SUMMARY
Genes for seven putative serine proteases (ChpA–ChpG) belonging to the trypsin subfamily and homologous to the virulence factor pat‐1 were identified on the chromosome of Clavibacter michiganensis subsp. michiganensis (Cmm) NCPPB382. All proteases have signal peptides indicating export of these proteins. Their putative function is suggested by two motifs and an aspartate residue typical for serine proteases. Furthermore, six cysteine residues are located at conserved positions. The genes are clustered in a chromosomal region of about 50 kb with a significantly lower G + C content than common for Cmm. The genes chpA, chpB and chpD are pseudogenes as they contain frame shifts and/or in‐frame stop codons. The genes chpC and chpG were inactivated by the insertion of an antibiotic resistance cassette. The chpG mutant was not impaired in virulence. However, in planta the titre of the chpC mutant was drastically reduced and only weak disease symptoms were observed. Complementation of the chpC mutant by the wild‐type allele restored full virulence. ChpC is the first chromosomal gene of Cmm identified so far that affects the interaction of the pathogen with the host plant.
INTRODUCTION
Clavibacter michiganensis subsp. michiganensis (Cmm) is a Gram‐positive, plant pathogenic actinomycete (family Microbacteriaceae) (Stackebrandt et al., 1997) causing bacterial wilt and canker of tomato plants (Solanum lycopersicum L.) (Davis et al., 1984; Strider, 1969). Infection by Cmm results in a systemic tracheobacteriosis with an in planta titre of 109–1010 bacteria per gram plant homogenate. The first symptom of disease is unilateral wilting, followed at later stages by canker lesions on the stem (Wallis, 1977). The wild‐type strain NCPPB382 carries two plasmids that are essential for virulence, pCM1 and pCM2 (Meletzus et al., 1993). Both plasmids carry a single virulence factor, the gene celA coding for an endo‐β‐1,4‐glucanase on plasmid pCM1 (Jahr et al., 2000), and the gene pat‐1 encoding a putative serine protease on plasmid pCM2 (Dreier et al., 1997). Loss of one plasmid, either pCM1 with celA or pCM2 with pat‐1, reduces virulence, i.e. development of disease symptoms is delayed. When both plasmids are lost a complete loss of virulence results and the typical wilting symptoms do not occur, although the bacteria are still able to colonize the host plant. Thus, the loss of the virulence factors converts Cmm into a non‐virulent endophyte of tomato (Dreier et al., 1997; Gartemann et al., 2003; Jahr et al., 2000; Meletzus et al., 1993). This shows that gene functions required for the colonization of the host plant are encoded by the chromosome.
Hybridization of total DNA of the wild‐type strain NCPPB382 against a pat‐1 probe led to the identification of three homologous genes, termed phpA/B, carried on plasmid pCM2, and the pseudogene chpA, located on the chromosome (Burger et al., 2005). It was shown that these genes are not involved in Cmm–tomato interaction. Clavibacter michiganensis subsp. sepedonicus is the causal agent of bacterial wilt and ring rot of potato and also carries several pat‐1 homologous genes (Holtsmark et al., 2008). Using quantitative reverse transcriptase (RT)‐PCR, five of these genes were analysed during infection of potato and in liquid culture. It was shown that during infection three pat‐1 homologous genes were downregulated and two were upregulated, implying an involvement of these genes in the infection process (Holtsmark et al., 2008).
In Cmm, analysis of the nucleotide sequence surrounding the chpA locus revealed that chpA maps in a cluster of six additional genes putatively encoding serine proteases with homology to Pat‐1. Due to the relationship of the chp genes to the virulence gene pat‐1 we considered it possible that these genes could be involved in the interaction of Cmm with the host plant. Here we present the classical genetic approach specifically to inactivate candidate genes that might be involved in the pathogenicity of Cmm and to analyse the phenotype of the corresponding mutants in comparison with the respective wild‐type reference strain.
RESULTS
Features of the Pat‐1 protein family
In a recent publication (Burger et al., 2005) we have reported on the chpA gene (chromosomal homology of pat‐1). chpA is a pseudogene, containing internal stop codons as well as two frame shifts. The hypothetical gene product is homologous to Pat‐1, indicating that the functional gene could encode a putative serine protease. Analysis of the nucleotide sequence around the chpA locus revealed six more pat‐1 homologous genes (chpB, chpC, chpD, chpE, chpF and chpG) in a region of about 50 kb of the chromosome (Fig. 1).
Figure 1.
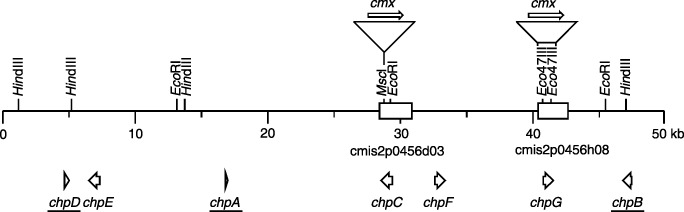
Physical map of the 50‐kb chromosomal region of Clavibacter michiganensis subsp. michiganensis (Cmm) carrying the chp genes. Pseudogenes are underlined. Boxes indicate the fragments used to construct plasmids for gene replacement or complementation. The insertion sites of the cassettes carrying the chloramphenicol exporter gene cmx are indicated.
Like chpA, the genes chpB and chpD are also pseudogenes. ChpB contains a frame shift 138 nt downstream of the ATG start codon and chpD has a frame shift at position 114 downstream of the ATG start codon and an in‐frame stop codon at position 547–549. For the multiple alignment of the Chp proteins the reading frames of ChpA, ChpB and ChpD were restored at the appropriate amino acid positions (Fig. 2). All Pat‐1 homologues have a putative signal peptide indicating that these proteins are secreted. Two motifs characteristic for serine proteases of the trypsin type ([LIVM][ST]A[STAG]HC and [DNSTAGC][GSTAPIMVQH]x(2)G[DE]SG[GS][SAPHV][LIVMFYWH][LIVMFYSTANQH]PROSITE, PDOC00124) are highly conserved. The histidine and serine residues indicated by bold letters in the motifs are those amino acids participating in catalysis. An aspartate as part of the catalytic triad is also conserved. Furthermore, six cysteine residues are located at conserved positions in all members of this protein family except in ChpC, which has only four of the conserved cysteine residues. The functional members of the Pat‐1 family have 277–286 amino acid residues and molecular weights of between 29.0 and 35.8 kDa. The G + C content of the genes varies between 51.9 and 65.5 mol% and thus is significantly lower than the average G + C content of 72.6 mol% for Cmm. Furthermore, the codon usage deviates from the normal codon usage of Cmm. The presence of rare codons as observed for pat‐1 and chpA (Burger et al., 2005) is a common feature of all chp genes. A putative sortase motif with homology to (LPXTG) (Mazmanian et al., 2001), which may be important for anchoring of the protein to the cell wall, was identified only in Pat‐1 and the ChpA pseudogene.
Figure 2.
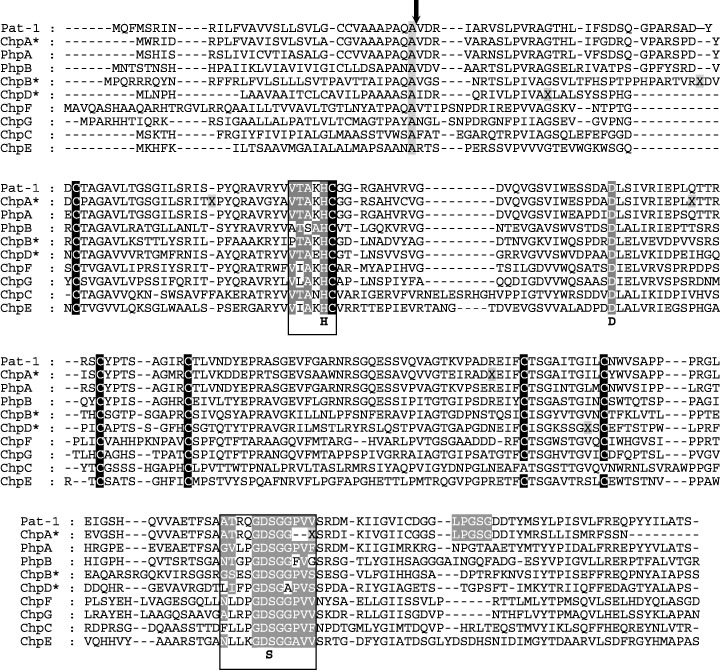
Multiple alignment of the Pat‐1 family members of Clavibacter michiganensis subsp. michiganensis (Cmm). Pseudogenes are marked with asteriks. They contain ‘X’ introduced to restore the reading frame. Conserved cysteines, the conserved aspartate of the catalytic triad, the two serine proteases motifs and the putative sortase motif (LPGSG) are also indicated. The three amino acid residues (H, D, S) of the catalytic triad are exhibited below the alignment. The arrow marks the hypothetical processing site (AQA↓V) although this motif is not conserved in all putative gene products.
Construction of chpC and chpG mutants
In order to assess a possible function for the chp genes in plant–microbe interaction, Cmm NCPPB382 was subjected to targeted homologous recombination for the generation of chp mutants. Thus far, we have been able to obtain mutations in the genes chpC and chpG.
A 1.9‐kb BsaAI DNA fragment containing the chloramphenicol resistance gene (cmx) from hybrid plasmid pEC70 (Tauch et al., 1998) was inserted into the internal MscI site of the chpC gene carried by plasmid cmis2p0456d03, a sequenced plasmid from the shotgun cloning of the genome project (Fig. 1). Both orientations of the cmx cassette relative to chpC were obtained (pIGCα and pIGCβ). In plasmid cmis2p0456h08, which carries the chpG gene (Fig. 1), an internal 640‐bp Eco47III fragment was replaced by a 1.9‐kb BsaAI fragment of pEC70 carrying the cmx gene. Both orientations of the cmx cassette relative to chpG were obtained (pIGGα and pIGGβ). The hybrid plasmids based on the Escherichia coli vector pSMART, which cannot replicate in Cmm, were introduced into the wild‐type strain NCPPB382 by electroporation. Clones in which recombination between the wild‐type genes and the inactivated gene on the plasmid had occurred (formation of a cointegrate) were selected with chloramphenicol in the medium. If a second crossover had occured at the other side flanking the cmx gene, the intact chromosomal chp gene was exchanged by the inactivated chp gene.
To detect such knock‐out mutants, total DNA of chloramphenicol‐resistant clones was isolated, hydrolysed with NcoI and BamHI and hybridized against a labelled pUC probe which shares homology with pSMART. Clones which gave a signal were interpreted as cointegrates and discarded. Clones which gave no signal with the pUC probe were further analysed by hybridization against chpC and chpG probes, respectively. DNA from the chpC mutant designated CMM101chpCβ gave 2.9‐kb and 10‐kb signals (NcoI digestion) or 2.6‐kb and 5.9‐kb signals (BamHI hydrolysis) against the chpC probe, as expected for a double crossover event with the cmx cassette integrated in the β‐orientation (Fig. 3). NcoI‐digested total DNA of the chpG mutant designated CMM101chpGβ gave signals of 1.1 and 2.9 kb in hybridizations against a cmx probe (data not shown) and of 1.1, 2.0 and 2.9 kb against the chpG probe, showing that the desired inactivation of chpG was achieved and that the cmx cassette was integrated in the opposite direction relative to chpG (Fig. 4).
Figure 3.
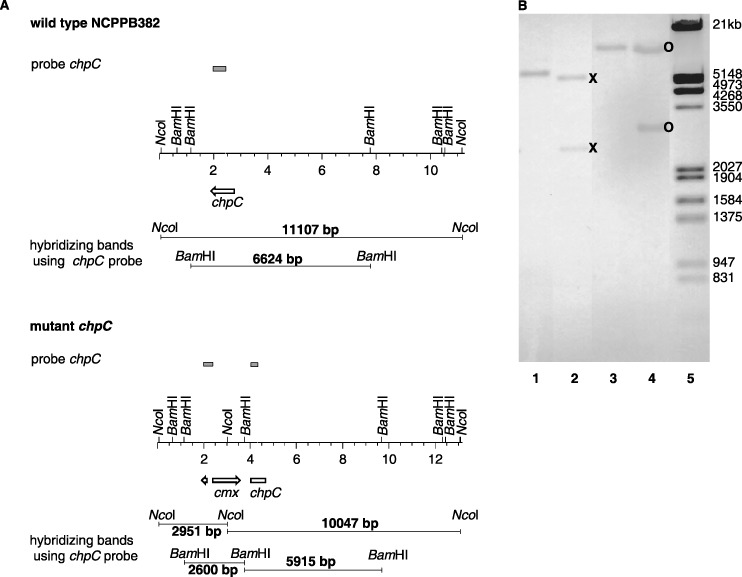
(A) Physical map of the chpC region in the wild‐type and in the chpC mutant. (B) Southern hybridization against the chpC probe. BamHI‐digested total DNA of the wild‐type (lane1) and the chpC mutant (lane 2), NcoI‐digested total DNA of the wild‐type (lane 3) and the chpC mutant (lane 4), digoxygenin‐11‐dUTP‐labelled EcoRI/HindIII‐digested λ marker (lane 5). The expected signals of 2.9 and 10 kb (NcoI digestion, ○) and 2.6 and 5.9 kb (BamHI hydrolysis,  ) were obtained for the chpC mutant.
) were obtained for the chpC mutant.
Figure 4.
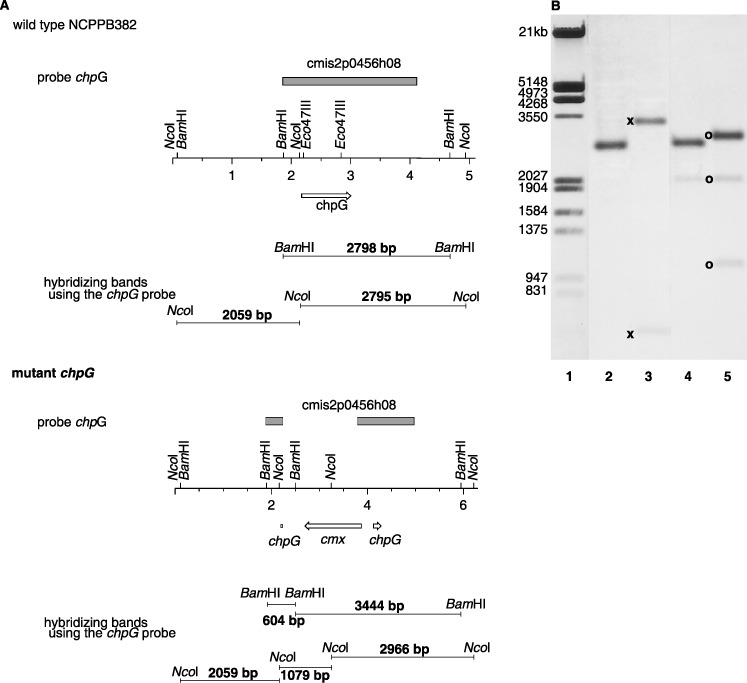
(A) Physical map of the chpG region in the wild‐type and in the mutant. (B) Southern hybridization against the chpG probe. BamHI‐digested total DNA of the wild‐type (lane 2) and the mutant (lane 3), NcoI‐digested total DNA of the wild‐type (lane 4) and the mutant (lane 5), digoxygenin‐11‐dUTP‐labelled EcoRI/HindIII‐digested λ marker (lane 1). The occurrence of signals of 1.1, 2.0 and 2.9 kb (NcoI digestion, ○) and 0.6 and 3.4 kb (BamHI hydrolysis, 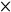 ) against the chpG probe demonstrated that the desired inactivation of chpG was achieved.
) against the chpG probe demonstrated that the desired inactivation of chpG was achieved.
In addition, BamHI‐digested total DNA was hybridized against the chpG probe. The hybridizing bands had sizes of 0.6 and 3.4 kb as expected for a double crossover (Fig. 4).
During electroporation of the wild‐type strain Cmm NCPPB382 we frequently observed loss of plasmid pCM2. Such derivatives, termed CMM101 (they still carry plasmid pCM1), are still virulent and able to colonize tomato plants efficiently.
Thus, the chpC and the chpG mutants were checked for the presence of both plasmids pCM1 and pCM2 via hybridization against specific celA (pCM1) and pat‐1 (pCM2) probes. As expected, only celA (pCM1) was found while pat‐1 gave no signal, indicating that plasmid pCM2 was lost in the mutants (data not shown). According to their plasmid status, which is identical to the plasmid curing derivative CMM101, carrying only plasmid pCM1 with the pathogenicity factor celA, the mutants were designated CMM101chpCβ and CMM101chpGβ.
Assessment of virulence of chpC and chpG mutants
In tests for the pathogenic phenotype of mutants CMM101chpCβ and CMM101chpG as shown in Fig. 5, 64 tomato plants were infected with one of the mutant strains or with CMM101 as a control. In these experiments CMM101chpCβ displayed very weak virulence; only seven of 64 plants showed symptoms with curled and wilting leaves 28 days post‐infection (dpi). In contrast to that, infection by the reference strain CMM101 as control, showed curled and wilting leaves in 32 of 64 plants already 15 to 16 dpi (Fig. 5). This comparison shows that virulence of the mutant strain CMM101chpCβ is drastically reduced as compared with the reference strain CMM101. The experiment was repeated four times with 32 plants in each experimental group. Although there are variations in the sensitivity of the tomato plants to infection and general vitality of the plants due to seasonal effects, the qualitative results were identical to the example shown in Fig. 5. The wilting indices as shown in Table 1 did vary by 1–2 days in different experiments. However, infection with the chpC mutant always resulted in only 2–5 plants with mild disease symptoms out of 32 plants.
Figure 5.
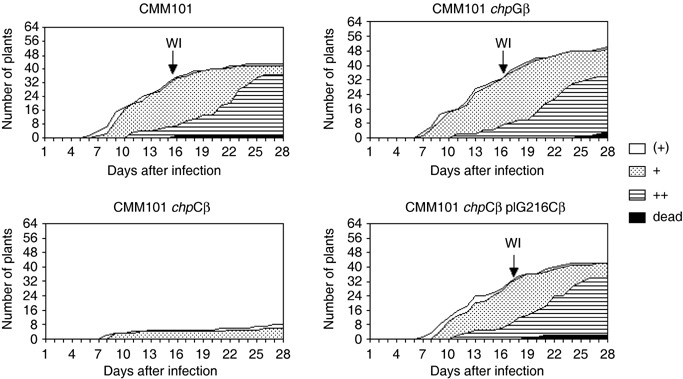
Diagram showing the wilting symptoms and wilting indices of control strain CMM101 compared with the chpC mutant (CMM101chpCβ), the chpG mutant (CMM101chpGβ) and the complemented chpC mutant (CMM101chpCβ−pIG216Cβ). Plants with beginning leaf curling just at the tips are indicated by a ‘(+)’, obviously curled leaves by a ‘+’ and strong wilting symptoms were recorded as ‘++’ (when at least two‐thirds of the leaves showed wilting symptoms). When plants showed severe wilting symptoms they were termed ‘dead’. The wilting index (WI) is indicated and defined as the number of days required for 50% of the plants to show clear wilting symptoms ‘+’.
Table 1.
Clavibacter michiganensis subsp. michiganensis (Cmm) strains and their plasmid status, wilting indices and titre.
| Strain | Plasmid(s) | Wilting index (d)* | Titre (cfu/g plant homogenate)† | SD | Reduction of titre‡ |
|---|---|---|---|---|---|
| Cmm NCPPB 382 | pCM1, pCM2 | 12 | 7.4 × 109 § | ±7.3 × 109 | – |
| CMM101 | pCM1 | 15–16 | 9.9 × 109 § | ±4.1 × 109 | – |
| CMM101chpCβ | pCM1 | – | 2.0 × 107 ¶ | ±3.2 × 107 | 495 |
| CMM101chpCβ pIG216Cβ | pCM1 pIG216Cβ | 17 | 1.9 × 109 ¶ | ±1.7 × 109 | 5.2 |
| CMM101chpGβ | pCM1 | 15–16 | 1.8 × 109 § | ±2.4 × 109 | 5.5 |
Time post‐infection when 50% of the infected plants show wilting symptoms, i.e. curled leaves; n = 64 plants.
After 28 days.
x‐fold reduction of bacterial titre compared with CMM101.
n = 10 plants.
n = 20 plants.
When the titre in planta was determined, we found that strain CMM101chpCβ did not colonize the plants as well as the reference strain CMM101 or wild‐type Cmm NCPPB382. The titre of CMM101chpCβ was reduced 500‐fold to 2 × 107 cfu/g plant homogenate as compared with 1 × 1010 cfu/g plant homogenate for CMM101 (Table 1). Obviously, the chpC mutant is not able to colonize the host plant as efficiently as the reference strain. Although the wilt‐inducing pathogenicity gene celA is present in the mutant strain, the titre in planta seems not to be high enough for the development of full virulence. To exclude the possibility that the mutation in chpC has an effect on general growth behaviour we determined the generation times in different media. The generation times of the chpC and the chpG mutants compared with CMM101 did not differ and were 2.5 h in rich medium and 4 h in minimal medium, respectively.
In contrast to CMM101chpCβ, the chpG mutant CMM101chpGβ was not affected in virulence and colonization of the host plant. When CMM101chpGβ was compared with the control strain CMM101 the virulence assays and titre in planta indicated no significant difference (Fig. 5, Table 1). However, when leaves of the non‐host plant Mirabilis jalapa were infiltrated with cell suspensions of the chpC and the chpG mutants we observed no induction of a hypersensitive reaction (HR) for the chpG mutant (data not shown). This indicates that the putative serine protease encoded by chpG is also involved in plant–microbe interaction.
Complementation of the chpC mutant with plasmid pIG216Cβ
To confirm that the phenotype of the CMM101chpCβ mutant was not caused by some other mutation in the chromosome, a merozygote containing the inactivated and the wild‐type chpC gene was constructed. A 2.5‐kb EcoRV fragment containing the complete chpC of cmis2p456d03 was cloned into the EcoRV‐digested E. coli–Cmm shuttle vector pHN216 (Laine et al., 1996), resulting in plasmid pIG216Cβ, which was introduced into the chpC mutant by electroporation. Plasmid DNA from neomycin‐resistant colonies was isolated and the presence of the plasmids was verified by NcoI digestion (data not shown). One clone carrying the hybrid plasmid pIG216Cβ designated CMM101chpCβ‐pIG216Cβ was chosen for assays on tomato plants.
In the virulence assay 64 tomato plants were infected with the merozygotic strain CMM101chpCβ‐pIG216Cβ and scored daily for disease symptoms. The virulence of strain CMM101chpCβ‐pIG216Cβ was almost identical to the control strain CMM101 with a wilting index of 17 days (Fig. 5). In addition, the titre in planta of the complemented chpC mutant was close to that of CMM101 (Table 1). This indicates that trans‐complementation of the chpC mutation by the wild‐type gene had occurred and no other mutation or rearrangement in the chromosome of Cmm was responsible for the phenotype of the CMM101chpCβ mutant.
DISCUSSION
One of the virulence factors of Cmm NCPPB382 is the putative serine protease Pat‐1, which is encoded by plasmid pCM2 (Dreier et al., 1997). The exact function of this serine protease and its substrate remains unknown. The first identified Pat‐1 homologues, phpA, phpB on plasmid pCM2 and chpA on the chromosome, were found by hybridizations against a pat‐1 probe (Burger et al., 2005). Six further proteins homologous to Pat‐1 (ChpB–ChpG) were found to be encoded in a 50‐kb region of the Cmm chromosome.
Most microbial proteases are secreted enzymes and can be classified based on the essential catalytic residue at their active site. They include aspartate proteases, cysteine proteases, metalloproteases and serine proteases. Pat‐1 and the homologous proteins may comprise a family of serine proteases belonging to the chymotrypsin subfamily S1A (Fig. 2). At the N‐terminus they all have a signal peptide indicating that these proteins may be secreted. Some of them also share the hypothetical processing site AQA↓V (Burger et al., 2005). Furthermore, several cysteine residues are conserved as is also the case in some other members of the serine protease subfamily S1A. It is possible that cysteines form disulphide bridges in the mature enzymes. As the chpA, chpB and chpD genes contain frame shifts or in‐frame stop codons, only truncated and possibly non‐functional proteins originate from these genes. The other genes chpC, chpE, chpF and chpG may encode functional serine proteases; however, when Cmm is grown in rich or minimal media the supernatant contains no protease activity when tested with common serine protease substrates, such as casein, azocasein and azocoll (Burger et al., 2005). This indicates that these putative serine proteases might have a specific activity or are only expressed in the plant. Holtsmark et al. (2008) recently demonstrated that pat‐1 homologous genes in Clavibacter michiganensis subsp. sepedonicus were also expressed in rich medium. However, depending on which pat‐1 homologous gene was tested the transcript level decreased or increased during infection of the host plant, indicating that expression of these genes is indeed affected in the interaction with the host tomato.
To elucidate the possible function of these serine proteases we examined whether they are involved in pathogenicity, as is the case for Pat‐1. Thus far, we have only succeeded to inactivate the genes chpC and chpG by insertion of an antibiotic resistance cassette and exchange of the wild‐type gene by the inactivated one. Inactivation of the chpG gene had no effect on virulence or colonization, while knock‐out of the chpC gene resulted in a significant reduction in virulence and bacterial growth in planta. The titre of CMM101chpCβin planta 28 dpi was only 2 × 107 cfu/g plant homogenate as compared with 9.9 × 109 cfu/g plant homogenate for CMM101 and only seven of 64 plants showed curled leaves, the mildest manifestation of disease. Thus, a correlation seems to exist between development of wilting symptoms and the bacterial titre in planta. Assays with several Cmm strains and mutants showed that only when titres of higher than 108 bacteria/g plant homogenate are reached is wilting of tomato plants observed (data not shown). Cmm grows in the xylem fluid and a slow bacterial growth in planta may be caused by a lack of nutrients, especially as Cmm is auxotrophic for methionine, thiamine and nicotinic acid. Thus, these compounds and sugars or organic acids have to be provided in the xylem in sufficient concentration to allow growth of Cmm. It is possible that the ChpC serine protease degrades or processes specific proteins of either plant or bacterial origin to enrich the xylem fluid with nutrients (perhaps methionine) or that ChpC generates a signal which induces the plant to secrete nutrients into the xylem fluid. In order to test this hypothesis, it will be necessary to study the composition of the xylem fluid in infected and non‐infected tomato plants.
Many plant pathogens producing proteases have been studied and the results suggest that proteases seem to play a role in virulence. However, often neither the exact mode of action nor the substrate or target of the proteases is known. An example is the interference with regulatory mechanisms of the plant by cysteine proteases of proteobacteria translocated into the plant cell via the type III secretion system. The proteases XopD, AvrXv4, AvrPphB and AvrRpt2 were shown to hydrolyse specific host target proteins (Hotson and Mudgett, 2004; Hotson et al., 2003). XopD of Xanthomonas campestris pv. vesicatoria disturbs plant signal transduction as it mimics plant‐specific SUMO (small ubiquitin‐like modifier) proteases responsible for the activation or degradation of proteins destined to be SUMOylated. In planta analysis showed that XopD targets numerous SUMO‐modified proteins and therefore may affect several plant signalling pathways leading to stress responses or defence reactions against pathogens (Hanania et al., 1999; Hotson et al., 2003). However, thus far we have no indication for a type III secretion system in Cmm. In spite of this, ChpC may still be a similar effector which is transported into the plant cell by a different transport system.
Filamentous fungi secrete a broad range of proteolytic enzymes during penetration and colonization of the plant tissue. The infection of potato tubers with Fusarium eumartii is accompanied by an accumulation of a serine protease activity (Olivieri et al., 1998), which suggests that this protease activity might be involved in plant–pathogen interaction (Olivieri et al., 2002). In fact, the extracellular serine protease of Fusarium eumartii belonging to the subtilase subfamily was shown to degrade potato pathogenesis‐related (PR) proteins as well as specific polypeptides in the intercellular washing fluid and cell‐wall proteins from potato tubers (Olivieri et al., 2002). It is possible that ChpC serine protease plays a similar role in the interaction with tomato plants.
Altogether, the importance of proteases as virulence factors remains unclear because in many cases the inactivation of proteases by deletion, mutation or disruption did not affect pathogenicity (Jaton‐Ogay et al., 1994). This seems to be the case for the putative serine protease ChpG, but for ChpC we can clearly show that it has an effect on the interaction of Cmm with tomato plants. One of the main goals for the future will be the identification of substrate molecules for the putative Chp serine proteases in order to obtain clues to their function in the Clavibacter–tomato interaction.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and media
Bacterial strains and plasmids used and constructed in this study are listed in Table 1. Strains of Cmm were grown at 25 °C on C‐medium (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, 5 g/L glucose, pH 7.2) or M9 medium (6 g/L Na2HPO4, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.5 g/L NaCl, 1 mm MgSO4, 0.01 mm CaCl2, 200 mg/L methionine, 200 mg/L thiamine, 20 mg/L nicotinic acid, 2 g/L glucose). E. coli was grown on TBY medium (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, pH 7.2) at 37 °C. When selecting for antibiotic resistance to E. coli, media contained 10 µg/mL chloramphenicol or 50 µg/mL kanamycin. After electroporation strains of Cmm were grown on SB medium (10 g/L tryptone, 5 g/L yeast extract, 4 g/L NaCl, 0.5 m sorbitol, 20 mm MgCl2, 20 mm CaCl2). Selection for chloramphenicol and/or neomycin resistance was achieved with SB medium supplemented with these antibiotics at 10 and 75 µg/mL, respectively.
DNA isolation, manipulation and transfer
Plasmid DNA of E. coli used for cloning and electroporation was isolated and purified with QIAprep spin columns as specified by the manufacturer (QIAGEN, Hilden, Germany). Preparation of total DNA for hybridization was done as described by Hopwood et al. (1985). Plasmid DNA of Cmm was isolated by the method of Birnboim and Doly (1979) with the following modifications. Ten‐millilitre cultures were harvested, washed with 2 mL 10% glycerol, and frozen at –20 °C overnight. The pellet was resuspended in 300 µL P1 buffer (50 mm Tris‐HCl, 10 mm EDTA, 100 µg/mL RNaseA, pH 8) with addition of 7 mg/mL lysozyme. Lysis was performed with 300 µL buffer P2 (200 mm NaOH, 1% SDS) for 5 min. Three hundred microlitres of buffer P3 (3 m KAc, pH 5.5) was added to neutralize the suspension. DNA hydrolysis with restriction endonucleases, ligation and transformation were carried out by standard procedures (Sambrook et al., 1989). Isolation of restriction fragments from agarose gels was done with the QIAquick Gel Extraction Kit as specified by the manufacturer (QIAGEN).
Sequence analysis
Amino acid sequence alignment was performed with the alignment programs ClustalX (Thompson et al., 1997) and GeneDoc (Nicholas et al., 1997). Signal sequences were predicted using SignalP (http://www.cbs.dtu.dk/services/SignalP, Nielson et al., 1997).
Construction of plasmids for gene replacement and complementation
The plasmid cmis2p0456d03 carrying the native complete chpC gene was used for the construction of a mutagenesis vector. Cmis2p0456d03 cannot replicate in Cmm. Plasmid pIGCβ was constructed by cloning the chloramphenicol resistance gene cmx of pEC70 (Tauch et al., 1998) as a 1.9‐kb BsaAI fragment into the unique MscI restriction site of cmis2p0456d03 within the chpC gene.
In plasmid cmis2p0456h08, which carries the native chpG gene, a 640‐bp Eco47III fragment containing the promoter region and part of chpG was replaced by the 1.9‐kb BsaAI fragment of pEC70 carrying the cmx gene (pIGGβ) (Fig. 1).
Plasmid pIG216Cβ used for complementation was constructed based on the E. coli–Cmm shuttle vector pHN216 (Laine et al., 1996). The 2.5‐kb EcoRV fragment of cmis2p0456d03 carrying the chpC gene was integrated into the vector pHN216 linearized by EcoRV.
Electroporation
Electroporation into E. coli was conducted according to the Gene Pulser manual (Bio‐Rad Laboratories, Krefeld, Germany). The electroporation into Cmm was performed as described by Kirchner et al. (2001).
Southern blot analysis
An internal 465‐bp fragment of chpC was generated by PCR (primer: chpC‐1: CCCCATCGGAACGGTTTATTGG and chpC‐2: GCTCTGCTCTGTGAGACGATG). Amplification of the 465‐bp fragment was done according to the following conditions: 4 min at 94 °C, 35 cycles of 1.5 min at 94 °C, 1.5 min at 64 °C and 1.5 min at 72 °C, and finally 10 min at 72 °C. DNA was labelled using the random primed DNA labelling kit (Roche Diagnostics, Mannheim, Germany). For chpG the 2.3‐kb EcoRV fragment of cmis2p0456h08 was used as a probe. The digoxygenin‐11‐dUTP‐labelled 1.9‐kb BsaAI fragment of pEC70 was used to detect the cmx gene. pUC18 DNA was labelled with digoxigenin‐11‐dUTP by nick translation (Sambrook et al., 1989). Digested chromosomal and plasmid DNA fragments were separated on 0.8–1.0% agarose gels and transferred to nylon membranes (porablot, Macherey & Nagel, Düren, Germany; or Nytrans, Schleicher & Schuell, Dassel, Germany) by capillary blots (Smith and Summers, 1980). Hybridizations were carried out at 68 °C overnight in a buffer containing 5× SSC (0.75 m NaCl plus 0.075 m sodium citrate), 0.02% sodium dodecyl sulphate, 0.1% Na lauroylsarcosyl, and 2% blocking reagent (Roche Diagnostics). The nylon membrane was washed twice with 0.1× SSC, 0.1% sodium dodecyl sulphate at 68 °C for 15 min. Detection was carried out as recommended by the manufacturer with anti‐DIG‐AP conjugate and NBT and BCIP as chromogenic substrates (Roche Diagnostics).
Virulence assay
Tomato plants (Lycopersicon esculentum cv. Moneymaker) used for the standard infection procedure were about 14 days old (two‐leaf stage). Fresh Cmm cultures, which were grown at 28 °C on rich medium, were harvested by centrifugation and adjusted to an optical density (580 nm) of 8–9 with sterile water. After removal of adhering soil from the roots, the plants were immersed for 15–20 min in the high‐titre bacterial suspension to achieve an effective infection, which under these conditions is about 70–90%. The infected plants were then planted into sterile soil. Growth of the plants took place at 25 °C with a day–night rhythm of 16 h day at 120 000 lx and 8 h night, and 40–50% relative air humidity. Each experimental group consisted of 64 plants. Disease symptoms were estimated by scoring infected plants for wilting symptoms. The wilting index is defined as the number of days required for 50% of the plants to display wilting symptoms (leaf curling) (Meletzus et al., 1993). Parallel to this, plants were examined daily over a period of up to 28 days for the development of wilting symptoms. Plants with beginning leaf curling just at the tips were assigned the score ‘(+)’, obviously curled leaves were ‘+’ and strong wilting symptoms were recorded as ‘++’ (when at least two‐thirds of the leaves showed wilting symptoms). When all leaves showed wilting and photosynthesis was no longer possible, the plants were termed ‘dead’.
Determination of bacterial titres in planta
Twenty‐eight days after infection 10–20 tomato plants were harvested by cutting the stem 1 cm above the soil. Single plants were frozen in liquid nitrogen, ground to powder in a sterile mortar and then suspended in buffer (7 g/L Na2HPO4·2H2O, 5 g/L NaCl, 3 g/L KH2PO4, pH 7) (1 mL buffer/g fresh weight). Appropriate dilutions were plated on selective medium and incubated at 25 °C for 3–5 days to determine the number of colony forming units (cfu). The mean of the 10–20 individual titre determinations was taken to calculate the in planta titre and the standard deviation.
ACKNOWLEDGEMENTS
We are indebted to E.‐M. Zellermann for excellent technical assistance. This study was supported by the Deutsche Forschungsgemeinschaft (DFG) (Ei 535/12‐1) and BMBF (FKZ:0313805A).
Nucleotide sequence data are to be found at GenBank as accession number AM711867 (complete chromosome).
REFERENCES
- Birnboim, H.C. and Doly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, A. , Gräfen, I. , Engemann, J. , Niermann, E. , Pieper, M. , Kirchner, O. , Gartemann, K.‐H. and Eichenlaub, R. (2005) Identification of homologues to the pathogenicity factor Pat‐1, a putative serine protease of Clavibacter michiganensis subsp. michiganensis . Microbiol. Res. 160, 417–427. [DOI] [PubMed] [Google Scholar]
- Davis, M.J. , Gillaspie, A.G. Jr. , Vidaver, A.K. and Harris, R.W. (1984) Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Bacteriol. 34, 107–117. [Google Scholar]
- Dreier, J. , Meletzus, D. and Eichenlaub, R. (1997) Characterization of the plasmid encoded virulence region pat‐1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis . Mol. Plant–Microbe Interact. 10, 195–206. [DOI] [PubMed] [Google Scholar]
- Gartemann, K.‐H. , Kirchner, O. , Engemann, J. , Gräfen, I. , Eichenlaub, R. and Burger, A. (2003) Clavibacter michiganensis subsp. michiganensis: first steps in the understanding of virulence of a gram‐positive phythopathogenic bacterium. J. Biotech. 106, 179–191. [DOI] [PubMed] [Google Scholar]
- Hanania, U. , Furman‐Matarasso, N. , Ron, M. and Avni, A. (1999) Isolation of a novel SUMO protein from tomato that suppresses EIX‐induced cell death. Plant J. 19, 533–541. [DOI] [PubMed] [Google Scholar]
- Holtsmark, I. , Takle, G.W. and Brurberg, M.B. (2008) Expression of putative virulence factors in the potato pathogen Clavibacter michiganensis subsp. sepedonicus during infection. Arch. Microbiol. 189, 131–139. [DOI] [PubMed] [Google Scholar]
- Hopwood, D.A. , Bibb, M.J. , Chater, K.F. , Kieser, T. , Bruton, C.J. , Kieser, H.M. , Lydiate, D.J. , Smith, C.P. , Ward, J.M. and Schrempf, H. (1985) Genetic Manipulations of Streptomyces. A Laboratory Manual. Norwich, UK: John Innes Foundation. [Google Scholar]
- Hotson, A. and Mudgett, M.B. (2004) Cysteine proteases in phytopathogenic bacteria: identification of plant targets and activation of innate immunity. Curr. Opin. Plant Biol. 7, 384–390. [DOI] [PubMed] [Google Scholar]
- Hotson, A. , Chosed, R. , Shu, H. , Orth, K. and Mudgett, M.B. (2003) Xanthomonas type III effector XopD targets SUMO‐conjugated proteins in planta . Mol. Microbiol. 50, 377–389. [DOI] [PubMed] [Google Scholar]
- Jahr, H. , Dreier, J. , Meletzus, D. , Bahro, R. and Eichenlaub, R. (2000) The endo‐(‐1,4‐glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant–Microbe Interact. 13, 703–714. [DOI] [PubMed] [Google Scholar]
- Jaton‐Ogay, K. , Paris, S. , Huerre, M. , Quadroni, M. , Falchetto, R. , Togni, G. , Latgé, J.‐P. and Monod, M. (1994) Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus . Mol. Microbiol. 14, 917–928. [DOI] [PubMed] [Google Scholar]
- Kirchner, O. , Gartemann, K.‐H. , Zellermann, E.‐M. , Eichenlaub, R. and Burger, A. (2001) A highly efficient transposon mutagenesis system for the tomato pathogen Clavibacter michiganensis subsp. michiganesis . Mol. Plant–Microbe Interact. 14, 1312–1318. [DOI] [PubMed] [Google Scholar]
- Laine, M.J. , Nakhei, H. , Dreier, J. , Lehtila, K. , Meletzus, D. , Eichenlaub, R. and Metzler, M.C. (1996) Stable transformation of the gram‐positive phytopathogenic bacterium Clavibacter michiganensis subsp. sepedonicus with several cloning vectors. Appl. Environ. Microbiol. 62, 1500–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian, S.K. , Ton‐That, H. and Schneewind, O. (2001) Sortase‐catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus . Mol. Microbiol. 40, 1049–1057. [DOI] [PubMed] [Google Scholar]
- Meletzus, D. , Bermpohl, A. , Dreier, J. and Eichenlaub, R. (1993) Evidence for plasmid‐encoded virulence factors in the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis NCPPB382. J. Bacteriol. 175, 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, K.B. , Nicholas, H.B. Jr. and Deerfield, D.W. II (1997) GeneDoc: analysis and visualization of genetic variation. Embnew. News, 4, 14 http://www.psc.edu/biomed/genedoc [Google Scholar]
- Nielson, H. , Engelbrecht, J. , Brunak, S. and Von Heijne, G. (1997) Identification of procaryotic and eucaryotic signal peptides and prediction of their cleavage site. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Olivieri, F. , Godoy, A.V. , Escande, A. and Casalongué, C.A. (1998) Analysis of intercellular washing fluids of potato tubers and dection of increased proteolytic activity upon fungal infection. Physiol. Plantarum, 104, 232–238. [Google Scholar]
- Olivieri, F. , Zanetti, M.E. , Oliva, C.R. , Covarrubias, A.A. and Casalongué, C.A. (2002) Characterization of an extracellular serine protease of Fusarium eumartii and its action on pathogenesis related proteins. Eur. J. Plant Pathol. 108, 63–72. [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: a Laboratory Manual. New York: Cold Spring Harbor Laboratory. [Google Scholar]
- Smith, G.E. and Summers, M.D. (1980) A bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl‐paper. Anal. Biochem. 109, 123–129. [DOI] [PubMed] [Google Scholar]
- Stackebrandt, E. , Rainey, F.A. and Ward‐Rainey, N.L. (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47, 479–491. [Google Scholar]
- Strider, D.L. (1969) Bacterial canker of tomato caused by Corynebacterium michiganense: a literature review and bibliography. North Carolina Agricultural Experiment Station Tech. Bull. 193. [Google Scholar]
- Tauch, A. , Zheng, Z. , Pühler, A. and Kalinowski, J. (1998) The Corynebacterium striatum resistance transposon Tn5564: genetic organization and transposition in Corynebacterium glutamicum . Plasmid, 40, 126–139. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis, F.M. (1977) Ultrastructural histopathology of tomato plants infected with Corynebacterium michiganense . Physiol. Plant Pathol. 13, 307–317. [Google Scholar]


