB in Nicotiana benthamiana. Myc::BSK8 and FLS2::HPB were transiently expressed in N. benthamiana leaves and a streptavidin bead‐based pulldown assay was performed. FLS2::HPB was pulled down with streptavidin beads. Nicotiana benthamiana leaves expressing only Myc::BSK8 were used as a negative control. Both pulldown samples and input samples were analysed by immunoblotting using anti‐haemagglutinin (HA) and anti‐Myc antibodies. This experiment was performed twice with similar results.
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Jeff Dangl (University of North Carolina, Chapel Hill, NC, USA) for the anti‐RIN4 antibody, rpm1 rps2 double mutant and RPM1‐Myc transgenic plants, David Mackey (Ohio State University, Columbus, OH, USA) for the fls2 mutant and Silke Robatzek (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for the anti‐FLS2 antibody. This work was supported by grant IOS‐0419648 (Arabidopsis 2010 program) to Jane Glazebrook and Fumiaki Katagiri, and grant MCB‐0918908 to Fumiaki Katagiri from the National Science Foundation. Yiping Qi was supported by a Hamm Memorial Graduate Student Fellowship and a PBS Doctoral Dissertation Fellowship from the University of Minnesota.
REFERENCES
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C.C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , Deen, H. , Geralt, M. , Hazari, N. , Hom, E. , Karnes, M. , Mulholland, C. , Ndubaku, R. , Schmidt, I. , Guzman, P. , Aguilar‐Henonin, L. , Schmid, M. , Weigel, D. , Carter, D.E. , Marchand, T. , Risseeuw, E. , Brogden, D. , Zeko, A. , Crosby, W.L. , Berry, C.C. and Ecker, J.R. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. (2005) Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6, 973–979. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J. and Staskawicz, B.J. (2003) Initiation of RPS2‐specified disease resistance in Arabidopsis is coupled to the AvrRpt2‐directed elimination of RIN4. Cell, 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Benschop, J.J. , Mohammed, S. , O'Flaherty, M. , Heck, A.J. , Slijper, M. and Menke, F.L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell Proteomics, 6, 1198–1214. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. , Kunkel, B.N. , Dahlbeck, D. , Brown, K.L. , Schmidt, R. , Giraudat, J. , Leung, J. and Staskawicz, B.J. (1994) RPS2 of Arabidopsis thaliana: a leucine‐rich repeat class of plant disease resistance genes. Science, 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C. , Nam, J. and Dangl, J.L. (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA, 95, 15 849–15 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Bauer, Z. , Regenass, M. , Boller, T. and Felix, G. (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell, 18, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Boller, T. and Robatzek, S. (2007a) Flagellin signalling in plant immunity. Adv. Exp. Med. Biol. 598, 358–371. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nurnberger, T. , Jones, J.D. , Felix, G. and Boller, T. (2007b) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gohre, V. , Spallek, T. , Haweker, H. , Mersmann, S. , Mentzel, T. , Boller, T. , de Torres, M. , Mansfield, J.W. and Robatzek, S. (2008) Plant pattern‐recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol. 18, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cells, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Grant, M.R. , Godiard, L. , Straube, E. , Ashfield, T. , Lewald, J. , Sattler, A. , de Torres, M. , Mansfield, J.W. and Robatzek, S. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science, 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Holt, B.F., 3rd , Belkhadir, Y. and Dangl, J.L. (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science, 309, 929–932. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kim, M.G. , Cunha, L. , McFall, A.J. , Belkhadir, Y. , DebRoy, S. , Dangl, J.L. and Mackey, D. (2005) Two Pseudomonas syringae type III effectors inhibit RIN4‐regulated basal defense in Arabidopsis. Cell, 121, 749–759. [DOI] [PubMed] [Google Scholar]
- Li, J. , Wen, J. , Lease, K.A. , Doke, J.T. , Tax, F.E. and Walker, J.C. (2002) BAK1, an Arabidopsis LRR receptor‐like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell, 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Lu, D. , Wu, S. , Gao, X. , Zhang, Y. , Shan, L. and He, P. (2010) A receptor‐like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA, 107, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Holt, B.F. , Wiig, A. and Dangl, J.L. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1‐mediated resistance in Arabidopsis. Cell, 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Mackey, D. , Belkhadir, Y. , Alonso, J.M. , Ecker, J.R. and Dangl, J.L. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2‐mediated resistance. Cell, 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Medzhitov, R. and Janeway, C.A., Jr (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell, 91, 295–298. [DOI] [PubMed] [Google Scholar]
- Mindrinos, M. , Katagiri, F. , Yu, G.L. and Ausubel, F.M. (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide‐binding site and leucine‐rich repeats. Cell, 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Nam, K.H. and Li, J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell, 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Qi, Y. and Katagiri, F. (2009) Purification of low‐abundance Arabidopsis plasma‐membrane protein complexes and identification of candidate components. Plant J. 57, 932–944. [DOI] [PubMed] [Google Scholar]
- Rixon, H.W. , Brown, G. , Aitken, J. , McDonald, T. , Graham, S. and Sugrue, R.J. (2004) The small hydrophobic (SH) protein accumulates within lipid‐raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85, 1153–1165. [DOI] [PubMed] [Google Scholar]
- Robatzek, S. , Chinchilla, D. and Boller, T. (2006) Ligand‐induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F. , Golstein, C. , Ade, J. , Stoutemyer, M. , Dixon, J.E. and Innes, R.W. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science, 301, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H. , Karlowski, W.M. , Pan, R. , Tzeng, Y.H. , Mayer, K.F. and Li, W.H. (2004) Comparative analysis of the receptor‐like kinase family in Arabidopsis and rice. Plant Cell, 16, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W. , Kim, T.W. , Oses‐Prieto, J.A. , Sun, Y. , Deng, Z. , Zhu, S. , Wang, R. , Burlingame, A.L. and Wang, Z.Y. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science, 321, 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. and Katagiri, F. (2010) Comparing signaling mechanisms engaged in pattern‐triggered and effector‐triggered immunity. Curr. Opin. Plant Biol. 13, 459–565. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, R.F. , Henk, A. , Mowery, P. , Holub, E. and Innes, R.W. (1998) A mutation within the leucine‐rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell, 10, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Li, W. , Xiang, T. , Liu, Z. , Laluk, K. , Ding, X. , Zou, Y. , Gao, M. , Zhang, X. , Chen, S. , Mengiste, T. , Zhang, Y. and Zhou, J.M. (2010) Receptor‐like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe, 7, 290–301. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Identification of BSK1 (A) and BSK8 (B) by liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) in the RPS2‐HPB pulldown samples. This figure shows original database search results viewed in the Scaffold viewer ( http://www.proteomesoftware.com/Scaffold/Scaffold_viewer.htm). BSK1 protein was identified with 100% probability and BSK8 protein was identified with 94% probability (top left panel) based on the identification of two BSK1 peptides and one BSK8 peptide at a 95% probability level (top right panel).
Fig. S2 Detection of BSK8::YFP::HA protein from the p35S::BSK8::YFP::HA transgenic Arabidopsis line #2. Leaf tissues of a control bsk8 plant (SALK_090812) and a p35S::BSK8::YFP::HA #2 plant (in SALK_090812 background) were flash‐frozen in liquid nitrogen and ground to a fine powder. Then, 2 × Laemmli buffer [4% sodium dodecylsulphate (SDS), 20% glycerol, 10% 2‐mercaptoethanol, 0.004% bromophenol blue, 0.125 M Tris‐HCl, pH 6.8] was added at a ratio of 1 g tissue to 2 mL buffer. The samples were boiled for 6 min and centrifuged at 16 000 g for 10 min. The supernatant was employed for immunoblotting using anti‐green fluorescent protein (GFP) antibody. This experiment was performed twice with similar results.
Fig. S3 Myc::BSK8 forms a protein complex with FLS2::HPB in Nicotiana benthamiana. Myc::BSK8 and FLS2::HPB were transiently expressed in N. benthamiana leaves and a streptavidin bead‐based pulldown assay was performed. FLS2::HPB was pulled down with streptavidin beads. Nicotiana benthamiana leaves expressing only Myc::BSK8 were used as a negative control. Both pulldown samples and input samples were analysed by immunoblotting using anti‐haemagglutinin (HA) and anti‐Myc antibodies. This experiment was performed twice with similar results.
Supporting info item
Supporting info item
Supporting info item


