SUMMARY
Tomato leaf curl New Delhi virus (ToLCNDV) infection causes significant yield loss in tomato. The availability of a conventional tolerance source against this virus is limited in tomato. To understand the molecular mechanism of virus tolerance in tomato, the abundance of viral genomic replicative intermediate molecules and virus‐directed short interfering RNAs (siRNAs) by the host plant in a naturally tolerant cultivar H‐88‐78‐1 and a susceptible cultivar Punjab Chhuhara at different time points after agroinfection was studied. We report that less abundance of viral replicative intermediate in the tolerant cultivar may have a correlation with a relatively higher accumulation of virus‐specific siRNAs. To study defence‐related host gene expression in response to ToLCNDV infection, the suppression subtractive hybridization technique was used. A library was prepared from tolerant cultivar H‐88‐78‐1 between ToLCNDV‐inoculated and Agrobacterium mock‐inoculated plants of this cultivar at 21 days post‐inoculation (dpi). A total of 106 nonredundant transcripts was identified and classified into 12 different categories according to their putative functions. By reverse Northern analysis and quantitative real‐time polymerase chain reaction (qRT‐PCR), we identified the differential expression pattern of 106 transcripts, 34 of which were up‐regulated (>2.5‐fold induction). Of these, eight transcripts showed more than four fold induction. qRT‐PCR analysis was carried out to obtain comparative expression profiling of these eight transcripts between Punjab Chhuhara and H‐88‐78‐1 on ToLCNDV infection. The expression patterns of these transcripts showed a significant increase in differential expression in the tolerant cultivar, mostly at 14 and 21 dpi, in comparison with that in the susceptible cultivar, as analysed by qRT‐PCR. The probable direct and indirect relationship of siRNA accumulation and up‐regulated transcripts with the ToLCNDV tolerance mechanism is discussed.
INTRODUCTION
Plants are routinely attacked by a spectrum of parasites, such as viruses, bacteria, fungi, nematodes and insects. Unlike animals, plants are sessile and lack a somatically adaptive immune system in their response to pathogens. Instead, plants have evolved various multifaceted mechanisms to protect themselves from various biotic and abiotic challenges (Thordal‐Christian, 2003). A large number of crop plants are susceptible to infection by viruses. Geminiviruses are an emerging group of plant viruses that affect horticultural crops in tropical and subtropical areas throughout the world. Tomato leaf curl disease (ToLCD) is one of the most important constraints to tomato production in the tropics and subtropics, particularly in South and Southeast Asia (Chakraborty, 2008; Czosnek and Laterrot, 1997; Green and Kalloo, 1994; Saikia and Muniyappa, 1989; Vasudeva and Samraj, 1948). The disease is caused by strains of Tomato leaf curl New Delhi virus (ToLCNDV), a species of the genus Begomovirus, family Geminiviridae, and is transmitted by whiteflies (Bemisia tabaci Genn.). The limited availability of a conventional resistance source has restricted the efforts to develop a cultivar with durable resistance. The majority of the begomoviruses within the Old World are monopartite in nature and often associated with satellite DNA‐β molecules (Nawaz‐ul‐Rehman et al., 2009). However, bipartite begomoviruses, such as ToLCNDV (possessing DNA‐A and DNA‐B of about 2.5–2.7 kb in size), are predominant in major tomato‐growing areas of the country (Chakraborty, 2008). DNA‐A encodes the components necessary for replication and DNA‐B encodes proteins for systemic movement inside the host. The viral genome multiplies by a combination of rolling circle and recombination‐dependent replication in the nucleus via a double‐stranded DNA intermediate (Gutierrez, 1999; Jeske et al., 2001; Preiss and Jeske, 2003). At least five species of Tomato leaf curl virus (ToLCV) have been reported in southern India (Kirthi et al., 2002), where ToLCD incidence in the summer season (February to June) in susceptible varieties increases rapidly to 100% and yield losses often exceed 90% (Saikia and Muniyappa, 1989). In northern India, ToLCNDV is the predominant species, resulting in severe ToLCD, either alone or in combination with other tomato‐infecting begomoviruses (Chakraborty, 2008).
Interactions between plants and pathogens induce a series of defence responses (Hammond‐Kosack and Jones, 1996). When a plant and a pathogen come into contact, close communications occur between the two organisms (Hammond‐Kosack and Jones, 2000). Plants are adapted to detect the presence of pathogens and to respond with antimicrobial defences and other stress responses. Systemic infection of plants by virus requires modifications that allow viral replication and movement. This is associated with the suppression of host gene defence responses and changes in host gene expression. It has been shown that the induction of these mechanisms involves a complex network of signal perception, amplification and transduction, in which several molecules and defence‐related genes participate (Xiong et al., 2001). Several studies have investigated the genes expressed during begomovirus–host interaction. In Arabidopsis, numerous defence‐associated genes have been identified and have shown to be coordinately regulated in response to infection with various viruses (Whitham et al., 2003). During these interactions, the plant defence system is strictly regulated which, in turn, determines the outcome (McDowell and Dangle, 2000). Undoubtedly, these diverse and complex interactions are a product of many factors, including differences in signal transduction between the interacting organisms and the relative dominance of the pathway involved (Takemoto and Hardham, 2004).
Begomoviruses have been shown to induce and to become the target of post‐transcriptional gene silencing (PTGS) in plants (Akbergenov et al., 2006; Chellappan et al., 2004; Lucioli et al., 2003). Symptom recovery in infected plants over time is correlated with the accumulation of short interfering RNAs (siRNAs) targeted against specific viruses (Rodriguez‐Negrete et al., 2008). siRNA‐mediated gene silencing has become an important method for the analysis of gene functions in eukaryotes and, at the same time, holds promise for the development of new approaches aimed at controlling plant viruses. The molecular basis of disease development following virus infection has been the subject of intense investigation (Vanitharani et al., 2005). The ability to silence viral genes by the generation of cellular double‐stranded RNA (dsRNA) targeted against viral genomes has resulted in the suppression of virus infection in animal as well as plant systems (Ding et al., 2004).
The subtractive complementary DNA (cDNA) hybridization technique is a powerful approach to gain preliminary insights by identifying and isolating cDNA of differentially expressed genes. Suppressive subtractive hybridization (SSH) is used to selectively amplify target cDNA fragments (differentially expressed) and simultaneously suppress the amplification of nontarget DNA. The method is based on the suppression polymerase chain reaction (PCR) effect. In order to identify the molecular mechanisms underlying tolerance or resistance to virus infections, a strategy combining SSH with reverse Northern analysis and quantitative real‐time polymerase chain reaction (qRT‐PCR) was used to analyse the difference in gene expression between tolerant and susceptible tomato cultivars, namely H‐88‐78‐1 and Punjab Chuhhara, respectively. To understand the molecular regulation of these processes, the relevant subsets of differentially expressed transcripts of interest were identified, cloned and studied in detail. Those transcripts expressed in the tolerant cultivar alone may act as stress sensors, transcriptional activators or signal transduction pathway components, and thus may behave as dominating factors for resistance towards viral infection. We identified a series of transcripts that may exhibit viral tolerance inducibility, including stress response factors, transporters, transcription factors and transcripts encoding proteins of unknown functions. In the present investigation, we studied the tolerant phenotypic character of tomato cultivar H‐88‐78‐1 in terms of viral DNA and siRNA accumulation and host response to ToLCNDV infection.
RESULTS AND DISCUSSION
Infectivity analysis for ToLCNDV tolerance in tomato cultivars
We screened seven tomato cultivars for tolerance and susceptibility to ToLCNDV infection under glasshouse conditions. Scoring was performed on the basis of the percentage infectivity at 21 days post‐inoculation (dpi), and was subsequently classified as tolerant (T), moderately tolerant (MT), susceptible (S) and highly susceptible (HS) (Table 1). A total of 75 plants from three independent experiments was tested for infectivity analysis for each cultivar. Leaf curl symptoms started to appear in the cultivar Punjab Chhuhara (highly susceptible to ToLCNDV) within 7 dpi, whereas, in the highly resistant cultivar H‐88‐78‐1, symptom initiation started at 15 dpi (Table 1). We scored the number of plants showing symptoms at 21 dpi (Fig. 1). The highest infectivity (92%) was observed in cultivar Punjab Chhuhara, followed by 86.7% in 15SB, both of which were grouped as highly susceptible cultivars (Table 1). Cultivar H‐86 was categorized as susceptible, with 58.6% of the inoculated plants showing symptoms, whereas hybrids TLBRH‐5 and TLBRH‐6 were considered to be moderately tolerant with 38.7% and 36% infection, respectively. Two cultivars, H‐88‐78‐1 and LA1777, showed lower infectivity (9.3% and 12%, respectively), and hence were grouped as tolerant cultivars (Table 1). Southern blotting of infected tomato genomic DNA with a part of ToLCNDV DNA‐A as probe showed a 2.4‐fold higher accumulation of viral replicative intermediate in Punjab Chhuhara in comparison with that in H‐88‐78‐1 at 21 dpi (Fig. 2A,B). Hence, H‐88‐78‐1 was selected as a tolerant cultivar for further studies.
Table 1.
Infectivity of Tomato leaf curl New Delhi virus in selected tomato cultivars by Agrobacterium‐mediated inoculation.
| Cultivar | Plants infected/inoculated | First symptom appearance (dpi) | Symptom severity* | Overall grade† |
|---|---|---|---|---|
| LA1777 | 9/75 | 12 | + | T |
| H‐88‐78‐1 | 7/75 | 15 | + | T |
| TLBRH‐5 | 29/75 | 10 | ++ | MT |
| TLBRH‐6 | 27/75 | 10 | ++ | MT |
| H‐86 | 44/75 | 9 | +++ | S |
| Punjab Chhuhara | 69/75 | 7 | ++++ | HS |
| 15SB | 65/75 | 7 | ++++ | HS |
Observations were made at 21 days post‐inoculation (dpi).
+, Least severe; ++, moderately severe; +++, severe; ++++, highly severe.
T, Tolerant (1–20%); MT, moderately tolerant (20.1–40%); S, susceptible (40.1–60%); HS, highly susceptible (60.1–100%).
Figure 1.

Phenotypic expression of tomato cultivars at 21 days post‐inoculation (dpi). (A) H‐88‐78‐1 inoculated with pCAMBIA2301 as mock. (B) H‐88‐78‐1 inoculated with Tomato leaf curl New Delhi virus (ToLCNDV). (C) Punjab Chhuhara inoculated with ToLCNDV.
Figure 2.
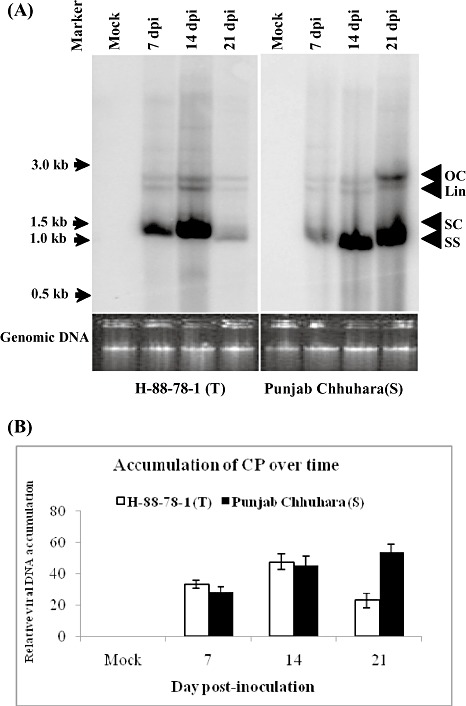
Infectivity analysis on tolerant (H‐88‐78‐1; T) and susceptible (Punjab Chhuhara; S) cultivars of tomato. (A) Southern blot of tomato genomic DNA hybridized with Tomato leaf curl New Delhi virus (ToLCNDV) coat protein (CP) gene as probe. (B) Relative accumulation of viral DNA in ToLCNDV‐infected tolerant cultivar H‐88‐78‐1 and susceptible cultivar Punjab Chhuhara at different time points. Viral replicative forms are indicated as open circular (OC), linear (Lin), supercoiled (SC) and single strand (SS). Bars indicate the standard deviations (± SD).
Accumulation of siRNA in tolerant cultivars
To study the molecular mechanism of tolerance in these tomato cultivars, the accumulation of viral gene‐specific siRNAs in the leaf tissues of H‐88‐78‐1 and Punjab Chhuhara was studied at different time points. To determine the association of antiviral defence with PTGS in terms of virus‐specific siRNA accumulation, total RNA was analysed by polyacrylamide gel electrophoresis (PAGE) to resolve low‐molecular‐weight RNAs. Northern blots with replication‐associated protein (Rep) gene as probe detected the accumulation of 21–23‐nucleotide‐long RNA products in virus‐infected samples, indicating that the ToLCNDV‐infected cultivar H‐88‐78‐1 was able to induce PTGS with the production of virus‐specific siRNAs (Fig. 3A). The blot was quantified for siRNA accumulation, and it was found that the amount of siRNAs increased gradually (>85‐fold at 21 dpi) with time in H‐88‐78‐1 (Fig. 3A,B). In infected cultivar H‐88‐78‐1, virus‐derived siRNA accumulation was 26‐fold at 7 dpi, 36‐fold at 14 dpi and reached a maximum of 90‐fold at 21 dpi (Fig. 3B). However, in the cultivar Punjab Chhuhara, a contrasting pattern in the rate of siRNA accumulation was observed. In Punjab Chhuhara, siRNA accumulation started at 7 dpi (43‐fold), when it was more abundant when compared with the tolerant cultivar; it then decreased at 14 dpi (32‐fold) and 21 dpi (25‐fold) (Fig. 3B). The increased level of siRNA at later times in cultivar H‐88‐78‐1 suggests that this accumulation may have a possible correlation with the lower level of viral replication in this cultivar. The induction of siRNA has been shown to down‐regulate viral DNA accumulation and gene expression in plant cells (Vanitharani et al., 2003). The accumulation of virus‐derived siRNAs has been reported in local and systemically infected leaf tissue of virus‐infected plants (Moissiard and Voinnet, 2004; Szittya et al., 2002), demonstrating the activation of virus‐induced gene silencing (VIGS), which results in reduced accumulation of the invading virus (Szittya et al., 2002). In our experiment, Southern blot analysis also indicated less accumulation of viral DNA‐A in the tolerant cultivar at 21 dpi (Fig. 2A,B). However, the intensity and efficacy of virus‐induced PTGS varies with intrinsic features of the viral genome and its interaction with the host (Moissiard and Voinnet, 2004; Voinnet, 2001, 2005; Waterhouse et al., 2001). Mechanisms other than the mere accumulation of siRNA are also likely to play a role in the natural resistance against virus infection, as transgene‐mediated siRNAs are also produced in susceptible plants (Ribeiro et al., 2007).
Figure 3.
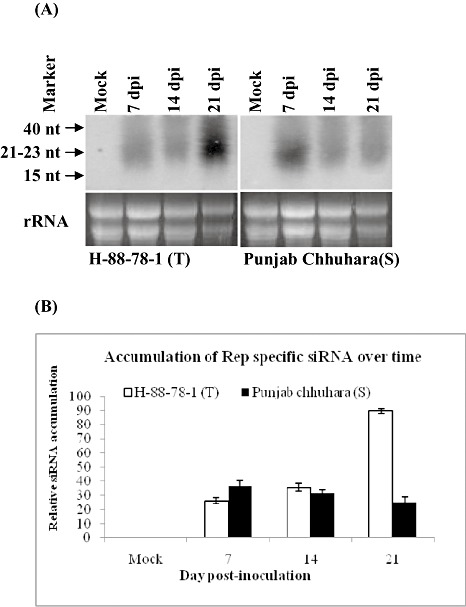
Identification of Tomato leaf curl New Delhi virus (ToLCNDV)‐derived short interfering RNA (siRNA) in tolerant (H‐88‐78‐1; T) and susceptible (Punjab Chhuhara; S) cultivars of tomato. (A) Northern hybridization showing the accumulation of virus‐derived siRNAs. (B) Identification of replication‐associated protein (Rep)‐specific siRNA accumulation in ToLCNDV‐infected tolerant cultivar H‐88‐78‐1 and susceptible cultivar Punjab Chhuhara at different time points. A plant infected with Agrobacterium tumefaciens harbouring pCAMBIA2301 alone was used as mock. Total RNA was separated by 15% polyacrylamide gel electrophoresis and hybridized with the ToLCNDV Rep gene fragment as probe. The sizes of the standard oligonucleotides and siRNAs are indicated. Bars indicate the standard deviations (± SD). Ethidium bromide‐stained ribosomal RNA is shown for equivalent loading.
Identification and classification of ToLCNDV‐responsive genes in the tolerant cultivar
A total of 400 clones was randomly picked from the forward subtracted library. After single‐pass sequencing and vector sequence removal, 259 expressed sequence tag (EST) sequences were obtained, 106 of which were nonredundant. All of these 106 clones were functionally annotated by Blastx against the GenBank nonredundant EST databases, and subsequently classified into 12 functional categories according to their putative function (Table 2). Transcripts assigned to the metabolism category accounted for the largest (32.07%) contribution, followed by photosynthetic transcripts (14.15%). Transcripts were also involved in other processes, including cell cycle/DNA and RNA processing (4.71%), signalling/receptor function (4.71%), hormone/hormonal regulation (4.71%), abiotic stress (1%), cell development/housekeeping (7.54%), cellular transport (6.60%), transcription/ translational factor (5.66%), defence response (3.77%), protein degradation (5.66%) and unknown function (9.43%) (Table 2). Our study listed the abundance of transcripts in the host plant in response to ToLCNDV infection. Similar studies have been reported on Tomato mottle taino virus (ToMoTV) and Cucumber mosaic virus (CMV) to identify the up‐regulated host transcripts during infection (Collazo et al., 2005; Ruiz‐Medrano et al., 2007).
Table 2.
Categorization of transcripts expressed in tolerant tomato cv. H‐88‐78‐1 after Tomato leaf curl New Delhi virus (ToLCNDV) infection according to their putative function and fold induction.
| GenBank accession ID | Similarity of transcripts | GenBank match | Organism | Obtained sequence length (bp)‡ | E‐value | Fold induction (± SD)* |
|---|---|---|---|---|---|---|
| CELL CYCLE AND DNA/RNA PROCESSING | ||||||
| GR979494 | RNA polymerase‐associated protein LEO1 | EEF47773.1 | Ricinus communis | 808 | 3.00E‐60 | 3.27 (± 0.33) |
| GR979429 | Histone h2b, putative | EEF29752.1 | R. communis | 337 | 1.00E‐19 | 1.43 (± 0.89) |
| GR979472 | Dead box ATP‐dependent RNA helicase | EEF41818.1 | R. communis | 539 | 2.00E‐48 | 3.37 (± 1.24) |
| GR979481 | Putative DNA2‐NAM7 helicase family protein | AAL31652.1 | Oryza sativa | 833 | 6.00E‐121 | 3.98 (± 0.93) |
| GR979485 | FtsH‐like protein Pftf precursor | AAD17230.1 | Nicotiana tabacum | 469 | 5.00E‐42 | −1.89 (± 0.37) |
| SIGNALLING /RECEPTOR | ||||||
| GR979482 | Serine/threonine protein kinase PBS1† | EEF43813.1 | R. communis | 607 | 4.00E‐65 | 4.01 (± 0.79) |
| GR979493 | Signal recognition particle 19 kDa protein | EEF43168.1 | R. communis | 496 | 1.00E‐26 | 2.34 (± 0.97) |
| GR979435 | Receptor protein kinase | EEF50514.1 | R. communis | 513 | 1.00E‐52 | 1.54 (± 0.41) |
| GR979412 | Chloroplast N receptor‐interacting protein 1 | ACA79924.1 | Nicotiana benthamiana | 309 | 2.00E‐31 | −1.82 (± 0.39) |
| GR979457 | ADP‐ribosylation factor 1 | ACG31280.1 | Zea mays | 288 | 1.00E‐49 | 3.07 (± 1.73) |
| HORMONE/HORMONAL REGULATION | ||||||
| GR979487 | Gibberellin‐regulated protein 1† | EEF35557.1 | R. communis | 618 | 6.00E‐11 | 4.15 (± 0.75) |
| GR979477 | Auxin influx transport protein | ABN81352.1 | Casuarina glauca | 679 | 4.00E‐123 | 2.26 (± 0.83) |
| GR979484 | Indole‐3‐acetic acid‐amido synthetase GH3 | EEF28642.1 | R. communis | 601 | 3.00E‐17 | 2.01 (± 0.13) |
| GR979403 | Similar to IBR3 | refNP_187337.2 | Vitis vinifera | 850 | 2.00E‐116 | 2.09 (± 0.93) |
| GR979430 | Cytokinin binding protein CBP57 | dbjBAA03710.1 | Nicotiana sylvestris | 214 | 9.00E‐30 | 1.06 (± 0.42) |
| ABIOTIC STRESS | ||||||
| GR979463 | ERD15 | ABB89735.1 | Capsicum annuum | 493 | 2.00E‐60 | −2.23 (± 0.13) |
| CELL DEVELOPMENT/HOUSEKEEPING | ||||||
| GR979431 | 60S ribosomal protein L13 | AAQ96375.1 | Solanum brevidens | 570 | 1.00E‐90 | 1.23 (± 0.76) |
| GR979459 | Similar to HSC70‐1 | ABX26256.1 | Panax quinquefolius | 258 | 9.00E‐38 | 1.75 (± 0.57) |
| GR979461 | Actin | ACJ04738.1 | Sedum alfredii | 479 | 8.00E‐82 | 1.03 (± 0.56) |
| GR979434 | α‐Tubulin | embCAD13178.1 | N. tabacum | 502 | 4.00E‐82 | 1.02 (± 0.36) |
| GR979424 | Cell wall protein | embCAA54561.1 | Solanum lycopersicum | 439 | 3.00E‐09 | 1.37 (± 0.41) |
| GR979420 | Ribosome biogenesis protein nop10 | EEF32551.1 | R. communis | 232 | 3.00E‐26 | 1.01 (± 0.31) |
| GR979417 | Senescence‐associated protein | AAZ23261.1 | N. tabacum | 308 | 1.00E‐36 | 1.57 (± 0.83) |
| GR979492 | Xyloglucan endotransglycosylase | AAZ08349.1 | S. lycopersicum | 386 | 2.00E‐73 | −1.87 (± 0.91) |
| METABOLISM | ||||||
| GR979471 | Phosphoglycerate kinase precursor | AAC26785.1 | Solanum tuberosum | 503 | 5.00E‐85 | 1.48 (± 0.77) |
| GR979401 | Aminomethyltransferase | spP54260.1 | S. tuberosum | 462 | 1.00E‐81 | 1.98 (± 0.39) |
| GR979392 | C‐4 sterol methyl oxidase 2 | AAQ83692.1 | N. benthamiana | 267 | 2.00E‐30 | 2.12 (± 0.93) |
| GR979425 | S‐Adenosylmethionine decarboxylase | ABQ42184.1 | S. lycopersicum | 500 | 6.00E‐26 | 2.33 (± 0.67) |
| GR979409 | Plastidic aldolase NPALDP1 | dbjBAA77604.1 | Nepenthes paniculata | 352 | 1.00E‐30 | 1.93 (± 0.13) |
| GR979456 | Triose phosphate isomerase | ABB02628.1 | S. tuberosum | 371 | 1.00E‐60 | 2.63 (± 0.29) |
| GR979444 | Glycine hydroxymethyltransferase | spP50433.1 | S. tuberosum | 845 | 6.00E‐151 | 2.29 (± 0.31) |
| GR979427 | Glyoxysomal malate dehydrogenase | AAU29200.1 | S. lycopersicum | 790 | 9.00E‐140 | 1.17 (± 0.69) |
| GR979447 | Plastid glutamine synthetase GS2 | AAG40236.2 | S. tuberosum | 217 | 1.00E‐34 | 2.23 (± 0.72) |
| GR979399 | Geranylgeranyl reductase | embCAA07683.1 | N. tabacum | 399 | 5.00E‐71 | 1.23 (± 0.77) |
| GR979448 | Serine‐pyruvate aminotransferase, putative | EEF31258.1 | R. communis | 447 | 8.00E‐68 | 1.14 (± 0.47) |
| GR979449 | Hydroxypyruvate reductase | dbjBAB44155.1 | R. communis | 293 | 3.00E‐46 | −1.19 (± 0.82) |
| GR979450 | Catalase | AAR14052.2 | S. tuberosum | 744 | 4.00E‐148 | −1.87 (± 0.59) |
| GR979468 | Alanine aminotransferase 2 | refNP_001151209.1 | Z. mays | 542 | 1.00E‐36 | 1.59 (± 0.87) |
| GR979466 | Phosphoribulokinase | refNP_001149223.1 | Z. mays | 716 | 2.00E‐112 | −1.22 (± 0.86) |
| GR979440 | Precursor of carboxylase h‐protein 4 | refXP_002326710.1 | Populus trichocarpa | 449 | 2.00E‐64 | 1.04 (± 0.34) |
| GR979453 | Aldolase | AAA33643.1 | Pisum sativum | 230 | 3.00E‐35 | 2.59 (± 0.11) |
| GR979421 | Catalase isozyme | ABY21246.1 | S. tuberosum | 196 | 1.00E‐30 | 1.15 (± 0.41) |
| GR979489 | β‐Mannosidase | AAL37714.1 | S. lycopersicum | 519 | 4.00E‐58 | −1.12 (± 0.69) |
| GR979452 | Glyceraldehyde‐3‐phosphate dehydrogenase | ABW89104.1 | Helianthus annuus | 576 | 6.00E‐75 | 2.67 (± 0.67) |
| GR979476 | PAPS‐reductase‐like protein | AAB05871.2 | Catharanthus roseus | 532 | 2.00E‐35 | 2.23 (± 0.46) |
| GR979478 | FAD5 (FATTY ACID DESATURASE 5) | refNP_566529.1 | Arabidopsis thaliana | 375 | 3.00E‐56 | 2.11 (± 0.63) |
| GR979483 | Acyl carrier protein | EEF35894.1 | A. thaliana | 790 | 4.00E‐42 | 1.86 (± 0.49) |
| GR979396 | Glycosyltransferase | refXP_002323701.1 | P. trichocarpa | 561 | 6.00E‐57 | 1.31 (± 0.34) |
| GR979469 | Benzoyl coenzyme A | AAT68601.1 | Petunia hybrida | 269 | 2.00E‐38 | −1.07 (± 0.63) |
| GR979389 | Glycine dehydrogenase | spO49954.1 | S. tuberosum | 407 | 8.00E‐21 | 2.78 (± 1.09) |
| GR979405 | 5‐Phosphoribosyl‐1‐pyrophosphate amidotransferase | AAR06289.1 | N. tabacum | 357 | 3.00E‐37 | 1.36 (± 0.25) |
| GR979410 | Carbonic anhydrase | embCAH60891.1 | S. lycopersicum | 468 | 2.00E‐62 | 1.91 (± 0.23) |
| GR979467 | Inorganic pyrophosphatase | embCAA58699.1 | N. tabacum | 592 | 2.00E‐35 | −1.87 (± 0.78) |
| GR979414 | Methionine synthase | AAF74983.1 | S. tuberosum | 430 | 8.00E‐57 | 1.56 (± 0.84) |
| GR979438 | Putative S‐adenosylmethionine synthetase | ACF17673.1 | C. annuum | 315 | 3.00E‐09 | −1.53 (± 0.13) |
| GR979436 | Ribulose bisphosphate carboxylase, small subunit | AAA34192.1 | S. lycopersicum | 226 | 6.00E‐24 | 2.57 (± 0.67) |
| GR979416 | S‐Adenosyl‐l‐homocysteine hydrolase | AAV31754.1 | N. tabacum | 735 | 9.00E‐133 | 2.98 (± 0.89) |
| GR979397 | Ribulose bisphosphate carboxylase/oxygenase activase | spO49074.1 | Lycopersicon pennellii | 287 | 3.00E‐23 | −1.17 (± 0.79) |
| CELLULAR TRANSPORT | ||||||
| GR979488 | ABC1 family protein | refNP_565214.1 | A. thaliana | 305 | 3.00E‐06 | 2.55 (± 0.54) |
| GR979395 | Similar to aquaporin | dbjBAD95790.1 | S. lycopersicum | 488 | 3.00E‐24 | 2.78 (± 0.78) |
| GR979445 | ATP‐dependent transporter, putative | EEF38635.1 | R. communis | 761 | 2.00E‐61 | 2.9 (± 0.12) |
| GR979454 | Nonspecific lipid transfer protein | embCAJ19705.1 | S. lycopersicum | 524 | 4.00E‐59 | 3.71 (± 0.44) |
| GR979462 | Vacuolar ATP synthase proteolipid subunit | EEF48309.1 | R. communis | 214 | 3.00E‐12 | 1.51 (± 0.56) |
| GR979437 | ATP synthase (γ subunit) | spP29790.1 | N. tabacum | 622 | 3.00E‐62 | 2.32 (± 0.09) |
| GR979455 | Water channel protein | dbjBAA20075.1 | Nicotiana excelsior | 591 | 9.00E‐99 | 2.87 (± 0.23) |
| TRANSCRIPTION/TRANSLATIONAL FACTOR | ||||||
| GR979443 | MRNA‐binding protein precursor | AAD21574.3 | S. lycopersicum | 423 | 2.00E‐75 | 2.38 (± 0.13) |
| GR979473 | Similar to elongation factor 2 | ACG42954.1 | V. vinifera | 763 | 1.00E‐116 | 2.62 (± 0.12) |
| GR979479 | Eukaryotic translation initiation factor 2 | ABB86256.1 | S. tuberosum | 307 | 4.00E‐130 | 1.34 (± 0.59) |
| GR979480 | Elongation factor 1‐α | AAR83865.1 | C. annuum | 837 | 1.00E‐17 | 2.69 (± 0.55) |
| GR979441 | AGO4‐2 | ABC61505.1 | N. benthamiana | 562 | 6.00E‐94 | 2.03 (± 0.67) |
| GR979390 | Basic helix‐loop‐helix (bHLH) family protein† | refNP_001119080.1 | A. thaliana | 359 | 9.00E‐22 | 4.63 (± 0.89) |
| PROTEIN DEGRADATION | ||||||
| GR979393 | 26S proteasome AAA‐ATPase subunit RPT4a† | dbjBAC23035.1 | S. tuberosum | 637 | 3.00E‐100 | 4.22 (± 0.13) |
| GR979460 | RUB1 (RELATED TO UBIQUITIN 1 | refNP_564379.2 | A. thaliana | 619 | 1.00E‐57 | 3.23 (± 0.77) |
| GR979415 | Ubiquitin‐conjugating enzyme E2† | refNP_564379.2 | S. lycopersicum | 357 | 7.00E‐54 | 5.55 (± 0.64) |
| GR979475 | Armadillo repeat motif‐containing protein† | AAK60564.1 | N. tabacum | 820 | 5.00E‐79 | 5.20 (± 0.98) |
| GR979474 | Ubiquitin‐associated uba/ubx domain‐containing protein | EEF34340.1 | R. communis | 681 | 8.00E‐26 | 3.29 (± 0.75) |
| GR979413 | Cysteine proteinase 3 | spQ40143 | S. lycopersicum | 555 | 2.00E‐31 | −2.57 (± 0.33) |
| DEFENCE RESPONSE | ||||||
| GR979439 | Cysteine protease TDI‐65† | spQ40143.1 | S. lycopersicum | 682 | 3.00E‐80 | 4.55 (± 0.83) |
| GR979391 | Pathogenesis‐related leaf protein 6† | spP04284.2 | S. lycopersicum | 738 | 3.00E‐92 | 5.66 (± 0.41) |
| GR979470 | Phosphoric diester hydrolase, putative | EEF40839.1 | R. communis | 341 | 2.00E‐36 | 1.36 (± 0.63) |
| GR979458 | PR8/chloroplast thiazole biosynthetic protein | AAZ93636.1 | N. tabacum | 485 | 2.00E‐23 | −1.78 (± 0.64) |
| PHOTOSYNTHESIS/ENERGY | ||||||
| GR979428 | Ferredoxin‐I | AAA34192.1 | S. lycopersicum | 571 | 5.00E‐62 | 2.97 (± 0.70) |
| GR979394 | Cytochrome b6‐f complex iron–sulphur subunit | spQ69GY7.1 | S. tuberosum | 404 | 5.00E‐67 | −1.13 (± 0.31) |
| GR979464 | Thioredoxin m(mitochondrial)‐type | EEF42142.1 | R. communis | 328 | 9.00E‐41 | −2.32 (± 0.63) |
| GR979442 | NADH‐ubiquinone oxidoreductase subunit‐like | ABB02646.1 | S. tuberosum | 343 | 2.00E‐59 | 3.79 (± 0.89) |
| GR979406 | Oxygen‐evolving enhancer protein 1 | spP23322.2 | S. lycopersicum | 736 | 4.00E‐109 | −1.85 (± 0.03) |
| GR979423 | Chlorophyll a–b‐binding protein 4 | spP14278.1 | S. lycopersicum | 553 | 9.00E‐103 | −1.88 (± 0.61) |
| GR979422 | Chlorophyll a–b‐binding protein | spP13869.1 | P. hybrida | 487 | 1.00E‐62 | 3.39 (± 0.24) |
| GR979426 | Photosystem I subunit XI | AAO85557.1 | Nicotiana attenuata | 271 | 1.00E‐33 | 2.98 (± 0.19) |
| GR979398 | Chlorophyll‐binding protein 1B | prf1204205B | S. lycopersicum | 694 | 2.00E‐82 | 2.89 (± 0.37) |
| GR979432 | Photosystem II 10 kDa polypeptide | spQ40163.1 | S. lycopersicum | 218 | 3.00E‐30 | 2.83 (± 0.57) |
| GR979433 | Photosystem II oxygen‐evolving complex protein 3 | AAU03361.1 | S. lycopersicum | 544 | 7.00E‐75 | 2.07 (± 0.13) |
| GR979451 | Chlorophyll a–b‐binding protein 3C‐like | ABB55370.1 | S. tuberosum | 744 | 4.00E‐118 | 3.31 (± 0.99) |
| GR979402 | Chlorophyll a–b‐binding protein 5 | spP14279 | S. lycopersicum | 612 | 1.00E‐117 | 1.57 (± 0.17) |
| GR979491 | Chloroplast pigment‐binding protein CP29 | ABG73415.1 | N. tabacum | 583 | 4.00E‐93 | 2.40 (± 0.77) |
| GR979465 | Lhca5 protein | ABN10530.1 | Gentiana lutea | 272 | 1.00E‐06 | –2.06 (± 0.21) |
| OTHERS/UNKNOWN FUNCTION | ||||||
| GR979446 | Hypothetical protein | ABK96267.1 | P. trichocarpa | 586 | 1.00E‐87 | 1.48 (± 0.14) |
| GR979411 | Unknown | ABB16969.1 | S. tuberosum | 306 | 1.00E‐36 | 1.89 (± 0.22) |
| GR979419 | Hypothetical protein | refXP_002274904.1 | V. vinifera | 691 | 5.00E‐64 | 2.22 (± 1.11) |
| GR979418 | Conserved hypothetical protein | EEF30430.1 | R. communis | 350 | 3.00E‐05 | −2.02 (± 0.97) |
| GR979404 | Hypothetical protein LOC100193383 | refNP_001131619.1 | Z. mays | 240 | 6.00E‐29 | 1.08 (± 0.03) |
| GR979407 | Unknown | ACJ84739.1 | Medicago truncatula | 518 | 1.00E‐44 | 1.37 (± 0.81) |
| GR979408 | Unknown protein | refNP_566336.1 | A. thaliana | 835 | 1.00E‐71 | 1.90 (± 0.60) |
| GR979486 | Hypothetical protein OsJ_18110 | EEE63300.1 | O. sativa | 536 | 9.00E‐06 | −1.98 (± 0.73) |
| GR979490 | Predicted protein | refXP_002298321.1 | P. trichocarpa | 190 | 1.00E‐12 | 1.26 (± 0.93) |
| GR979400 | Unknown | ABB16993.1 | S. tuberosum | 570 | 5.00E‐57 | −2.00 (± 0.13) |
Fold inductions are presented as the expression ratio ToLCNDV stressed to mock of each transcript to that of tubulin with SDs. Negative values of fold induction show the down‐regulation of transcripts. Standard deviations (± SD) were calculated from three independent experiments. The transcripts are listed according to their possible functions. The E‐value is used to indicate the significance of sequence similarity.
Transcripts selected for quantitative real‐time polymerase chain reaction (qRT‐PCR).
Obtained length of transcript in base pairs (bp).
Identification of differentially expressed clones by reverse Northern analysis
The effective signal intensities of the spots were calculated by subtracting the normalized intensity of the negative control. Reverse Northern hybridization analysis revealed that, of the 106 transcripts, 34 (32.07%) were up‐regulated (more than 2.5‐fold induction) in response to ToLCNDV infection (Table 2). Of these, eight transcripts showed more than four fold induction after ToLCNDV infection in comparison with control at 21 dpi (Fig. 4A, B). These identified transcripts were involved in the defence response, transcription process, proteolysis and hormone signalling.
Figure 4.
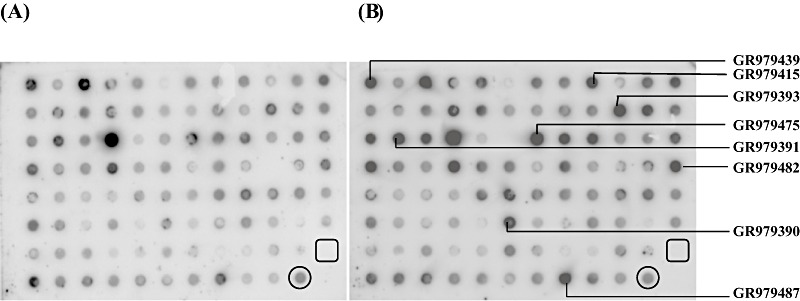
Reverse Northern blot of transcripts obtained from subtracted cDNA library. DNA array hybridization was performed on a nylon membrane. Segments of representative identical nylon membranes containing cDNA spots from the subtracted cDNA library generated with Tomato leaf curl New Delhi virus (ToLCNDV)‐infected tolerant cultivar H‐88‐78‐1. The figure shows hybridization with 32P‐labelled cDNA probes prepared from equal amounts of poly(A+) RNA of control (A) and virus‐infected (B) H‐88‐78‐1. Circles indicate α‐tubulin and squares indicate neomycin phosphotransferase II (NPTII) spots.
qRT‐PCR of the identified differentially expressed clones
To validate the changes in mRNA abundance, as detected by reverse Northern analysis, qRT‐PCR was performed to evaluate quantitatively the relative abundance of eight transcripts in both H‐88‐78‐1 and Punjab Chhuhara. All of these transcripts exhibited strong induction at 14 and 21 dpi in the tolerant cultivar in response to ToLCNDV infection. It was interesting to note that the accumulation of different transcripts occurred with different kinetics. Several transcripts were induced after 7 dpi, whereas the expression of some had declined by 21 dpi. Transcripts encoding the serine/threonine kinase PBS1 (GR979482), cysteine protease TDI‐65 (GR979439), basic helix–loop–helix (bHLH) family protein (GR979390) and 26S proteasome AAA‐ATPase subunit RPT4a (GR979393) were highly up‐regulated at 21 dpi, whereas the ubiquitin‐conjugating enzyme E2 (GR979415) and Armadillo repeat motif (ARM)‐containing proteins (GR979475) showed a late response, with induction at 28 dpi in the tolerant cultivar (Fig. 5A–H).
Figure 5.
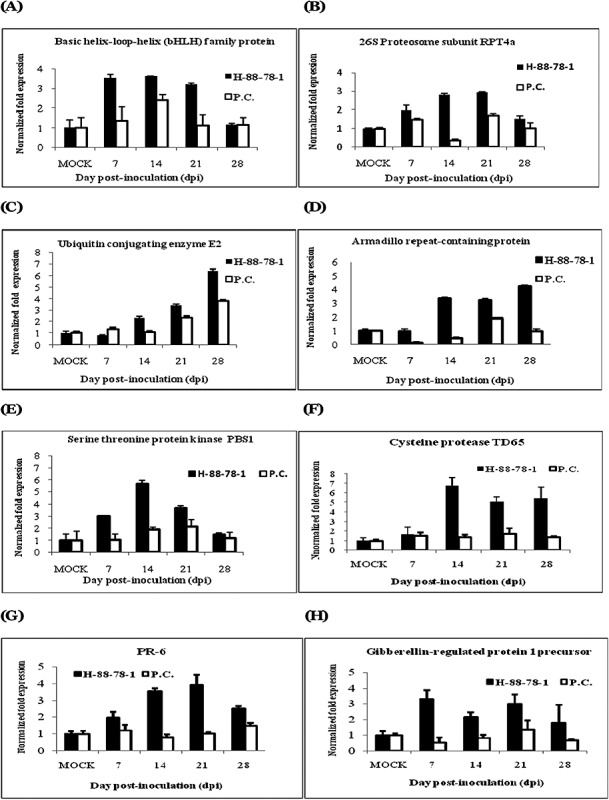
Quantitative real‐time polymerase chain reaction (qRT‐PCR) analyses of the expression of selected transcripts in response to Tomato leaf curl New Delhi virus (ToLCNDV) infection in tolerant (H‐88‐78‐1) and susceptible (Punjab Chhuhara) cultivars. Expression patterns of basic helix–loop–helix (bHLH) (A), 26S proteasome AAA‐ATPase subunit RPT4a (B), ubiquitin‐conjugating enzyme E2 (C), Armadillo repeat motif‐containing protein (D), serine/threonine protein kinase (E), cysteine protease (F), pathogenesis‐related leaf protein 6 (G) and gibberellin‐regulated protein 1 (H). Bars indicate the standard deviations (± SD).
Transcript GR979390 encodes a bHLH transcription factor. The expression level of this transcript in the susceptible cultivar was normal, whereas, in the tolerant cultivar, the relative expression level was 3.5‐fold at 7 dpi and remained constant until 21 dpi, suggesting that its induction might suppress the susceptibility to ToLCNDV infection (Fig. 5A). It is relevant to note that Arabidopsis bHLH (AtMYC2) functions as a transcriptional activator in abscisic acid (ABA) signalling (Abe et al., 2003). ABA regulates processes related to abiotic and biotic stress tolerance and disease resistance (Finkelstein et al., 2002; Ton et al., 2009). Although ABA has emerged as an important regulator of biotic defence responses, its role against viruses is yet to be resolved fully. ABA promotes resistance in some plant–pathogen interactions, whereas it increases susceptibility in Arabidopsis, soybean, potato, etc. (Asselbergh et al., 2008). An elevated level of ABA in tobacco was positively correlated with increased disease resistance (Fraser, 1982; Whenham et al., 1986). The increased level of this transcription factor may lead to the activation of ABA signalling, which, in turn, may regulate plant defence for enhanced tolerance to ToLCNDV.
Transcript GR979393 showed similarity with the putative 26S proteasome AAA‐ATPase subunit RPT4a of Solanum tuberosum. Putative 26S proteasome AAA‐ATPase subunit RPT4a is a base subcomplex of the proteasome regulatory particle. This subunit uses ATP hydrolysis to assist during the recognition and/or hydrolysis of substrate by 26S proteasome (Groll and Huber, 2003; Vierstra et al., 1999). Differential expression of the 26S proteasome subunit increased substantially until 21 dpi in the tolerant cultivar H‐88‐78‐1. There was more than 2.5‐fold induction in the tolerant cultivar H‐88‐78‐1 in comparison with the mock‐inoculated control. In the susceptible cultivar, it tended to increase only up to 1.7‐fold (Fig. 5B). Previously, it has been suggested that the polyubiquitination of the movement protein (MP) and subsequent degradation by the 26S proteasome may play a substantial role in the regulation of virus spread (Reichel and Beachy, 2000). Intriguingly, proteasomes purified from sunflower also possess endonuclease activity capable of cleaving Tobacco mosaic virus (TMV) RNA (Ballut et al., 2003).
Transcript GR979415 was homologous to the ubiquitin‐conjugating enzyme E2 of Solanum lycopersicum. This enzyme interacts with ubiquitin ligase (E3), a catalyst that mediates the final transfer of ubiquitin to the ε‐amino group of the target protein. Hence, it is an important component of the ubiquitin‐mediated proteolysis pathway. Enhanced expression (approximately two fold) of this transcript was detected at 14 dpi in the tolerant relative to the susceptible cultivar, which steadily increased until 28 dpi (Fig. 5C). The enhanced level of the ubiquitin‐conjugating enzyme E2 may be responsible for the increased processing of ubiquitin‐mediated proteolysis of viral proteins. The importance of ubiquitinization in plant defence has also been documented in other recent findings (Craig et al., 2009; Devoto et al., 2003; Glickman and Ciechanover, 2002).
Transcript GR979475 corresponds to the ARM‐containing protein of Nicotiana tabacum. ARM is approximately 40 amino acids in length, is typically present in a variable number of tandem copies and functions in protein–protein interactions (Hammond‐Kosack and Jones, 1996; Hatzfeld, 1999). In our experiment, the basal expression of this transcript was more than three fold greater in the tolerant cultivar at 14 dpi and reached a maximum at 28 dpi (4.25‐fold), whereas, in the susceptible cultivar, its expression declined to less than that of the mock‐inoculated control (Fig. 5D). This suggests that ARM‐containing proteins may play a unique role in the ubiquitin‐mediated pathway of protein degradation in the tolerant cultivar H‐88‐78‐1. Recently, the role of the ARM‐containing U‐box ligase in plant defence has been described (Dreher and Callis, 2007). In these proteins, the U‐box functions as the E2 interaction domain, whereas the ARM domain might contribute directly or indirectly to substrate recognition. Several other ARM‐containing proteins are induced by elicitors in several plant species and appear to participate in the plant defence response in both basal and resistance gene‐mediated pathways (Dreher and Callis, 2007).
Transcript GR979482 showed similarity with the serine/threonine protein kinase PSB1 of Ricinus communis. Protein kinases play a central role in signalling during pathogen recognition and the subsequent activation of the plant defence mechanism (Romeis, 2001; Swiderski and Innes 2001). This transcript was induced at all time points in the tolerant cultivar, with maximum mRNA accumulation at 14 dpi (greater than five fold), whereas, in the susceptible cultivar, its expression was lower (Fig. 5E). Some recent studies have indicated the involvement of the serine/threonine protein kinase OXI1 during downstream signalling after the oxidative burst following pathogen attack (Legaz et al., 1998; Rentel et al., 2004). An increased level of this transcript has suggested that, possibly, the serine/threonine kinase plays an important role in ToLCNDV tolerance.
Transcript GR979439 corresponds to the cysteine protease TDI‐65 of Solanum lycopersicum. Recently, proteolytic enzymes and processes have been implicated or shown to play a regulatory role in the plant defence response (Bernoux et al., 2008; Krüger et al., 2002; Lam and Del Pozo, 2000; Nishimura and Somerville, 2002). The protease that cleaves its targets in a caspase‐like manner (such as cysteine proteases in animal systems) has been found recently to be active early in the Nicotiana–TMV interaction (Chichkova et al., 2004). The expression of this transcript reached six fold at 14 dpi, followed by more than four fold induction at 21 and 28 dpi. The susceptible cultivar failed to up‐regulate its expression during all time points of infection (Fig. 5F). This elevation in the level of transcript expression suggests that, in the tolerant cultivar, the early defence response, generated by the cysteine protease, is capable of suppressing the viral infection.
Transcript GR979391 encodes pathogenesis‐related leaf protein 6 (PR‐6). A cysteine proteinase inhibitor class of PR‐6 proteins is induced by pathogen attack (Vidyasekaran, 2002). PR‐6 exhibited a rapid up‐regulation as early as 7 dpi (two fold) and reached a maximum level at 21 dpi (3.9‐fold) in comparison with the mock‐inoculated control in the tolerant cultivar (Fig. 5G). Some PR proteins release elicitor molecules from the host cell wall surface, which may stimulate the biosynthesis of phenolic compounds, such as phytoalexins (Vidyasekaran, 2002). Plants possess both preformed and inducible mechanisms to resist pathogen invasion. The significance of PR proteins in the inhibition of pathogen infection has been reviewed in a recent article, where they were shown to induce the defence response to suppress pathogenesis (Van Loon et al., 2006).
The transcript GR979487 encodes gibberellin regulatory protein 1, which is a hormone‐regulated protein which may function as a defence gene. Gibberellic acid (GA) is a growth‐promoting hormone that positively regulates processes such as normal plant growth and development (Swain and Singh, 2005). The gibberellin regulatory protein transcript showed up to 3.5‐fold induction at early stages of ToLCNDV infection (7 dpi) in the tolerant cultivar in comparison with the mock‐inoculated control (Fig. 5H). An antimicrobial peptide Snakin‐2, resembling gibberellin regulatory protein 1, was induced locally by wounding and responded to pathogen infection in a GA‐regulated manner in potato (Herzog et al., 1995; Lobo et al., 2002). Hence, it is suggested that gibberellin regulatory protein 1 of cultivar H‐88‐78‐1 may provide tolerance by assisting the plant to overcome ToLCNDV infection to yield normal growth and development. ToLCNDV infection in the susceptible tomato cultivar may have affected GA biosynthesis; as a result, plant growth was affected.
In summary, gene expression profiles were investigated in one tolerant and one susceptible cultivar following inoculation with ToLCNDV. In both cases, differences were found in symptom severity and in the accumulation of the viral genomic DNA component and corresponding virus‐specific siRNAs at different time points (7, 14 and 21 dpi). Our limited knowledge of host gene expression during geminivirus infection in planta is a result of the absence of a well‐characterized, compatible, virus–host system suitable for transcriptome profiling studies. Our study reveals that the changes in host gene expression that occur during ToLCNDV interaction are associated with the tolerant characteristics of cultivar H‐88‐78‐1. A strong correlation of siRNA accumulation with ToLCNDV tolerance was observed in cultivar H‐88‐78‐1. A recent study has indicated the presence of a strong RNAi pathway in a soybean cultivar conferring immunity against Mungbean yellow mosaic India virus (Yadav et al., 2009). Plants have evolved two antiviral mechanisms based on RNA degradation: RNAi‐ and 20S‐mediated degradation (Ballut et al., 2005). 20S‐mediated RNA degradation can be considered as the first component of the plant antiviral defence, targeting nonhost RNAs, a less fine‐tuned mechanism than RNAi (Dielen et al., 2009). In the past few years, the ubiquitin proteasomal system (UPS) has emerged as an essential protagonist in plant–pathogen interactions. Viral proteins themselves are the target of UPS (Reichel and Beachy, 2000). In our study, most of the up‐regulated transcripts (GR979393, GR979460, GR979415, GR979475 and GR979474) identified were involved in UPS. The induction of these transcripts at different stages of ToLCNDV infection suggests that they function in providing tolerance to ToLCNDV infection in the tolerant cultivar H‐88‐78‐1. It should be highlighted that, among the various components of protein degradation, defence signalling transcripts are over‐expressed in response to pathogen attack. Over‐expression of transcripts, such as bHLH transcription factors (GR979390) and gibberellin‐regulated protein 1 (GR979487), during ToLCNDV infection advocates coordination among hormone signalling and defence response pathways (Asselbergh et al., 2008; Herzog et al., 1995; Lobo et al., 2002). Further, characterization and functional analysis of these identified genes can lead to a better understanding of host defence during interaction with begomoviruses. With the ongoing effort to sequence the tomato genome, the identified transcripts may play an important role in breeding for tomato improvement and the development of cultivars with improved disease resistance.
EXPERIMENTAL PROCEDURES
Construction of infectious ToLCNDV clone
Tandem repeat constructs of ToLCNDV genomic components (DNA‐A and DNA‐B) were available from the Molecular Virology Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi, India (Chakraborty et al., 2008). These constructs were recloned into pCAMBIA2301 at the SmaI site and confirmed by restriction digestion using appropriate enzymes.
Agroinoculation
Tandem repeat constructs of DNA‐A and DNA‐B of ToLCNDV were introduced into Agrobacterium tumefaciens strain EHA105 by the freeze–thaw method. Agrobacterium tumefaciens harbouring DNA‐A and DNA‐B was grown overnight in yeast extract broth (YEB) (pH 7.0) at 28 °C. The cultures were centrifuged at 5000 g for 10 min and the cell pellets were suspended in YEB (pH 5.6) with 100 mm acetosyringone (Sigma, USA). Tomato seeds were germinated in vermiculite to the two‐leaf stage. Agroinoculation was performed by the stem inoculation method, in which wounds were made around the growing nodal region of the tomato stem by pricking three to four times with a 30‐gauge needle. About 20 µL of Agrobacterium suspension containing an equimolar concentration of A. tumefaciens harbouring DNA‐A and DNA‐B were mixed and applied to the wounds. For mock inoculation, Agrobacterium harbouring pCAMBIA2301 alone was used. The plants were kept in a glasshouse at 25 °C for symptom development until 45 dpi.
Southern hybridization
The leaf samples (inoculated as well as systemic) from ToLCNDV‐infected and mock‐inoculated plants were collected at 7, 14 and 21 dpi. Total DNA from symptomatic leaves of tomato cultivars (five plants per treatment) were isolated by the cetyltrimethylammonium bromide (CTAB) method (Porebski et al., 1997). For Southern hybridization, 5 µg of total DNA was electrophoresed on 1% agarose gel in TBE [Tris‐borate EDTA; 45 mm Tris‐borate, 1 mm EDTA (pH 8)]. Samples were transferred to hybond‐N+ membrane (Amersham Bioscience, USA) and hybridized with a α32P‐deoxycytidine triphosphate (dCTP)‐labelled DNA‐A‐specific coat protein (CP) sequence amplified using 5′‐ACAGAAAACCCAGAATGTACAGAA‐3′ and 5′‐CAACATTAAGGCATTTTCAGTATG‐3′. Radiolabelled probe was prepared and hybridized by a high prime DNA labelling kit (Roche, Indianapolis, IN, USA) according to the manufacturer's protocol.
Detection of virus‐specific siRNA
Total RNA from infected as well as mock‐inoculated tomato leaves was isolated using TRIzol reagent (Sigma).Total RNA (50 µg) was resuspended in formamide RNA loading dye and denatured at 95 °C for 2 min. Samples were electrophoresed on 15% polyacrylamide (19 : 1) gel containing 7 m urea in 0.5 × TBE, and transferred to nylon membrane according to the procedure described by Sambrook and Russell (2001). The Rep gene of ToLCNDV was PCR amplified using specific primer pairs (5′‐CTCAAAGGTGTATAGCAATGATGC‐3′ and 5′‐AAAGTCGAATCTGTTATTT‐3′) for probe preparation. Probes were labelled with α32P‐dCTP using a high prime DNA labelling kit (Roche). Hybridization with radiolabelled probe was performed overnight at 42 °C. The blot was scanned in a Phosphor‐imager (Typhoon‐9210, GE Healthcare, NJ, USA) and quantified using Bio‐Rad Quantity One software (USA).
Construction of subtracted cDNA library
Frozen leaves were ground in liquid nitrogen and total RNA was isolated using TRIzol reagent (Sigma) according to the manufacturer's instructions. RNA integrity was examined by electrophoresing samples on denaturing formaldehyde–1.2% agarose–ethyl bromide gel. The quantity and quality of isolated total RNA was examined spectrophotometrically. Messenger RNA (mRNA) was purified from total RNA using the MagneSphere mRNA Purification Kit (Promega, Madison, USA) according to the manufacturer's protocol. A forward subtraction was performed between 21‐day‐old ToLCNDV‐infected H‐88‐78‐1 as tester and mock‐infected H‐88‐78‐1 (with Agrobacterium harbouring vector pCAMBIA2301 only) as driver. Purified mRNA (2 µg) was used for reverse transcription and first‐strand cDNA thus prepared was used for SSH with the Clontech PCR Select‐cDNA Subtraction kit (BD Biosciences, Clontech, CA, USA). In brief, driver and tester cDNAs were RsaI digested, extracted with phenol–chloroform, ethanol precipitated and resuspended in water. Tester cDNA was split into two pools and each was ligated to a different adapter (provided with the cDNA subtraction kit). Unsubtracted tester control cDNA was ligated to both adapters. Two rounds of hybridization and PCR amplification were carried out to normalize and enrich the differentially expressed cDNAs according to the manufacturer's protocol, with the following changes: the primary PCR was performed for 30 cycles (94 °C, 30 s; 65 °C, 30 s; 72 °C, 90 s) and the secondary PCR was performed for 16 cycles (94 °C, 30 s; 66 °C, 30 s; 72 °C, 90 s). Products of the secondary PCR were purified and cloned into pGEMT‐easy vector (Promega) and transformed into DH5αEscherichia coli competent cells.
DNA sequencing and data analysis
Sequences of the recombinant plasmids were determined with an automated sequencer (ABI Sequencer, Version No.3770, Applied Biosystems, USA) using M13 forward and reverse primers. Nucleotides and translated sequences were compared with nonredundant sequences of the GenBank database using the blast sequence alignment program (Altschul et al., 1997). ESTs of more than 100 nucleotides in length and an E‐value of less than 1E‐05 were considered to be significant. Further functional classification was performed according to the MIPS data base (http://mips.helmholtz‐muenchen.de/proj/funcatDB/search_main_frame.html).
Reverse Northern hybridization
Individual clones of the subtracted cDNA library were amplified in a 96‐well PCR plate using M13 forward and reverse primers in a 50‐µL reaction at an annealing temperature of 60 °C for 30 cycles. The products were analysed on agarose gel to confirm the insert size, quality and quantity. Purified PCR products were denatured by adding an equal volume of 0.6 m sodium hydroxide. An equal volume of each denatured PCR product (about 100 ng) of above 300 bp in size was spotted onto two Hybond N+ membranes (Amersham Bioscience) using a BIO‐DOT dot‐blot apparatus (Bio‐Rad) in 96‐well formats to prepare two identical arrays. In addition, a PCR product of α‐tubulin cDNA (produced using primer sequences 5′‐GGTACTGGTCTTCAAGGTTTC‐3′ and 5′‐TTGTCATAGATAGCCTCATTGT‐3′) was spotted as internal control to normalize the signals of two different blots corresponding to ToLCNDV‐treated and mock‐treated samples. A PCR product of the neomycin phosphotransferase II (NPTII) gene from the vector pCAMBIA 1305.1 using primer sequences (5′‐TTTTCTCCCAATCAGGCTTG‐3′ and 5′‐TCAGGCTCTTTCACTCCATC‐3′) was also spotted as a negative control to subtract the background noise. The membranes were neutralized with neutralization buffer (0.5 m Tris‐Cl, pH 7.4, 1.5 m NaCl) for 3 min, washed with 2% standard saline citrate (SSC) and cross‐linked using UV cross‐linker (Stratagene, La Jolla, CA, USA).
qRT‐PCR analysis
For eight highly up‐regulated transcripts, the differential expression was confirmed using qRT‐PCR. Total RNA was isolated at 7, 14, 21 and 28 dpi from the leaves of ToLCNDV‐ and mock‐inoculated tolerant (H‐88‐78‐1) and susceptible (Punjab Chuhhara) tomato cultivars by TRIzol reagent (Sigma). Total RNA (2 µg) was used to synthesize first‐strand cDNA from each sample using Superscript reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the supplier's manual. The primers used for qRT‐PCR were designed from the sequences of selected transcripts using Primer Express Version 3.0 (Table 3). qRT‐PCR was performed on a Step One Real‐Time PCR System (Applied Biosystems) using Power SYBR Green dye (Applied Biosystems). PCR was performed for each sample in triplicate; α‐tubulin, a constitutively expressed protein, was used as internal control. The amount of transcript of each gene, normalized to the internal control α‐tubulin, was analysed using the 2‐ΔΔCt method (Livak and Schmittgen, 2001). The amount of transcript of each target gene under the mock condition was designated as 1.0. The PCR conditions were kept as 95 °C for 10 min, 95 °C for 15 s, 60 °C for 1 min for 40 cycles, 95 °C for 15 s and 60 °C for 1 min. The experiment was repeated three times to check the reproducibility.
Table 3.
Primers used for quantitative real‐time polymerase chain reaction (qRT‐PCR) analysis.
| Primer ID | Amplicon length (bp) | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|---|
| 26S proteasome AAA‐ATPase subunit RPT4a | 61 | GGGCTGATATGCGGAATGTT | TCACGCTCTGCACGAATTG |
| Armadillo repeat motif‐containing protein | 65 | GCGATCTCTTGCACGCAAA | TGGATAGCTCCAAAAGCAGATG |
| Basic helix–loop–helix protein | 66 | CACCGGATGGGTCACAATC | TCTTCCTCCTATTCCTCTCTGATACAA |
| Cysteine protease | 56 | TCCGAAGGCCCCAATAGG | CACTGGGAGTGAAGGCAATGA |
| Gibberellin‐regulated protein 1 | 63 | CATTTTCATCCTCCTCATACAGGTT | TTGCCTGCCTCTATCTGTTCAGT |
| Pathogenesis‐related leaf protein 6 | 61 | AGGGCAGCCGTGCAATT | ATTGGCTGGTAGCGTGGTTATAG |
| Serine/threonine protein kinase | 63 | TGCATTGCAAACAGCAACAA | CCAAGAGATCCTTCACCAATGAG |
| α‐Tubulin | 62 | AACCTGTGAGATAAGGCGATTGA | GGCGCTCATTGGACATTGA |
| Ubiquitin‐conjugating enzyme E2 | 60 | TCAGCTGGACCCATGATTGTT | TGCTGGCCCAGTAGCTGAA |
ACKNOWLEDGEMENTS
We are grateful to the Director, National Institute of Plant Genome Research (NIPGR) for providing facilities, to the Council for Scientific and Industrial Research, Government of India for providing a Senior Research Fellowship to Mr Neeraj K Rai, and to the Department of Biotechnology, Government of India for providing financial support (Grant no. BT/PR/5274/AGR/16/464/2004). We thank Mr Rajeev K. Yadav, Ms Swati Puranik and Ms Kajal Kumari (NIPGR, New Delhi, India) for help with the manuscript. We would like to thank the reviewers for their critical comments.
REFERENCES
- Abe, H. , Urao, T. , Ito, T. , Seki, M. , Shinozaki, K. and Shinozaki, K.Y. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell, 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbergenov, R. , Si‐Ammour, A. , Blevins, T. , Amin, I. , Kutter, C. , Vanderschuren, H. , Zhang, P. , Gruissem, W. , Meins, Jr., F. , Hohn, T. and Pooggin, M.M. (2006) Molecular characterization of geminivirus‐derived small RNAs in different plant species. Nucleic Acids Res. 34, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh, B. , Vleesschauwer, D.D. and Hofte, M. (2008) Global switches and fine‐tuning—ABA modulates plant pathogen defense. Mol. Plant–Microbe Interact. 21, 709–719. [DOI] [PubMed] [Google Scholar]
- Ballut, L. , Petit, F. , Mouzeyar, S. , Le Gall, O. , Candresse, T. , Schmid, P. , Nicolas, P. and Badaovi, S. (2003) Biochemical identification of proteosome‐associated endonuclease activity in sunflower. Biochim. Biophys. Acta, 1645, 30–39. [DOI] [PubMed] [Google Scholar]
- Ballut, L. , Drucker, M. , Pugniere, M. , Cambon, F. , Blanc, S. , Roquet, F. , Candresse, T. , Schmid, H.‐P. , Nicolas, P. , Gall, O.L. and Badaoui, S. (2005) HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. J. Gen. Virol. 86, 2595–2603. [DOI] [PubMed] [Google Scholar]
- Bernoux, M. , Timmers, T. , Jauneau, A. , Briere, C. , De Wit, P.J.G.M. , Marco, Y. and Deslandes, L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1‐R‐mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell, 20, 2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S. (2008) Tomato leaf curl viruses from India (Geminiviridae) In: Encyclopedia of Virology (Mahy B.W.J. and Van Regenmortel M.H.V., eds), pp. 124–133. Oxford: Elsevier. [Google Scholar]
- Chakraborty, S. , Vanitharani, R. , Chattopadhyay, B. and Fauquet, C.M. (2008) Supervirulent pseudorecombination and asymmetric synergism between genomic components of two distinct species of begomovirus associated with severe tomato leaf curl disease in India. J. Gen. Virol. 89, 818–828. [DOI] [PubMed] [Google Scholar]
- Chellappan, P. , Vanitharani, R. , Pita, J. and Fauquet, C.M. (2004) Short interfering RNA accumulation correlates with host recovery in DNA virus‐infected hosts, and gene silencing targets specific viral sequences. J. Virol. 78, 7465–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkova, N.V. , Kim, S.H. , Titova, E.S. , Kalkum, M. , Morozov, V.S. , Rubtsov, Y.P. , Kalinina, N.O. , Taliansky, M.E. and Vartapetian, A.B. (2004) A plant caspase like protease activated during the hypersensitive response. Plant Cell, 16, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo, C. , Romos, P.L. , Chacon, O. , Borroto, C.J. , Lopez, Y. , Pozol, M. , Thomma, B.H.P. , Hein, I. and Borras‐Hidalgo, O. (2005) Phenotypical and molecular characterization of the Tomato mottle taino virus–Nicotiana megalosiphon interaction. Physiol. Mol. Plant Pathol. 67, 231–236. [Google Scholar]
- Craig, A. , Ewan, R. , Mesmar, J. , Gudipati, V. and Sadanandom, A. (2009) E3 ubiquitin ligases and plant innate immunity. J. Exp. Bot. 60, 1123–1132. [DOI] [PubMed] [Google Scholar]
- Czosnek, H. and Laterrot, H. (1997) A worldwide survey of Tomato yellow leaf curl viruses . Arch. Virol. 142, 1391–1406. [DOI] [PubMed] [Google Scholar]
- Devoto, A. , Muskett, P.R. and Shirasu, K. (2003) Role of ubiquitination in the regulation of plant defense against pathogens. Curr. Opin. Plant Biol. 6, 307–311. [DOI] [PubMed] [Google Scholar]
- Dielen, A.S. , Badaoui, S. , Candresse, T. and German‐Retana, S. (2009) The ubiquitin/26S proteasome system in plant–pathogen interactions: a never‐ending hide‐and‐seek game. Mol. Plant Pathol. 11, 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W. , Li, H. , Lu, R. , Li, F. and Li, W.X. (2004) RNA silencing: a conserved antiviral immunity of plants and animals. J. Virus Res. 102, 109–115. [DOI] [PubMed] [Google Scholar]
- Dreher, K. and Callis, J. (2007) Ubiquitin, hormones and biotic Stress. Ann. Bot. 99, 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. , Gampala, S.S.L. and Rock, C.D. (2002) Abscisic acid signaling in seed and seedling. Plant Cell, 14, S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, R.S.S. (1982) Are ‘pathogenesis related’ proteins involved in acquired systemic resistance of tobacco plants to Tobacco mosaic virus . J. Gen. Virol. 58, 305–313. [Google Scholar]
- Glickman, M.H. and Ciechanover, A. (2002) The ubiquitin proteosome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428. [DOI] [PubMed] [Google Scholar]
- Green, S.K. and Kalloo, G. (1994) Leaf curl and yellowing viruses of pepper and tomato: an overview. Technical Bulletin No. 21, Asian Vegetable Research and Development Centre, Taiwan, p. 51.
- Groll, M. and Huber, R. (2003) Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell Biol. 35, 606–616. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (1999) Geminivirus DNA replication. Cell. Mol. Life Sci. 56, 313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Jones, J.D.G. (1996) Resistance gene‐dependent plant defense responses. Plant Cell, 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Jones, J.D.G. (2000) Responses to plant pathogens In: Biochemistry and Molecular Biology of Plants (Buchanan B., Gruissem R. and Jones R., eds), pp. 1102–1156. Rockville, MD: American Society of Plant Physiologists. [Google Scholar]
- Hatzfeld, M. (1999) The armadillo family of structural proteins. Int. Rev. Cytol. 186, 179–224. [DOI] [PubMed] [Google Scholar]
- Herzog, M. , Dorne, A.M. and Grellet, F. (1995) GASA a gibberellin regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol. Biol. 27, 743–752. [DOI] [PubMed] [Google Scholar]
- Jeske, H. , Lutgemeier, M. and Preiss, W. (2001) DNA forms indicate rolling circle and recombination‐dependent replication of Abutilon mosaic virus . EMBO J. 20, 6158–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirthi, N. , Maiya, S.P. , Murthy, M.R.N. and Savithri, H.S. (2002) Evidence of recombination among the tomato leaf curl virus strains/species from Bangalore, India. Arch. Virol. 147, 255–272. [DOI] [PubMed] [Google Scholar]
- Krüger, J. , Thomas, C.M. , Golstein, C. , Dixon, M.S. , Smoker, M. , Tang, S. , Mulder, L. and Jones, J.D.G. (2002) A tomato cysteine protease required for Cf‐2‐dependent disease resistance and suppression of autonecrosis. Science, 296, 744–747. [DOI] [PubMed] [Google Scholar]
- Lam, E. and Del Pozo, O. (2000) Caspase‐like protease involvement in the control of plant cell death. Plant Mol. Biol. 44, 417–428. [DOI] [PubMed] [Google Scholar]
- Legaz, M.E.R. , De Armas, D. Pinon, C. and Vicente, C. (1998) Relationships between phenolics‐conjugated polyamines and sensitivity of sugarcane to smut (Ustilago scitaminea). J. Exp. Bot. 49, 1723–1728. [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lobo, M.B. , Segura, A. , Mozeno, M. , Lopez, G. , Olmedo, G. and Molina, A. (2002) Snakin‐2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 128, 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucioli, A. , Noris, E. , Brunetti, A. , Tavazza, R. , Ruzza, V. , Castillo, A.G. , Bejarano, R. , Accotto, G.P. and Tavazza, M. (2003) Tomato yellow leaf curl Sardinia virus rep‐derived resistance to homologous and heterologous geminiviruses occurs by different mechanisms and is overcome if virus‐mediated transgene silencing is activated. J. Virol. 77, 6785–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M. and Dangle, J.L. (2000) Signal transduction in the plant immune response. Trends Biochem. Sci. 25, 79–82. [DOI] [PubMed] [Google Scholar]
- Moissiard, G. and Voinnet, O. (2004) Viral suppression of RNA silencing in plants. Mol. Plant Pathol. 5, 71–82. [DOI] [PubMed] [Google Scholar]
- Nawaz‐ul‐Rehman, M.S. , Mansoor, S. , Briddon, R.W. and Fauquet, C.M. (2009) Maintenance of an Old World betasatellite by a New World helper begomovirus and possible rapid adaptation of the betasatellite. J. Virol. 83, 9347–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M. and Somerville, S. (2002) Resisting attack. Science, 295, 2032–2033. [DOI] [PubMed] [Google Scholar]
- Porebski, S. , Bailey, L.G. and Baurn, B.R. (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. [Google Scholar]
- Preiss, W. and Jeske, H. (2003) Multitasking in replication is common among geminiviruses. J. Virol. 77, 2972–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel, C. and Beachy, R.N. (2000) Degradation of Tobacco mosaic virus movement protein by the 26S proteasome. J. Virol. 74, 3330–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel, M.C. , Lecourieux, D. , Ouaked, F. , Usher, S.L. , Petersen, L. , Okamoto, H. , Knight, H. , Peck, S.C. , Grierson, C.S. , Hirt, H. and Knight, M.R. (2004) OXI1 kinase is necessary for oxidative burst‐mediated signalling in Arabidopsis . Nature, 427, 858–861. [DOI] [PubMed] [Google Scholar]
- Ribeiro, S.G. , Lohuis, H. , Goldbach, R. and Prins, M. (2007) Tomato chlorotic mottle virus is a target of RNA silencing but the presence of specific short interfering RNAs does not guarantee resistance in transgenic plants. J. Virol. 81, 1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Negrete, E.A. , Carrillo‐Tripp, J. and Rivera‐Bustamante, R.F. (2008) RNA silencing against Geminivirus: complementary action of PTGS and TGS in host recovery. J. Virol. 83, 1332–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis, T. (2001) Protein kinases in the plant defense response. Curr. Opin. Plant Biol. 4, 807–814. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Medrano, R. , Moya, J.H. , Xoconnostle‐Cazares, B. and Lucas, W.J. (2007) Influence of cucumber mosaic virus infection on the mRNA population present in the phloem translocation stream of pumpkin plants. Funct. Plant Biol. 34, 292–301. [DOI] [PubMed] [Google Scholar]
- Saikia, A.K. and Muniyappa, V. (1989) Epidemiology and control of Tomato leaf curl virus in Southern India. Trop. Agric. 66, 350–354. [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd edn, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Swain, S.M. and Singh, D.P. (2005) Tale tales from sly dwarves: novel functions of gibberellins in plant development. Trends Plant Sci. 10, 123–129. [DOI] [PubMed] [Google Scholar]
- Swiderski, M.R. and Innes, R.W. (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26, 101–112. [DOI] [PubMed] [Google Scholar]
- Szittya, G. , Molnar, A. , Silhavy, D. , Hornyik, C. and Burgyan, J. (2002) Short defective interfering RNAs of Tombusviruses are not targeted but trigger post‐transcriptional gene silencing against their helper virus. Plant Cell, 14, 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, D. and Hardham, A.R. (2004) The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physol. 136, 3864–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal‐Christian, H. (2003) Fresh insights into processes of non‐host resistance. Curr. Opin. Plant. Biol. 6, 351–357. [DOI] [PubMed] [Google Scholar]
- Ton, J. , Victor, F. and Brigitte, M.M. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Vanitharani, R. , Chellappan, P. and Fouquet, C.M. (2003) Short interfering RNA‐mediated interference of gene expression and viral DNA accumulation in cultured plant cells. Proc. Natl. Acad. Sci. USA, 100, 9632–9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitharani, R. , Chellappan, P. and Fouquet, C.M. (2005) Geminiviruses and RNA silencing. Trends Plant Sci. 10, 144–151. [DOI] [PubMed] [Google Scholar]
- Vasudeva, R.S. and Samraj, J. (1948) Leaf curl disease of tomato. Phytopathology, 38, 364–369. [Google Scholar]
- Vidyasekaran, P. (2002) Bacterial disease resistance in plants In: Molecular Biology and Biotechnological Application (Vidyasekaran P., ed.), pp. 185–293. New York: The Howarth Press. [Google Scholar]
- Vierstra, R.D. , Fu, H. , Girod, P.A. , Doelling, J.H. , Nocker, S.V. , Hochstrasser, M. and Finley, D. (1999) Structure and functional analysis of the 26S proteasome subunit from plants. Mol. Biol. Rep. 26, 137–146. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet. 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M. , Wang, M.B. and Lough, T. (2001) Gene silencing as an adaptive defense against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Whenham, R.J. , Fraser, R.S.S. , Brown, L.P. and Payne, J.A. (1986) Tobacco mosaic virus induced increase in absisic acid concentration in tobacco leaves: intracellular location in light and dark green areas, and relationship to symptom development. Planta, 168, 592–598. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Quan, S. , Chang, H.S. , Cooper, B. , Estes, B. , Zhu, T. , Wang, X. and Yu‐Ming, H. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. [DOI] [PubMed] [Google Scholar]
- Xiong, L. , Lee, M.W. , Qi, M. and Yang, Y. (2001) Identification of defense‐related rice genes by suppression subtractive hybridization and differential screening. Mol. Plant–Microbe Interact. 14, 685–692. [DOI] [PubMed] [Google Scholar]
- Yadav, R.K. , Shukla, R.K. and Chattopadhyay, D. (2009) Soybean cultivar resistant to Mungbean yellow mosaic India virus infection induces viral RNA degradation earlier than the susceptible cultivar. Virus Res. 144, 89–95. [DOI] [PubMed] [Google Scholar]


