SUMMARY
Taxonomy: Papaya ringspot virus (PRSV) is an aphid‐transmitted plant virus belonging to the genus Potyvirus, family Potyviridae, with a positive sense RNA genome. PRSV isolates belong to either one of two major strains, P or W. The P strains infect both papaya and cucurbits whereas the W strains infect only cucurbits.
Geographical distribution: PRSV‐P is found in all major papaya‐growing areas.
Physical properties: Virions are filamentous, non‐enveloped and flexuous measuring 760–800 × 12 nm. Virus particles contain 94.5% protein and 5.5% nucleic acid. The protein component consists of the virus coat protein (CP), which has a molecular weight of about 36 kDa as estimated by Western blot analysis. Density of the sedimenting component in purified PRSV preparations is 1.32 g/cm3 in CsCl.
Genome: The PRSV genome consists of a unipartite linear single‐stranded positive sense RNA of 10 326 nucleotides with a 5′ terminus, genome‐linked protein, VPg.
Transmission: The virus is naturally transmitted via aphids in a non‐persistent manner. Both the CP and helper component (HC‐Pro) are required for vector transmission. This virus can also be transmitted mechanically, and is typically not seed‐transmitted.
Hosts: PRSV has a limited number of hosts belonging to the families Caricaceae, Chenopodiaceae and Cucurbitaceae. Propagation hosts are: Carica papaya, Cucurbita pepo and Cucumis metuliferus cv. accession 2459. Local lesion assay hosts are: Chenopodium quinoa and Chenopodium amaranticolor.
Control: Two transgenic papaya varieties, Rainbow and SunUp, with engineered resistance to PRSV have been commercially grown in Hawaii since 1998. Besides transgenic resistance, tolerant varieties, cross‐protection and other cultural practices such as isolation and rogueing of infected plants are used to manage the disease.
Virus code: 00.057.0.01.045.
Virus accession number: 57010045.
Useful link: http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/57010045.htm
INTRODUCTION
Papaya ringspot virus (PRSV), a member of the aphid‐transmitted genus Potyvirus, is the cause of a destructive disease and a major limiting factor for papaya and cucurbit cultivation worldwide (Purcifull et al., 1984). PRSV is grouped into papaya‐infecting type‐P (PRSV‐P) and non‐papaya ‐infecting type‐W (PRSV‐W). PRSV‐P isolates infect plants in the families Caricaceae, Cucurbitaceae and Chenopodiaceae whereas isolates of PRSV‐W infect plants in the families Cucurbitaceae and Chenopodiaceae. PRSV‐P and PRSV‐W are serologically indistinguishable and are transmitted non‐persistently by several species of aphids. PRSV‐P infection is typically characterized by the production of ringspot symptoms on fruit of infected papaya trees (Jensen, 1949). In addition to ringspots, PRSV produces a range of other symptoms such as leaf mosaic and chlorosis, water‐soaked oily streaks on the petiole and upper part of the trunk, distortion of young leaves that sometimes results in shoestring‐like symptoms that resemble mite damage, stunting of infected plants, and flower abortion (Fig. 1). Consequently, fruit production can be severely decreased, and fruit sugar levels reduced by 50% or more. Symptoms on cucurbits are similar to those on papaya. PRSV is able to infect members of the family Chenopodiaceae such as Chenopodium amaranticolor and Chenopodium quinoa but the disease symptoms are restricted to local lesions on the leaves (Purcifull et al., 1984). Infection assays on these ‘local lesion hosts’ are used as supplementary information for evaluation of PRSV pathogenicity. The virus can be propagated and maintained for experimental studies on Carica papaya, Cucurbita pepo and Cucumis metuliferus, although in the case of Cucumis metuliferus it will proliferate on some accessions but not others (cv. Accession 2459 but not, for example, PI 292190) (Yeh et al., 1984). Recently, Momordica charantia (a member of the family Cucurbitaceae) is reported to be a climbing weed host reservoir for PRSV‐P in Jamaica with infected plants exhibiting vein clearing symptoms (Chin et al., 2007).
Figure 1.
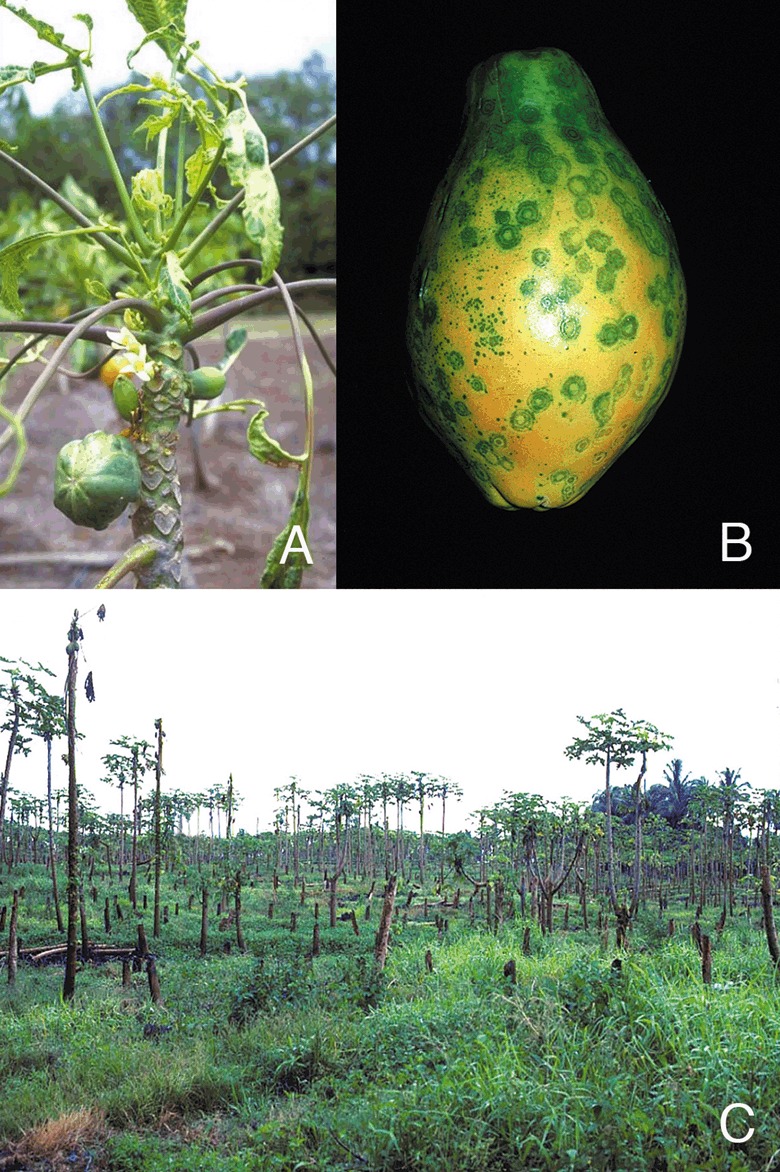
Symptoms of PRSV on papaya. (A) PRSV‐infected papaya tree, (B) ringspot symptoms on fruit, and (C) PRSV‐infected papaya orchard in Puna area of Hawaii in 1994.
The devastating nature of PRSV on commercial crops is exemplified by the near destruction of the Hawaiian papaya industry in the 1990s (Ferreira et al., 2002; Gonsalves, 1998, 2006; Gonsalves et al., 2006, 2007a). This pathogen profile reviews the current knowledge on the biology of PRSV, pathogenesis, genome, isolate/sequence diversity and development of disease control strategies. An overview on the control of PRSV by development of genetically engineered virus resistance in papaya has been included as it is the most successful approach to combat PRSV and it has been commercially implemented in Hawaii. However, PRSV remains a threat to the economic production of papaya and cucurbits worldwide (Fermin and Gonsalves, 2003; Fermin et al., 2004; Fuchs and Gonsalves, 2007; Gonsalves, 1998, 2006; Gonsalves and Fermin, 2004; Gonsalves and Ferreira, 2003; Gonsalves et al., 2004a,2004b, 2006, 2007a, 2007b; Suzuki et al., 2007; Tripathi et al., 2006).
GENOME ORGANIZATION
Papaya ringspot virus belongs to the genus Potyvirus, family Potyviridae. The virus consists of non‐enveloped flexuous filamentous particles which consist of a positive‐sense, single‐stranded, unipartite RNA genome encapsidated by the genome‐encoded coat protein (CP). The genomic RNA of PRSV is 10 326 nucleotides long followed by a tract of polyA sequence (An) (Fig. 2). An open reading frame starting at nucleotide 86 and ending at nucleotide 10 120 encodes a polyprotein of 3344 amino acids from which all the proteins of the virus are derived. As with other potyviruses, several proteins are produced through a combination of cotranslational, post‐translational, autoproteolytic and transproteolytic processing by three virus‐encoded endoproteases, P1, HC‐Pro and NIa (Adams et al., 2005; Yeh and Gonsalves, 1985; Yeh et al., 1992). The viral genes [name (product size)], listed in the order of their occurrence (5′ to 3′), in the PRSV genome are: P1 (63k), helper component (HC‐Pro, 52k), P3 (46k), cylindrical inclusion protein (CI, 72k), 6K (6k), nuclear inclusion protein a (NIa, 48k), nuclear inclusion protein b (NIb, 59k) and coat protein (CP, 35k) (Fig. 2) (Quemada et al., 1990; Wang and Yeh, 1997; Yeh and Gonsalves, 1985; Yeh et al., 1992) . The P1 protein of PRSV is 18–34k larger than that found in other potyviruses. Several of the virus‐encoded proteins have been identified by the non‐virion complexes they form in infected plant cells, including the HC‐Pro (also known as the amorphous inclusion or AI protein), the CI associated with cylindrical or pinwheel inclusions, and NIa and NIb proteins associated with nuclear inclusion bodies.
Figure 2.
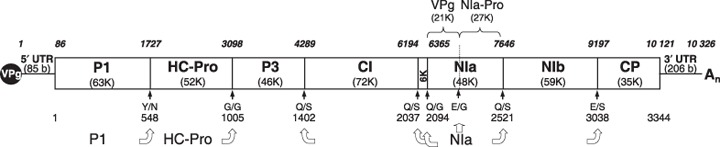
PRSV genome map of the Hawaiian isolate HA (GenBank accession number S46722). Numbers at the top indicate nucleic acid position. Numbers at the bottom indicate the amino acid position in the polyprotein. Molecular weights of the individual viral proteins are shown below the viral protein name in parentheses. The 5′ and 3′ untranslated regions (UTR) are marked along with their lengths (in bases, b) in parentheses. Amino acids flanking the cleavage sites of proteases P1, HC‐Pro and NIa (shown with block arrows) are indicated below black arrows marking the relative cleavage site position. The solid black circle labelled VPg represents the genome‐linked protein.
Interestingly, two cleavage sites at the N‐terminus of the CP have been predicted based on PRSV sequence analysis. The first, VFHQ/SKNE (Quemada et al., 1990), produces a CP of 33k, smaller than the 36k estimated from authentic CP (Gonsalves and Ishii, 1980), and an NIb protein containing 537 amino acids, which is about 20 amino acids larger than those of other potyviruses. The second, VYHE/SRGTD (Yeh et al., 1992), produces a CP and Nb of similar size to other potyviruses. There is no firm evidence to suggest that only one cleavage site is used for processing. Heterogeneous CP products would be expected if both sites were used in polyprotein processing. This may explain why purified CP preparations that have been stored frequently contain the major 36k form as well as CPs that are 2–5k smaller.
The 85‐base, 5′ untranslated region (UTR) of the PRSV genome is rich in A and U residues, similar to other potyviruses, but is 63–121 bases shorter. The 5′ UTR shows sequence similarity to the 5′ UTR of a number of other potyviruses, suggesting that this region may serve a common functional role (Yeh et al., 1992).
The virus‐encoded VPg protein that is derived from the autoproteolytic processing of the NIa protease in potyviruses is a covalently bound component in a number of plant and animal viral genomes. The role of the VPg is related to priming of RNA synthesis during replication of the viral genome and possibly packaging of the virion. Evidence from studies of similar positive‐strand viruses suggest that viral genome replication occurs via intermediate complexes, which include the virus‐encoded NIb (RNA‐dependent RNA polymerase or RdRp) and minus‐strand templates (Riechmann et al., 1992).
HOST SPECIFICITY DETERMINANTS, PATHOGENESIS AND VECTOR TRANSMISSION
As with other potyviruses, PRSV is typically stylet‐borne and is transmitted by many species of aphids (mainly Myzus persicae and Aphis gossypii) in a non‐persistent manner. The acquisition and transmission of infectious PRSV virion particles occurs during the brief period when the aphid superficially probes into the plant. Two virus‐encoded proteins, CP and HC‐Pro, are required for this process (Maia et al., 1996; Peng et al., 1998; Pirone, 1991; Pirone and Blanc, 1996).
The development of a system to produce infectious viral transcripts from recombinant PRSV in vitro (Chiang and Yeh, 1997) has provided a unique opportunity to begin to identify viral‐encoded gene segments involved in various viral functions, including pathogenicity, host range and vector transmission (Chen et al., 2001b; Lee et al., 2001), as well as to probe the mechanisms involved in induced virus resistance caused by cross‐protection and transgene‐induced post‐transcriptional gene silencing (Chiang et al., 2001; You et al., 2005). The results of bioassay studies with in vitro transcripts of recombinant PRSV‐W with its CP gene replaced with that of a CP gene from PRSV‐P demonstrated that the CP gene is not a determinant for infection of papaya (Chen et al., 2001b). Instead, similar experiments with PRSV‐W containing PRSV‐P segments from the region including the NIa gene and a portion of the NIb gene were required for papaya infection. Mutations in the PRSV‐P genome at two amino acid positions in the NIa‐Pro region, which are conserved within isolates of the same host range type but differ between PRSV‐P and PRSV‐W, 2309 (K→D) and 2487 (I→V), demonstrated that these two residues are critical for conferring PRSV pathogenicity of papaya (Chen et al., 2003).
In order to identify the genetic determinants for pathogenicity across host species, recombinants were generated between a severe PRSV‐P strain (PRSV HA) and a nitrous acid mutant derived from it (PRSV HA5‐1). Mutations in the P1 and HC‐Pro genes of the severe strain engineered to match those found in the mild strain resulted in the attenuation of both PRSV HA symptoms in papaya as well as reduced local lesion formation on Chenopodium quinoa (Chiang et al., 2007; Lee et al., 2001). The results also indicated that of the two genes, HC‐Pro was the major determinant for local lesion formation in Chenopodium quinoa.
Understanding the interactions occurring between the host and viral pathogen and between multiple viruses and their host during mixed infections is of fundamental biological interest as well as a key to developing strategies to control or manage disease. In the case of PRSV, the use of cross‐protection, a tool which has been used to protect a plant from the severe symptoms of a virulent virus by prior inoculation with an attenuated form of the same virus, has provided a clear demonstration that these biological interactions exist. Although cross‐protection is not clearly understood at the molecular level, studies suggest sequence‐specific RNA‐mediated natural defence, which is similar to post‐transcriptional gene silencing (PTGS) (Ratcliff et al., 1999). Experiments to evaluate the efficacy of cross‐protection against virulent PRSV‐P strains differing in geographical origins suggest that cross‐protection is more effective against challenge strains of the virus that are related more closely related to the virus used for cross‐protection (Gonsalves and Garnsey, 1989; Yeh and Gonsalves, 1994), consistent with the notion that sequence specificity plays a role in the efficacy of protection. Further evidence for the involvement of viral homology and the importance of extent and position of this homology in the phenomenon of strain‐specific cross‐protection was provided in a study using the original and recombinant versions of the attenuated PRSV‐P HA5‐1 on Cucumis metuliferus followed by infection with virulent strains PRSV‐W or PRSV‐P. While the attenuated PRSV mutant HA5‐1 provided 90–100% protection against the severe parental strain PRSV HA under greenhouse and field conditions, it provided only 20–30% protection against the PRSV‐W from Taiwan (Yeh and Gonsalves, 1984, 1994). By contrast, the recombinant HA5‐1 carrying both the heterologous CP and the 3′ UTR from PRSV‐W significantly enhanced the protection against PRSV‐W in cucurbits (You et al., 2005). In particular, the heterologous 3′ UTR was found to be critical for enhancement of the protection against PRSV‐W in Cucumis metuliferus. Interestingly, the same recombinant HA5‐1 virus carrying the heterologous PRSV‐W CP or 3′ UTR reduced the effectiveness of protection against PRSV HA in papaya, indicating that both the CP and the 3′ UTR are also important in cross‐protection in papaya (You et al., 2005).
SEQUENCE DIVERSITY AND EVOLUTION
Knowledge of sequence diversity among isolates of a virus and their distribution has the potential to deepen our understanding of viral origins, development, dispersion and disease etiology. This information would be useful in developing effective virus disease management programmes. With PRSV, most studies have focused on examining sequence variation in the CP gene. Initially data from the USA and Australia (Bateson et al., 1994; Quemada et al., 1990) suggested that there was little sequence variation among the CP gene from PRSV isolates within these countries. However, recent sequence data of PRSV CP genes of isolates from India (Jain et al., 2004) and Mexico (Silva‐Rosales et al., 2000) suggest greater sequence variation among the local populations of PRSV isolates in other countries. A phylogenetic tree illustrating the relationship of PRSV isolates from different countries utilizing published CP gene sequences (Bateson et al., 2002; Jain et al., 2004) is shown in Fig. 3. Reported CP sequence diversity at the amino acid (10%) and nucleic acid (14%) levels were highest among the Asian populations of PRSV isolates (Jain et al., 2004). Interestingly, this variation is considerably less than that found among isolates of other potyviruses, such as Yam mosaic virus (YMV), where the nucleotide sequence diversity was reportedly as high as 28%. The PRSV isolates collected within India were as different from each other in CP gene sequence (0–11%) as they were to the CP gene sequences of isolates collected in Bangladesh (9–11%), other Asian countries (4–14%), Australia (5–11%) and the Americas (5–11%). A study by Bateson et al. (2002) also described the high levels of diversity in PRSV isolates from the Indian subcontinent and proposed that they probably represented the oldest population of PRSV, and based on their basal position according to phylogenetic analysis, the origin of PRSV might have been South Asia.
Figure 3.
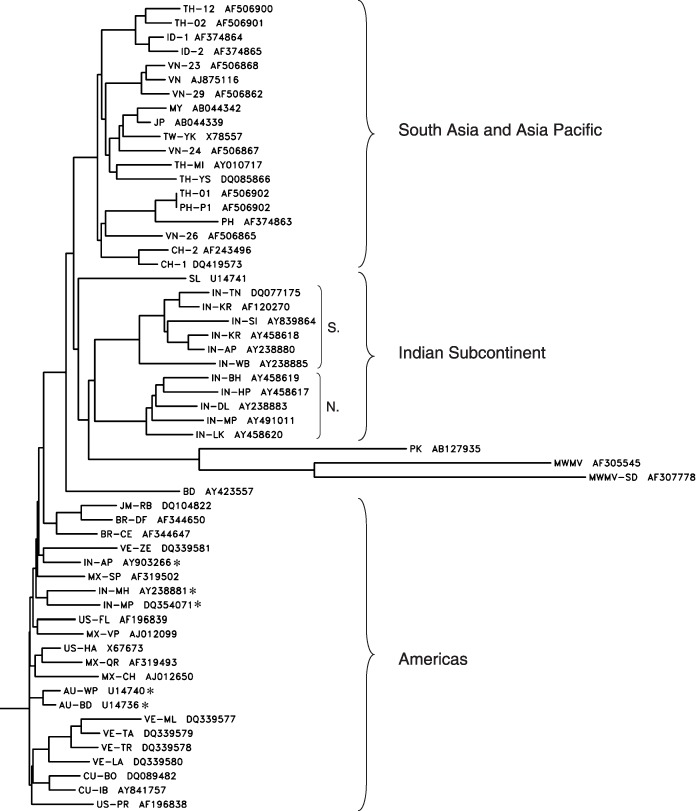
Neighbour‐joining tree showing the relationships between CP gene sequences of PRSV isolates from various countries. The tree was constructed with Clustal W and Drawgram programs using CP sequences of PRSV isolates from AU (Australia), BD, (Bangladesh), BR (Brazil), CH (China), CU (Cuba), ID (Indonesia), IN (India), MX (Mexico), MY (Malaysia), JM (Jamaica), JP (Japan), PH (Philippines), SL (Sri Lanka), TW (Taiwan), TH (Thailand), US (USA), VE (Venezuela) and VN (Vietnam). A CP sequence from a PRSV‐W type isolate from Pakistan (PK) and Moroccan watermelon mosaic virus (MWMV) CP genes were used as outgroup sequences. N. and S. denote North and South India, respectively. Asterisks denote PRSV CP sequences of isolates that are not from the Americas, but cluster with sequences of PRSV from countries of that region.
Some of the variation observed between PRSV‐P isolates was due to differences in the CP gene length, which varied between 840 nucleotides encoding 280 amino acids for the KA2 isolate from India (Jain et al., 2004) and 870 nucleotides encoding 290 amino acids for the VNW‐38 isolate from central Vietnam (Bateson et al., 2002). All described differences in CP length were confined to the first 50 amino acids at the amino terminal end of the CP, a region of noted variability due to changes in the number of so‐called EK repeats (Bateson et al., 2002; Jain et al., 2004). The sequence differences in this region of the virus genome lead mainly to conservative amino acid substitutions, which probably preserves CP function. At present, variation in length of the CP gene has been described from among PRSV populations within countries such as India (Jain et al., 2004) and Vietnam (Bateson et al., 2002) but not from Thailand (Bateson et al., 2002).
An interesting question regarding PRSV is the origin of types P and W. A number of viral diseases on cucurbits were initially associated with Watermelon mosaic virus 1 (WMV‐1) but not with any viral diseases of papaya. Later, serological and molecular characterization showed that WMV‐1 was virtually identical to PRSV. Did type P originate from W, or conversely, did they evolve independently? A sequence study of PRSV‐P and PRSV‐W from Australia suggests that the recent outbreak of PRSV‐P came from the population of PRSV‐W already present in Australia rather than as a new introduction (Bateson et al., 1994, 2002). This proposal is also supported by the diversity of cucurbit‐infecting potyviruses and virus isolates that are phylogenetically closer to PRSV.
CONTROL STRATEGIES: CONVENTIONAL TO GENETICALLY ENGINEERED RESISTANCE
PRSV disease management practices include quarantine, eradication, avoidance by planting papaya in areas isolated from the virus, continual rogueing of infected plants, use of tolerant lines to lower the economic losses caused by PRSV, cross‐protection and transgenic resistance. The Hawaiian papaya story is described here because it involves all of the above efforts and ultimately transgenic resistance in papaya was most successful in controlling the virus.
Development of PRSV‐resistant cultivars through conventional breeding met with limited success because of difficulties in overcoming intergeneric reproductive barriers of wild, related species of papaya (Gonsalves et al., 2006; Manshardt, 1992; Manshardt and Wenslaff, 1989). In addition, partial loss of tolerance in back‐crosses with the commercial papaya parent also limits the usefulness of this approach. As a control strategy for PRSV in papaya, cross‐protection was initiated in 1979 and used briefly in Hawaii, Florida, Mexico, Taiwan and Thailand (Wang et al., 1987; Yeh and Gonsalves, 1994; Yeh et al., 1988). Cross‐protection involves the use of a mild virus strain to protect plants against economic damage caused by the severe strain of the same virus or related virus (Gonsalves and Garnsey, 1989; Yeh and Gonsalves, 1984). Mild strain HA5‐1 was selected from severe strain HA after nitric acid mutagenesis and used to cross‐protect papayas in Hawaii (Yeh and Gonsalves, 1984). Although used for about 5 years, relatively severe virus symptoms occurred on certain cultivars, such as Sunrise, and it was thus not widely adopted (Ferreira et al., 1992; Mau et al., 1989; Pitz et al., 1994). In Taiwan, cross‐protection was evaluated extensively, but it proved to be only marginally effective. The value of the mild strain HA5‐1 was more fully appreciated when it was used later as the source of the CP gene for genetically engineering resistance. Use of the CP gene from the mild PRSV strain would probably mitigate against the impact of potential viral recombination possibly associated with CP gene transformation.
Although other laboratories have engineered resistance against PRSV using the CP (Bau et al., 2003; Cheng et al., 1996; Davis and Ying, 2004; Lines et al., 2002) or the replicase (Chen et al., 2001a) genes of PRSV, we will emphasize here the Hawaii transgenic papaya case because the resultant Rainbow and SunUp cultivars are the only transgenic papayas to be deregulated and commercialized, and to be sold and consumed in the mainland US and Canada (Gonsalves, 1998, 2006; Gonsalves et al., 2004b, 2007a; Suzuki et al., 2007; Tripathi et al., 2006).
Hawaii transgenic papaya case
Although first reported in 1945 (Jensen, 1949), PRSV was probably observed as early as 1937 (Parris, 1938) on the island of Oahu in Hawaii. PRSV appeared to cause a relatively mild disease of papaya until the middle 1950s when a strain of the virus causing yellow mosaic disease was reported (Ishii and Holtzmann, 1963). This new strain was initially described as a new papaya virus (Papaya mosaic virus) but shown later to be a different strain of PRSV, suggesting that the new yellow mosaic strain had either evolved from the original milder strain of the virus or that a second more severe strain was introduced in the mid‐1950s. Interestingly, within 10 years of its occurrence, the new yellow mosaic strain of PRSV quickly became widespread and severely affected production. On Oahu papaya acreage declined from a peak of just over 243 ha in 1956 to less than 16 ha in 1968.
With the decline in papaya production on Oahu due to both PRSV and encroaching urbanization, the Puna district on the island of Hawaii became the major production centre. By the early 1990s, production averaged about 25 000 tonnes of fruit annually, and represented about 95% of Hawaii's papaya production (Ferreira et al., 1992).
By 1971 PRSV had become established on the island of Hawaii in home‐grown papaya in the Hilo and Kona districts; it quickly became established in Keaau also, about 6.5–8 km from Hilo. A strict quarantine was imposed prohibiting the movement of papaya seedlings between the Hilo/Keaau region and commercial production areas in Puna. In effect, a PRSV‐free buffer of about 30 km kept commercial production relatively isolated from PRSV. In this buffer zone, only occasional backyard or homeowner plants were noted, and remained PRSV free.
In 1992, PRSV was observed in Puna (Ferreira et al., 2002). Within 3 years, commercial production in Puna was no longer possible (Fig. 1C), except for a few relatively isolated areas.
By the autumn of 1995, growers attempted to ‘outrun’ the virus by relocating production along the Hamakua coast, just north of Hilo, in former areas of sugarcane production where PRSV was absent. In this way, papaya production on the island of Hawaii was partially spared the total impact of PRSV, as production declined from 23 600 tonnes to 11 600 tonnes from 1992 to 1999.
As the PRSV epidemic played out in Puna, efforts to develop a solution were well underway, having been initiated about 6 years earlier. The concept of pathogen‐derived resistance (PDR), conceived in the mid‐1980s (Sanford and Johnston, 1985), offered a new approach for controlling virus diseases. In 1986, Gonsalves’ laboratory at Cornell University in collaboration with the Upjohn Company and the University of Hawaii started work to engineer papaya with the CP gene of the mild PRSV strain HA5‐1. The CP gene construct was originally designed with the concept that protein expression was required for resistance. The PRSV CP is produced by post‐translational protease cleavage of a PRSV polyprotein and does not contain a native translation signal specific to CP or even a 5′ UTR. Therefore, a chimeric PRSV CP transgene construct was made utilizing the translation signals found in the leader sequence (5′ untranslated RNA translational enhancer and initial 16 amino acid coding sequences) of the Cucumber mosaic virus (CMV) CP gene fused in‐frame to the structural sequence of the PRSV CP including the Q/S protein cleavage site and 70 nucleotides of the non‐coding region. This was accomplished by cloning the PRSV HA5‐1 CP structural sequence from plasmid pPRV117 between the CaMV 35S double enhancer promoter—translational leader sequence and CaMV 35S terminator of a CMV expression cassette (Quemada et al., 1990, 1991). This PRSV CP expression cassette was finally cloned into pGA482GG, a modified version of the Agrobacterium transformation vector pGA482 (An et al., 1988) that contained the nptII (neomycin phosphotransferase II) gene behind a nopaline synthase promoter and a uidA (GUS or β glucuronidase) gene behind a CaMV 35S promoter, used for kanamycin selection and colorimetric screening of transformants, respectively (An, 1986; Ling et al., 1991).
The red‐fleshed cultivars Sunrise and Sunset, and the yellow‐fleshed Kapoho were transformed with PRSV HA5‐1 CP biolistically (Fitch et al., 1990). Nine selected transgenic lines from original transformants, six Sunset and three Kapoho, were screened for resistance with PRSV HA from which the mild strain HA5‐1 had been derived. R0‐micropropagated plants of the first transformant line designated 55‐1 showed excellent resistance to PRSV HA (Fig. 4A) (Fitch et al., 1992).
Figure 4.

Evaluation of transgenic papaya for PRSV resistance. (A) R0‐transgenic papaya line 55‐1 (left) and nontransgenic control (right) six months after inoculation with PRSV HA in the greenhouse. (B) Aerial photograph of the 1‐acre plot of Rainbow papaya in Puna (Hawaii) 28 months after transplanting. The Rainbow block was surrounded by non‐transgenic susceptible Sunrise plants, which are severely infected by PRSV.
In 1991, the first field assessment of line 55‐1 was made (Lius et al., 1997). Nearly all (95%) of the non‐transgenic plants and those of a transgenic line that lacked the CP gene were infected by 77 days after the start of the field trial, whereas none of the line 55‐1 plants was ever infected. PRSV was not recovered from line 55‐1 plants except for two plants, which showed virus symptoms on side shoots but none on the leaves of the main canopy. Plants of line 55‐1 grew normally and produced fruit with total soluble solids of about 13%, within the expected range for fruit from uninfected plants. Thus, by mid‐1993, the trial had provided convincing evidence that line 55‐1 would be useful for controlling PRSV in Hawaii (Gonsalves, 1998).
Given the imposing presence of PRSV on the island of Hawaii and the performance of line 55‐1 and its R1 derivatives in the greenhouse and field trials, an active attempt to utilize this new transgenic germplasm for the development of commercial cultivars was made. Because the major cultivar grown in Puna was the yellow‐fleshed Kapoho, an F1 hybrid of Kapoho and a homozygous selection of line 55‐1 (SunUp) was made to produce a yellow‐fleshed, hemizygous cultivar, later named Rainbow (Manshardt, 1998). The resulting Rainbow cultivar bore pear‐shaped fruit with yellow‐orange flesh as anticipated and together with SunUp was ready to be tested in field trials in Puna in late 1995.
The results of the 1995 field trial were excellent and clearly demonstrated the potential value of transgenic papaya (Fig. 4B) for reclaiming papaya production in Puna (Ferreira et al., 2002; Gonsalves et al., 2004b). Of the non‐transgenic controls plants, 50% showed virus symptoms within 4.5 months after transplanting, and all were infected by 11 months. The growth differences between the transgenic and non‐transgenic trees were remarkable (Fig. 5); transgenic plants grew vigorously, with dark green leaves and full fruit columns, whereas non‐transgenic plants were stunted, with yellow and mosaic leaves and very sparse fruit columns. Fruit production data of Rainbow and SunUp in field tests showed that the yields were at least three times higher than the industry average while maintaining the percentage soluble solids above the minimum requirement for commercial fruits (Ferreira et al., 2002).
Figure 5.

View of PRSV disease progress in the field test in Puna (Hawaii) at 18 and 23 months (top and bottom, respectively). In each view the susceptible Sunrise variety is shown on the left and resistant transgenic Rainbow on the right.
The US governmental agencies involved with deregulation of a transgenic product are the Animal and Plant Health Inspection Services (APHIS), Food and Drug Administration (FDA) and Environmental Protection Agency (EPA). By 1997, approvals from all three agencies were granted and in April 1998, licence agreements from all parties were obtained, allowing the commercial cultivation of papaya 55‐1 or its derivatives in the State of Hawaii. The deregulation of Hawaiian transgenic papaya has been previously reviewed (Gonsalves, 1998; Gonsalves et al., 2007a; Suzuki et al., 2007). At present, transgenic papaya is widely sold in Hawaii and exported to the mainland US and Canada. The transgenic papaya has clearly demonstrated a great impact on controlling the destructive disease caused by PRSV, enabling an increase in non‐transgenic papaya production as well as transgenic papaya production and diversification of papaya cultivars in Hawaii. This is perhaps a good model of engineered resistance in papaya for disease control, and biotechnology and technology transfer to other regions.
Although just over 70% of the acreage grown in Puna is transgenic (Rainbow), a significant amount of the non‐transgenic (susceptible) Kapoho cultivar continues to be grown for sale in Hawaii's most important market in Japan. The use of the transgenic cultivars ensures that a significant amount of non‐transgenic papayas can continue to be produced in areas previously devastated by PRSV.
Potential threat to transgenic resistance
Since its release in 1998, the transgenic papaya has had great socio‐economic impact on the Hawaiian papaya industry (Gonsalves, 1998, 2006; Gonsalves et al., 2007a). This success can largely be attributed to both the stability/durability of the transgenic resistance, and the desirable horticultural and fruit quality attributes of the cultivars, Rainbow and SunUp.
However, because the PRSV population in Hawaii is relatively homogeneous, the papaya industry in Hawaii is in a precarious position should resistance breakdown occur due to the emergence or arrival of divergent PRSV strains.
Greenhouse inoculation studies by Tennant et al. (1994) showed that the resistance of R1 transgenic papaya of line 55‐1 was narrow. R1 plants were resistant to isolates from Hawaii but susceptible to PRSV strains occurring in other countries. Rainbow, which like the 55‐1 R1 plants is hemizygous for the CP transgene, shared a similar narrow resistance, whereas SunUp (homozygous) showed good resistance to isolates from Hawaii as well as to isolates from Jamaica and Brazil (Table 1). These results suggest that doubling transgene dosage will broaden transgenic resistance (Tennant et al., 2001).
Table 1.
Coat protein (CP) nucleotide sequence homologies of PRSV isolates to PRSV HA5‐1 and summary of reactions of different isolates to Rainbow and SunUp papaya.
| PRSV isolates | % Homology to transgene CP | Reaction to isolates | |||||
|---|---|---|---|---|---|---|---|
| N | Core | C | 3′ncr | Overall | Rainbow | SunUp | |
| Hawaii‐HA | 99.3 | 99.8 | 100 | 100 | 99.8 | R | R |
| Hawaii‐OA | 97.3 | 98.0 | 100 | 95.7 | 97.9 | sR | R |
| Hawaii‐KA | 95.3 | 97.1 | 98.3 | 93.6 | 96.7 | sR | R |
| Hawaii‐KE | 95.3 | 97.1 | 98.3 | 93.6 | 96.7 | sR | R |
| Jamaica‐JA | 89.3 | 95.0 | 91.5 | 69.6 | 92.5 | S | R |
| Brazil‐BR | 84.4 | 93.9 | 98.3 | 73.3 | 91.6 | S | R |
| Thailand‐TH | 83.7 | 90.7 | 91.5 | 89.4 | 89.5 | S | sR |
N = 199 nucleotides of the amino terminus, core = 641 nucleotides of the core region, C = 59 nucleotides of the carboxy terminus, and 3′ncr = 35 nucleotides of the non‐coding regions following the stop codon. R = resistant; sR = susceptible at young stages and resistant at older stages; S = susceptible. Table is modified from Tennant et al. (2001).
Additional work with line 63‐1 (Tennant et al., 2005) further confirms the relationship between transgene dosage and broader resistance (Souza et al., 2005). Line 63‐1 is similar to line 55‐1, but had double insertion of the CP gene, and was resistant to a range of isolates of PRSV not only from Hawaii (HA) but also from Jamaica (JA), Thailand (TH) and Brazil (BR) (Souza et al., 2005; Tennant et al., 2005).
Comparison of the CP gene sequences from the various PRSV isolates suggested that resistance is positively correlated with the degree of homology between the CP of the infecting virus and the transgene. PRSV isolates from Hawaii showed 97–100% sequence homology to the transgene CP, whereas the isolates from elsewhere showed 89–93%CP sequence homology to the transgene. Among all isolates of PRSV from outside of Hawaii, the CP gene of the Thailand PRSV had the least homology to the transgene CP (Table 1) and in turn had the most severe symptom expression.
CP expression data from tested transgenic papaya plants were consistently lower in homozygous SunUp than hemizygous Rainbow. Nuclear run‐on experiments were done to determine whether the lower expression levels were due to decreased transcription or PTGS. These results showed clearly and consistently higher transcription in homozygous (SunUp) than in hemizygous (Rainbow) plants. In contrast, Northern assays showed that the steady‐state levels of the transgene transcript in SunUp were lower or equal to levels in Rainbow. In another study by Souza et al. (2005), resistant plants of line 63‐1 had lower CP expression levels than susceptible plants. These results strongly suggest that the mechanism of transgenic resistance is sequence homology‐dependent and RNA‐mediated via PTGS (Tennant et al., 2001). This requirement for transgene homology limits the applicability of transgenic resistance in different geographical regions that may harbour PRSV strains which are molecularly diverse.
Although there appears to be a strong relationship between transgene and challenge virus CP gene homology for resistance to be expressed, other factors also influence the host's response. Experiments on the infectivity of recombinant PRSV HA with different segments of the CP gene and/or its 3′ UTR replaced with the cognate segment from PRSV isolates that can overcome transgenic resistance suggest that position effects and homology within the CP gene are also important (Chiang et al., 2001).
Studies with recombinant PRSV transcripts carrying other heterologous PRSV segments showed that the HC‐Pro gene of the challenge strain plays a role in determining virus pathogenicity and virulence on transgenic papaya. In addition, HC‐Pro acts as a suppressor of the plant's gene silencing defence mechanism, which in turn influences effectiveness of resistance (Bau et al., 2004b; Tripathi et al., 2003, 2004; Yeh et al., 2005).
Other studies have shown that the absence of transgene homology with the challenge virus CP gene does not always correlate with ability of a PRSV strain to breakdown resistance in transgenic papaya (Chen et al., 2002; Ruanjan et al., 2007; Tripathi et al., 2004; Yeh et al., 2003). Agro‐transformation of papaya with the CP gene of a native Taiwanese isolate, PRSV YK (Cheng et al., 1996), showed good resistance in several field trials from 1996 to 1999 (Bau et al., 2003, 2004a). Several lines were highly resistant to the homologous strain (PRSV YK) and also to three different geographical PRSV isolates from Hawaii, Thailand and Mexico (Bau et al., 2003). Interestingly, during the fourth field test in 1999, PRSV infection of this line was observed and the PRSV strain isolated from infected transgenic line was designated PRSV 5‐19 (Chen et al., 2002; Tripathi et al., 2004). Further characterization of the virus revealed that the nucleotide identity between the transcript of the CP transgene and PRSV 5‐19 RNA was less divergent than that between the CP transgene and other geographical strains of PRSV that were not able to overcome the transgenic resistance (Tripathi et al., 2004).
This observation suggests that the breakdown of the transgenic resistance was not correlated with the sequence divergence between the infecting virus and the transgene. To study the potential role of silencing suppressor HC‐Pro in the resistance breakdown, an infectious viral recombinant was constructed by replacing the HC‐Pro region of PRSV YK with that of PRSV 5‐19 and the resistance against the recombinant was evaluated in transgenic papaya lines. These results demonstrated that the heterologous HC‐Pro region of 5‐19 alone was sufficient to breakdown the transgene‐linked resistance, even though the sequences of the transgene transcript shared 100% identity with the genome of the infecting virus (Yeh et al., 2005). This resistance breakdown by the PRSV 5‐19 of PRSV was most probably due to the involvement of the HC‐Pro gene as a silencing suppressor and/or virulence enhancer (Tripathi et al., 2004; Yeh et al., 2005).
The above observations show that resistance in transgenic papaya can be overcome by PRSV with distant homology to the transgene, or by PRSV strains with HC‐Pro that can sufficiently suppress the silencing mechanism of transgenic papaya. It would therefore be important to develop transgenic papaya that could mitigate against the impact of these PRSV strains.
CONCLUSION AND FUTURE PROSPECTS
Papaya ringspot virus belongs to one of the largest and most economically important plant virus groups. Utilizing the concept of CP‐mediated resistance, transgenic papaya cultivars Rainbow and SunUp were developed and released in a timely manner to save Hawaii's papaya industry from damage caused by PRSV. Evidence suggests that transgene conferred PRSV resistance is mediated by RNA via homology‐dependent post‐transcriptional gene silencing or PTGS. While the transgenic resistance in Rainbow and SunUp has proven durable for nearly 10 years in Hawaii, resistance breakdown could occur with the emergence of new viral strains locally or by the introduction of divergent strains from regions where the virus exists, particularly from localities where the suppression of the gene silencing mechanism by HC‐Pro has been demonstrated. This potential for resistance breakdown suggests that it is important to monitor the PRSV population for its diversity and the arrival or emergence of new and more virulent strains in nature. The recent increase in cumulative data on PRSV gene sequences, including analyses on isolate variability and phylogenetic relationships between different geographical isolates will be helpful for devising effective PRSV management strategies such as design for the most effective transgene for specific regions.
In fact, evidence that transgenes could only protect against closely related virus strains stimulated the development of a next generation of transgenics that used a new approach of combining different transgene sequences to produce plants that are resistant to multiple viruses or virus isolates. Broad‐spectrum resistance to diverse strains of PRSV will benefit small‐scale farming worldwide. Advances in our basic knowledge of pathogenicity and infectivity determinants of PRSV, the mechanism of gene silencing and gene silencing suppression in relation to transgenic resistance and its breakdown should also contribute to transgene design and other viral resistance strategies.
However, technology is not the major hurdle in commercializing virus‐resistant transgenic plants for other regions. Although Rainbow and SunUp papaya have had a great socio‐economic impact in Hawaii and the fruit has been consumed in Hawaii, the US mainland and Canada without any reported adverse effects on human health or the environment, other issues remain as major concerns limiting the commercialization of transgenic plants in many countries. In fact, several countries have developed papaya resistant to PRSV, but deregulation and governmental acceptance are the biggest and last remaining challenges limiting its application in the field and real adoption by all farmers.
REFERENCES
- Adams, M.J. , Antoniw, J.F. and Beaudoin, F. (2005) Review: overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 6, 471–487. [DOI] [PubMed] [Google Scholar]
- An, G. (1986) Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol. 81, 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, G. , Ebert, P.R. , Mitra, A. and Ha, S.B. (1988) Binary vectors In: Plant Molecular Biology Manual (Gelvin S. and Schilperoort R.A., eds), pp. 1–19. Boston: Kluwer Academic Publishers. [Google Scholar]
- Bateson, M. , Henderson, J. , Chaleeprom, W. , Gibbs, A. and Dale, J. (1994) Papaya ringspot potyvirus: isolate variability and origin of PRSV type P (Australia). J. Gen. Virol. 75, 3547–3553. [DOI] [PubMed] [Google Scholar]
- Bateson, M.F. , Lines, R.E. , Revill, P. , Chaleeprom, W. , Ha, C.V. , Gibbs, A.J. and Dale, J.L. (2002) On the evolution and molecular epidemiology of the potyvirus Papaya ringspot virus . J. Gen. Virol. 83, 2575–2585. [DOI] [PubMed] [Google Scholar]
- Bau, H.J. , Cheng, Y.H. , Yu, T.A. , Yang, J.S. , Liou, P.C. , Hsiao, C.H. , Lin, C.Y. and Yeh, S.D. (2004a) Field evaluation of transgenic papaya lines carrying the coat protein gene of Papaya ringspot virus in Taiwan. Plant Dis. 85, 594–599. [DOI] [PubMed] [Google Scholar]
- Bau, H.J. , Cheng, Y.H. , Yu, T.A. , Yang, J.S. and Yeh, S.D. (2003) Broad‐spectrum resistance to different geographic strains of Papaya ringspot virus in coat protein gene transgenic papaya. Phytopathology, 93, 112–120. [DOI] [PubMed] [Google Scholar]
- Bau, H.J. , Tripathi, S. , Chen, L.F. , Cheng, Y.H. , Yu, T.A. , Yang, J.S. and Yeh, S.D. (2004b) Transgenic papaya for control of papaya ringspot virus and analysis of viral factors involved in resistance breakdown. In: 10th SCBA International Symposium Vol. W‐21‐04. Beijing, China. [Google Scholar]
- Chen, G. , Ye, C.M. , Huang, J.C. , Yu, M. and Li, B.J. (2001a) Cloning of the papaya ringspot virus (PRSV) replicase gene and generation of PRSV‐resistant papayas through the introduction of the PRSV replicase gene. Plant Cell Rep. 20, 272–277. [Google Scholar]
- Chen, K.C. , Wang, C.H. , Liu, F.L. , Su, W.C. and Yeh, S.D. (2001b) Construction of in vitro infectious clone of a type W strain of Papaya ringspot virus and analysis of host determinant for papaya infection. Plant Pathol. Bull. 10, 215–216. [Google Scholar]
- Chen, K.C. , Wang, C.H. , Liu, F.L. , Su, W.C. and Yeh, S.D. (2003) The NIa gene of Papaya ringspot virus is the host determinant for papaya infection In: 7th International Congress of Plant Molecular Biology. Barcelona, Spain. [Google Scholar]
- Chen, L.F. , Bau, H.J. and Yeh, S.D. (2002) Identification of viruses capable of breaking transgenic resistance of papaya conferred by the coat protein gene of papaya ringspot virus. Acta Hortic. 575, 465–474. [Google Scholar]
- Cheng, Y.H. , Yang, J.S. and Yeh, S.D. (1996) Efficient transformation of papaya by coat protein gene of papaya ringspot virus mediated by Agrobacterium following liquid‐phase wounding of embryogenic tissues with carborundum. Plant Cell Rep. 16, 127–132. [DOI] [PubMed] [Google Scholar]
- Chiang, C.H. , Lee, C.Y. , Wang, C.H , Jan, F.J. , Lin, S.S. , Chen, T.C. , Raja, J. and Yeh, S.D. (2007) Genetic analysis of an attenuated Papaya ringspot virus strain applied for cross‐protection. Eur. J. Plant Pathol. 118, 333–348. [Google Scholar]
- Chiang, C.H. , Wang, J.J. , Jan, F.J. , Yeh, S.D. and Gonsalves, D. (2001) Comparative reactions of recombinant papaya ringspot viruses with chimeric coat protein (CP) genes and wild‐type viruses on CP‐transgenic papaya. J. Gen. Virol. 82, 2827–2836. [DOI] [PubMed] [Google Scholar]
- Chiang, C.H. and Yeh, S.D. (1997) Infectivity assays of in vitro and in vivo transcripts of papaya ringspot potyvirus. Bot. Bull. Acad. Sinica, 38, 153–163. [Google Scholar]
- Chin, M , Ahmad, M.H. and Tennant, P. (2007) Momordica charantia is a weed host reservoir for Papaya ringspot virus type P in Jamaica. Plant Dis. 91, 1518. [DOI] [PubMed] [Google Scholar]
- Davis, M.J. and Ying, Z. (2004) Development of papaya breeding lines with transgenic resistance to Papaya ringspot virus. Plant Dis. 88, 352–358. [DOI] [PubMed] [Google Scholar]
- Fermin, D. and Gonsalves, D. (2003) Papaya: engineering resistance against papaya ringspot virus by native, chimeric and synthetic transgenes In: Virus and Virus‐like Diseases of Major Crops in Developing Countries (Loebenstein G. and Thottappilly G., eds), pp. 497–518. Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Fermin, G. , Tennant, P. , Gonsalves, C. , Lee, D. and Gonsalves, D. (2004) Comparative development and impact of transgenic papayas in Hawaii, Jamaica, and Venezuela In: Transgenic Plants: Methods and Protocols (Pena L., ed.), pp. 399–430. Totowa, NJ: The Human Press Inc. [DOI] [PubMed] [Google Scholar]
- Ferreira, S.A. , Mau, R.F.L. , Manshardt, R. , Pitz, K.Y. and Gonsalves, D. (1992) Field evaluation of papaya ringspot virus cross protection In: Proceedings of the 28th Annual Hawaii Papaya Industry Association Conference, pp. 14–19. Honolulu: USA. [Google Scholar]
- Ferreira, S.A. , Pitz, K.Y. , Manshardt, R. , Zee, F. , Fitch, M. and Gonsalves, D. (2002) Virus coat protein transgenic papaya provides practical control of papaya ringspot virus in Hawaii. Plant Dis. 86, 101–105. [DOI] [PubMed] [Google Scholar]
- Fitch, M.M.M. , Manshardt, R.M. , Gonsalves, D. , Slightom, J.L. and Sanford, J.C. (1990) Stable transformation of papaya via microprojectile bombardment. Plant Cell Rep. 9, 189–194. [DOI] [PubMed] [Google Scholar]
- Fitch, M.M.M. , Manshardt, R.M. , Gonsalves, D. , Slightom, J.L. and Sanford, J.C. (1992) Virus resistant papaya derived from tissues bombarded with the coat protein gene of papaya ringspot virus. Bio/Technology, 10, 1466–1472. [Google Scholar]
- Fuchs, M. and Gonsalves, G. (2007) Safety of virus‐resistant transgenic plants two decades after their introduction: Lesson from realistic field risk assessment studies. Annu. Rev. Phytopathol. 47, 173–202. [DOI] [PubMed] [Google Scholar]
- Gonsalves, D. (1998) Control of papaya ringspot virus in papaya: a case study. Annu. Rev. Phytopathol. 36, 415–437. [DOI] [PubMed] [Google Scholar]
- Gonsalves, D. (2006) Transgenic papaya: development, release, impact, and challenges. Adv. Virus Res. 67, 317–354. [DOI] [PubMed] [Google Scholar]
- Gonsalves, D. and Fermin, G. (2004) The use of transgenic papaya to control papaya ringspot virus in Hawaii and transfer of this technology to other countries In: Handbook of Plant Biotechnology (Christau P. and Klee H., eds), pp. 1165–1182. London, UK: John Wiley & Sons. [Google Scholar]
- Gonsalves, D. and Ferreira, S. (2003) Transgenic papaya: a case for managing risks of papaya ringspot virus in Hawaii. Plant Health Progress doi: 10.1094/PHP-2003-1113-03-RV [DOI] [Google Scholar]
- Gonsalves, D. , Ferreira, S.A. , Suzuki, J.Y. and Tripathi, S. (2007a) Papaya In: A Compendium of Transgenic Crop Plants (Kole C. and Hall T.C., eds). London: Blackwell Publishing; (in press). [Google Scholar]
- Gonsalves, D. and Garnsey, S.M. (1989) Cross protection techniques for control of plant virus diseases in the tropics. Plant Dis. 73, 592–597. [Google Scholar]
- Gonsalves, D. , Gonsalves, C. , Ferreira, S. , Pitz, K. , Fitch, M. , Manshardt, R. and Slightom, J. (2004b) Transgenic virus resistant papaya: from hope to reality for controlling papaya ringspot virus in Hawaii. APSnet Feature Story for July 2004. http://www.apsnet.org/online/feature/ringspot
- Gonsalves, D. and Ishii, M. (1980) Purification and serology of papaya ringspot virus. Phytopathology, 70, 1028–1032. [Google Scholar]
- Gonsalves, C. , Lee, D.R. and Gonsalves, D. (2004a) Transgenic virus‐resistant papaya: the Hawaiian ‘Rainbow’ was rapidly adopted by farmers and is of major importance in Hawaii today. APSnet feature story for August–September 2004. http://www.apsnet.org/online/feature/rainbow.
- Gonsalves, D. , Suzuki, J.Y. , Tripathi, S. and Ferreira, S.A. (2007b) Papaya ringspot virus (Potyviridae) In: Encyclopedia of Virology (Mahy B.W.J. and Van Regenmortel M.H.V., eds). Oxford: Elsevier Ltd; (in press). [Google Scholar]
- Gonsalves, D. , Vegas, A. , Prasartsee, V. , Drew, R. , Suzuki, J.Y. and Tripathi, S. (2006) Developing papaya to control Papaya ringspot virus by transgenic resistance, intergeneric hybridization, and tolerance breeding. In: Plant Breeding Reviews Vol. 26 (Janick J., ed.), pp. 35–73. Hoboken, NJ: John Wiley and Sons, Inc. [Google Scholar]
- Ishii, M. and Holtzmann, O.V. (1963) Papaya mosaic disease in Hawaii. Plant Dis. Rep. 47, 947–951. [Google Scholar]
- Jain, R.K. , Sharma, J. , Sivakumar, A.S. , Sharma, P.K. , Byadgi, A.S. , Verma, A.K. and Varma, A. (2004) Variability in the coat protein gene of Papaya ringspot virus isolates from multiple locations in India. Arch. Virol. 149, 2435–2442. [DOI] [PubMed] [Google Scholar]
- Jensen, D.D. (1949) Papaya virus diseases with special reference to papaya ringspot. Phytopathology, 39, 191–211. [Google Scholar]
- Lee, C.Y. , Chiang, C.H. and Yeh, S.D. (2001) Analyses of the roles of P1 and HC‐Pro genes of Papaya ringspot virus for the attenuated symptoms on papaya plants and local lesion formation on Chenoposium quinoa . Plant Pathol. Bull. 10, 211–212. [Google Scholar]
- Lines, R.E. , Persley, D. , Dale, J.L. , Drew, R. and Bateson, M.F. (2002) Genetically engineered immunity to Papaya ringspot virus in Australian papaya cultivars. Mol. Breeding, 10, 119–129. [Google Scholar]
- Ling, K. , Namba, S. , Gonsalves, C. , Slightom, J.L. and Gonsalves, D. (1991) Protection against detrimental effects of potyvirus infection in transgenic tobacco plants expressing the papaya ringspot virus coat protein gene. Bio/Technology, 9, 752–758. [DOI] [PubMed] [Google Scholar]
- Lius, S. , Manshardt, R.M. , Fitch, M.M.M. , Slightom, J.L. , Sanford, J.C. and Gonsalves, D. (1997) Pathogen‐derived resistance provides papaya with effective protection against papaya ringspot virus. Mol. Breeding, 3, 161–168. [Google Scholar]
- Maia, I.G. , Haenni, A.L. and Bernardi, F. (1996) Potyviral HC‐Pro: a multifunctional protein. J. Gen. Virol. 77, 1335–1341. [DOI] [PubMed] [Google Scholar]
- Manshardt, R.M. (1992) Papaya. In: Biotechnology of Perennial Fruit Crops, Vol. 21 (Hammerschlag F.A. and Litz R.E., eds), pp. 489–511. Wallingford, UK: CAB International. [Google Scholar]
- Manshardt, R.M. (1998) ‘UH Rainbow’ papaya. Univ. Hawaii Coll. Trop. Agric. Hum. Resources , New Plant for Hawaii‐1, 2.
- Manshardt, R.M. and Wenslaff, T.F. (1989) Interspecific hybridization of papaya with other Carica species. J. Am. Soc. Hortic. Sci. 114, 689–694. [Google Scholar]
- Mau, R.F.L. , Gonsalves, D. and Bautista, R. (1989) Use of cross protection to control papaya ringspot virus at Waianae. In: Proc. 25th Annu. Hawaii Papaya Ind. Assoc. Conf ., pp. 77–84. Hawaii, USA. [Google Scholar]
- Parris, G.K. (1938) A new disease of papaya in Hawaii. Proc. Am. Soc. Hortic. Sci. 36, 263–265. [Google Scholar]
- Peng, Y.H. , Kadoury, D. , Gal‐On, A. , Huet, H. , Wang, Y. and Raccah, B. (1998) Mutations in the HC‐Pro gene of zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J. Gen. Virol. 79, 897–904. [DOI] [PubMed] [Google Scholar]
- Pirone, T.P. (1991) Viral genes and gene products that determine insect transmissibility. Semin. Virol. 2, 81–87. [Google Scholar]
- Pirone, T.P. and Blanc, S. (1996) Helper‐dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34, 227–247. [DOI] [PubMed] [Google Scholar]
- Pitz, K. , Ferreira, S. , Mau, R. and Gonsalves, D. (1994) Papaya cross protection: the near‐commercialization experience on Oahu In: Proc. 30th Annu. Hawaii Papaya Ind. Assoc. Conf., pp. 4–6. Maui, USA. [Google Scholar]
- Purcifull, D. , Edwardson, J. , Hiebert, E. and Gonsalves, D. (1984) Papaya ringspot virus. CMI/AAB Descr. Plant Viruses , No. 209. (No. 84 Revis., July 1984), 8.
- Quemada, H.D. , Gonsalves, D. and Slightom, J.L. (1991) Expression of coat protein gene from cucumber mosaic virus strain C in tobacco: protection against infections by CMV strains transmitted mechanically or by aphids. Phytopathology, 81, 794–802. [Google Scholar]
- Quemada, H. , L’Hostis, B. , Gonsalves, D. , Reardon, I.M. , Heinrikson, R. , Hiebert, E.L. , Sieu, L.C. and Slightom, J.L. (1990) The nucleotide sequences of the 3′‐terminal regions of papaya ringspot virus strains w and p. J. Gen. Virol. 71, 203–210. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F.G. , MacFarlane, S.A. and Baulcombe, D.C. (1999) Gene silencing without DNA: RNA‐mediated cross‐protection between viruses. Plant Cell, 11, 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L. , Lain, S. and Garcia, J.A. (1992) Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73, 1–16. [DOI] [PubMed] [Google Scholar]
- Ruanjan, P. , Kertbundit, S. and Juricek, M. (2007) Post‐transcriptional gene silencing is involved in resistance of transgenic papayas to papaya ringspot virus. Biol. Plantarum, 51, 517–520. [Google Scholar]
- Sanford, J.C. and Johnston, S.A. (1985) The concept of parasite‐derived resistance—Deriving resistance genes from the parasite's own genome. J. Theor. Biol. 113, 395–405. [Google Scholar]
- Silva‐Rosales, L. , Becerra‐Leor, N. , Ruiz‐Castro, S. , Teliz‐Ortiz, D. and Noa‐Carrazana, J.C. (2000) Coat protein sequence comparisons of three Mexican isolates of papaya ringspot virus with other geographical isolates reveal a close relationship to American and Australian isolates. Arch. Virol. 145, 835–843. [DOI] [PubMed] [Google Scholar]
- Souza, M.T., Jr , Tennant, P.F. and Gonsalves, D. (2005) Influence of coat protein transgene copy number on resistance in transgenic line 63‐1 against Papaya ringspot virus isolates. HortScience, 40, 2083–2087. [Google Scholar]
- Suzuki, J.Y. , Tripathi, S. and Gonsalves, D. (2007) Virus‐resistant transgenic papaya: commercial development and regulatory and environmental issues In: Biotechnology and Plant Disease Management (Punja Z.K., DeBoer S. and Sanfacon H., eds), pp. 336–361. Willingford, UK: CAB International. [Google Scholar]
- Tennant, P. , Fermin, G. , Fitch, M.M. , Manshardt, R.M. , Slightom, J.L. and Gonsalves, D. (2001) Papaya ringspot virus resistance of transgenic Rainbow and SunUp is affected by gene dosage, plant development, and coat protein homology. Eur. J. Plant Pathol. 107, 645–653. [Google Scholar]
- Tennant, P.F. , Gonsalves, C. , Ling, K.S. , Fitch, M. , Manshardt, R. , Slightom, J.L. and Gonsalves, D. (1994) Differential protection against papaya ringspot virus isolates in coat protein gene transgenic papaya and classically cross‐protected papaya. Phytopathology, 84, 1359–1366. [Google Scholar]
- Tennant, P. , Souza, M.T., Jr, Gonsalves, D. , Fitch, M.M. , Manshardt, R.M. and Slightom, J.L. (2005) Line 63‐1: a new virus‐resistant transgenic papaya. HortScience, 40, 1196–1199. [Google Scholar]
- Tripathi, S. , Bau, H.J. , Chen, L.F. and Yeh, S.D. (2004) The ability of Papaya ringspot virus strains overcoming the transgenic resistance of papaya conferred by the coat protein gene is not correlated with higher degrees of sequence divergence from the transgene. Eur. J. Plant Pathol. 110, 871–882. [Google Scholar]
- Tripathi, S. , Chen, L.F. , Bau, H.J. and Yeh, S.D. (2003) In addition to transgene divergence potyviral HC‐Pro gene plays an important role in breaking down coat protein gene‐mediated transgene resistance. In: 7th International Congress of Plant Molecular Biology , p. 367.
- Tripathi, S. , Suzuki, J. and Gonsalves, D. (2006) Development of genetically engineered resistant papaya for Papaya ringspot virus in a timely manner—A comprehensive and successful approach. In: Plant–Pathogen Interactions: Methods and Protocols Vol. 354 (Ronald P., ed.), pp. 197–240. Totowa, NJ: The Humana Press, Inc. [DOI] [PubMed] [Google Scholar]
- Wang, C.H. and Yeh, S.D. (1997) Divergence and conservation of the genomic RNAs of Taiwan and Hawaii strains of papaya ringspot potyvirus. Arch. Virol. 142, 271–285. [DOI] [PubMed] [Google Scholar]
- Wang, H.L. , Yeh, S.‐D. , Chiu, R.J. and Gonsalves, D. (1987) Effectiveness of cross‐protection by mild mutants of papaya ringspot virus for control of ringspot disease of papaya in Taiwan. Plant Dis. 71, 491–497. [Google Scholar]
- Yeh, S.D. and Gonsalves, D. (1984) Evaluation of induced mutants of papaya ringspot virus for control by cross protection. Phytopathology, 74, 1086–1091. [Google Scholar]
- Yeh, S.D. and Gonsalves, D. (1985) Translation of Papaya ringspot virus RNA in vitro: detection of a possible polyprotein that is processed for capsid protein, cylindrical‐inclusion protein, and amorphous‐inclusion protein. Virology, 143, 260–271. [DOI] [PubMed] [Google Scholar]
- Yeh, S.D. and Gonsalves, D. (1994) Practices and perspective of control of papaya ringspot virus by cross protection. In: Advances in Disease Vector Research Vol. 10 (Harris K.F., ed.), pp. 237–257. New York: Springer. [Google Scholar]
- Yeh, S.D. , Gonsalves, D. and Provvidenti, R. (1984) Comparative studies on host range and serology of papaya ringspot virus and watermelon mosaic virus 1. Phytopathology, 74, 1081–1085. [Google Scholar]
- Yeh, S.D. , Gonsalves, D. , Wang, H.L. , Namba, R. and Chiu, R.J. (1988) Control of papaya ringspot virus by cross protection. Plant Dis. 72, 375–380. [Google Scholar]
- Yeh, S.D. , Jan, F.J. , Chiang, C.H. , Doong, P.J. , Chen, M.C. , Chung, P.H. and Bau, H.J. (1992) Complete nucleotide sequence and genetic organization of papaya ringspot virus RNA. J. Gen. Virol. 73, 2531–2541. [DOI] [PubMed] [Google Scholar]
- Yeh, S.D. , Tripathi, S. , Bau, H.J. and Chen, L.F. (2003) Identification and variability analyses of virus strains capable of breaking transgenic resistance of papaya conferred by the coat protein gene of Papaya ringspot virus. In: Abstracts 7th International Congress of Plant Molecular Biology , p. 367 Barcelona, Spain. [Google Scholar]
- Yeh, S.D. , You, B.J. , Tripathi, S. , Chen, L.F. and Bau, H.J. (2005) Transgenic resistance overcome by potyviral Hc‐Pro gene in a transgene sequence‐homology independent manner In: Microbes in Changing World. San Francisco, USA: International Union of Microbiological Societies. [Google Scholar]
- You, B.J. , Chiang, C.H. , Chen, L.F. , Su, W.C. and Yeh, S.D. (2005) Engineered mild strain of Papaya ringspot virus for broader cross protection in cucurbits. Phytopathology, 95, 533–540. [DOI] [PubMed] [Google Scholar]


