SUMMARY
To identify positive regulators of cell death in plants, we performed a high‐throughput screening, employing potato virus X‐based overexpression in planta of a cDNA library derived from paraquat‐treated Nicotiana benthamiana leaves. The screening of 30 000 cDNA clones enabled the identification of an ADP‐ribosylation factor 1 (ARF1) that induces cell death when overexpressed in N. benthamiana. Overexpression of the guanosine diphosphate (GDP)‐locked mutant of ARF1 did not trigger cell death, suggesting that ARF1 guanosine triphosphatase (GTPase) activity is necessary for the observed cell death‐inducing activity. The ARF1 transcript level increased strongly following treatment with Phytophthora infestans elicitor INF1, as well as inoculation with a non‐host pathogen Pseudomonas cichorii in N. benthamiana. In addition, ARF1 was induced in the interaction between the N gene and tobacco mosaic virus (TMV) in Nicotiana tabacum. By contrast, inoculation with the virulent pathogen Pseudomonas syringae pv. tabaci did not affect ARF1 expression in N. benthamiana. Virus‐induced gene silencing of ARF1 in N. benthamiana resulted in a stunted phenotype, and severely hampered non‐host resistance towards P. cichorii. In addition, ARF1 silencing partially compromised resistance towards TMV in N. benthamiana containing the N resistance gene. By contrast, and in accordance with the ARF1 gene expression profile, silencing of ARF1 transcription did not alter the susceptibility of N. benthamiana towards the pathogen P. syringae pv. tabaci. These results strongly implicate ARF1 in the non‐host resistance to bacteria and N gene‐mediated resistance in N. benthamiana.
INTRODUCTION
Plants typically defend against a wide range of parasites by preventing them from entering attacked cells. This non‐host resistance (plant immunity) is based on a two‐layer protection, which includes pre‐ and postinvasive defence components (Lipka et al., 2005). On infection by host pathogens, by contrast, resistant host plants induce a complex set of signalling pathways, ultimately leading to the hypersensitive response (HR), a rapid death of invaded cells, which is associated with the restriction of pathogen growth (Heath, 2000). These host resistance processes can also occur during non‐host resistance, and the extent to which these resistance responses overlap is an active area of research (for a review, see Mysore and Ryu, 2004).
High‐throughput functional screening, i.e. the transient expression of a large number of genes from hosts or pathogens to elicit defence reactions, has been proven to be instrumental in the identification of regulators and components of these signalling pathways (Karrer et al., 1998; Nasir et al., 2005; Qutob et al., 2002; Takahashi et al., 2007; Takken et al., 2000; Torto et al., 2003). In this study, we performed a high‐throughput screening for cell death‐inducing factors, and identified an ADP‐ribosylation factor 1 (ARF1) as a positive regulator of cell death. ARFs constitute a highly conserved family belonging to the Ras superfamily of guanosine triphosphate (GTP)‐binding proteins or guanosine triphosphatases (GTPases) (Kahn et al., 1992). Like all members of the Ras superfamily, ARFs function as molecular switches between an ‘inactive’ guanosine diphosphate (GDP)‐bound and an ‘active’ GTP‐bound state. Guanine exchange factors (GEFs) convert inactive cytosolic GDP‐ARF into active, membrane‐associated GTP‐ARF, and GTPase‐activating proteins (GAPs) reform GDP‐ARF (Kjeldgaard et al., 1996). ARF1s in plants are well known to play a critical role in the formation of transport vesicles by recruiting cytosolic coat proteins to sites of vesicle budding (Kirchhausen, 2000). It has been shown that ARF1 specifically recruits coat protein I (COPI) to transport vesicles, thereby mediating retrograde vesicle trafficking from the Golgi to the endoplasmic reticulum (Kirchhausen, 2000). In addition to interaction with coat proteins, it has been shown in animals that ARF1 is able to interact with other effectors to assist its role in vesicle trafficking (for a review, see Donaldson et al., 2005; D'Souza‐Schorey and Chavrier, 2006; Nie et al., 2003; Randazzo et al., 2000). Indeed, ARF1 is known to interact with lipid‐modifying enzymes, and stimulates the activity of phospholipase D (PLD; Brown et al., 1993; Cockcroft et al., 1994) and phosphatidylinositol 4‐phosphate 5‐kinase (PIP5‐kinase; Jones et al., 2000), leading to the production of phosphatidic acid (PA) and phosphatidylinositol 4,5‐biphosphate (PIP2). It has been suggested that changes in specific phospholipids may mediate ARF1 action in membrane traffic. Similarly, ARF1 has been shown to regulate a dynamic pool of actin, thereby also facilitating the formation and/or dissociation of nascent vesicles from donor membranes (D'Souza‐Schorey and Chavrier, 2006).
Recently, it has become clear that ARF1 also plays a critical role in the pathogenesis of bacteria (Moss and Vaughan, 1991; Nagai et al., 2002) and viruses (Richards et al., 2002) in mammals. However, it remains elusive whether such a role can be attributed to ARF1 in plants. Nomura et al. (2006) reported that the Arabidopsis ARF‐GEF AtMIN7 is specifically targeted by HopM1, a Pseudomonas syringae virulence factor. However, to our knowledge, a direct association of ARF1 with disease response in plants has been reported only once for a rice ARF1: RARF1 (Lee et al., 2003). Although the authors did not report any loss‐of‐function data, they showed a rapid pathogen‐induced increase in gene expression of RARF1. In addition, the expression of pathogenesis‐related genes and enhanced disease resistance to a fungal pathogen was observed in transgenic Nicotiana tabacum plants expressing RARF1.
In this report, we provide evidence for a role of ARF1 in disease response in Nicotiana benthamiana. The ARF1 transcript level strongly increased following several types of biotic stress. In accordance with gene expression data, loss‐of‐function analysis of ARF1 seriously hampered non‐host resistance to Pseudomonas cichorii and partially compromised N gene‐mediated resistance towards tobacco mosaic virus (TMV) in N. benthamiana.
RESULTS
Functional screening for plant cell death‐inducing factors in N. benthamiana identifies an ARF1
The potato virus X (PVX)‐based binary expression vector pSfinx (Takken et al., 2000) was employed to screen for cell death‐inducing genes in N. benthamiana. Therefore, a directional cDNA library was constructed from mRNA isolated from N. benthamiana leaves treated with paraquat to generate reactive oxygen species and thus to induce defence‐related genes. A screening of 30 000 cDNA clones by the inoculation of N. benthamiana leaves allowed the identification of around 240 independent clones causing cell death within 3 weeks after inoculation. Clones, for which the cell death phenotype was subsequently confirmed by infiltration of a liquid Agrobacterium tumefaciens culture containing the pSfinx vector, were sequenced and analysed for gene annotation. The majority of these cDNA clones coded for proteins involved in protein degradation, such as ubiquitin‐like proteins, subunits of the proteasome complex and other peptidases. Amongst others, several classes of cDNA were identified that coded for protein kinases and soluble N‐ethylmaleimide‐sensitive factor (NSF) attachment proteins. Clone pSfinx:8‐51 was investigated in more detail, as it caused cell death in N. benthamiana leaves as rapidly as 7 days after inoculation, as well as within 4 days after infiltration (Fig. 1). blastx analysis (Altschul et al., 1997) of the 928‐bp insert of clone pSfinx:8‐51 (GenBank accession DQ531849) revealed the presence of a full‐length cDNA showing significant homology (E = 1 × e−97) to an ARF1 cDNA from Arabidopsis (GenBank accession NP_182239), as well as to ARF1 cDNAs from several dicots and monocots. The predicted open reading frame (ORF) of NbARF1 encodes a 181‐amino acid protein with very high (96%) sequence identity to other known ARF1 proteins.
Figure 1.

Overexpression of NbARF1 causes cell death in Nicotiana benthamiana. Agrobacterium tumefaciens clones transformed with the pSfinx vector containing no insert (pSfinx, left) or the NbARF1 cDNA (pSfinx:8‐51, right) were infiltrated in the right half of the leaf of an 8‐week‐old N. benthamiana plant. A photograph of the leaf phenotype was taken 4 days after infiltration.
ARF1 is a member of a gene family and its expression is induced by challenging with INF1, TMV and the non‐host pathogen P. cichorii
In order to characterize the genomic organization of the ARF genes in N. benthamiana, Southern hybridization was performed with the full insert sequence of pSfinx:8‐51. The hybridization pattern showed several bands with diverse signal intensities, suggesting that NbARF1 might be a member of a gene family in N. benthamiana (Fig. 2a, right panel). In an attempt to design a probe specific for the ARF1 gene identified in this study, a 399‐bp fragment harbouring the 3′‐end of NbARF1 ORF and the 3′‐untransformed region (3′‐UTR) (designated as ‘3′‐ORF‐UTR’) was used as probe (Fig. 2a, left panel). The number of fragments detected by this probe was two (BamHI and EcoRV) or three (EcoRI), suggesting that there is a maximum of two gene copies showing homology to this fragment. We further tested a 215‐bp fragment corresponding to the NbARF1 3′‐UTR (‘3′‐UTR’), but the hybridization pattern was the same as that obtained by ‘3′‐ORF‐UTR’ (data not shown). To study NbARF1 expression in relation to the defence response in N. benthamiana, Northern blot analysis was carried out with the probe corresponding to the NbARF1‘3′‐ORF‐UTR’ using RNA extracted from leaf material collected at defined time intervals after infiltration with the HR‐inducing elicitor INF1 (Kamoun et al., 1998), and after inoculation with the non‐host bacterial pathogen P. cichorii (Hikichi et al., 1998) or the host pathogen P. syringae pv. tabaci (Taguchi et al., 2001). In addition, total RNA was extracted from N gene‐containing N. tabacum leaves inoculated with TMV. A low basal NbARF1 gene expression was detected, which was not significantly affected by water infiltration (Fig. 2b). However, on infiltration with the elicitor INF1, NbARF1 transcription was strongly induced (Fig. 2b). Although a strong induction of NbARF1 gene expression was detected 6 h after infiltration with P. cichorii (Fig. 2c), no increase in NbARF1 expression was observed following challenge of N. benthamiana leaves with P. syringae pv. tabaci (Fig. 2d). To investigate whether ARF1 expression is involved in R gene‐mediated defence, N. tabacum harbouring the N gene was employed. The N gene from N. tabacum confers a gene‐for‐gene resistance towards most members of the Tobamovirus family by causing HR (Whitham et al., 1994). We used a synchronous HR‐inducing system based on a temperature shift. Nicotiana tabacum harbouring the N gene is permissive of TMV at high temperature above 26 °C (Weststeijn, 1981). Thus, incubation at 30 °C of TMV‐infected, N gene‐harbouring N. tabacum leaves results in the multiplication and spread of the virus without lesion formation. Shifting TMV‐infected leaves from 30 °C to 20 °C induces synchronous HR formation in the infected region. We attempted a temperature shift experiment using N gene‐harbouring N. benthamiana inoculated with TMV, but failed to produce synchronous HR induction. As the probe for hybridization, we used the ‘3‐ORF‐UTR’ fragment of NbARF1. It should be noted that the NbARF1 ORF shares 96% nucleotide sequence identity with NtARF1 (data not shown). A slight but clear induction in NtARF1 expression was observed after temperature shift in the leaves inoculated with TMV, but not in mock‐inoculated leaves (Fig. 2e). These results suggest the involvement of ARF1 in the N gene‐mediated defence response towards TMV, as well as in defence reactions towards the non‐host pathogen P. cichorii, but not towards the pathogen P. syringae pv. tabaci.
Figure 2.
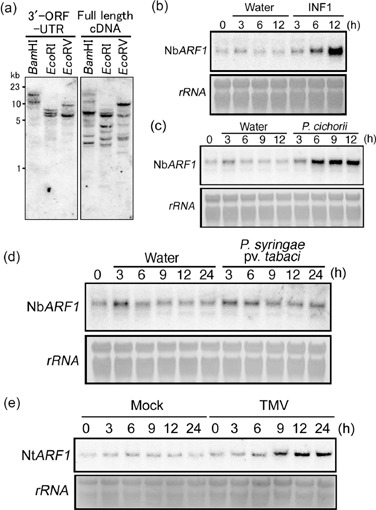
Genomic organization of the ARF1 gene and analysis of ARF1 gene expression in Nicotiana spp. (a) Genomic Southern analysis suggests that NbARF1 is a member of a small gene family in Nicotiana benthamiana. Ten micrograms of total DNA were digested to completion with BamIHI, EcoRI and EcoRV, respectively, and hybridized with the ‘3′‐ORF‐UTR’ fragment (left) or full pSfinx:8‐51 insert (full‐length cDNA; right). (b–e) Northern analysis of ARF1 gene expression following biotic stresses in N. benthamiana and Nicotiana tabacum. Ten micrograms of total RNA were separated in a denaturing agarose gel, and rRNAs were stained by methylene blue on the membranes as loading controls. The probe used for Northern blotting was the ‘3′‐ORF‐UTR’ as used in (a). NbARF1 gene expression was induced in N. benthamiana following infiltration with a solution containing 100 nm of the Phytophthora infestans elicitor INF1 (b), inoculation with the non‐host pathogen Pseudomonas cichorii (OD600 = 0.1) (c), but not following inoculation with the virulent pathogen Pseudomonas syringae pv. tabaci (OD600 = 0.1) (d). The NtARF1 transcript level increased in N gene‐harbouring N. tabacum inoculated with tobacco mosaic virus following a temperature shift (e).
Inducible expression of ARF1 protein triggers cell death and ion leakage
To confirm a correlation between ARF1 protein expression and cell death, a glucocorticoid‐inducible GVG expression vector (Aoyama and Chua, 1997) derivative, GVG‐ARF1‐HA, was constructed by cloning a fusion between the NbARF1 ORF and a single epitope of influenza haemagglutinin (HA) into the basic GVG vector. Following transient transformation by infiltration of Agrobacterium harbouring an empty GVG vector or GVG‐ARF1‐HA, gene expression was induced by infiltrating leaves with the glucocorticoid hormone dexamethasone (DEX). Cell death occurred 24 h after DEX treatment in ARF1‐HA‐expressing plants, whereas no cell death was observed in control plants infiltrated with Agrobacterium harbouring empty GVG (Fig. 3a). In addition, complete desiccation of the ARF1‐HA‐expressing leaf was observed 48 h after induction (Fig. 3a). The presence of the ARF1 protein was confirmed by immunoblot analysis using an anti‐HA antibody. High quantities of ARF1‐HA protein were detected at 6, 9 and 12 h after induction in ARF1‐HA‐transformed leaves (Fig. 3b). The attenuated expression of ARF1‐HA detected 24 h after DEX induction can probably be explained by the fact that cell death was already advanced at that time point (Fig. 3b). These results demonstrate that overexpression of ARF1 protein causes cell death in N. benthamiana. To further characterize cellular events on ARF1‐HA overexpression, ion leakage was analysed. Ion leakage became clearly detectable following DEX treatment in GVG‐ARF1‐HA‐transformed leaves, and was significantly higher than that in control leaves (Fig. 3c). Apparently, membranes became permeable on ARF1‐HA overexpression.
Figure 3.
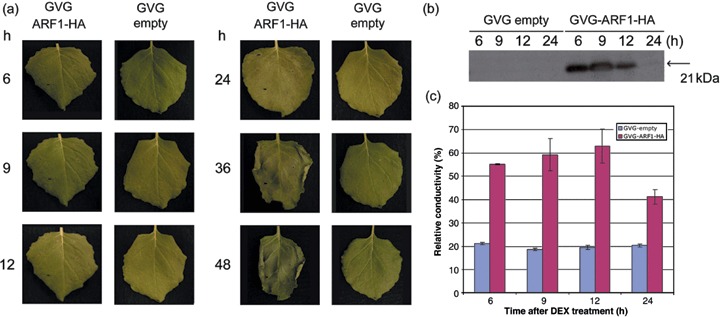
Correlation between NbARF1 protein expression and cell death in Nicotiana benthamiana. Leaves of an 8‐week‐old N. benthamiana plant were transiently transformed with Agrobacterium tumefaciens containing the empty GVG vector or GVG‐ARF1‐HA. Forty‐eight hours later, leaves were infiltrated with 30 µm of dexamethasone (DEX) to trigger ARF1‐HA protein expression. (a) Photographs of leaf phenotypes taken at different time points (6–48 h) after DEX induction. (b) Western blot analysis of ARF1‐HA protein at the indicated time points (6–24 h) by employing the anti‐HA antibody. (c) Ion leakage in ARF1‐HA‐expressing N. benthamiana leaves compared with control (empty GVG) leaves measured at the same time points as in (b).
Functional domains of ARF1
As described above, a common feature of small GTPases, such as ARF1, is the regulated binding and hydrolysis of guanine nucleotides. It is known that ‘inactive’ GDP‐bound or ‘active’ GTP‐bound ARF1 mutants can be generated by specifically mutating crucial amino acid residues in the ARF1 protein. Replacement of the threonine residue in the consensus sequence GLDAAGKT (motif P) with asparagine (T31N mutant) results in an inactive GDP‐locked ARF1 mutant. By contrast, substitution of the glutamine residue in the consensus sequence DVGGQ (motif G) by leucine (Q71L mutant) impairs intrinsic GTP hydrolysis, thereby generating a constitutively active GTP‐locked ARF1 mutant (Dascher and Balch, 1994; Kahn et al., 1995; Zhang et al., 1994). In addition to these two well‐described mutants, another ARF1 mutant was also obtained by deletion of the glycine residue at position 2 (G2Δ mutant), which is necessary for myristoylation of the ARF1 protein (Franco et al., 1996; Kahn et al., 1995). We hypothesize that the G2Δ mutant is incapable of being tethered to plasma membrane, thereby losing its normal function. In order to investigate whether the G2, T31 and Q71 amino acid residues play a role in the cell death‐inducing ability of NbARF1, pTA7001‐derived GVG vectors were constructed containing the GDP‐locked ARF1 mutant (ARF1T31N‐HA), GTP‐locked ARF1 mutant (ARF1Q71L‐HA) and G2 deletion mutant (ARF1G2Δ‐HA). A pTA7001‐derived vector containing the wild‐type (WT) ARF1 (ARF1WT‐HA) was employed as a positive control. Following transient transformation of N. benthamiana by infiltration with Agrobacterium harbouring these GVG vectors, ARF1 gene expression was induced by infiltrating the leaves with DEX. In all cases, the proteins of the expected size were successfully expressed, as shown by Western blot analysis with anti‐HA antibody (Fig. 4b). Rapid cell death occurred following DEX induction in the ARF1WT‐HA‐ and ARF1Q71L‐HA producing plants (Fig. 4a). No clear difference in timing and severity of cell death was found between the ARF1WT‐HA‐ and ARF1Q71L‐HA‐overexpressing leaves. By contrast, overexpression of ARF1G2Δ‐HA and ARF1T31N‐HA did not cause rapid cell death (Fig. 4a). If observations were continued for a longer time, cell death became slightly visible in leaves producing these mutants, being less severe in the G2Δ mutant than in the T31N mutant. These results suggest that GTPase activity of ARF1 properly tethered to plasma membrane is necessary to cause rapid cell death in N. benthamiana leaves.
Figure 4.
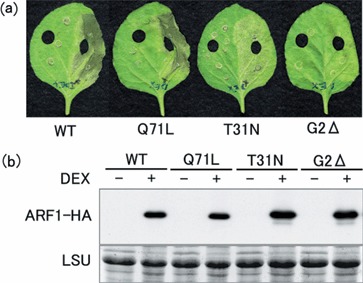
Cell death caused by mutant versions of ARF1. Whole leaves of Nicotiana benthamiana were infiltrated with agrobacterium carrying plasmids coding for ARF1 wild‐type protein (WT) or mutated versions Q71L, T31N and G2D, respectively. Forty‐eight hours after infiltration, the right halves of the leaves were infiltrated with dexamethasone (DEX) for the induction of protein expression. (a) Photographs were taken after another 48 h. (b) Samples for Western analysis were taken 6 h after DEX treatment. ARF1 proteins (ARF1‐HA) were detected with an anti‐HA‐tag antibody after the blot had been stained with amido black to visualize the large subunit of rubisCO protein (LSU) to confirm equal loading.
Virus‐induced gene silencing of ARF1
Loss‐of‐function analysis of ARF1 was performed by virus‐induced gene silencing (VIGS). Initially, a 375‐bp region from the NbARF1 ORF was amplified and cloned in antisense orientation in a tobacco rattle virus (TRV)‐derived expression vector (pTV:00; Ratcliff et al., 2001). However, on infiltrating this construct into N. benthamiana, plant growth was arrested and the plants died within 3 weeks (results not shown), probably because overall silencing of related genes coding for ARFs occurred in these plants, thereby blocking the critical roles of ARF1 in cell survival and development. In a second attempt, a 399‐bp region was amplified from NbARF1 comprising the full 3′UTR and 188 bp of the ORF (‘3′‐ORF‐UTR’, the same fragment as used in Fig. 2). From Southern blot analysis using this fragment (Fig. 2a), we hypothesize that, at most, two genes including NbARF1 were targeted for silencing. When the generated pTV:ARF1 construct was infiltrated into N. benthamiana, plants showed a severe stunted phenotype 4 weeks later (Fig. 5a). Silencing of NbARF1 gene expression was confirmed by real‐time polymerase chain reaction (PCR) in the seventh and eighth leaves above the inoculated leaf. Transcript levels of NbARF1 in pTV:ARF1‐infiltrated plants were estimated to be reduced by 80% when compared with pTV:00 control plants (Fig. 5b). To study the effect of ARF1 silencing on plant defence, P. cichorii, a non‐host bacterial pathogen, was inoculated onto N. benthamiana leaves silenced for ARF1. Growth of P. cichorii, determined in three independent experiments 20 h after inoculation, was significantly higher in ARF1‐silenced plants than in pTV:00‐infiltrated control plants (Fig. 6a). This result suggests a crucial role for ARF1 in non‐host resistance towards a bacterial pathogen. To examine the effect of ARF1 silencing on the defence reaction towards the host pathogen P. syringae pv. tabaci, bacterial growth was determined 48 h after infiltration in pTV:00‐ and pTV:ARF1‐innoculated plants. However, no difference in growth was observed between these plants (Fig. 6b). These results indicate that ARF1 silencing affects non‐host resistance, but not the basal resistance to a host pathogen. To investigate the role of ARF1 in R gene‐mediated resistance, N gene‐containing N. benthamiana was infiltrated with a green fluorescent protein (GFP)‐expressing TMV (TocJ/GFP; Hori and Watanabe, 2003), which allowed the easy detection of viral spreading. As expected, very small GFP spots were observed following infection with TMV on pTV:00‐infiltrated, N gene‐containing N. benthamiana (Fig. 6c). However, the lesion size in ARF1‐silenced, N gene‐containing N. benthamiana was consistently larger than that in non‐silenced plants in three independent experiments (Fig. 6c). The average size of randomly chosen GFP spots of ARF1‐silenced plants was significantly (P < 0.05) larger than that of control plants (Fig. 6d). This result indicates a partial loss of N gene‐mediated resistance towards TMV in ARF1‐silenced plants.
Figure 5.
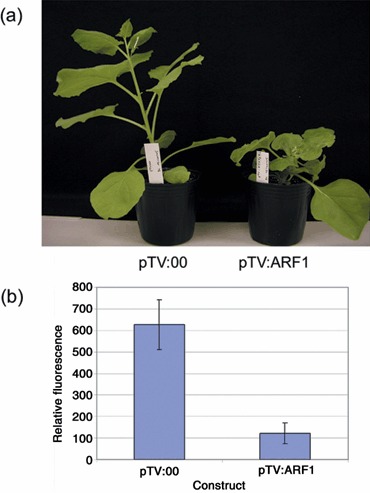
Virus‐induced gene silencing of NbARF1 in Nicotiana benthamiana. The two youngest leaves of a 4‐leaf stage N. benthamiana plantlet were infiltrated with pTV:ARF1 or with pTV:00 as a control treatment. Observations were done 4 weeks after inoculation. (a) Growth of ARF1 silenced plants (pTV:ARF1, right) was stunted compared to control plants (pTV:00, left). (b) NbARF1 transcript level was reduced by 80% in ARF1 silenced plants compared to control plants as quantified by real time PCR.
Figure 6.
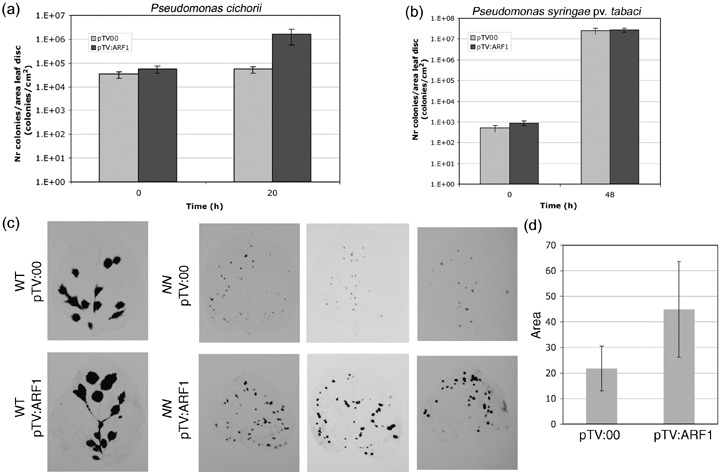
Virus‐induced gene silencing of NbARF1 in Nicotiana benthamiana compromises defence responses. (a) ARF1‐silenced (pTV:ARF1) or control (pTV:00) N. benthamiana leaves were infiltrated with Pseudomonas cichorii (OD600 = 0.01). The growth of P. cichorii in ARF1‐silenced or control N. benthamiana leaves was determined at the indicated time points (0 and 20 h). (b) ARF1‐silenced or control N. benthamiana leaves were infiltrated with Pseudomonas syringae pv. tabaci (OD600 = 0.0001). The growth of P. syringae pv. tabaci was determined at the indicated time points (0 and 48 h). (c) Wild‐type (WT) and N gene‐harbouring (NN) N. benthamiana plants, each inoculated with pTV:ARF1 or pTV:00, were infected with green fluorescent protein (GFP)‐expressing tobacco mosaic virus (TMV, TocJ/GFP). The spread of TMV was observed in three leaves, 11 days after inoculation, by measuring GFP fluorescence, indicated as black regions. The same results were obtained in three independent experiments. (d) Bar graph showing the sizes of randomly selected spots of GFP fluorescence (n = 113 for each bar) from the leaves of control (pTV:00) and ARF1‐silenced (pRV:ARF1) plants shown in (c). The difference is statistically significant (P < 0.05).
DISCUSSION
Functional screening of plant genes identifies an ARF
The aim of this study was the identification of cell death‐causing factors by functional in vivo screening of a cDNA library in N. benthamiana. To enable screening for a broad range of downstream factors involved in plant cell death, cDNA was derived from N. benthamiana leaves under chemically induced oxidative stress. By employing the PVX‐based binary expression vector pSfinx (Takken et al., 2000), we identified a cDNA encoding an ARF1 (NbARF1). Members of these small GTPases are present in various eukaryotic organisms, including yeast (Stearns et al., 1990), mammals (Monaco et al., 1990; Tsuchiya et al., 1991) and several higher plants, such as Arabidopsis thaliana (Regad et al., 1993), rice (Higo et al., 1994), potato (Szopa and Müller‐Röber, 1994), maize (Verwoert et al., 1995), carrot (Kiyosue and Shinozaki, 1995) and wheat (Kobayashi‐Uehara et al., 2001). Genomic Southern hybridization analysis suggested that several copies of NbARF1 genes could be present in the N. benthamiana genome (Fig. 2a), indicating that ARF genes are organized in a small multigene family in plants (Higo et al., 1994; Kiyosue and Shinozaki, 1995; Kobayashi‐Uehara et al., 2001; Regad et al., 1993). We observed that transiently overexpressed GFP‐tagged NbARF1 was localized on (endo)membranes as well as in the cytosol (results not shown), corroborating previous reports (Ritzenthaler et al., 2002; Takeuchi et al., 2002) of the primary function of ARF1 in vesicle trafficking in plants (Contreras et al., 2004; Lee et al., 2002; Memon, 2004; Molendijk et al., 2004; Pimpl et al., 2003). Importantly, we showed that, although the WT and GTP‐locked mutant of ARF1 caused rapid cell death, the GDP‐locked mutant (ARF1T31N‐HA) and a mutant that could not be tethered to the plasma membrane (ARF1G2Δ‐HA) failed to cause rapid cell death (Fig. 4). These results suggest that the GTPase function of ARF1 is involved in the observed cell death phenotype. In addition to NbARF1, we identified several ARF1‐like cDNAs that caused cell death after overexpression, although the effect was not as strong as with NbARF1 (data not shown). This finding suggests that the enhancement of the function of ARF1 and ARF1‐related proteins may generally lead to cell death.
ARF1 plays a critical role in plant defence
NbARF1 transcripts were detected at low basal level (Fig. 2b), suggesting housekeeping functions for ARF1 in N. benthamiana, in accordance with previous reports in plants (Kobayashi‐Uehara et al., 2001; Xu and Scheres, 2005). However, infiltration with the HR‐inducing elicitor INF1 induced NbARF1 (Fig. 2b). Furthermore, although NbARF1 expression was strongly induced in N. benthamiana following inoculation with the non‐host pathogen P. cichorii (Fig. 2c), no increase in NbARF1 expression was observed following inoculation with the pathogen P. syringae pv. tabaci (Fig. 2d). This is in contrast with the induction of RARF1 observed in rice following inoculation with virulent and avirulent strains of Magnaporthe grisea, although the transcript level was more rapidly induced in the incompatible reaction than in the compatible interaction (Lee et al., 2003). Finally, the NtARF1 transcript level was clearly increased in N gene‐containing N. tabacum inoculated with TMV (Fig. 2e).
A second observation of the role of ARF1 in defence was demonstrated by loss‐of‐function analysis via VIGS. Initial attempts, employing a 375‐bp region of the NbARF1 ORF, failed, probably because silencing was targeted to most members of this conserved gene family, resulting in a phenotype which was too severe. Gebbie et al. (2005) similarly failed to generate homozygous Arabidopsis lines harbouring antisense suppression constructs for ARF1. VIGS of the NbARF1 transcript level was only successful by employing a smaller region derived from the ORF, together with the full 3′UTR (3’‐ORF‐UTR), which resulted in a stunted phenotype (Fig. 5a). From the result of Southern blot analysis using the same 3′‐ORF‐UTR fragment as probe (Fig. 2a), we judged that, at most, two genes were targeted for silencing. We attempted VIGS of a single ARF1 gene using a 215‐bp fragment corresponding to the ARF1 3′‐UTR (3′‐UTR), but failed to cause gene silencing (data not shown). A stunted phenotype, as obtained here, was also observed in Arabidopsis following stable antisense suppression of ARF1 (Gebbie et al., 2005); this can probably be attributed to reduced cell expansion, as vesicle trafficking is crucial for the delivery of materials to the plasma membrane. In addition, reduced cell division was observed in ARF1‐suppressed plants of Arabidopsis (Gebbie et al., 2005), as well as in Arabidopsis plants containing a mutation in GNOM, a GEF of ARF1 (Geldner et al., 2003).
Silencing of NbARF1 severely compromised the plant defence against P. cichorii, indicating that ARF1 plays a role in non‐host resistance (Fig. 6a). Furthermore, R gene‐mediated resistance was partially compromised in NbARF1‐silenced, N gene‐containing N. benthamiana, as HR was delayed following inoculation with TMV (Fig. 6c,d). Although these results further support a role for ARF1 in defence, silencing of NbARF1 did not affect the susceptibility of N. benthamiana towards the virulent bacterial pathogen P. syringae pv. tabaci (Fig. 6b), indicating a differential involvement of ARF1 in different types of defence. A specific role of ARF1 in non‐host and R gene‐mediated resistance, but not in the basal resistance against the host pathogen, is interesting and requires future research.
To our knowledge, a direct role for ARF1 GTPases in plant defence has been described only once for RARF1 isolated from rice. Constitutive overexpression of RARF1 in N. tabacum triggered the formation of spontaneous lesions, induced pathogenesis‐related genes and resulted in increased resistance towards the oomycete pathogen Phytophthora parasitica var. nicotianae (Lee et al., 2003). However, the authors did not suggest a possible molecular mechanism to explain these observations. Recently, however, Nomura et al. (2006) described the identification of a P. syringae virulence factor, HopM1, which mediated the destruction of Arabidopsis AtMIN7 via the host proteasome. AtMIN7 was identified as an ARF‐GEF, and the authors observed that the pathogenesis of P. syringae in Arabidopsis was promoted following the destruction of AtMIN7 by HopM1. These results indicate that P. syringae has evolved a mechanism to eliminate a vesicle traffic pathway as an effective strategy to overcome host immunity (Nomura et al., 2006). The finding of Nomura et al. (2006) corroborates our observation. ARF‐GEF is needed to convert ARF‐GDP to ARF‐GTP, resulting in the activation of ARF function. Therefore, it is reasonable that the loss of function of ARF‐GEF and ARF both result in a decrease in plant resistance.
In addition to ARF GTPases, another family of GTPases belonging to the Ras superfamily of GTPases, namely the Rho GTPases, has recently been shown to play a key role in disease resistance and response to abiotic stress in plants (Agrawal et al., 2003; Gu et al., 2004). The best‐characterized Rho GTPase in plants in connection with defence is OsRac1 from rice (Kawasaki et al., 1999, 2006; Ono et al., 2001; Suharsono et al., 2002). Similar to our results, Moeder et al. (2005) observed a differential role of OsRac1 in defence towards a non‐host bacterial pathogen compared with a virulent bacterial pathogen. Stable overexpression of Rac1‐T24N, a dominant‐negative mutant of OsRac1, in N. tabacum severely compromised resistance towards the non‐host pathogen P. syringae pv. maculicola, whereas it did not affect susceptibility towards the virulent pathogen P. syringae pv. tabaci. In addition, and further corroborating our NbARF1 data in relation to viral defence, Moeder et al. (2005) observed reduced HR development following TMV infection of N gene‐containing N. tabacum expressing the dominant‐negative Rac1‐T24N. Although it is well known in mammals that ARF1 and Rac1 GTPases can interact with each other via an Arfaptin protein (D'Souza‐Schorey et al., 1997; Shin and Exton, 2001; Tarricone et al., 2001), it remains elusive whether ARF1 can interact with Rho GTPases in plants via an as yet unidentified protein.
In conclusion, we have provided strong evidence that ARF1 is involved in cell death signalling, as its overexpression causes cell death. Our data further implicate a role for ARF1 in plant defence, as the expression of NbARF1 and NtARF1 is induced on challenge with the non‐host pathogen P. cichorii and TMV, respectively. Corroborating these data, non‐host resistance and N gene‐mediated resistance are compromised following the silencing of NbARF1.
EXPERIMENTAL PROCEDURES
Plant material and paraquat treatment
WT and N gene‐containing N. benthamiana, as well as N gene‐containing N. tabacum, plants were grown under glasshouse conditions at 23 °C without supplemental light. Fully developed leaves of approximately 8‐week‐old N. benthamiana plants were treated by application of a solution containing 50 µm paraquat (Sigma‐Aldrich Japan, Tokyo, Japan), 10 mm phosphate buffer (pH 7.2) and 0.1% (v/v) Tween 20 with a soft paintbrush. Following 2 h in the dark, the plants were exposed to continuous light with an irradiance of 300 µE. The leaves were harvested after 3, 8 and 24 h, frozen in liquid nitrogen and used for total RNA isolation. One treated leaf was kept attached to the plant and monitored for cell death after a longer exposure to light.
cDNA library construction in pSfinx vector and functional screening
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN Japan, Tokyo, Japan). mRNA was isolated from total RNA by the mRNA Purification Kit (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK). mRNA was subsequently used for double‐stranded cDNA synthesis with asymmetrical SfiI sites, employing the Creator SMART™ cDNA Library Construction Kit (Clontech Laboratories, Mountain View, CA, USA). Thereafter, the cDNA was cloned in sense orientation into the pSfinx vector (Takken et al., 2000), and ultracompetent Escherichia coli DH5α (TOYOBO, Osaka, Japan) cells were transformed with the recombinant vector. Cells were subsequently plated onto Luria–Bertani (LB) agar containing 50 µg/mL kanamycin to high density. After forming colonies for 12 h at 37 °C, approximately 80 000 colonies were collected and pooled for plasmid isolation (QIAprep Spin Kit, QIAGEN Japan). Plasmids were then employed to transform Agrobacterium strain LBA4404. More than 30 000 individual colonies were transferred from agar plates to 384‐well microtitre plates filled with LB agar medium, and kept for further use. For functional screening, A. tumefaciens clones were transferred to 96‐well microtitre plates harbouring liquid LB medium, and subsequently cultured for 48 h at 28 °C. Liquid‐cultured cells were lifted by a tooth‐pick, and inoculated onto N. benthamiana leaves as described by Takken et al. (2000). Up to five expanded leaves per plant were inoculated with 96 colonies per leaf. Putative positive clones that induced cell death around the inoculation site were rescreened by infiltrating a liquid culture of A. tumefaciens cells harbouring the corresponding pSfinx clone into a fully expanded leaf of an approximately 8‐week‐old N. benthamiana plant.
Southern blot and Northern blot analysis
For Southern analysis, total DNA was prepared from N. benthamiana leaves employing the DNeasy Plant Kit (QIAGEN Japan), digested with the restriction enzymes BamHI, EcoRI and EcoRV, respectively, and separated by electrophoresis on a 1% (w/v) agarose gel before transfer to nylon membrane (Hybond N+, GE Healthcare). Total RNA was isolated from N. benthamiana and N. tabacum by the method of Nagy et al. (1988). Aliquots (10 µg each) were separated on formaldehyde–1% (w/v) agarose gels, and blotted onto membranes (Hybond N+, GE Healthcare). Hybridizations were accomplished with the full insert sequence of the original pSfinx:8‐51 clone (928 bp), the 3’‐ORF‐UTR fragment (399 bp) or the 3’‐UTR fragment (215 bp).
These fragments were PCR amplified, and labelled using the TAKARA BcaBEST Labelling Kit (TaKaRa, Kyoto, Japan) and [32P]dCTP (3000 Ci/mmol; GE Healthcare). Following hybridization, the membranes were washed under high stringency conditions [0.1 × standard saline citrate (SSC) and 0.1% (w/v) sodium dodecyl sulphate (SDS)] at 60 and 68 °C for Northern and Southern blots, respectively. Detection was performed using a BAS2000 bio‐imaging auto‐analyser (Fuji Photo Film, Tokyo, Japan).
Inducible expression of HA‐tagged ARF1 and its mutant versions
The coding sequence for a single HA tag and a diglycine linker (5′‐GGGGGTTATCCATACGATGTTCCAGATTATGCT‐3′; GGYPYDVPDYA) was fused in frame to the 3′‐end of the NbARF1 ORF by PCR. The resulting cDNA was directionally cloned in the XhoI and SpeI sites of a GVG vector, pTA7001 (Aoyama and Chua, 1997), to create GVG‐ARF1‐HA. This binary vector was subsequently transferred to A. tumefaciens GV3101 by chemical transformation. In order to establish transient transformation, A. tumefaciens cells were infiltrated into fully expanded leaves of approximately 8‐week‐old N. benthamiana plants. Forty‐eight hours after infiltration, gene expression was induced by infiltrating the same leaves with DEX [30 µm in 0.1% (v/v) ethanol]. A series of pTA7001‐derived vectors was constructed containing mutant versions of ARF1: a G2Δ mutant of ARF1 (GVG‐ARF1G2Δ‐HA), T31N mutant of ARF1 (GVG‐ARF1T31N‐HA) and Q71L mutant of ARF1 (GVG‐ARF1Q71L‐HA). These were generated by overlap extension PCR (Ho et al., 1989).
Immunoblot analysis
For the detection of ARF1‐HA protein, leaf samples were ground under liquid nitrogen and extracted with 10 mm phosphate buffer (pH 7.0) containing 0.1% (v/v) Triton‐X100 (2 mL per gram of tissue). After centrifugation, the supernatants were used for electrophoresis in the presence of 0.4% (w/v) SDS on 12% (w/v) polyacrylamide slab gels (Laemmli, 1970), followed by electrophoretic transfer to poly(vinylidene difluoride) (PVDF) membranes (Millipore, Billerico, MA, USA). After binding of the anti‐HA antibody (Roche, Mannheim, Germany), the localization of antigens was detected by a secondary antibody (Promega, Madison, WI, USA), and visualized by chemiluminescence using the ECL system (GE Healthcare).
Construction of silencing vector and inoculation of N. benthamiana
A 399‐bp ARF1 cDNA fragment (‘3′‐ORF‐UTR’) was amplified by PCR from the original pSfinx plasmid harbouring NbARF1 with the primer pair 5′‐CTGTGCTGCTTGTTTTTGCT‐3′ and 5′‐CTTCGTTTACAAATTTATG‐3′, annealing between positions 470 and 868 of the ARF1 gene. The generated PCR product was then cloned in the antisense orientation into the KpnI and SpeI sites of pTV:00 (Ratcliff et al., 2001) to generate pTV:ARF1. Virus infection on N. benthamiana was performed as described by Ratcliff et al. (2001) with some minor modifications. Briefly, liquid cultures of A. tumefaciens GV3101 harbouring pTV:00, pTV:ARF1 or pBintra6 (Ratcliff et al., 2001) were grown to saturation in liquid LB medium. Cultures were centrifuged and thereafter resuspended in 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES)–KOH (pH 5.6), 10 mM MgCl2 and 150 µm acetosyringone [optical density at 600 nm (OD600) of 0.9 and 0.8 for pBintra6 and pTV:00/pTV:ARF1, respectively]. Cultures were subsequently incubated at room temperature for 3 h. For TRV infections, separate cultures containing pBintra6 and pTV:00 (or pTV:ARF1) were mixed in a 1 : 1 ratio. The mixture was subsequently infiltrated into the two youngest leaves of N. benthamiana (four‐leaf stage). Four weeks after infiltration, post‐transcriptional gene silencing was confirmed by real‐time PCR employing a primer pair (5′‐AATGACAGAGACCGTGTTGTTGA‐3′ and 5′‐ACAGCATCCCGAAGCTCATC‐3′), annealing between positions 397 and 473 of the NbARF1 ORF.
Inoculation with P. cichorii and P. syringae pv. tabaci and determination of growth kinetics
Inoculation with bacteria and determination of growth kinetics were essentially performed as described by Sharma et al. (2003). Briefly, a culture of P. cichorii SPC9001 (Hikichi et al., 1998) or P. syringae pv. tabaci (Taguchi et al., 2001) was grown overnight at 28 °C in liquid LB medium containing rifampicin (20 µg/mL). Following centrifugation, bacterial cells were washed and resuspended in 10 mm MgCl2 (OD600 of 0.01 and 0.0001 for P. cichorii and P. syringae pv. tabaci, respectively). The bacterial cell suspension was thereafter infiltrated into N. benthamiana leaves using a needle‐less syringe. At the indicated post‐inoculation time points, small leaf discs (5 mm in diameter) were punched out of the infiltrated areas of four plants. The leaf discs were subsequently homogenized in 750 µL of 10 mm MgCl2, and serial dilutions were plated onto LB plates supplemented with rifampicin. Following incubation at 28 °C for 24 h, the colonies were counted to determine the increase in the number of bacteria.
Inoculation with GFP‐expressing TMV and determination of growth
The inoculation of N. benthamiana leaves with a GFP‐expressing TMV (TocJ/GFP) was performed as described by Hori and Watanabe (2003). Eleven days after inoculation, viral growth was determined by measuring GFP fluorescence in detached inoculated leaves with a Fluorimager (Fluorimager 595, Molecular Dynamics, Sunnyvale, CA, USA). The lesion size was calculated using APS Assess Software (http://www.apsnet.org/press/assess/).
Temperature shift experiment of TMV in N. tabacum
A temperature shift experiment was performed by rub‐inoculating 8‐week‐old mature detached leaves of N gene‐containing N. tabacum with TMV (10 µg/mL) in 10 mm phosphate buffer (pH 7.0) using carborandum. Mock inoculations were performed by rubbing N. tabacum leaves with phosphate buffer and carborandum alone. Following incubation at 30 °C for 48 h under continuous light, the temperature was reduced to 20 °C, allowing the leaves to initiate HR, which is induced on recognition of TMV. Leaf samples were taken at the indicated time points and subsequently employed for total RNA isolation.
Sequence data from this article have been deposited with the GenBank data libraries under accession number DQ531849.
Supporting information
Figure S1. Southern blot analysis of NbARF1 (For reviewing purpose only)
Figure S2. Northern blot analysis of NbARF1 (For reviewing purpose only)
Figure S3. Result of RT‐PCR of NbARF1 after attempt of VIGS with NbARF1 3’‐UTR fragment (For reviewing purpose only)
This material is available as part of the online article from:
http://www.blackwell‐synergy.com/doi/full/10.1111/j.1364‐3703.2007.00440.x
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
All work was carried out at the Iwate Biotechnology Research Center, Iwate, Japan.
We acknowledge Dr Matthieu Joosten (Wageningen University, Wageningen, The Netherlands) for providing the pSfinx vector, and Dr David Baulcombe (Sainsbury Laboratory, John Innes Centre, Norwich, UK) for pTV:00 and pBintra6. We thank Dr Nam‐Hai Chua (Rockefeller University, New York, USA) for pTA7001 and Dr Yu‐ichiro Watanabe (University of Tokyo, Tokyo, Japan) for TMV‐GFP. We are also grateful to Dr Yuki Ichinose (Okayama University, Okayama, Japan) and Dr Kazumi Suzuki (Iwate Biotechnology Research Center, Kitakami, Japan) for provision of P. syringae pv. tabaci and P. cichorii SPC9001, respectively. Many thanks are due to Ms Akiko Hirabuchi and Ms Hiroe Utsushi for excellent technical assistance. This work was supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science (JSPS), and by grants from the ‘Program for Promotion of Basic Research Activities for Innovative Biosciences’ (Japan) and the ‘Iwate University 21st Century COE Program: Establishment of Thermo‐Biosystem Research Program’.
REFERENCES
- Agrawal, G.K. , Iwahashi, H. and Rakwal, R. (2003) Small GTPase ‘Rop’: molecular switch for plant defense responses. FEBS Lett. 546, 173–180. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T. and Chua, N.‐H. (1997) A glucocorticoid‐mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Brown, H.A. , Gutowski, S. , Moomaw, C.R. , Slaughter, C. and Sternweis, P.C. (1993) ADP‐ribosylation factor, a small GTP‐dependent regulatory protein, stimulates phospholipase D activity. Cell, 75, 1137–1144. [DOI] [PubMed] [Google Scholar]
- Cockcroft, S. , Thomas, G.M.H. , Fensome, A. , Geny, B. , Cunningham, E. , Gout, I. , Hiles, I. , Totty, N.F. , Truong, O. and Hsuan, J.J. (1994) Phospholipase D: a downstream effector of ARF in granulocytes. Science, 263, 523–526. [DOI] [PubMed] [Google Scholar]
- Contreras, I. , Ortiz‐Zapater, E. and Aniento, F. (2004) Sorting signals in the cytosolic tail of membrane proteins involved in the interaction with plant ARF1 and coatomer. Plant J. 38, 685–698. [DOI] [PubMed] [Google Scholar]
- Dascher, C. and Balch, W.E. (1994) Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 269, 1437–1448. [PubMed] [Google Scholar]
- Donaldson, J.G. , Honda, A. and Weigert, R. (2005) Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta, 1744, 364–373. [DOI] [PubMed] [Google Scholar]
- D'Souza‐Schorey, C. , Boshans, R.L. , McDonough, M. , Stahl, P.D. and Van Aelst, L. (1997) A role for POR1, a Rac1‐interacting protein, in ARF6‐mediated cytoskeletal rearrangements. EMBO J. 16, 5445–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza‐Schorey, C. and Chavrier, P. (2006) ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358. [DOI] [PubMed] [Google Scholar]
- Franco, M. , Chardin, P. , Chabre, M. and Paris, S. (1996) Myristoylation‐facilitated binding of the G protein ARF1GDP to membrane phospholipids is required for its activation by a soluble nucleotide exchange factor. J. Biol. Chem. 271, 1573–1578. [DOI] [PubMed] [Google Scholar]
- Gebbie, L.K. , Burn, J.E. , Hocart, C.H. and Williamson, R.E. (2005) Genes encoding ADP‐ribosylation factors in Arabidopsis thaliana L. Heyn.; genome analysis and antisense suppression. J. Exp. Bot. 414, 1079–1091. [DOI] [PubMed] [Google Scholar]
- Geldner, N. , Anders, N. , Wolters, H. , Keicher, J. , Kornberger, W. , Muller, P. , Delbarre, A. , Ueda, T. , Nakano, A. and Jürgens, G. (2003) The Arabidopsis GNOM ARF‐GEF mediates endosomal recycling, auxin transport, and auxin‐dependent plant growth. Cell, 112, 219–230. [DOI] [PubMed] [Google Scholar]
- Gu, Y. , Wang, Z. and Yang, Z. (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr. Opin. Plant Biol. 7, 527–536. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000) Hypersensitive response‐related death. Plant Mol. Biol. 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Higo, H. , Kishimoto, N. , Saito, A. and Higo, K. (1994) Molecular cloning and characterization of a cDNA encoding a small GTP‐binding protein related to mammalian ADP‐ribosylation factor from rice. Plant Sci. 100, 41–49. [Google Scholar]
- Hikichi, Y. , Suzuki, K. , Toyoda, K. , Horikoshi, M. , Hirooka, T. and Okuno, T. (1998) Successive observation of growth and movement of genetically lux‐marked Pseudomonas cichorii and the response of host tissues in the same lettuce leaf. Ann. Phytopathol. Soc. Jpn. 64, 519–525. [Google Scholar]
- Ho, S.N. , Hunt, H.D. , Horton, R.M. , Pullen, J.K. and Pease L.R. (1989) Site‐directed mutagenesis by overlap extension using the polymerase chain‐reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hori, K. and Watanabe, Y. (2003) Construction of a Tobamovirus vector that can systematically spread and express foreign gene products in Solanaceous plants. Curr. Top. Microbiol. 20, 129–136. [Google Scholar]
- Jones, D.H. , Morris, J.B. , Morgan, C.P. , Kondo, H. , Irvine, R.F. and Cockcroft, S. (2000) Type I phosphatidylinositol 4‐phosphate 5‐kinase directly interacts with ADP‐ribosylation factor 1 and is responsible for phosphatidylinositol 4,5‐biphosphate synthesis in the Golgi compartment. J. Biol. Chem. 275, 13 962–13 966. [DOI] [PubMed] [Google Scholar]
- Kahn, R.A. , Clark, J. , Rulka, C. , Stearns, T. , Zhang, C.‐J. , Randazzo, P.A. , Terui, T. and Cavenagh, M. (1995) Mutational analysis of Saccharomyces cerevisiae ARF1. J. Biol. Chem. 270, 143–150. [DOI] [PubMed] [Google Scholar]
- Kahn, R.A. , Der, C.J. and Bokoch, G.M. (1992) The Ras superfamily of GTP‐binding proteins: guidelines on nomenclature. FASEB J. 6, 2512–2513. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Van West, P. , Vleeshouwers, V.G. , De Groot, K.E. and Govers, F. (1998) Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell, 10, 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer, E.E. , Beachy, R.N. and Holt, C.A. (1998) Cloning of tobacco genes that elicit the hypersensitive response. Plant. Mol. Biol. 36, 681–690. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T. , Henmi, K. , Ono, E. , Hatakeyama, S. , Iwano, M. , Satoh, H. and Shimamoto, K. (1999) The small GTP‐binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA, 96, 10 922–10 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T. , Koita, H. , Nakatsubo, T. , Hasegawa, K. , Wakabayashi, K. , Takahashi, H. , Umemura, K. , Umezawa, T. and Shimamoto, K. (2006) Cinnamoyl‐CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA, 103, 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen, T. (2000) Three ways to make a vesicle. Nat. Rev. Mol. Cell Biol. 1, 187–198. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T. and Shinozaki, K. (1995) Cloning of a carrot cDNA for a member of the family of ADP‐ribosylation factors (ARFs) and characterization of the binding of nucleotides by its product after expression in E. coli . Plant Cell Physiol. 36, 849–856. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard, M. , Nyborg, J. and Clark, B.F.C. (1996) The GTP binding motif: variations on a theme. FASEB J. 10, 1347–1368. [PubMed] [Google Scholar]
- Kobayashi‐Uehara, A. , Shimosaka, E. and Handa, H. (2001) Cloning and expression analysis of cDNA encoding an ADP‐ribosylation factor from wheat: tissue‐specific expression of wheat ARF. Plant Sci. 160, 535–542. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, M.H. , Min, M.K. , Lee, Y.J. , Jin, J.B. , Shin, D.H. and Kim, D.H. (2002) ADP‐ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis . Plant Physiol. 129, 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.Y. , Hong, J.K. , Kim, C.Y. , Chun, H.J. , Park, H.C. , Kim, J.C. , Yun, D.‐J. , Chung, W.K. , Lee, S.‐H. , Lee, S.Y. , Cho, M.J. and Lim, C.O. (2003) Over‐expressed rice ADP‐ribosylation factor 1 (RARF1) induces pathogenesis‐related genes and pathogen resistance in tobacco plants. Physiol. Plant. 119, 573–581. [Google Scholar]
- Lipka, V. , Dittgen, J. , Bednarek, P. , Bhat, R. , Wiermer, M. , Stein, M. , Landtag, J. , Brandt, W. , Rosahl, S. , Scheel, D. , Llorente, F. , Molina, A. , Parker, J. , Somerville, S. and Schulze‐Lefert, P. (2005) Pre‐ and postinvasion defenses contribute to nonhost resistance in Arabidopsis . Science, 310, 1180–1183. [DOI] [PubMed] [Google Scholar]
- Memon, A.R. (2004) The role of ADP‐ribosylation factor and SAR1 in vesicular trafficking in plants. Biochim. Biophys. Acta, 1664, 9–30. [DOI] [PubMed] [Google Scholar]
- Moeder, W. , Yoshioka, K. and Klessig, D.F. (2005) Involvement of the small GTPase Rac in the defense responses of tobacco to pathogens. Mol. Plant–Microbe Interact. 2, 116–124. [DOI] [PubMed] [Google Scholar]
- Molendijk, A.J. , Ruperti, B. and Palme, K. (2004) Small GTPases in vesicle trafficking. Curr. Opin. Plant Biol. 7, 694–700. [DOI] [PubMed] [Google Scholar]
- Monaco, L. , Murtagh, J.J. , Newman, K.B. , Tsai, S.‐C. , Moss, J. and Vaughan, M. (1990) Selective amplification of an mRNA and related pseudogene for a human ADP‐ribosylation factor, a guanine nucleotide‐dependent protein activator of cholera toxin. Proc. Natl. Acad. Sci. USA, 87, 2206–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, J. and Vaughan, M. (1991) Activation of cholera‐toxin and Escherichia coli heat‐labile enterotoxins by ADP‐ribosylation factors, a family of 20‐kDa guanine nucleotide‐binding proteins. Mol. Microbiol. 5, 2621–2627. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S. and Ryu, C.‐M. (2004) Nonhost resistance: how much do we know? Trends Plant Sci. 9, 97–104. [DOI] [PubMed] [Google Scholar]
- Nagai, H. , Kagan, J.C. , Zhu, X. , Kahn, R.A. and Roy, C.R. (2002) A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science, 295, 679–682. [DOI] [PubMed] [Google Scholar]
- Nagy, F. , Kay, S.A. and Chua, N.‐H. (1988) Analysis of gene expression in transgenic plants In Plant Molecular Biology Manual (Gelvin S.B. and Schilperoort R.A., eds), pp. 1–29. Dordrecht: Kluwer. [Google Scholar]
- Nasir, K.H.B. , Takahashi, Y. , Ito, A. , Saitoh, H. , Matsumura, H. , Kanzaki, H. , Shimizu, T. , Ito, M. , Fujisawa, S. , Sharma, P.C. , Ohme‐Takagi, M. , Kamoun, S. and Terauchi, R. (2005) High‐throughput in planta expression screening identifies a class II ethylene‐responsive element binding factor‐like protein that regulates plant cell death and non‐host resistance. Plant J. 43, 491–505. [DOI] [PubMed] [Google Scholar]
- Nie, Z. , Hirsch, D.S. and Randazzo, P.A. (2003) Arf and its many interactors. Curr. Opin. Cell. Biol. 15, 396–404. [DOI] [PubMed] [Google Scholar]
- Nomura, K. , DebRoy, S. , Lee, Y.H. , Pumplin, H. , Jones, J. and He, S.Y. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science, 313, 220–223. [DOI] [PubMed] [Google Scholar]
- Ono, E. , Wong, H.N. , Kawasaki, T. , Hasegawa, M. , Kodama, O. and Shimamoto, K. (2001) Essential role of the small GTPase Rac in disease resistance in rice. Proc. Natl. Acad. Sci. USA, 98, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl, P. , Hanton, S.L. , Taylor, J.P. , Pinto‐daSilva, L.L. and Denecke, J. (2003) The GTPase ARF1p controls the sequence‐specific vacuolar sorting route to the lytic vacuole. Plant Cell, 15, 1242–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Kamoun, S. and Gijzen, M. (2002) Expression of a Phytophthora sojae necrosis‐inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32, 361–373. [DOI] [PubMed] [Google Scholar]
- Randazzo, P.A. , Nie, Z. , Miura, K. and Hsu, V.W. (2000) Molecular aspects of the cellular activities of ADP‐ribosylation factors. Sci. STKE, 2000, re1. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F. , Martin‐Hernandez, A.M. and Baulcombe, D.C. (2001) Tobacco Rattle Virus for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Regad, F. , Bardet, C. , Tremousaygue, D. , Moisan, A. , Lescure, B. and Axelos, M. (1993) cDNA cloning and expression of an Arabidopsis GTP‐binding protein of the ARF family. FEBS Lett. 316, 133–136. [DOI] [PubMed] [Google Scholar]
- Richards, A.A. , Stang, E. , Pepperkok, R. and Parton, R.G. (2002) Inhibitors of COP‐mediated transport and cholera toxin action inhibit Simian Virus 40 infection. Mol. Biol. Cell, 13, 1750–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler, C. , Nebenführ, A. , Movafeghi, A. , Stussi‐Garaud, C. , Behnia, L. , Staehelin, L.A. and Robinson, D.G. (2002) Reevaluation of the effects of Brefeldin A on plant cells using tobacco bright yellow 2 cells expressing Golgi‐targeted green fluorescent protein and COPI antisera. Plant Cell, 14, 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P.C. , Ito, A. , Shimizu, T. , Terauchi, R. , Kamoun, S. and Saitoh, H. (2003) Virus‐induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1‐induced hypersensitive response (HR) in Nicotiana benthamiana . Mol. Gen. Genomics, 269, 583–591. [DOI] [PubMed] [Google Scholar]
- Shin, O.‐H. and Exton, J.H. (2001) Differential binding of Arfaptin 2/POR1 to ADP‐ribosylation factors and Rac1. Biochem. Biophys. Res. Commun. 285, 1267–1273. [DOI] [PubMed] [Google Scholar]
- Stearns, T. , Kahn, R.A. , Botstein, D. and Hoyt, M.A. (1990) ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol. Cell. Biol. 10, 6690–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suharsono, U. , Fujisawa, Y. , Kawasaki, T , Iwasaki, Y. , Satoh, H. and Shimamoto, K. (2002) The heterotrimeric G protein α subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA, 99, 13 307–13 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szopa, J. and Müller‐Röber, B. (1994) Cloning and expression analysis of an ADP‐ribosylation factor from Solanum tuberosum L. Plant Cell Rep. 14, 180–183. [DOI] [PubMed] [Google Scholar]
- Taguchi, F. , Tanaka, R. , Kinoshita, S. , Ichinose, Y. , Imura, Y. , Andi, S. , Toyoda, K. , Shiraishi, T. and Yamada, T. (2001) Harpinpst from Pseudomonas syringae pv. tabaci is defective and deficient in its expression and HR‐inducing activity. J. Gen. Plant Pathol. 67, 116–123. [Google Scholar]
- Takahashi, Y. , Nasir, K.H.B. , Ito, A. , Kanzaki, H. , Matsumura, H. , Saitoh, H. , Fujisawa, S. , Kamoun, S. and Terauchi, R. (2007) High‐throughput screen of cell death‐inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1‐induced cell death signaling and non‐host resistance to Pseudomonas cichorii . Plant. J. 49, 1030–1040. [DOI] [PubMed] [Google Scholar]
- Takeuchi, M. , Ueda, T. , Yahara, N. and Nakano, A. (2002) ARF1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31, 499–515. [DOI] [PubMed] [Google Scholar]
- Takken, F.L.W. , Luderer, R. , Gabriels, S.J.E.J. , Westerick, N. , Lu, R. , De Wit, P.J.G.M. and Joosten, M.H.A.J. (2000) A functional cloning strategy, based on a binary PVX‐expression vector, to isolate HR‐inducing cDNAs of plant pathogens. Plant J. 24, 275–283. [DOI] [PubMed] [Google Scholar]
- Tarricone, C. , Xiao, B. , Justin, N. , Walker, P.A. , Rittinger, K. , Gamblin, S.J. and Smerdon, S.J. (2001) The structural basis of Arfaptin‐mediated cross‐talk between Rac and Arf signaling pathways. Nature, 411, 215–219. [DOI] [PubMed] [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A.R. , Van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome. Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, M. , Price, S.R. , Tsai, S.‐C. , Moss, J. and Vaughan, M. (1991) Molecular identification of ADP‐ribosylation factor mRNAs and their expression in mammalian cells. J. Biol. Chem. 266, 2772–2777. [PubMed] [Google Scholar]
- Verwoert, I.I.G.S. , Brown, A. , Slabas, A.R. and Stuitje, A.R. (1995) A Zea mays GTP‐binding protein of the ARF family complements an Escherichia coli mutant with a temperature‐sensitive malonyl‐coenzyme A: acyl carrier protein transacylase. Plant Mol. Biol. 27, 629–633. [DOI] [PubMed] [Google Scholar]
- Weststeijn, E.A. (1981) Lesion growth and virus localization in leaves of Nicotiana tabacum cv. Xanthi nc. after inoculation with tobacco mosaic virus and incubation alternately at 22 °C and 32 °C. Physiol. Plant Pathol. 18, 357–368. [Google Scholar]
- Whitham, S. , Dinesh‐Kumar, S.P. , Choi, D. , Hehl, R. , Corr, C. and Baker, B. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the interleukin‐1 receptor. Cell, 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Xu, J. and Scheres, B. (2005) Dissection of Arabidopsis ADP‐ribosylation factor 1 function in epidermal cell polarity. Plant Cell, 17, 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.J. , Rosenwald, A.G. , Willingham, M.C. , Skuntz, S. , Clark, J. and Kahn, R.A. (1994) Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo . J. cell Biol. 124, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Southern blot analysis of NbARF1 (For reviewing purpose only)
Figure S2. Northern blot analysis of NbARF1 (For reviewing purpose only)
Figure S3. Result of RT‐PCR of NbARF1 after attempt of VIGS with NbARF1 3’‐UTR fragment (For reviewing purpose only)
This material is available as part of the online article from:
http://www.blackwell‐synergy.com/doi/full/10.1111/j.1364‐3703.2007.00440.x
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


