SUMMARY
Arbuscular mycorrhizal (AM) symbiosis occurs between fungi of the phylum Glomeromycota and most terrestrial plants. However, little is known about the molecular symbiotic signalling between AM fungi (AMFs) and non‐leguminous plant species. We sought to further elucidate the molecular events occurring in tomato, a non‐leguminous host plant, during the early, pre‐symbiotic stage of AM symbiosis, i.e. immediately before and after contact between the AMF (Glomus intraradices) and the host. We adopted a semi‐synchronized AMF root infection protocol, followed by genomic‐scale, microarray‐based, gene expression profiling at several defined time points during pre‐symbiotic AM stages. The microarray results suggested differences in the number of differentially expressed genes and in the differential regulation of several functional groups of genes at the different time points examined. The microarray results were validated and one of the genes induced during contact between AMF and tomato, the expansin‐like EXLB1, was functionally analysed. Expansins, encoded by a large multigene family, facilitate plant cell expansion. However, no biological or biochemical function has yet been established for plant‐originated expansin‐like proteins. EXLB1 transcripts were localized early during the association to cells that may perceive the fungal signal, and later during the association in close proximity to sites of AMF hypha–root colonization. Moreover, in transgenic roots, we demonstrated that a reduction in the steady‐state level of EXLB1 transcript was correlated with a reduced rate of infection, reduced arbuscule expansion and reduced AMF spore formation.
INTRODUCTION
Arbuscular mycorrhizal (AM) symbiosis occurs between fungi of the phylum Glomeromycota (Schüssler et al., 2001) and most terrestrial plants (Gianinazzi‐Pearson, 1996). These fungi are found in virtually all major ecosystems, forming a mutualistic association with most vascular plants. This mutualism is a bidirectional event: the fungus supplies phosphate and other mineral nutrients required by the plant from inaccessible portions of the rhizosphere and, in return, obtains organic carbon fixed by the photosynthesizing plant (Smith and Read, 1997). AM symbiosis is usually most beneficial to the plant host; it serves to increase its supply of mineral nutrients, especially under nutrient‐limiting conditions, improves rooting and plant establishment, improves the uptake of ions with low mobility and accelerates budding and flowering (Smith and Read, 1997).
Numerous studies have examined the biological, chemical and molecular characteristics of AM symbiosis, and these have been discussed in a number of recent reviews (for example, Balestrini and Lanfranco, 2006; Bucher, 2007; Franken et al., 2007; Javot et al., 2007; Krajinski and Frenzel, 2007; Liu et al., 2007; Maeda et al., 2006; Paszkowski, 2006; Reinhardt, 2007). These and other studies have suggested the existence of molecular ‘symbiotic signalling’ between the AM fungus (AMF) and the host plant, long before the first colonization structures appear on the root epidermis (reviewed by Paszkowski, 2006). This ‘pre‐contact’ molecular dialogue is orchestrated by a set of chemicals, which includes secondary metabolites, flavonoids and isoflavonoids (Harrison, 2005; Vierheilig and Piché, 2002). Recently, strigolactones, a sesquiterpene lactone, have been suggested to be the active factors triggering AMF branching (Akiyama et al., 2005).
In parallel, as part of AM symbiotic signalling, AMFs release mobile, diffusible signals which are perceived by the plant's genetic machinery, triggering the activation of signal transduction pathways (Kosuta et al., 2003; Olah et al., 2005). Although their exact chemical identity is unknown, empirical evidence strongly suggests that these signalling moieties possess functional properties similar to rhizobial Nod factors (Navazio et al., 2007). Once the fungal hypha comes into contact with a host root, an appressorium (i.e. a contact structure) is formed by the fungus, and a pre‐penetration apparatus (PPA) is formed by the host plant (Genre et al., 2005; Siciliano et al., 2007).
Genetic studies and the genomic‐based identification of genes involved in the host plant response to mycorrhization have focused mainly on the leguminous model plant species Medicago truncatula (for example, Barker et al., 1990; Brechenmacher et al., 2004; Cook, 1999; reviewed by Krajinski and Frenzel, 2007) and Lotus japonicus (for example, Akiyama et al., 2005; Demchenko et al., 2004; Handberg and Stougaard, 1992; Kistner et al., 2005; Küster et al., 2007; Maeda et al., 2006). The choice of these two legumes makes sense, as a large volume of information is already available on well‐developed rhizobial genetics to which AM symbiosis shows a high degree of similarity (Borisov et al., 2007; Gherbi et al., 2008; Hirsch and Kapulnik, 1998; Kistner et al., 2005; Limpens et al., 2003; van Rhijn et al., 1997). As a result, although most terrestrial plants are non‐leguminous, much less is known about the AM association with them. Because of this lack of information, tomato (Solanum lycopersicum L.; for example, Balestrini et al., 2007; Barker et al., 1998; 2001, 2003; Herrera‐Medina et al., 2007; Nagy et al., 2005), maize (Zea mays L.; for example, Xia et al., 2007; reviewed by Paszkowski, 2006) and rice (Oryza sativa L.; for example, Gao et al., 2007; Glassop et al., 2007; Güimil et al., 2005; Paszkowski et al., 2002; reviewed by Paszkowski, 2006) are now being worked on by various research groups as model species.
We sought to further elucidate the molecular events occurring in tomato, a non‐leguminous host plant, during its interaction with the model AMF Glomus intraradices. We focused on the early stages of AM symbiosis—the phases pre‐ and post‐AMF‐host contact—about which little is known. During these stages, even before contact is formed, a pivotal role may exist for ‘symbiotic signalling’ between the host and the fungus, which may affect the gene expression of the plant host (reviewed by Gianinazzi‐Pearson et al., 2007; Koltai et al., 2009).
One of the obstacles in studying AM symbiosis, especially for gene expression profiling, is the difficulty in synchronizing the initial developmental events leading to the establishment of the symbiosis; this synchronization is important for the accurate elucidation of the physiological and genetic events during the early stages of the interaction (Weidmann et al., 2004). Hence, we first semi‐synchronized AMF root infection, and semi‐synchronized time points were selected for genomic‐scale, microarray‐based, gene expression profiling. The microarray results were validated and one of the genes induced during contact between AMF and tomato, the expansin‐like EXLB1, was taken for functional tests.
Expansins are encoded by a large multigene family (Chen et al., 2001 and references therein; Cosgrove, 1999), and are considered to be plant cell wall‐loosening factors (McQueen‐Mason et al., 1992). By disrupting the hydrogen bonds between cellulose and hemicellulose polymers in the plant cell wall (McQueen‐Mason and Cosgrove, 1995), they facilitate cell expansion in vivo (Cosgrove, 1999); cell expansion, in turn, is one of the processes controlling cell growth. In plants, four expansin protein families have been recognized: EXPA, EXPB, EXLA and EXLB. Multiple family members of EXPA and EXPB have been shown to induce rapid extension or stress relaxation of plant cells in vitro (Kende et al., 2004). The two expansin‐like families, EXLA (EXPANSIN‐LIKE A) and EXLB (EXPANSIN‐LIKE B), possess expansin domains, but their amino acid sequences are divergent from EXPA and EXPB. No biological or biochemical function has yet been established for any member of the plant EXLA or EXLB families (Kende et al., 2004; Sampedro and Cosgrove, 2005). However, a nematode‐originated expansin‐like protein has been found to be synthesized and excreted by cyst‐nematodes during plant infection and, importantly, to be active in plant cell wall loosening (Qin et al., 2004). In addition, an expansin‐like protein has been suggested to be involved in AMF‐induced cell wall expansion, and has been shown to be an early host marker for successful mycorrhization (Siciliano et al., 2007).
Plant expansins have been shown to be involved in several types of plant–microbe interaction, including those with nematodes (Gal et al., 2006; Wieczorek et al., 2006) and with nitrogen‐fixing rhizobia (Giordano and Hirsch, 2004). Recently, expansins have been localized to cucumber root cells inoculated with AMF (Balestrini et al., 2005); however, no functional tests were performed to demonstrate the involvement of expansin in the AM symbiosis.
In this study, partial transcriptional silencing of EXLB was correlated with a reduction in AMF spore formation in transgenic roots. These results suggest a role for plant expansins in the early stages of the AMF–host interaction, by affecting the ability of the fungus to proliferate.
RESULTS
Verification and determination of G. intraradices infection rate on tomato seedlings
Following the synchronization of fungal penetration and colonization in tomato roots, a time‐course experiment was carried out to determine the level of synchronization; synchronization was performed using the ‘infection bio‐indicator’ (IBI) process (detailed in Experimental procedures). The time points chosen for further analysis are illustrated in Fig. 1. Root samples were evaluated daily and microscopic observation of the trypan blue‐stained root systems showed no signs of fungal contact in the tomato root system within the first 24 and 48 h post‐inoculation (hpi). The first sign of fungal contact was seen on root systems sampled at 72 hpi: 0.15 ± 0.04 entry points/cm length were counted from five individual replicate plants (20 root segments were examined from each individual plant). At 96 hpi, an increment in entry points was observed: 0.52 ± 0.07 entry points/cm were counted for each of the five replicates (20 root segments examined per plant). The control treatment did not show any signs of infection at any of the time points examined.
Figure 1.
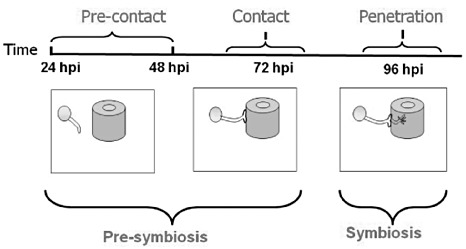
Illustration of time points examined during arbuscular mycorrhizal fungus (AMF)–tomato interaction. Spores are represented by circles, roots by cylinders. hpi, hours post‐inoculation.
Differential expression of genes during symbiosis of G. intraradices with tomato
Microarray experiments were performed to profile tomato gene expression during G. intraradices symbiosis. Differentially expressed genes (expressed above 1.33‐ or below 0.75‐fold in inoculated vs. non‐inoculated roots) were identified during the course of the pre‐ and post‐contact phase of G. intraradices with the roots (Supplementary files S1 and S2, see Supporting Information).
At 24 hpi, only 11 genes were found to be altered in their expression (Fig. 2) relative to the other time points. At 48 hpi, 89 genes showed altered expression (Fig. 2) relative to 24 hpi. These results suggest that plant genes are induced, or repressed, even before contact is established between the fungus and the host.
Figure 2.
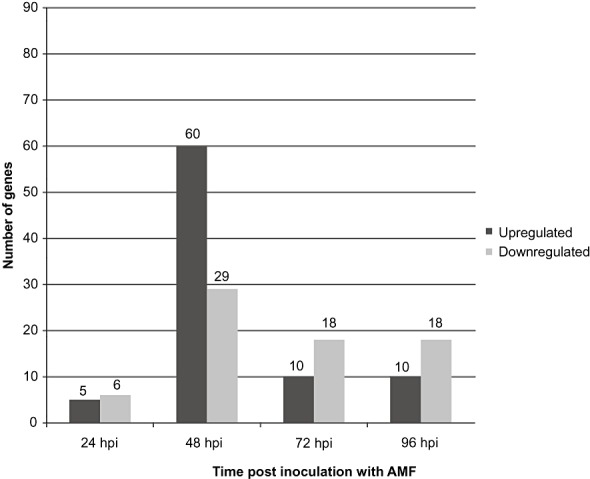
Total number of up‐ and down‐regulated genes as determined from microarray experiments at each time point examined during arbuscular mycorrhizal fungus (AMF)–tomato interaction. hpi, hours post‐inoculation. The number of genes is presented above each column.
At 72 hpi, contact is established between the fungus and the host, and appressoria are formed. At 96 hpi, the fungus penetrates, and fungal structures are formed inside the tomato roots. During each of these time points, microarray analysis suggested that only 28 genes exhibited altered expression (Fig. 2).
Thirteen microarray‐derived results of gene expression at selected time points (Table 1) were validated by quantitative real‐time polymerase chain reaction (qPCR). The qPCR results for these genes demonstrated a pattern of expression that tended to agree with that of the microarray analysis (Table 1), validating the microarray data.
Table 1.
Results of quantitative polymerase chain reaction (qPCR) and microarray analyses.
| SGN accession number/time (hpi) of sample taken | Gene annotation and biological function based on family build | Fold change by qPCR | Fold change by microarray data |
|---|---|---|---|
| 24 hpi | |||
| SGN‐U148923 | Cupin 1; Molecular Function: nutrient reservoir activity (GO:0045735); Cupin, RmlC‐type | 3.03* | 2.91 |
| 48 hpi | |||
| SGN‐U144150 | 4‐Hydroxyphenylpyruvate dioxygenase; Molecular Function: 4‐hydroxyphenylpyruvate dioxygenase activity (GO:0003868); Biological Process: aromatic amino acid family metabolism (GO:0009072); glyoxalase/bleomycin resistance protein/dioxygenase | 1.88* | 1.79 |
| SGN‐U144012 | Expansin 45, endoglucanase‐like; Barwin‐related endoglucanase; rare lipoprotein A (EXLB1 homologue) | 2.02* | 2.58 |
| SGN‐U148120 | Proteinase inhibitor I3, Kunitz legume; Molecular Function: endopeptidase inhibitor activity (GO:0004866); Kunitz inhibitor ST1‐like | 2.04* | 2.47 |
| SGN‐U144291 | Late embryogenesis abundant protein 3; Biological Process: response to stress (GO:0006950) | 1.25 | 1.20 |
| 72 hpi | |||
| SGN‐U148923 | Cupin 1; Molecular Function: nutrient reservoir activity (GO:0045735); Cupin, RmlC‐type | 11.19* | 8.20 |
| SGN‐U144536 | Fatty acid desaturase subdomain; Biological Process: fatty acid desaturation (GO:0006636); Cellular Component: membrane (GO:0016020); Molecular Function: oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water (GO:0016717); Fatty acid desaturase; Molecular Function: oxidoreductase activity (GO:0016491); Fatty acid acyl‐CoA desaturase; Biological Process: fatty acid biosynthesis (GO:0006633); Molecular Function: oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen (GO:0016705) | 1.82* | 2.05 |
| SGN‐U146006 | Heavy metal transport/detoxification protein; Biological Process: metal ion transport (GO:0030001); Molecular Function: metal ion binding (GO:0046872) | 1.67* | 2.30 |
| SGN‐U143331 | Serine‐type endopeptidase inhibitor activity (GO:0004867) | 2.86* | 8.53 |
| 96 hpi | |||
| SGN‐U148923 | Cupin 1; Molecular Function: nutrient reservoir activity (GO:0045735); Cupin, RmlC‐type | 2.78* | 3.86 |
| SGN‐U143331 | Proteinase inhibitor type II CEVI57 precursor | 1.46 | 1.11 |
| SGN‐U152045 | Cytochrome P450; Molecular Function: monooxygenase activity (GO:0004497), Molecular Function: iron ion binding (GO:0005506); Biological Process: electron transport (GO:0006118); Molecular Function: haem binding (GO:0020037) | 2.35* | 1.90 |
| SGN‐U144410 | Early light‐inducible protein, chlorophyll a/b‐binding family protein/early light‐induced protein (ELIP) | 0.57 | 0.50 |
Values of fold change represent expression fold change of genes in plants inoculated with mycorrhiza vs. non‐inoculated controls. SGN gene annotation is based on Tomato 200607 #1 build. hpi, hours post‐inoculation.
Values were found to be significant at a one‐tailed t‐test (P≤ 0.05).
Genes that were significantly regulated at the different examined time points during the pre‐ and post‐contact phases of G. intraradices with the roots were classified on the basis of biological function gene ontology. Classification identified several groups of up‐ and down‐regulated genes. Genes within the different gene groups and their relative expression values are listed in Supplementary file S3 (see Supporting Information) and presented in Fig. 3. Among the most abundant functional groups were those containing genes involved in molecular transport and transcription regulation, stress response, protein degradation and nucleic acid metabolism.
Figure 3.
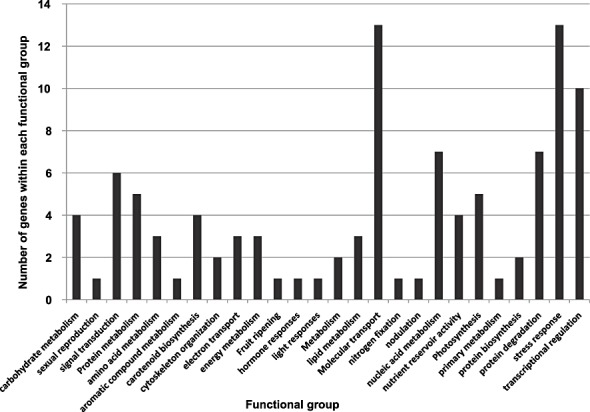
Functional groups and number of genes that were up‐ and down‐regulated within each functional group. Genes were determined from microarray experiments at all time points examined during the arbuscular mycorrhizal fungus (AMF)–tomato interaction.
Several genes were differentially expressed at more than one time point. These included SGN‐U144362 (expressed protein), which was up‐regulated at 48 hpi and down‐regulated at 72 hpi; SGN‐U145445 (unknown protein), which was down‐regulated at 48 and 96 hpi; SGN‐U146006 (heavy metal‐associated, domain‐containing protein), which was up‐regulated at 24, 48 and 72 hpi; SGN‐U148923 (germin‐like protein), which was up‐regulated at 24, 48, 72 and 96 hpi; and SGN‐U155544 (ribulose bisphosphate carboxylase small chain 2A), which was up‐regulated at 24 and 96 hpi.
Functional assessment of expansin association during infection of G. intraradices on tomato
One of the genes that was up‐regulated during the early events of G. intraradices infection, at 48 hpi, was an expansin‐like gene (EXLB1, SGN accession no. SGN‐U144012; Table 1 and Supplementary files S2 and S3). Although expansins have previously been shown to be involved in AMF infection in cucumber cells (Balestrini et al., 2005; Siciliano et al., 2007), to our knowledge, no functional tests have been performed demonstrating the involvement of expansin in the infection process. Moreover, no role has been demonstrated for any plant expansin‐like gene. To further characterize EXLB1 transcription, we quantified EXLB1 transcription during the various early time points of the AMF–tomato interaction. EXLB1 transcription was found to be induced to some extent at 24, 48 and 72 hpi, whereas, during the later time points (i.e. 96 hpi), EXLB1 transcription was reduced in AMF‐inoculated vs. non‐inoculated plants (Fig. 4).
Figure 4.
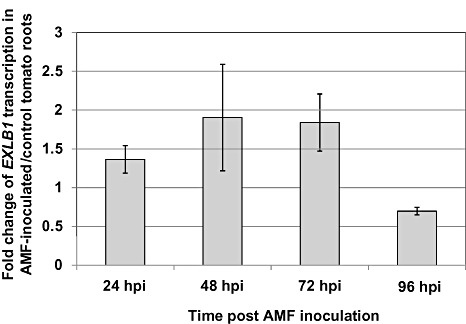
Relative transcription of EXLB1 determined by quantitative polymerase chain reaction in tomato roots inoculated with arbuscular mycorrhizal fungus (AMF) vs. non‐inoculated control. hpi, hours post‐inoculation.
Second, in situ localization of EXLB1 transcripts was performed at several time points early during the interaction (24–96 hpi), and later following mycorrhizal penetration and establishment [4 weeks post‐inoculation (wpi)].
At 96 hpi, EXLB1 transcripts could only be localized to the epidermal cells of the differentiated parts of young roots. No signal could be detected at the earlier time points (not shown) or in non‐infected controls (Fig. 5). It should be noted that the discrepancy between qPCR and in situ results of EXLB1 transcription may be caused by two factors: (i) incomplete synchronization, which results in the accumulation of events; for example, assessment of the 48‐hpi time point also examines the 0‐ and 24‐hpi time points (but not those at 72 or 96 hpi); full synchronization of the mycorrhizal process currently cannot be achieved; hence, both qPCR and in situ analyses detect, at each time point, mycorrhizal‐associated events that occur at that time point and beforehand; (ii) the qPCR method is highly sensitive and averages all events, whereas in situ localization examines individual events; to summarize, the local signal obtained in the in situ experiments may reflect events that occur at 48 or 72 hpi, rather than at 96 hpi.
Figure 5.
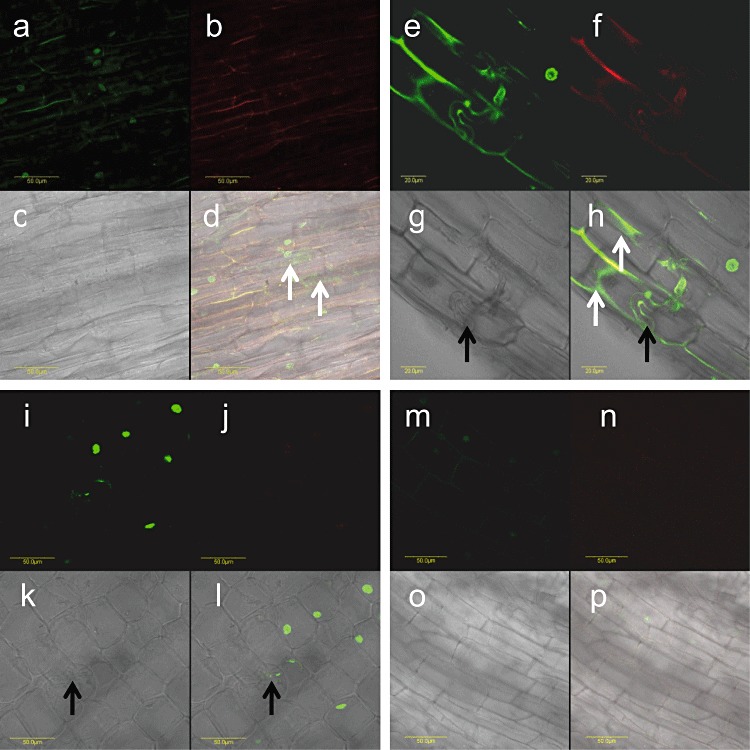
In situ localization of EXLB1 transcripts in longitudinal sections of tomato roots inoculated with arbuscular mycorrhizal fungi (AMF). (a–d, e–h) EXLB1 transcript localization in AMF‐inoculated root tissue at 96 h post‐inoculation (hpi) (a–d) and 4 weeks post‐inoculation (wpi) (e–h). (i–l) AMF‐infected control in which no primers were added to the polymerase chain reaction. (m–p) AMF non‐infected control (with primers). (a, e, i, m) Detection of signal. (b, f, j, n) Detection of autofluorescence. (c, g, k, o) Bright field of the section. (d, h, l, p) Overlay of signal detection, autofluorescence detection and bright field. Green signal in cell cytoplasm indicates EXLB1 transcripts (marked by white arrows); red and yellow signals represent autofluorescence; green signal in nuclei (both plant and fungus cells) represents non‐specific genomic DNA amplification. Black arrows indicate fungal hyphae. Bars represent 50 µm (a–d, i–p); bars represent 20 µm (e–h).
At 4 wpi, the results suggest that EXLB1 transcripts are localized to root cortical cells, which are in close proximity to the colonizing fungal hyphae (Fig. 5). No signal was detected in root parts distant from the AMF infection site (not shown). These results suggest a local induction of EXLB1 transcription on AMF–tomato interaction. qPCR results confirmed the in situ observation: EXLB1 expression was induced 1.7‐fold (±0.2) in AMF‐infected roots relative to non‐infected roots at 4 wpi.
Next, we further elucidated the role of EXLB1 during AM symbiosis by performing functional tests. Transgenic roots with an expansin antisense construct have been shown previously to contain less expansin transcript (Gal et al., 2006). The steady‐state level of EXLB1 transcript was shown by qPCR to be significantly reduced in these transgenic expansin antisense roots, when compared with transgenic control roots expressing green fluorescent protein (GFP) (Fig. 6a). Hence, to further elucidate the role of EXLB1 during AM symbiosis, we examined the symbiotic structures generated on transgenic expansin antisense roots and GFP controls. The symbiotic structures (e.g. arbuscules) were present in both expansin antisense roots and GFP controls (Fig. 6b). However, some arbuscules in expansin antisense roots seemed to be smaller and not fully expanded, whereas, in the GFP control, arbuscules were larger and better expanded within the root cells. Moreover, the rate of infection was reduced in expansin antisense roots by more than twofold in comparison with that in GFP controls (13.3 ± 1.7% and 31.7 ± 13.6%, respectively).
Figure 6.
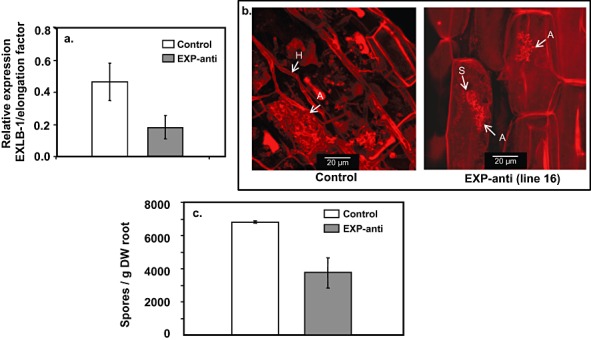
(a) Relative expression of EXLB1 vs. elongation factor in transgenic tomato roots inoculated with arbuscular mycorrhizal fungus (AMF). Transgenic roots contain either expansin antisense (EXP‐anti) or green fluorescent protein (GFP) (control) constructs. All 14 generated transgenic lines were analysed. Average and standard deviations are presented for the eight lines containing EXP‐anti and for the six lines containing GFP (control) constructs. (b) Examples of stained AMF symbiotic structures. Hyphae (H), spores (S) and arbuscules (A) are marked. (c) Number of spores per gram dry weight (DW) of transgenic roots inoculated with AMF. Transgenic roots contain either EXP‐anti or GFP (control) constructs. All 14 generated transgenic lines were analysed. Average and standard deviations are presented for the eight lines containing EXP‐anti and for the six lines containing GFP (control) constructs.
In addition, the spore production of AMF was measured to reflect the success of the symbiotic process on the transgenic expansin antisense roots. Fewer spores were produced by G. intraradices inoculated on expansin antisense roots compared with control G. intraradices infection of GFP‐transformed roots (Fig. 6c). Hence, a reduction in expansin expression may repress, to some extent, G. intraradices spore production.
DISCUSSION
The association between AMFs and their hosts has been shown to occur before contact is established, and even before any morphological differences are apparent within the host or fungus. In this study, we sought to elucidate some of the molecular events occurring in the host immediately pre‐ and post‐contact with the fungus. We first performed a genomic‐scale screening for tomato genes whose expression was altered as a result of the symbiotic association. We then performed an in‐depth study, consisting of functional tests, for an expansin‐like gene that was found to be induced on contact between the host and fungus.
Global profiling of host gene expression during the AM association suggested that, as the pre‐contact phase of the symbiotic association progressed, the number of differentially expressed genes increased, when compared with non‐inoculated plants. Hence, plant genes are induced, or repressed, even before contact is established between the fungus and the host. However, shortly after contact is established, once the appressorium is formed and the fungus penetrates, fewer genes are differentially expressed. Hence, it may be that fewer genes need to be expressed during this contact stage as a result of the plant's ‘readiness’ for the fungus following gene expression during previous stages of the AM association. Alternatively, gene expression may be repressed during this stage of the association, perhaps in order to repress defence‐ and stress‐associated host responses.
The ontology of biologically functional groups and the annotation of up‐ and down‐regulated genes at 24, 48, 72 and 96 hpi suggested that several functional groups were differentially expressed during the early time points of the AMF–plant interaction. These groups may provide some insight into the physiological and metabolic processes triggered by the presence of AMF in plants. However, these insights require additional experimental investigation and are mentioned only briefly. Interestingly, few genes were differentially expressed at more than one time point. This suggests that, at each of the examined time points, a different set of genes takes a leading role, facilitating the notion of a dynamic and developing process of AMF–tomato interaction during the early time points of recognition and contact.
Among the genes induced in our experiments during the early time points were those associated with signal transduction and, in particular, a receptor kinase. Two key players of AMF–plant interactions have been suggested to be the receptor‐like kinase DMI2 and the calcium‐ and calmodulin‐dependent protein kinase DMI3, the former acting upstream and the latter downstream of the calcium spike associated with early AMF infection of plants (2002, 2004; Balestrini and Lanfranco, 2006; Endre et al., 2002; Genre et al., 2005; Geurts et al., 2005; Lévy et al., 2004; Messinese et al., 2007; Oldroyd et al., 2005). Although no homology was found between the protein kinase identified in this study and DMI 2 or DMI3, it may be that the differentially expressed genes associated with signal transduction in our system, and especially the receptor kinase, are involved in signal transduction early on in the AMF–tomato association.
One of the functional groups which included many of the differentially expressed genes was molecular transport. Molecular transport‐associated genes include those that putatively encode proteins involved in sugar, lipid or ion transport. The elaborate molecular transport taking place between plant hosts and fungi has been well characterized (for example, Hause and Fester, 2005). Such activity during the early stages of a plant–fungus association probably reflects the metabolic activity of the plant, which is induced or repressed on early encounter with the fungus. Interestingly, in a recently published paper by Guether et al. (2009), which examined the response of Lotus japonicus to the AMF Gigaspora margarita 4 and 28 days post‐infection, several nutrient transporters were identified. In our system of G. intraradices–tomato, only one potassium transporter, potassium channel TORK1, was identified as being differentially expressed; phosphate transporters, ammonium and amino acid transporters were not differentially expressed. Our results are probably dependent on the fact that we considered early stages of the interaction and, during these time points, nutrients are probably not yet being exchanged between host and fungus, leading to a different result from that of Guether et al. (2009) in which a later stage was considered.
Another functional group that included many of the differentially expressed genes was the stress‐responsive group. Among the stress‐responsive genes were several protease inhibitors. These were found to be up‐regulated, whereas several proteases were found to be down‐regulated, suggesting a reduction in protein degradation during the early AMF–plant association (including during appressorium formation). The finding of the induction of protease inhibitors is in agreement with the results of Guether et al. (2009). However, our results are in contrast with the findings of Roussel et al. (2001), which suggested that serine protease is induced in pea roots during appressorium formation, 6 days post‐inoculation (dpi). Interestingly, proteases have been suggested to be induced in a broad range of mycorrhiza‐inoculated plants. Hence, they have been suggested to play a role in symbiotic processes, especially on elongation of internal hyphae and vesicle formation (Takeda et al., 2007). It may be that, during the early time points examined in the present work, i.e. pre‐contact and during appressorium formation and penetration, the inhibition of protein degradation, rather than protein induction, is needed for successful fungal colonization. As plant proteases have been suggested to digest not only plant proteins but also microbe proteins (Takeda et al., 2007), repression of protein degradation may prevent the degradation of fungal proteins before and on contact with the plant host.
One nodulation‐related gene and two germin‐like genes were induced in our system. Both nodulation genes and germins (i.e. oxalate oxidases and homopentameric glycoproteins; Lane et al., 1993) are known to be associated with rhizobium symbiosis, and have been identified previously in AMF–M. truncatula associations, before contact is established between the plant and the fungus, and during appressorium and PPA formation (Brechenmacher et al., 2004; Dean et al., 1999; Genre et al., 2005; Gucciardo et al., 2007; Kosuta et al., 2003; Massoumou et al., 2007). Notably, one of the germin‐like genes was induced during all examined time points in our system, further suggesting an important role for this gene in AMF–tomato associations.
Multiple genes and several proteins were identified by Siciliano et al. (2007) and Amiour et al. (2006) in association with induced expression in Medicago during PPA formation. These studies were performed in wild‐type Medicago and in a mycorrhiza‐defective Medicago mutant that does not support the formation of a PPA (Amiour et al., 2006; Genre et al., 2005). The work by Siciliano et al. (2007) identified 107 M. truncatula genes up‐regulated during early contact with Gigaspora margarita, 15 of which were confirmed to be induced during the interaction and were involved in signal transduction, defence responses and cell wall modifications. Functional groups that include signal transduction and cell wall modification genes were also identified in our experimental tomato–AMF system. Special attention should be given to the identification of an expansin‐like gene by Siciliano et al. (2007), which is discussed further below.
One of the genes induced during pre‐contact between fungus and host in our system was the expansin‐like gene EXLB1: its induction was identified most markedly at 48 and 72 hpi. Expansins have been found to be induced in M. truncatula roots 5 days after inoculation with Glomus mosseae (Weidmann et al., 2004), and in M. truncatula roots inoculated with Glomus versiforme 10, 17, 22, 31 and 38 dpi (Liu et al., 2004). Expansins have been localized to mycorrhiza‐colonized cucumber root cells (Balestrini et al., 2005), but no functional tests were performed to demonstrate the involvement of expansin in AMF symbiosis.
A biological role has been demonstrated for an expansin‐like gene from a plant‐parasitic nematode during the pathogenic nematode–plant interaction process (Qin et al., 2004). Although this expansin‐like gene is of nematode origin, it has been suggested to play a role in the nematode–plant interaction (Qin et al., 2004). Therefore, expansin‐like proteins may be a common component in the establishment of both parasitism and symbiosis. Moreover, an expansin‐like gene was found to be significantly up‐regulated in the early phase of infection by AMF (Siciliano et al., 2007). Its transcripts were localized to wild‐type M. truncatula epidermal cells during Gigaspora margarita appressorium development (Siciliano et al., 2007). The expansin‐like protein was suggested to be involved in cell wall expansion, and was shown to be an early host marker for successful mycorrhization (Siciliano et al., 2007).
Therefore, we chose the expansin‐like protein EXLB1 for further analysis of its involvement in AM symbiosis. It may be that, early on in the association, before any physical contact is made between the fungus and host, host root epidermal cells perceive a signal, perhaps originating from the fungus, and initiate a cellular response that includes the induction of EXLB1 transcription. This assumption is based on the following results: at an early time point of the AMF–tomato association, at 96 hpi, EXLB1 transcription was localized to epidermal cells of differentiated parts of young roots. During the later stages of the AMF–tomato interaction, at 4 wpi, the induction of the EXLB1 transcript was localized only to root cortical cells that were in close proximity to the invading fungal hyphae. Hence, transcription of plant EXLB1 is induced locally, rather than systemically. During the later stages of fungal colonization, once the symbiosis is well established, the induction of EXLB1 transcription may still be needed locally for successful AMF colonization.
To further examine the requirement of EXLB1 expression for AM symbiosis, the symbiosis was evaluated in transgenic roots in which the steady‐state transcript level of this gene is reduced. In these roots, a reduced rate of infection, reduced arbuscule expansion and reduced AMF spore formation were recorded in comparison with controls. Hence, on reduction of expansin expression, including that of EXLB1, the fungus seems to be inhibited in the utilization of its full reproductive potential. We cannot exclude, however, the possibility that the reduced expression of another expansin (such as Le‐ExpA‐5; Gal et al., 2006) led to the observed phenotype of inhibited spore formation. Nevertheless, no expansin other than EXLB1 was found to be induced in our system during the AMF–tomato interaction.
What is the mechanism that associates cell wall expansion with fungus development and its ability to complete its life cycle? A reduction in the level of EXLB1 expression results in a reduced level of mycorrhizal infection and restricted symbiotic structures, and hence in a reduced level of spore formation (as detected in the transgenic lines). Perhaps these extracellular proteins are needed to loosen the cell wall during both the early events of AMF–plant penetration and during later stages of development of arbuscules and symbiotic association (Balestrini et al., 2005).
Moreover, it has been suggested that expansin‐like genes may be involved in the construction of the interface surrounding the infection hyphae (Balestrini and Lanfranco, 2006; Siciliano et al., 2007). The early induction of EXLB1 found in our study further supports the suggestion (Balestrini and Lanfranco, 2006; Siciliano et al., 2007) of host ‘readiness’ for its symbiont, perhaps even before contact is established.
To summarize, our study and those of others have established expansin‐like proteins as an important component of AM symbiosis, as part of the symbiotic signalling taking place between host and fungus during pre‐symbiosis and symbiosis. The recruitment of plant expansin‐like proteins is an important factor for successful symbiosis, one that allows the utilization of the full reproductive potential of a fungus, suggesting the host's ability to control fungal penetration. Alternatively, this recruitment of plant expansin‐like proteins may reflect the ability of the fungus to redirect the host's genetic programming in order to establish a fungal interface. Moreover, as expansin‐like proteins have a role in the pathogenic process of plant‐parasitic nematodes, these proteins may be a common component in host colonization for the establishment of either beneficial symbiosis or harmful parasitism. As the expansin‐like gene was induced even before contact was established between the fungus and the host, its induction may be a result of the mobile, diffusible signal suggested to be secreted by the fungus and perceived by the plant, redirecting gene expression in the host plant.
EXPERIMENTAL PROCEDURES
Plant and fungal material
Surface‐sterilized Solanum lycopersicum var. VF36 seeds were allowed to germinate on agar medium containing 10 g Difco agar and 10 g sucrose in 1 L of distilled water. Following 48 h of incubation at room temperature, the seedlings were ready to be planted. Glomus intraradices (isolate BEG 141) inoculum was produced as described previously (David‐Schwartz et al., 2001), and inoculum particles were screened to exclude those larger than 0.5 cm in diameter. For mock inoculation, the same inoculum was autoclaved (120 °C for 60 min, twice) before application.
Tomato plant growth and inoculation
Short‐term (24–96 hpi) experiments were carried out under controlled glasshouse conditions. Both experiments were performed in sand as a rooting matrix, amended with 0.15 g of both K2SO4 and super‐phosphate (final concentration, 15 mg/L of potassium and phosphorus), and autoclaved at 121 °C for 1 h. After inoculation and planting, the plants were grown under controlled glasshouse conditions at 28/22 °C (day/night) and a photoperiod of 12 h, under natural illumination supplemented and extended with incandescent illumination (6 µE/s cm2 at plant level; March–April). Plants were irrigated daily with UV‐treated water, which was supplemented twice weekly with inorganic phosphate (Pi)‐free nutrient‐modified Johnson solution (Johnson et al., 1957).
To semi‐synchronize the infection process for the short‐term experiments, opaque plastic trays (single cell volume, 200 mL) were used as containers, and inoculation was carried out by mixing the inocula with the sand in each cell to a final concentration of 300 propagules per cell. Following inoculum application, the trays were watered with the Pi‐free nutrient solution (see above) and incubated in a controlled glasshouse chamber until planting. A subportion of the trays, serving as IBIs, were planted with 4‐week‐old tomato seedlings and were sampled daily for AMF root colonization. When the infection rate of the IBI plants exceeded 10% (about 2 weeks after planting), the rest of the trays were planted with 4‐week‐old tomato seedlings, which were prepared separately, and designated as time 0 for the experiment.
For the long‐term (4 wpi) experiment, 3‐L pots were filled with the above‐described sand, and inoculum (live or autoclaved) was placed in a layer 3 cm below the soil surface.
Root sampling and infection evaluation
At the sampling times, plants were removed from the medium, and their roots were separated, washed, dried and immediately frozen in liquid nitrogen. The samples were kept frozen at −80 °C until use. To evaluate the root systems for inoculation, at each time point examined, roots from five individual replicate plants were pooled, washed, cleared and then stained with trypan blue (Phillips and Hayman, 1970). Stained roots were examined for mycorrhizal colonization under a dissecting microscope, and the percentage colonization was estimated using the gridline‐intersection method described by Giovannetti and Mosse (1980).
Isolation of total RNA and cDNA synthesis
Total RNA was extracted from tomato roots using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) and digested with DNase I enzyme (Qiagen) using the on‐column digestion method, according to the manufacturer's instructions.
For cDNA synthesis, 1 µg of total RNA and 0.1 µm of random hexamer primers were heated for 5 min at 65 °C and snap‐chilled on ice. The following components were added to this reaction mixture: 0.2 mm dNTP mixture (Fermentas, Glen Burnie, MD, USA), M‐MuLV‐reverse transcriptase (RT) buffer (1 × final concentration), 40 U RNase inhibitor (Takara, Otsu, Shiga, Japan), 200 U M‐MuLV‐RT enzyme (Fermentas) and diethylpyrocarbonate‐treated water to a reaction volume of 25 µL. The reaction was incubated at 42 °C for 60 min.
Microarray analysis
Microarray chip description
The tomato microarray chip (TOM1) was developed and printed by Cornell University (Ithaca, NY, USA): it contains 12 500 clones, selected at random from a number of different cDNA libraries derived from a range of tissues, including leaf, root, fruit and flower.
Microarray experiment and data analysis
Total RNA was extracted separately from mycorrhizal and mock‐inoculated tomato roots at 12, 24, 36 and 48 hpi using an RNeasy Plant Mini Kit (Qiagen), and treated with DNase I (Qiagen). Total RNA (3 µg) was taken for mRNA amplification using the MessageAmp™ II amplification kit (Ambion, Austin, TX, USA). The amplified RNA (aRNA) was then used for cDNA synthesis, with 1 mm aminoallyl‐dUTP included for labelling. Traces of aRNA were removed by alkaline hydrolysis using 10 µL NaOH (1 m), incubated at 70 °C for 15 min and neutralized by 10 µL HCl (1 m). The cDNA was then ethanol precipitated using glycogen as a carrier, and the resultant pellet obtained post‐centrifugation was air dried at room temperature.
The aminoallyl‐incorporated cDNA was labelled with cyanine‐3/cyanine‐5 (Cy3/Cy5) fluorescent dye using the LabelStar Array Kit (Amersham, Uppsala, Sweden). Post‐labelling, the fluorescently labelled cDNA was cleaned of excess dye and nucleotides with a QIAquick PCR Purification Kit (Qiagen). The amount of incorporated fluorescent dye was calculated by measuring the absorbance at A 550/A 260 and A 650/A 260. The samples were stored at −20 °C until further use.
Pre‐hybridization was performed essentially as described by Bar‐Or et al. (2005). Each chip was hybridized by mixing two samples of Cy3‐ and Cy5‐labelled probes (each 100 pm); the volume was reduced to 8.5 µL using a Microcon YM30 filter (Millipore, Cambridge, UK). The following reagents were then added: 1.5 µL 20 × SSPE [3.6 m NaCl, 0.2 m NaH2PO4‐H2O, 0.02 m ethylenediaminetetraacetic acid (EDTA), pH to 7.7], 1 µL poly dA (15 µg/mL), 1 µL yeast tRNA (4 mg/mL), 24 µL hybridization buffer [62.8% formamide, 0.8% sodium dodecylsulphate (SDS), 4 × Denhardt's solution and 5 × SSPE]. The probe solution was applied to the slide using HybriSlip covers (Shleicher & Schuell, Dassel, Germany) after denaturation at 98 °C for 3 min and centrifugation for 1 min at 13 000 g. The slides were wrapped in aluminium foil and incubated at 42 °C overnight. Post‐incubation washes were carried out as described by Bar‐Or et al. (2005).
Quantification and data analysis
The slides were scanned for fluorescence emission using a DNA microarray scanner (Agilent Technologies, Santa Clara, CA, USA). The spot intensities were quantified using Imagene 6 software (BioDiscovery, El Segundo, CA, USA) and the channel ratio was determined using the median‐of‐ratio method. Filtration on flags (to filter out absent signals) and on errors [to filter out genes with high (above 1.4) standard deviation] was applied. Data normalization was performed using the overall background by applying per‐spot and per‐chip normalization, and the default Lowess function (35% smoothing; GeneSpring 5.1; Silicon Genetics, Redwood City, CA, USA). Filtration on confidence was performed based on a one‐sample t‐test (P≤ 0.05), and the resulting gene lists were filtered for differentially expressed genes (above 1.33‐ and below 0.75‐fold change in inoculated vs. non‐inoculated samples). For each examined treatment (i.e. time point and infection vs. mock inoculation with mycorrhiza), three biological and two technical replicates (i.e. dye swap) were performed. The ontology of biologically functional groups and the annotation of up‐ and down‐regulated genes were determined on the basis of The Solanaceae Genomics Networks (http://www.sgn.cornell.edu/).
qPCR
qPCR was set up using the components supplied in the SYBR Green I Kit (Eurogentec SA, Seraing, Belgium) and specific gene primers (Table 2). The reaction mixture consisted of the following components (final concentration): 1 × reaction buffer, 2.5 mm MgCl2, 200 µm dNTP mixture, 0.4 µm forward and reverse primers, 0.2 U of Hot GoldStar™ enzyme (Eurogentec SA), 0.75 µL of SYBR Green I (1 : 2000 dilution in dimethylsulphoxide), 1 µL of the template and, finally, PCR‐grade water to a final volume of 25 µL. qPCR analysis was carried out on an ABI PRISM 7000 instrument (Perkin‐Elmer Biosystems, Norwalk, CT, USA) according to the following programme: 10 min at 95 °C, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s. The primers used for qPCR are presented in Table 2. The threshold cycle (C t) was calculated by the ABI PRISM 7000 instrument software. The level of expression of the target genes was calculated relative to that of the reference mRNA, S. lycopersicum elongation factor α (Table 1); elongation factor α was validated in preliminary experiments to be constitutively expressed in AMF‐infected and non‐infected roots (data not shown). Means and standard deviations were calculated for three biological and two technical replicates for each examined treatment. Values of the expression ratios of treatment vs. control were analysed for significance (P≤ 0.05) in a one‐tailed t‐test.
Table 2.
Primers used for quantitative polymerase chain reaction.
| Reverse primer sequence 5′–3′ | Forward primer sequence 5′–3′ | SGN accession number |
|---|---|---|
| CGTTCTGCCTTAAGTTTGTCAAGC | GAGGTTCGAGAAGGAAGCTGCT | X53043 (GenBank accession number) |
| GGCTGCTGAT AACCCTGGAC | CTGATGTTCAGATTCCTTGAACAAA | SGN‐U144012 |
| TTGCGACACA AACAGTCAAC AAA | GGTGGGATAACAACTAAATGAAACGT | SGN‐U144536 |
| GGAGATACGA CGCATGAACTGAA | CGTTTCTCTTGGTCTCTTGGCAT | SGN‐U144150 |
| CAATTTCCTTAGCATGAGTTTCTGG | TGGGAGTGTTAGAAGCAATGTGA | SGN‐U144291 |
| TTCGTTAGCGTCCACCGG | TGTCAAGCATTAGGAACACA | SGN‐U148120 |
| CTTAGAAAATCCCACTCAGTTGAATCA | GGGTACCCCTACTTATTGTGAATGG | SGN‐U144536 |
| TCCAATGGGCTTTCCATGAG | ATAGGGCAGAAGTTCTCCATACAGG | SGN‐U152045 |
| CGAACTCCCT TGATATTTAG CAGC | AATGGCACGTCCTTTCAACAT | SGN‐U146006 |
| TCCCAAACCCAAGATGACCA | CTTACCTACTACTTGTTGTTGGATTATTGC | SGN‐U143331 |
| TGGGTAGTCCACTTAAGTTGTCACA | CTATACAATACTAGTACTTCTTCAC | SGN‐U144410 |
In situ localization of gene transcripts
The localization of EXLB1 transcripts was performed essentially as described in Gal et al. (2006) with a few modifications. Briefly, plants were grown and infected with AMF as described above. At 24, 48, 72 and 96 hpi, roots were taken out of the pots, rinsed and fixed in formalin–acetic acid–alcohol (FAA) (Gal et al., 2006). For the 4‐wpi analysis, tomato roots were inoculated with the ‘whole inoculum’ fraction of AMF (i.e. inoculum containing spores, hyphae and infected root segments) and allowed to grow for 4 weeks under glasshouse conditions, as described previously by David‐Schwartz et al. (2001). The inoculated roots, containing various stages of AMF colonization, were fixed in FAA and sectioned using a Vibratome microtome (Series 1000 Sectioning System, Technical Products International, O'Fallon, MO, USA). Sections were inserted into 0.2‐mL tubes and reverse transcription (RT) and PCR were performed essentially as described in Gal et al. (2006) with the following modifications: for RT, the final concentration of dNTPs was 0.2 mm and the final concentration of gene‐specific primer was 0.1 µm. The primers used for RT and amplification of EXLB1 were as follows: forward, 5′‐GCAACTATCATGGCTACCTTGC‐3′; reverse, 5′‐CTACAATGGCTGCTGATAACCC‐3′. Only the reverse primer was used for RT; both primers were used for PCR. In the controls, no primers were added to the PCR mixture. Following PCR, excised tissues were transferred to glass slides containing 10 × phosphate‐buffered saline, and immediately observed using a confocal microscope (Olympus IX81,Tokyo, Japan) to detect the fluorescence signal (excitation and emission wavelengths of 488 nm and 505–560 nm, respectively). First, control sections were observed and the BA505I filter and argon 488‐nm laser beam were modified such that no fluorescence signal was detected in the control. The same microscopic setting was used to detect the signal in the experiment. Autofluorescence was detected using excitation and emission wavelengths of 488 nm and ≥660 nm, respectively. Liquid left in the PCR tubes following tissue transfer was subjected to gel electrophoresis on an ethidium bromide‐stained agarose gel to verify the size of the in situ‐amplified fragment, its presence in the experiment and its absence in the controls. Each experiment and control were repeated twice, with 20 different sections of five different plants observed in each repeat.
Construction and propagation of transgenic roots
Transgenic roots were constructed and propagated as described in Gal et al. (2006). Briefly, tomato (S. lycopersicum cv. VF36) cotyledons were transformed with a 35S promoter‐LeEXPA5‐antisense construct, using Agrobacterium rhizogenes strain A15834 as the vector. Emerging roots were excised and transferred to Gamborg medium (Gamborg et al., 1968) containing 50 µg/mL kanamycin (Duchefa, Haarlem, the Netherlands) for the selection of emerging roots bearing the transgenic construct. Control root cultures expressing GFP were prepared similarly using the original pBIN m‐gfp5‐ER vector expressing GFP‐sense under the control of the 35S promoter. The ubiquitous expression of GFP in these controls was verified by microscopic observation (DMLB microscope, Leica, Heidelberg, Germany).
Analysis of transgenic roots for steady‐state levels of EXLB1 transcript and G. intraradices symbiosis
Fourteen independent transgenic lines, eight containing expansin antisense (EXP‐anti) and six containing GFP (control) constructs, were taken for further analysis. To determine the steady‐state level of EXLB1 transcript, qPCR was performed as described above. For the establishment of AMF symbiosis on the transgenic roots, root cultures were grown on solid M‐medium (Bécard and Fortin, 1988) in an environmentally controlled incubator, in constant darkness, at 25 °C. After establishment of each clone (in 10‐cm Petri dishes), inoculation was carried out in vitro by placing 10–15 sterile spores in several places along the roots. Spores were taken from carrot root organ culture, as described by St‐Arnaud et al. (1996). Cultures were incubated in an inverted position in the dark at 27 °C. Cultures were sampled for the number and organization of intracellular structures of mycorrhizal establishment for 5 wpi. Roots were stained with acid fuchsin, as described in Floss et al. (2008).
After 3–4 months, the number of spores in each plate was counted, and then calculated per gram of root dry weight. It should be noted that spore production may be the most straightforward measure of the full reproductive potential of a fungus, suggesting the host's ability to control fungal penetration. Two replicate plates were set up for each clone.
Supporting information
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This work was supported by an Israeli Science Foundation Grant (number 522/02‐1) to HK and YK.
REFERENCES
- Akiyama, K. , Matsuzaki, K. and Hayashi, H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature, 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Amiour, N. , Recorbet, G. , Robert, F. , Gianinazzi, S. and Dumas‐Gaudot, E. (2006) Mutations in DMI3 and SUNN modify the appressorium‐responsive root proteome in arbuscular mycorrhiza. Mol. Plant–Microbe Interact. 19, 988–997. [DOI] [PubMed] [Google Scholar]
- Ané, J.M. , Lévy, J. , Thoquet, P. , Kulikova, O. , De Billy, F. , Penmetsa, V. , Kim, D.J. , Debellé, F. , Rosenberg, C. , Cook, D.R. , Bisseling, T. , Huguet, T. and Dénarié, J. (2002) Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol. Plant–Microbe Interact. 15, 1108–1118. [DOI] [PubMed] [Google Scholar]
- Ané, J.M. , Kiss, G.B. , Riely, B.K. , Penmetsa, R.V. , Oldroyd, G.E. , Ayax, C. , Lévy, J. , Debellé, F. , Baek, J.M. , Kalo, P. , Rosenberg, C. , Roe, B.A. , Long, S.R. , Dénarié, J. and Cook, D.R. (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science, 303, 1364–1367. [DOI] [PubMed] [Google Scholar]
- Balestrini, R. and Lanfranco, L. (2006) Fungal and plant gene expression in arbuscular mycorrhizal symbiosis. Mycorrhiza, 16, 509–524. [DOI] [PubMed] [Google Scholar]
- Balestrini, R. , Cosgrove, D.J. and Bonfante, P. (2005) Differential location of a‐expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta, 220, 889–899. [DOI] [PubMed] [Google Scholar]
- Balestrini, R. , Gómez‐Ariza, J. , Lanfranco, L. and Bonfante, P. (2007) Laser microdissection reveals that transcripts for five plant and one fungal phosphate transporter genes are contemporaneously present in arbusculated cells. Mol. Plant–Microbe Interact. 20, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Bar‐Or, C. , Kapulnik, Y. and Koltai, H. (2005) A broad characterization of the transcriptional profile of the compatible tomato response to the plant parasitic root knot nematode Meloidogyne javanica . Eur. J. Plant Pathol. 111, 181–192. [Google Scholar]
- Barker, D.G. , Bianchi, S. , Blondon, F. , Dattée, Y. , Duc, G. , Flament, P. , Gallusci, P. , Génier, G. , Guy, P. , Muel, X. , Tourneur, J. , Dénarié, J. and Huguet, T. (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium–legume symbiosis. Plant Mol. Biol. Rep. 8, 40–49. [Google Scholar]
- Barker, S.J. , Stummer, B. , Gao, L. , Dispain, I. , O'Connor, P.J. and Smith, S.E. (1998) A mutant in Lycopersicon esculentum Mill. with highly reduced VA mycorrhizal colonisation: isolation and preliminary characterisation. Plant J. 15, 791–797. [DOI] [PubMed] [Google Scholar]
- Bécard, G. and Fortin, J.A. (1988) Early events of vesicular–arbuscular mycorrhiza formation on Ri T‐DNA transformed roots. New Phytol. 108, 211–218. [DOI] [PubMed] [Google Scholar]
- Borisov, A. , Danilova, T. , Koroleva, T. , Kuznetsova, E. , Madsen, L. , Mofett, M. , Naumkina, T. , Nemankin, T. , Ovchinnikova, E. , Pavlova, Z. , Petrova, N. , Pinaev, A. , Radutoiu, S. , Rozov, S. , Rychagova, T. , Shtark, O. , Solovov, I. , Stougaard, J. , Tikhonovich, I. , Topunov, A. , Tsyganov, V. , Vasil'chikov, A. , Voroshilova, V. , Weeden, N. , Zhernakov, A. and Zhukov, V. (2007) Regulatory genes of garden pea (Pisum sativum L.) controlling the development of nitrogen‐fixing nodules and arbuscular mycorrhiza: a review of basic and applied aspects. Appl. Biochem. Microbiol. 43, 237–243. [PubMed] [Google Scholar]
- Brechenmacher, L. , Weidmann, S. , Van Tuinen, D. , Chatagnier, O. , Gianinazzi, S. , Franken, P. and Gianinazzi‐Pearson, V. (2004) Expression profiling of up‐regulated plant and fungal genes in early and late stages of Medicago truncatula–Glomus mosseae interactions. Mycorrhiza, 14, 253–262. [DOI] [PubMed] [Google Scholar]
- Bucher, M. (2007) Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 173, 11–26. [DOI] [PubMed] [Google Scholar]
- Chen, F. , Dahal, P. and Bradford, K.J. (2001) Two tomato expansin genes show divergent expression and localization in embryos during seed development and germination. Plant Physiol. 127, 928–936. [PMC free article] [PubMed] [Google Scholar]
- Cook, D.R. (1999) Medicago truncatula: a model in the making! Curr. Opin. Plant Biol. 2, 301–304. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999) Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 391–417. [DOI] [PubMed] [Google Scholar]
- David‐Schwartz, R. , Badani, H. , Wininger, S. , Levy, A. , Galili, G. and Kapulnik, Y. (2001) Identification of a novel genetically controlled step in mycorrhizal colonization: plant resistance to infection by fungal spores but not extra‐radical hyphae. Plant J. 27, 561–569. [DOI] [PubMed] [Google Scholar]
- David‐Schwartz, R. , Gadkar, V. , Wininger, S. , Bendov, R. , Galili, G. , Levy, A. and Kapulnik, Y. (2003) Isolation of a pre‐mycorrhizal infection (pmi2) mutant of tomato, resistant to arbuscular mycorrhizal fungal colonization. Mol. Plant–Microbe Interact. 16, 382–388. [DOI] [PubMed] [Google Scholar]
- Dean, R.M. , Rivers, R.L. , Zeidel, M.L. and Roberts, D.M. (1999) Purification and functional reconstitution of soybean nodulin 26. An aquaporin with water and glycerol transport properties. Biochemistry, 38, 347–353. [DOI] [PubMed] [Google Scholar]
- Demchenko, K. , Winzer, T. , Stougaard, J. , Parniske, M. and Pawlowski, K. (2004) Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol. 163, 381–392. [DOI] [PubMed] [Google Scholar]
- Endre, G. , Kereszt, A. , Kevei, Z. , Mihacea, S. , Kalo, P. and Kiss, G.B. (2002) A receptor kinase gene regulating symbiotic nodule development. Nature, 417, 962–966. [DOI] [PubMed] [Google Scholar]
- Floss, D.S. , Hause, B. , Lange, P.R. , Küster, H. , Strack, D. and Walter, M.H. (2008) Knock‐down of the MEP pathway isogene 1‐deoxy‐D‐xylulose 5‐phosphate synthase 2 inhibits formation of arbuscular mycorrhiza‐induced apocarotenoids, and abolishes normal expression of mycorrhiza‐specific plant marker genes. Plant J. 56, 86–100. [DOI] [PubMed] [Google Scholar]
- Franken, P. , Donges, K. , Grunwald, U. , Kost, G. , Rexer, K.H. , Tamasloukht, M. , Waschke, A. and Zeuske, D. (2007) Gene expression analysis of arbuscule development and functioning. Phytochemistry, 68, 68–74. [DOI] [PubMed] [Google Scholar]
- Gal, T.Z. , Aussenberg, E.R. , Burdman, S. , Kapulnik, Y. and Koltai, H. (2006) Expression of a plant expansin is necessary for completion of the root knot nematode life cycle. Planta, 4, 1–8. [DOI] [PubMed] [Google Scholar]
- Gamborg, O.L. , Miller, R.A. and Ojima, K. (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. [DOI] [PubMed] [Google Scholar]
- Gao, X. , Kuyper, T.W. , Zou, C. , Zhang, F. and Hoffland, E. (2007) Mycorrhizal responsiveness of aerobic rice genotypes is negatively correlated with their zinc uptake when nonmycorrhizal. Plant Soil, 290, 283–291. [Google Scholar]
- Genre, A. , Chabaud, M. , Timmers, T. , Bonfante, P. and Barker, D.G. (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell, 17, 3489–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts, R. , Fedorova, E. and Bisseling, T. (2005) Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8, 346–352. [DOI] [PubMed] [Google Scholar]
- Gherbi, H. , Markmann, K. , Svistoonoff, S. , Estevan, J. , Autran, D. , Giczey, G. , Auguy, F. , Péret, B. , Laplaze, L. , Franche, C. , Parniske, M. and Bogusz, D. (2008) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc. Natl. Acad. Sci. USA, 105, 4928–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi‐Pearson, V. (1996) Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell, 8, 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianinazzi‐Pearson, V. , Séjalon‐Delmas, N. and Genre, A. (2007) Plants and arbuscular mycorrhizal fungi: cues and communication in the early steps of symbiotic interactions. Adv. Bot. Res. 46, 181–219. [Google Scholar]
- Giordano, W. and Hirsch, A.M. (2004) The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover–Sinorhizobium meliloti interaction. Mol. Plant–Microbe Interact. 17, 613–622. [DOI] [PubMed] [Google Scholar]
- Giovannetti, M. and Mosse, B. (1980) An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. [Google Scholar]
- Glassop, D. , Smith, S.E. and Smith, F.W. (2007) Rice phosphate transporters associated with phosphate uptake in rice roots colonised with arbuscular mycorrhizal fungi. Can. J. Bot. 85, 644–651. [Google Scholar]
- Gucciardo, S. , Wisniewski, J.P. , Brewin, N.J. and Bornemann, S. (2007) A germin‐like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. J. Exp. Bot. 58, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Guether, M. , Balestrini, R. , Hannah, M. , He, J. , Udvardi, M.K. and Bonfante, P. (2009) Genome‐wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus . New Phytol. 182, 200–212. [DOI] [PubMed] [Google Scholar]
- Güimil, S. , Chang, H.S. , Zhu, T. , Sesma, A. , Osbourn, A. , Roux, C. , Ioannidis, V. , Oakeley, E.J. , Docquier, M. , Descombes, P. , Briggs, S.P. and Paszkowski, U. (2005) Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. USA, 102, 8066–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg, K. and Stougaard, J. (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 2, 487–496. [Google Scholar]
- Harrison, M.J. (2005) Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 59, 19–42. [DOI] [PubMed] [Google Scholar]
- Hause, B. and Fester, T. (2005) Molecular and cell biology of arbuscular mycorrhizal symbiosis. Planta, 221, 184–196. [DOI] [PubMed] [Google Scholar]
- Herrera‐Medina, M.J. , Steinkellner, S. , Vierheilig, H. , Ocampo Bote, J.A. and García Garrido, J.M. (2007) Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 175, 554–564. [DOI] [PubMed] [Google Scholar]
- Hirsch, A.M. and Kapulnik, Y. (1998) Signal transduction pathways in mycorrhizal associations: comparisons with the Rhizobium–legume symbiosis. Fungal Genet. Biol. 23, 205–212. [DOI] [PubMed] [Google Scholar]
- Javot, H. , Pumplin, N. and Harrison, M.J. (2007) Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ. 30, 310–322. [DOI] [PubMed] [Google Scholar]
- Johnson, C.M. , Stout, P.R. , Beyer, J.C. and Carlson, A.B. (1957) Comparative chlorine requirements of different species. Plant Soil, 8, 337–353. [Google Scholar]
- Kende, H. , Bradford, K. , Brummell, D. , Cho, H.T. , Cosgrove, D. , Fleming, A. , Gehring, C. , Lee, Y. , McQueen‐Mason, S. , Rose, J. and Voesenek, L.A. (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 55, 311–314. [DOI] [PubMed] [Google Scholar]
- Kistner, C. , Winzer, T. , Pitzschke, A. , Mulder, L. , Sato, S. , Kaneko, T. , Tabata, S. , Sandal, N. , Stougaard, J. , Webb, K.J. , Szczyglowski, K. and Parniske, M. (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell, 17, 2217–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai, H. , Gadkar, V. and Kapulnik, Y. (2009) Biochemical and practical views of arbuscular mycorrhizal fungus–host association in horticultural crops In: Horticultural Reviews (Janick J., ed.) San Francisco, CA: John Wiley & Sons, Inc. (in press). [Google Scholar]
- Kosuta, S. , Chabaud, M. , Lougnon, G. , Gough, C. , Dénarié, J. , Barker, D.G. and Bécard, G. (2003) A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis‐specific MtENOD11 expression in roots of Medicago truncatula . Plant Physiol. 131, 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajinski, F. and Frenzel, A. (2007) Towards the elucidation of AM‐specific transcription in Medicago truncatula . Phytochemistry, 68, 75–81. [DOI] [PubMed] [Google Scholar]
- Küster, H. , Vieweg, M.F. , Manthey, K. , Baier, M.C. , Hohnjec, N. and Perlick, A.M. (2007) Identification and expression regulation of symbiotically activated legume genes. Phytochemistry, 68, 8–18. [DOI] [PubMed] [Google Scholar]
- Lane, B.G. , Dunwell, J.M. , Ray, J.A. , Schmitt, M.R. and Cuming, A.C. (1993) Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268, 12 239–12 242. [PubMed] [Google Scholar]
- Lévy, J. , Bres, C. , Geurts, R. , Chalhoub, B. , Kulikova, O. , Duc, G. , Journet, E.P. , Ané, J.M. , Lauber, E. , Bisseling, T. , Dénarié, J. , Rosenberg, C. and Debellé, F. (2004) A putative Ca2+ and calmodulin‐dependent protein kinase required for bacterial and fungal symbioses. Science, 303, 1361–1364. [DOI] [PubMed] [Google Scholar]
- Limpens, E. , Franken, C. , Smit, P. , Willemse, J. , Bisseling, T. and Geurts, R. (2003) LysM domain receptor kinases regulating rhizobial Nod factor‐induced infection. Science, 302, 630–633. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Blaylock, L. and Harrison, M.J. (2004) cDNA arrays as tools to identify mycorrhiza‐regulated genes: identification of mycorrhiza induced genes that encode or generate signaling molecules implicated in the control of root growth. Can. J. Bot. 82, 1177–1185. [Google Scholar]
- Liu, J. , Maldonado‐Mendoza, I. , Lopez‐Meyer, M. , Cheung, F. , Town, C.D. and Harrison, M.J. (2007) Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 50, 529–544. [DOI] [PubMed] [Google Scholar]
- Maeda, D. , Ashida, K. , Iguchi, K. , Chechetka, S.A. , Hijikata, A. , Okusako, Y. , Deguchi, Y. , Izui, K. and Hata, S. (2006) Knockdown of an arbuscular mycorrhiza‐inducible phosphate transporter gene of Lotus japonicus suppresses mutualistic symbiosis. Plant Cell Physiol. 47, 807–817. [DOI] [PubMed] [Google Scholar]
- Massoumou, M. , Van Tuinen, D. , Chatagnier, O. , Arnould, C. , Brechenmacher, L. , Sanchez, L. , Selim, S. , Gianinazzi, S. and Gianinazzi‐Pearson, V. (2007) Medicago truncatula gene responses specific to arbuscular mycorrhiza interactions with different species and genera of Glomeromycota. Mycorrhiza, 17, 223–234. [DOI] [PubMed] [Google Scholar]
- McQueen‐Mason, S. and Cosgrove, D.J. (1995) Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 107, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen‐Mason, S. , Durachko, D.M. and Cosgrove, D.J. (1992) Two endogenous proteins that induce cell wall expansion in plants. Plant Cell, 4, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese, E. , Mun, J.‐H. , Yeun, L.H. , Jayaraman, D. , Rougé, P. , Barre, A. , Lougnon, G. , Schornack, S. , Bono, J.‐J. and Cook, D.R. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium‐ and calmodulin‐dependent protein kinase of Medicago truncatula . Mol. Plant–Microbe Interact. 20, 912–921. [DOI] [PubMed] [Google Scholar]
- Nagy, R. , Karandashov, V. , Chague, V. , Kalinkevich, K. , Tamasloukht, M. , Xu, G. , Jakobsen, I. , Levy, A.A. , Amrhein, N. and Bucher, M. (2005) The characterization of novel mycorrhiza‐specific phosphate transporters from Solanum esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 42, 236–250. [DOI] [PubMed] [Google Scholar]
- Navazio, L. , Moscatiello, R. , Genre, A. , Novero, M. , Baldan, B. , Bonfante, P. and Mariani, P. (2007) Diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol. 144, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah, B. , Briere, C. , Bécard, G. , Dénarié, J. and Gough, C. (2005) Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J. 44, 195–207. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E. , Harrison, M.J. and Udvardi, M. (2005) Peace talks and trade deals. Keys to long‐term harmony in legume–microbe symbioses. Plant Physiol. 137, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski, U. (2006) A journey through signaling in arbuscular mycorrhizal symbioses. New Phytol. 172, 35–46. [DOI] [PubMed] [Google Scholar]
- Paszkowski, U. , Kroken, S. , Roux, C. and Briggs, S.P. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA, 99, 13 324–13 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J.M. and Hayman, D.S. (1970) Improved procedure for clearing roots and staining parasitic and vesicular–arbuscular fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55, 158–161. [Google Scholar]
- Qin, L. , Kudla, U. , Roze, E.H. , Goverse, A. , Popeijus, H. , Nieuwland, J. , Overmars, H. , Jones, J.T. , Schots, A. , Smant, G. , Bakker, J. and Helder, J. (2004) Plant degradation: a nematode expansin acting on plants. Nature, 427, 30. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D. (2007) Programming good relations—development of the arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 10, 98–105. [DOI] [PubMed] [Google Scholar]
- Van Rhijn, P. , Fang, Y. , Galili, S. , Shaul, O. , Atzmon, N. , Wininger, S. , Eshed, Y. , Lum, M. , Li, Y. , To, V. , Fujishige, N. , Kapulnik, Y. and Hirsch, A.M. (1997) Expression of early nodulin genes in alfalfa mycorrhizae indicates that signal transduction pathways used in forming arbuscular mycorrhizae and Rhizobium‐induced nodules may be conserved. Proc. Natl. Acad. Sci. USA, 94, 5467–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel, H. , Van Tuinen, D. , Franken, P. , Gianinazzi, S. and Gianinazzi‐Pearson, V. (2001) Signalling between arbuscular mycorrhizal fungi and plants: identification of a gene expressed during early interactions by differential RNA display analysis. Plant Soil, 232, 13–19. [Google Scholar]
- Sampedro, J. and Cosgrove, D.J. (2005) The expansin superfamily. Genome Biol. 6, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüssler, A. , Schwarzott, D. and Walker, C. (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421. [Google Scholar]
- Siciliano, V. , Genre, A. , Balestrini, R. , Cappellazzo, G. , DeWit, P.J. and Bonfante, P. (2007) Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol. 144, 1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.E. and Read, D.J. (1997) Mycorrhizal Symbiosis. London: Academic Press. [Google Scholar]
- St‐Arnaud, M. , Hamel, C. , Vimard, B. , Caron, M. and Fortin, J.A. (1996) Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol. Res. 100, 328–332. [Google Scholar]
- Takeda, N. , Kistner, C. , Kosuta, S. , Winzer, T. , Pitzschke, A. , Groth, M. , Sato, S. , Kaneko, T. , Tabata, S. and Parniske, M. (2007) Proteases in plant root symbiosis. Phytochemistry, 68, 111–121. [DOI] [PubMed] [Google Scholar]
- Vierheilig, H. and Piché, Y. (2002) Signalling in arbuscular mycorrhiza: facts and hypotheses. Adv. Exp. Med. Biol. 505, 23–39. [DOI] [PubMed] [Google Scholar]
- Weidmann, S. , Sanchez, L. , Descombin, J. , Chatagnier, O. , Gianinazzi, S. and Gianinazzi‐Pearson, V. (2004) Fungal elicitation of signal transduction‐related plant genes precedes mycorrhiza establishment and requires the dmi3 gene in Medicago truncatula . Mol. Plant–Microbe Interact. 17, 1385–1393. [DOI] [PubMed] [Google Scholar]
- Wieczorek, K. , Golecki, B. , Gerdes, L. , Heinen, P. , Szakasits, D. , Durachko, D.M. , Cosgrove, D.J. , Kreil, D.P. , Puzio, P.S. , Bohlmann, H. and Grundler, F.M.W. (2006) Expansins are involved in the formation of nematode‐induced syncytia in roots of Arabidopsis thaliana . Plant J. 48, 98–112. [DOI] [PubMed] [Google Scholar]
- Xia, Y.S. , Chen, B.‐D. , Christie, P. , Smith, A.F. , Wang, Y.‐S. and Li, W.‐L. (2007) Arsenic uptake by arbuscular mycorrhizal maize (Zea mays L.) grown in an arsenic‐contaminated soil with added phosphorus. J. Environ. Sci. 19, 1245–1251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


