SUMMARY
Fumonisins comprise a class of carcinogenic mycotoxins produced by Fusarium verticillioides during colonization of maize kernels. In previous work, we identified ZFR1, which is predicted to encode a Zn(II)2Cys6 zinc finger transcription factor required for fumonisin B1 (FB1) production during growth on kernels. In this study, we characterized the role of ZFR1 in colonizing maize kernels and inducing FB1 biosynthesis. The ZFR1 deletion strain (Δzfr1) grew approximately 2.5‐fold less than the wild‐type on endosperm tissue and a variety of other carbon sources, including glucose and amylopectin. However, the Δzfr1 strain displayed higher α‐amylase activity and expression of genes involved in starch saccharification than the wild‐type, thus indicating that the reduced growth of the Δzfr1 strain was not due to inhibition of amylolytic enzymes. In the wild‐type strain, expression of six genes encoding putative sugar transporters was significantly greater on endosperm tissue than on germ tissue, and expression of at least three of the six genes was negatively affected by disruption of ZFR1. Intriguingly, disruption of FST1 had no effect on growth, kernel colonization or kernel pH but decreased FB1 production by approximately 82% on maize kernels. Based on these findings, we hypothesize that ZFR1 controls FB1 biosynthesis by regulating genes involved in the perception or uptake of carbohydrates.
INTRODUCTION
Fusarium verticillioides (Sacc.) Nirenberg (teleomorph: Gibberella moniliformis Wineland, synonym Gibberella fujikuroi mating population A) is a ubiquitous fungal pathogen that causes ear, kernel, seedling and stem rots of maize (Zea mays). During colonization of maize kernels, F. verticillioides produces a group of structurally related polyketide mycotoxins collectively referred to as fumonisins. FB1, the predominant fumonisin produced by F. verticillioides (Vismer et al., 2004), is a potent inhibitor of ceramide synthase (Desai et al., 2002) and thus disrupts sphingolipid biosynthesis. Ingestion of FB1 results in leukoencephalomalacia in horses and has been implicated as a cause of oesophageal cancer and birth defects in humans (Hendricks, 1999; Joffe, 1986). Because of health concerns related to consumption of FB1, regulatory agencies throughout the world have established guidelines regarding acceptable levels of FB1 in grains and food products.
FB1 biosynthesis is influenced by components of the maize kernel (Shim et al., 2003). One such component is starch, which changes dramatically in the kernel as it approaches maturity (Ingle et al., 1965). Immature kernels lacking starch do not support FB1 biosynthesis or expression of fumonisin biosynthetic (FUM) genes, but as kernels approach maturity and accumulate starch, they support high levels of FB1 production (Bluhm and Woloshuk, 2005; Warfield and Gilchrist, 1999). Growth of F. verticillioides is comparable throughout each stage of kernel development (Bluhm and Woloshuk, 2005), indicating that immature kernels either repress FB1 biosynthesis or lack the ability to induce FUM gene expression. In mature kernels, the starch‐rich tissue of the endosperm supports 10‐ to 20‐fold higher levels of FB1 biosynthesis than germ tissue (Shim et al., 2003), and disruption of the α‐amylase gene of F. verticillioides results in low levels of FB1 biosynthesis on endosperm fractions (Bluhm and Woloshuk, 2005). Analyses of FB1 production in maize kernel mutants and in defined liquid media indicate that amylopectin, a component of starch, supports high levels of FB1 biosynthesis (Bluhm and Woloshuk, 2005).
In a previous study, we identified ZFR1, a gene encoding a putative zinc‐finger transcription factor required for wild‐type levels of FB1 production on maize kernels (Flaherty and Woloshuk, 2004). In the present study, we investigated the molecular mechanisms through which ZFR1 regulates colonization of kernel tissues and FUM gene transcription. We evaluated the role of ZFR1 in regulating four distinct aspects of kernel colonization: growth in the presence of specific carbohydrates found in maize kernels, hydrolysis of starch, perception of glucose, and expression of putative mono‐ and oligo‐saccharide transporters. From these experiments, we identified FST1, a putative sugar transporter regulated by ZFR1 that is required for FB1 biosynthesis but not growth during kernel colonization. The discovery that ZFR1 regulates sugar transporter genes that in turn affect fumonisin biosynthesis during kernel colonization reveals a possible linkage between the perception of starch and the induction of fumonisin biosynthesis and provides new insight into the role of sugar transport proteins in plant pathogenesis.
RESULTS
Growth of the Δzfr1 mutant on carbohydrates found in maize kernels
Although visual assessments indicated that the Δzfr1 strain grew at approximately wild‐type levels on whole kernels, ergosterol analysis indicated the mutant grew 20% less than the wild‐type strain (Table 1). On endosperm fractions of kernels, growth of the Δzfr1 strain was approximately 60% less than wild‐type (Table 1). In contrast, the Δzfr1 strain grew greater than twice as much as wild‐type in germ fractions of kernels (Table 1), indicating that the growth phenotype of the mutant is strongly influenced by kernel tissue. Growth of the Δzfr1 strain was substantially less than wild‐type when provided with glucose, maltose, dextrin or amylopectin as the sole carbon source (Fig. 1). Together, these observations suggested that the Δzfr1 strain is impaired in the perception or utilization of carbohydrates.
Table 1.
Growth of the Δzfr1 strain on germ and endosperm fractions of maize kernels.
| Fungal strain | Growth (ergosterol)* | ||
|---|---|---|---|
| Whole kernel | Germ | Endosperm | |
| Wild type | 110 ± 11 | 160 ± 17 | 100 ± 9 |
| Δzfr1 | 90 ± 7 | 370 ± 33 | 40 ± 5 |
Measurements were taken after 7 days of growth. Data were reported as micrograms of ergosterol per gram of kernel tissue ± the standard error of the mean for three samples.
Figure 1.
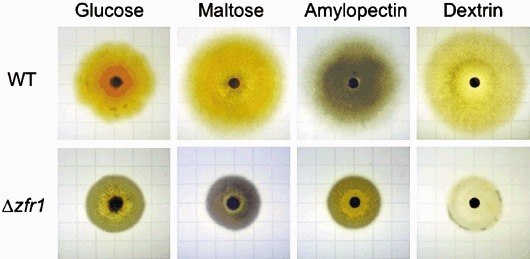
Growth of the wild‐type (WT) and Δzfr1 strains on minimal media containing either glucose, maltose, amylopectin or dextrin as the sole carbon source. Each square of the background grid represents 14 × 14 mm. Pictures were taken after 4 days of growth.
Glucokinase activity in the Δzfr1 strain
In many organisms, hexokinases regulate carbohydrate metabolism by phosphorylating glucose and other hexose sugars and thus function as sugar sensors. To determine whether impairment of hexokinase activity accounted for the reduced growth of the Δzfr1 strain in the presence of carbohydrates, we measured the expression of putative hexokinase‐encoding genes and glucokinase activity. Homology‐based searches of the F. verticillioides genome revealed the presence of at least two genes encoding putative hexokinases (HXK1 and HXK2). HXK1 and HXK2 were expressed at low levels in both the wild‐type and Δzfr1 strains on germ tissue (Table 2). However, on the starch‐rich tissue of the endosperm, expression of HXK1 and HXK2 increased by approximately 120‐ and 27‐fold, respectively, in the wild‐type, and by approximately 45‐ and 18‐fold, respectively, in the Δzfr1 strain (Table 2). Consistent with these findings, glucokinase activity was substantially higher in both strains grown on endosperm tissue as compared with germ tissue with no significant differences observed between the two strains (Table 2). From these observations, we concluded that disruption of ZFR1 has a negligible effect on carbohydrate perception by hexokinases.
Table 2.
Glucokinase expression and activity in the wild‐type and Δzfr1 strains grown on germ and endosperm fractions of maize kernels.
| Fungal strain | HXK1 expression* | HXK2 expression | Glucokinase activity† | |||
|---|---|---|---|---|---|---|
| Germ | Endosperm | Germ | Endosperm | Germ | Endosperm | |
| Wild‐type | 1.0 (0.7–1.3) | 120 (100–150) | 1.0 (0.7–1.3) | 27.3 (27.0–27.7) | 2.0 ± 0.2 | 131 ± 4.9 |
| Δzfr1 | 1.5 (1.3–1.9) | 45 (40–50) | –0.7 (–0.3 to –1.1) | 18.2 (18.0–18.3) | 1.7 ± 0.4 | 119 ± 3.7 |
As analysed by qPCR. Expression of HXK1 and HXK2 was normalized to TUB2 and was calculated relative to expression in wild‐type growing on germ tissue. Data represent fold differences in expression. The relative expression of each gene was calculated as 2ΔΔCt. The range of expression, in parentheses, for each gene = (2ΔΔCt–s–2ΔΔCt+s), where s = the standard deviation of the ΔΔCt value.
Data are reported as 1 × 10−4 units of glucokinase activity per gram total protein ± the standard error of the mean for three samples.
Expression of genes encoding amylolytic enzymes in the Δzfr1 strain
To test the possibility that the Δzfr1 strain is impaired in its ability to hydrolyse starch, we measured the expression of six genes predicted to encode enzymes involved in starch saccharification. In both the wild‐type and the Δzfr1 mutant, expression of all six genes was low when grown on germ tissue (Fig. 2A). Surprisingly, on endosperm tissue at least three amylolytic genes were upregulated in the Δzfr1 strain compared with the wild‐type, including the α‐amylase AMY1 (three‐fold higher), a putative glucoamylase designated GMY1 (eight‐fold higher) and a putative maltase designated MLT1 (two‐fold higher) (Fig. 2A).
Figure 2.
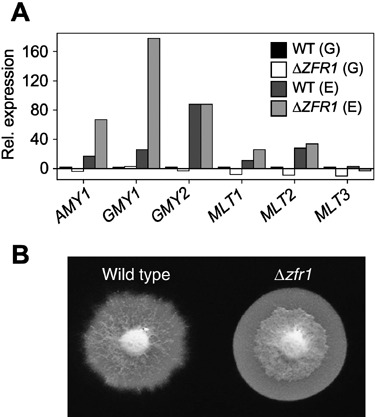
Expression and activity of amylolytic genes in the wild‐type and Δzfr1 strains. (A) Expression of an a‐amylase (AMY1), two glucoamylases (GMY1 and GMY2), and three maltases (MLT1, MLT2 and MLT3) was compared between the strains after 6 days of growth on germ (G) and endosperm (E) tissues of mature maize kernels. For each gene, expression was normalized to TUB2 expression and calculated as fold‐changes in expression relative to expression in wild‐type growing on germ tissue. (B) Amylase activity was measured by staining starch plates with iodine after 3 days of growth. Unstained areas are indicative of starch saccharification.
To determine whether the increased levels of expression of amylolytic genes in the Δzfr1 strain corresponded to higher levels of starch hydrolysis, we grew the wild‐type and Δzfr1 on plates containing amylose. By staining plates with an iodine solution, the amount of starch hydrolysis was determined qualitatively; unstained areas of the plate result from enzymatic degradation of amylose. Consistent with higher levels of amylolytic enzyme activity, the amount of clearing beneath mycelia of the Δzfr1 strain was much greater than beneath wild‐type colonies (Fig. 2B). These results indicated that the Δzfr1 strain is able to perceive and hydrolyse starch at levels equal to or greater than the wild‐type strain.
Regulation of putative sugar transporter genes by kernel tissue and zfr1
Observations that the Δzfr1 strain was impaired in growth on carbohydrates despite high levels of amylolytic and hexokinase activity suggested that ZFR1 may regulate sugar uptake. In fungi, sugar uptake is facilitated in large part by membrane‐bound transporters belonging to the major facilitator superfamily (MFS) (Pao et al., 1998). The genome of F. verticillioides contains at least 381 genes predicted to encode MFS proteins, of which 147 share significant amino acid similarity with known sugar transporters. Using microarrays described previously (Sagaram et al., 2006), we were able to measure the expression profiles of 50 of the 147 putative sugar transporter genes in the wild‐type strain during growth on germ and endosperm tissues. Surprisingly, although endosperm tissues of kernels are rich in carbohydrates, only six putative sugar transporters were more highly expressed during growth on endosperm tissue (Fig. 3A). To verify the expression data obtained from microarrays, we arbitrarily selected three putative sugar transporter genes (FST1, FST2 and FST4) for expression analysis by quantitative PCR (qPCR). Expression of FST1, FST2 and FST4 was two‐ to three‐fold higher in the wild‐type strain during growth on endosperm tissue compared with germ tissue (Fig. 3B). To determine if ZFR1 regulates the expression of sugar transporters, we measured the expression of FST1, FST2 and FST4 on endosperm tissue. On germ tissue, transcription of FST1, FST2 and FST4 was reduced in the Δzfr1 strain by approximately two‐ to five‐fold (Fig. 3B). On endosperm tissue, expression of FST1 and FST2 was reduced by approximately two‐fold in the Δzfr1 strain, and expression of FST4 was reduced by approximately 20‐fold (Fig. 3B). These results showed that ZFR1 regulated the expression of sugar transporters during kernel colonization.
Figure 3.
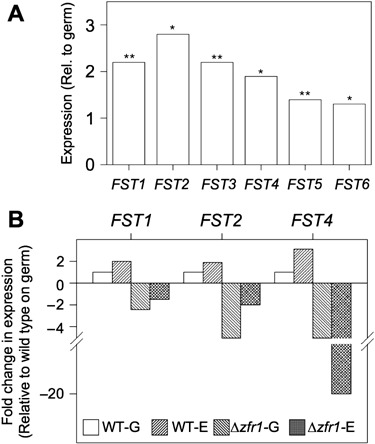
Expression of putative sugar transporter genes in the wild‐type strain during colonization of maize kernels. (A) Microarray analysis of six sugar transporter genes differentially expressed in germ and endosperm tissues. For each gene, expression was normalized to TUB2 by dividing the spot intensity of the gene by the intensity recorded for TUB2. Measurements were taken after 4 days of growth on kernel tissues. Asterisks indicate a P value < 0.05 (*) or < 0.01 (**). (B) qPCR verification of microarray results and analysis of sugar transporter expression in the Δzfr1 strain. Expression of FST1, FST2 and FST4 was measured in wild‐type growing on germ (WT‐G) and endosperm (WT‐E) and in the Δzfr1 strain growing on germ (Δzfr1‐G) and endosperm (Δzfr1‐E). For each gene, expression was normalized to TUB2 expression and calculated as fold‐changes in expression relative to expression in wild‐type growing on germ tissue.
Growth and FB1 biosynthesis in a Δfst1 strain
To determine the function of FST1 during kernel colonization, we disrupted the gene through double homologous recombination (Fig. 4). Growth of the Δfst1 strain was similar to the wild‐type and complemented strains when provided glucose or maltose as the primary carbon source (Fig. 5), thus suggesting that the Δfst1 strain was not significantly impaired in the uptake or utilization of either carbohydrate. On whole kernels of maize, growth of the Δfst1 strain was slightly higher than either the wild‐type or complemented strain (Table 3), which indicates that FST1 is dispensable for kernel colonization. Additionally, disruption of FST1 had no effect on kernel acidification after colonization (Table 3).
Figure 4.
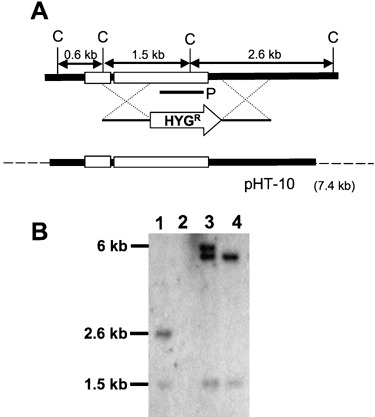
Disruption and complementation of FST1. (A) A hygromycin‐resistance cassette (HYGR) was inserted into the FST1 gene by homologous recombination to create a Δfst1 strain. For complementation, protoplasts of Δfst1 were co‐transformed with pHT‐10 (constructed by cloning the FST1 locus into the pGEM‐T‐Easy vector; dashed lines represent vector sequence) and pKS‐GEN, a vector harbouring a geneticin resistance cassette. For Southern analysis, genomic DNA was digested with ClaI (C) and probed with a 473‐bp region of FST1 (P). (B) The presence or absence of FST1 was verified by Southern analysis in the wild‐type (lane 1), Δfst1 (lane 2), FST‐COMP1 (lane 3) and FST‐COMP2 (lane 4) strains. Integration of pHT‐10 was ectopic in the transformants, and the FST‐COMP1 strain appeared to contain two copies of the complementation construct and thus was not used in subsequent experiments.
Figure 5.
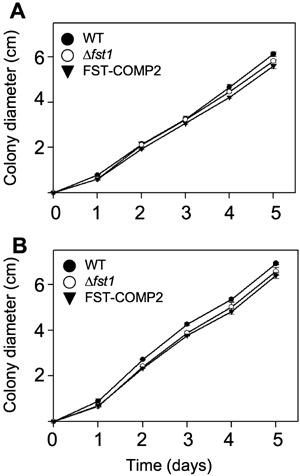
Growth of the wild‐type, Δfst1, and FST‐COMP2 strains with either glucose (A) or maltose (B) as the sole carbon source. Radial growth was measured for three individual colonies of each strain. Error bars represent the standard error of the mean.
Table 3.
Growth, FB1 production, and change in kernel pH of the Δfst1 strain.*
| Strain | FB1 † | Ergosterol‡ | pH (ΔpH)§ |
|---|---|---|---|
| Wild type | 111 ± 9 | 293 ± 15 | 4.8 ± 0.1 (–0.7) |
| Δfst1 | 20 ± 8 | 381 ± 88 | 5.0 ± 0.3 (–0.5) |
| FST‐COMP2 | 100 ± 6 | 300 ± 58 | 5.0 ± 0.2 (–0.5) |
All measurements were taken from three experimental replicates after 10 days of growth on whole kernels of maize.
As measured in micrograms of FB1 per gram of maize, ± standard error.
As measured in micrograms of ergosterol per gram of maize, ± standard error.
ΔpH = [(the pH of the uninoculated material)–(the average pH after 10 days of growth)].
To determine if FST1 affects fumonisin biosynthesis during kernel colonization, we measured production of FB1 by the wild‐type, Δfst1 and complemented strains after 10 days of growth on mature maize kernels. Despite robust growth on kernels, the Δfst1 strain produced approximately five‐fold less FB1 than the wild‐type strain (Table 3). Production of FB1 was restored in the complemented strain, thus confirming that the reduction in FB1 biosynthesis is fully attributable to disruption of FST1 (Table 3).
DISCUSSION
The discovery and characterization of ZFR1 represented an important advance in understanding the genetic regulation of fumonisin biosynthesis during maize kernel colonization. However, the manner in which ZFR1 regulates fumonisin biosynthesis is not fully understood. In many fungi, genes encoding enzymes that produce a specific class of secondary metabolites are physically grouped into clusters, and frequently these clusters contain a gene encoding a pathway‐specific transcriptional regulator. Although ZFR1 is predicted to encode a transcriptional regulator, it is not located within the FUM gene cluster, and recently a cryptic gene (FUM21) was discovered within the FUM cluster that functions as a pathway‐specific regulator of FUM gene transcription (Brown et al., 2007). Therefore, the regulation of fumonisin biosynthesis by ZFR1 during kernel colonization is most probably an indirect consequence of other changes in the transcriptome of the fungus. Consistent with this hypothesis, the Δzfr1 strain grew less than the wild‐type on endosperm fractions of mature kernels as well as various carbohydrates such as glucose and maltose. Initially, this finding suggested that the Δzfr1 strain was defective in the perception or utilization of starch, or possibly its amylolytic breakdown products such as maltose and glucose, in the endosperm tissue. However, our observations indicate that the Δzfr1 mutant perceives extracellular starch in the kernel environment. Expression of amylolytic genes in the Δzfr1 strain was substantially higher when grown on endosperm tissue than on germ tissue; unexpectedly, amylase activity was higher in the mutant than in the wild‐type. The elevated levels of gene expression and amylolytic activity suggest that the Δzfr1 mutant is able to perceive and hydrolyse extracellular starch, but either cannot perceive or fully utilize the products of hydrolysis. Additionally, disruption of ZFR1 had little effect on overall hexokinase activity or on the induction of HXK1 and HXK2 on endosperm tissue, thus leading us to conclude that the phenotypes of the Δzfr1 mutant are not caused by aberrant hexokinase expression or activity during kernel colonization. However, we cannot completely dismiss the possibility that HXK1 and HXK2 function as sugar sensors in F. verticillioides or that ZFR1 is involved in regulating their expression. To clarify further the roles of HXK1 and HXK2 in fumonisin biosynthesis and kernel colonization, functional studies of these two genes are currently underway.
Although the molecular aspects of glucose sensing and uptake have been studied extensively in yeast, less is known about these pathways in filamentous fungi. Twenty sugar transporters have been characterized in Saccharomyces cerevisae (Boles and Hollenberg, 1997), all of which belong to the sugar porter (SP) subfamily of the MFS. In yeast and filamentous fungi, glucose transport is facilitated by both constitutive low‐affinity and glucose‐repressible high‐affinity systems that probably rely on high‐ and low‐affinity glucose transporters such as HGT‐1 (Xie et al., 2004). In addition to transporting sugars across the plasma membrane, some members of the SP subfamily function as sugar sensors, including SNF3 and RGT2 in yeast, and RCO‐3 in Neurospora crassa (Madi et al., 1994). A domain search of the genome of F. verticillioides reveals at least 147 putative genes with significant similarity to sugar transporters, none of which has been characterized as to function or substrate specificity. Of the 50 putative sugar transporter genes of F. verticillioides available for microarray analysis, six showed higher levels of expression on endosperm tissue, including genes with high homology to hexose transporters, maltose transporters and other oligosaccharide transporters. Intriguingly, and at least three of these putative transporters induced on endosperm tissue were down‐regulated in the Δzfr1 mutant. Based on these observations, we conclude that ZFR1 is required for the expression of a crucial subset of the genes involved in perceiving or acquiring sugars. The exact regulatory mechanism through which ZFR1 functions, however, is not clear. ZFR1 may interact directly with the promoters of genes encoding sugar transport proteins, or conversely may play a more global role in regulating carbohydrate metabolism. It is also important to note that the microarray contained probes corresponding to only 34% of the putative sugar transporter genes in F. verticillioides and that 12% of the 50 genes that were represented had increased gene expression, thus making it likely that other sugar transporter genes also show increased expression on maize kernel endosperm. Future experiments will focus on identifying additional sugar transport genes involved in the regulation of secondary metabolism as well as determining the effect of ZFR1 on the transcriptome of F. verticillioides during kernel colonization.
With respect to fumonisin biosynthesis, we hypothesize that one or more sugar transporter homologues of F. verticillioides sense specific molecules involved in pathway induction. Several lines of evidence suggest that FST1 plays such a role during kernel colonization. Expression of FST1 is induced by endosperm tissue, the fraction of maize kernels that supports the highest level of FB1 biosynthesis. Disruption of FST1 significantly reduced FB1 biosynthesis on maize kernels, but had negligible effects on growth or kernel acidification during colonization. Also, the Δfst1 strain displayed no reduction in growth in defined media containing glucose or maltose as the sole carbon source, thus indicating that its role in uptake is negligible or compensated for by other transporters. Together, these observations suggest that disruption of FST1 does not cause an overall reduction in sugar uptake, but rather a deficiency in specific aspects of sugar signalling. In earlier work, we found that amylopectin induces high levels of fumonisin biosynthesis during kernel colonization (Bluhm and Woloshuk, 2005). Our current effort is to test the hypothesis that FST1 functions to sense specific breakdown products of starch during kernel colonization and activates as yet undescribed signal transduction pathways that induce fumonisin biosynthesis.
This study has shown that the uptake of sugars into the fungal cell is likely to be a central component of kernel colonization and the induction of fumonisin biosynthesis in F. verticillioides, and that ZFR1 may affect fumonisin biosynthesis by regulating sugar uptake or perception. A thorough analysis of genes involved in sugar sensing and transport is anticipated to shed new light on pathogenesis and the accumulation of mycotoxins in maize kernels. For example, breeders of maize have documented a lack of correlation between disease severity of F. verticillioides and fumonisin accumulation; often, asymptomatic kernels or ears contain relatively high levels of fumonisins. Further studies are needed to determine if the regulation of sugar transporters, possibly by ZFR1, plays a role in this phenomenon.
CONCLUSIONS
The regulation of fumonisin biosynthesis during maize kernel colonization is complex and poorly understood at the molecular level. The evidence presented in this study indicates that Δzfr1 strains display reduced growth on maize kernel endosperm tissue and a variety of carbohydrates including starch and breakdown products of starch.
Based on our findings, we hypothesize that ZFR1 is required for the expression of FST1 and other sugar transporters during kernel colonization. Our observation that FST1 is required for fumonisin biosynthesis but not growth during kernel colonization suggests that FST1 functions as a carbohydrate sensor and thus a key linkage between primary and secondary metabolism. This study provides new information on the function of sugar transporters in mycotoxigenesis and could provide novel approaches to control the accumulation of fumonisins in maize.
EXPERIMENTAL PROCEDURES
Fungal strains and culture conditions
F. verticillioides strain 7600 (Fungal Genetics Stock Center, University of Kansas Medical School, Kansas City, KS) was used as wild‐type in this study. The mutant strain Δzfr1 was described previously (Flaherty and Woloshuk, 2004). Mutant strain Δfst1 and complemented strains FST‐COMP1 and FST‐COMP2 were generated for this study as described below. Cultures were maintained on potato dextrose agar (PDA; B&D, Sparks, MD). Cracked maize kernels and kernel fractions were prepared as described by Shim et al. (2003). Radial growth was measured on BSAL‐agar media [0.1 m KH2PO4, pH 4.5; 0.01 m MgSO4; 0.1 m NaCl; 0.1% bovine serum albumin; 2% (w/v) of a primary carbon source consisting of either glucose, maltose, amylopectin or dextrin].
Quantification of fungal growth
Radial growth measurements were performed as described by Bluhm and Woloshuk (2006). The ergosterol content of kernels or liquid cultures was analysed as a quantitative measure of fungal growth (Bluhm and Woloshuk, 2005; Seitz et al., 1979). Ground kernels (100–400 mg) were extracted overnight at room temperature in 2 : 1 chloroform/methanol. Liquid cultures (10 mL) were pelleted by centrifugation, dried on sterile filter paper, ground in liquid nitrogen with a mortar and pestle, and extracted in 1 mL of 2 : 1 chloroform/methanol. Ergosterol analysis was performed on an HPLC system with UV detection at 282 nm (Beckman Coulter, Fullerton, CA) and a 4.6 U ODS column (200 × 4.6 mm, Beckman Coulter). Compounds were eluted with 100% methanol at a flow rate of 1.0 mL/min. Quantities were determined by comparing peak areas of samples to a standard curve generated from HPLC‐grade ergosterol (Sigma, St. Louis, MO).
Nucleic acid isolation and analysis
Bacterial plasmids were isolated with a Qiagen miniprep DNA purification system (Qiagen, Valencia, CA). Fungal genomic DNA was collected from cultures grown in YEPD media as described by Woloshuk et al. (1994). For Southern blot analysis, genomic DNA was digested with ClaI, fractionated by electrophoresis through 0.7% agarose gels and transferred to nylon membranes (Nytran; Schleicher and Schuell, Keene, NH). Probes were radiolabelled with the Rediprime II random prime labelling system (Amersham Pharmacia Biotech Inc., Piscataway, NJ) and hybridizations were performed as described by Flaherty et al. (2003). For analyses of gene expression, total RNA was extracted from maize kernels as follows: ground colonized kernels (0.5–1 g) were suspended in 10 mL water‐equilibrated phenol (pH 4.0) to which 10 mL Tris‐HCl (pH 8.0) was added. Phases were separated by centrifugation (10 min at 12 000 g, 4 °C) and the aqueous phase was subsequently extracted with 10 mL phenol/chloroform (1 : 1) and 10 mL chloroform/isoamyl alcohol (24 : 1). RNA was precipitated overnight at –20 °C after the addition of ethanol to a final volume of 70%. Precipitated RNA was pelleted by centrifugation (30 min at 12 000 g, 4 °C), air‐dried briefly and purified with the RNeasy RNA purification kit (Qiagen).
Identification of genes predicted to function in sugar sensing, amylolysis or sugar transport
Homology‐based analyses of expressed sequence tags (ESTs) (http://compbio.dfci.harvard.edu/tgi) and genomic sequence of F. verticillioides (http://www.broad.mit.edu) were used to identify two genes predicted to function in sugar sensing (HXK1, GenBank accession no. EU247513; HXK2, GenBank accession no. EU247514) and six genes involved in starch hydrolysis (AMY1, GenBank accession no. DQ143884; GMY1, GenBank accession no. EU247508; GMY2, GenBank accession no. EU247509; MLT1, GenBank accession no. EU247510; MLT2, GenBank accession no. EU247511; and MLT3, GenBank accession no. EU247512). The genome of F. verticillioides is also predicted to contain 381 genes encoding MFS proteins, 147 of which are homologous to sugar transporters in yeast and other filamentous fungi. Of these 147 genes, 50 were represented on microarrays including FST1 (GenBank accession no. EU152990), FST2 (GenBank accession no. EU152991), FST3 (GenBank accession no. EU152992), FST4 (GenBank accession no. EU152993), FST5 (GenBank accession no. EU152994) and FST6 (GenBank accession no. EU152995).
Determination of glucokinase activity
Germ and endosperm fractions of maize kernels (Beck's 5322) prepared as described above were inoculated with wild‐type or the Δzfr1 strain of F. verticillioides. After 6 days, kernels were harvested as described above. Glucokinase activity was determined essentially as described by Olsson et al. (2003). Briefly, ground colonized kernels were extracted for 10 min in buffer AT (50 mm Tris‐HCl, pH 7.8, 5 mm MgCl2, 1 mm EDTA, 1% Trition X‐100). Total protein concentration for each extract was determined with Bradford reagent (Sigma) and volumes were subsequently adjusted for each sample to achieve identical concentrations in assay buffer consisting of 100 mm HEPES‐KOH, pH 7.8, 10 mm MgCl2, 2 mm ATP, 0.8 mm NADP, 0.2 U glucose‐6‐phosphate dehydrogenase (Sigma) and 10 mm glucose (Sigma). After incubation at 30 °C for 20 min, the A340 was measured for each sample. One unit of glucokinase activity was determined as the formation of 1 µmol NADPH/min. The concentration of NADPH was calculated based on an extinction coefficient of A = 6200/mol. To determine amylase activity, strains were grown on starch plates (0.3 m NaNO3, 6 mm K2HPO4, 4 mm MgSO4, 7 mm KCl, 66 µm FeSO4 and 2% amylose) and stained with an iodine solution (0.5% iodine and 5% potassium iodine) to visualize α‐amylase activity as described by Fakhoury and Woloshuk (1999).
Microarray analysis of gene expression
Germ and endosperm fractions of mature maize kernels (Beck's 5322) were separated as described by Shim et al. (2003). Five grams of each fraction were placed in 20 mL scintillation vials (RPI, Mt. Prospect, IL), autoclaved and inoculated with 1 × 105 conidia of wild‐type F. verticillioides. After 4 days of growth, vials were flash‐frozen in liquid nitrogen and total RNA was extracted from samples. RNA samples were labelled and hybridized to microarrays by NimbleGen (Madison, WI). Oligonucleotide probes of 24 bp were derived from analysis of the entire suite of ESTs available from the F. verticillioides sequencing project (Brown, et al., 2005). Each of the 11 126 unique ESTs is represented twice at randomly assigned positions on the microarray to provide technical replication. Data were normalized by robust multiarray average (Bolstad et al., 2003; Irizarry et al., 2003) included in NimbleScan software. For each EST, expression was normalized to TUB2 by dividing the spot intensity of each EST by the intensity recorded for TUB2. The experiment was conducted with two biological replicates. For each gene, the statistical significance of changes in gene expression was calculated with a t‐test for equal and unequal variance using PROC TTEST from SAS. The array platform and experimental data were submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE8535.
Quantitative PCR (qPCR) analysis of gene expression
Primers used in this study are listed in Table 4; primers for TUB2 were described previously by Bluhm and Woloshuk (2005). RNA was extracted from ground maize kernels and cDNA was generated as described by Flaherty et al. (2003). Reactions were performed in an ABI 7700 Sequence Detection System (Applied Biosystems) and data were collected with Sequence Detector Software version 1.7 (Applied Biosystems). Each reaction contained 10 µL of QuantiTect SYBR®‐green PCR Master mix (Qiagen, Valencia, CA), forward and reverse primers (500 nm of each), cDNA template and nuclease‐free water to a final volume of 20 µL. PCR cycling conditions were 2 min at 50 °C (one cycle); 10 min at 95 °C (one cycle); 15 s at 95 °C followed by 1 min at 60 °C (40 cycles). Expression of genes was measured in triplicate. To verify that the efficiencies of the target and reference reactions were approximately equal, reactions were performed using the primers for each gene or TUB2 with serial dilutions of cDNA as template. After verifying that the efficiencies of the primers were acceptable, expression levels were calculated by the comparative Ct method (Applied Biosystems) with TUB2 as the endogenous reference for normalization.
Table 4.
Primers used in this study.
| Primer name* | Sequence (5′→3′) |
|---|---|
| FST1‐1 | TCCTGCCTGATGATCTTGGTCTG |
| FST1‐2 | TTTGTGCTCACCGCCTGGACTCGAACTGATAACTGACAC |
| FST1‐3 | GAATAGATGCCGACCGGGAACATCCTGGCTCTGAAT |
| FST1‐4 | ACCGTCACGTGCGCTACTTACTTC |
| FST1‐5 | CCGTCACGTGCGCTACTTACTTCA |
| FST1‐nF | AACTCTGGTATGCCGCTTGGTG |
| FST1‐nR | TCTGTCATCTTGCCTGCTGGTAAA |
| FST1‐ns5 | TGCTCTTCTCTTCCTCGCCTCTTT |
| FST1‐ns3 | AACACCAGCCCAATTCTCCCTAAC |
| FST1‐cF | CATGGGAATAGCAGTGCCTTTGAC |
| HYG5 | GTCCAGGCGGTGAGCACAAA |
| HYG3 | TTCCCGGTCGGCATCTATTC |
| HYG13 | AATACGAGGTCGCCAACATCTTCT |
| FST1‐pF | TCCTCGCCTCTTTCATCGTCACCT |
| FST1‐pR | TGCCAAACCGTCAAACATCTCCT |
| AMY1rtF | GGCATGGGATTTGATGCTATTTGG |
| AMY1rtR | GTCATGAGCAGATTTGACAAGACTC |
| GMY1rtF | CTGGTGTTATTCCTCCCTCTTGGG |
| GMY1rtR | GAGAGCCGTCCTTCTTAACATTGATGTAC |
| GMY2rtF | ACAAGTCTCTCCAGCGCAAGATC |
| GMY2rtR | CAGAAGCTTTGCATGAAGCAGAGAAC |
| MLT1rtF | AGCCTCTTTGACGAGACTGAATCAG |
| MLT1rtR | TCGCAGACTTCCTCTCTGACGAA |
| MLT2rtF | GACGCTCATTTCCAAGGAAGTGC |
| MLT2rtR | TGACATCGTGAACTTGTTTTCTGACTGC |
| MLT3rtF | TCCAATTTCGGTGGCGGTAGTG |
| MLT3rtR | TGAAGCATAAATGGCCCGACGC |
| HXK1rtF | GAGAACCAAGATATCTCTCTGCTACGC |
| HXK1rtR | TAAGGTTCTCCCAGGGGTCTTC |
| HXK2rtF | ATCTCAACTCGAGCACGAACGACTG |
| HXK2rtR | GCCGCAACGGACATGATGGA |
| FST1rtF | CTTCTGATGCTCTTCTCTTCCTCGC |
| FST1rtR | TCTGGTATATCTCACCAATGAACGCGAT |
| FST2rtF | GGTATCTCTGAAAAGGAGGAGCCAG |
| FST2rtR | TGGCATCGCCTTTGAACTG |
| FST4rtF | TCATGAACTTCTTCATCCAGGTTGCTC |
| FST4rtR | AAAGGCGAGCTCGACGGCAAT |
The rtF or rtR extension indicates the forward or reverse primer, respectively, for quantification of gene expression by real‐time PCR.
Identification and disruption of FST1
A disruption vector for FST1 was constructed by the double‐joint PCR strategy (Yu et al., 2004). DNA fragments corresponding to the 5′ (1.0 kb) and 3′ (1.6 kb) regions of FST1 were amplified from genomic DNA of F. verticillioides with primer pairs FST1‐1/FST1‐2 and FST1‐3/FST1‐4, respectively, and a 1.4‐kb hygromycin B phosphotransferase (HYGR) gene cassette was amplified from pCB1003 (Carroll et al., 1994) with primers HYG5/HYG3. A 3.6‐kb fusion product was amplified from the three individual templates with nested primers FST1‐nF/FST1‐nR and was cloned into the pGEM‐T‐Easy vector (Promega) to create pFST1‐KO. Protoplasts of F. verticillioides were transformed as described previously (Proctor et al., 1999). Transformants were screened by PCR with primers FST1‐ns5/FST1‐ns3 and HYG13/FST1‐5 to identify strains disrupted in FST1. One isolate that tested positive for disruption of FST1 by PCR was confirmed by Southern analysis and designated Δfst1. For complementation of Δfst1, we amplified 4.4‐kb DNA fragment, containing a 1.8‐kb coding region of FST1, by primers FST1‐cF/ FST1‐4, and cloned the product into pGEM‐T‐Easy to generate pHT‐10. Protoplasts of the Δfst1 strain were co‐transformed with pHT‐10 and pKS‐GEN, a vector created by cloning the geneticin resistance cassette from pSM334 (Flaherty et al., 2003) into the XbaI site of pBluescript‐KS. Geneticin‐resistant colonies were screened by PCR with primers FST1‐ns5/FST1‐ns3 and confirmed by Southern analysis for the presence of FST1.
FB1 extraction and analysis
Maize kernels were ground and extracted as described by Bluhm and Woloshuk (2005), and 5 mL of the supernatant was passed over an equilibrated C18 column (Agilent Technologies, Palo Alto, CA) as described by Rice et al. (1995). Samples were derivatized with O‐pthaldedealdehyde as described by Rice et al. (1995) and analysed with an HPLC system (Shimadzu, Kyoto, Japan) equipped with a C18 column (5 U, 150 × 4.6 mm; Alltech, Deerfield, IL) and a fluorescence detector (335 nm excitation, 440 nm emission). For each sample, FB1 was measured by comparing peak areas to a standard curve generated by analysis of HPLC‐grade FB1 (Sigma).
ACKNOWLEDGEMENTS
We thank Drs Larry Dunkle, Jin‐Rong Xu and Guri Johal for helpful discussions and reviews of this manuscript and Aiden Thompson for technical assistance. Support for this research was provided by USDA‐NRI grant # 2005‐35201‐16233. This report constitutes Journal Publication 2007‐18152 of the Purdue University Agriculture Experiment Station. Disclaimer: names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the products, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
REFERENCES
- Bluhm, B.H. and Woloshuk, C.P. (2005) Amylopectin induces fumonisin B1 production by Fusarium verticillioides during colonization of maize kernels. Mol. Plant–Microbe Interact. 18, 1333–1339. [DOI] [PubMed] [Google Scholar]
- Bluhm, B.H. and Woloshuk, C.P. (2006) Fck1, a c‐type cyclin‐dependent kinase, interacts with Fcc1 to regulate development and secondary metabolism in Fusarium verticillioides . Fungal Genet. Biol. 43, 146–154. [DOI] [PubMed] [Google Scholar]
- Boles, E. and Hollenberg, C.P. (1997) The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21, 85–111. [DOI] [PubMed] [Google Scholar]
- Bolstad, B. , Irizarry, R. , Astrand, M. and Speed, T. (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics, 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Brown, D.W. , Butchko, R.A.E. , Busman, M. and Proctor, R.H. (2007) The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production. Eukaryot. Cell, 6, 1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.W. , Cheung, F. , Proctor, R.H. , Butchko, R.A.E. , Zheng, L. , Lee, Y. , Utterback, T. , Smith, S. , Feldblyum, T. , Glenn, A.E. , Plattner, R.D. , Kendra, D.F. , Town, C.D. and Whitelaw, C.A. (2005) Analysis of 87 000 expressed sequence tags reveals alternatively spliced introns in multiple genes of the fumonisin gene cluster. Fungal Genet. Biol. 42, 848–861. [DOI] [PubMed] [Google Scholar]
- Carroll, A.M. , Sweigard, J.A. and Valent, B. (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41, 22. [Google Scholar]
- Desai, K. , Sullards, M.C. , Allegood, J. , Wang, E. , Schmelz, E.M. , Hartl, M. , Humpf, H.U. , Liotta, D.C. , Peng, Q. and Merrill, A.H. (2002) Fumonisins and fumonisin analogs as inhibitors of ceramide synthase and inducers of apoptosis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids, 1585, 188–192. [DOI] [PubMed] [Google Scholar]
- Fakhoury, A.M. and Woloshuk, C.P. (1999) Amy1, the α‐amylase of Aspergillus flavus: Involvement in aflatoxin biosynthesis in maize kernels. Phytopathology, 89, 908–914. [DOI] [PubMed] [Google Scholar]
- Flaherty, J.E. and Woloshuk, C.P. (2004) Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster‐type gene, ZFR1 . Appl. Environ. Microbiol. 70, 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty, J.E. , Pirttila, A.M. , Bluhm, B.H. and Woloshuk, C.P. (2003) PAC1, a pH regulatory gene from Fusarium verticillioides . Appl. Environ. Microbiol. 69, 5222–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks, K. (1999) Fumonisins and neural tube defects in South Texas. Epidemiology, 10, 198–200. [DOI] [PubMed] [Google Scholar]
- Ingle, J. , Beitz, D. and Hageman, R.H. (1965) Changes in composition during development and maturation of maize seeds. Plant Physiol. 50, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A. , Hobbs, B. , Collins, F. , Beazer‐Barclay, Y.D. , Antonellis, K.J. , Scherf, U. and Speed, T.R. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Joffe, A.Z. (1986) Fusarium Species: Their Biology and Toxicology. New York: Wiley and Sons. [Google Scholar]
- Madi, L. , Ebbole, D.J. , White, B.T. and Yanofsky, C. (1994) Mutants of Neurospora crassa that alter gene‐expression and conidia development. Proc. Natl Acad. Sci. USA, 91, 6226–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson, T. , Thelander, M. and Ronne, H. (2003) A novel type of chloroplast stromal hexokinase is the major phosphorylating enzyme in the moss Physcomitrella patens . J. Biol. Chem. 278, 44439–44447. [DOI] [PubMed] [Google Scholar]
- Pao, S.S. , Paulsen, I.T. and Saier Jr., M.H. (1998) Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor, R.H. , Desjardins, A.E. , Plattner, R.D. and Hohn, T.M. (1999) A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27, 100–112. [DOI] [PubMed] [Google Scholar]
- Rice, L.G. , Ross, P.F. , Dejong, J. , Plattner, R.D. and Coats, J.R. (1995) Evaluation of a liquid chromatographic method for the determination of fumonisins in corn, poultry feed, and Fusarium culture material. J. AOAC Int. 78, 1002–1009. [PubMed] [Google Scholar]
- Sagaram, U.S. , Butchko, R.A.E. and Shim, W.‐B. (2006) The putative monomeric G‐protein GBP1 is negatively associated with fumonisin B1 production in Fusarium verticillioides . Mol. Plant Pathol. 7, 381–389. [DOI] [PubMed] [Google Scholar]
- Seitz, L.M. , Sauer, D.B. , Burroughs, R. , Mohr, H.E. and Hubbard, J.D. (1979) Ergosterol as a measure of fungal growth. Phytopathology, 69, 1202–1203. [Google Scholar]
- Shim, W.‐B. , Flaherty, J.E. and Woloshuk, C.P. (2003) Comparison of fumonisin B1 biosynthesis in maize germ and degermed kernels by Fusarium verticillioides . J. Food Protect. 66, 2116–2122. [DOI] [PubMed] [Google Scholar]
- Vismer, H.F. , Snijman, P.W. , Marasas, W.F.O. and Van Schalkwyk, D.J. (2004) Production of fumonisins by Fusarium verticillioides strains on solid and in a defined liquid medium—effects of L‐methionine and inoculum. Mycopathologia, 158, 99–106. [DOI] [PubMed] [Google Scholar]
- Warfield, C.Y. and Gilchrist, D.G. (1999) Influence of kernel age on fumonisin B1 production in maize by Fusarium moniliforme . Appl. Environ. Microbiol. 65, 2853–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshuk, C.P. , Foutz, K.R. , Brewer, J.F. , Bhatnagar, D. , Cleveland, T.E. and Payne, G.A. (1994) Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60, 2408–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. , Wilkinson, H.H. , Correa, A. , Lewis, Z.A. , Bell‐Pedersen, D. and Ebbole, D.J. (2004) Transcriptional response to glucose starvation and functional analysis of a glucose transporter of Neurospora crassa . Fungal Genet. Biol. 41, 1104–1119. [DOI] [PubMed] [Google Scholar]
- Yu, J.‐H. , Hamari, Z. , Han, K.‐H. , Seo, J.‐A. , Reyes‐Domínguez, Y. and Scazzocchio, C. (2004) Double‐Joint PCR (DJ‐PCR): a PCR‐based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. [DOI] [PubMed] [Google Scholar]


