SUMMARY
Microbial plant pathogens impose a continuous threat to global food production. Similar to animals, an innate immune system allows plants to recognize pathogens and swiftly activate defence. To activate a rapid response, receptor‐mediated pathogen perception and subsequent downstream signalling depends on post‐translational modification (PTM) of components essential for defence signalling. We discuss different types of PTMs that play a role in mounting plant immunity, which include phosphorylation, glycosylation, ubiquitination, sumoylation, nitrosylation, myristoylation, palmitoylation and glycosylphosphatidylinositol (GPI)‐anchoring. PTMs are rapid, reversible, controlled and highly specific, and provide a tool to regulate protein stability, activity and localization. Here, we give an overview of PTMs that modify components essential for defence signalling at the site of signal perception, during secondary messenger production and during signalling in the cytoplasm. In addition, we discuss effectors from pathogens that suppress plant defence responses by interfering with host PTMs.
INTRODUCTION
Plants are continuously challenged by a variety of organisms, such as viruses, bacteria, fungi, oomycetes, nematodes and insects. Microbes that manage to circumvent structural barriers such as the cell wall and the cuticle are generally not able to invade a plant because of the activation of a primary defence response resulting in non‐host resistance. Most of the microbes that are able to evade or suppress the primary defence response are recognized by the plant via the effector proteins that they secrete, which results in the activation of a secondary defence response that in most cases involves a hypersensitive response (HR). Eventually, only a small subset of microbes has evolved into successful pathogens that are able to suppress and/or circumvent both the primary and the secondary defence responses of the plant (Bent and Mackey, 2007; Chisholm et al., 2006; De Wit, 2007; Jones and Dangl, 2006; Nürnberger et al., 2004). These pathogens cause disease, and can result in severe crop losses.
The primary and secondary defence responses of plants leading to resistance rely on the swift activation of signal transduction cascades, whereby cellular changes caused by the secondary defence response are generally most pronounced (Jones and Dangl, 2006). Research on the molecular aspects of recognition and subsequent defence signalling was initiated by the proposition of the gene‐for‐gene hypothesis by Flor (1942). Since then, many sophisticated pathogen recognition mechanisms have been discovered that initiate highly complex signalling cascades, eventually leading to host genotype‐specific resistance. Thus far, the main focus of molecular phytopathologists has been the identification and functional analysis of resistance (R) proteins and their cognate pathogen effectors, the so‐called race‐specific elicitors (Bent and Mackey, 2007; Takken and Tameling, 2007). In addition, transcriptional changes that occur upon pathogen recognition have been extensively studied by microarray and cDNA‐AFLP experiments (Eulgem, 2005; Wise et al., 2007), and the role of individual genes in resistance has been studied by transient/stable knockdown and knockout studies (Baulcombe, 1999; Burch‐Smith et al., 2004; Glazebrook et al., 1997).
Initial plant defence responses occur extremely fast upon recognition of a pathogen (Laxalt and Munnik, 2002; Nürnberger and Scheel, 2001; Wojtaszek, 1997), which implies the involvement of post‐translational modifications (PTMs) of pre‐existing proteins in signal transduction cascades. A definite role for PTMs in defence signal transduction became apparent with the discovery of protein phosphorylation events in parsley cells upon elicitor treatment (Dietrich et al., 1990), and with the observation that activated mitogen‐activated protein kinases (MAPKs), which require phosphorylation for activation, are involved in the primary resistance response of parsley to Phytophthora sojae (Ligterink et al., 1997). Furthermore, some receptors contain kinase domains themselves, which enable them to phosphorylate downstream substrates (Martin et al., 2003; Van Ooijen et al., 2007). In recent years, the general importance of PTMs in signal transduction cascades has become clear (Thurston et al., 2005; Xing et al., 2002) and its relevance for successful plant defence signalling was further confirmed by reports describing direct manipulation of PTMs by pathogens in order to suppress plant immune responses (Kim et al., 2005b; Mudgett, 2005; Shan et al., 2007). In this review we will discuss different types of host protein PTMs that play a role in plant defence signalling. In addition, we discuss effectors from pathogens that specifically interfere with host PTMs to suppress plant defence responses, thereby underlining the importance of PTMs in defence signalling.
POST‐TRANSLATIONAL MODIFICATIONS, HOW DO THEY OCCUR?
Single genes can give rise to a diversity of RNA transcripts because of gene splicing and each of these transcripts is translated into a protein that can subsequently be proteolytically processed and/or post‐translationally modified. PTMs are responsible for a major increase in complexity from genome to proteome. For example, the human genome, containing approximately 30 000 open reading frames, is predicted to give rise to approximately 1.8 million different protein species (Jensen, 2004; Kersten et al., 2006). PTMs are involved in protein regulation and are therefore often reversible, rapid, controlled and highly specific but they usually affect only a small percentage of the total pool of a specific protein (Johnson, 2004). Furthermore, PTMs are catalysed by specific enzymes that in turn are often also regulated by PTMs (Peck, 2006). Currently, more than 300 types of PTMs have been described (Jensen, 2004), but here we focus on the major PTMs that have been implicated in defence signalling.
Phosphorylation
Phosphorylation is the most predominant covalent modification of proteins and implies the reversible attachment of a phosphate group to an amino acid residue. Phosphorylation has been described to play a major role in defence signalling cascades (de la Fuente van Bentem and Hirt, 2007; Peck, 2003; Thurston et al., 2005; Xing et al., 2002). Four types of phosphorylation occur, of which N‐, S‐ and acyl‐phosphorylation are very uncommon. O‐phosphorylation is the most common type and is here further referred to as phosphorylation. Phosphorylation mainly occurs on the hydroxyl group of hydroxyamino acids such as serine, threonine and tyrosine but can also occur on unusual residues such as hydroxy‐proline (Reinders and Sickmann, 2005). Phosphorylation is executed by protein kinases that transfer a phosphoryl (PO3) group from ATP to the hydroxyl group in the polar rest (R‐) group of the amino acid residue, resulting in a phosphoester (R–O–PO3) bond. Dephosphorylation occurs by protein phosphatases that hydrolyse the phosphoester bond, thereby releasing the phosphoryl group and restoring the hydroxyamino acid into its unphosphorylated state (Sickmann and Meyer, 2001). Generally, only a small percentage of the total pool of a certain protein in the cell is phosphorylated and a transient change of only a few percent can be sufficient to activate signalling. The opposite activity of kinases and phosphatases balances phosphorylation‐based signalling cascades, rendering them highly dynamic (Reinders and Sickmann, 2005).
Ubiquitination
Another highly dynamic PTM that is implicated in defence signalling is ubiquitination. Ubiquitination refers to a three‐step enzymatic cascade to covalently attach a small conserved polypeptide, ubiquitin, to a protein. First, the C‐terminal glycine of ubiquitin, which is maturated by deubiquitination enzymes (DUBs), forms together with the thiol group (SH) of a cysteine in the active site of the ubiquitin‐activating enzyme (E1), a thioester (RE1–S–CO–RUb). Subsequently, the activated ubiquitin is transferred to a cysteine residue of the ubiquitin‐conjugating enzyme (E2). Finally, the ubiquitin‐ligase protein (E3), which interacts with the ubiquitinated E2 enzyme, initiates attachment of the ubiquitin moiety to the target protein by an isopeptide bond between the C‐terminal glycine of ubiquitin and the ɛ‐amino group of a lysine residue of the target protein (Vierstra, 2003). The target protein often requires phosphorylation prior to binding to the E3 complex. To form a polyubiquitinated protein, these three steps are repeated so that each new ubiquitin moiety is attached to a lysine residue of the previous ubiquitin moiety. Polyubiquitination can lead to lysine (K) 48‐ and K63‐linked chains, depending on which lysine in the ubiquitin moiety is targeted for ubiquitination, and on the E2 conjugating enzyme. Proteins modified with a K48‐chain are normally targeted to the 26S proteasome for degradation, whereas K63‐chains are involved in endocytosis of the protein, its activation or modification of its activity (Angot et al., 2007). Some proteins are only monoubiquitinated and this may also trigger a change in the localization and/or activity of the protein (Haglund et al., 2003). Eventually, DUBs are capable of removing the covalently bound ubiquitin moieties thereby changing the fate of the protein, but they also recycle ubiquitin moieties from ubiquitinated proteins processed by the proteasome (Kerscher et al., 2006; Vierstra, 2003).
Sumoylation
Similar to ubiquitination, proteins can be decorated with a small ubiquitin‐related modifier (SUMO) moiety during defence signalling (Miura et al., 2007; Novatchkova et al., 2004). Sumoylation has been reported in cell‐cycle activity, DNA repair, nuclear localization, enzymatic activity and stability of proteins and in the modulation of transcription factor activity (Miura et al., 2007). Similar to ubiquitin, SUMO is processed to expose its C‐terminal glycine that is subsequently attached to a lysine residue of a target protein, via conjugation machinery similar to that for ubiquitination. However, sumoylation differs from ubiquitination as it has only one universal E2‐conjugating enzyme that does not always require an E3‐ligase to transfer SUMO to the targeted protein. Furthermore, a weak consensus motif for sumoylation has been identified in target proteins and normally only mono‐sumoylation occurs although poly‐sumoylation has been reported. Finally, the cysteine proteases required for SUMO maturation and desumoylation belong to a distinct family of ubiquitin‐like protein proteases (ULPs) (Chosed et al., 2006).
S‐nitrosylation
S‐nitrosylation of proteins is another mechanism to regulate cellular processes and although not very well described, this modification is regarded as influential as protein phosphorylation (Lindermayr et al., 2006). Protein S‐nitrosylation occurs on cysteine residues, mainly via two mechanisms. Proteins can either become S‐nitrosylated via an oxygen‐dependent reaction where nitrosonium (NO+) reacts with a thiolate group (R–S−) of the cysteine in the protein, or nitric oxide (NO) can be transferred from a nitrosothiol (SNO) to the thiol group (SH) of the cysteine (transnitrosylation). SNOs consist of small molecules, like glutathione, with a thiol group (GSH), that react with NO resulting in S‐nitrosoglutathione (GSNO), and are suggested to be the NO reservoirs and NO donors in the cell (Lindermayr et al., 2006). Although reports on S‐nitrosylation during plant–pathogen interactions are rare, the production of NO and its signalling function during plant–pathogen interactions are well described (Romero‐Puertas et al., 2004). The presence of GSNO reductase activity in plants, which releases NO from GSNO, indicates that the formation of SNOs could play an important role in NO signalling (Lindermayr et al., 2005).
Glycosylation
Covalent linkage of an oligosaccharide side chain to a protein is referred to as protein glycosylation. The two most predominant types are N‐glycosylation and O‐glycosylation. Here, we only consider N‐glycosylation, which can affect the asparagine residue in the sequence motif asparagine–X–serine/threonine (X can be any amino acid except proline) and which refers to the oligosaccharide side chain attachment to the asparagine residue. N‐glycosylation starts co‐translationally at the endoplasmic reticulum (ER) by the transfer of an oligosaccharide precursor, Glc3Man9GlcNAc2, onto the amide nitrogen of the asparagine residue. Subsequently, the oligosaccharide matures by the removal of glucose and mannose residues or by the attachment of new sugar residues to generate glycans and complex‐type glycans (Saint‐Jore‐Dupas et al., 2007). Glycosylation occurs quite frequently and can affect the biological activity and the function of proteins, and has been reported to occur also on resistance proteins (Van der Hoorn et al., 2005).
N‐myristoylation and S‐palmitoylation
Next to the attachment of sugars to proteins, proteins can also be modified co‐translationally (N‐myristoylation) or post‐translationally (S‐palmitoylation) with fatty acids. N‐myristoylation, also referred to as myristoylation, is the modification of a protein with myristate, a hydrophobic 14‐carbon fatty acid. Catalysed by N‐myristoyltransferase, myristate is in general covalently and irreversibly attached through amide linkage to the N‐terminal glycine exposed after removal of the initial methionine residue of the target protein by aminopeptidases. Myristoylation targets proteins to a membrane and thereby promotes interactions between these proteins and membrane‐associated protein complexes (Farazi et al., 2001). Protein myristoylation plays an important role in defence signalling in tomato against Pseudomonas syringae (Andriotis and Rathjen, 2006). S‐palmitoylation, also referred to as S‐acylation, is the thioesterification of palmitate (a 16‐carbon fatty acid) to a cysteine residue in a protein. S‐palmitoylation is catalyzed by palmityl acyltransferases (PAT) or occurs via a spontaneous autoacylation in the presence of long‐chain acyl‐coenzyme As (CoAs) and lipid vesicles. S‐palmitoylation supports initial plasma‐membrane binding of proteins (Smotrys and Linder, 2004), including proteins required for the perception of pathogen elicitors, and might play a role in protein trafficking (Kim et al., 2005a).
GPI‐anchoring
GPI‐anchoring implies the attachment of a glycosylphosphatidylinositol (GPI) to anchor cell‐surface proteins to the plasma membrane, where they can play a role in elicitor perception. GPI is synthesized at the ER via the sequential linkage of sugars and other components to phosphatidylinositol (PI). GPI transamidases recognize and cleave the C‐terminal GPI attachment signal peptide of the target and mediate attachment to the GPI anchor. The GPI‐anchored protein is subsequently secreted via the Golgi apparatus and attached to the plasma membrane (Maeda et al., 2006).
PTMS OF HOST PROTEINS INVOLVED IN SIGNAL PERCEPTION
Pathogen recognition is mediated by a group of protein receptors which can be divided in a few major classes. Two classes account for the receptor‐like proteins (RLPs) and the receptor‐like kinases (RLKs) that are localized in the plasma membrane and contain extracellular leucine‐rich repeats (LRRs). The RLPs lack a cytoplasmic signalling domain, while RLKs have a cytoplasmic kinase domain. Two other classes are formed by receptors that are cytoplasmically localized and that contain a nucleotide‐binding (NB) site and LRRs. One class is referred to as TIR‐NB‐LRRs as these NB‐LRRs contain an N‐terminal domain similar to the Drosophila Toll receptor and the interleukin 1 receptor (TIR). The other class is referred to as CC‐NB‐LRRs, as N‐terminal domain structures in which frequently coiled‐coil (CC) motifs are predicted are found in addition to the NB‐LRR domains (Martin et al., 2003; Van Ooijen et al., 2007). It has become clear that the primary (non‐host) defence response elicited by microbe‐associated molecular patterns (MAMPs) and the secondary (host genotype‐specific) defence response induced by race‐specific elicitors are in fact mediated by very similar receptors (Gómez‐Gómez and Boller, 2000; Zipfel et al., 2006). These receptors are today referred to as pattern recognition receptors (PRRs) and R proteins, respectively (Bent and Mackey, 2007; Jones and Dangl, 2006).
Signal perception by RLKs
The best studied model system in Arabidopsis for primary defence signalling is the perception of bacterial flagellin, or its 22‐amino‐acid conserved epitope, flg22, by the membrane‐bound PRR FLS2. FLS2 is an RLK and autophosphorylation of its kinase domain seems to be required for binding of flg22 and might affect the stability of the FLS2‐flg22 complex (Gómez‐Gómez et al., 2001). Mutation of four, probably not autophosphorylated, phosphorylation sites in the C‐terminal region of the protein did not affect flg22 binding but abolished or reduced downstream signalling. Mutation of one of these sites also significantly reduced FLS2 internalization by endocytosis (Robatzek et al., 2006). FLS2 endocytosis might be triggered by ubiquitination as the required conserved (PEST) motif is present in the cytoplasmic region of the FLS2 protein, and FLS2 endocytosis is followed by its degradation (Fig. 1; Robatzek et al., 2006). It has been found recently that the FLS2 receptor specifically binds to one of the somatic embryogenesis receptor kinases, SERK3, also referred to as BRI1‐associated receptor kinase 1 (BAK1), in a ligand‐dependent manner (Chinchilla et al., 2007; Heese et al., 2007). Upon perception of brassinosteroids (BRs), which are plant steroid hormones, BAK1 forms a heterodimer with the plasma membrane receptor kinase BRI1 (BRASSINOSTEROID‐INSENSITIVE 1). Both BAK1 and BRI1 display BR‐dependent phosphorylation (Wang et al., 2005), which enhances the interaction and complex formation between the two proteins that are subsequently internalized via endocytosis (reviewed by Karlova and de Vries, 2006). Possibly, the ligand‐dependent FLS2‐BAK1 complex formed in vivo is internalized in a similar way to the BRI1‐BAK1 complex (Fig. 1; Chinchilla et al., 2007). The kinase‐associated protein phosphatase (KAPP) might negatively regulate FLS2 signalling as it binds and dephosphorylates FLS2 (Gómez‐Gómez et al., 2001). Recently, a highly homologous receptor that recognizes an 18‐amino‐acid fragment of the bacterial elongation factor Tu (EF‐Tu) was identified (Zipfel et al., 2006). Just like FLS2, this EF‐Tu receptor (EFR) requires BAK1 for downstream signalling and upon stimulation both PRRs induce the transcription of a similar set of genes, including a large amount of additional RLKs. Furthermore, they induce a common set of responses including downstream MAPK activation and extracellular alkalization (Chinchilla et al., 2007; Zipfel et al., 2006). Phosphorylation of the EFR receptor itself has not yet been reported, but the homology to the FLS2 signalling cascade suggests a role for EFR‐mediated phosphorylation upon EF‐Tu perception (reviewed by Nürnberger and Kemmerling, 2006). In addition, an RLK referred to as RPG1 confers resistance of barley to Puccinia graminis f. sp. tritici. RPG1 contains two tandem kinase domains of which only the C‐terminal domain is functional and displays autophosphorylation required for resistance. In accordance with FLS2 signalling, RPG1 appears to be degraded in a proteasome‐dependent way upon inoculation with an avirulent strain, which implies that RPG1 becomes ubiquitinated (Nirmala et al., 2006, 2007). Furthermore, the rice RLK Xa21, which mediates recognition of the effector AvrXa21 from Xanthomonas oryzae pv. oryzae (Song et al., 1995), has a kinase domain that autophosphorylates on several serine and threonine residues and that stabilizes the protein and probably protects it from proteolytic cleavage (Liu et al., 2002; Xu et al., 2006). Xa21 phosphorylates the Xa21‐binding protein 3 (XB3) that binds in vivo to the receptor and which is required for its accumulation. XB3 is a RING finger‐containing protein that can function as an E3 ubiquitin ligase and it is hypothesized that XB3 is phosphorylated by Xa21 upon pathogen recognition. XB3 subsequently ubiquitinates a downstream component, which could be a negative regulator of defence signalling that is targeted for degradation (Fig. 1; Wang et al., 2006). However, referring back to FLS2, XB3 might also mediate Xa21 ubiquitination and degradation. It is tempting to speculate that RLK‐mediated signalling is initiated by phosphorylation and formation of a ligand‐dependent protein complex that internalizes and is subsequently degraded in a proteasome‐dependent manner.
Figure 1.
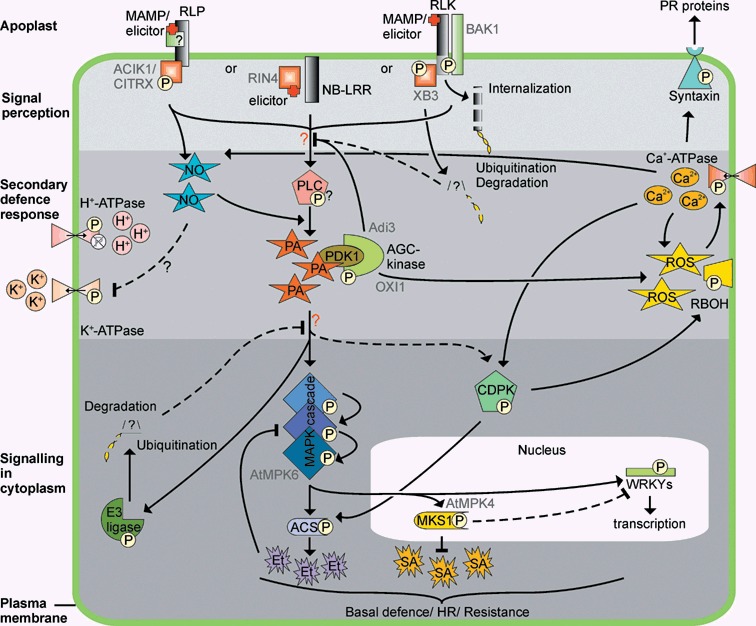
Defence‐related signal transduction cascades that depend on post‐translational modifications. Receptors mediate recognition of microbe‐associated molecular patterns (MAMPs) and race‐specific elicitors (elicitor), but they require additional proteins for their function. Proteins with nucleotide‐binding and leucine‐rich repeat domains (NB‐LRR) recognize their cognate elicitors intracellularly, while receptor‐like proteins (RLP) and receptor‐like kinases (RLK) are probably activated outside the cell. RLPs require additional proteins that bind the cytoplasmically localized part of the protein to mediate downstream signalling, while RLKs require their kinase domain to autophosphorylate and form complexes with additional proteins. RLKs might become ubiquitinated, after which they are internalized and targeted for proteasome‐mediated degradation. Signalling downstream from the receptor eventually leads to the formation of secondary messengers such as phosphatidic acid (PA), possibly via phospholipase C (PLC) phosphorylation, and nitric oxide (NO). The concentrations of ions such as H+, K+ and Ca2+ are controlled by (de)phosphorylation of the respective ATPase while the production of ROS is stimulated upon phosphorylation of the NADPH oxidases (RBOH). The secondary messengers also mediate phosphorylation of proteins such as calcium‐dependent protein kinases (CDPK), or syntaxins, which might promote the release of pathogenesis‐related (PR) proteins into the apoplast. Mitogen‐activated protein kinase (MAPK) cascades are activated by phosphorylation of the individual components, which eventually leads to the phosphorylation of, amongst others WRKY transcription factors, 1‐aminocyclopropane‐1‐carboxylic acid synthase (ACS) and MAP kinase substrate 1 (MKS1), which influence the production of ethylene (Et) and salicylic acid (SA), respectively. Also, E3‐ligases are activated, which might result in the ubiquitination and subsequent degradation of negative regulators of the signalling cascades, thereby providing a positive feedback loop. In addition, negative feedback loops are required to prevent an uncontrolled hypersensitive response (HR). For example, MAPK (MPK)‐mediated ethylene production negatively regulates MAPK activation. The secondary messengers influence each other and fine‐tune the downstream signal while proteins modified by secondary messengers might inhibit receptor‐mediated signals. Eventually, a balanced signal will lead to increased (basal) resistance and possibly a HR. Phosphorylation states as presented in this figure represent the active state of the protein. Protein names indicated in grey might be specific for a particular plant–pathogen interaction. ACIK1, Avr9/Cf‐9‐induced kinase 1; CITRX, Cf‐9‐interacting thioredoxin; RIN4, RPM1‐interacting protein 4; BAK1, BRASSINOSTEROID‐INSENSITIVE 1; XB3, Xa21‐binding protein 3; Adi3, AvrPto‐dependent Pto‐interacting protein 3; PDK1, 3‐phosphoinositide‐dependent protein kinase‐1; AGC‐kinase, protein kinase A, G and C family; OXI1, oxidative signal‐inducible 1.
Signal perception by RLPs
RLPs, which lack a kinase domain and thus lack autophosphorylation, are represented by, for example, the so‐called Cf proteins of tomato plants mediate resistance to Cladosporium fulvum (Rivas and Thomas, 2005). One of the family members is Cf‐9, which is highly glycosylated, a feature required for its stability and for a full Cf‐9‐mediated HR. Probably, Cf‐9 N‐glycosylation is required for a stable structural conformation and/or interactions with the cell wall (Piedras et al., 2000; Van der Hoorn et al., 2005). Cf‐9 has no signalling domain but the cytoplasmic C‐terminus interacts with a thioredoxin (CITRX; for Cf‐9‐interacting thioredoxin) that accelerates the Cf‐9/Avr9‐induced HR upon transcriptional knockdown by virus‐induced gene silencing (VIGS) (Rivas et al., 2004). In addition, the Avr9/Cf‐9‐induced kinase 1 (ACIK1), which encodes a cytoplasmic serine/threonine kinase, compromises the Cf‐9/Avr9‐ and Cf‐4/Avr4‐induced HR and resistance upon VIGS (Durrant et al., 2000; Rowland et al., 2005). Interestingly, ACIK1 binds and phosphorylates CITRX and binds the C‐terminus of Cf‐9 with CITRX as an adaptor protein, thereby forming a complex that can mediate downstream signalling (Nekrasov et al., 2006). However, it remains difficult to understand how the downstream signalling from Cf‐9 actually takes place given that ACIK1 is a positive regulator and CITRX a negative regulator of Cf‐9/Avr9‐induced defence signalling, and the catalytic domains are not required for the interaction between the different proteins (Nekrasov et al., 2006). We hypothesize that Cf‐9, CITRX and ACIK1 form a complex under normal conditions in unchallenged plants. Upon elicitation by Avr9, ACIK1 phosphorylates CITRX, which destabilizes the complex and releases CITRX and ACIK1 into the cytoplasm where they can activate downstream signalling components resulting in a balanced defence response (Fig. 1). In addition to race‐specific elicitor recognition, RLPs also mediate MAMP‐induced defence responses. The MAMP xylanase from Trichoderma viride triggers signalling through the ethylene‐inducing xylanase (EIX) PRR, which is an RLP (Ron and Avni, 2004). Chitin, a major component from fungal cell walls, is a MAMP that triggers signalling by the chitin oligosaccharide elicitor‐binding protein CEBiP, which is an RLP that is highly glycosylated, just like Cf‐9 (Kaku et al., 2006). However, for these PRRs it remains to be elucidated how the perceived signal is transferred further downstream to the cytoplasm.
Signal perception by NB‐LRRs
Resistance to Pseudomonas species is in most cases conferred by NB‐LRRs. The interaction between tomato and Pseudomonas syringae pv. tomato (Pst) is a well‐studied model system. Resistance to Pst expressing the elicitor genes AvrPto and/or AvrPtoB requires the Pto and the Prf genes (Kim et al., 2002; Salmeron et al., 1994). Pto encodes a serine/threonine protein kinase (Loh and Martin, 1995) and, originally, Pto was reported as the AvrPto‐matching R protein (Martin et al., 1993). However, further analysis revealed Prf as a CC‐NB‐LRR protein, which is capable of signalling in the absence of Pto, while Pto is incapable of signalling in the absence of Prf (Salmeron et al., 1996). It was also shown that Prf and Pto interact in vivo and that Prf accumulates to higher levels in the presence of Pto (Mucyn et al., 2006). Therefore, Prf is now classified as the R protein that activates downstream signalling (Van Ooijen et al., 2007). Still, Pto plays an important role in AvrPto and AvrPtoB perception as Pto specifically binds both elicitors and several other Pto‐interacting (Pti) proteins (Kim et al., 2002; Sessa et al., 2000b; Tang et al., 1996). Pti1 represents a serine/threonine kinase which is phosphorylated by the Pto kinase, and this phosphorylation is required for Pto/Pti1 interaction (Sessa et al., 2000a). In vitro, Pto autophosphorylates at eight sites, three of which are required for HR development and AvrPto binding, and one is only required for HR development, indicating that Pto kinase activity is required for the AvrPto/Prf‐dependent HR elicitation (reviewed by Pedley and Martin, 2003). Further research revealed two additional phosphorylation sites in the activation loop of Pto required for AvrPto binding. Substitution of these residues by aspartic acid (D), which mimics the negative charge introduced by phosphorylation, resulted in a Prf‐dependent and AvrPto‐independent HR in tomato (Rathjen et al., 1999). To complicate Pto‐mediated AvrPto perception further, Pto was also found to be myristoylated at the N‐terminus, which negatively regulates its kinase activity (Andriotis and Rathjen, 2006). A model summarizing these results has been proposed: Pto is myristoylated to suppress its kinase activity and to be targeted to a cellular membrane, most likely the plasma membrane, where it binds to Prf. AvrPto targets the complex and causes displacement of the myristoylated N terminus of Pto, which results in derepression of the kinase domain, Pto phosphorylation and activation, and subsequent signalling via Prf (Andriotis and Rathjen, 2006; Balmuth and Rathjen, 2007). Furthermore, AvrPto and phosphorylated Pto form a complex with AvrPto‐dependent Pto‐interacting protein 3 (Adi3). Adi3 is a member of the AGC family of protein kinases (protein kinase A, G and C family) and negatively regulates Pto‐AvrPto‐induced host cell death when phosphorylated by 3‐phosphoinositide‐dependent protein kinase‐1 (PDK1) or Pto. In contrast to the AvrPto‐dependent Pto/Adi3 interaction, Adi3 phosphorylation by Pto is independent of AvrPto and is not required for Pto/AvrPto/Adi3 complex formation. Therefore, it is hypothesized that Adi3‐mediated negative regulation is released when bound to the Pto‐AvrPto complex (Devarenne et al., 2006). Possibly, phosphorylated Adi3 negatively regulates elicitor‐independent Pto signalling under normal conditions to avoid activation of defence responses. Upon elicitation, Adi3 is dephosphorylated and binds to Pto, which leads to Pto‐mediated signalling. As described in other defence signalling cascades, secondary messengers such as phosphatidic acid might be produced (see below) that bind and possibly activate PDK1 (Testerink et al., 2004). PDK1 might subsequently phosphorylate Adi3 to negatively regulate Pto signalling again, thereby forming a negative feedback loop (Fig. 1).
Interactions between Pseudomonas syringae and Arabidopsis are also intensively studied and several intracellular NB‐LRRs have been described to mediate recognition of elicitors from different P. syringae strains (Nimchuk et al., 2003). The R proteins RPS2 and RPS5 provide resistance to P. syringae pathovars expressing AvrRpt2 or AvrPphB, respectively, whereas RPM1 provides resistance to P. syringae pathovars expressing AvrRpm1 or AvrB (reviewed by Belkhadir et al., 2004b). In a yeast two‐hybrid screen, two RPM1‐interacting proteins (RINs), RIN2 and RIN3, were identified which represent RING‐finger ubiquitin E3 ligases and which also weakly interact with RPS2. These RINs seem to enhance the RPM1‐ and RPS2‐mediated HR; however, they do not restrict bacterial growth in the plant. Although RIN2 and RIN3 encode proteins that show E3 ligase activity in vitro, a target protein that might serve as a negative regulator of the HR and is degraded remains to be identified (Kawasaki et al., 2005). Another protein that physically interacts with RPM1 and RPS2 is RIN4, a protein that negatively regulates RPM1‐ and RPS2‐mediated resistance (Belkhadir et al., 2004a). RIN4 is C‐terminally palmitoylated, which is required for RIN4 localization to the plasma membrane and its functioning (Belkhadir et al., 2004a; Day et al., 2005; Kim et al., 2005a). It is hypothesized that RIN4 is bound to RPM1 and RPS2 under normal conditions to regulate defence signalling negatively and that RIN4 is released from the complex upon R protein triggering (Fig. 1). In addition, defence signalling by RPM1 or RPS2 requires the non‐race‐specific disease resistance 1 (NDR1) protein, which is glycosylated and C‐terminally processed. NDR1 is thought to undergo GPI modification at its processed C‐terminus and this GPI‐anchor places the protein on the outer surface of the plasma membrane with a short part of the N‐terminus in the cytoplasm, where it binds the C‐terminal half of RIN4 (Coppinger et al., 2004; Day et al., 2006). Upon inoculation with an AvrRpt2‐producing Pst strain, RIN4 is cleaved by the cysteine protease activity of the AvrRpt2 effector, after which the negative regulation of RPS2 by RIN4 is released (Fig. 1; Takemoto and Jones, 2005). A C‐terminal membrane‐embedded RIN4 fragment is not degraded after cleavage and positively regulates RPS2‐mediated signalling by its interaction with NDR1 (Day et al., 2006). Probably, RIN4‐mediated RPM1 activation by AvrRpm1/AvrB elicitation occurs via a different mechanism, as RIN4 degradation abolishes RPM1 signalling, and RPM1 activation depends on RIN4 phosphorylation (Kim et al., 2005a,b). In addition to RPM1 and RPS2, also RPS5‐mediated resistance depends on NDR1 but RPS5 does not require RIN4 (Coppinger et al., 2004). Instead, RPS5‐mediated resistance to P. syringae depends on a serine/threonine protein kinase PBS1 that binds to RPS5 in unchallenged plants (Ade et al., 2007; Swiderski and Innes, 2001). Similar to AvrRpt2‐mediated cleavage of RIN4 and the subsequent activation of RPS2, the cysteine protease AvrPphB cleaves PBS1, which activates RPS5. PBS1 requires a functional kinase domain that is probably involved in autophosphorylation; however, neither the phosphorylation nor the elimination of PBS1 is sufficient to activate RPS5. Therefore, it is hypothesized that a phosphorylated cleavage product of PBS1 is required for RPS5 activation (Shao et al., 2003).
PTMS OF PLASMA MEMBRANE‐LOCALIZED HOST PROTEINS INVOLVED IN DOWNSTREAM SIGNALLING
In addition to the above described complexes that are at least partially localized and/or bound to the plasma membrane, several other post‐translationally modified membrane‐localized proteins exist that are not directly involved in signal perception but play a role in downstream responses.
Transport of secondary messengers over the plasma membrane by ATPases
Secondary messengers are transported over membranes by pumps that are driven by the hydrolysis of ATP and are referred to as ATPases. An important subclass of ATPases is formed by the H+‐ATPases that mediate the generation of electrochemical gradients across the plasma membrane, which is the energy source for most transport proteins (Palmgren, 2001). H+‐ATPases require phosphorylation on a threonine residue in the N‐terminus for their activity and are inactivated by dephosphorylation of this site. However, a plasma membrane H+‐ATPase from Arabidopsis was also inactivated by phosphorylation on a serine residue by the PKS5 serine/threonine protein kinase. This phosphorylation event prevents interaction with a 14‐3‐3 protein and therefore inhibits the activity of the H+‐ATPase (Fig. 1; Fuglsang et al., 2007). Furthermore, plasma membrane‐bound H+‐ATPases are dephosphorylated upon recognition of the Avr5 elicitor of C. fulvum by Cf‐5 tomato suspension cells (Vera‐Estrella et al., 1994). In addition to H+‐ATPases, Ca2+‐ATPases also seem to be regulated via phosphorylation. For example, in closing Vicia guard cells, Ca2+‐ATPases become phosphorylated, which enhances Ca2+ import in the cell (Köhler and Blatt, 2002). Furthermore, elicitation of the plasma membrane of Cf‐5 tomato protoplasts with the Avr5 elicitor activates a Ca2+‐ATPase by G‐protein‐dependent phosphorylation (Fig. 1; Gelli et al., 1997). Also, K+ channel activity seems to depend on phosphorylation. The stimulation of the K+ outward channels and the suppression of the K+ inward channels upon elicitation of transgenic Cf‐9‐expressing Nicotiana tabacum cells with Avr9 is completely blocked by broad‐range protein kinase inhibitors (Blatt et al., 1999). K+ channels might also be nitrosylated as NO blocks outward K+ channels in guard cells (Fig. 1; Sokolovski and Blatt, 2004).
Syntaxins and other membrane‐bound proteins in defence signalling
To identify plasma membrane‐bound proteins in Arabidopsis that are (de)phosphorylated upon defence signalling, 32P pulse‐labelled suspension‐cultured cells were elicited with flg22 and plasma membrane proteins were analysed by two‐dimensional gel electrophoresis (Nühse et al., 2003). This revealed several differentially phosphorylated proteins such as the syntaxin AtSyp122. Syntaxins are part of the SNARE complex and play a central role in exocytosis as they mediate vesicle fusion to the plasma membrane (Fasshauer, 2003). Phosphorylation of AtSyp122 is Ca2+‐dependent, which leads to the hypothesis that a Ca2+ influx stimulates exocytosis of defence proteins and other compounds via syntaxins (Fig. 1). In agreement with this hypothesis, the same phosphoproteomics screen revealed a second syntaxin, AtSyp132, of which the N. benthamiana orthologue, NbSyp132, contributes to the exocytosis of pathogenesis‐related (PR) proteins into the apoplast upon Pto/AvrPto‐induced defence signalling (Kalde et al., 2007). Furthermore, NbSyp132 contributes to basal‐ and salicylate‐associated defence against bacterial pathogens in plants (Kalde et al., 2007). Another plasma membrane‐localized syntaxin, Syp121 or PEN1, is required for resistance to powdery mildew in barley but does not play a role in Pto‐mediated resistance to Pst (Fig. 1; Collins et al., 2003; Kalde et al., 2007). The orthologue NtSyp121 is phosphorylated upon Cf‐9/Avr9‐activated signalling, which appeared to be specific as this syntaxin is not phosphorylated upon elicitation with flg22 (Heese et al., 2005).
Recent technical advances in phosphoproteomics now enable phosphopeptide or phosphoprotein purification and their immediate analysis by mass spectrometry. A non‐quantitative analysis of phosphorylated plasma membrane‐bound proteins from flg22‐elicited Arabidopsis cells revealed over 300 phosphorylation sites although it remains unclear to what extent these phosphorylation sites play a role during signalling cascades (Nühse et al., 2004). Recently, quantitative phosphoproteome studies of flg22‐ or xylanase‐treated Arabidopsis cells revealed several differentially phosphorylated proteins. Some of these proteins, such as calcium‐dependent protein kinases (CDPKs) and ATPases, have already been described to be regulated by phosphorylation, but for other proteins like auxin efflux carriers and respiratory burst oxidase protein D, phosphorylation‐mediated regulation is novel (Benschop et al., 2007; Nühse et al., 2007). Further functional analysis of the identified phosphoproteins will reveal new insights into defence‐related signalling cascades.
PTMS LEADING TO THE FORMATION OF HOST SECONDARY MESSENGERS
When a microbe is recognized by the plant, defence signalling cascades are activated. Thus far, it is unclear how signals are transferred from the receptor to one or more downstream pathways such as the MAPK pathway. Studies using suspension‐cultured cells indicate that in intact plants, secondary messengers are produced upon elicitation and they are thought to play a role in amplifying and transferring the signal downstream into the signalling cascade (Laxalt and Munnik, 2002).
NO signalling
Elicitation of tomato cells with xylanase results in the production of the secondary messenger NO (Fig. 1; Laxalt et al., 2007). In Arabidopsis, NO is synthesized by the NO synthase enzyme, AtNOS1 (Guo et al., 2003), or results from the reduction of nitrate by nitrate reductase (NR) (Romero‐Puertas et al., 2004). However, other mechanisms to generate NO are also likely to exist (Neill et al., 2007). To transfer a signal, the highly reactive NO molecules can modify a variety of target proteins by S‐nitrosylation. An extensive study in Arabidopsis led to the identification of many proteins that can be modified by S‐nitrosylation, of which some proteins such as superoxide dismutases and Hsp90 have been reported in defence signalling as well (Lindermayr et al., 2005).
PA signalling
Another secondary messenger is the phospholipid‐derived molecule phosphatidic acid (PA), which is produced upon signal perception via the phospholipase C or D (PLC/PLD) pathway (Bargmann and Munnik, 2006; Laxalt and Munnik, 2002; Testerink and Munnik, 2005). Most elicitors reported to induce PA production stimulate PLC‐mediated formation of PA via the phosphorylation of the intermediate diacylglycerol (DAG) by DAG kinase (DGK), although some elicitors also activate the PLD pathway (Andersson et al., 2006; De Jong et al., 2004; Van der Luit et al., 2000). The PLCs might be activated by upstream kinases that have been activated as a result of receptor triggering, as an Arabidopsis PLC was reported to be phosphorylated upon flagellin elicitation (Fig. 1; Nühse et al., 2007). In soybean, PA generated upon wounding has been shown to activate the MAPK cascade as the addition of exogenous PA to suspension‐cultured cells specifically activates a MAPK (Fig. 1). PA formation in wound‐induced leaves can be blocked with PLD inhibitors (Lee et al., 2001). Furthermore, PA stimulates the oxidative burst upon elicitation (Andersson et al., 2006; De Jong et al., 2004). In xylanase‐treated tomato suspension cells, PA is produced via the PLC/DGK pathway, which is activated by a xylanase‐triggered NO accumulation. How NO exactly activates the PLC/DGK pathway remains unclear, although NO might act directly on PLC and/or DGK by protein S‐nitrosylation (Fig. 1). NO might also affect the PLC/DGK pathway indirectly via the MAPK signalling cascade, via altered Ca2+ levels or via a change in redox potential in the cell (Laxalt et al., 2007; Lee et al., 2001). In Arabidopsis, PA targets have been identified and include heat shock protein 90, serine/threonine kinases and phosphatases (Testerink et al., 2004). Another target is the previously described phosphoinositide‐dependent kinase PDK1 (Fig. 1, see above) (Anthony et al., 2006). PDK1 interacts with the OXI1 kinase (oxidative signal‐inducible 1; also referred to as AGC2‐1) and subsequently phosphorylates and activates OXI1, which is involved in oxidative burst‐mediated signalling in Arabidopsis (Fig. 1; Anthony et al., 2004, 2006; Rentel et al., 2004). OXI1, in turn, phosphorylates the serine/threonine kinase PTI1‐2, which has high sequence homology to the tomato Pti1 kinase. The signalling pathway PDK1/OXI1/PTI1‐2 was shown to be specific for lipid signalling, whereas reactive oxygen species (ROS) and flagellin signals converge further downstream in the OXI1/PTI1‐2 pathway, independently of PDK1 (Anthony et al., 2006). As the AGC kinase Adi3 is also phosphorylated by PDK1 (see above), we suggest that PDK1 functions as a spider in the web for transferring receptor‐mediated PA signals to downstream signalling cascades via AGC kinases (Fig. 1). To balance the signalling cascade, PA signals are attenuated by PA kinase (PAK), which converts PA into the lipid DAG pyrophosphate (DGPP) (Munnik et al., 1996). However, because DGPP accumulation is associated with PA‐induced signalling, DGPP itself might also function as a secondary messenger. The observation that DGPP is broken down by the DGPP phosphatase (DPP) might confirm this hypothesis (reviewed by Van Schooten et al., 2006).
ROS signalling
ROS are important secondary messengers responsible for the oxidative burst. Upon pathogen recognition, the plant responds with a bi‐phasic production of ROS (Lamb and Dixon, 1997). ROS can be produced inside the plant cell in several organelles; however, a membrane‐bound respiratory oxidative burst protein (RBOHD; an NADPH oxidase) is considered as the source of ROS upon elicitation by pathogens (Torres and Dangl, 2005). The Arabidopsis RBOHD protein is heavily phosphorylated at seven different amino acid residues and differentially phosphorylated at three residues upon elicitation with flg22 or xylanase (Benschop et al., 2007; Nühse et al., 2007). In accordance, another member from the RBOH family, RBOHB, is phosphorylated by calcium‐dependent protein kinases (CDPKs) in potato, which causes a subsequent oxidative burst (Fig. 1; Kobayashi et al., 2007). Upon signal‐induced phosphorylation, the activated oxidase converts O2 into 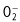 , which subsequently forms the stable component hydrogen peroxide (H2O2), which is removed by catalases or peroxidases when the signal is transferred further down. Besides a signalling role, H2O2 also has direct antimicrobial effects, cross‐links cell walls and activates transcription of defence‐related genes (Lamb and Dixon, 1997).
, which subsequently forms the stable component hydrogen peroxide (H2O2), which is removed by catalases or peroxidases when the signal is transferred further down. Besides a signalling role, H2O2 also has direct antimicrobial effects, cross‐links cell walls and activates transcription of defence‐related genes (Lamb and Dixon, 1997).
Calcium signalling
A secondary messenger that links several defence‐related processes is the ubiquitous messenger calcium (Ca2+) (Lecourieux et al., 2006). Ca2+ is imported to and exported from the cell and the vacuole by Ca2+‐ATPases that are regulated via phosphorylation, and stimulates the production of NO and ROS upon recognition of an avirulent pathogen (Fig. 1; Delledonne, 2005). Strikingly, H2O2 also stimulates rapid Ca2+ influxes upon elicitation, which reveals a role for Ca2+ signalling up‐ and downstream of ROS (Fig. 1; Lamb and Dixon, 1997). These data imply that secondary messengers produced via pathways that rely on PTMs connect several components of the defence signalling cascades, but also influence each other to balance the downstream responses.
PTMS OF HOST PROTEINS IN THE CYTOPLASM
The MAPK cascade
A major conserved signalling cascade that is activated by a large range of biotic and abiotic stress stimuli in plants is the MAPK cascade (Fig. 1; Pedley and Martin, 2005; Zhang et al., 2006). MAPK cascades consist of three functionally linked protein kinases that transfer the stress signals. A stress signal causes phosphorylation and activation of the most upstream MAPK kinase kinase (MAPKKK). Subsequently, the MAPKKK phosphorylates and activates a MAPK kinase (MAPKK), which in turn does the same with a MAPK (MPK). The MAPK then phosphorylates downstream target(s) thereby transferring the signal further downstream (Fig. 1). The Arabidopsis genome encodes 20 MAPKs, ten MAPKKs and 60 putative MAPKKKs and, in addition, it contains several protein phosphatases that control the cascade by dephosphorylating the MAPK cascade components (Ichimura et al., 2002; Martín et al., 2005). For example, the AtMPK6 protein, activated upon most stress stimuli, is controlled by the phosphatases ABI1, AP2C1, MKP1 and MKP2 in Arabidopsis (Lee and Ellis, 2007; Leung et al., 2006; Schweighofer et al., 2007; Ulm et al., 2002). The AtMPK4 protein negatively regulates defence responses upon phosphorylation, which implies also that the activation of protein phosphatases can mediate the transfer of stress‐related signals (Ichimura et al., 2006; Suarez‐Rodriguez et al., 2007). It is interesting to note that most stress stimuli mainly activate AtMPK6, ‐3 and ‐4 and their orthologues in other plant species during stress‐related signalling. Therefore, stress‐related signalling cascades are considered to converge in the MAPK cascades, after which the signal is transferred into different downstream pathways (Pedley and Martin, 2005; Zhang et al., 2006). In Cf‐4 tomato, three highly homologous MAPKs, LeMPK1, ‐2 (both are orthologous to AtMPK6) and LeMPK3 (the AtMPK3 orthologue), are activated upon Avr4‐elicitation. These LeMPKs appeared to have different phosphorylation specificities and a different role in defence signalling, suggesting that the signal can eventually be transferred to different substrates and possibly different downstream signalling cascades (Stulemeijer et al., 2007). Thus far, only a few MAPK targets have been described. The AtMPK6 protein phosphorylates 1‐aminocyclopropane‐1‐carboxylic acid synthase 6 and 2 (ACS6/2), which are key enzymes in ethylene biosynthesis, and WRKY transcription factors upon flg22 elicitation (Fig. 1; Asai et al., 2002; Liu and Zhang, 2004; Menke et al., 2005). The AtMPK4 protein phosphorylates MAP kinase substrate 1 (MKS1), which negatively regulates salicylic acid‐dependent resistance upon phosphorylation (Fig. 1; Andreasson et al., 2005). Furthermore, AtMPK3 was recently found to phosphorylate the transcription factor VIP1, which is involved in regulating the expression of the PR1 pathogenesis‐related gene (Djamei et al., 2007).
CDPK‐mediated signalling
CDPKs contain a calmodulin‐like domain with Ca2+ binding sites and represent another class of kinases. In the absence of Ca2+, the kinase domain of CDPKs is not phosphorylated, which points to a direct regulation by Ca2+(Fig. 1; Ludwig et al., 2004). Tobacco NtCDPK2 was the first CDPK reported to be involved in plant defence signalling in transgenic Cf‐9 tobacco upon elicitation with the Avr9 effector. NtCDPK2 is required for HR development and is activated by phosphorylation (Fig. 1). Furthermore, NtCDPK2 enhances ethylene production, which subsequently negatively regulates the MAPK signalling cascade (Fig. 1). In addition, a tomato CDPK phosphorylates tomato ACS2 (Tatsuki and Mori, 2001), the orthologue of which in Arabidopsis was shown to be phosphorylated by AtMPK6 (Liu and Zhang, 2004). This observation suggests that two kinase signalling cascades, both leading to an ethylene‐dependent cell death, can cross‐talk to fine‐tune the final outcome (Ludwig et al., 2005). Finally, the potato CDPK, StCDPK5, phosphorylates StRBOHB thereby regulating the oxidative burst (Fig. 1; Kobayashi et al., 2007).
Ubiquitination in defence signalling
In recent years, several proteins with E3 ubiquitin ligase activity that play a role in defence signalling have been reported, indicating that ubiquitination is important for resistance of plants to pathogens (Fig. 1). An extensive transcriptional analysis of Cf‐9 transgenic tobacco cells elicited with Avr9 revealed two genes, ACRE189 and ACRE276, the encoded proteins of which possess in vitro E3 ligase activity and which are required for Cf‐9‐ and Cf‐4‐mediated defence signalling (Durrant et al., 2000; Yang et al., 2006). The closest orthologue of ACRE276 in Arabidopsis, PUB17, is also required for RPM1‐ and RPS4‐mediated resistance to Pseudomonas syringae pv. tomato expressing the elicitors AvrB or AvrRPS4, respectively (Yang et al., 2006). ACRE74, which encodes another tobacco E3 ligase (NtCMPG1), is also required for Cf‐9/Avr9‐induced signalling in addition to defence responses induced by Pto/AvrPto and the Phytophthora infestans elicitor Inf1 (González‐Lamothe et al., 2006). Furthermore, a functional tomato E3 ligase, ATL6, is transcriptionally upregulated upon elicitation with a cell‐wall protein fraction from Pythium oligandrum (Hondo et al., 2007). In addition to E3 ubiquitin ligase activity, transient‐induced gene silencing (TIGS) of the ubiquitin‐encoding gene itself and subsequent complementation studies in powdery mildew‐inoculated resistant barley suggest a role for K48‐linked polyubiquitination in defence signalling. Although K48‐linked polyubiquitination normally results in proteasome‐mediated protein degradation, here the polyubiquitination event but not the subsequent degradation is required for the defence response (Dong et al., 2006). Finally, the Arabidopsis E1 ubiquitin‐activating enzyme UBA1 is required for defence responses induced upon recognition of the AvrRpt2 effector (Goritschnig et al., 2007).
Sumoylation in defence signalling
In addition to ubiquitination, sumoylation also plays a role in defence signalling, although evidence for this remains scarce. Thus far, there are only two reports that show an increase in protein sumoylation upon exposure to abiotic stress conditions such as heat shock, H2O2, ethanol and the defensive compound against herbivores, canavanine (Kurepa et al., 2003; Saracco et al., 2007). However, overexpression of SUMO in tobacco appears to block HR development upon xylanase infiltration (Hanania et al., 1999) and a SUMO E3 ligase, SIZ1, was reported to regulate salicylic acid‐mediated innate immunity in Arabidopsis (Lee et al., 2007). SIZ1 also appears to negatively regulate systemic‐acquired resistance and the expression of PR genes. The best evidence for the importance of sumoylation in defence signalling originates from the observation that pathogen effectors interfere with the host sumoylation cascade. The Xanthomonas campestris effector XopD is injected into the host cell upon infection and encodes an active cysteine protease with plant‐specific SUMO substrate specificity. XopD specifically desumoylates host proteins, thereby most likely interfering with the host defence signalling cascade upon infection (Hotson et al., 2003). Another effector from X. campestris, AvrXv4, requires its protease activity to reduce the amount of SUMO‐conjugated proteins in the host cell, which leads to suppression of localized cell death in inoculated plants (Roden et al., 2004). The effector AvrBsT, which also possesses protease activity, requires its catalytic domain to induce cell death in N. benthamiana (reviewed by Hotson and Mudgett, 2004; Orth et al., 2000). Additionally, some effectors seem to interact with proteins from the host sumoylation machinery. Xylanase interacts with SUMO in a yeast‐two‐hybrid system (Hanania et al., 1999) and the replication protein RepAC1 from geminiviruses interacts with the SUMO E3 ligase SCE1 from N. benthamiana (Castillo et al., 2004). However, the biological relevance of these observations remains to be elucidated. Yet, if sumoylation did not play any role in defence signalling, the various effectors mentioned above would not enhance virulence for the pathogen and they would probably have been eliminated from the population during evolution to avoid recognition by resistant plants.
EFFECTORS OF PATHOGENS MODIFY PTMS IN DEFENCE SIGNALLING
It has become apparent from the information given above that plants depend on rapid PTMs in signalling cascades to defend themselves against intruding pathogens. An active defence response is triggered by the recognition of elicitors that are secreted by the invading pathogen. Therefore, the intriguing question remains as to why pathogens still secrete elicitors that induce avirulence. Increasing evidence is accumulating that these elicitors act as effectors that specifically interfere with the host defence mechanisms to increase the virulence of pathogens in the absence of the cognate R protein (Abramovitch et al., 2006a; Alfano and Collmer, 2004; He et al., 2007; Mudgett, 2005). To reach this goal, as mentioned above for sumoylation effectors regularly modify the PTM status of host proteins, thereby targeting primary and/or secondary defence responses. Here, we describe the virulence function of some effectors of P. syringae.
The RIN4 protein is targeted by two effectors from Pst, AvrRpm1 and AvrB, which indirectly induce RIN4 phosphorylation, thereby enhancing the negative regulation of the primary defence response by RIN4, which leads to increased host susceptibility and pathogen virulence (Kim et al., 2005b; Mackey et al., 2002). The effector HopAI1 dephosphorylates AtMPK6 and AtMPK3 in the MAPK cascade through phosphothreonine lyase activity, which is an alternative cleavage of the phosphate from the threonine residue (Zhang et al., 2007). Similarly, the HopPtoD2 effector functions as a protein tyrosine phosphatase downstream of the host MAPKKs (Espinosa et al., 2003). As MAPK cascades are activated in most stress‐related responses, the position of interference is strategic given that the effectors might interfere in many signalling cascades. Even more intriguing is the abuse of the MAPK cascade by Agrobacterium, the T‐DNA of which hitch‐hikes with a phosphorylated AtMPK3 substrate, the transcription factor VIP1, into the nucleus thereby circumventing the defence response (Djamei et al., 2007). Effectors not only modify protein phosphorylation but can also modify protein ubiquitination. The effector AvrPtoB, the N‐terminal part of which is recognized by the Fen kinase, has a C‐terminal E3 ubiquitin ligase domain. AvrPtoB ubiquitinates the Fen kinase and subsequently targets it for degradation, thereby abolishing the recognition of its own N‐terminal region (Abramovitch et al., 2006b; Janjusevic et al., 2006; Rosebrock et al., 2007). AvrPtoB will probably not prove to be the only effector that mediates ubiquitination of host proteins, as a screen of the available bacterial genomes revealed several new putative effectors that are predicted to mimic subunits of the ubiquitination pathway (Angot et al., 2007).
CONCLUDING REMARKS
Rapid PTMs of proteins in defence signalling are essential tools for plants to respond swiftly to pathogen invasion. In this review, we have given an overview of PTMs that modify components essential for defence signalling at the site of signal perception, during secondary messenger production and during signalling in the cytoplasm. PTMs regulate protein localization and activity and provide complex mechanisms to balance responses in the cell without the prerequisite of protein synthesis. As recent technological developments allow high‐throughput analysis of modified proteins, we expect that many previously unidentified components of defence signalling cascades, which are not transcriptionally regulated, will be revealed in the coming years.
NOTE
Recently it was shown that AvrPto of Pseudomonas syringae inhibits plant immunity triggered by diverse MAMPs. The bacterial effector suppresses early defense gene transcription and intercepts MAPK signaling upstream of MAPKKK at the plasma membrane, linked to the MAMP receptor (He et al., 2006). Xiang and co‐workers (Xiang et al., 2008) studied the mechanism behind this observation and found that AvrPto binds the MAMP‐triggered receptor‐kinases FLS2 and EFR. Targeting of these receptors is required for the virulence function of AvrPto and inhibits FLS2 and EFR kinase activity. As a result of this, MAPK cascade activation is inhibited and MAMP‐induced immune responses are suppressed.
ACKNOWLEDGEMENTS
We thank Wladimir I. L. Tameling for critically reading the manuscript, and Bart P. H. J. Thomma and Pierre J. G. M. de Wit for helpful comments. I.J.E.S. and M.H.A.J.J. are supported by the Dutch Organization for Scientific Research (NWO; VIDI grant to M.H.A.J.J.).
REFERENCES
- Abramovitch, R.B. , Anderson, J.C. and Martin, G.B. (2006a) Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch, R.B. , Janjusevic, R. , Stebbins, C.E. and Martin, G.B. (2006b) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl Acad. Sci. USA, 103, 2851–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade, J. , DeYoung, B.J. , Golstein, C. and Innes, R.W. (2007) Indirect activation of a plant nucleotide binding site‐leucine‐rich repeat protein by a bacterial protease. Proc. Natl Acad. Sci. USA, 104, 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, J.R. and Collmer, A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. [DOI] [PubMed] [Google Scholar]
- Andersson, M.X. , Kourtchenko, O. , Dangl, J.L. , Mackey, D. and Ellerström, M. (2006) Phospholipase‐dependent signalling during the AvrRpm1‐ and AvrRpt2‐induced disease resistance responses in Arabidopsis thaliana . Plant J. 47, 947–959. [DOI] [PubMed] [Google Scholar]
- Andreasson, E. , Jenkins, T. , Brodersen, P. , Thorgrimsen, S. , Petersen, N.H.T. , Zhu, S. , Qiu, J.‐L. , Micheelsen, P. , Rocher, A. , Petersen, M. , Newman, M.‐A. , Bjorn Nielsen, H. , Hirt, H. , Somssich, I. , Mattsson, O. and Mundy, J. (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriotis, V.M.E. and Rathjen, J.P. (2006) The Pto kinase of tomato, which regulates plant immunity, is repressed by its myristoylated N terminus. J. Biol. Chem. 281, 26578–26586. [DOI] [PubMed] [Google Scholar]
- Angot, A. , Vergunst, A. , Genin, S. and Peeters, N. (2007) Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, R.G. , Henriques, R. , Helfer, A. , Meszaros, T. , Rios, G. , Testerink, C. , Munnik, T. , Deak, M. , Koncz, C. and Bögre, L. (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, R.G. , Khan, S. , Costa, J. , Pais, M.S. and Bögre, L. (2006) The Arabidopsis protein kinase PTI1–2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J. Biol. Chem. 281, 37536–37546. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Balmuth, A. and Rathjen, J.P. (2007) Genetic and molecular requirements for function of the Pto/Prf effector recognition complex in tomato and Nicotiana benthamiana . Plant J. 51, 978–990. [DOI] [PubMed] [Google Scholar]
- Bargmann, B.O.R. and Munnik, T. (2006) The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 9, 515–522. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1999) Fast forward genetics based on virus‐induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y. , Nimchuk, Z. , Hubert, D.A. , Mackey, D. and Dangl, J.L. (2004a) Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell, 16, 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y. , Subramaniam, R. and Dangl, J.L. (2004b) Plant disease resistance protein signaling: NBS‐LRR proteins and their partners. Curr. Opin. Plant Biol. 7, 391–399. [DOI] [PubMed] [Google Scholar]
- Benschop, J.J. , Mohammed, S. , O’Flaherty, M. , Heck, A.J.R. , Slijper, M. and Menke, F.L.H. (2007) Quantitative phospho‐proteomics of early elicitor signalling in Arabidopsis. Mol. Cell Proteomics, 6, 1198–1214. [DOI] [PubMed] [Google Scholar]
- Bent, A. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Blatt, M.R. , Grabov, A. , Brearley, J. , Hammond‐Kosack, K. and Jones, J.D.G. (1999) K+ channels of Cf‐9 transgenic tobacco guard cells as targets for Cladosporium fulvum Avr9 elicitor‐dependent signal transduction. Plant J. 19, 453–462. [DOI] [PubMed] [Google Scholar]
- Burch‐Smith, T.M. , Anderson, J.C. , Martin, G.B. and Dinesh‐Kumar, S.P. (2004) Applications and advantages of virus‐induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. [DOI] [PubMed] [Google Scholar]
- Castillo, A.G. , Kong, L.J. , Hanley‐Bowdoin, L. and Bejarano, E.R. (2004) Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 78, 2758–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nurnberger, T. , Jones, J.D.G. , Felix, G. and Boller, T. (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host‐microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Chosed, R. , Mukherjee, S. , Lois, L.M. and Orth, K. (2006) Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem. J. 398, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N.C. , Thordal‐Christensen, H. , Lipka, V. , Bau, S. , Kombrink, E. , Qiu, J.L. , Hückelhoven, R. , Steins, M. , Freialdenhoven, A. , Somerville, S.C. and Schulze‐Lefert, P. (2003) SNARE‐protein‐mediated disease resistance at the plant cell wall. Nature, 425, 973–977. [DOI] [PubMed] [Google Scholar]
- Coppinger, P. , Repetti, P.P. , Day, B. , Dahlbeck, D. , Mehlert, A. and Staskawicz, B.J. (2004) Overexpression of the plasma membrane‐localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana . Plant J. 40, 225–237. [DOI] [PubMed] [Google Scholar]
- Day, B. , Dahlbeck, D. and Staskawicz, B.J. (2006) NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell, 18, 2782–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, B. , Dahlbeck, D. , Huang, J. , Chisholm, S.T. , Li, D. and Staskawicz, B.J. (2005) Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell, 17, 1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, C.F. , Laxalt, A.M. , Bargmann, B.O. , De Wit, P.J.G.M. , Joosten, M.H.A.J. and Munnik, T. (2004) Phosphatidic acid accumulation is an early response in the Cf‐4/Avr4 interaction. Plant J. 39, 1–12. [DOI] [PubMed] [Google Scholar]
- De Wit, P.J.G.M. (2007) How plants recognize pathogens and defend themselves. Cell Mol. Life Sci . PMID: 17876517. [DOI] [PMC free article] [PubMed]
- Delledonne, M. (2005) NO news is good news for plants. Curr. Opin. Plant Biol. 8, 390–396. [DOI] [PubMed] [Google Scholar]
- Devarenne, T.P. , Ekengren, S.K. , Pedley, K.F. and Martin, G.B. (2006) Adi3 is a Pdk1‐interacting AGC kinase that negatively regulates plant cell death. EMBO J. 25, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, A. , Mayer, J.E. and Hahlbrock, K. (1990) Fungal elicitor triggers rapid, transient, and specific protein phosphorylation in parsley cell suspension cultures. J. Biol. Chem. 265, 6360–6368. [PubMed] [Google Scholar]
- Djamei, A. , Pitzschke, A. , Nakagami, H. , Rajh, I. and Hirt, H. (2007) Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science, 318, 453–456. [DOI] [PubMed] [Google Scholar]
- Dong, W. , Nowara, D. and Schweizer, P. (2006) Protein polyubiquitination plays a role in basal host resistance of barley. Plant Cell, 18, 3321–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E. , Rowland, O. , Piedras, P. , Hammond Kosack, K.E. and Jones, J.D.G. (2000) cDNA‐AFLP reveals a striking overlap in race‐specific resistance and wound response gene expression profiles. Plant Cell, 12, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa, A. , Guo, M. , Tam, V.C. , Fu, Z.Q. and Alfano, J.R. (2003) The Pseudomonas syringae type III‐secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol. Microbiol. 49, 377–387. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78. [DOI] [PubMed] [Google Scholar]
- Farazi, T.A. , Waksman, G. and Gordon, J.I. (2001) The biology and enzymology of protein N‐myristoylation. J. Biol. Chem. 276, 39501–39504. [DOI] [PubMed] [Google Scholar]
- Fasshauer, D. (2003) Structural insights into the SNARE mechanism. Biochim. Biophys. Acta, 1641, 87–97. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1942) Inheritance of pathogenicity in Melampsora lini. Phytopathology, 32, 653–669. [Google Scholar]
- De La Fuente van Bentem, S. and Hirt, H. (2007) Using phosphoproteomics to reveal signalling dynamics in plants. Trends Plant Sci. 12, 404–411. [DOI] [PubMed] [Google Scholar]
- Fuglsang, A.T. , Guo, Y. , Cuin, T.A. , Qiu, Q. , Song, C. , Kristiansen, K.A. , Bych, K. , Schulz, A. , Shabala, S. , Schumaker, K.S. , Palmgren, M.G. and Zhu, J.‐K. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+‐ATPase by preventing interaction with 14‐3‐3 protein. Plant Cell, 19, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelli, A. , Higgins, V.J. and Blumwald, E. (1997) Activation of plant plasma membrane Ca2+‐permeable channels by race‐specific fungal elicitors. Plant Physiol. 113, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. , Rogers, E.E. and Ausubel, F.M. (1997) Use of Arabidopsis for genetic dissection of plant defense responses. Annu. Rev. Genet. 31, 547–569. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis . Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. , Bauer, Z. and Boller, T. (2001) Both the extracellular leucine‐rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell, 13, 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- González‐Lamothe, R. , Tsitsigiannis, D.I. , Ludwig, A.A. , Panicot, M. , Shirasu, K. and Jones, J.D.G. (2006) The U‐box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell, 18, 1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritschnig, S. , Zhang, Y. and Li, X. (2007) The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 49, 540–551. [DOI] [PubMed] [Google Scholar]
- Guo, F.Q. , Okamoto, M. and Crawford, N.M. (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science, 302, 100–103. [DOI] [PubMed] [Google Scholar]
- Haglund, K. , Di Fiore, P.P. and Dikic, I. (2003) Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 28, 598–604. [DOI] [PubMed] [Google Scholar]
- Hanania, U. , Furman‐Matarasso, N. , Ron, M. and Avni, A. (1999) Isolation of a novel SUMO protein from tomato that suppresses EIX‐induced cell death. Plant J. 19, 533–541. [DOI] [PubMed] [Google Scholar]
- He, P. , Shan, L. , Lin, N.C. , Martin, G.B. , Kemmerling, B. , Nürnberger, T. and Sheen, J. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell, 125, 563–575. [DOI] [PubMed] [Google Scholar]
- He, P. , Shan, L. and Sheen, J. (2007) Elicitation and suppression of microbe‐associated molecular pattern‐triggered immunity in plant‐microbe interactions. Cell Microbiol. 9, 1385–1396. [DOI] [PubMed] [Google Scholar]
- Heese, A. , Hann, D.R. , Gimenez‐Ibanez, S. , Jones, A.M.E. , He, K. , Li, J. , Schroeder, J.I. , Peck, S.C. and Rathjen, J.P. (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. USA, 104, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese, A. , Ludwig, A.A. and Jones, J.D.G. (2005) Rapid phosphorylation of a syntaxin during the Avr9/Cf‐9‐race‐specific signaling pathway. Plant Physiol. 138, 2406–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondo, D. , Hase, S. , Kanayama, Y. , Yoshikawa, N. , Takenaka, S. and Takahashi, H. (2007) The LeATL6‐associated ubiquitin/proteasome system may contribute to fungal elicitor‐activated defense response via the jasmonic acid‐dependent signaling pathway in tomato. Mol. Plant–Microbe. Interact. 20, 72–81. [DOI] [PubMed] [Google Scholar]
- Hotson, A. and Mudgett, M.B. (2004) Cysteine proteases in phytopathogenic bacteria: identification of plant targets and activation of innate immunity. Curr. Opin. Plant Biol. 7, 384–390. [DOI] [PubMed] [Google Scholar]
- Hotson, A. , Chosed, R. , Shu, H. , Orth, K. and Mudgett, M.B. (2003) Xanthomonas type III effector XopD targets SUMO‐conjugated proteins in planta. Mol. Microbiol. 50, 377–389. [DOI] [PubMed] [Google Scholar]
- Ichimura, K. , Casais, C. , Peck, S.C. , Shinozaki, K. and Shirasu, K. (2006) MEKK1 is required for MPK4 activation and regulates tissue‐specific and temperature‐dependent cell death in Arabidopsis. J. Biol. Chem. 281, 36969–36976. [DOI] [PubMed] [Google Scholar]
- Ichimura, K. , Shinozaki, K. , Tena, G. , Sheen, J. , Henry, Y. , Champion, A. , Kreis, M. , Zhang, S. and Hirt, H. (2002) Mitogen‐activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Janjusevic, R. , Abramovitch, R.B. , Martin, G.B. and Stebbins, C.E. (2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science, 311, 222–226. [DOI] [PubMed] [Google Scholar]
- Jensen, O.N. (2004) Modification‐specific proteomics: characterization of post‐translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 8, 33–41. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl Acad. Sci. USA, 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde, M. , Nuhse, T.S. , Findlay, K. and Peck, S.C. (2007) The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis‐related protein 1. Proc. Natl Acad. Sci. USA 104, 11850–11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova, R. and De Vries, S.C. (2006) Advances in understanding brassinosteroid signaling. Sci. STKE, 2006, pe36. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T. , Nam, J. , Boyes, D.C. , Holt III, B.F. , Hubert, D.A. , Wiig, A. and Dangl, J.L. (2005) A duplicated pair of Arabidopsis RING‐finger E3 ligases contribute to the RPM1‐ and RPS2‐mediated hypersensitive response. Plant J. 44, 258–270. [DOI] [PubMed] [Google Scholar]
- Kerscher, O. , Felberbaum, R. and Hochstrasser, M. (2006) Modification of proteins by ubiquitin and ubiquitin‐like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180. [DOI] [PubMed] [Google Scholar]
- Kersten, B. , Agrawal, G.K. , Iwahashi, H. and Rakwal, R. (2006) Plant phosphoproteomics: a long road ahead. Proteomics, 6, 5517–5528. [DOI] [PubMed] [Google Scholar]
- Kim, H.S. , Desveaux, D. , Singer, A.U. , Patel, P. , Sondek, J. and Dangl, J.L. (2005a) The Pseudomonas syringae effector AvrRpt2 cleaves its C‐terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl Acad. Sci. USA, 102, 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.G. , Da Cunha, L. , McFall, A.J. , Belkhadir, Y. , DebRoy, S. , Dangl, J.L. and Mackey, D. (2005b) Two Pseudomonas syringae type III effectors inhibit RIN4‐regulated basal defense in Arabidopsis. Cell, 121, 749–759. [DOI] [PubMed] [Google Scholar]
- Kim, Y.J. , Lin, N.‐C. and Martin, G.B. (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell, 109, 589–598. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M. , Ohura, I. , Kawakita, K. , Yokota, N. , Fujiwara, M. , Shimamoto, K. , Doke, N. and Yoshioka, H. (2007) Calcium‐dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell, 19, 1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, B. and Blatt, M.R. (2002) Protein phosphorylation activates the guard cell Ca2+ channel and is a prerequisite for gating by abscisic acid. Plant J. 32, 185–194. [DOI] [PubMed] [Google Scholar]
- Kurepa, J. , Walker, J.M. , Smalle, J. , Gosink, M.M. , Davis, S.J. , Durham, T.L. , Sung, D.Y. and Vierstra, R.D. (2003) The small ubiquitin‐like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and ‐2 conjugates is increased by stress. J. Biol. Chem. 278, 6862–6872. [DOI] [PubMed] [Google Scholar]
- Lamb, C. and Dixon, R.A. (1997) The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Laxalt, A.M. and Munnik, T. (2002) Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 5, 332–338. [DOI] [PubMed] [Google Scholar]
- Laxalt, A.M. , Raho, N. , Ten Have, A. and Lamattina, L. (2007) Nitric oxide is critical for inducing phosphatidic acid accumulation in xylanase‐elicited tomato cells. J. Biol. Chem. 282, 21160–21168. [DOI] [PubMed] [Google Scholar]
- Lecourieux, D. , Ranjeva, R. and Pugin, A. (2006) Calcium in plant defence‐signalling pathways. New Phytol. 171, 249–269. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Nam, J. , Park, H.C. , Na, G. , Miura, K. , Jin, J.B. , Yoo, C.Y. , Baek, D. , Kim, D.H. , Jeong, J.C. , Kim, D. , Lee, S.Y. , Salt, D.E. , Mengiste, T. , Gong, Q. , Ma, S. , Bohnert, H.J. , Kwak, S.‐S. , Bressan, R.A. , Hasegawa, P.M. and Yun, D.‐J. (2007) Salicylic acid‐mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49, 79–90. [DOI] [PubMed] [Google Scholar]
- Lee, J.S. and Ellis, B.E. (2007) Arabidopsis MAPK phosphatase MKP2 positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 mitogen‐activated protein kinases. J. Biol. Chem. 282, 25020–25029. [DOI] [PubMed] [Google Scholar]
- Lee, S. , Hirt, H. and Lee, Y. (2001) Phosphatidic acid activates a wound‐activated MAPK in Glycine max . Plant J. 26, 479–486. [DOI] [PubMed] [Google Scholar]
- Leung, J. , Orfanidi, S. , Chefdor, F. , Mészáros, T. , Bolte, S. , Mizoguchi, T. , Shinozaki, K. , Giraudat, J. and Bögre, L. (2006) Antagonistic interaction between MAP kinase and protein phosphatase 2C in stress recovery. Plant Sci. 171, 596–606. [Google Scholar]
- Ligterink, W. , Kroj, T. , Zur Nieden, U. , Hirt, H. and Scheel, D. (1997) Receptor‐mediated activation of a MAP Kinase in pathogen defense of plants. Science, 276, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Lindermayr, C. , Saalbach, G. and Durner, J. (2005) Proteomic identification of S‐nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr, C. , Saalbach, G. , Bahnweg, G. and Durner, J. (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S‐nitrosylation. J. Biol. Chem. 281, 4285–4291. [DOI] [PubMed] [Google Scholar]
- Liu, G.Z. , Pi, L.Y. , Walker, J.C. , Ronald, P.C. and Song, W.Y. (2002) Biochemical characterization of the kinase domain of the rice disease resistance receptor‐like kinase XA21. J. Biol. Chem. 277, 20264–20269. [DOI] [PubMed] [Google Scholar]
- Liu, Y. and Zhang, S. (2004) Phosphorylation of 1‐aminocyclopropane‐1‐carboxylic acid synthase by MPK6, a stress‐responsive mitogen‐activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell, 16, 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, Y.‐T. and Martin, G.B. (1995) The Pto bacterial resistance gene and the Fen insecticide sensitivity gene encode functional protein kinases with serine/threonine specificity. Plant Physiol. 108, 1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, A.A. , Romeis, T. and Jones, J.D.G. (2004) CDPK‐mediated signalling pathways: specificity and cross‐talk. J. Exp. Bot. 55, 181–188. [DOI] [PubMed] [Google Scholar]
- Ludwig, A.A. , Saitoh, H. , Felix, G. , Freymark, G. , Miersch, O. , Wasternack, C. , Boller, T. , Jones, J.D.G. and Romeis, T. (2005) Ethylene‐mediated cross‐talk between calcium‐dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl Acad. Sci. USA 102, 10736–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Holt B.F., III, Wiig, A. and Dangl, J.L. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1‐mediated resistance in Arabidopsis . Cell, 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Maeda, Y. , Ashida, H. , Kinoshita, T. and Minoru, F. (2006) CHO glycosylation mutants: GPI anchor. Methods Enzymol. 416, 182–205. [DOI] [PubMed] [Google Scholar]
- Martin, G.B. , Bogdanove, A.J. and Sessa, G. (2003) Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- Martin, G.B. , Brommonschenkel, S.H. , Chunwongse, J. , Frary, A. , Ganal, M.W. , Spivey, R. , Wu, T. , Earle, E.D. and Tanksley, S.D. (1993) Map‐based cloning of a protein kinase gene conferring disease resistance in tomato. Science, 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Martín, H. , Flández, M. , Nombela, C. and Molina, M. (2005) Protein phosphatases in MAPK signalling: we keep learning from yeast. Mol. Microbiol. 58, 6–16. [DOI] [PubMed] [Google Scholar]
- Menke, F.L.H. , Kang, H.G. , Chen, Z. , Jeong, M.P. , Kumar, D. and Klessig, D.F. (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR‐like cell death in tobacco. Mol. Plant–Microbe. Interact. 18, 1027–1034. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Jin, J.B. and Hasegawa, P.M. (2007) Sumoylation, a post‐translational regulatory process in plants. Curr. Opin. Plant Biol. 10, 1–8. [DOI] [PubMed] [Google Scholar]
- Mucyn, T.S. , Clemente, A. , Andriotis, V.M.E. , Balmuth, A.L. , Oldroyd, G.E.D. , Staskawicz, B.J. and Rathjen, J.P. (2006) The tomato NBARC‐LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell, 18, 2792–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531. [DOI] [PubMed] [Google Scholar]
- Munnik, T. , De Vrije, T. , Irvine, R.F. and Musgrave, A. (1996) Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G‐protein activation in plants. J. Biol. Chem. 271, 15708–15715. [DOI] [PubMed] [Google Scholar]
- Neill, S. , Bright, J. , Desikan, R. , Hancock, J. , Harrison, J. and Wilson, I. (2007) Nitric oxide evolution and perception. J. Exp. Bot. doi:10.1093. [DOI] [PubMed] [Google Scholar]
- Nekrasov, V. , Ludwig, A.A. and Jones, J.D.G. (2006) CITRX thioredoxin is a putative adaptor protein connecting Cf‐9 and the ACIK1 protein kinase during the Cf‐9/Avr9‐ induced defence response. FEBS Lett. 580, 4236–4241. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z. , Eulgem, T. , Holt III, B.F. and Dangl, J.L. (2003) Recognition and response in the plant immune system. Annu. Rev. Genet. 37, 579–609. [DOI] [PubMed] [Google Scholar]
- Nirmala, J. , Brueggeman, R. , Maier, C. , Clay, C. , Rostoks, N. , Kannangara, C.G. , Von Wettstein, D. , Steffenson, B.J. and Kleinhofs, A. (2006) Subcellular localization and functions of the barley stem rust resistance receptor‐like serine/threonine‐specific protein kinase Rpg1. Proc. Natl Acad. Sci. USA, 103, 7518–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirmala, J. , Dahl, S. , Steffenson, B.J. , Kannangara, C.G. , Von Wettstein, D. , Chen, X. and Kleinhofs, A. (2007) Proteolysis of the barley receptor‐like protein kinase RPG1 by a proteasome pathway is correlated with Rpg1‐mediated stem rust resistance. Proc. Natl Acad. Sci. USA, 104, 10276–10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova, M. , Budhiraja, R. , Coupland, G. , Eisenhaber, F. and Bachmair, A. (2004) SUMO conjugation in plants. Planta, 220, 1–8. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S. , Boller, T. and Peck, S.C. (2003) A plasma membrane syntaxin is phosphorylated in response to the bacterial elicitor flagellin. J. Biol. Chem. 278, 45248–45254. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S. , Bottrill, A.R. , Jones, A.M.E. and Peck, S.C. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse, T.S. , Stensballe, A. , Jensen, O.N. and Peck, S.C. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell, 16, 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger, T. and Kemmerling, B. (2006) Receptor protein kinases—pattern recognition receptors in plant immunity. Trends Plant Sci. 11, 519–522. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T. and Scheel, D. (2001) Signal transmission in the plant immune response. Trends Plant Sci. 6, 372–379. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Orth, K. , Xu, Z. , Mudgett, M.B. , Bao, Z.Q. , Palmer, L.E. , Bliska, J.B. , Mangel, W.F. , Staskawicz, B.J. and Dixon, J.E. (2000) Disruption of signaling by Yersinia effector YopJ, a ubiquitin‐like protein protease. Science, 290, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Palmgren, M.G. (2001) Plant plasme membrane H+‐ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Peck, S.C. (2003) Early phosphorylation events in biotic stress. Curr. Opin. Plant Biol. 6, 334–338. [DOI] [PubMed] [Google Scholar]
- Peck, S.C. (2006) Phosphoproteomics in Arabidopsis: moving from empirical to predictive science. J. Exp. Bot. 57, 1523–1527. [DOI] [PubMed] [Google Scholar]
- Pedley, K.F. and Martin, G.B. (2003) Molecular basis of Pto‐mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243. [DOI] [PubMed] [Google Scholar]
- Pedley, K.F. and Martin, G.B. (2005) Role of mitogen‐activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 8, 541. [DOI] [PubMed] [Google Scholar]
- Piedras, P. , Rivas, S. , Droge, S. , Hillmer, S. and Jones, J.D. (2000) Functional, c‐myc‐tagged Cf‐9 resistance gene products are plasma‐membrane localized and glycosylated. Plant J. 21, 529–536. [DOI] [PubMed] [Google Scholar]
- Rathjen, J.P. , Chang, J.H. , Staskawicz, B.J. and Michelmore, R.W. (1999) Constitutively active Pto induces a Prf‐dependent hypersensitive response in the absence of avrPto. EMBO J. 18, 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders, J. and Sickmann, A. (2005) State‐of‐the‐art in phosphoproteomics. Proteomics, 5, 4052–4061. [DOI] [PubMed] [Google Scholar]
- Rentel, M.C. , Lecourieux, D. , Ouaked, F. , Usher, S.L. , Petersen, L. , Okamoto, H. , Knight, H. , Peck, S.C. , Grierson, C.S. , Hirt, H. and Knight, M.R. (2004) OXI1 kinase is necessary for oxidative burst‐mediated signalling in Arabidopsis. Nature, 427, 858–861. [DOI] [PubMed] [Google Scholar]
- Rivas, S. and Thomas, C.M. (2005) Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum . Annu. Rev. Phytopathol. 43, 395–436. [DOI] [PubMed] [Google Scholar]
- Rivas, S. , Rougon‐Cardoso, A. , Smoker, M. , Schauser, L. , Yoshioka, H. and Jones, J.D. (2004) CITRX thioredoxin interacts with the tomato Cf‐9 resistance protein and negatively regulates defence. EMBO J. 23, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Robatzek, S. , Chinchilla, D. and Boller, T. (2006) Ligand‐induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis . Genes Dev. 20, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden, J. , Eardley, L. , Hotson, A. , Cao, Y. and Mudgett, M.B. (2004) Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant–Microbe. Interact. 17, 633–643. [DOI] [PubMed] [Google Scholar]
- Romero‐Puertas, M.C. , Perazzolli, M. , Zago, E.D. and Delledonne, M. (2004) Nitric oxide signalling functions in plant‐pathogen interactions. Cell Microbiol. 6, 795–803. [DOI] [PubMed] [Google Scholar]
- Ron, M. and Avni, A. (2004) The receptor for the fungal elicitor ethylene‐inducing xylanase is a member of a resistance‐like gene family in tomato. Plant Cell, 16, 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock, T.R. , Zeng, L. , Brady, J.J. , Abramovitch, R.B. , Xiao, F. and Martin, G.B. (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature, 448, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, O. , Ludwig, A.A. , Merrick, C.J. , Baillieul, F. , Tracy, F.E. , Durrant, W.E. , Fritz‐Laylin, L. , Nekrasov, V. , Sjolander, K. , Yoshioka, H. and Jones, J.D.G. (2005) Functional analysis of Avr9/Cf‐9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf‐9‐dependent disease resistance in tomato. Plant Cell, 17, 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint‐Jore‐Dupas, C. , Faye, L. and Gomord, V. (2007) From planta to pharma with glycosylation in the toolbox. Trends Biotechnol. 25, 317–323. [DOI] [PubMed] [Google Scholar]
- Salmeron, J.M. , Barker, S.J. , Carland, F.M. , Mehta, A.Y. and Staskawicz, B.J. (1994) Tomato mutants altered in bacterial disease resistance provide evidence for a new locus controlling pathogen recognition. Plant Cell, 6, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron, J.M. , Oldroyd, G.E.D. , Rommens, C.M.T. , Scofield, S.R. , Kim, H.‐S. , Lavelle, D.T. , Dahlbeck, D. and Staskawicz, B.J. (1996) Tomato Prf is a member of the leucine‐rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell, 86, 123–133. [DOI] [PubMed] [Google Scholar]
- Saracco, S.A. , Miller, M.J. , Kurepa, J. and Vierstra, R.D. (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145, 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer, A. , Kazanaviciute, V. , Scheikl, E. , Teige, M. , Doczi, R. , Hirt, H. , Schwanninger, M. , Kant, M. , Schuurink, R. , Mauch, F. , Buchala, A. , Cardinale, F. and Meskiene, I. (2007) The PP2C‐type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell, 19, 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa, G. , D’Ascenzo, M. and Martin, G.B. (2000a) The major site of the pti1 kinase phosphorylated by the pto kinase is located in the activation domain and is required for pto‐pti1 physical interaction. Eur. J. Biochem. 267, 171–178. [DOI] [PubMed] [Google Scholar]
- Sessa, G. , D’Ascenzo, M. and Martin, G.B. (2000b) Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto‐Pto‐mediated hypersensitive response. EMBO J. 19, 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, L. , He, P. and Sheen, J. (2007) Intercepting host MAPK signaling cascades by bacterial type III effectors. Cell Host Microbe, 1, 167–174. [DOI] [PubMed] [Google Scholar]
- Shao, F. , Golstein, C. , Ade, J. , Stoutemyer, M. , Dixon, J.E. and Innes, R.W. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science, 301, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Sickmann, A. and Meyer, H.E. (2001) Phosphoamino acid analysis. Proteomics, 1, 200–206. [DOI] [PubMed] [Google Scholar]
- Smotrys, J.E. and Linder, M.E. (2004) Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73, 559–587. [DOI] [PubMed] [Google Scholar]
- Sokolovski, S. and Blatt, M.R. (2004) Nitric oxide block of outward‐rectifying K+ channels indicates direct control by protein nitrosylation in guard cells. Plant Physiol. 136, 4275–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y. , Wang, G.L. , Chen, L.L. , Kim, H.S. , Pi, L.Y. , Holsten, T. , Gardner, J. , Wang, B. , Zhai, W.X. , Zhu, L.H. , Fauquet, C. and Ronald, P. (1995) A receptor kinase‐like protein encoded by the rice disease resistance gene, Xa21 . Science, 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Stulemeijer, I.J.E. , Stratmann, J.W. and Joosten, M.H.A.J. (2007) The tomato MAP kinases LeMPK1, ‐2 and ‐3 are activated during the Cf‐4/Avr4‐induced HR and have distinct phosphorylation specificities. Plant Physiol. 144, 1481–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez‐Rodriguez, M.C. , Adams‐Phillips, L. , Liu, Y. , Wang, H. , Su, S.H. , Jester, P.J. , Zhang, S. , Bent, A.F. and Krysan, P.J. (2007) MEKK1 is required for flg22‐induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski, M.R. and Innes, R.W. (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26, 101–112. [DOI] [PubMed] [Google Scholar]
- Takemoto, D. and Jones, D.A. (2005) Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol. Plant–Microbe. Interact. 18, 1258–1268. [DOI] [PubMed] [Google Scholar]
- Tameling, W.I. and Takken, F.L.W. (2007) Resistance proteins: scouts of the plant innate immune system. doi 10.1007/s10658-007-9187-8 [DOI]
- Tang, X. , Frederick, R.D. , Zhou, J. , Halterman, D.A. , Jia, Y. and Martin, G.B. (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science, 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tatsuki, M. and Mori, H. (2001) Phosphorylation of tomato 1‐ aminocyclopropane‐1‐carboxylic acid synthase, LE‐ACS2, at the C‐terminal region. J. Biol. Chem. 276, 28051–28057. [DOI] [PubMed] [Google Scholar]
- Testerink, C. and Munnik, T. (2005) Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375. [DOI] [PubMed] [Google Scholar]
- Testerink, C. , Dekker, H.L. , Lim, Z.Y. , Johns, M.K. , Holmes, A.B. , Koster, C.G. , Ktistakis, N.T. and Munnik, T. (2004) Isolation and identification of phosphatidic acid targets from plants. Plant J. 39, 527–536. [DOI] [PubMed] [Google Scholar]
- Thurston, G. , Regan, S. , Rampitsch, C. and Xing, T. (2005) Proteomic and phosphoproteomic approaches to understand plant‐pathogen interactions. Physiol. Mol. Plant Pathol. 66, 3–11. [Google Scholar]
- Torres, M.A. and Dangl, J.L. (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. [DOI] [PubMed] [Google Scholar]
- Ulm, R. , Ichimura, K. , Mizoguchi, T. , Peck, S.C. , Zhu, T. , Wang, X. , Shinozaki, K. and Paszkowski, J. (2002) Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L. , Wulff, B.B.H. , Rivas, S. , Durrant, M.C. , Van der Ploeg, A. , De Wit, P.J.G.M. and Jones, J.D.G. (2005) Structure‐function analysis of Cf‐9, a receptor‐like protein with extracytoplasmic leucine‐rich repeats. Plant Cell, 17, 1000–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Luit, A.H. , Piatti, T. , Van Doorn, A. , Musgrave, A. , Felix, G. , Boller, T. and Munnik, T. (2000) Elicitation of suspension‐cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 123, 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen, G. , Van den Burg, H.A. , Cornelissen, B.J.C. and Takken, F.L.W. (2007) Structure and function of resistance proteins in Solanaceous plants. Annu. Rev. Phytopathol. 45, 43–72. [DOI] [PubMed] [Google Scholar]
- Van Schooten, B. , Testerink, C. and Munnik, T. (2006) Signalling diacylglycerol pyrophosphate, a new phosphatidic acid metabolite. Biochim. Biophys. Acta, 1761, 151–159. [DOI] [PubMed] [Google Scholar]
- Vera‐Estrella, R. , Barkla, B.J. , Higgins, V.J. and Blumwald, E. (1994) Plant defense response to fungal pathogens. Activation of host‐plasma membrane H+‐ATPase by elicitor‐induced enzyme dephosphorylation. Plant Physiol. 104, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8, 135–142. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Goshe, M.B. , Soderblom, E.J. , Phinney, B.S. , Kuchar, J.A. , Li, J. , Asami, T. , Yoshida, S. , Huber, S.C. and Clouse, S.D. (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID‐INSENSITIVE1 receptor kinase. Plant Cell, 17, 1685–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.S. , Pi, L.Y. , Chen, X. , Chakrabarty, P.K. , Jiang, J. , De Leon, A.L. , Liu, G.Z. , Li, A. , Benny, U. , Oard, J. , Ronald, P.C. and Song, W.Y. (2006) Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21‐mediated disease resistance. Plant Cell, 18, 3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, R.P. , Moscou, M.J. , Bogdanove, A.J. and Whitham, S.A. (2007) Transcript profiling in host‐pathogen interactions. Annu. Rev. Phytopathol. 45, 329–369. [DOI] [PubMed] [Google Scholar]
- Wojtaszek, P. (1997) Oxidative burst: An early plant response to pathogen infection. Biochem. J. 322, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, T. , Zong, N. , Zou, Y. , Wu, Y. , Zhang, J. , Xing, W. , Li, Y. , Tang, X. , Zhu, L. , Chai, J. and Zhou, J.M. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80. [DOI] [PubMed] [Google Scholar]
- Xing, T. , Ouellet, T. and Miki, B.L. (2002) Towards genomic and proteomic studies of protein phosphorylation in plant‐pathogen interactions. Trends Plant Sci. 7, 224–230. [DOI] [PubMed] [Google Scholar]
- Xu, W.H. , Wang, Y.S. , Liu, G.Z. , Chen, X. , Tinjuangjun, P. , Pi, L.Y. and Song, W.Y. (2006) The autophosphorylated Ser686, Thr688, and Ser689 residues in the intracellular juxtamembrane domain of XA21 are implicated in stability control of rice receptor‐like kinase. Plant J. 45, 740–751. [DOI] [PubMed] [Google Scholar]
- Yang, C.‐W. , Gonzalez‐Lamothe, R. , Ewan, R.A. , Rowland, O. , Yoshioka, H. , Shenton, M. , Ye, H. , O'Donnell, E. , Jones, J.D.G. and Sadanandom, A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U‐BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell, 18, 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. , Long, C. , Lan, L. , Chai, J. , Chen, S. , Tang, X. and Zhou, J.M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Liu, Y. , Yang, T. , Zhang, L. , Xu, S. , Xue, L. and An, L. (2006) Diverse signals converge at MAPK cascades in plant. Plant Physiol. Biochem. 44, 274–283. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D.G. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]


