SUMMARY
Clubroot disease of Brassicaceae is caused by an obligate biotrophic protist, Plasmodiophora brassicae. During root gall development, a strong sink for assimilates is developed. Among other genes involved in sucrose and starch synthesis and degradation, the increased expression of invertases has been observed in a microarray experiment, and invertase and invertase inhibitor expression was confirmed using promoter::GUS lines of Arabidopsis thaliana. A functional approach demonstrates that invertases are important for gall development. Different transgenic lines expressing an invertase inhibitor under the control of two root‐specific promoters, Pyk10 and CrypticT80, which results in the reduction of invertase activity, showed clearly reduced clubroot symptoms in root tissue with highest promoter expression, whereas hypocotyl galls developed normally. These results present the first evidence that invertases are important factors during gall development, most probably in supplying sugars to the pathogen. In addition, root‐specific repression of invertase activity could be used as a tool to reduce clubroot symptoms.
INTRODUCTION
Clubroot disease within the family Brassicaceae is caused by the obligate biotrophic protist Plasmodiophora brassicae. During disease development, the root is transformed into a large gall, thus also influencing the aerial part of the plant (Siemens et al., 2002). Gall development is controlled, among other factors, by two plant hormones, i.e. auxin and cytokinin (Ludwig‐Müller and Schuller, 2008). The pathogen is completely dependent on its host for nutritional purposes. Therefore, the root gall is transformed into a strong metabolic sink accumulating sugars and starch (Keen and Williams, 1969). Reallocation of assimilates from leaves to infected hypocotyls has been described in this phase of infection (Keen and Williams, 1969). Amyloplasts accumulate in infected host roots, but remain restricted to host cells (Mithen and Magrath, 1992), such that starch hydrolysis has to take place in the host cytoplasm and plastids. A change in partitioning of carbon, with a higher root to shoot ratio, is observed in infected plants (Evans and Scholes, 1995). In addition, the amount of starch in the leaves of infected plants is lower than in control plants, and both soluble and storage carbohydrates accumulate in infected tissues (Evans and Scholes, 1995). Brodmann et al. (2002) found that glucose and fructose, but not sucrose, increased in infected hypocotyls and roots, suggesting the induction of invertase‐mediated active sink metabolism, because starch also increased simultaneously in clubroots at that time point. This is in accordance with observations that young vegetative secondary plasmodia of the pathogen are found near the vascular bundle (Kobelt et al., 2000). In a study using the Affymetrix ATH1 microarray, many genes associated with sugar transport and metabolism were differentially regulated (Siemens et al., 2006). Although the former indicated a strong change in trafficking of metabolites between different cells and compartments, the large number of genes involved in starch synthesis, which were upregulated, demonstrated the importance of assimilate accumulation in infected roots.
In higher plants, the growth and metabolism of sink tissues are mainly sustained by sucrose synthesized in source leaves and transported through the phloem into sink tissues. The use of sucrose in sink tissues requires cleavage of the glycosidic bond, catalysed by both sucrose synthase and invertases. Sucrose synthase cleaves sucrose into uridine diphospho‐ (UDP‐)glucose and UDP‐fructose, whereas invertases hydrolyse sucrose into hexose monomers. Three types of invertase isoenzyme are distinguished on the basis of solubility, subcellular localization, pH optima and isoelectric point (Roitsch and González, 2004). Cell wall‐bound invertases have been shown to play a crucial function in carbohydrate partitioning and supply of photoassimilates to sink tissues (Goetz et al., 2001; Hirsche et al., 2008; Tang et al., 1999; Weschke et al., 2003). Both vacuolar and cell wall invertases have been shown to play a key role in the active growth of young roots (Eschrich, 1980; Godt and Roitsch, 2006; Tang et al., 1999; Tymowska‐Lalanne and Kreis, 1998), with a predominant role of vacuolar invertase in root elongation, especially in the lower part of the roots, where cell division and elongation occur (Seergeva et al., 2006). Vacuolar invertase could be a key regulator of cell expansion because of the doubled osmotic potential generated by sucrose cleavage in the vacuole. In addition, it could play a role in the stimulation of phloem unloading to parenchyma cells (Seergeva et al., 2006; Sturm and Tang, 1999).
In addition, cell wall invertases could play a key function in providing an enhanced flow of utilizable carbohydrates to pathogens. Hexoses generated by the increased invertase activity, induced by pathogen attack, act, in turn, as signalling molecules, repressing photosynthetic genes (reviewed in Berger et al., 2007). The repression of cell wall invertase impaired and delayed defence reactions to Phytophthora nicotianae in leaves of tobacco (Essmann et al., 2008) and to Xanthomonas campestris in leaves of tomato (Kocal et al., 2008).
In addition, in symbiotic interactions, such as in arbuscular mycorrhizal roots, increased expression and activity of cell wall invertases at stages with high carbohydrate demand support a key function of this enzyme in increasing sink strength by phloem unloading (Schaarschmidt et al., 2006). The observed reduction in mycorrhiza formation in tobacco roots with diminished apoplastic root invertase activity demonstrates the role of invertases in supplying hexoses to the mycorrhizal root (2007a, 2007b). In tomato, the reduced mycorrhizal colonization in plants affected in the synthesis of jasmonic acid is related to a reduction in the expression and activity of cell wall invertase, suggesting that one of the mechanisms by which jasmonic acid modulates mycorrhiza formation may be through the influence of invertase activity in carbon partitioning in the plant (Tejeda‐Sartorius et al., 2008). Invertases are part of a regulation network with cross‐links to nearly all classes of phytohormones, which is important because of the increased demand of carbohydrates during hormone‐promoted growth (Proels and Roitsch, 2009; Roitsch and González, 2004).
Currently, different transgenic approaches based on altered sucrose metabolism by ectopic expression or antisense repression of an invertase have addressed the control of source–sink relationships and the effect of carbohydrate metabolism on disease development. Antisense expression of a vacuolar or cell wall invertase in tap roots of carrot resulted in an increased leaf to root ratio, with reduced tap root development in cell wall invertase antisense lines (Tang et al., 1999). The hexoses generated by the constitutive expression of a yeast invertase in the cell wall of tobacco and Arabidopsis resulted in enhanced resistance to tobacco mosaic virus (Herbers et al., 1996). The reduction of invertase activity by the expression of an antisense construct or the generation of single knock‐out plants presents the limitation of repression of a specific invertase isoform, leading to a lack of clear effects, as has been described for resistance against Pseudomonas syringae or Alternaria brassicicola in Arabidopsis thaliana single knock‐out plants for cell wall invertases (Berger et al., 2007). Proteinaceous invertase inhibitors constitute, in addition to transcriptional regulation, one of the main mechanisms regulating plant invertases (Rausch and Greiner, 2004). The use of an invertase inhibitor has proven to be a useful tool to circumvent the limitations of antisense inhibition for the prevention of cold‐induced sweetening of potato tubers (Greiner et al., 1999) and to elucidate the role of invertases in cytokinin‐mediated delay of senescence (Balibrea Lara et al., 2004).
In this article, the role of invertases in the development of clubroot disease was analysed. In an attempt to modify photoassimilate allocation to the root, root invertase activity was reduced by tissue‐specific expression of an invertase inhibitor. Reduced invertase activity in the root led to reduced clubroot symptoms, mainly in the root tissue, as a result of the root‐specific promoters used in this study, whereas hypocotyl galls developed normally.
RESULTS
Expression of invertases during clubroot disease
The root galls caused by the obligate biotrophic pathogen P. brassicae are dependent on nutrients from the host plant (reviewed in Ludwig‐Müller et al., 2009). Therefore, data obtained in previously conducted microarray experiments on galls of the compatible clubroot interaction in Arabidopsis thaliana ecotype Col‐0 (Siemens et al., 2006) have been re‐examined for differentially expressed genes associated with sugar metabolism, and presented in a MAPMAN graph (Figs S1 and S2, see Supporting Information). The two time points of the analysis reflect the establishment of the gall without visible symptoms and the pathogen mostly at the stage of young vegetative plasmodia (TP1), and the fully grown gall with mature plasmodia and resting spores of the pathogen (TP2), where nutrients from the host are needed for the growth of the pathogen. It was shown that, in general, sugar metabolism genes, including invertases, were upregulated during gall formation.
As invertases are key enzymes for phloem unloading, there is reason to believe that they might be important for the nutrition of the obligate biotrophic pathogen; therefore promoter::GUS lines (GUS, β‐glucuronidase) for four cell wall and two vacuolar invertases, as well as for three invertase inhibitors, were generated and analysed for expression during clubroot development (Fig. 1). In contrast with expression analyses, promoter::GUS lines would also provide information on the spatial expression of invertase genes in the galls. For the earlier time point, we chose the first visible GUS staining in the majority of young galls at 13 days after inoculation (dai). Quantitative real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) indicated expression in control roots of cell wall invertases AtcwINV5 and AtcwINV1 and vacuolar invertases Atßfruct4 and Atßfruct3, but not AtcwINV4 and AtcwINV2 (data not shown). GUS expression in galls could be shown for the cell wall invertase line pAtcwINV1::GUS (Fig. 1E,F), vacuolar invertases pAtßfruct3::GUS (Fig. 1A,B) and pAtßfruct4::GUS (Fig. 1C,D), and the invertase inhibitor pAtC/VIF1::GUS (Fig. 1G,H). In the other studied lines, pAtcwINV4::GUS, pAtcwINV5::GUS, pAtcwINV2::GUS, pAtC/VIF2::GUS and pAtC/VIF3::GUS, GUS expression in roots could not be visualized by staining under the growth conditions optimal for gall development, and these lines are therefore not shown [for more information on the promoter::GUS lines, see Table S1 (Supporting Information)].
Figure 1.
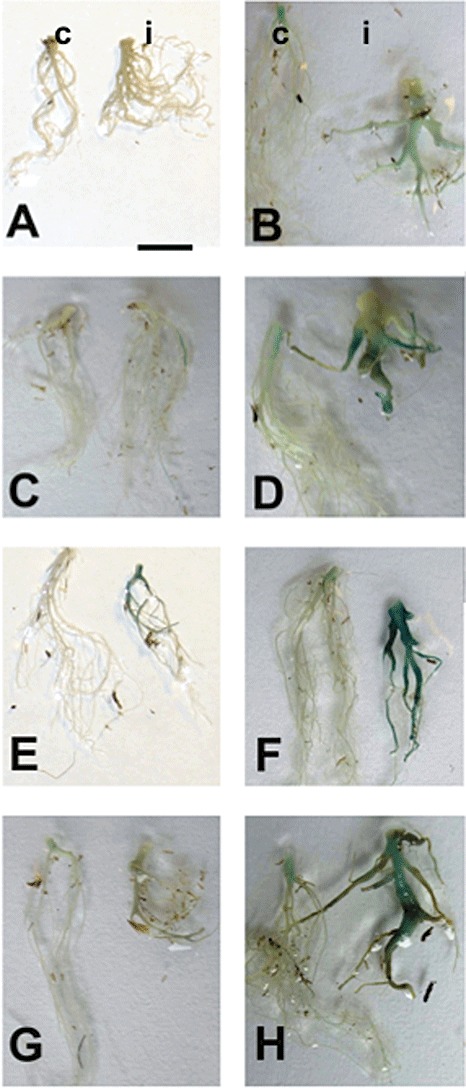
Expression pattern analysis of selected invertase and invertase inhibitor genes in Plasmodiophora brassicae‐infected (i) and control (c) roots using promoter::GUS fusions and X‐Gluc staining of whole roots (GUS, β‐glucuronidase; X‐Gluc, 5‐bromo‐4‐chloro‐3‐indoyl‐β‐d‐glucuronide). The bar indicates 1 cm. The first symptoms became macroscopically visible at 12–14 days after inoculation (dai); therefore, in the left‐hand panels, 13‐dai plants are shown, which equals the first time point of the microarray experiment (for more details, see text). In the right‐hand panels, 23‐dai plants are shown. The expression pattern is shown for promoter::GUS lines of vacuolar invertases pAtßfruct3::GUS (A, B) and pAtßfruct4::GUS (C, D), the cell wall invertase pAtcwINV1::GUS (E, F) and the invertase inhibitor pAtC/VIF1::GUS (G, H).
The gene for extracellular invertase, AtcwINV1, was slightly upregulated in young galls (Fig. 1E,F), when the symptoms started to become visible, and strongly upregulated in developed galls. The same pattern was observed for the expression of the gene for vacuolar invertase, Atßfruct4 (Fig. 1C,D), although the intensity of GUS staining was lower for this invertase. In contrast, the line pAtßfruct3::GUS showed very weak expression level at 13 dai (Fig. 1A,B). Upregulation in galls could only be shown by GUS staining at TP2. The invertase inhibitor reporter line pAtC/VIF1::GUS revealed a weak expression level at 13 dai, but, at 23 dai, the expression was strong in the upper part of the root and hypocotyl of control plants and highly upregulated in infected roots (Fig. 1G,H).
Functional approaches to reduce invertase activity in roots
The fact that several invertases were upregulated according to promoter::GUS studies (Fig. 1) made the use of knockout lines for single genes not feasible. Therefore, in an attempt to modify the sink strength of the roots and to study the effect on the whole plant and disease development, we expressed different invertase inhibitors under the control of root‐specific promoters. Two different root‐specific promoters were used, the promoter C of the root‐specific β‐glucosidase PYK10 gene (Nitz et al., 2001) and a cryptic promoter described by Mollier et al. (2000). Both promoters have been shown to confer root‐specific expression by the use of GUS reporter plants. The fragment C of the Pyk10 promoter is active at different intensities throughout the whole plant in young developmental stages but, from 5 days after germination (dag) onwards, expression is restricted to the whole root, except for the elongation zone, with no detectable expression in the upper part of the plant at day 14 (Nitz et al., 2001). The 2.1‐kb fragment of the cryptic promoter directed expression of the GUS reporter gene mainly to the root, with the exception of the elongation zone of the root tip, with maximal expression at 7 and 15 dag, more localized to the end of the roots and secondary roots from 26 dag onwards (Mollier et al., 2000). Two different Arabidopsis invertase inhibitors, with high homology to vacuolar (AtC/VIF1) and cell wall (AtC/VIF2) invertase inhibitors from tobacco, were used for the reduction of invertase activity in the root. Recent in vitro evidence has shown that AtC/VIF1 specifically inhibits vacuolar invertase, whereas AtC/VIF2 inhibits both acid invertase activities, although with a 10‐fold higher activity for vacuolar than for cell wall invertase (Link et al., 2004). From the different independent transformants obtained for pPyk10::AtC/VIF2, pCrypticT80::AtC/VIF2 and pCrypticT80::AtC/VIF1, several lines were chosen for further analysis of the effect of invertase inhibitor activity on plant phenotype. These lines were shown to have a statistically significant increased expression of the corresponding transgene compared with the wild‐type by Northern blot analysis (Fig. 2). Lines 1 and 2 for pPyk10::AtC/VIF2 and pCrypticT80:AtC/VIF1 were homozygous lines obtained from the same heterozygous parental line; therefore, only one of the two lines is shown. The transcript analysis is presented for all three lines in the case of pCrypticT80::AtC/VIF2.
Figure 2.
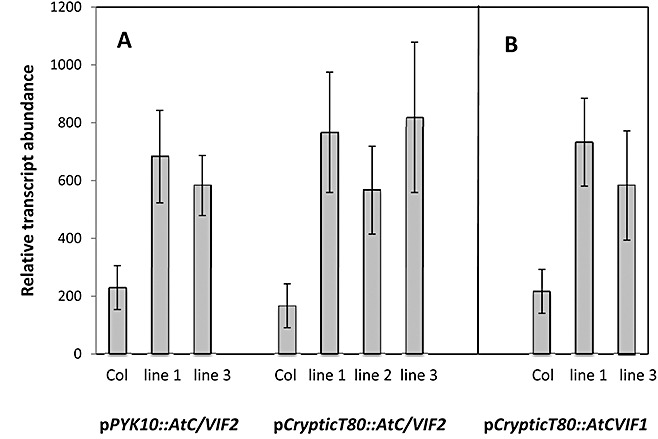
Analysis of transgene expression. RNA was isolated from roots of individual plants grown for 40 days. (A) Hybridization of RNA isolated from wild‐type Col‐0 pPyk10::AtC/VIF2 and pCrypticT80::AtC/VIF2 transgenic plants with a probe for AtC/VIF2. (B) Hybridization with an AtC/VIF1 probe of RNA isolated from wild‐type Col‐0 and pCrypticT80::AtC/VIF1 transgenic plants. Lines 1 and 2 for pPyk10::AtC/VIF2 and pCrypticT80:AtC/VIF1 were homozygous lines obtained from the same heterozygous parental line; therefore, only one of the two lines is shown here as an example. Data were similar for the second line. Transcript analysis is presented for three lines in the case of pCrypticT80::AtC/VIF2. At least three independent RNA samples per line were analysed and the mean values ± ;standard error are presented as the relative transcript abundance (arbitrary units). Expression of the transgene was normalized on the expression of rRNA.
The growth conditions affected the phenotype of the plants, and only the combination of long‐day conditions and cultivation in Hoagland solution produced the phenotype referred to in Fig. 3 (data not shown). Expression of the transgene resulted in enhanced growth of transgenic plants during early development, although no apparent difference was observed in the time of flowering compared with wild‐type control plants (Fig. 3). Transgenic plants did not show a consistent alteration in root fresh weight, but rather a tendency to an increased shoot weight because of greater branching (data not shown), and therefore an altered shoot to root ratio. In addition, the seed weight per plant showed a marked increase in comparison with wild‐type plants. In soil‐grown plants, greater branching was not observed and therefore the differences in shoot phenotypes were not so evident (data not shown); noninfected root phenotypes were similar to the wild‐type (Table 1), which is important for the evaluation of infection experiments.
Figure 3.
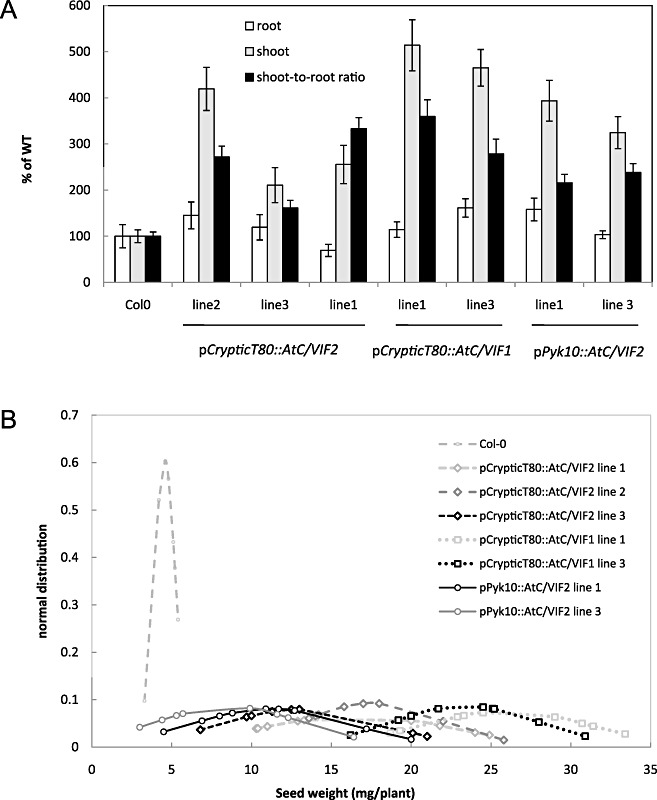
Characterization of transgenic lines expressing two invertase inhibitors under two root‐specific promoters. The phenotype of the selected Arabidopsis transgenic plants was examined after 7 weeks of growth in long‐day conditions. (A) Shoot and root fresh weights were quantified and the shoot to root ratio was determined for at least eight plants of each independent line. Values are referred to those of control plants (WT), considered as 100%. (B) Total seed weight per plant was determined in each individual plant after siliques were dried and the seeds collected. The normal distribution of seed weight was calculated for each plant and the results are plotted for all transgenic lines. The experiment was repeated three times.
Table 1.
Overexpression of an invertase inhibitor under the control of two different root‐specific promoters (Pyk10, CrypticT80) leads to a reduction of the disease index after infection with Plasmodiophora brassicae in different independent lines.
| Line | Club fresh weight (mg) | Root fresh weight (mg) | Root index Ri/Rni | Disease index |
|---|---|---|---|---|
| Col | 47.0 ± 11.6 | 10.7 ± 2.1 | 4.3 ± 0.5 | 100 |
| Ws‐0 | 43.7 ± 5.3 | 10.2 ± 1.7 | 4.1 ± 0.4 | 97 |
| pPyk10::invertase inhibitor AtC/VIF2 | ||||
| pPyk10::AtC/VIF2 line 1 | 30.3 ± 5.9* | 10.6 ± 1.7 | 2.8 ± 0.6* | 82 |
| pPyk10::AtC/VIF2 line 2 | 34.5 ± 5.1* | 10.8 ± 1.9 | 3.5 ± 0.5* | 79 |
| pPyk10::AtC/VIF2 line 3 | 36.6 ± 4.7* | 10.4 ± 0.5 | 3.8 ± 0.8* | 81 |
| pCrypticT80::invertase inhibitor AtC/VIF2 | ||||
| pCrypticT80::AtC/VIF2 line 1 | 29.2 ± 5.1* | 10.4 ± 1.2 | 2.8 ± 0.6* | 88 |
| pCrypticT80::AtC/VIF2 line 2 | 33.5 ± 11.8* | 9.8 ± 1.9 | 3.4 ± 0.6* | 74 |
| pCrypticT80::AtC/VIF2 line 3 | 44.8 ± 6.2 | 10.5 ± 0.2 | 4.4 ± 0.6 | 89 |
| pCrypticT80::invertase inhibitor AtC/VIF1 | ||||
| pCrypticT80::AtC/VIF1 line 1 | 33.7 ± 9.3* | 10.7 ± 1.7 | 2.8 ± 0.8* | 89 |
| pCrypticT80::AtC/VIF1 line 2 | 30.9 ± 3.2* | 9.5 ± 1.3 | 3.3 ± 0.5* | 78 |
| pCrypticT80::AtC/VIF1 line 3 | 30.0 ± 7.2* | 12.0 ± 1.6 | 2.4 ± 0.9* | 83 |
The mean values and standard deviations of the club weight, root weight and root index (Ri/Rni), and disease indices, of the different host–pathogen combinations are given. Only experiments were presented with n > 35. For the susceptible wild‐type Col, with DI > 95, all lines showed an infection rate of 100%. Inoculation (106 spores/mL with single‐spore isolate e3) was performed at 14 days after germination.
Significant difference compared with ecotype by Student's t‐test from at least eight experiments.
In order to analyse the effect of ectopic invertase inhibitor expression in the root, in situ staining for invertase activity in transgenic plants was compared with that of wild‐type control plants. This analysis prevents the described protective effect of sucrose on the inhibition of invertase activity in in vitro tests, usually performed at high concentrations of sucrose not reflecting the actual concentration of this sugar in tissue. The results showed a strongly reduced staining in the roots of transgenic plants (Fig. 4A–F), which expressed either one of the invertase inhibitors compared with control wild‐type Col‐0 and Wassilewskija plants (Fig. 4G,H). Staining in transgenic control plants, transformed with the empty binary plasmid, was similar to that of wild‐type plants (data not shown), thus confirming that the absence of staining for invertase activity in the transgenic lines is caused by the activity of the inhibitor. As invertase enzyme activity, and not expression, was downregulated in this experiment, we did not carry out analysis on RNA or protein levels in infected roots.
Figure 4.
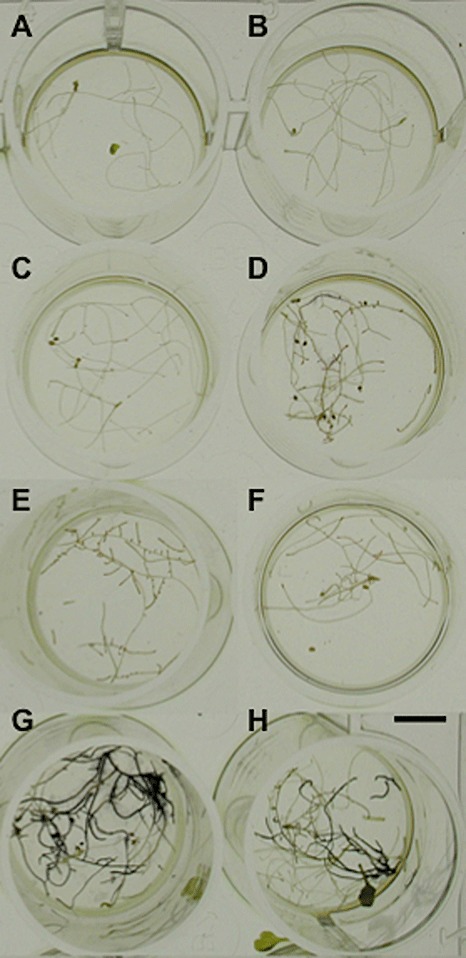
In situ staining of invertase activity in roots. Staining of invertase activity in roots of Col‐0 (G) and Ws‐0 (H) wild‐type plants is shown in comparison with roots from line 2 (A), line 1 (B) and line 3 (C) of pCrypticT80::AtC/VIF1, line 1 (D) and line 2 (E) of pPyk10::AtC/VIF2, and line 1 of pCrypticT80::AtC/VIF2 (F) transgenic plants. Roots shown correspond to 28‐day‐old grown plants. The results obtained were similar after 22 and 28 days of growth. The bar represents 1 cm.
In addition, acid invertase activity, corresponding to extracellular and vacuolar invertases, was determined in the roots and shoots of the selected transgenic lines (pCrypticT80::AtC/VIF1‐1 and pCrypticT80::AtC/VIF1‐2) and compared with that in wild‐type control plants (Fig. 5). No significant effect was observed on invertase activity in shoots. In roots, extracellular invertase activity was reduced in both transgenic lines.
Figure 5.
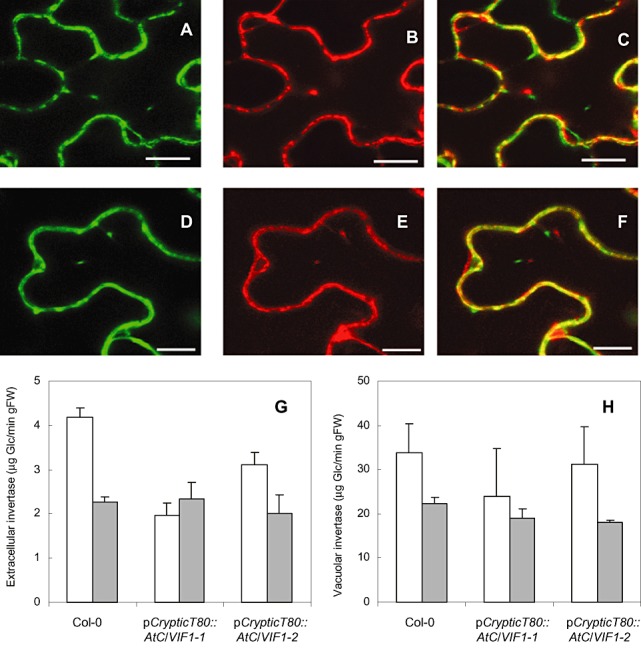
Subcellular localization of the invertase inhibitors used in this study. XFP fusions were transiently expressed in tobacco epidermal cells and analysed via confocal laser scanning microscopy. (A–C) AtC/VIF2 localization in tobacco epidermal cells; scale bar, 10 µm. (A) AtC/VIF2::YFP fluorescence. (B) Cell wall invertase::RFP fluorescence. (C) Overlap of (A) and (B). (D–F) AtC/VIF1 localization in tobacco epidermal cells; scale bar, 5 µm. (D) AtC/VIF1::YFP fluorescence. (E) Cell wall invertase::RFP fluorescence. (F) Overlap of (D) and (E). (G, H) Determination of extracellular (G) and vacuolar (H) invertase activity in wild‐type Col and two transgenic lines (pCryptic::AtC/VIF1). White histograms, roots; grey histograms, shoots.
Subcellular localization of invertase inhibitors AtC/VIF1 and AtC/VIF2
In addition to measuring the activities of vacuolar and extracellular invertases in transgenic lines, we obtained further evidence that extracellular invertase is the target of the two invertase inhibitors used to downregulate invertase activity in this study (Fig. 5). Both invertase inhibitors, as well as a cell wall marker (invertase), were clearly localized in the cell wall in transient expression assays using Nicotiana benthamiana epidermal cells, as shown by confocal laser scanning microscopy. Together with invertase activity showing reduction only for the extracellular and not vacuolar form, this is good evidence that extracellular (cell wall) invertase is the target downregulated in our transgenic lines.
Enhanced root‐specific expression of invertase inhibitors causes a decrease in disease symptoms
The clubroot disease symptoms of different transgenic lines overproducing invertase inhibitors AtC/VIF2 and AtC/VIF1 specifically in the root system under the control of two different root‐specific promoters (Pyk10, CrypticT80) were compared with those of the susceptible ecotypes Col‐0 and Ws‐0 (Table 1). The CrypticT80 promoter caused root‐specific expression and was also induced in clubroot galls, as shown in a promoter reporter line (Fig. S3, see Supporting Information). The expression pattern caused by the Pyk10 promoter revealed an elevated expression in the root system, but only weak expression in the hypocotyl (Fig. S4, see Supporting Information). Although expression was lower in infected roots at a later time point, the expression level was estimated to be sufficiently high to drive root‐specific invertase inhibitor expression in older galls. Attempts to determine invertase activity in situ in infected roots failed because of the high hydrogen peroxide levels present in root galls, which interfered with the enzyme assay (data not shown).
As several invertases were upregulated during gall formation (Fig. 1), mutation of a single invertase gene did not result in any visible phenotype concerning gall development (data not shown). Overexpression of any invertase inhibitor gene/protein should result in the repression of invertase activity, and thus in a lower sucrose import into infected cells. As we used invertase inhibitors localized to the cell wall, these will affect apoplastic invertases. Resistance to clubroot is defined by the reduction in diseased root weight, which is correlated with gall size and a reduction in resting spores, and a lower disease index (Siemens et al., 2002). Ecotypes Col‐0 and Ws‐0 showed the typical disease indices (DI > 95) and root indices (Ri/Rni > 4, roots of infected plants/roots of noninfected plants) of susceptible lines (Table 1). The plants showed clubs, which developed over the whole root and hypocotyl, and also extended in part into the rosette ground (Fig. 6). The overproduction of AtC/VIF2 and AtC/VIF1 caused an apparent tolerance to P. brassicae, indicated by reduced swellings of the root part, resulting in lower disease indices and lower root indices. All analysed overproducers of invertase inhibitor genes showed the different phenotypes presented in Fig. 6. The plants showed clear disease symptoms but, in comparison with ecotype Col‐0, large clubs were induced less frequently and infected roots showed greater variation of disease symptoms. The transgenic plants revealed reduced galls, either as hypocotyl galls with slight infection of the root system (Fig. 6D,E) or as slim galls in the hypocotyl and main root (Fig. 6G,H). In contrast with hypocotyl galls, strong gall development was rarely observed at lateral roots (Fig. 6F) but, in all lines, infected lateral roots could also grow into small galls (Fig. 6I). The disease indices and root indices indicate the susceptibility of the lines, and the mean root indices and disease indices in transgenic plants were significantly lower than those of Col‐0 and Ws‐0. In summary, the root indices of the investigated transgenic lines provided an indication of increased tolerance caused by the overexpression of invertase inhibitor genes.
Figure 6.
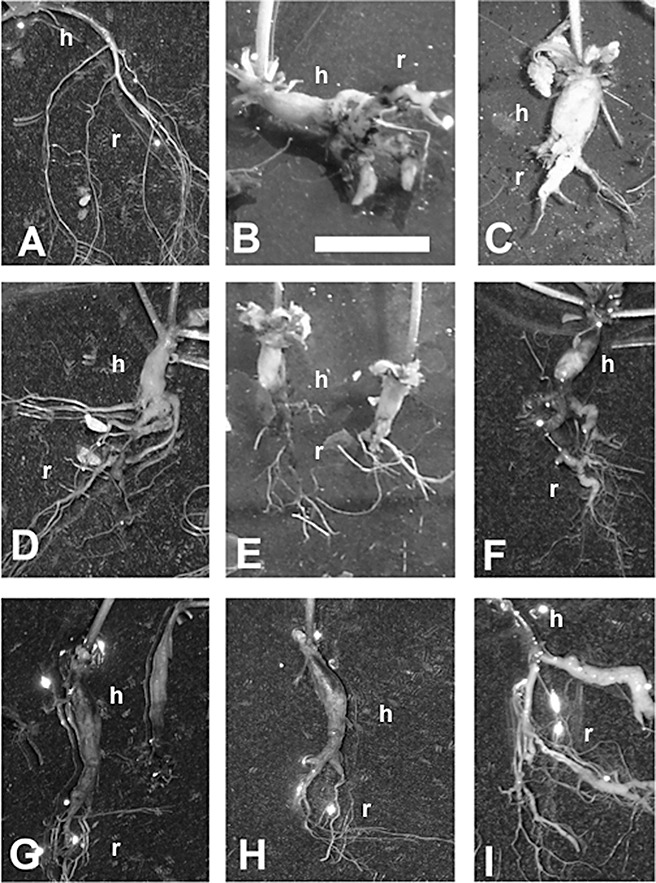
Phenotypes of transgenic plants after inoculation with Plasmodiophora brassciae and under control conditions. Shown are noninfected control (A) and infected roots of different lines at 28 days after inoculation (dai) (B–I). The bar indicates 1 cm. Clubroots of susceptible ecotypes Ws‐0 (B) and Col‐0 (C) are shown in comparison with clubroots of transgenic lines showing: hypocotyl gall but only slight infection of the root system [(D) pPyk10::AtC/VIF2 line 3 and (E) pPyk10::AtC/VIF2 line 1], weak hypocotyl and more pronounced root galls [(F) pCrypticT80::AtC/VIF2 line 2], hypocotyl and main root infected but the galls are slim [(G) pCrypticT80::AtC/VIF1 line 2 and (H) pCrypticT80::AtC/VIF2 line 1] and weak infection at lateral roots proving that root infection can also be developed [(I) pCrypticT80::AtC/VIF1 line 3] (h, hypocotyl; r, root). Photographs of these lines were selected because of their representative gall phenotypes.
DISCUSSION
Invertase gene expression is upregulated in root galls of Arabidopsis after P. brassicae infection
Clubroot disease of Brassicaceae, caused by the obligate biotrophic pathogen P. brassicae, results in large root galls (clubs). For the nutrition of the pathogen during gall formation, the partitioning of assimilates into the roots has been proposed (Ludwig‐Müller et al., 2009). Strong indications for the global involvement of sugar metabolism and transport originate from microarray analysis (Siemens et al., 2006). In this study, we re‐examined the data for genes involved in sugar metabolism, generated promoter–reporter gene fusion lines of invertase and invertase inhibitor genes to verify the regulation of invertases and invertase inhibitors during clubroot disease (1, 3) and produced transgenic lines with decreased invertase activity by the expression of two invertase inhibitor genes under the control of two root‐specific promoters (2, 4, 5) to investigate their role in gall formation (Fig. 6, Table 1).
Invertases are key enzymes for sucrose metabolism and thus assimilate partitioning. They are encoded by small gene families in plants with different expression patterns and regulation, reflecting specific roles of the different isoenzymes in the regulation of plant growth and development. A number of studies have reflected the great impact of constitutively increasing (Büssis et al., 1997; von Schaewen et al., 1990; 1991, 1997) or decreasing (Balibrea Lara et al., 2004; Goetz et al., 2001; Sturm and Tang, 1999; Tang et al., 1999) invertase activity in different cell compartments or specific developmental stages, resulting in marked alterations of plant phenotype not only because of the role of the enzymes in the control of assimilate partitioning, but also because of the control of sugar composition (hexose to sucrose ratio) and the crucial signalling role of sugar molecules (Koch, 1996; Rolland et al., 2006).
Both soluble sugars (hexoses and sucrose) and starch accumulated in the galls of infected Arabidopsis (Brodmann et al., 2002; Evans and Scholes, 1995; Mithen and Magrath, 1992), consistent with an upregulation of expression of sucrose (sucrose synthase) and starch (starch synthase) biosynthetic genes (Siemens et al., 2006; Supporting Information). The visual display of MAPMAN‐generated data in the current study organized the genes for starch/sucrose metabolism in contextual groups, which facilitates the discovery of general trends. This tool can be used to visualize the differences in sugar metabolism between an early time point (10 dai; TP1) and a later time point where galls are fully developed (23 dai; TP2) (Fig. S1A,B). At TP1, genes for starch synthesis, glycolysis and tricarboxylic acid (TCA) appeared to be consistently upregulated (also adenosine‐5′‐diphosphate glucose pyrophosphorylase and sucrose phosphatases), whereas sucrose synthase genes [sucrose‐UDP glucosyltransferase (EC 2.4.1.13)] were mostly downregulated. At TP2, starch synthesis appeared to be further upregulated but, in part, degradation was also upregulated, reflecting the mixture of processes in the cells infected with different stages of the pathogen (Figs S1 and S2). In addition, several invertase genes were upregulated. The mechanism underlying an accumulation of soluble sugars and starch in clubroot galls remains to be elucidated, but may well be associated with increased cytokinin concentration in infected tissues (Devos et al., 2005; Siemens et al., 2006), as cytokinins induce both cell wall invertases and hexose transporters, enhance metabolic sinks and lead to increased starch synthesis (Balibrea Lara et al., 2004; Devos et al., 2006; Ehness and Roitsch, 1997; Siemens et al., 2006).
The verification of microarray data was addressed by the generation of promoter–reporter gene fusion lines of invertases and invertase inhibitors and the monitoring of GUS activity during clubroot development (Fig. 1). The data using promoter::GUS lines for several invertases partially confirmed the data obtained from microarray analysis. In particular, during later infection, when galls were clearly visible, invertase expression was upregulated (Fig. 1). In the current study, we therefore aimed to alter assimilate partitioning to the root in order to analyse the regulatory role of carbon availability on the development of clubroot disease in Arabidopsis (see below).
Extracellular invertase activity is suppressed in transgenic plants
With this aim, we decreased invertase activity specifically in the root by the overexpression of two different invertase inhibitor genes under the control of two different root‐specific promoters, and analysed the effect on plant morphology and gall development. It should be noted that we did not imply, at this stage, a role for invertase inhibitors in clubroot formation; rather, we used these tools to downregulate invertase activity. Root specificity of the two promoters has been reported (Mollier et al., 2000; Nitz et al., 2001). The Pyk10 promoter was shown to be active in control roots and root galls (Fig. S4). As, for the gene driven by the CrypticT80 promoter, no information is available, web‐based gene expression tools could not be used. The promoter activity in galls was therefore verified using a promoter::GUS line (Fig. S3). As plant hormone homeostasis is altered in clubroots, the possibility of hormone induction was investigated for PYK10 using the eFP Browser (http://www.bar.utoronto.ca; Winter et al., 2007). No influence of any hormone on PYK10 expression was observed (Fig. S4D).
Invertase inhibitor activity in transgenic lines was confirmed by histochemical staining for invertase activity in whole roots, with markedly reduced staining in transgenic plants relative to control plants (Fig. 4). In addition, extracellular invertase activity in root extracts, but not leaves, of transgenic plants was reduced (Fig. 5), and the two invertase inhibitors used for the downregulation of invertase activity in a root‐specific manner were localized to the cell wall (Fig. 5). These data confirm that mainly extracellular invertase is targeted by the inhibitors used to downregulate activity. Although vacuolar invertases also play a role in development (Roitsch and González, 2004), the role of extracellular invertases lies in phloem unloading and the determination of the sink strength of a tissue (Roitsch et al., 2003), and thus highlights the importance for the redirection of assimilates to infected roots (see also below). The induction of extracellular invertases would allow the pathogen to withdraw carbohydrates for nutrition. This concept has been mainly described for leaf pathogens (reviewed in Berger et al., 2007; Biemelt and Sonnewald, 2006), but we have shown here that root pathogens can also benefit from carbohydrate reallocation. However, it should be noted that hexoses might also act as defence signals, among others (e.g. Herbers et al., 1996), and that a role for invertase in plant defence is possible.
Invertase suppression in clubs increases tolerance to P. brassicae
Consistent with the data showing increased invertase expression, we were able to modulate gall formation using root‐specific promoters (pPyk10 and pCrypticT80) to drive the expression of invertase inhibitors. The transgenic plants were tolerant to clubroot, i.e. root gall formation was reduced and galls of the hypocotyl were mostly unaltered. This is a result of the specific activity of the promoters in the root (Figs S3 and S4). This approach has the advantage of not changing the overall assimilate partitioning of the plants, where shoot development is promoted, and has only a slight effect on root development (Fig. 3). Thus, the reduction in sink strength of the roots by the inhibition of invertase activity apparently favours partitioning to the shoot. Although the transgenic lines in the infection experiments did not show large differences in shoot biomass, as did controls, it cannot be ruled out completely that other factors, such as more vigorous plants, could have contributed to the tolerance to clubroot observed in this study. However, the similar expression patterns of promoters in the area of the gall in which size reduction was observed argue against this. Future studies should therefore involve the measurement of carbohydrates to substantiate this hypothesis. Moreover, it cannot be ruled out that possible pleiotropic effects of the genetic engineering of root invertase activity change the performance of the plant, i.e. modifications in shoot/root carbon allocations could result in high seed production (Fig. 3), but less reallocation of carbon to the roots could be a disadvantage under abiotic stress. These relationships require further investigation, if possible under agronomically important growth conditions.
The upregulation of invertase has been found in several plant–pathogen interactions (Berger et al., 2007), as described here for clubroot disease. Increased invertase activity in clubroots could also be a cause of increased starch synthesis after the uptake of glucose and fructose in infected cells. Several groups have shown an increase in soluble sugars, as well as starch, in root galls (Brodmann et al., 2002; Evans and Scholes, 1995; Mithen and Magrath, 1992). Starch granules could be remobilized by mature plasmodia of the pathogen during the process of resting spore production. However, mutants defective in ADP glucose pyrophosphorylase and starch accumulation (adg1‐1, adg2‐1) were not more tolerant to P. brassicae infection than wild‐type plants (Siemens et al., 2002), indicating that starch accumulation is not a prerequisite for club development, or that the defect in one metabolic pathway could be substituted by another. In addition, the starch‐overproducing mutant sex1‐1 was tested, for which a more sensitive reaction to P. brassicae could be expected, but again this mutant did not show any differences to the wild‐type (Siemens et al., 2002). Therefore, the reduction in gall formation when invertase activity is reduced indicates that hexoses are metabolized directly by the pathogen, and the accumulation of starch granules is more a by‐product of the plant to cope with the access to hexoses induced by the pathogen during early club development.
It has been shown that, typically, invertase activity is abundant and does not limit the carbon availability to a given tissue (Miller and Chourey, 1992) or in symbiotic interactions (Schaarschmidt et al., 2007b). Thus, it is expected that the increase in sink strength by the ectopic overexpression of heterologous (Heyer et al., 2004; Sonnewald et al., 1997; Tomlinson et al., 2004) or orthologous (Von Schweinichen and Büttner, 2005) invertases should not affect the Plasmodiophora–plant interaction. In contrast with the transgenic Arabidopsis plants generated in this study, transgenic pPyk10::AtC/VIF2 tobacco plants showed no apparent alteration of vegetative growth (Schaarschmidt et al., 2007b), which might be explained by a different expression pattern of the Arabidopsis Pyk10 promoter in other species because of the occurrence of specific transcription factors (Nitz et al., 2001).
In conclusion, our data reveal a role for assimilate partitioning and phloem unloading via extracellular invertases in gall development, which would be expected for an obligate biotrophic pathogen, such as P. brassicae. The claim made from the reduced gall size can be substantiated by showing that mainly extracellular invertase activity is reduced in the roots of transgenic lines, because of the cell wall localization of both invertase inhibitors (Fig. 5). Thus, earlier speculations on the importance of the reallocation of carbohydrates to galls (Evans and Scholes, 1995) and the increase in various sugars in infected roots (Brodmann et al., 2002) are confirmed by the functional approach presented in this study. In addition, this approach is promising for the suppression of gall formation, and could be improved in the future by using promoters expressed in roots and hypocotyls.
EXPERIMENTAL PROCEDURES
Plant material
Ecotypes Columbia (Col‐0), Cape Verde Islands (Cvi‐0) and Wassilewskija (Ws‐0) of A. thaliana were originally provided by the Arabidopsis Seed Stock Centre (NASC, Nottingham, UK). Brassica rapa cv. Granaat (ECD05) plants were used for the propagation of the P. brassicae isolate.
Transgenic plants generated and used in this investigation are listed in Table S1. They were designated with the promoter name and gene name. For A. thaliana sequences, the AGI number was used to designate the promoter (marked by p) or coding sequences, or standard abbreviations for the coded proteins were employed (see Table S1).
Pathogen material
Plasmodiophora brassicae isolate e3 is a single‐spore isolate (Fähling et al., 2003; Graf et al., 2004). Clubroot galls were stored at −20 °C until required. Resting spores were extracted by homogenizing mature clubroot galls of Chinese cabbage, followed by filtering through gauze (pore width, 25 µm) and two centrifugation steps (2500 g, 10 min). For propagation of the P. brassicae isolate, B. rapa cv. Granaat (ECD05) plants were inoculated with 4 mL of spore suspension (107 spores/mL) 4 days after sowing and cultivated for 8 weeks in a glasshouse.
Plant growth
For in vitro culture, Arabidopsis seeds were sterilized by incubation in 70% ethanol for 2 min, and subsequently in a solution containing 8% NaOCl and 0.01% Tween‐20 for 10 min. Seeds were finally washed five times with sterile distilled water.
Seeds were initially sown on glucose‐containing Murashige and Skoog (MS) plates, supplemented with antibiotic when needed. Seedlings were transplanted after 14 days, watered with Hoagland solution (Gibeaut et al., 1997) and grown under long‐day conditions (16 h light) in a growth chamber with a constant temperature of 23 °C for 35–45 additional days.
For phytopathological analysis, plants were cultivated under a controlled environment in soil (21 °C, 16 h light, 100 µmol photons/s/m2).
Phytopathological analysis and histological examinations
Fourteen‐day‐old A. thaliana plants cultivated under a controlled environment (21 °C, 16 h light, 100 µmol photons/s/m2) were routinely inoculated by injecting the soil around each plant with 2 mL of a resting spore suspension of the pathogen with a standard concentration of 106 spores/mL, according to Siemens et al. (2002).
For all experiments, the highly susceptible ecotype Cvi‐0 was inoculated as the susceptible control, and only tests in which Cvi‐0 was scored with an infection rate of 100% and a disease index above 90 were considered for analysis.
Disease symptoms were assessed at 28 dai using a scale consisting of five classes and by estimating the Ri/Rni indices according to Siemens et al. (2002). Scoring of disease symptoms was performed using a scale consisting of five classes according to Klewer et al. (2001): 0 (no symptoms); 1 (very small clubs, mainly on lateral roots that do not impair the main root); 2 (small clubs covering the main root and few lateral roots); 3 (medium‐sized to bigger clubs, also including the main root and hypocotyl, fine roots are partly unaffected, plant growth may be impaired); 4 (severe clubs in lateral, main root, hypocotyls or rosette, fine roots completely destroyed, plant growth is affected). DI was calculated using a five‐grade scale according to the formula, DI = (1n 1+ 2n 2+ 3n 3+ 4n 4)100/4N t, where n 1–n 4 are the numbers of plants in the indicated classes and N t is the total number of plants tested. For each isolate and line combination, at least 35 Arabidopsis plants were analysed in triplicate. These data were analysed with the Kruskal–Wallis test and, subsequently, the mean rank differences were compared (Siegel and Castellan, 1988).
For quantitative estimation, the plants were cut at the top of the hypocotyl into shoots and roots. The infected roots were measured as galls, ignoring occasionally remaining noninfected lateral roots. The root fresh weight was used to calculate the root index Ri/Rni according to Siemens et al. (2002). For each isolate and line combination, at least 35 Arabidopsis plants were analysed in triplicate and compared by Student's t‐test. Root sections were prepared according to Kobelt et al. (2000).
Plasmid construction for root‐specific expression of invertase inhibitors
The complete cDNA of AtC/VIF1 (At1g47960) was amplified by RT‐PCR from total RNA isolated from A. thaliana leaves using the primers AtvInh‐1 and Atvinh‐2 containing Acc65I/XhoI and XbaI/ApaI restriction sites, respectively. A list of the primers used is given in Table S2 (see Supporting Information). Complete cDNA corresponding to AtC/VIF2 (At5g64620) was amplified from cDNA using the primers Atcwinh‐1 and Atcwinh‐2 containing Acc65I/XhoI and XbaI/ApaI restriction sites, respectively. The corresponding PCR products were cloned into vector pBluescript KS+ using Acc65I and XbaI restriction sites, generating pMCG3 and pMCG2 plasmids, respectively. Both cDNAs were cloned into the binary plasmid vector pBIN‐Hyg‐Tx (Gatz and Lenk, 1998) by restriction of the previous clones with Acc65I/XbaI, and cloning into the corresponding sites of plasmid pTF2‐6 (Bonfig et al., 2007), to generate plasmids pMCG4 (AtC/VIF2) and pMCG5 (AtC/VIF1).
A 1.4‐kb root‐specific promoter of PYK10, corresponding to fragment C of the complete promoter described by Nitz et al. (2001), was amplified from Arabidopsis genomic DNA using nested PCRs with two pairs of primers: first pyk10‐Forw and pyk10‐Rev and, subsequently pyk10‐F2 and pyk10‐C, containing Acc65I restriction sites at the 5′ end. The PCR product was cloned into the plasmid pTF2‐6, containing the tobacco cell wall invertase inhibitor between the Acc65I/XbaI sites of the pBIN‐Hyg‐Tx binary plasmid. Corresponding transcriptional fusions between the Pyk10 promoter and AtC/VIF1 and AtC/VIF2 were generated by ligation of pMCG5 and pMCG4 binary plasmids, linearized by Acc65I, with an Acc65I fragment corresponding to the promoter, generating pMCG7‐5 and pMCG6‐1 plasmids, respectively.
The cryptic promoter T80, conferring root‐specific expression (Mollier et al., 2000), was amplified from plasmid X7‐KS, generously provided by M. Simon (INRA, Versailles, France), using primers crypF and crypR containing an Acc65I restriction site at the 5′ end. The corresponding fusions between the cryptic promoter and AtC/VIF1 and AtC/VIF2 were generated by ligation of the linearized pMCG7‐5 and pMCG6‐1 plasmids, restricted with Acc65I, with the Acc65I‐digested PCR product, to generate pMCG12‐1 and pMCG11‐9 plasmids, respectively.
Plasmid construction for promoter::GUS plants
The promoter regions were amplified from genomic DNA with the listed primers containing the GATEWAY cloning (Invitrogen, Karlsruhe, Germany) attB1 and attB2 sites (Table S2). GATEWAY cloning into the vector pBGWFS7 (http://www.psb.rug.ac.be/gateway/list‐of‐constructs.html) was performed following the manufacturer's instructions, and the correct insertion of the promoter region was confirmed by sequencing. The resulting vectors consisted of the promoter in front of an egfp/uidA‐gene fusion.
Plasmid construction for subcellular localization studies in N. benthamiana
Invertase inhibitor sequences were amplified from Arabidopsis leaf cDNA with aatB flanked primers P1/P2 (AtC/VIF2) and P3/P4 (AtC/VIF1), respectively (for primer sequences, see Table S2). The PCR product was recovered using a PCR purification kit and cloned into the pDONR201 vector (Invitrogen) using the BP reaction, and subsequently into the pB7YGW2 vector (http://www.psb.ugent.be/gateway/index.php) using the LR reaction, according to the manufacturer's instructions (Invitrogen, http://www.invitrogen.com), except that BP and LR reactions were downscaled to one‐quarter of the reaction volume.
Transformation of A. thaliana plants
After mobilizing the constructs in Agrobacterium tumefaciens, A. thaliana ecotype Columbia plants were transformed using the floral dip method (Clough and Bent, 1998). Transformed seeds were selected on MS plates supplemented with 50 mg/L hygromycin and 1% glucose, or were screened for glufosinate (BASTA) resistance on soil. Transgenic plants were further checked to contain the corresponding constructs by PCR with specific primers. Control plants corresponding to the empty binary plasmid were also obtained.
Transformation of N. benthamiana leaves
Agrobacterium‐mediated transformation of N. benthamiana leaves was performed using Agrobacterium tumefaciens cells (strain C58C1) containing the appropriate construct. Bacteria were grown overnight in 30 mL yeast extract and beef (YEB) medium supplemented with carbenicillin (50 µg/mL), rifampicin (100 µg/mL) and spectinomycin (50 µg/mL) until reaching the stationary phase. After centrifugation at 3000 g for 30 min at ambient temperature, the cells were suspended in 10–15 mL of infiltration buffer [10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.9, 150 µm acetosyringone] and incubated with gentle agitation for 2 h. The cell suspensions were adjusted to an optical density (OD) of 1.0 with infiltration buffer, mixed when appropriate and infiltrated into the lower epidermis of 8–12‐week‐old N. benthamiana leaves with a 1‐mL syringe. For colocalization with cell wall marker, the invertase inhibitor fusion constructs (AtC/VIF1::YFP, AtC/VIF2::YFP) were mixed in equal volumes with a cell wall marker (cell wall invertase::RFP).
Microscopy and image analysis
Confocal laser scanning images were taken with an LSM 510 Meta inverted microscope (Zeiss, Oberkochen, Germany). Yellow fluorescent protein (YFP) fluorescence was excited with a 514‐nm laser line, and emitted fluorescence was collected using a 530–600‐nm band‐pass filter. Red fluorescent protein (RFP) fluorescence was excited with the 543‐nm laser line and was collected with a 560‐nm long‐pass filter.
Reporter gene assay
For the analysis of GUS activity, tissue samples of transformants were treated with GUS staining buffer {100 mm Na2HPO4/NaH2PO4, pH 7.0, 10 mm sodium ethylenediaminetetraacetate (Na2EDTA), 0.5 mm K3[Fe(CN)6], 0.5 mm K4[Fe(CN)6]} and 0.08% 5‐bromo‐4‐chloro‐3‐indolyl (X‐Gluc; Duchefa, Haarlem, the Netherlands) for 16 h at 37°C (Jefferson, 1987). Green tissues were bleached with ethanol before examination.
Invertase in situ staining
Roots were fixed in 2% paraformaldehyde with 2% polyvinylpyrrolidone 40, 0.001 m dithiothreitol (DTT), pH 7.0, at 4 °C, as described in Sergeeva and Vreugdenhil (2002).
For the in situ staining of invertase activity, roots of plants, grown as described previously, after 22 and 28 days of growth, were incubated in a solution containing 38 mm sodium phosphate, pH 6.0, 25 U/mL glucose oxidase (GOD), 0.03% nitroblue tetrazolium (NBT), 0.0168% phenazine methosulphate (PMS) and 2.4% sucrose until the appearance of dark staining. In control reactions, sucrose was omitted and replaced with phosphate buffer. After the incubation period, roots were rinsed in water and photographed.
Invertase activity determination
The enzyme extraction and activity determination of extracellular and vacuolar invertases were performed as described previously (Balibrea Lara et al., 2004) at pH 4.5. Glucose liberated in the assay was determined using a glucose test kit (Roche, Indianapolis, IN, USA). In each case, control reactions using the same volume of water instead of sucrose were performed. Samples of roots and shoots corresponded to plants grown for 14 days on 1% glucose MS plates, and then transplanted and watered with Hoagland solution, and grown for an additional 40 days under long‐day conditions. Pools of roots and shoots of 10 independent plants were made. Three pools of plants of each line were used for activity determinations. The activity of each sample was determined at least in triplicate.
RNA extraction
Total RNA was isolated with Tritidy (Applichem, Darmstadt, Germany), as described in Bonfig et al. (2007), for Northern analysis, or with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) for quantitative PCR, following the manufacturer's instructions. For quantitative PCR, to eliminate residual genomic DNA present in the preparation, the samples were treated with RNase‐free DNaseI (Promega, Mannheim, Germany) and the RNA was subsequently bound to RNeasy Spin columns (Qiagen) for purification.
Transcript estimation by quantitative real‐time PCR
After elution with RNase‐free water, 2 µg of RNA were transcribed into first‐strand cDNA using the Omniscript RT Kit from Qiagen with an oligo dT primer. Samples treated similarly, but without reverse transcriptase, were used as a negative control in PCR in order to exclude contamination with genomic DNA.
Real‐time PCR was performed using Platinum Taq‐DNA Polymerase (Invitrogen) and SYBR‐Green as fluorescent reporter in a Biorad iCycler (Bio‐Rad Laboratories GmbH, München, Germany). Amplification of genomic DNA was circumvented by designing primers spanning an intron. Primer sequences are given in Table S2. A serial dilution of flower cDNA was used as standard curve to optimize the amplification efficiency for gene‐specific and actin primers. Each reaction was performed in triplicate, and the specificity of amplification products was confirmed by melting curve and gel electrophoresis analysis. Relative expression levels were calculated and normalized with respect to Act2/8 mRNA according to the method of Muller et al. (2002).
Transcript estimation by Northern blot analysis
Ten micrograms of RNA were loaded onto 1.2% agarose formaldehyde gels and blotted onto nitrocellulose membranes. Filters were hybridized with radioactively labelled DNA corresponding to the gene of interest, washed with decreasing concentrations of standard saline citrate (SSC) and exposed to a screen up to 24 h. The screen was imaged with a PhosphorImager (BAS; Fuji, Tokyo, Japan). At least three independent RNA samples per line were analysed, and the mean values ± SE are presented as relative transcript abundance. The transcripts were normalized on rRNA expression.
Re‐evaluation of sugar metabolism‐associated genes from transcriptome analysis
Transcriptome analysis has been described in Siemens et al. (2006). The complete dataset is available at Array Express of the European Bioinformatics Institute (http://www.ebi.ac.uk/arrayexpress/), experiment no E‐MEXP‐254. For this microarray experiment, two distinct developmental stages of clubroot disease were analysed by gene expression profiling on the basis of three independent hybridizations for each time point and treatment (Siemens et al., 2006). The data were further re‐analysed for the genes involved in sugar metabolism using MAPMAN software (http://gabi.rzpd.de/projects/MapMan/) for visualization and statistical analysis (Thimm et al., 2004; Usadel et al., 2005).
The annotation of the transcripts presented in the tables was performed using the Affymetrix probe set‐IDs for the respective genes, and annotation and AGI numbers from the Salk Institute Genomic Analysis Laboratory database SIGnAL (http://signal.salk.edu/tabout.htm) were retrieved. The annotation of highly regulated genes was constantly updated using The Gene Indices (TGI; http://compbio.dfci.harvard.edu/tgi) and TAIR (http://www.arabidopsis.org/) databases.
Supporting information
Fig. S1 Major CHO metabolism, glycolysis and tricarboxylic acid (TCA) cycle during clubroot development. Expression data were extracted from microarray data (M‐EXP‐254) and analysed using MAPMAN software. (A) TP1: 10 days after inoculation (dai). (B) TP2: 23 dai. Blue, upregulated genes; red, downregulated genes. Wilcoxon rank test with Benjamini Hochberg correction indicates that the functional classes major CHO synthesis (P value: 0.009), glycolysis (P value: 0.0002) and TCA cycle (P‐value: 0.01) are consistently upregulated at TP1. At TP2, the respective values indicate a consistent upregulation of the functional classes major CHO synthesis/metabolism (P value: 0.00005) and starch and sugar metabolism (P value: 0.002).
Fig. S2 Sugar (left) and starch (right) metabolism during two different time points of clubroot development. Expression data were extracted from a microarray (M‐EXP‐254) and analysed using MAPMAN software. (A) TP1: 10 days after inoculation (dai). (B) TP2: 23 dai.
Fig. S3 Expression of β‐glucuronidase (GUS) under the control of the crypticT80 promoter. (A) Control roots. (B) Roots infected with Plasmodiophora brassicae. The crypticT80 promoter is highly induced in root but not hypocotyl galls (r, root; h, hypocotyl). The photographs were taken by Cornelia Horn, Institute of Botany, Technische Universität Dresden, Germany.
Fig. S4 Expression levels of PYK10 (At3g09260). (A) Root expression according to microarray data (E‐MEXP‐254). (B–D) Expression according to Arabidopsis eFP Browser (http://www.bar.utoronto.ca; Winter et al., 2007). (B, C) Tissue‐specific expression shows high levels only in the hypocotyls and roots of young seedlings and the roots of older seedlings, where the expression is mainly in the outer cell files. (D) Induction by hormones; no prominent induction by any of the tested hormones, at least at the seedling stage, was observed. dai, days after inoculation; dag, days after germination.
Table S1 Description of the plant material used in this investigation.
Table S2 List of primers used in this study.
Supporting info item
REFERENCES
- Balibrea Lara, M.E. , Gonzalez Garcia, M. , Fatima, T. , Ehneß, R. , Lee, T.K. , Proels, R. , Tanner, W. and Roitsch, T. (2004) Extracellular invertase is an essential component of cytokinin mediated delay of senescence. Plant Cell, 16, 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S. , Sinha, A.K. and Roitsch, T. (2007) Plant physiology meets phytopathology: plant primary metabolism and plant–pathogen interactions. J. Exp. Bot. 58, 4019–4026. [DOI] [PubMed] [Google Scholar]
- Biemelt, S. and Sonnewald, U. (2006) Plant–microbe interactions to probe regulation of plant carbon metabolism. J. Plant Physiol. 163, 307–318. [DOI] [PubMed] [Google Scholar]
- Bonfig, K.B. , Berger, S. , Fatima, T. , González, M.‐C. and Roitsch, T. (2007) Metabolic control of seedling development by invertases. Funct. Plant Biol. 34, 508–516. [DOI] [PubMed] [Google Scholar]
- Brodmann, D. , Schuller, A. , Ludwig‐Müller, J. , Aeschbacher, R.A. , Wiemken, A. , Boller, T. and Wingler, A. (2002) Induction of trehalase in Arabidopsis plants infected with the trehalose‐producing pathogen Plasmodiophora brassicae . Mol. Plant–Microbe Interact. 15, 693–700. [DOI] [PubMed] [Google Scholar]
- Büssis, D. , Heineke, D. , Sonnewald, U. , Willmitzer, L. , Raschke, K. and Heldt, H.‐W. (1997) Solute accumulation and decreased photosynthesis in leaves of potato plants expressing yeast‐derived invertase either in the apoplast, vacuole or cytosol. Planta, 202, 126–136. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Devos, S. , Vissenberg, K. , Verbelen, J.‐P. and Prinsen, E. (2005) Infection of Chinese cabbage by Plasmodiophora brassicae leads to a stimulation of plant growth: impacts on cell wall metabolism and hormone balance. New Phytol. 166, 241–250. [DOI] [PubMed] [Google Scholar]
- Devos, S. , Laukens, K. , Deckers, P. , Van Der Straeten, D. , Beeckman, T. , Inze, D. , van Onckelen, H. , Witters, E. and Prinsen, E. (2006) A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis. Mol. Plant–Microbe Interact. 19, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Ehness, R. and Roitsch, T. (1997) Co‐ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Eschrich, W. (1980) Free space invertase, its possible role in phloem unloading. Ber. Deutsch. Bot. Ges. 93, S363–S378. [Google Scholar]
- Essmann, J. , Schmitz‐Thom, I. , Schön, H. , Sonnewald, S. , Weis, E. and Scharte, J. (2008) RNA interference‐mediated repression of cell wall invertase impairs defense in source leaves of tobacco. Plant Physiol. 147, 1288–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J. and Scholes, J.D. (1995) How does clubroot (Plasmodiophora brassicae) alter the regulation of carbohydrate metabolism in Arabidopsis thaliana? Asp. Appl. Biol. 42, 125–132. [Google Scholar]
- Fähling, M. , Graf, H. and Siemens, J. (2003) Pathotype‐separation of Plasmodiophora brassicae by the host plant. J. Phytopathol. 151, 425–430. [Google Scholar]
- Gatz, C. and Lenk, I. (1998) Promoters that respond to chemical inducers. Trends Plant Sci. 3, 352–358. [Google Scholar]
- Gibeaut, D.M. , Hulett, J. , Cramer, G.R. and Seemann, J.R. (1997) Maximal biomass of Arabidopsis thaliana using a simple, low‐maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 115, 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt, D. and Roitsch, T. (2006) The developmental and organ specific expression of sucrose cleaving enzymes in sugar beet suggest a transition between apoplasmic and symplasmic phloem unloading at the seedling stage. Plant Physiol. Biochem. 44, 656–665. [DOI] [PubMed] [Google Scholar]
- Goetz, M. , Godt, D.E. , Guivarc'h, A. , Kahmann, U. , Chriqui, D. and Roitsch, T. (2001) Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl. Acad. Sci. USA, 98, 6522–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, H. , Fähling, M. and Siemens, J. (2004) Chromosome polymorphism of the obligate biotrophic parasite Plasmodiophora brassicae . J. Phytopathol. 152, 86–91. [Google Scholar]
- Greiner, S. , Rausch, T. , Sonnewald, U. and Herbers, K. (1999) Ectopic expression of a tobacco invertase inhibitor homolog prevents cold‐induced sweetening of potato tubers. Nat Biotechnol. 17, 708–711. [DOI] [PubMed] [Google Scholar]
- Herbers, K. , Meuwly, P. , Frommer, W.B. , Metraux, J.P. and Sonnewald, U. (1996) Systemic acquired resistance mediated by the ectopic expression of invertase: possible hexose sensing in the secretory pathway. Plant Cell, 8, 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer, A.G. , Raap, M. , Schroer, B. , Marty, B. and Willmitzer, L. (2004) Cell wall invertase expression at the apical meristem alters floral, architectural, and reproductive traits in Arabidopsis thaliana . Plant J. 39, 161–169. [DOI] [PubMed] [Google Scholar]
- Hirsche, J. , Engelke, T. and Roitsch, T. (2008) Interspecies compatibility of anther specific promoters for generating male sterile plants by metabolic engineering. Theor. Appl. Genet. 118, 235–245. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Keen, N.T. and Williams, P.H. (1969) Translocation of sugars into infected cabbage tissues during clubroot development. Plant Physiol. 44, 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klewer, A. , Luerßen, H. , Graf, H. and Siemens, J. (2001) Restriction fragment length polymorphism markers to characterize Plasmodiophora brassicae single‐spore isolates with different virulence patterns. J. Phytopathol. 149, 121–127. [Google Scholar]
- Kobelt, P. , Siemens, J. and Sacristan, M.D. (2000) Histological characterisation of the incompatible interaction between Arabidopsis thaliana and the obligate biotrophic pathogen Plasmodiophora brassicae . Mycol. Res. 104, 220–225. [Google Scholar]
- Kocal, N. , Sonnewald, U. and Sonnewald, S. (2008) Cell wall‐bound invertase limits sucrose export and is involved in symptom development and inhibition of photosynthesis during compatible interaction between tomato and Xanthomonas campestris pv vesicatoria . Plant Physiol. 148, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, K.E. (1996) Carbohydrate‐modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Link, M. , Rausch, T. and Greiner, S. (2004) In Arabidopsis thaliana, the invertase inhibitors AtC/VIF1 and 2 exhibit distinct target enzyme specificities and expression profiles. FEBS Lett. 573, 105–109. [DOI] [PubMed] [Google Scholar]
- Ludwig‐Müller, J. and Schuller, A. (2008) What can we learn from clubroots: alterations in host roots and hormone homeostasis caused by Plasmodiophora brassicae . Eur. J. Plant Pathol. 121, 291–302. [Google Scholar]
- Ludwig‐Müller, J. , Prinsen, E. , Rolfe, S. and Scholes, J. (2009) Metabolism and plant hormone action during the clubroot disease. J. Plant Growth Regul. 28, 229–244. [Google Scholar]
- Miller, E.M. and Chourey, P.S. (1992) The maize invertase‐deficient miniature2 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell, 4, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen, R. and Magrath, R. (1992) A contribution to the life history of Plasmodiophora brassicae: secondary plasmodia development in root galls of Arabidopsis thaliana . Mycol. Res. 96, 877–885. [Google Scholar]
- Mollier, P. , Hoffmann, B. , Orsel, M. and Pelletier, G. (2000) Tagging of a cryptic promoter that confers root‐specific GUS expression in Arabidopsis thaliana . Plant Cell Rep. 19, 1076–1083. [DOI] [PubMed] [Google Scholar]
- Muller, P.Y. , Janovjak, H. , Miserez, A.R. and Dobbie, Z. (2002) Processing of gene expression data generated by quantitative real‐time RT‐PCR. Biotechniques, 32, 1372–1379. [PubMed] [Google Scholar]
- Nitz, I. , Berkefeld, H. , Puzio, P.S. and Grundler, F.M.W. (2001) Pyk10, a seedling and root specific gene and promoter from Arabidopsis thaliana . Plant Sci. 161, 337–346. [DOI] [PubMed] [Google Scholar]
- Proels, R.K. and Roitsch, T. (2009) Regulation of source/sink relations by extracellular invertase Lin6 of tomato: a pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by diurnal rhythm. J. Exp. Bot. 60, 1555–1567. [DOI] [PubMed] [Google Scholar]
- Rausch, T. and Greiner, S. (2004) Plant protein inhibitors of invertases. Biochim. Biophys. Acta, 1696, 253–261. [DOI] [PubMed] [Google Scholar]
- Roitsch, T. and González, M.‐C. (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 9, 606–613. [DOI] [PubMed] [Google Scholar]
- Roitsch, T. , Balibrea, M.E. , Hofmann, M. , Proels, R. and Sinha, A.K. (2003) Extracellular invertase: key metabolic enzyme and PR protein. J. Exp. Bot. 54, 513–524. [DOI] [PubMed] [Google Scholar]
- Rolland, F. , Baena‐Gonzalez, E. and Sheen, J. (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. [DOI] [PubMed] [Google Scholar]
- Schaarschmidt, S. , Roitsch, T. and Hause, B. (2006) Arbuscular mycorrhiza induces gene expression of the apoplastic invertase LIN6 in tomato (Lycopersicon esculentum) roots. J. Exp. Bot. 57, 4015–4023. [DOI] [PubMed] [Google Scholar]
- Schaarschmidt, S. , Kopka, J. , Ludwig‐Müller, J. and Hause, B. (2007a) Regulation of arbuscular mycorrhization by apoplastic invertases: enhanced invertase activity in the leaf apoplast affects the symbiotic interaction. Plant J. 51, 390–405. [DOI] [PubMed] [Google Scholar]
- Schaarschmidt, S. , González, M.‐C. , Roitsch, T. , Strack, D. , Sonnewald, U. and Hause, B. (2007b) Regulation of arbuscular mycorrhization by carbon. The symbiotic interaction cannot be improved by increased carbon availability accomplished by root‐specifically enhanced invertase activity. Plant Physiol. 143, 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen, A. , Stitt, M. , Schmidt, R. , Sonnewald, U. and Willmitzer, L. (1990) Expression of a yeast‐derived invertase in the cell wall of tobacco and Arabidopsis plants leads to accumulation of carbohydrate and inhibition of photosynthesis and strongly influences growth and phenotype of transgenic tobacco plants. EMBO J. 9, 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva, L.I. and Vreugdenhil, D. (2002) In situ staining of activities of enzymes involved in carbohydrate metabolism in plant tissues. J. Exp. Bot. 53, 361–370. [DOI] [PubMed] [Google Scholar]
- Seergeva, L.I. , Keurentjes, J.J.B. , Bentsink, L.N. , Vonk, J. , van de Plas, L.H.W. , Koornneef, M. and Vreugdenhil, D. (2006) Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Plant Physiol. 103, 2994–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, S. and Castellan, N.J. (eds) (1988) Nonparametric Statistics for the Behavioral Sciences. New York: McGraw‐Hill Book Company. [Google Scholar]
- Siemens, J. , Nagel, M. , Ludwig‐Müller, J. and Sacristán, M.D. (2002) The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: parameters for disease quantification and screening of mutant lines. J. Phytopathol. 150, 592–605. [Google Scholar]
- Siemens, J. , Keller, I. , Sarx, J. , Kunz, S. , Schuller, A. , Nagel, W. , Schmülling, T. , Parniske, M. and Ludwig‐Müller, J. (2006) Transcriptome analysis of Arabidopsis clubroots indicates a key role for cytokinins in disease development. Mol. Plant–Microbe Interact. 19, 480–494. [DOI] [PubMed] [Google Scholar]
- Sonnewald, U. , Brauer, M. , von Schaewen, A. , Stitt, M. and Willmitzer, L. (1991) Transgenic tobacco plants expressing yeast‐derived invertase in either the cytosol, vacuole or apoplast: a powerful tool for studying sucrose metabolism and sink/source interactions. Plant J. 1, 95–106. [DOI] [PubMed] [Google Scholar]
- Sonnewald, U. , Hajirezaei, M.R. , Kossmann, J. , Heyer, A. , Trethewey, R.N. and Willmitzer, L. (1997) Increased potato tuber size resulting from apoplastic expression of a yeast invertase. Nat. Biotechnol. 15, 794–797. [DOI] [PubMed] [Google Scholar]
- Sturm, A. and Tang, G.‐Q. (1999) The sucrose‐cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 4, 1360–1385. [DOI] [PubMed] [Google Scholar]
- Tang, G.‐Q. , Lüscher, M. and Sturm, A. (1999) Antisense repression of vacuolar and cell wall invertase in transgenic carrot alters early plant development and sucrose partitioning. Plant Cell, 11, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejeda‐Sartorius, M. , Martínez de la Vega, O. and Délano‐Frier, J.P. (2008) Jasmonic acid influences mycorrhizal colonization in tomato plants by modifying the expression of genes involved in carbohydrate partitioning. Physiol. Plant. 133, 339–353. [DOI] [PubMed] [Google Scholar]
- Thimm, O. , Blaesing, O. , Gibon, Y. , Nagel, A. , Meyer, S. , Krüger, P. , Selbig, J. , Müller, L.A. , Rhee, S.Y. and Stitt, M. (2004) MAPMAN: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Tomlinson, K.L. , McHugh, S. , Labbe, H. , Grainger, J.L. , James, L.E. , Pomeroy, K.M. , Mullin, J.W. , Miller, S.S. , Dennis, D.T. and Miki, B.L.A. (2004) Evidence that the hexose‐to‐sucrose ratio does not control the switch to storage product accumulation in oilseeds: analysis of tobacco seed development and effects of overexpressing apoplastic invertase. J. Exp. Bot. 55, 2291–2303. [DOI] [PubMed] [Google Scholar]
- Tymowska‐Lalanne, Z. and Kreis, M. (1998) Expression of the Arabidopsis thaliana invertase gene family. Planta, 207, 259–265. [DOI] [PubMed] [Google Scholar]
- Usadel, B. , Nagel, A. , Thimm, O. , Redestig, H. , Blaesing, O.E. , Palacios‐Rojas, N. , Selbig, J. , Hannemann, J. , Piques, M.C. , Steinhauser, D. , Scheible, W.R. , Gibon, Y. , Morcuende, R. , Weicht, D. , Meyer, S. and Stitt, M. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Schweinichen, C. and Büttner, M. (2005) Expression of a plant cell wall invertase in roots of Arabidopsis leads to early flowering and an increase in whole plant biomass. Plant Biol. 7, 469–475. [DOI] [PubMed] [Google Scholar]
- Weschke, W. , Panitz, R. , Gubatz, S. , Wang, Q. , Radchuk, R. , Weber, H. and Wobus, U. (2003) The role of invertases and hexose transporters in controlling sugar ratios in maternal and filial tissues in barley caryopses during early development. Plant J. 33, 395–411. [DOI] [PubMed] [Google Scholar]
- Winter, D. , Vinegar, B. , Nahal, H. , Ammar, R. , Wilson, G.V. and Provart, N.J. (2007) An ‘Electronic Fluorescent Pictograph’ Browser for exploring and analyzing large‐scale biological data sets. PLoS ONE 2, e718. doi: 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Major CHO metabolism, glycolysis and tricarboxylic acid (TCA) cycle during clubroot development. Expression data were extracted from microarray data (M‐EXP‐254) and analysed using MAPMAN software. (A) TP1: 10 days after inoculation (dai). (B) TP2: 23 dai. Blue, upregulated genes; red, downregulated genes. Wilcoxon rank test with Benjamini Hochberg correction indicates that the functional classes major CHO synthesis (P value: 0.009), glycolysis (P value: 0.0002) and TCA cycle (P‐value: 0.01) are consistently upregulated at TP1. At TP2, the respective values indicate a consistent upregulation of the functional classes major CHO synthesis/metabolism (P value: 0.00005) and starch and sugar metabolism (P value: 0.002).
Fig. S2 Sugar (left) and starch (right) metabolism during two different time points of clubroot development. Expression data were extracted from a microarray (M‐EXP‐254) and analysed using MAPMAN software. (A) TP1: 10 days after inoculation (dai). (B) TP2: 23 dai.
Fig. S3 Expression of β‐glucuronidase (GUS) under the control of the crypticT80 promoter. (A) Control roots. (B) Roots infected with Plasmodiophora brassicae. The crypticT80 promoter is highly induced in root but not hypocotyl galls (r, root; h, hypocotyl). The photographs were taken by Cornelia Horn, Institute of Botany, Technische Universität Dresden, Germany.
Fig. S4 Expression levels of PYK10 (At3g09260). (A) Root expression according to microarray data (E‐MEXP‐254). (B–D) Expression according to Arabidopsis eFP Browser (http://www.bar.utoronto.ca; Winter et al., 2007). (B, C) Tissue‐specific expression shows high levels only in the hypocotyls and roots of young seedlings and the roots of older seedlings, where the expression is mainly in the outer cell files. (D) Induction by hormones; no prominent induction by any of the tested hormones, at least at the seedling stage, was observed. dai, days after inoculation; dag, days after germination.
Table S1 Description of the plant material used in this investigation.
Table S2 List of primers used in this study.
Supporting info item


