SUMMARY
Bacterial pathogens employ the type III secretion system to secrete and translocate effector proteins into their hosts. The primary function of these effector proteins is believed to be the suppression of host defence responses or innate immunity. However, some effector proteins may be recognized by the host and consequently trigger a targeted immune response. The YopJ/HopZ/AvrRxv family of bacterial effector proteins is a widely distributed and evolutionarily diverse family, found in both animal and plant pathogens, as well as plant symbionts. How can an effector family effectively promote the virulence of pathogens on hosts from two separate kingdoms? Our understanding of the evolutionary relationships among the YopJ superfamily members provides an excellent opportunity to address this question and to investigate the functions and virulence strategies of a diverse type III effector family in animal and plant hosts. In this work, we briefly review the literature on YopJ, the archetypal member from Yersinia pestis, and discuss members of the superfamily in species of Pseudomonas, Xanthomonas, Ralstonia and Rhizobium. We review the molecular and cellular functions, if known, of the YopJ homologues in plants, and highlight the diversity of responses in different plant species, with a particular focus on the Pseudomonas syringae HopZ family. The YopJ superfamily provides an excellent foundation for the study of effector diversification in the context of wide‐ranging, co‐evolutionary interactions.
INTRODUCTION
The recognition of conserved pathogen‐associated molecular patterns (PAMPs) by the host induces a form of innate immunity, termed ‘PAMP‐triggered immunity’ (PTI) in the plant pathology literature (Chisholm et al., 2006; Jones and Dangl, 2006). Examples of well‐characterized bacterial PAMPs that can induce plant PTI include flagellin, a structural component of the bacterial flagella, and Ef‐Tu, a component of the protein translational machinery (Boller and Felix, 2009). Successful phytopathogens must suppress PTI to establish infections and proliferate in the host tissue, and many bacterial pathogens are able to accomplish this via the type III secretion system (T3SS) and its translocated type III secreted effector (T3SE) proteins (Galan and Wolf‐Watz, 2006). Plants have responded evolutionarily to this challenge through a second and more directed tier of immunity, termed ‘effector‐triggered immunity’ (ETI), in which certain effectors (or, more typically, the actions of these effectors) induce a defence response that is often accompanied by a localized programmed cell response, termed the ‘hypersensitive response’ (HR). ETI relies on resistance (R) proteins, which characteristically have either a coiled‐coil (CC) or toll‐like‐receptor (TIR) domain linked to a nucleotide‐binding site‐leucine‐rich repeat (NBS‐LRR) (Dangl and Jones, 2001). However, it should be noted that cell death is not an ETI‐specific response, as some PAMPs can induce HR‐like responses (Thomma et al., 2011). In the grand tradition of the classic arms race, some pathogens have been able to counter ETI, either by losing the defence‐eliciting T3SE or via the action of other T3SEs that disrupt ETI signalling.
The YopJ superfamily of T3SEs is one of the largest and most widely distributed bacterial effector families. Yersinia pestis, the causal agent of the black plague, contains the archetypal member YopJ. YopJ homologues are also found in the animal pathogens Salmonella (AvrA), Vibrio (VopJ) and Aeromonas (AopP), which cause infections ranging from gastroenteritis to typhoid fever and other life‐threatening diseases. In plant pathogens, YopJ homologues are found in species of Pseudomonas (the HopZ family), Xanthomonas (AvrRxv, AvrXv4, AvrBsT and XopJ), Erwinia (ORFB) and Ralstonia (PopP1 and PopP2), and the plant symbiont Rhizobium (Y4LO) (Fig. 1) (Ma et al., 2006).
Figure 1.
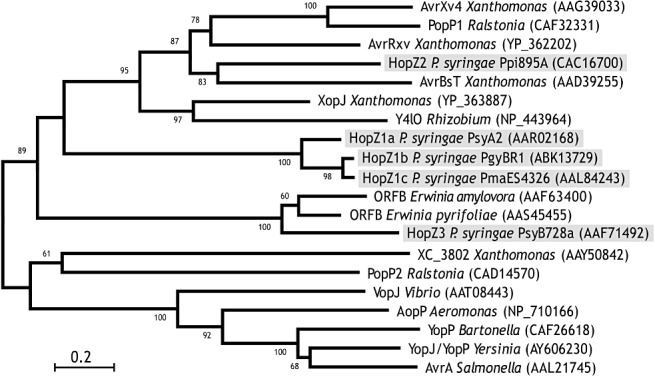
Phylogenetic relationships of the YopJ superfamily of type III secreted effector (T3SE) proteins. Neighbour‐joining tree of YopJ family of T3SE proteins. Bootstrap support is indicated above each node, with only values above 60% being shown. Accession numbers for each protein are presented in parentheses following the protein name and species. Modified from Ma et al. (2006). The scale bar indicates the evolutionary distances computed using the JTT substitution matrix (in numbers of amino acid substitutions per site).
In this article, we begin with a brief summary of an extensive body of work on YopJ, followed by a more detailed discussion of the functions of YopJ homologues from phytopathogens, with particular emphasis on the Pseudomonas syringae HopZ family (summarized in Table 1 and Fig. 2). Despite the extensive sequence and host‐specific diversity within the YopJ superfamily, there appears to be significant similarity in molecular function, suggesting that YopJ may provide a valuable framework to study evolutionary diversification of this type III effector superfamily.
Table 1.
The YopJ superfamily in plant‐associated bacteria.
| Allele | Pathovar of origin | Enzymatic activity | Phenotype in host | Localization | Reference |
|---|---|---|---|---|---|
| YopJ | Yersinia pestis, Y. pseudotuberculosis | Acetyltransferase, SUMO protease, ubiquitin protease | Inhibition of MAPK and NFκB signalling pathways; macrophage cell death; blocking both innate and adaptive immunity | ? | Monack et al. (1997); Mukherjee et al. (2006); 1999, 2000); Sweet et al. (2007) |
| HopZ1a | Pseudomonas syringae pv. syringae | Protease, acetyltransferase | HR in Arabidopsis *, †, Glycine max (soybean) † , § , Oryza sativa (rice) † , Sesamum indicum (sesame) † , Nicotiana benthamiana † , § , †† | Membrane | Lewis et al. (2008); Ma et al. (2006); Morgan et al. (2010); Zhou et al. (2009); A. Lee et al., unpublished data |
| HopZ1b | P. syringae pv. glycinea | Protease?, acetyltransferase? | Weak HR in Arabidopsis † , ‡‡ , HR in N. benthamiana § , †† , promotes P. syringae virulence in G. max (soybean) ‡ | Membrane | Lewis et al. (2008); Ma et al. (2006); Zhou et al. (2009); A. Lee et al., unpublished data |
| HopZ1c | P. syringae pv. maculicola | Protease, acetyltransferase? | ? | Membrane | Lewis et al. (2008); Ma et al. (2006); A. Lee et al., unpublished data |
| HopZ2 | P. syringae pv. pisi | Protease, acetyltransferase? | HR in Phaseolus vulgaris (common bean) † , promotes P. syringae virulence in Arabidopsis ‡ | Membrane | Arnold et al. (2001); Lewis et al. (2008); Ma et al. (2006); A. Lee et al., unpublished data |
| HopZ3 | P. syringae pv. syringae | Protease, acetyltransferase? | HR in Nicotiana tabacum †† and Phaseolus vulgaris (common bean) †† , reduces P. syringae growth and disease symptoms in Phaseolus vulgaris § , ¶ and N. benthamiana § , ¶ , suppresses HR induced by AvrPto1 †† , HopAA1 †† , HopM1 †† , HopAE1 †† and HopZ1b**, †† in N. benthamiana, promotes P. syringae virulence in Arabidopsis § , ¶ | Soluble | Lewis et al. (2008); Ma et al. (2006); Vinatzer et al. (2006); Zhou et al. (2009); A. Lee et al., unpublished data |
| AvrBsT | Xanthomonas campestris pv. vesicatoria | ? | HR in Capsicum annuum (pepper) † , ¶ , †† , C. pubescens (tree chilli) † , N. benthamiana † and Arabidopsis Pi‐0 † , suppresses AvrBs1 HR in pepper † , ¶ , †† | Nucleus | Cunnac et al. (2007); Escolar et al. (2002); Minsavage et al. (1990); Orth et al. (2000); Szczesny et al. (2010) |
| AvrRxv | X. campestris pv. vesicatoria | ? | HR in Phaseolus vulgaris (common bean) † , G. max (soybean) † , Vigna sinensis (cowpea) † , Medicago sativa (alfalfa) † , Zea mays (corn) † , Gossypium hirsutum (cotton) † and Solanum esculentum (tomato) † , § , ¶ , †† | Cytoplasm or membrane | Bonshtien et al. (2005); Ciesiolka et al. (1999); 1988, 1993) |
| AvrXv4 | X. campestris pv. vesicatoria | SUMO protease | HR in Solanum pennellii (wild tomato) † and N. benthamiana † , †† | Cytoplasm | Astua‐Monge et al. (2000); Roden et al. (2004) |
| XopJ | X. campestris pv. vesicatoria | ? | HR in N. benthamiana †† and N. clevelandii †† , suppresses PTI in Arabidopsis ‡‡ | Membrane | Bartetzko et al. (2009); Thieme et al. (2007) |
| PopP1 | Ralstonia solanacearum | ? | HR in Petunia § , ¶ | Cytoplasm (predicted) | Lavie et al. (2002) |
| PopP2 | R. solanacearum | Acetyltransferase | HR in Arabidopsis § , ¶ | Nucleus | Deslandes et al. (2003); Narusaka et al. (2009); Tasset et al. (2010) |
| Y4LO | Rhizobium species | ? | Contributes to symbiosome differentiation ¶ | ? | Yang et al. (2009) |
HR, hypersensitive response; MAPK, mitogen‐activated protein kinase; NFκB, nuclear factor kappa B; PTI, pathogen‐associated molecular pattern (PAMP)‐triggered immunity.
Effector expressed in natural strain.
Cloned effector expressed in virulent strain lacking any homologue.
Cloned effector expressed in nonhost strain lacking any homologue.
Cloned effector expressed in strain knocked out for same allele.
Strain knocked out for effector.
Cloned effector expressed in virulent strain with different homologue.
Effector expressed by Agrobacterium‐mediated transient expression.
Effector expressed in transgenic plant.
Figure 2.
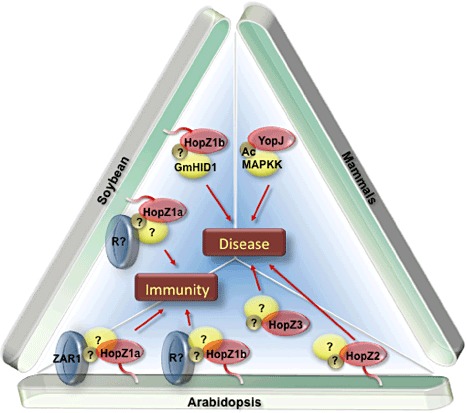
Functional diversification of the YopJ superfamily of type III secreted effector (T3SE) proteins. The HopZ family of Pseudomonas syringae T3SE proteins demonstrates the remarkable functional diversification of the YopJ superfamily in plants. In Arabidopsis thaliana ecotype Col‐0, HopZ1a and HopZ1b are recognized by two independent resistance (R) proteins resulting in a hypersensitive response (HR) (Lewis et al., 2010). Both recognition events require an intact myristoylation sequence and catalytic cysteine for recognition, indicating that it is likely that the enzymatic activity of these effectors on membrane‐bound host targets is recognized by their corresponding R proteins (Lewis et al., 2008). HopZ2 promotes the growth of P. syringae in Arabidopsis. This activity requires an intact myristoylation sequence and catalytic cysteine, indicating that HopZ2 targets a membrane‐associated protein(s) to promote P. syringae virulence (Lewis et al., 2008). HopZ3 also promotes the growth of P. syringae, but does not possess a myristoylation sequence, suggesting that its targets are soluble proteins (Vinatzer et al., 2006). In soybean, HopZ1a induces an HR in cultivars OAC Bayfield and Williams 82, which requires an intact myristoylation sequence and catalytic cysteine (Zhou et al., 2009). HopZ2 and HopZ3 also trigger an HR in soybean cultivars OAC Bayfield and Williams 82 (not shown; R. L. Morgan and W. Ma, unpublished data). HopZ1b can promote the growth of P. syringae in soybean cultivar OAC Bayfield, and it has recently been demonstrated that this virulence activity partially requires the 2‐hydroxyisoflavanone dehydratase (GmHID1), which interacts directly with HopZ1b (Zhou et al., 2011). Although all members of the HopZ family possess the catalytic triad present in the archetypical member YopJ, their biochemical function on host targets remains to be demonstrated. YopJ can acetylate and inactivate eukaryotic mitogen‐activated protein kinase kinases (MAPKKs), thereby suppressing host immune responses (Mittal et al., 2006; Mukherjee et al., 2006; Mukherjee and Orth, 2008). Ac, acetyl group.
YOPJ FUNCTIONS AND PHENOTYPES
The Yersinia pestis and Y. pseudotuberculosis T3SE YopJ blocks the mammalian innate immune response by inhibiting the mitogen‐activated protein kinase (MAPK) and the nuclear factor kappa B (NFκB) signalling pathways (Mukherjee et al., 2006; 1999, 2000). Furthermore, YopJ promotes apoptosis in macrophages by preventing the activation of the NFκB pathway (Cornelis and Wolf‐Watz, 1997; Monack et al., 1997; Zhang et al., 2005). YopJ inhibits these signalling pathways by binding directly to the MAPK kinases (MAPKKs) and the inhibitor of the NFκB complex, IκBβ, but not to the upstream MAPK kinase kinases (MAPKKKs) or the downstream MAPKs (Orth et al., 1999). The interaction between YopJ and MAPKKs inhibits the phosphorylation of MAPKKs, thus blocking the signal transduction that leads to cytokine production and anti‐apoptotic factor expression (2006, 2007; 1999, 2000).
YopJ contains a conserved catalytic triad [histidine (His), glutamate (Glu) and cysteine (Cys)], and mutations in these catalytic residues disrupt YopJ's ability to inhibit the MAPK and NFκB pathways (Orth et al., 2000). Given that the predicted secondary structure and catalytic domain of YopJ are similar to those of the adenovirus protease (AVP), it was initially hypothesized that YopJ was a cysteine protease (Orth et al., 2000). Indeed, experiments have demonstrated de‐sumoylating and de‐ubiquitinating activity for YopJ (Orth et al., 2000; Sweet et al., 2007). However, direct targets of these protease activities remain to be identified. In addition, using liquid chromatography‐tandem mass spectrometry, Mukherjee et al. (2006) showed that YopJ acetylates MKK6 on serine (Ser) and threonine (Thr) residues in its activation loop. An in vitro acetylation assay further demonstrated that YopJ requires its catalytic Cys to acetylate MKK6 (Mukherjee et al., 2006; Mukherjee and Orth, 2008). In the presence of acetyl‐CoA and YopJ, MKK6 or IκBβ could no longer be phosphorylated by the upstream MAPKKK, thus providing an elegant mechanistic explanation for how YopJ inhibits the MAPK and NFκB signalling pathways. Given the similarity to cysteine proteases, YopJ has been proposed to modify its substrates via a ‘ping–pong mechanism’, whereby YopJ first forms an acetyl‐enzyme covalent intermediate before transferring the acetyl group to the Ser/Thr [or lysine (Lys)] residues of its substrate (Mukherjee et al., 2007).
Mittal et al. (2006) provided additional evidence that YopJ acetylates other MAPKKs (mitogen‐activated protein kinase/extracellular signal‐regulated kinase kinase 1/2, MEK1/2) in their activation loop, which consequently blocks the phosphorylation of these Ser residues in the kinase. Interestingly, Mittal et al. (2006) also observed that, in addition to acetylating its host target MEK2, YopJ appears to autoacetylate. However, autoacetylation does not appear to be required for the acetylation of MEK by YopJ (Mittal et al., 2010). The autoacetylation site of YopJ and its functional consequences remain to be determined. One possibility is that autoacetylation represents the acetyl‐enzyme covalent intermediate predicted from the ping–pong mechanism of transfer in which the catalytic Cys would be acetylated (Mukherjee et al., 2007). YopJ also appears to require a eukaryotic activating factor, inositol hexakisphosphate (IP6), for full activation of its acetyltransferase activity (Mittal et al., 2010).
The MAPK signalling pathway is highly conserved across eukaryotes and therefore, perhaps not surprisingly, YopJ can also disrupt the MAPK signalling pathways in Saccharomyces cerevisiae (Hao et al., 2008; Yoon et al., 2003). Expression of wild‐type YopJ, but not the catalytically inactive form, in yeast disrupts the MAPK pathways used for pheromone perception (mating pathway) and high osmolarity growth (HOG pathway). Similar to the observation in the mammalian system, YopJ disrupts the mating pathway and the HOG pathway in yeast by preventing the phosphorylation of the MAPK Fus3p and Hog1p, respectively (Yoon et al., 2003). Specifically, YopJ blocks the HOG pathway by binding and acetylating Pbs2, a conserved MAPKK, to prevent the activation and phosphorylation of the kinase (Hao et al., 2008). YopJ cannot block the MAPK pathway downstream of the activated MAPKK, Pbs2, as phospho‐mimic mutants of Pbs2 can grow in the presence of high osmolarity and YopJ. Using a suppressor screen to identify Pbs2 mutants that are insensitive to YopJ inhibition of the HOG pathway, Hao et al. (2008) identified a conserved hydrophobic region in the kinase domain of Pbs2 required for YopJ binding. If YopJ is unable to bind to this hydrophobic α‐helix in Pbs2 and other MAPKKs, it cannot acetylate these targets. This represents an elegant use of a heterologous system to understand how YopJ interacts with and modifies its targets, the MAPKKs.
The YopJ superfamily provides an excellent opportunity to investigate the diversification of a T3SE family. How can an effector family effectively promote the virulence of pathogens on hosts from two separate kingdoms? Studies on the phytopathogenic members of the YopJ superfamily are beginning to reveal the mechanisms by which an effector family can diversify not only between animal and plant hosts, but also between different plant hosts. Below, we provide an overview of the YopJ family members from phytopathogens on various plant hosts, which highlights the remarkable functional diversity of this T3SE family.
THE P. SYRINGAE HOPZ FAMILY
The HopZ family of P. syringae comprises three major forms: HopZ1, HopZ2 and HopZ3. Under pressure from the host immune system, HopZ1 diversified into at least three allelic forms (HopZ1a, HopZ1b and HopZ1c) (Ma et al., 2006). HopZ1a induces a strong defence response in a wide variety of plant species, including Arabidopsis thaliana, soybean, sesame, Nicotiana benthamiana and rice (Lewis et al., 2008; Ma et al., 2006). HopZ1b is weakly recognized in Arabidopsis and strongly recognized in N. benthamiana, whereas HopZ1c does not induce defence responses in any host tested so far (Lewis et al., 2008; Zhou et al., 2009). As HopZ1a is most similar to the ancestral HopZ allele in P. syringae, it has been hypothesized that members of HopZ1 diversified under pressure from the host immune response (Ma et al., 2006). HopZ2 is more closely related to Xanthomonas YopJ homologues than the Pseudomonas HopZs, and was putatively acquired by horizontal gene transfer, whereas HopZ3, which is quite divergent from HopZ1 and HopZ2, was putatively acquired by horizontal gene transfer from Erwinia species (Ma et al., 2006). All of the members of the HopZ family, except HopZ3, contain a consensus myristoylation site (Gly2), which is required for membrane localization (Lewis et al., 2008; Zhou et al., 2009).
Based on their relatedness to YopJ and the conservation of the catalytic triad, members of the HopZ family of effector proteins were originally hypothesized to be cysteine proteases (Ma et al., 2006; Orth et al., 2000). The HopZ proteins display weak protease activity using a fluorescence‐based assay with modified casein as a generic substrate (Ma et al., 2006); however, the host targets of this activity remain to be identified. More recently, our data have suggested that HopZ1a can autoacetylate in the presence of eukaryotic cofactors (A. Lee et al., unpublished data). This is reminiscent of YopJ, suggesting that the HopZ family may also be cofactor‐activated acetyltransferases (2006, 2010).
Functions of the HopZ family in Arabidopsis
HopZ1a is recognized by the CC NBS‐LRR R protein ZAR1 in Arabidopsis (Lewis et al., 2010). Recognition of HopZ1a depends on its catalytic Cys residue (Cys216) as well as the consensus myristoylation site (Gly2), which is required for membrane localization. These data suggest that ZAR1 recognizes the membrane‐localized enzymatic activity of HopZ1a (Lewis et al., 2008; Ma et al., 2006).
In the absence of ZAR1 recognition in zar1 mutant Arabidopsis plants, HopZ1a demonstrates a catalytic Cys‐dependent virulence function by promoting the growth of a nonhost P. syringae pv. cilantro 0788–9 strain (Pci0788‐9; Lewis et al., 2010), that natively carries no HopZ allele, although it is closely related to a virulent, HopZ1c‐bearing P. syringae pv. maculicola strain (Ma et al., 2006). These results suggest that ZAR1 may have evolved to monitor and respond to an ancestral virulence function of HopZ1 (Lewis et al., 2010). HopZ1a also appears to suppress ETI induced by the T3SEs AvrRpt2, AvrRpm1 and AvrRps4, as shown by a competitive index measure (2009, 2010). AvrRpt2, AvrRpm1 and AvrRps4 are recognized by different R proteins and require different R gene signalling components relative to HopZ1a to trigger ETI (Aarts et al., 1998; Austin et al., 2002; Century et al., 1995; Feys and Parker, 2000; Lewis et al., 2010; Macho et al., 2010; Muskett et al., 2002; Tornero et al., 2002). It remains to be determined how HopZ1a may suppress ETI from these diverse effector proteins.
Although similar to HopZ1a (72.1% amino acid identity, 80.7% similarity), HopZ1b is only weakly recognized in Arabidopsis ecotype Col‐0 and causes an HR in approximately 25% of leaves (Lewis et al., 2008). The recognition of HopZ1b depends on its catalytic Cys residue (Cys212) and, surprisingly, is independent of ZAR1 (2008, 2010). Therefore, at least two resistance proteins have evolved in Arabidopsis to recognize members of the HopZ family. HopZ1c is almost identical to HopZ1b (97% amino acid identity up to the insertion mutation in hopZ1c that results in a 19‐amino‐acid frameshift and premature stop codon), but is truncated at the C‐terminus, resulting in a protein that is about one‐third smaller than HopZ1b. HopZ1c retains the catalytic residues, but does not have any observable functions in Arabidopsis (or any other plant host) so far (Lewis et al., 2008; Zhou et al., 2009).
HopZ2 is more similar to the Xanthomonas homologues of the YopJ superfamily than to the HopZ1 alleles (Fig. 1; Ma et al., 2006). HopZ2 was originally identified because of its avirulence function in Phaseolus vulgaris (common bean) cultivars (formerly AvrPpiG; Arnold et al., 2001). In Arabidopsis, HopZ2 has a virulence function and promotes the growth of the nonhost Pci0877‐9 strain (Lewis et al., 2008). Virulence again depends on the catalytic Cys of HopZ2 (Cys229) and localization to the membrane, indicating that HopZ2 virulence targets are membrane localized (Lewis et al., 2008).
HopZ3 is most similar to Erwinia members of the YopJ superfamily, which remain uncharacterized at this time (formerly HopPsyV; Deng et al., 2003; Ma et al., 2006; Fig. 1). HopZ3 differs from the HopZ1 and HopZ2 alleles in that it is not membrane associated (Lewis et al., 2008) and the hopZ3 operon contains a predicted type III chaperone (Ma et al., 2006). HopZ3 promotes in planta multiplication of P. syringae pv. syringae strain B728a (PsyB728a) in the nonhost Arabidopsis (Vinatzer et al., 2006).
Functions of the HopZ family in soybean
The cellular functions of HopZ1a and HopZ1b were investigated in Glycine max (soybean) after the discovery that all the P. syringae pv. glycinea strains, which were largely isolated from soybean, produce functional HopZ1b (Ma et al., 2006). Bacterial growth assays demonstrated that HopZ1b can promote P. syringae growth in soybean. For example, the kidney bean isolate P. syringae pv. phaseolicola strain 1302A (Pph1302A) carries no hopZ homologue and multiplies to only a low level in the soybean cultivar OAC Bayfield, but can grow to a 10‐fold higher population density in soybean when transformed with a hopZ1b allele (Zhou et al., 2009).
Although the wild‐type P. syringae pv. glycinea strain BR1 (PgyBR1), carrying the endogenous hopZ1b, is virulent on the soybean cultivars OAC Bayfield and Williams 82, PgyBR1 expressing HopZ1a triggers HR in both cultivars (Ma et al., 2006; Zhou et al., 2009). As discussed earlier, both the HR‐triggering activity of HopZ1a and the pathogen growth‐promoting activity of HopZ1b require the catalytic Cys residue, indicating that the enzymatic activities of HopZ1a and HopZ1b are important for their cellular functions. Domain shuffling experiments demonstrated that a central domain upstream of the conserved catalytic Cys residue of HopZ1a and HopZ1b determines the allelic specificity (Morgan et al., 2010). Within this domain, a single substitution of Cys141 found in HopZ1a with Lys found at the corresponding position in HopZ1b (Lys137) abolishes HopZ1a‐triggered HR in soybean. As this position is under strong positive selection, the Cys141/Lys137 mutation might represent a key step during HopZ1 evolution to evade host recognition. Protein structure modelling analysis indicates that the Cys141/Lys137 residue may play a role in substrate binding (Morgan et al., 2010). Indeed, characterization of HopZ1a‐ and HopZ1b‐interacting proteins in soybean revealed both common and distinct proteins associating with these two alleles (Zhou et al., 2011). These data suggest that sequence diversification allows altered substrate‐binding specificity, which then leads to different cellular functions of HopZ1 alleles.
HopZ1a and HopZ1b both interact with 2‐hydroxyisoflavanone dehydratase (GmHID1), which is an enzyme involved in soybean isoflavone biosynthesis (Zhou et al., 2011). Silencing of GmHID1 leads to increased susceptibility of soybean to P. syringae infection, as well as a compromised HopZ1b‐mediated increase in P. syringae growth, indicating that this enzyme plays a role in host defence and HopZ1b‐mediated virulence. Furthermore, isoflavone levels in soybean leaves inoculated with P. syringae pv. glycinea strains producing functional HopZ1b are significantly lower than in leaves inoculated with P. syringae pv. glycinea carrying the catalytic mutant of HopZ1b. These data, taken together, suggest that HopZ1 suppresses plant defence by inhibiting isoflavone production (Zhou et al., 2011).
HopZ2 and HopZ3 both trigger HR in soybean cultivars OAC Bayfield and Williams 82 (R. L. Morgan and W. Ma, unpublished data); yet, it remains to be determined whether the predicted catalytic residues are required for HopZ2‐ and HopZ3‐triggered HR in soybean. HopZ1c does not have any observable phenotype in soybean so far.
Functions of the HopZ family in N. benthamiana
The functions of the HopZ effector proteins have been investigated in N. benthamiana using Agrobacterium‐mediated transient expression. Both HopZ1a and HopZ1b trigger a catalytic Cys‐dependent HR‐like programmed cell death in N. benthamiana (Lewis et al., 2008; Ma et al., 2006; Zhou et al., 2009). Moreover, the C‐terminal domain downstream of the catalytic core, which is lacking in HopZ1c, is also required for HopZ1b to induce HR in N. benthamiana (H. B. Zhou and W. Ma, unpublished data). This C‐terminal domain might be important for maintaining the protein structure necessary for the enzymatic activity of HopZ1b.
Although both HopZ1a and HopZ1b trigger HR‐like cell death in N. benthamiana, they may be differentially recognized. The N‐terminal myristoylation enhances the HopZ1b‐triggered HR, but does not affect the HopZ1a‐triggered HR. The cell death symptoms elicited by the mutant HopZ1a(G2A) were indistinguishable from those of the wild‐type HopZ1a, whereas HopZ1b(G2A) triggered a weaker cell death symptom, which was also delayed compared with the wild‐type HopZ1b (Zhou et al., 2009).
HopZ2 also triggers an HR‐like programmed cell death when it is transiently expressed in N. benthamiana. Similar to HopZ1a and HopZ1b, the catalytic Cys residue is also required for this activity (Lewis et al., 2008; Ma et al., 2006). To date, it is not known whether the potential myristoylation site, which is conserved between HopZ1 and HopZ2, is important for the HopZ2‐triggered HR in this host.
Transient expression of HopZ3 in N. benthamiana blocks the HR‐like cell death elicited by effectors AvrPto1, HopAA1, HopM1 and HopAE1 (Vinatzer et al., 2006). HopZ1b‐triggered cell death is also suppressed by HopZ3 (Zhou et al., 2009). Furthermore, the catalytic Cys residue of HopZ3 is required to block HR triggered by HopZ1b, indicating that HR suppression by HopZ3 depends on its enzymatic activity. Although it is possible that HopZ3 suppresses the HopZ1b‐triggered HR by competing for binding to similar substrates, it is more likely that HopZ3 targets a common signal transduction component downstream of the initial recognition of these different effectors. This study also demonstrated that HopZ3 can induce an HR‐like cell death response in Phaseolus vulgaris (snap bean) and Nicotiana tabacum (tobacco) using Agrobacterium‐mediated transient expression. Interestingly, HopZ3 negatively contributes to the epiphytic growth of PsyB728a in N. benthamiana despite a lack of cell death induction (Vinatzer et al., 2006). It will be interesting to investigate the potential role of HopZ3 in preinvasive immunity.
Much of the data obtained for the HopZ family in N. benthamiana have relied on transient expression using strong promoters. As this approach results in the over‐expression of these proteins relative to the levels delivered by bacteria, it will be important to demonstrate that the functions ascribed using transient expression translate to functions when delivered by the T3SS.
THE XANTHOMONAS YOPJ HOMOLOGUES
Xanthomonas campestris pathovars contain several YopJ homologues, including AvrBsT, AvrRxv, AvrXv4 and XopJ. This list is likely to expand as more Xanthomonas species and pathovars are characterized and/or sequenced. Unlike the HopZ family in P. syringae, a single Xanthomonas pathovar can carry multiple, different YopJ homologues (Szczesny et al., 2010). As in P. syringae, the Xanthomonas YopJ homologues display extensive functional divergence.
AvrBsT was originally described because of its avirulence function in Capsicum annuum (pepper) isogenic lines carrying the Bs1 R gene (Minsavage et al., 1990). It also causes an HR in C. pubescens (Escolar et al., 2002) and N. benthamiana (Orth et al., 2000). The avirulence function in N. benthamiana (and presumably other species) depends on its catalytic triad (Orth et al., 2000). AvrBsT can also cause an HR in the Arabidopsis ecotype Pi‐0 when it is expressed and delivered by the P. syringae T3SS (Cunnac et al., 2007). AvrBsT‐induced resistance is impaired in R gene signalling mutants, suggesting that the Pseudomonas‐delivered AvrBsT is recognized in an R gene‐mediated fashion in Arabidopsis (Cunnac et al., 2007). Unlike HopZ1a‐induced resistance, AvrBsT‐mediated resistance is dependent on certain known signalling components (i.e. NDR1, EDS1, PAD4, SID2, NPR1), which suggests that the R gene that recognizes AvrBsT differs from ZAR1, which recognizes HopZ1a (Cunnac et al., 2007; Lewis et al., 2010). Using P. syringae to deliver AvrBsT, Cunnac et al. (2007) designed a genetic screen based on the natural genetic variation in AvrBsT‐induced resistance between the resistant Pi‐0 and susceptible Col‐0 ecotypes, and found that the AvrBsT‐induced HR in Pi‐0 was caused by a recessive mutation in SOBER1. SOBER1 demonstrates phospholipase activity in vitro (Kirik and Mudgett, 2009). Functional SOBER1 abrogates the AvrBsT‐induced HR in Arabidopsis and prevents the accumulation of phosphatidic acid (Cunnac et al., 2007; Kirik and Mudgett, 2009). In other systems, homologues to SOBER1 have been implicated in cellular signalling as lipid secondary messengers (Cunnac et al., 2007). Kirik and Mudgett (2009) propose that SOBER1 phospholipase activity suppresses downstream phospholipid stress signalling in response to AvrBsT.
A virulence function has not yet been demonstrated for AvrBsT. Knocking out AvrBsT in X. campestris pv. vesicatoria strain 85–10 does not change the virulence of this strain in tomato; however, this strain also contains two other YopJ homologues, AvrRxv and XopJ, which may confer virulence (Szczesny et al., 2010).
AvrBsT is able to suppress ETI induced by AvrBs1, a non‐YopJ effector protein (Szczesny et al., 2010). Other YopJ homologues in X. campestris pv. vesicatoria strain 85–10 (AvrRxv and XopJ) cannot suppress the AvrBs1‐induced HR, suggesting that their functions have diverged. The ETI suppression function of AvrBsT depends on its catalytic Cys residue (Cys222), and occurs within the plant cell, as demonstrated by Agrobacterium‐mediated transient expression of AvrBsT and AvrBs1 in pepper leaves. The ETI suppression activity of AvrBsT may indicate that AvrBsT and AvrBs1 target a conserved host protein. AvrBsT interacts with an SnRK1 kinase in yeast two‐hybrid assays and by bimolecular fluorescence. The AvrBs1 HR is reduced when SnRK1 is silenced, but it remains to be determined whether SnRK1 is required for AvrBsT‐mediated suppression of the AvrBs1 HR (Szczesny et al., 2010).
AvrRxv from X. campestris pv. vesicatoria induces an HR in some cultivars of Phaselous vulgaris (common bean), G. max (soybean), Vigna sinensis (cowpea), Medicago sativa (alfalfa), Zea mays (corn) and Gossypium hirsutum (cotton) (Whalen et al., 1988), as well as Solanum esculentum (tomato) (Wang et al., 1994; Whalen et al., 1993). Loss of the HR was observed when each residue of the catalytic core (His180, Glu200, Cys244) was independently mutated, indicating that the catalytic function of AvrRxv is necessary for recognition (Bonshtien et al., 2005; Whalen et al., 2008). AvrRxv does not contain a consensus myristoylation sequence and appears to localize to the cytoplasm or the membrane (Bonshtien et al., 2005). AvrRxv interacts with a 14‐3‐3 protein in yeast two‐hybrid assays and in vitro (Whalen et al., 2008). 14‐3‐3 proteins function in protein–protein interactions in diverse cellular activities. It remains to be determined how the AvrRxv–14‐3‐3 interaction contributes to avirulence in tomato.
AvrXv4 from X. campestris pv. vesicatoria has an avirulence function in Solanum pennellii (wild tomato) (Astua‐Monge et al., 2000) and N. benthamiana, which is dependent on its catalytic Cys (Cys219) and His (His155) residues (Roden et al., 2004). In susceptible hosts, AvrXv4 makes a small contribution to virulence (Roden et al., 2004). AvrXv4 has small ubiquitin modifier (SUMO) protease activity and causes a reduction in sumoylated proteins in N. benthamiana and C. annuum (Roden et al., 2004). The specific targets of AvrXv4 activity and the role of sumoylation in pathogenicity remain unanswered; however, it is likely that the targets are cytoplasmic, where AvrXv4 is localized in plant cells (Roden et al., 2004).
XopJ causes an HR in N. benthamiana and N. clevelandii (Thieme et al., 2007). Induction of the HR depends on the plasma membrane localization of XopJ, which probably occurs by myristoylation (Thieme et al., 2007). In Agrobacterium‐mediated transient expression experiments in N. benthamiana, XopJ is found at the plasma membrane and in vesicles that colocalize with a Golgi marker, suggesting that it trafficks through the secretory pathway. Interestingly, catalytic mutants of XopJ are only found at the plasma membrane (Bartetzko et al., 2009). In susceptible hosts, the enzymatic activity of XopJ is necessary for XopJ to block secretion to the apoplast (Bartetzko et al., 2009). In addition, XopJ can partially suppress PTI, and it is possible that the block in secretion contributes to the impairment of PTI.
THE RALSTONIA POPP FAMILY
Ralstonia solanacearum, the bacterial wilt pathogen, has at least two T3SEs that are part of the YopJ superfamily. PopP1 is believed to have been acquired by horizontal gene transfer and has an avirulence function in Petunia (Lavie et al., 2002). Interestingly, the Rhizobium homologue Y4LO and the Xanthomonas XopJ effector protein are part of the same clade. A second R. solanacearum homologue, PopP2, forms a clade with an uncharacterized Xanthomonas homologue (Ma et al., 2006). PopP2 causes an HR in Arabidopsis and is recognized by the cooperative action of the RRS1 and RPS4 R proteins (Deslandes et al., 2002; Narusaka et al., 2009). RRS1 is an atypical TIR‐NBS‐LRR R protein with a C‐terminal WRKY domain found in plant transcription factors (Deslandes et al., 2002), whereas RPS4 is a TIR‐NBS‐LRR R protein originally characterized as recognizing the unrelated P. syringae effector AvrRps4 (Gassmann et al., 1999). Recognition of PopP2 requires its catalytic activity (Tasset et al., 2010) and occurs by direct interaction between PopP2 and RRS1 in the nucleus (Deslandes et al., 2003). RPS4 has also been shown to be nuclear localized, which is necessary for AvrRps4‐triggered defence responses (Wirthmueller et al., 2007). However, it is unknown whether RPS4 interacts directly or indirectly with PopP2 and where this interaction occurs.
Tasset et al. (2010) demonstrated that PopP2 is an acetyltransferase that is autoacetylated at K383, a modification necessary for RRS1 recognition. PopP2 appears to stabilize the RRS1 protein and to prevent its degradation by the proteasome (Tasset et al., 2010). In addition, RRS1 resistance requires RD19, a cysteine protease, whose expression is induced on infection with R. solanacearum (Bernoux et al., 2008). RD19 is relocalized to the nucleus when co‐expressed with RRS1 (Bernoux et al., 2008). RD19 interacts with RRS1 but not PopP2 (Bernoux et al., 2008). As a result, when PopP2 is knocked out in R. solanacearum, the strain is highly virulent in Arabidopsis (Narusaka et al., 2009).
THE RHIZOBIUM Y4LO HOMOLOGUE
Rhizobium species are usually considered to exist in symbiosis with their plant hosts, where they contribute to nitrogen fixation, and the Rhizobium T3SS can contribute to nodulation in some host plants (Marie et al., 2001). A YopJ homologue, Y4LO, has been identified in Rhizobium species (Astua‐Monge et al., 2000; Ciesiolka et al., 1999; Ma et al., 2006). Y4LO is most closely related to the XopJ homologue from Xanthomonas (Ma et al., 2006). Y4LO can be delivered by the P. syringae T3SS into plants (J. Chang, Oregon State University, Corvallis, OR, USA, personal communication). In Tephrosia vogelii, Phaseolus vulgaris and Vigna sinensis, Y4LO contributes to the production of effective nitrogen‐fixing nodules and normal nitrogen content in the plant (Yang et al., 2009). Strains of Rhizobium with Y4LO disruptions still form nodules; however, differentiation into the symbiosome, with the subsequent ability to fix nitrogen, does not appear to complete normally (Yang et al., 2009).
CONCLUSIONS AND FUTURE DIRECTIONS
The YopJ superfamily shows remarkable diversification for a T3SE family, with homologues in animal pathogens, plant pathogens and plant symbionts (1, 2; Table 1). In plants, members of the YopJ superfamily have been demonstrated to contribute to either disease or immunity in a host‐dependent manner (Fig. 2). In animals, members of the YopJ superfamily have well‐characterized, disease‐promoting roles; however, it remains to be established whether they can also limit bacterial virulence as observed in plants. Interestingly, the YopJ homologue AvrA has been suggested to limit virulence in vertebrates, indicating that the YopJ superfamily may also display avirulence activities outside of the plant kingdom (Collier‐Hyams et al., 2002).
All of the YopJ homologues described so far require a common catalytic triad, despite different enzymatic activities. YopJ and PopP2 are acetyltransferases and HopZ1a also shows evidence of this function (Mittal et al., 2006; Mukherjee et al., 2006; Tasset et al., 2010; A. Lee et al., unpublished data). Other members are SUMO proteases (AvrXv4), and weak protease activity has been shown for the HopZ family (Ma et al., 2006; Roden et al., 2004). However, the host targets of these enzymatic activities are still unknown in many cases. It will be critical to demonstrate enzymatic activity on host substrates to confirm their function. It is possible that these effector proteins are bifunctional enzymes with distinct enzymatic functions encoded by the same catalytic triad, as observed with cysteine proteases and acetyltransferases (Mukherjee et al., 2007). Neofunctionalization of enzymatic function might be more likely in strains of Xanthomonas, where multiple YopJ homologues are found in a single strain.
The targets of phytopathogenic YopJ homologues remain unidentified and are of great interest. Will the plant YopJ homologues also target kinases, as does YopJ? MAP kinases are targeted by several unrelated effector proteins, for example HopAI1 and HopF2, to suppress PTI (Lewis et al., 2009; Wang et al., 2010; Zhang et al., 2007). A SnRK kinase has been shown to interact with AvrBsT. However, the enzymatic activity of AvrBsT and the resultant modification of its targets remain to be determined (Szczesny et al., 2010). It remains to be seen whether disruption of kinase signalling is a general immune suppression technique of the YopJ superfamily. Given the diverse subcellular localization patterns of the YopJ homologues, they probably employ an array of molecular strategies to promote immunosuppression and pathogenesis.
ACKNOWLEDGEMENTS
This article is based on presentations given at the 8th International Conference on Pseudomonas syringae Pathovars and Related Pathogens held at Oxford University, UK, in September 2010. We thank Brenden Hurley, Dr David Mackey and an anonymous reviewer for providing thoughtful and constructive comments. This publication was supported by Natural Sciences and Engineering Research Council (NSERC) discovery grants to DD and DSG; an NSERC graduate scholarship to AL; the Canadian Foundation for Innovation (DD); a Canada Research Chair in Plant–Microbe Systems Biology (DD) and Comparative Genomics (DSG); the Centre for the Analysis of Genome Evolution and Function (DD and DSG); the National Science Foundation (NSF) (IOS#0847870) and United States Department of Agriculture—Research Support Allocation Process (USDA‐RSAP; WM).
REFERENCES
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis . Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, D.L. , Jackson, R.W. , Fillingham, A.J. , Goss, S.C. , Taylor, J.D. , Mansfield, J.W. and Vivian, A. (2001) Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi . Microbiology, 147, 1171–1182. [DOI] [PubMed] [Google Scholar]
- Astua‐Monge, G. , Minsavage, G.V. , Stall, R.E. , Vallejos, C.E. , Davis, M.J. and Jones, J.B. (2000) Xv4‐avrxv4: a new gene‐for‐gene interaction identified between Xanthomonas campestris pv. vesicatoria race T3 and the wild tomato relative Lycopersicon pennellii . Mol. Plant–Microbe Interact. 13, 1346–1355. [DOI] [PubMed] [Google Scholar]
- Austin, M.J. , Muskett, P. , Kahn, K. , Feys, B.J. , Jones, J.D.G. and Parker, J.E. (2002) Regulatory role of SGT1 in early R gene‐mediated plant defenses. Science, 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Bartetzko, V. , Sonnewald, S. , Vogel, F. , Hartner, K. , Stadler, R. , Hammes, U.Z. and Bornke, F. (2009) The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall‐associated defense responses. Mol. Plant–Microbe Interact. 22, 655–664. [DOI] [PubMed] [Google Scholar]
- Bernoux, M. , Timmers, T. , Jauneau, A. , Briere, C. , de Wit, P. , Marco, Y. and Deslandes, L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1‐R‐mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell, 20, 2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bonshtien, A. , Lev, A. , Gibly, A. , Debbie, P. , Avni, A. and Sessa, G. (2005) Molecular properties of the Xanthomonas AvrRxv effector and global transcriptional changes determined by its expression in resistant tomato plants. Mol. Plant–Microbe Interact. 18, 300–310. [DOI] [PubMed] [Google Scholar]
- Century, K.S. , Holub, E.B. and Staskawicz, B.J. (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA, 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Ciesiolka, L.D. , Hwin, T. , Gearlds, J.D. , Minsavage, G.V. , Saenz, R. , Bravo, M. , Handley, V. , Conover, S.M. , Zhang, H. , Caporgno, J. , Phengrasamy, N.B. , Toms, A.O. , Stall, R.E. and Whalen, M.C. (1999) Regulation of expression of avirulence gene avrRxv and identification of a family of host interaction factors by sequence analysis of AvrBsT. Mol. Plant–Microbe Interact. 12, 35–44. [DOI] [PubMed] [Google Scholar]
- Collier‐Hyams, L.S. , Zeng, H. , Sun, J. , Tomlinson, A.D. , Bao, Z.Q. , Chen, H. , Madara, J.L. , Orth, K. and Neish, A.S. (2002) Cutting Edge: Salmonella AvrA effector inhibits the key proinflammatory, anti‐apoptotic NF‐kappa B pathway. J. Immunol. 169, 2846–2850. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R. and Wolf‐Watz, H. (1997) The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23, 861–867. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Wilson, A. , Nuwer, J. , Kirik, A. , Baranage, G. and Mudgett, M.B. (2007) A conserved carboxylesterase is a SUPPRESSOR OF AVRBST‐ELICITED RESISTANCE in Arabidopsis . Plant Cell, 19, 688–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D.G. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Deng, W.L. , Rehm, A.H. , Charkowski, A.O. , Rojas, C.M. and Collmer, A. (2003) Pseudomonas syringae exchangeable effector loci: sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J. Bacteriol. 185, 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Theulieres, F. , Hirsch, J. , Feng, D.X. , Bittner‐Eddy, P. , Beynon, J. and Marco, Y. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1‐R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA, 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, L. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolar, L. , van den Ackerveken, G. , Pieplow, S. , Rossier, O. and Bonas, U. (2002) Type III secretion and in planta recognition of the Xanthomonas avirulence proteins AvrBs1 and AvrBsT. Mol. Plant Pathol. 2, 287–296. [DOI] [PubMed] [Google Scholar]
- Feys, B.J. and Parker, J.E. (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Galan, J.E. and Wolf‐Watz, H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature, 444, 567–573. [DOI] [PubMed] [Google Scholar]
- Gassmann, W. , Hinsch, M.E. and Staskawicz, B.J. (1999) The Arabidopsis RPS4 bacterial resistance gene is a member of the TIR‐NBS‐LRR family of disease resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Hao, Y.H. , Wang, Y. , Burdette, D. , Mukherjee, S. , Keitany, G. , Goldsmith, E. and Orth, K. (2008) Structural requirements for Yersinia YopJ inhibition of MAP kinase pathways. PLoS ONE, 3, e1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kirik, A. and Mudgett, M.B. (2009) SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc. Natl. Acad. Sci. USA, 106, 20 532–20 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie, M. , Shillington, E. , Eguiluz, C. , Grimsley, N. and Boucher, C. (2002) PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host‐specificity factor and modulates aggressiveness of Ralstonia solanacearum . Mol. Plant–Microbe Interact. 15, 1058–1068. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D. , Abada, W. , Ma, W.B. , Guttman, D.S. and Desveaux, D. (2008) The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana . J. Bacteriol. 190, 2880–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.D. , Desveaux, D. and Guttman, D.S. (2009) The targeting of plant cellular systems by injected type III effector proteins. Semin. Cell Dev. Biol. 20, 1055–1063. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D. , Wu, R. , Guttman, D.S. and Desveaux, D. (2010) Allele‐specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 6, e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W.B. , Dong, F.F.T. , Stavrinides, J. and Guttman, D.S. (2006) Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2, e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. , Ruiz‐Albert, J. , Tornero, P. and Beuzon, C.R. (2009) Identification of new type III effectors and analysis of the plant response by competitive index. Mol. Plant Pathol. 10, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. , Guevara, C.M. , Tornero, P. , Ruiz‐Albert, J. and Beuzon, C.R. (2010) The Pseudomonas syringae effector protein HopZ1a suppresses effector‐triggered immunity. New Phytol. 187, 1018–1033. [DOI] [PubMed] [Google Scholar]
- Marie, C. , Broughton, W.J. and Deakin, W.J. (2001) Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4, 336–342. [DOI] [PubMed] [Google Scholar]
- Minsavage, G.V. , Dahlbeck, D. , Whalen, M.C. , Kearney, B. , Bonas, U. , Staskawicz, B.J. and Stall, R.E. (1990) Gene‐for‐gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria–pepper interactions. Mol. Plant–Microbe Interact. 3, 41–47. [Google Scholar]
- Mittal, R. , Peak‐Chew, S.Y. and McMahon, H.T. (2006) Acetylation of MEK2 and I kappa B Kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc. Natl. Acad. Sci. USA, 103, 18 574–18 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, R. , Peak‐Chew, S.Y. , Sade, R.S. , Vallis, Y. and McMahon, H.T. (2010) The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J. Biol. Chem. 285, 19 927–19 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack, D.M. , Mecsas, J. , Ghori, N. and Falkow, S. (1997) Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc. Natl. Acad. Sci. USA, 94, 10 385–10 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, R.L. , Zhou, H.B. , Lehto, E. , Nguyen, N. , Bains, A. , Wang, X.Q. and Ma, W.B. (2010) Catalytic domain of the diversified Pseudomonas syringae type III effector HopZ1 determines the allelic specificity in plant hosts. Mol. Microbiol. 76, 437–455. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S. and Orth, K. (2008) In vitro signaling by MAPK and NF kappa B pathways inhibited by Yersinia YopJ. Methods Enzymol. 438, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S. , Keitany, G. , Li, Y. , Wang, Y. , Ball, H.L. , Goldsmith, E.J. and Orth, K. (2006) Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science, 312, 1211–1214. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S. , Hao, Y.H. and Orth, K. (2007) A newly discovered post‐translational modification: the acetylation of serine and threonine residues. Trends Biochem. Sci. 32, 210–216. [DOI] [PubMed] [Google Scholar]
- Muskett, P.R. , Kahn, K. , Austin, M.J. , Moisan, L.J. , Sadanandom, A. , Shirasu, K. , Jones, J.D.G. and Parker, J.E. (2002) Arabidopsis RAR1 exerts rate‐limiting control of R gene‐mediated defenses against multiple pathogens. Plant Cell, 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka, M. , Shirasu, K. , Noutoshi, Y. , Kubo, Y. , Shiraishi, T. , Iwabuchi, M. and Narusaka, Y. (2009) RRS1 and RPS4 provide dual Resistance‐gene system against fungal and bacterial pathogens. Plant J. 60, 218–226. [DOI] [PubMed] [Google Scholar]
- Orth, K. , Palmer, L.E. , Bao, Z.Q. , Stewart, S. , Rudolph, A.E. , Bliska, J.B. and Dixon, J.E. (1999) Inhibition of the mitogen‐activated protein kinase kinase superfamily by a Yersinia effector. Science, 285, 1920–1923. [DOI] [PubMed] [Google Scholar]
- Orth, K. , Xu, Z.H. , Mudgett, M.B. , Bao, Z.Q. , Palmer, L.E. , Bliska, J.B. , Mangel, W.F. , Staskawicz, B. and Dixon, J. (2000) Disruption of signaling by Yersinia effector YopJ, a ubiquitin‐like protein protease. Science, 290, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Roden, J. , Eardley, L. , Hotson, A. , Cao, Y.Y. and Mudgett, M.B. (2004) Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant–Microbe Interact. 17, 633–643. [DOI] [PubMed] [Google Scholar]
- Sweet, C.R. , Conlon, J. , Golenbock, D.T. , Goguen, J. and Silverman, N. (2007) YopJ targets TRAF proteins to inhibit TLR‐mediated NF‐kappa B, MAPK and IRF3 signal transduction. Cell. Microbiol. 9, 2700–2715. [DOI] [PubMed] [Google Scholar]
- Szczesny, R. , Buttner, D. , Escolar, L. , Schulze, S. , Seiferth, A. and Bonas, U. (2010) Suppression of the AvrBs1‐specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1‐related kinase. New Phytol. 187, 1058–1074. [DOI] [PubMed] [Google Scholar]
- Tasset, C. , Bernoux, M. , Jauneau, A. , Pouzet, C. , Briere, C. , Kieffer‐Jacquinod, S. , Rivas, S. , Marco, Y. and Deslandes, L. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1‐R‐mediated immunity in Arabidopsis . PLoS Pathog. 6, e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme, F. , Szczesny, R. , Urban, A. , Kirchner, O. , Hause, G. and Bonas, U. (2007) New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N‐myristoylation motif. Mol. Plant–Microbe Interact. 20, 1250–1261. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P. , Nurnberger, T. and Joosten, M.H. (2011) Of PAMPs and effectors: the blurred PTI–ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P. , Merritt, P. , Sadanandom, A. , Shirasu, K. , Innes, R.W. and Dangl, J.L. (2002) RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell, 14, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer, B.A. , Teitzel, G.M. , Lee, M.W. , Jelenska, J. , Hotton, S. , Fairfax, K. , Jenrette, J. and Greenberg, J.T. (2006) The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non‐host plants. Mol. Microbiol. 62, 26–44. [DOI] [PubMed] [Google Scholar]
- Wang, J.F. , Stall, R.E. and Vallejos, C.E. (1994) Genetic analysis of a complex hypersensitive reaction to bacterial spot in tomato. Phytopathology, 84, 126–132. [Google Scholar]
- Wang, Y.J. , Li, J.F. , Hou, S.G. , Wang, X.W. , Li, Y.A. , Ren, D.T. , Chen, S. , Tang, X.Y. and Zhou, J.M. (2010) A Pseudomonas syringae ADP‐ribosyltransferase inhibits Arabidopsis mitogen‐activated protein kinase kinases. Plant Cell, 22, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, M.C. , Stall, R.E. and Staskawicz, B.J. (1988) Characterization of a gene from a tomato pathogen determining hypersensitive resistance in non‐host species and genetic analysis of this resistance in bean. Proc. Natl. Acad. Sci. USA, 85, 6743–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen, M.C. , Wang, J.F. , Carland, F.M. , Heiskell, M.E. , Dahlbeck, D. , Minsavage, G.V. , Jones, J.B. , Scott, J.W. , Stall, R.E. and Staskawicz, B.J. (1993) Avirulence gene avrRxv from Xanthomonas campestris pv. vesicatoria specifies resistance on tomato line Hawaii 7998. Mol. Plant–Microbe Interact. 6, 616–627. [DOI] [PubMed] [Google Scholar]
- Whalen, M.C. , Richter, T. , Zakhareyvich, K. , Yoshikawa, M. , Al‐Azzeh, D. , Adefioye, A. , Spicer, G. , Mendoza, L.L. , Morales, C.Q. , Klassen, V. , Perez‐Baron, G. , Toebe, C.S. , Tzovolous, A. , Gerstman, E. , Evans, E. , Thompson, C. , Lopez, M. and Ronald, P.C. (2008) Identification of a host 14‐3‐3 protein that interacts with Xanthomonas effector AvrRxv. Physiol. Mol. Plant Pathol. 72, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller, L. , Zhang, Y. , Jones, J.D.G. and Parker, J.E. (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1‐dependent defense. Curr. Biol. 17, 2023–2029. [DOI] [PubMed] [Google Scholar]
- Yang, F.J. , Cheng, L.L. , Zhang, L. , Dai, W.J. , Liu, Z. , Yao, N. , Xie, Z.P. and Staehelin, C. (2009) Y4LO of Rhizobium sp strain NGR234 is a symbiotic determinant required for symbiosome differentiation. J. Bacteriol. 191, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yoon, S. , Liu, Z.C. , Eyobo, Y. and Orth, K. (2003) Yersinia effector YopJ inhibits yeast MAPK signaling pathways by an evolutionarily conserved mechanism. J. Biol. Chem. 278, 2131–2135. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Cui, H. , Chen, L.J. , Li, H.T. , Zou, Y. , Long, C.Z. , Lan, L.F. , Chai, J.J. , Chen, S. , Tang, X.Y. and Zhou, J.M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Ting, A.T. , Marcu, K.B. and Bliska, J.B. (2005) Inhibition of MAPK and NF‐kappa B pathways is necessary for rapid apoptosis in macrophages infected with Yersinia . J. Immunol. 174, 7939–7949. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Lin, J. , Johnson, A. , Morgan, R.L. , Zhong, W. and Ma, W. (2011) Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe, 9, 177–186. [DOI] [PubMed] [Google Scholar]
- Zhou, H.B. , Morgan, R.L. , Guttman, D.S. and Ma, W.B. (2009) Allelic variants of the Pseudomonas syringae type III effector HopZ1 are differentially recognized by plant resistance systems. Mol. Plant-Microbe Interact. 22, 176–189. [DOI] [PubMed] [Google Scholar]


