SUMMARY
The translocation of Melon necrotic spot virus (MNSV) within tissues of inoculated and systemically infected Cucumis melo L. ‘Galia’ was studied by tissue‐printing and in situ hybridization techniques. The results were compatible with the phloem vascular components being used to spread MNSV systemically by the same assimilate transport route that runs from source to sink organs. Virus RNAs were shown to move from the inoculated cotyledon toward the hypocotyl and root system via the external phloem, whereas the upward spread through the stem to the young tissues took place via the internal phloem. Virus infection was absent from non‐inoculated source tissues as well as from both shoot and root apical meristems, but active sink tissues such as the young leaves and root system were highly infected. Finally, our results suggest that the MNSV invasion of roots is due to virus replication although a destination‐selective process is probably necessary to explain the high levels of virus accumulation in roots. This efficient invasion of the root system is discussed in terms of natural transmission of MNSV by the soil‐borne fungal vector.
INTRODUCTION
Once a plant virus has been transmitted to a specific host plant by an appropriate vector, it begins its infectious cycle through an intracellular multiplication step in the initially infected cell accompanied by genome replication and plant defence suppression (Brodersen and Voinnet, 2006; Burgyan, 2006; Dunoyer and Voinnet, 2005). This process is followed by intracellular translocation of either the virus particles or the non‐virion nucleoprotein complexes towards the cell periphery to colonize the neighbouring cells progressively by crossing the cell wall through the symplastic connections known as plasmodesmata (PD) (Ruiz‐Medrano et al., 2004; Scholthof, 2005). For this local spreading, virus‐encoded movement proteins (MP), which can bind nucleic acids, target PD and modify their size exclusion limit (Haupt et al., 2005; Herranz and Pallas, 2004; Herranz et al., 2005; Lucas, 2006; Navarro et al., 2006; Peremyslov et al., 2004; Prokhnevsky et al., 2005; Wright et al., 2007). However, systemic infection of the plant prior to the full establishment of antiviral defence response in young growing parts is not feasible as a result of the time‐consuming cell‐to‐cell movement. Therefore, viruses make use of a pre‐existing rapid long‐distance translocation pathway of the plant by entering the vascular system through both major and minor leaf veins (Cheng et al., 2000; Nelson and van Bel, 1998; Ueki and Citovsky, 2007). Although some viruses may travel through the xylem (Dubois et al., 1994; Moreno et al., 2004; Opalka et al., 1998; Verchot et al., 2001), most viruses move systemically through phloem, which is a complex symplasmic domain that controls photoassimilate partitioning throughout the plant as well as inter‐organ signalling and coordination (Andrianifahanana et al., 1997; Barker and Harrison, 1986; Cheng et al., 2000; Derrick and Barker, 1992, 1997; Mas and Pallás, 1996). The phloem proper consists of two main cell types: the cells of the conducting sieve element (SE) and the companion cells (CC), connected to SE by specific branched PD (Sauer, 1997). However, virus entry into the SE–CC complex is constrained by both the bundle sheath (BS)–vascular parenchyma (VP) and VP–CC boundaries (Ding et al., 1996; Goodrick et al., 1991; Thompson and Garcia‐Arenal, 1998; Wintermantel et al., 1997). In addition, a discontinuous vascular movement model for Potato virus A (PVA) has recently been proposed, by which vascular transport of this virus would proceed in repeated movement cycles, each including virus loading to SE for transport over a short distance and unloading to CC and other phloem cells for replication (Germudson and Valkonen, 2006). Together, these results indicate that mechanisms controlling cell‐to‐cell and systemic movement differ.
It has been established that the long‐distance trafficking of viruses follows the non‐selective pressure‐driven stream of photosynthetic assimilates from source to sink organs (Leisner and Turgeon, 1993; Oparka and Santa Cruz, 2000; Santa Cruz, 1999). In plant families such as Solanaceae and Cucurbitaceae, which contain two different types of phloem, movement towards roots occurs through external phloem while the internal phloem is used for the translocation towards shoots (Andrianifahanana et al., 1997; Barker and Harrison, 1986; Cheng et al., 2000; Derrick and Barker, 1992, 1997). Moreover, accumulation of some viruses in specific tissues (Al‐Kaff and Covey, 1996) suggests that their final destination could also be regulated in a similar way to that of the 16‐kDa Cucurbita maxima phloem protein 1 (CmPP16‐1) and CmPP16‐2. These two plant paralogues of viral MPs are targeted to roots via interaction with unidentified phloem proteins (Aoki et al., 2005). Finally, the viral unloading at distal young organs takes place in the major veins probably due to the immature status of the minor veins (Cheng et al., 2000; Silva et al., 2002). The exact destination of the virus will depend on specific cell boundaries (Foster et al., 2002; Itaya et al., 2002; Zhu et al., 2002).
Regardless of the advances made in elucidating the molecular mechanisms involved in the cell‐to‐cell movement of viruses, our understanding of the factors controlling long‐distance movement is limited (for a complete review of plant virus movement see Ueki and Citovsky, 2007; Waigmann et al., 2004). Therefore, more studies are required in different virus–host plant systems. In the present study, we examined the systemic spread of Melon necrotic spot virus (MNSV) in its natural host Cucumis melo L. Galia. MNSV has been classified as belonging to the genus Carmovirus within the family Tombusviridae (Hibi and Furuki, 1985; Riviere and Rochon, 1990). This virus has a single‐stranded RNA genome 4.3 kb long (Riviere and Rochon, 1990) which codes for five functionally characterized proteins (Genovés et al., 2006; Navarro et al., 2006). MNSV has a range of host plants largely restricted to species of the family Cucurbitaceae. Characteristic symptoms of diseased plants include necrotic spots on leaves and stem necrosis. MNSV is naturally transmitted in soil by the zoospores of the chytrid fungus Olpidium bornovanus (Lange and Insunza, 1977), which occurs by attachment of virus particles to the outer surface of zoospores (Furuki, 1981). MNSV can also be seed‐transmitted (Campbell et al., 1996).
Studies previously performed in our laboratory showed that melon roots act as a reservoir for MNSV (Gosálvez et al., 2003). In the present study, a more accurate imaging of MNSV‐infected melon tissues using cell biology techniques demonstrates that MNSV moves through the phloem of melon plants by using the external and internal phloem for movement to roots and from roots to shoot apex, respectively. Moreover, roots were confirmed as a specific accumulation site of the virus, while evidence for their replication in this location is also provided.
RESULTS
The accumulation of MNSV in different parts of melon plants, including cotyledons, leaves, stems and roots, was previously studied in our laboratory by dot‐blot hybridization (Gosálvez et al., 2003). These previous results showed that the virus exits the inoculated cotyledon as early as 2 days post‐inoculation (dpi) given that the viral signal was already detected in the stem at this time. Moreover, the viral infection reached both uninoculated leaves and root tissues 4 dpi, when chlorotic to necrotic spot lesions become visible on cotyledons. Interestingly, the highest amount of viral RNA, apart from the inoculated cotyledons, was detected in roots 5–6 dpi (Gosálvez et al., 2003).
In the present study, a more accurate macroscopic study of MNSV movement in systemically infected melon plants was carried out using tissue‐printing RNA–RNA hybridization and a digoxigenin‐labelled RNA probe, complementary to 463 nt of the MNSV coat protein gene. Mechanical inoculation of MNSV virions into cotyledons was performed before the first leaf emerged. Thus, samples were taken 7–8 dpi when at least two leaves had already appeared (see schematic diagram in Fig. 1). At this point, the first leaf was fully expanded (named hereafter developed leaf) whereas the second one was in an immature young stage (named hereafter young leaf). Several MNSV‐infected plants showing a similar developmental stage were divided into seven different sections (as indicated in Fig. 1), including the developed leaf and its petiole, the hypocotyl and stem, the shoot tip, the young leaf and its petiole, one of the inoculated cotyledons and its corresponding petiole, and the roots. They were then simultaneously analysed by tissue‐printing hybridization.
Figure 1.
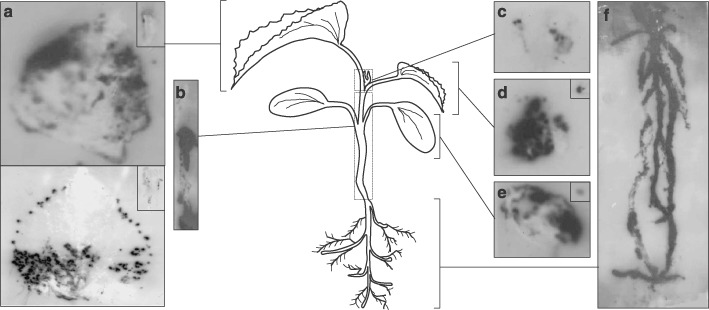
Analysis of the spatial distribution of MNSV in infected melon plants (8 dpi) by tissue printing hybridization of (a) first systemic leaves (developed leaves), (b) hypocotyl and main stem, (c) shoot tip, (d) second systemic leaf (young leaf), (e) MNSV‐inoculated cotyledon and (f) roots. Either longitudinal or cross‐section impressions of the petioles from both developed or young leaves and cotyledons, respectively, are displayed in the insets located in the top right of the corresponding panel. A diagram of the melon plant in the developmental stage when the samples were taken is displayed at the centre of the figure to better understand the origin of each panel. Tissue‐printing hybridization was performed using a digoxigenin‐labelled RNA probe for detecting MNSV vRNA as described in the Experimental Procedures. Five different plants were analysed but only those results clearly representative are shown.
In fully developed leaves, the hybridization signal corresponding to viral RNAs (vRNA) appeared to be associated with the symptomatic areas, which, interestingly, were asymmetrically distributed either in the entire leaf (Fig. 1a, top) or in the basal half‐leaf next to the petiole and at the hydathodes (Fig. 1a, bottom). Moreover, in this case, the hybridization signals (infection foci) were mainly concentrated on the left side of the leaf that was closest to one of the inoculated cotyledons (Fig. 1a, bottom). Longitudinal sections of petioles from these developed leaves showed a low‐intensity hybridization signal (Fig. 1a, insets). In some cases, neither symptoms nor hybridization signals were observed in these types of leaves (data not shown). This situation has already been observed in other virus–host interactions in which fully expanded leaves were not susceptible to viral infection (e.g. Mas and Pallás, 1996). In contrast, the highest hybridization signal was uniformly distributed over the surface of the young leaf (Fig. 1d) as well as on the cross‐sectioned petiole (Fig. 1d, inset) in all the analysed plants.
Similar results were obtained with the tissue‐blots from both the inoculated cotyledons and the cross‐section of the corresponding petiole (Fig. 1e and inset, respectively), which also showed a strong hybridization signal coinciding with the local lesions. Unexpectedly, hybridization signals in the vascular system of both leaves and cotyledons were not observed.
The tissue‐blots of longitudinal sections of the stem from the shoot tip (not included) to the hypocotyl (included) showed only a strong vRNA signal located below the cotyledon insertion point, while, again, no signal was observed in the upper part of the stem (Fig. 1b), even after extended infection periods of 17 dpi (data not shown). In the shoot tip, flanked by the petioles of the leaves, the viral‐specific signal was low and heterogeneous, although some strong spots were also observed (Fig. 1c). Finally, in agreement with previous results (Gosálvez et al., 2003) the roots showed the strongest hybridization signal, which was uniformly distributed (Fig. 1f). No signal was observed in the tissue‐printing hybridization performed on samples taken from healthy melon (data not shown).
Differential MNSV accumulation and replication in roots
To study whether the high levels of MNSV RNA accumulation in the root system originated from viral replication or were due to the existence of a root tropism, evidence for MNSV replication in this organ was investigated. Considering that both complementary vRNA (cvRNA) and double‐strand RNAs (dsRNAs) generally accompany RNA virus replication, their presence on MNSV‐infected roots was analysed. For this purpose, two RNA probes containing either cvRNA or vRNA sequences were used. Cotyledons (used as controls of a tissue where virus replication is active) as well as roots from MNSV‐infected melon plants were collected 10 dpi and used for tissue‐printing hybridization or dsRNA extraction followed by Northern blot analysis. The specificity and efficiency of the probes was checked by analysing transcripts of both polarities by dot‐blot assay, which confirmed that both riboprobes recognized only the corresponding complementary RNAs and at the same level of sensitivity (data not shown). In the tissue‐printing hybridization, the viral signals in symptomatic cotyledons revealed a differential distribution of vRNAs and cvRNAs, indicating the absence of self‐hybridization in either riboprobe (Fig. 2a, compare vRNA vs. cvRNA signals in the cotyledons panel). The vRNAs were uniformly distributed all around each infection focus/local lesion while cvRNAs were mainly detected in specific areas and also around the lesions. Interestingly, both vRNAs and cvRNAs were also detected in MNSV‐infected roots. Unlike cotyledons, no clear difference was found between the signal distribution of each virus strand, although the spots were not completely coincident (Fig. 2a, compare vRNA vs. cvRNA signals in the roots panel), suggesting that replication occurs in both primary and secondary roots. The ratio between cvRNA and vRNA was similar in both organs, as deduced by hybridization signal intensity. No signal was observed from the tissue‐printing hybridization analysis performed on samples taken from healthy plants using either riboprobe matching to vRNA (Fig. 2a) or to cvRNA (data not shown). In addition, dsRNAs from cotyledon and root samples were extracted, resolved by polyacrylamide gel electrophoresis and transferred to a nylon membrane. As MNSV produces dsRNAs corresponding to their genomic and both subgenomic RNAs, the subsequent Northern blot assay demonstrated the presence and the identity of all three species of RNA in cotyledon but also in root samples (Fig. 2b), although here the hybridization signal intensity was five‐fold lower than in dsRNAs from cotyledons.
Figure 2.
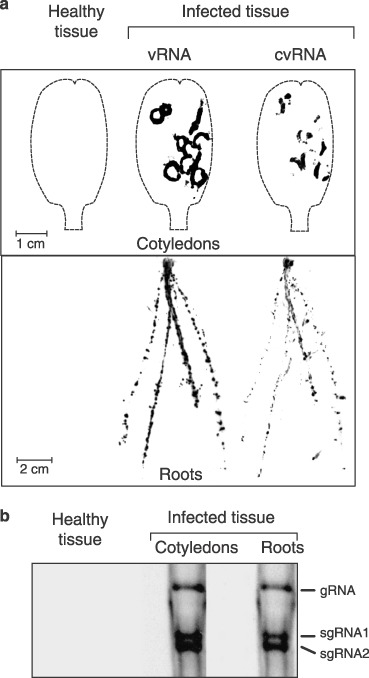
(a) Detection of either vRNA or cvRNA in both cotyledon and roots from MNSV‐infected melon plants (10 dpi) by tissue printing hybridization. Impressions of healthy tissues hybridized using the vRNA complementary riboprobe are shown. The contour of the cotyledons is shown by a broken line. (b) Northern‐blot analysis of the dsRNAs isolated either from the cotyledons or from the roots of MNSV‐infected melon plants and hybridized against an MNSV vRNA complementary riboprobe. A negative control from healthy tissue (cotyledons and roots mix) is shown. The position of the MNSV gRNA and both sgRNAs is indicated on the right.
Localization of MNSV in source and transport tissues
Besides the macroscopic tissue‐printing hybridization study, a fine microscopic imaging of MNSV RNA accumulation at the cellular level was also achieved by light microscopy in situ hybridization, using the MNSV cvRNA ribroprobe to reveal their translocation route in melon plants. Samples from both infected and uninfected melon plants showing a similar developmental stage to that described above (Fig. 1) were processed. MNSV infection of melon plants was achieved by mechanical inoculation of MNSV virions over the upper surface of the cotyledons and cross‐sections from inoculated cotyledons, including both symptomatic and asymptomatic areas, were analysed. Consistent with the results obtained by the tissue‐printing analysis, in situ hybridization showed that MNSV vRNAs (blue signal) were specifically located in the areas around and enclosed by the local lesions (Fig. 3a,b) where epidermal (E), palisade (Pp) and mesophyll (M) cells, as well as vascular parenchyma (VP) and bundle sheath (BS) cells were infected. However, vascular bundles (VB) from major veins showed no viral hybridization signals in phloem (companion or intermediate cells and sieve elements, CC and SE, respectively) components or xylem vessels (Fig. 3c,d). Although virus loading was not addressed here, this observation clearly suggests that virus must enter the phloem through minor veins, as observed in other virus–host combinations (for a review see Nelson and van Bel, 1998). The viral signal was regularly distributed in the abaxial half of the cotyledon within the symptomatic area, whereas some groups of non‐infected cells were observed in the adaxial half (Fig. 3a,b), suggesting that cell‐to‐cell movement of vRNAs was more efficient on the abaxial face. This situation is most probably the consequence of the difference between the number and size of the different cell types located on each side. We therefore investigated the pathway for the phloem‐dependent movement of MNSV in the stem of melon plants.
Figure 3.
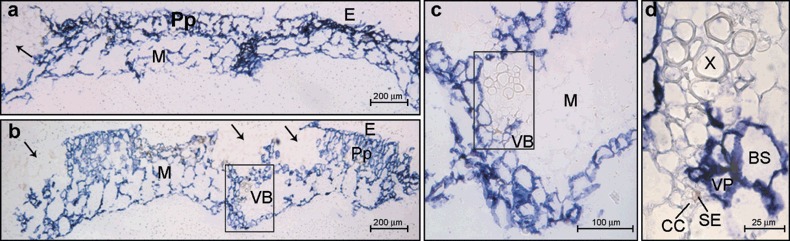
Microscopic imaging of the cellular localization of Melon necrotic virus (MNSV) RNAs by in situ RNA–RNA hybridization with a cvRNA riboprobe in cotyledons of systemically infected melon plants (6 dpi). The blue colour indicates the presence of target viral RNAs. (a,b) Cross‐sections of symptomatic and asymptomatic areas from two different cotyledons showing the viral‐specific blue signal in: epidermal cells, E; palisade parenchyma, Pp; and mesophyll cells, M. Black arrows point to the uninfected areas of the adaxial face of the cotyledon. The area inside the black box in panel (b) is enlarged in panel (c) to show a closer view of the cotyledon vascular bundle (VB). Panel (d) is a higher magnification of the same vascular bundle from panel (c) showing that vascular parenchyma (VP) and bundle sheath (BS) cells were infected but not the companion cells–sieve elements (CC–SE) complex.
Members of the families Solanaceae and Cucurbitaceae have a distinctive vascular anatomy, consisting of two types of phloem: fascicular and extrafascicular. The fascicular phloem is located in both the external (eP) and the internal (iP) phloem surrounding the xylem, and forming the bicollateral bundle VB (Hayward, 1938; McCauley and Evert, 1998a,b; Turgeon, 1989) (Fig. 4a,b). The extrafascicular phloem consists of both longitudinal strands of SE–CC complexes bordering the eP and iP (bundle‐associated extrafascicular phloem) and an anastomosing network in the cortex (cortical extrafascicular phloem) (Crafts, 1932; Fischer, 1884; Golecki et al., 1999). Thus, the route followed by the MNSV RNAs towards the sink tissues (roots, young leaf and apical shoot) was investigated by analysing cross‐sections of the stem taken at different distances, up and down, from the insertion point of the inoculated cotyledon. The internodal cross‐sections of the stem just above the cotyledon node revealed a poor virus‐specific signal mainly restricted to groups of cortex cells as well as to the internal fascicular phloem of some of the bicollateral vascular bundles (Fig. 4c–e).
Figure 4.
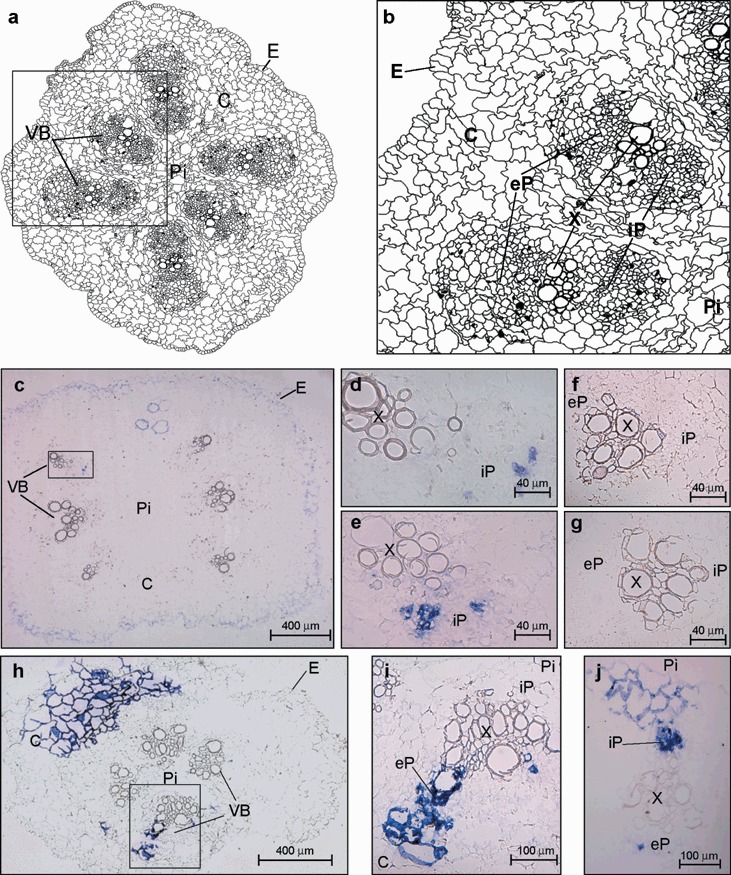
(a) Schematic representation of a cross‐section of the melon stem showing six vascular bundles (VB), the pith (Pi), the cortex (C) and the epidermis (E). (b) Diagram from the area enclosed by the black box in (a) showing an enlarged view of the vascular anatomy. The components of VB are designated as external, eP, and internal, iP, fascicular phloem, respectively and X, xylem. (c–j) Microscopic imaging of the cellular localization of the Melon necrotic spot virus (MNSV) RNAs by in situ RNA–RNA hybridization with a cvRNA riboprobe in transport tissues of systemically infected melon plants (6 dpi). The blue colour indicates the presence of target viral RNAs. (c) Internodal cross‐section of the stem region above the cotyledonary node showing MNSV RNA‐specific signals in cortical (C), and vascular bundle (VB) cells. The VB enclosed in the black box in panel (c) is enlarged in panel (d). (e) A closer view of another vascular bundle from a different cross‐section of the stem. (f,g) Two vascular bundles from different stem cross‐sections of healthy melon plant. (h) Hypocotyl cross‐section with only four vascular bundles showing the MNSV RNA‐specific signals in cortex (C) and vascular bundle (VB) cells. Panel (i) is an enlarged image of the vascular bundle enclosed by the black box in panel (h). (j) Vascular bundle from a hypocotyl cross‐section at 10 dpi where iP shows high hybridization levels.
The infected cells in the cross‐section from the hypocotyl (below the cotyledonary node) were located in both the cortex and the fascicular phloem but in this case in the eP (6 dpi) (Fig. 4h,i). Signal intensity increased as the analysed hypocotyl section approached the root and, interestingly, in later infection stages (10 dpi), the viral‐specific signal inside the VB of the hypocotyl region of the stem was mainly located in the iP (Fig. 4j). As virus‐specific signal was first detected in the hypocotyl rather than in the stem parts above the cotyledon node, the differential phloem location strongly suggests that MNSV root‐ward movement occurs through eP while iP is used for the shoot‐ward translocation.
Localization of MNSV in roots
MNSV infection of roots was also observed by in situ hybridization of both longitudinal and cross‐sections of the primary root tips as well as on lateral root primordia. Roots of dicotyledonous plants have centrally located xylem with a variable number of extensions projecting outward. The phloem tissue lies between these radiating arcs of xylem, generating a central structure known as the vascular cylinder (VC), which is enclosed by the innermost layer of the cortex (C), termed the endodermis (E). Unlike the above results obtained with the other organs, which showed a specific signal for distinct tissue areas, longitudinal sections of the primary root tip showed MNSV infection homogeneously distributed through all visible tissues (C and E), excluding some discrete cells and the root cap (RC) at 6 dpi (Fig. 5a,b). To determine better where the boundary of the infection at the primary meristem was located, a longitudinal section of the root tip, which this time included the central axis, was taken. It was seen that the non‐infected sector extended approximately 500 µm from the root ending, and, as before, included the root cap but also the tissues located in the zone of cell division (procambium, PC, ground meristem, GM, protoderm, PD, and apical meristem, Me) (Fig. 5c). Moreover, in order to study the virus invasion of the vascular cylinder, cross‐sections were taken from both the cell elongation zone and the cell division zone (Fig. 5d,e, respectively). In the former, the viral‐specific signal was homogeneously distributed in the whole section, including the epidermis, cortex and the phloem inside the vascular cylinder but not the xylem. In the zone of cell division, the label was limited to both the vascular cylinder (only inside the phloem components) and the surrounding cortex cells.
Figure 5.
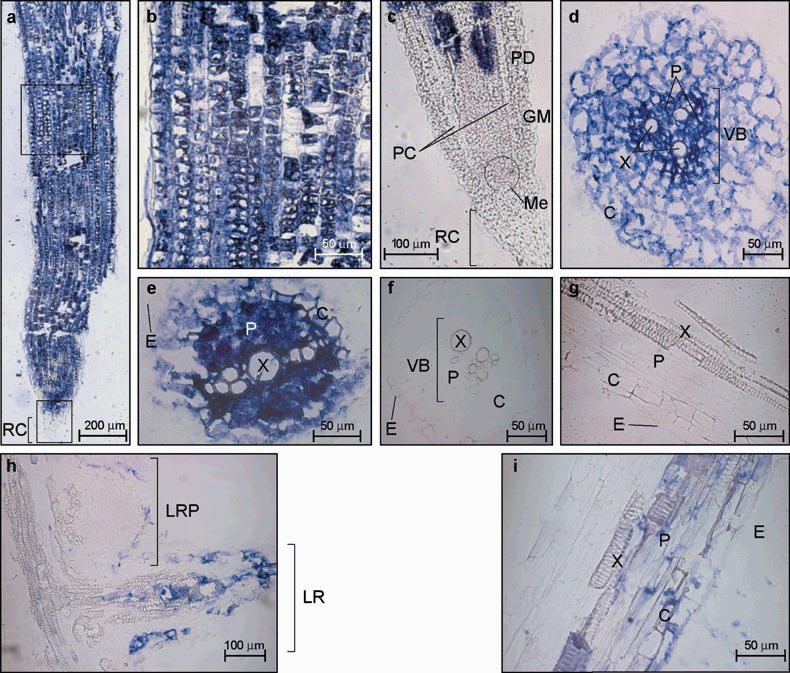
Microscopic imaging of the cellular localization of the Melon necrotic spot virus (MNSV) RNAs by in situ RNA–RNA hybridization with a cvRNA riboprobe in both the primary root tip (panels a–e) and lateral roots (panels h and i) of systemically infected melon plants. The blue colour indicates the presence of target viral RNAs. (a) Longitudinal section of a primary root tip. RC, root cap. (b) Enlarged image of the distal root‐tip sector enclosed by the upper black box from the longitudinal section of panel (a) showing discrete uninfected cells. (c) Longitudinal section through the central axis of a primary root tip (lower black box in panel (a)). MNSV invasion was not allowed at RC and primary meristem including protoderm (PD), ground meristem (GM), procambium (PC) and apical meristem (Me). (d) Cross‐section of the zone of cell elongation of primary root tip. (e) Cross‐section of the zone of cell division of primary root tip. (f,g) Cross‐section and longitudinal section of a primary root tip from healthy tissue. (h) Longitudinal section of both a lateral root primordial (LRP) and an elongated lateral root (LR). (i) Enlarged image of a longitudinal section of a lateral root. VB, vascular cylinder; E, epidermis; C, cortex; X, xylem; P, phloem.
Finally, it was hypothesized that viruses might also be delivered to developing lateral roots. The structure of the lateral roots is similar to that of the main root and the advance of virus infection through them might be expected to resemble that of the primary root. However, Tobacco mosaic virus (TMV) surprisingly replicates within the initial meristem of the lateral root primordia of Nicotiana benthamiana plants although its progress is quickly suppressed by gene silencing mechanisms, thus preventing replication in lateral roots (Valentine et al., 2002). Therefore, in situ hybridization of a longitudinal section of the primary root, including lateral roots, was performed to study the spread of MNSV within these organs (Fig. 5h). A section was taken 20 dpi to allow the putative establishment of the plant defence mechanisms. The elongation of a lateral root begins with the formation of a meristematic region in the outer boundary of the vascular cylinder known as the pericycle. Unlike the case of TMV infection of N. benthamiana, the root primordium and so root meristematic cells of melon plants showed no MNSV infection (see the lateral root in the upper part of Fig. 5h). As the lateral root grows it is pushed through the endodermis and cortex until it ruptures the surface and begins to elongate (Fig. 5h). At that more advanced development stage, MNSV was not only detected inside the vascular cylinder but also within some cortical and epidermic cells and the differentiating tracheary elements of the xylem in the lateral root (see the lower part of Fig. 5h,i) of melon lateral roots.
Localization of MNSV in aerial sink tissues
After long‐distance trafficking through the phloem, viruses are unloaded into growing sink tissues such as the shoot tip and young leaves. The shoot tip of melon plants is flanked by the petioles of both developed and young leaves (see the plant diagram in 1, 6) and consists of several shoot apical meristem units and immature leaves (Fig. 6c). A general view of a shoot tip from an infected melon plant after in situ hybridization is shown in Fig. 6(a,a′). The specific viral signal was heterogeneously distributed among the different parts of the shoot tip. Interestingly, an intense signal was observed in the stem pith just below each apical meristem (Me) and leaf primordium (LP) but a viral signal was always absent from the Me (Fig. 6a,b) even in later infection stages (10 dpi, Fig. 6d,e). During the last infection phase, the virus‐specific signal was more homogeneously distributed along the shoot tip, including cortex, stem pith and VB where MNSV was located exclusively in the phloem components (Fig. 6a–f).
Figure 6.
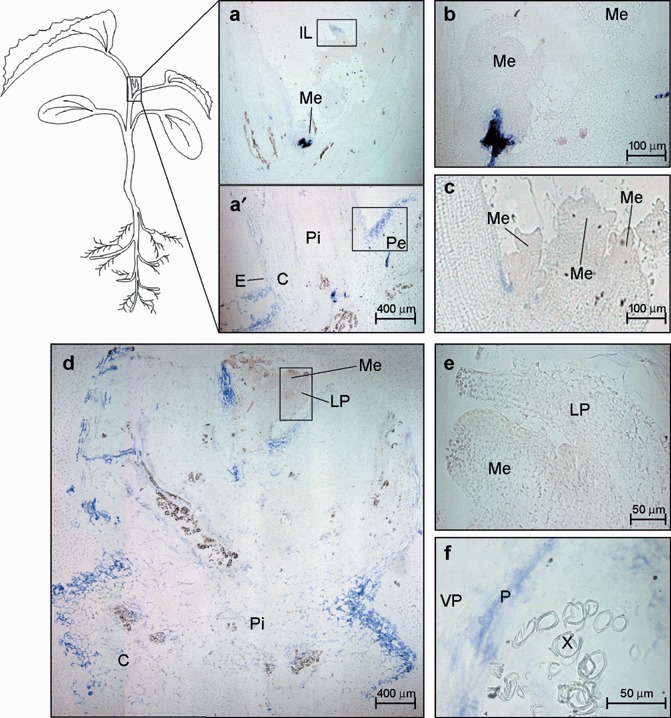
Microscopic imaging of the cellular localization of the Melon necrotic spot virus (MNSV) RNAs by in situ RNA–RNA hybridization with a cvRNA riboprobe in the shoot tip of systemically infected melon plants. The blue colour indicates the presence of target viral RNAs. (a,a′) Longitudinal section of the apical shoot tip showing the cortex (C), the stem pith (Pi), the insertion point of the young leaf petiole (Pe), an immature leaf (IL) and an apical meristem (Me) at 6 dpi. The meristem in panel (a) is enlarged in panel (b). The areas enclosed by both the lower and upper black boxes are enlarged in Fig. 7 in panels (a) and (d), respectively. (c) Longitudinal section of the multi‐meristematic apical shoot tip from a healthy melon plant. (d) Longitudinal section of the apical shoot tip at 10 dpi. Enlarged images of the apical meristem and the leaf primordia (LP) (black box) as well as a vascular bundle showing the MNSV infection into phloem (P) and vascular parenchyma (VP) are displayed on panels (e) and (f), respectively.
The virus‐specific signal in longitudinal sections of the young leaf petiole at the stem insertion point was mainly found in the epidermis and adjacent cortex cells and in some of the vascular components, including phloem and VP cells (Fig. 7a,b). The results were similar in the more distal longitudinal sections but no infected epidermis cell was observed (Fig. 7c). By contrast, in very young leaves, the specific viral signal was located in both the epidermis and mesophyll cells (Fig. 7d), as well as in the protophloem of the VB (Fig. 7d,e). Moreover, in the more distal cross‐sections from the stem insertion point of the developed leaf petiole (cross‐sections were taken at the leaf insertion point) the viral RNA signal was homogenously distributed throughout the epidermis (E), the components of both iP and eP, as well as in the vascular parenchyma (VP) cells and some groups of cortical cells close to the vascular bundle (VB) (Fig. 7f,g). Tissue‐printing of the corresponding developed leaf is shown in the lower part of Fig. 1(a).
Figure 7.
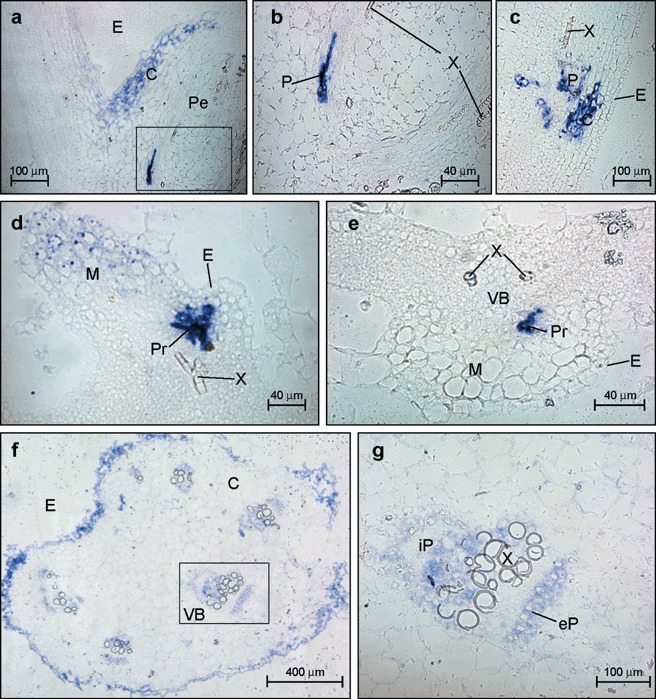
Microscopic imaging of the cellular localization of the Melon necrotic spot virus (MNSV) RNAs by in situ RNA–RNA hybridization with a cvRNA riboprobe in both the petiole of a young leaf (panels a–c), an immature leaf (panels d and e) and a petiole of a developed leaf (panels f and g) of systemically infected melon plants. The blue colour indicates the presence of target viral RNAs. (a) Longitudinal section of the young leaf petiole (Pe) at the stem insertion point showing MNSV infection (blue signal) at the epidermis (E) and adjacent cortical (C) cells. (b) A closer view of a vascular bundle (VB) showing MNSV infection on phloem (P) but not in xylem (X) components. (c) Longitudinal section of the young leaf petiole (Pe) at distal region from their stem insertion point. (d,e) Longitudinal and cross‐sections, respectively, of an immature leaf showing MNSV infection on immature epidermis and mesophyll cells (M) as well as in the protophloem (Pr). X, xylem. (f) Cross‐section of a developed leaf petiole taken close to the leaf insertion point. (g) A closer view of the vascular bundle inside the box in panel (f). The components of VB are designated as external, eP, and internal, iP, fascicular phloem, respectively.
DISCUSSION
In the present study, both macroscopic and microscopic images of the pathway used by the MSNV RNAs to systemically invade its natural host were obtained. As stated above, MNSV is transmitted by soil fungi. Here, we studied the route of viral trafficking after mechanical inoculation, which apparently could not reflect the natural pathway following transmission by the fungus. However, previous evidence indicates that this should not be the case. The resistance phenotype of different melon cultivars was similar after either mechanical or fungal transmission (see below), indicating that the results obtained here can be extrapolated, with some limitations, to a natural infection. One of these limitations would be the potential involvement of xylem tissue in viral trafficking after virus acquisition from the fungal vector. Viral RNAs were detected in xylem cells of young developing root tissue and at the hydathodes of primary leaves and, thus, the potential contribution of these vascular elements to the systemic movement of MNSV after fungal transmission cannot be ruled out. Data from tissue‐printing hybridization analysis and those from in situ hybridization were compatible with a route existing throughout the phloem vascular components for the systemic spreading of MNSV, parallel to the assimilate transport route from source to sink organs, similarly to that previously reported (Andrianifahanana et al., 1997; Cheng et al., 2000; Roberts et al., 1997). As revealed by the in situ hybridization studies, MNSV RNAs reached the vein of the inoculated cotyledon after cell‐to‐cell movement through all non‐vascular cell types. The accumulation of MNSV RNAs was detected in BS and VP cells but not in the CC–SE complex of major veins, suggesting that loading occurred specifically in minor veins or that the transit of the virus through these phloem components was too transitory to be detected by the techniques employed. Interestingly, the absence of viral infection in the CC–SE has previously been reported for other virus–host combinations such as Cucumber green mottle mosaic virus (CGMMV) in cucumber (Moreno et al., 2004) and Sunn‐hemp mosaic virus (SHMV) in Phaseolus vulgaris (Ding et al., 1998), the VP–CC boundary acting as an important barrier for systemic movement (Ueki and Citovsky, 2007; Waigmann et al., 2004).
Direct evidence of long‐distance MNSV movement through the phloem was obtained by analysis of the transport tissue in stem and hypocotyl. MNSV RNAs were localized in the CC–SE complex of the eP in the hypocotyl sections in early stages of infection, whereas at later stages viral RNAs were observed in the iP. In addition, cross‐sections of the stem above the cotyledon internode revealed the presence of viral RNAs in the iP. This differential distribution clearly reflects phloem specialization for the downward and upward transport of virus, as previously described for other virus–host interactions (Andrianifahanana et al., 1997; Barker and Harrison, 1986; Cheng et al., 2000; Derrick and Barker, 1992, 1997). However, unlike the results described for Pepper mottle virus (PepMoV) (Andrianifahanana et al., 1997) and TMV (Cheng et al., 2000) the downward movement of MNSV seems to be faster than its upward movement. At the beginning of the infection no transport between hypocotyl eP and iP was observed, suggesting a reduced or strictly controlled communication between both types of phloem. This new barrier to the advance of MNSV systemic infection may be related to the restriction of the long‐distance movement of the virus observed in varieties of melon such as C. melo L. cv. Doublon carrying the resistance controlled by the two dominant genes Mnr 1 and Mnr 2 (Mallor et al., 2003). Following mechanical inoculation of the cotyledons, plants of cv. Doublon showed local necrotic lesions in cotyledons but failed to develop any systemic symptoms. Additionally, MNSV infection was unable to spread beyond the base of the hypocotyl of these resistant plants when inoculation was performed on roots by means of O. bornovanus (Mallor et al., 2006). Our results shed some light on a possible mechanism to explain this type of resistance, i.e. in this resistant variety there should be a blockage of either iP loading or eP to iP transport of MNSV. This would also be valid for other virus–host systems, e.g. Capsicum annuum L. cv Avelar plants that allow downward movement of PepMoV through eP but restrict the upward movement through iP, resulting in virus‐free young tissues (Guerini and Murphy, 1999).
The unloading of MNSV RNAs at active sink organs occurs in an early infection stage given that these viruses consistently accumulated in both young leaves and roots. However, analysis of fully developed leaves revealed a variable pattern in the infection that included three different states whereby either the entire or only the basal portion of the leaf is infected while in other older leaves no virus signal was detected. The ability of the growing young leaves and photosynthetically active developed leaves to act as either sink or source tissue, respectively, accounts for the difference noted between them. Young leaves are strong sink tissues that allow the unloading of photoassimilates (Schaffer et al., 1996) as well as MNSV in the entire leaf, as observed in all the samples analysed. However, developed leaves have undergone a sink‐to_source transition characterized by an end to the import of both photoassimilates and virus as a result of the loss of symplastic continuity between the phloem and the surrounding tissue and due to the beginning of the export towards new sinks (Ding et al., 1988). Before this sink‐to‐source conversion is completed, however, an intermediate stage where both sink and source regions are simultaneously located in the same organ takes place. In this scenario, as occurs in different plant–virus systems (Cheng et al., 2000; Mas and Pallas, 1996; Roberts et al., 1997) the differing ability of MNSV to invade a leaf is related to the development stage of this organ (sink, sink‐to‐source or source) at the moment that the infection reaches it. Therefore, MNSV moves via the phloem with photoassimilates towards sink regions that will be easily infected, while source areas will not become infected because of their symplastic isolation. Interestingly, even though MNSV unloading from the vascular system did not occur in the apical portion of leaves undergoing the sink–source transition, discrete spots were frequently observed at the margins of these source areas. This situation is most probably due to the virus exiting from the vein endings. In these areas, a direct connection between the SE–CC complex and some mesophyll cells could exist, as has been described in Phaseolus vulgaris and C. annuum (de Morretes, 1962) or Beta vulgaris and Beta syringa (Esau, 1967, 1977), and so the complex is not symplastically isolated given the absence of the VP–BS/SE–CC boundary.
As well as source regions, shoot apical meristem (SAM) also evades the infection of many viruses such as TMV (Cheng et al., 2000), Potato leafroll virus (PLRV) (Faccioli et al., 1988), Carnation mottle virus (CarMV), Carnation vein mottle potyvirus (CVMV), Carnation latent carlavirus (CLV), Carnation etch ring caulimovirus (CERV) (Gosálvez‐Bernal et al., 2006), and others (Mori et al., 1982; reviewed in Hull, 2002). The reason why this tissue fails to allow virus replication remains unclear, although recent results indicating that RNA silencing mechanisms could be involved are contributing to clarify this point (Foster et al., 2002; Schwach et al., 2005). In fact, some viruses such as CMV may colonize shoot apical meristem cells at an early stage (7 dpi), although this tissue was seen to recover from infection (14–18 dpi) as a result of RNA silencing (Mochizuki and Ohki, 2004). An alternative explanation that cannot be ruled out is the existence of a plamodesmal selectivity operating in the virus exclusive of the meristem (Gisel et al., 1999; Kim et al., 2005). The same situation would occur at the root tips. In this case, the restriction would rely on the existence of a post‐phloem domain in roots that restricts the entrance of large macromolecules into the root tip, as has been demonstrated by the use of GFP fusions in Arabidopsis (Stadler et al., 2005). The root tips of plants infected with one of several viruses, such as Tomato bushy stunt virus (TBSV; Appiano and D’Agostino, 1983) and Clover yellow vein mosaic virus (ClYMV; Smith and Schlegel, 1964), have been found to be virus‐free. Despite the ability of MNSV to infect plants systemically, viral RNAs were excluded from both shoot and root apical meristem regions, suggesting that the defence mechanism was established faster than viral infection in these plant tissues.
The last step in the virus cycle involves its accumulation in organs or tissues, where it can be easily acquired by the corresponding vectors. As MNSV is transmitted by a soil‐borne fungal vector between roots (Furuki, 1981; Lange and Insunza, 1977), the observed accumulation of MNSV to high titres in roots seems advantageous for transmission. High titres in roots have also been observed for other viruses that are not fungus‐transmitted, but in that case viral accumulation was temporal and time‐linked to a specific developmental stage, i.e. the formation of the inflorescence bud (Lunello et al., 2007). MNSV RNAs accumulated strongly in all types of root cells but not in the root meristem. A stronger, more invasive infection of the root system (including colonization of the meristem cells) has been described for members of the genus Tobravirus (MacFarlane and Popovich, 2000) transmitted by plant‐parasitic nematodes that feed on roots (Ploeg et al., 1996). Interestingly, replication of TMV in roots could be prevented not only in the root meristem but also at a short distance before the lateral root tip by gene‐silencing mechanisms originated in the corresponding active meristem (Valentine et al., 2002). Given this scenario, the root defence mechanisms may operate at different degrees of intensity depending on the virus–host system in question. Nematodes destroy the root cells upon which they feed, although 5% of perforated cells remained alive and fully functional for the establishment of virus (Karanastasi et al., 2003). Therefore, the efficient transmission of tobraviruses would require faster suppression of the plant defence response than of MNSV, which is transmitted by a non‐destructive vector. Interestingly, the concentration of dsRNAs, which are considered as initiators of a silencing signal that generates a systemic post transcriptional gene silencing (PTGS) response, was approximately five‐fold lower than in cotyledons, even though the vRNA/cvRNA ratio is similar in both organs. The low accumulation of these viral molecules could be related to a viral mechanism against gene silencing but may also indicate reduced virus replication in this tissue. Another possibility is that replication in roots is favoured but insufficient to maintain the presence of high levels of MNSV RNAs, which, alternatively, could be achieved by a root tropism of viral infection. Whether this root destination‐selective movement is controlled by a host or a viral‐specific factor such as the tobravirus 2b helper protein (Valentine et al., 2004) remains unclear.
EXPERIMENTAL PROCEDURES
Virus source
The MNSV‐Al isolate used was originally obtained from a melon plant infected in the field (Cucumis melo L.) collected in Murcia (Spain) and kindly provided by Dr F. Botella (Miguel Hernandez University, Alicante, Spain) (Gosálvez et al., 2003). MNSV‐infected leaf tissue was homogenized in 30 mm phosphate buffer (pH 7.0) containing 20 mm mercaptoethanol, and the crude extract was used as inoculum. Subsequently, the virus was purified, essentially as described by Díez et al. (1998) for carmoviruses, and used to propagate infection by serial mechanical transmissions to melon plants (Cucumis melo L. ‘Galia’).
Plant material and bioassays
A total of 13 experiments including inoculation and sampling were carried out. Each experiment consisted of growing in 12 different pots a total of 36 melon plants (three plants per pot), from which ten pots (30 plants) were mechanically inoculated at the stage with no true leaf present, while the other two pots (six plants) were either mock‐inoculated or kept healthy as controls.
Bioassays including inoculated, mock‐inoculated and healthy plants were performed in a climatic chamber with a 16/8‐h day–night period, and temperatures of 25 and 18 °C, respectively, with an irradiance of 210 µmol photons/m2/s.
Sampling was done at different days post inoculation (dpi) from the same plant for the tissue‐printing as well as for the microscopy studies from the different organs studied (cotyledons, young and mature leaves, petioles, stems, roots and shoot tips).
Synthesis of digoxigenin‐labelled RNA probes
Digoxigenin‐labelled RNA probes were synthesized from the recombinant plasmid pMNSV‐CP containing the partial sequence of the MNSV coat protein (CP) gene placed between the T3 and T7 RNA polymerase promoters (Gosálvez et al., 2003). This clone was linearized and then used as template in in vitro transcriptions in the presence of either T3 or T7 RNA polymerase and DIG‐11‐UTP to obtain non‐isotopic viral riboprobes containing either viral RNA (vRNA) or complement to viral RNA (cvRNA), respectively (Pallás et al., 1998a), as recommended by the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). Both types of RNA transcripts were recovered by ethanol precipitation and resuspended in sterile water for use in later experiments.
Tissue‐printing hybridization
Melon leaves, cotyledons, petioles, stems and roots, as well as longitudinal sections of the shoot tips, were sampled at different days post inoculation (from 4 to 20 dpi) and directly pressed onto nylon membranes (Roche Molecular Biochemicals) as previously described (Más and Pallás, 1995). A total of 17 tissue‐printing experiments were performed, which included, in each experiment, all the organs analysed (cotyledons, leaves, petioles, stems, roots and shoot tips). The membranes were air‐dried and nucleic acids were bound by using a UV cross‐linker (700 × 100 µJ/cm2). Viral RNA detection was carried out as previously described (Pallás et al., 1998b), using the riboprobes described above (cvRNA or vRNA). Mock‐inoculated or healthy plants were used in each of the experiments and treated similarly.
dsRNA extraction and Northern blot assay
Inoculated cotyledons or roots from symptomatic melon plants were used to isolate double‐stranded RNAs (dsRNAs) by means of standard procedures (Astruc et al., 1996; Covelli et al., 2004) and subsequently purified by chromatography through CF11 cellulose (Whatman). Purified dsRNAs were electrophoresed through polyacrylamide 5% TAE 1× gel and transferred to positively charged nylon membranes (Roche Molecular Biochemicals) for Northern blot assay. Membranes were air‐dried and nucleic acids were bound by using a UV cross‐linker (700 × 100 µJ/cm2). Specific viral dsRNA was detected as described previously (Pallás et al., 1998a,b) by using the cvRNA digoxigenin‐labelled riboprobe described above.
Tissue fixation and embedding
Different organs of both healthy and MNSV‐infected melon plants, including cotyledons, young and developed leaves and their corresponding petioles, roots and stems, as well as shoot tips were sampled (from 4 to 20 dpi) and then fixed. Briefly, samples were vacuum‐infiltrated for 2 min in a freshly made mixture of 4%p‐formaldehyde and 2.5% glutaraldehyde in 0.2 m phosphate buffer (pH 7.2) at 4 °C for 4 h, essentially as described (García‐Castillo et al., 2001, 2003). After fixation, they were washed in 0.2 m phosphate buffer (pH 7.2), dehydrated in a tertiary butyl alcohol series, infiltrated and embedded in paraffin (Paraplast Plus, Sherwood Medical Co., St Louis, MO). Individual blocks of paraffin were sectioned using a Reichert‐Jung 2030 rotary microtome set to 8 or 10 µm. The sections were adhered to glass slides precoated with APTES (3‐amino‐propyl‐triethoxy‐silane) (Sigma), by heating them at 40 °C on a hot plate. The slides were allowed to stand overnight before further processing to ensure that the sections were tightly adhered to the slide.
To obtain some preliminary information about the anatomy of the melon organs we used a general staining for paraffin‐embedded plant material (Gerlach, 1969), performed on sections from the material sampled as described above under Plant material and bioassays.
In situ hybridization
In situ hybridization (ISH) was performed as previously described (García‐Castillo et al., 2001, 2003, Más et al., 2000). A total of 17 ISH experiments were performed, which included, in each experiment, slides containing sections from all the organs analysed (cotyledons, leaves, petioles, stems, roots and shoot tips), and from the different time‐points studied. Briefly, tissue sections on slides were de‐waxed with xylene (twice for 10 min each) and dehydrated with a series of decreasing ethanol concentrations from absolute ethanol to water. They were then treated with proteinase K (1 µg/mL, in 100 mm Tris‐HCl, 50 mm EDTA, pH 8.0) for 30 min at 37 °C, followed by a wash with PBS (135 mm NaCl, 1.5 mm KH2PO4, 8 mm Na2HPO4, 2.7 mm KCl, pH 7.2), for 2 min. Thereafter, proteinase K was blocked by glycine (2 mg/mL) in PBS and two further PBS washes of 30 s each. Hybridization was performed with 100 µL of hybridization solution consisting of 50% deionized formamide, 6× SSC, 3% SDS, yeast tRNA 0.1 mg/mL and a probe concentration of 200 ng/mL. The dig‐RNA probe used was always the cvRNA probe described above under Synthesis of digoxigenin‐labelled RNA probes. Hybridization was performed at 55 °C overnight. Hybridized probes were detected by antibody incubation using sheep anti‐digoxigenin conjugated to alkaline phosphatase (Roche Molecular Biochemicals) diluted 1 : 500 in 0.5% bovine serum albumin (BSA) in TBS buffer for 90 min. For colorigenic detection, slides were incubated with nitroblue tetrazolium and 5‐bromo‐4‐cloro‐3‐indolyl phosphate (NBT/BICP) (Roche Molecular Biochemicals) in equilibration buffer (100 mm Tris‐HCl, 100 mm NaCl, 50 mm MgCl2, pH 9.5). Thus, the hybridization signal is seen as blue areas in the sections. Slides were then dehydrated through an ethanol series of increasing concentration, immersed in xylene and mounted using Merckoglass (Merck, Darmstad, Germany). Sections were examined using a DMRB LEITZ microscope (Leica Microsistemas S.A., Barcelona, Spain) and photographed with a Leica DC 500 digital camera. Controls were made by performing, in each of the experiments, ISH on sections taken from healthy or mock‐inoculated plants, as well as on infected samples incubated without the probe. We did not use any contrasting dye after the ISH because the colorigenic detection procedure used (NBT/BCIP) is not compatible with any dye. This fact makes it more difficult to image and analyse those areas from the tissue that do not contain the virus, but did help us to decide, together with the controls performed, if a particular ISH experiment worked correctly.
ACKNOWLEDGEMENTS
This work was supported by grants BIO2002‐04099‐C02‐01 and BIO05‐7331 from DGICYT and GV04B‐183 from Generalitat Valenciana. B.G.‐B. was the recipient of a fellowship from the Instituto de Fomento‐Fundación Seneca from Comunidad de Murcia. A.G. and J.A.N. are recipients of a PhD fellowship and an I3P contract from the Spanish Ministerio de Educacion y Ciencia and the Consejo Superior de Investigaciones Científicas, respectively.
REFERENCES
- Al‐Kaff, N.S. and Covey, S.N. (1996) Unusual accumulations of Cauliflower mosaic virus in local lesions, dark green leaf tissue, and roots of infected plants. Mol. Plant–Microbe Interact., 9, 357–363. [Google Scholar]
- Andrianifahanana, M. , Lovins, K. , Dute, R. , Sikora, E. and Murphy, J.F. (1997) Pathway for phloem‐dependent movement of Pepper mottle potyvirus in the stem of Capsicum annuum . Phytopathology, 87, 892–898. [DOI] [PubMed] [Google Scholar]
- Aoki, K. , Suzui, N. , Fujimaki, S. , Dohmae, N. , Yonekura‐Sakakibara, K. , Fujiwara, T. , Hayashi, H. , Yamaya, T. and Sakakibara, H. (2005) Destination‐selective long‐distance movement of phloem proteins. Plant Cell, 17, 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appiano, A. and D’Agostino, G. (1983) Distribution of Tomato bushy stunt virus in root tips of systemically infected Gomphrena globosa . J. Ultrastruct. Res. 85, 239–248. [DOI] [PubMed] [Google Scholar]
- Astruc, N. , Marcos, J.F. , Macquaire, G. , Candresse, T. and Pallas, V. (1996) Studies on the diagnosis of hop stunt viroid in fruit trees: identification of new hosts and application of a nucleic acid extraction procedure based on non‐organic solvents. Eur. J. Plant Pathol. 102, 837–846 [Google Scholar]
- Barker, H. and Harrison, B.D. (1986) Restricted distribution of Potato leaf roll virus antigen in resistant potato genotypes and its effect on transmission of the virus by aphids. Ann. Appl. Biol. 109, 595–604. [Google Scholar]
- Brodersen, P. and Voinnet, O. (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22, 268–280. [DOI] [PubMed] [Google Scholar]
- Burgyan, J. (2006) Virus induced RNA silencing and suppression: defence and counter defence. J. Plant Pathol. 88, 233–244. [Google Scholar]
- Campbell, R.N. , Wipf‐Scheibel, C. and Lecoq, H. (1996) Vector‐assisted seed transmission of Melon necrotic spot virus in melon. Phytopathology, 86, 1294–1298. [Google Scholar]
- Cheng, N.H. , Lin, C.L. , Carter, S.A. and Nelson, R.S. (2000) Vascular invasion routes and systemic accumulation patterns of Tobacco mosaic virus in Nicotiana benthamiana . Plant J. 23, 349–362. [DOI] [PubMed] [Google Scholar]
- Covelli, L. , Coutts, R.H.A. , Di Serio, F. , Citir, A. , Acikgoz, S. , Hernandez, C. , Ragozzino, A. and Flores, R. (2004) Cherry chlorotic rusty spot and Amasya cherry diseases are associated with a complex pattern of mycoviral‐like double‐stranded RNAs. I. Characterization of a new species in the genus Chrysovirus . J. Gen. Virol. 85, 3389–3397. [DOI] [PubMed] [Google Scholar]
- Crafts, A.S. (1932) Phloem anatomy, exudation and transport of organic nutrients in cucurbits. Plant Physiol. 7, 183–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrick, P.M. and Barker, H. (1992) The restricted distribution of Potato leafroll luteovirus antigen in potato plants with transgenic resistance resembles that in clones with one type of host gene‐mediated resistance. Ann. Appl. Biol. 120, 451–457. [Google Scholar]
- Derrick, P.M. and Barker, H. (1997) Short and long distance spread of Potato leafroll luteovirus: effects of host genes and transgenes conferring resistance to virus accumulation in potato. J. Gen. Virol. 78, 243–251. [DOI] [PubMed] [Google Scholar]
- Díez, J. , Marcos, J.F. and Pallás, V. (1998) Carmovirus isolation and RNA extraction In: Methods in Molecular Biology, Volume 81: Plant Virology Protocols: From Virus Isolation to Transgenic Resistance (Foster G. and Taylor S., eds), pp. 211–217. Totowa, NJ: Humana Press Inc. [DOI] [PubMed] [Google Scholar]
- Ding, B. , Parthasarathy, M.V. , Niklas, K. and Turgeon, R. (1988) A morphometric analysis of the phloem‐unloading pathway in developing tobacco‐leaves. Planta, 176, 307–318. [DOI] [PubMed] [Google Scholar]
- Ding, X.S. , Carter, S.A. , Deom, C.M. and Nelson, R.S. (1998) Tobamovirus and potyvirus accumulation in minor veins of inoculated leaves from representatives of the Solanaceae and Fabaceae. Plant Physiol. 116, 125–136. [Google Scholar]
- Ding, X.S. , Shintaku, M.H. , Carter, S.A. and Nelson, R.S. (1996) Invasion of minor veins of tobacco leaves inoculated with tobacco mosaic virus mutants defective in phloem‐dependent movement. Proc. Natl Acad. Sci. USA, 93, 11155–11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, F. , Sangwan, R.S. and Sangwannorreel, B.S. (1994) Spread of Beet necrotic yellow vein virus in infected seedlings and plants of sugar‐beet (Beta‐vulgaris ). Protoplasma, 179, 72–82. [Google Scholar]
- Dunoyer, P. and Voinnet, O. (2005) The complex interplay between plant viruses and host‐RNA‐silencing pathways. Curr. Opin. Plant Biol. 8, 415–423. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1967) Minor veins in Beta leaves: structure related to function. Proc. Am. Philos. Soc. 111, 219–233. [Google Scholar]
- Esau, K. (1977) Anatomy of Seed Plants. New York: John Wiley and Sons. [Google Scholar]
- Faccioli, G. , Rubies‐Autonell, C. and Resca, R. (1988) Potato leafroll virus distribution in potato meristem tips and production of virus‐free plants. Potato Res. 31, 511–520. [Google Scholar]
- Fischer, A. (1884) Untersuchungen über das Siebröhrensystem der Cucurbitaceen. Berlin: Gebrüder Borntraeger. [Google Scholar]
- Foster, T.M. , Lough, T.J. , Emerson, S.J. , Lee, R.H. , Bowman, J.L. , Forster, R.L. and Lucas, W.J. (2002) A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell, 14, 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuki, I. (1981) Epidemiological Studies on Melon Necrotic Spot. Technical Bulletin 14. Shizuokaken, Japan: Shizuoka Agricultural Experiment Station. [Google Scholar]
- García‐Castillo, S. , Marcos, J.F. , Pallás V. and Sánchez‐Pina, M.A. (2001) Influence of the plant growing conditions on the translocation routes and systemic infection of Carnation mottle virus in Chenopodium quinoa plants. Physiol. Mol. Plant Pathol. 58, 229–238. [Google Scholar]
- García‐Castillo, S. , Sánchez‐Pina, M.A. and Pallás, V. (2003) Spatio‐temporal analysis of the RNAS, coat and movement (p7) proteins of Carnation Mottle Virus (CarMV) in Chenopodium quinoa plants. J. Gen. Virol. 84, 745–749. [DOI] [PubMed] [Google Scholar]
- Genovés, A. , Navarro, J.A. and Pallás, V. (2006) Functional analysis of the five Melon necrotic spot virus genome‐encoded proteins. J. Gen. Virol. 87, 2371–2380. [DOI] [PubMed] [Google Scholar]
- Gerlach, D. (1969) A rapid safranin‐crystal violet‐light green staining sequence for paraffin sections of plant materials. Stain Technol. 44, 210–211. [PubMed] [Google Scholar]
- Germudson, A. and Valkonen, J.P.T. (2006) P1‐and VPg‐transgenic plants show similar resistance to Potato virus A and may compromise long distance movement of the virus in plant sections expressing RNA silencing‐based resistance. Virus Res. 116, 208–213. [DOI] [PubMed] [Google Scholar]
- Gisel, A. , Barella, S. , Hempel, F.D. and Zambryski, P.C. (1999) Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana apices. Development, 126, 1879–1889. [DOI] [PubMed] [Google Scholar]
- Golecki, B. , Schulz, A. and Thompson, G.A. (1999) Translocation of structural P proteins in the phloem. Plant Cell, 11, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrick, B.J. , Kuhn, C.W. and Hussey, R.S. (1991) Restricted systemic movement of Cowpea chlorotic mottle virus in soybean with nonnecrotic resistance. Phytopathology, 81, 1426–1431. [Google Scholar]
- Gosálvez, B. , Navarro, J.A. , Lorca, A. , Botella, F. , Sánchez‐Pina, M.A. and Pallás, V. (2003) Detection of Melon necrotic spot virus in water samples and melon plants by molecular methods. J. Virol. Methods, 113, 87–93. [DOI] [PubMed] [Google Scholar]
- Gosálvez‐Bernal, B. , García‐Castillo, S. , Pallás, V. and Sánchez‐Pina, M.A. (2006) Distribution of carnation viruses in the shoot tip: exclusion from the shoot apical meristem. Physiol. Mol. Plant Pathol. 69, 43–51. [Google Scholar]
- Guerini, M.N. and Murphy, J.F. (1999) Resistance of Capsicum annum ‘Avelar’ to Pepper mottle potyvirus and alleviation of this resistance by co‐infection with Cucumber mosaic cucumovirus are associated with virus movement. J. Gen. Virol. 80, 2785–2792. [DOI] [PubMed] [Google Scholar]
- Haupt, S. , Cowan, G.H. , Ziegler, A. , Roberts, A.G. , Oparka, K.J. and Torrance, L. (2005) Two plant‐viral movement proteins traffic in the endocytic recycling pathway. Plant Cell, 17, 164–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward, H.E. (1938) The Structure of Economic Plants. New York: Macmillan Company. [Google Scholar]
- Herranz, M.C. and Pallas V. (2004) RNA‐binding properties and mapping of the RNA‐binding domain from the movement protein of Prunus necrotic ringspot virus . J. Gen. Virol. 85, 761–768. [DOI] [PubMed] [Google Scholar]
- Herranz, M.C. , Sanchez‐Navarro, J.A. , Sauri, A. , Mingarro, I. and Pallas, V. (2005) Mutational analysis of the RNA‐binding domain of the Prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell‐to‐cell movement. Virology, 339, 31–41. [DOI] [PubMed] [Google Scholar]
- Hibi, T. and Furuki, I. (1985) Melon Necrotic Spot Virus . CMI: AAB Descriptions of Plants Viruses no. 302. In: (eds) CMI/AAB Descriptions of plant viruses. Warwick, UK: Association of Applied Biologists.
- Hull, R. (2002) Mathews, Plant Virology, 4th edn. San Diego: Academic Press. [Google Scholar]
- Itaya, A. , Ma, F.S. , Qi, Y.J. , Matsuda, Y. , Zhu, Y.L. , Liang, G.Q. and Ding, B. (2002) Plasmodesma‐mediated selective protein traffic between ‘symplasmically isolated’ cells probed by a viral movement protein. Plant Cell, 14, 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanastasi, E. , Wyss, U. and Brown, D.J.F. (2003) An in vitro examination of Paratrichodorus anemones (Nematoda: Trichodoridae), with comments on the ability of the nematode to acquire and transmit Tobravirus particles. Nematology, 5, 421–434. [Google Scholar]
- Kim, I. , Kobayashi, K. , Cho, E. and Zambryski, P.C. (2005) Sub‐domains for transport via plasmodesmata corresponding to the apical‐basal axis are established during Arabidopsis embryogenesis. Proc. Natl Acad. Sci. USA, 102, 11945–11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, L. and Insunza, V. (1977) Root inhabiting Olpidium species: the O. radicale complex. Trans. British Mycol. Soc. 69, 377–384. [Google Scholar]
- Leisner, S.M. and Turgeon, R. (1993) Movement of virus and photoassimilate in the phloem: a comparative analysis. Bioassays, 15, 741–748. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. (2006) Plant viral movement proteins: agents for cell‐to‐cell trafficking of viral genomes. Virology, 344, 169–184. [DOI] [PubMed] [Google Scholar]
- Lunello, P. , Mansilla, C. , Sánchez, F. and Ponz, F. (2007) A developmentally linked, dramatic, and transient loss of virus from roots of Arabidopsis thaliana plants infected by either of two RNA viruses. Mol. Plant–Microbe Interact. 20, 1589–1595. [DOI] [PubMed] [Google Scholar]
- MacFarlane, S.A. and Popovich, A.H. (2000) Efficient expression of foreign proteins in roots from tobravirus vectors. Virology, 267, 29–35. [DOI] [PubMed] [Google Scholar]
- Mallor, C. , Alvarez, J.M. and Luis‐Arteaga, M. (2003) Inheritance of resistance to systemic symptom expression of Melon necrotic spot virus (MNSV) in Cucumis melo L. ‘Doublon’. Euphytica, 134, 319–324. [Google Scholar]
- Mallor, C. , Luis‐Arteaga, M. , Alvarez, J.M. , Montaner, C. and Floris, E. (2006) Resistance to Melon necrotic spot virus in Cucumis melo L. ‘Doublon’ artificially inoculated by the fungus vector Olpidium bornovanus . Crop Prot. 25, 426–431. [Google Scholar]
- Más, P. and Pallás, V. (1995) Non‐isotopic tissue‐printing hybridisation: a new technique to study long‐distance plant virus movement. J. Virol. Methods, 52, 317–326. [DOI] [PubMed] [Google Scholar]
- Mas, P. and Pallás, V. (1996) Long‐distance movement of Cherry leaf roll virus in infected tobacco plants. J. Gen. Virol. 77, 531–540. [DOI] [PubMed] [Google Scholar]
- Más, P. , Sánchez‐Pina, M.A. , Balsalobre, J.M. and Pallás, V. (2000) Subcellular localisation of Cherry leaf roll virus coat protein and genomic RNAs in tobacco leaves. Plant Sci. 153, 113–124. [DOI] [PubMed] [Google Scholar]
- McCauley, M.M. and Evert, R.F. (1988a) The anatomy of the leaf of potato, Solanum tuberosum L. ‘russet Burbank’. Bot. Gazette, 149, 179–195. [Google Scholar]
- McCauley, M.M. and Evert, R.F. (1988b) Morphology and vasculature of the leaf of potato (Solanum‐tuberosum ). Amer. J. Bot. 75, 377–390. [Google Scholar]
- Mochizuki, T. and Ohki, S.T. (2004) Shoot meristem tissue of tobacco inoculated with Cucumber mosaic virus is infected with the virus and subsequently recovers from infection by RNA silencing. J. Gen. Plant Pathol. 70, 363–366. [Google Scholar]
- Moreno, I.M. , Thompson, J.R. and García‐Arenal, F. (2004) Analysis of the systemic colonization of cucumber plants by Cucumber green mottle mosaic virus . J. Gen. Virol. 85, 749–759. [DOI] [PubMed] [Google Scholar]
- Mori, K. , Hosokawa, D. and Watanabe, M. (1982) Studies on multiplication and distribution of viruses in plants by the use of fluorescent antibody technique: I. Multiplication and distribution of viruses in shoot apices. Ann. Phytopathol. Soc. Jpn, 48, 433–443. [Google Scholar]
- De Morretes, B.L. (1962) Terminal phloem in vascular bundles of leaves of Capsicum annuum and Phaseolus vulgaris . Am. J. Bot. 49, 560–567. [Google Scholar]
- Navarro, J.A. , Genoves, A. , Climent, J. , Sauri, A. , Martinez‐Gil, L. , Mingarro, I. and Pallas, V. (2006) RNA‐binding properties and membrane insertion of Melon necrotic spot virus (MNSV) double gene block movement proteins. Virology, 356, 57–67. [DOI] [PubMed] [Google Scholar]
- Nelson, R.S. and Van Bel, A.J.E. (1998) The mystery of virus trafficking into, through and out of vascular tissue. Prog. Bot. 59, 476–533. [Google Scholar]
- Opalka, N. , Brugidou, C. , Bonneau, C. , Nicole, M. , Beachy, R.N. , Yeager, M. and Fauquet, C. (1998) Movement of Rice yellow mottle virus between xylem cells through pit membranes. Proc. Natl Acad. Sci. USA, 95, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J. and Santa Cruz, S. (2000) The great escape: phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 323–347. [DOI] [PubMed] [Google Scholar]
- Pallás, V. , Más, P. and Sánchez‐Navarro, J.A. (1998a) Detection of plant RNA viruses by non‐isotopic dot‐blot hybridisation In: ‘Methods in Molecular Biology’. Vol. 81: Plant Virology Protocols: From Virus Isolation to Transgenic Resistance (Foster G.D. and Taylor S.C., eds), pp. 461–468. Totowa, NJ: Humana Press Inc. [DOI] [PubMed] [Google Scholar]
- Pallás, V. , Sánchez‐Navarro, J.A. , Más, P. , Cañizares, M.C. , Aparicio, F. and Marcos, J.F. (1998b) Molecular diagnostic techniques and their potential role in stone fruit certification schemes. Options Méditer. 19, 191–208. [Google Scholar]
- Peremyslov, V.V. , Pan, Y.W. and Dolja, V.V. (2004) Movement protein of a closterovirus is a type III integral transmembrane protein localized to the endoplasmic reticulum. J. Virol. 78, 3704–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploeg, A.T. , Zoon, F.C. , Maas, P.W.T. (1996) Transmission efficiency of five tobravirus strains by Paratrichodorus teres . Eur. J. Plant Pathol. 102, 123–126. [Google Scholar]
- Prokhnevsky, A.I. , Peremyslov, V.V. and Dolja, V.V. (2005) Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. J. Virol. 7, 14421–14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere, C.J. and Rochon, D.M. (1990) Nucleotide‐sequence and genomic organization of Melon necrotic spot virus . J. Gen. Virol. 1, 1887–1896. [DOI] [PubMed] [Google Scholar]
- Roberts, A.G. , Santa‐Cruz, S. , Roberts, I.M. , Prior, D.A.M. , Turgeon, R. and Oparka, K.J. (1997) Phloem unloading in sink leaves of Nicotiana benthamiana: comparison of a fluorescent solute with a fluorescent virus. Plant Cell, 9, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Medrano, R. , Xoconostle‐Cazares, B. and Kragler, F. (2004) The plasmodesmatal transport pathway for homoeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 7, 641–650. [DOI] [PubMed] [Google Scholar]
- Santa Cruz, S. (1999) Perspective: phloem transport of viruses and macromolecules—what goes in must come out. Trends Microbiol. 7, 237–241. [DOI] [PubMed] [Google Scholar]
- Sauer, N. (1997) Sieve elements and companion cells‐extreme division of labour. Trends Plant Sci. 2, 285–286. [Google Scholar]
- Schaffer, A.A. , Pharr, D.M. and Madore, M.A. (1996) Cucurbits In: Photoassimilate Distribution in Plants and Crops. Source‐Sink Relationships (Zamsky E. and Schaffer A.A., eds), pp. 729–758. New York: Marcel Dekker, Inc. [Google Scholar]
- Scholthof, H.B. (2005) Plant virus transport: motions of functional equivalence. Trends Plant Sci. 10, 376–382. [DOI] [PubMed] [Google Scholar]
- Schwach, F. , Vaistij, F.E. , Jones, L. and Baulcombe, D.C. (2005) An RNA‐dependent RNA polymerase prevents meristem invasion by Potato Virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, M.S. , Wellink, J. , Goldbach, R.W. and Van Lent, J.W.M. (2002) Phloem loading and unloading of Cowpea mosaic virus in Vigna unguiculata . J. Gen. Virol. 83, 1493–1504. [DOI] [PubMed] [Google Scholar]
- Smith, S.H. and Schlegel, D.E. (1964) The distribution of Clover yellow mosaic virus in Vicia faba root tips. Phytopathology, 54, 1273–1274. [Google Scholar]
- Stadler, R. , Wright, K.M. , Lauterbach, C. , Amon, G. , Gahrtz, M. , Feuerstein, A. , Oparka, K.J. and Sauer, N. (2005) Expression of GFP‐fusions in Arabidopsis companion cells reveals non‐specific protein trafficking into sieve elements and identifies a novel post‐phloem domain in roots. Plant J. 41, 319–331. [DOI] [PubMed] [Google Scholar]
- Thompson, J.R. and Garcia‐Arenal, F. (1998) The bundle sheath‐phloem interface of Cucumis sativus is a boundary to systemic infection by Tomato aspermy virus . Mol. Plant–Microbe Interact. 11, 109–114. [Google Scholar]
- Turgeon, R. (1989) The sink_source transition in leaves. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 119–138. [Google Scholar]
- Ueki, S. and Citovsky, V. (2007) Spread throughout the plant: systemic transport of viruses In: Viral Transport in Plants (Waigmann E. and Heinlein M., eds), pp. 86–117. Berlin: Springer. [Google Scholar]
- Valentine, T. , Roberts, I.M. and Oparka, K.J. (2002) Inhibition of Tobacco mosaic virus replication in lateral roots is dependent on an activated meristem‐derived signal. Protoplasma, 219, 184–196. [DOI] [PubMed] [Google Scholar]
- Valentine, T. , Shaw, J. , Blok, V.C. , Phillips, M.S. , Oparka, K.J. and Lacomme, C. (2004) Efficient virus‐induced gene silencing in roots using a modified Tobacco rattle virus vector . Plant Physiol. 136, 3999–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. , Driskel, B.A. , Zhu, Y. , Hunger, R.M. and Littlefield, L.J. (2001) Evidence that soilborne wheat mosaic virus moves long distance through the xylem in wheat. Protoplasma, 218, 57–66. [DOI] [PubMed] [Google Scholar]
- Waigmann, E. , Ueki, S. , Trutnyeva, K. and Citovsky, V. (2004) The ins and out of non‐destructive cell‐to‐cell and systemic movement of plant viruses. Curr. Rev. Plant Sci. 23, 195–250. [Google Scholar]
- Wintermantel, W.M. , Banerjee, N. , Oliver, J.C. , Paolillo D.J. and Zaitlin, M. (1997) Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase‐mediated resistance. Virology, 231, 248–257. [DOI] [PubMed] [Google Scholar]
- Wright, K.M. , Wood, N.T. , Robert, A.G. , Chapman, S. , Boevink, P. , MacKenzie, K.M. and Oparka, K.J. (2007) Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic, 8, 21–31. [DOI] [PubMed] [Google Scholar]
- Zhu, Y.L. , Qi, Y.J. , Xun, Y. , Owens, R. , Ding, B. (2002) Movement of potato spindle tuber viroid reveals regulatory points of phloem‐mediated RNA traffic. Plant Physiol. 130, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]


