SUMMARY
Forward genetic screens are efficient tools for the dissection of complex biological processes, such as fungal pathogenicity. A transposon tagging system was developed in the vascular wilt fungus Fusarium oxysporum f. sp. lycopersici by inserting the novel modified impala element imp160::gfp upstream of the Aspergillus nidulans niaD gene, followed by transactivation with a constitutively expressed transposase. A collection of 2072 Nia+ revertants was obtained from reporter strain T12 and screened for alterations in virulence, using a rapid assay for invasive growth on apple slices. Seven strains exhibited reduced virulence on both apple slices and intact tomato plants. Five of these were true revertants showing the re‐insertion of imp160::gfp within or upstream of predicted coding regions, whereas the other two showed either excision without re‐insertion or no excision. Linkage between imp160::gfp insertion and virulence phenotype was determined in four transposon‐tagged loci using targeted deletion in the wild‐type strain. Knockout mutants in one of the genes, FOXG_00016, displayed significantly reduced virulence, and complementation of the original revertant with the wild‐type FOXG_00016 allele fully restored virulence. FOXG_00016 has homology to the velvet gene family of A. nidulans. The high rate of untagged virulence mutations in the T12 reporter strain appears to be associated with increased genetic instability, possibly as a result of the transactivation of endogenous transposable elements by the constitutively expressed transposase.
INTRODUCTION
Fusarium oxysporum is a ubiquitous soil‐borne ascomycete that causes vascular wilt disease on more than 100 different plant species, provoking severe losses in important crops such as banana, cotton, melon and tomato (Gordon and Martyn, 1997). This fungus is also increasingly being recognized as an emerging human pathogen which poses a lethal threat to immunocompromised individuals (Nucci and Anaissie, 2007). Its remarkably broad host range and the array of molecular tools available makes F. oxysporum an attractive model for studying different aspects of fungal infection (Di Pietro et al., 2003). Recently, a single isolate of the tomato pathogenic forma specialis (f. sp.) lycopersici was established as a multi‐host model for the dissection of virulence on both plant and mammalian hosts (Ortoneda et al., 2004). The complete genome sequence of this isolate is available (http://www.broad.mit.edu/annotation/fgi/), setting the basis for genome‐wide studies on pathogenicity determinants in F. oxysporum.
Forward genetic analysis provides an unbiased approach for the dissection of complex biological processes, such as fungal pathogenicity. Classical strategies using ultraviolet (UV) or chemical mutagenesis have been replaced by insertional mutagenesis techniques, including random restriction enzyme‐mediated integration (REMI) (Brown and Holden, 1998), Agrobacterium tumefaciens‐mediated transformation (ATMT) (Michielse et al., 2005) and transposon tagging (Daboussi and Capy, 2003). REMI is based on the integration of a defined DNA carrying a selectable marker by the in vivo action of a restriction enzyme. However, a substantial fraction (30%–50%) of REMI virulence mutants are untagged by the transforming DNA, and integration is not completely random, showing a bias for highly transcribed genomic regions (Brown and Holden, 1998). In spite of these limitations, REMI has been used successfully to identify novel pathogenicity factors in fungi (Balhadere and Talbot, 2001; Clergeot et al., 2001; Dufresne et al., 2000; Kimura et al., 2001; Sweigard et al., 1998; Urban et al., 1999), including several virulence determinants in F. oxysporum, such as a mitochondrial carrier protein (Inoue et al., 2002), a class V chitin synthase (Madrid et al., 2003), an F‐box protein (Duyvesteijn et al., 2005) and a Zn(II)2Cys6 family transcriptional regulator (Imazaki et al., 2007).
ATMT has been used to produce large‐scale T‐DNA insertions in plants (Alonso et al., 2003; Jeon et al., 2000) and, more recently, in fungi (Jeon et al., 2007). The main advantage of this technique is the possibility to transform intact fungal cells, obviating the need for protoplasting. ATMT generates a high percentage of transformants with single‐copy integrated DNA, which facilitates subsequent identification of the tagged genes, although small deletions at the insertion site are frequently observed (de Groot et al., 1998; Meng et al., 2007; Mullins et al., 2001). ATMT has been used for insertional mutagenesis in different fungal pathogens (Blaise et al., 2007; Choi et al., 2007; Idnurm et al., 2004; Walton et al., 2005), but most extensively in the rice blast fungus Magnaporthe grisea (Betts et al., 2007; Jeon et al., 2007; Meng et al., 2007). Although ATMT was originally thought to produce mainly random insertions (de Groot et al., 1998; Mullins et al., 2001), recent studies in M. grisea have detected a significant bias towards promoter regions and against open reading frames (ORFs), as well as a non‐random distribution among chromosomes (Choi et al., 2007; Meng et al., 2007).
Transposable elements (TEs) are ubiquitous DNA segments in prokaryotic and eukaryotic organisms which have the ability to move and replicate within genomes (Kidwell and Lisch, 1997). Class I retroelements transpose by reverse transcription of an RNA intermediate, whereas class II TEs transpose directly from DNA to DNA by a ‘cut‐and‐paste’ mechanism (Finnegan, 1992). Class II TEs are characterized by small inverted repeats (10–200 bp) bordering an internal transposase‐encoding ORF that is necessary for their transposition. The first TEs in filamentous fungi were identified in F. oxysporum (Daboussi and Langin, 1992) and, since then, all major classes of eukaryotic TEs have been found in this organism (Daboussi and Capy, 2003).
TEs have been used for insertional mutagenesis in plants, bacteria, insects and fungi (Daboussi and Capy, 2003; Kempken and Kuck, 2000), including Saccharomyces cerevisiae (Weil and Kunze, 2000), Tolypocladium inflatum (Kempken and Kuck, 2000), M. grisea (Villalba et al., 2001), F. oxysporum (Hua‐Van et al., 2001), Aspergillus nidulans (Li Destri Nicosia et al., 2001), Aspergillus fumigatus (Firon et al., 2003) and Ustilago maydis (Ladendorf et al., 2003). A major advantage of transposon tagging is that insertional mutants can be obtained rapidly without the need for repeated transformation, once a reporter strain is generated in which transposition of the TE can be monitored conveniently (Daboussi and Capy, 2003). The element most frequently used for insertional mutagenesis in fungi is impala160, a class II TE of the Tc1‐mariner superfamily, which was originally identified by transposon trapping in F. oxysporum (Langin et al., 1995). Characteristically for Tc1‐mariner TEs, impala160 inserts into a TA site which is duplicated on insertion, leaving a footprint after excision (Hua‐Van et al., 1998; Langin et al., 1995). The impala160 TE has been shown to transpose efficiently in different Fusarium species (Hua‐Van et al., 2001), A. nidulans (Li Destri Nicosia et al., 2001), A. fumigatus (Firon et al., 2003) and M. grisea (Villalba et al., 2001).
The aim of this study was to test the feasibility of large‐scale transposon mutagenesis as a tool for the genome‐wide identification of virulence determinants in the sequenced multi‐host model strain F. oxysporum f. sp. lycopersici 4287. A two‐component system (Hua‐Van et al., 2001) was used, based on a novel modified version of the impala160 element and a transposase constitutively expressed in trans, to generate a collection of 2072 transposon insertion strains. Screening of this collection using a rapid infection assay on apple slices identified a number of virulence mutants, but only one of these showed linkage between virulence phenotype and transposon insertion. The tagged gene showed homology to the fungal velvet family, providing the first evidence for a role of Velvet in virulence.
RESULTS
Establishment of a two‐component transposon tagging system in F. oxysporum f. sp. lycopersici strain 4287
A two‐component transposon tagging system was established in the tomato pathogenic F. oxysporum f. sp. lycopersici strain 4287 (Fig. S1, see ‘Supporting Information’). Plasmid pPtrap10B contained a novel modified impala element, imp160::gfp, in which the transposase gene was replaced with the egfp coding region, inserted into the promoter of the A. nidulans niaD gene (see ‘Experimental procedures’ for details). This plasmid was co‐transformed with vector pHEO62, carrying both the hygromycin resistance cassette and the imp160 transposase gene fused to the constitutive A. nidulans gpdA promoter (Hua‐Van et al., 2001), into the nitrate reductase (nit1) mutant CO6 derived from strain 4287 (Garcia‐Pedrajas and Roncero, 1996). CO6 has been shown previously to be fully virulent in tomato plants (Fig. S3, see ‘Supporting Information’). As a negative control, pPtrap10B was co‐transformed with vector pAN7‐1 carrying only the hygromycin resistance cassette (Punt et al., 1987). From a total of 44 hygromycin‐resistant transformants carrying the pHEO62 vector, 14 co‐transformants had also integrated the pPtrap10B construct, as determined by polymerase chain reaction (PCR) with green fluorescent protein (GFP)‐specific primers and by Southern analysis with the A. nidulans niaD gene probe (data not shown). As expected, all 14 co‐transformants exhibited a nia− phenotype when grown on selective minimal medium (MM) with nitrate as the sole nitrogen source. Of these, 10 gave rise to Nia+ revertants at frequencies between less than 10 and more than 100 per plate. One of the co‐transformants, named T12, produced Nia+ revertants at a very high rate (20–100 revertants per plate; Fig. S1C, see ‘Supporting Information’). By contrast, none of the six control co‐transformants carrying pPtrap10B and pAN7‐1 gave rise to Nia+ revertants, indicating that a high frequency of revertants was associated with the presence of the constitutively expressed imp160 transposase in vector pHEO62.
To confirm that the Nia+ phenotype was caused by transposition of the imp160::gfp element, 12 revertants obtained from co‐transformant T12 were analysed by Southern blot hybridization with the A. nidulans niaD probe. In all revertants tested, the imp160::gfp element had excised from the A. nidulans niaD promoter, as shown by a shift in size of the hybridizing EcoRI fragment from 3.8 kb in strain T12 to 2.7 kb in the revertants (Fig. 1A). To determine the frequency of re‐insertion of the imp160::gfp element, the same Southern blot was hybridized with the gfp probe. In nine of the 12 Nia+ revertants, single hybridizing bands of different sizes were detected, suggesting that re‐insertion of the imp160::gfp element had occurred in 75% of the revertants at single, apparently random sites of the F. oxysporum genome (Fig. 1B). Random insertion without apparent sequence preference (except for the TA nucleotide) is a hallmark of the Tc1‐mariner superfamily (Plasterk et al., 1999), and has been reported previously for impala160 transposition events in F. oxysporum (Hua‐Van et al., 2001), A. nidulans (Li Destri Nicosia et al., 2001), A. fumigatus (Firon et al., 2003) and M. grisea (Villalba et al., 2001).
Figure 1.
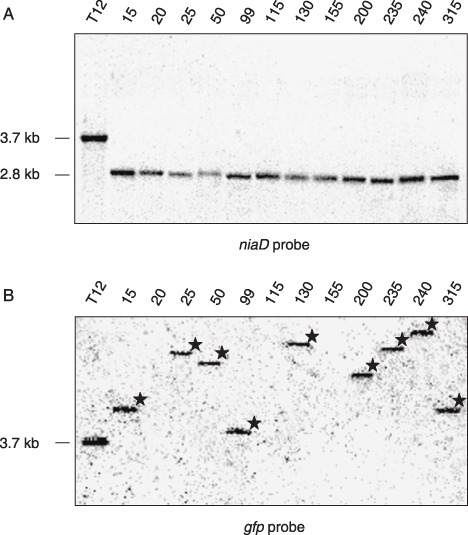
Southern blot analysis revealing the excision and re‐insertion pattern of the modified impala element. Genomic DNA of reporter strain T12 and different Nia+ revertants was treated with EcoRI and hybridized with the Aspergillus nidulans niaD probe (A) or the green fluorescent protein (gfp) probe (B). The hybridization pattern with niaD indicates a 100% excision rate in the Nia+ revertants, whereas the gfp probe shows re‐insertion of the imp160::gfp element at different genomic loci in nine of the 12 revertants studied (indicated by asterisks).
Transposon mutagenesis and virulence phenotype screening
A collection of 2072 independent F. oxysporum Nia+ revertants was obtained from co‐transformant T12 by direct selection on MM with nitrate as the sole nitrogen source (Fig. S1C, see ‘Supporting Information’). Based on the results shown above, we expected revertants in this setting to result predominantly from the excision of imp160::gfp, followed by re‐integration of the element in approximately 75% of cases. The revertants were screened for changes in virulence, using a rapid phenotypic assay for invasive growth on apple slices (Fig. S2, see ‘Supporting Information’). Microconidial suspensions of the revertants were spotted onto the surface of apple slices and incubated at 28 °C and 100% humidity. Inoculations were carried out in duplicate, and both positive (strain T12) and negative (mutant Δfmk1 lacking a mitogen‐activated protein kinase; Di Pietro et al., 2001) controls were routinely included. After 4 days, co‐transformant T12 had successfully invaded the surrounding fruit tissue, producing a brown halo of maceration and aerial mycelium on the fruit surface (Figs S2B and 3C, see ‘Supporting Information’). By contrast, the negative control Δfmk1 failed to grow invasively and to macerate the tissue. Of the 2072 revertants tested, 14 were less invasive than the wild‐type in the apple fruit assay. Seven of these revertants also showed significantly reduced virulence in a tomato root infection assay (Fig. S3, see ‘Supporting Information’). Revertants 5A11, 17A1 and 6C3 had a moderate virulence reduction, 6G5, 6F7 and 6F9 had a strong virulence reduction, and 7A1 was non‐pathogenic, similar to the Δfmk1 mutant.
Characterization of transposon insertion sites in virulence mutants
The genomic regions flanking the insertion site of the imp160:: gfp element were isolated in six revertants (5A11, 6C3, 6G5, 6F7, 7A1, 17A1) using inverse PCR. No flanking sequences could be recovered from strain 6F9, and no hybridizing signal was obtained in a Southern blot analysis with the gfp probe, suggesting that imp160::gfp had been lost in this revertant (data not shown). In the other six revertants, both flanks were obtained, indicating the absence of deletions at the transposon insertion site. Amplified fragments were cloned into pGEM‐T and sequenced. In revertant 6C3, the flanking sequences were identical to those in the original co‐transformant T12 (A. nidulans niaD promoter), suggesting that the Nia+ phenotype in this revertant is not caused by transposition of the imp160::gfp element, but rather by reversion of the nia−mutation of the endogenous nit1 gene. In the other five revertants, analysis of the flanking sequences confirmed the transposition of imp160::gfp to a different location in the genome. The sites of insertion were mapped by comparing the obtained sequences with the F. oxysporum genome database, and are listed in Table 1. In two revertants, the imp160::gfp element had inserted into different regions of chromosome 1, whereas in the other three revertants the element had inserted into chromosome 2, 5 and 10, respectively.
Table 1.
Insertion sites of the imp160::gfp element in the revertants with reduced virulence phenotype.
| Revertant | Closest predicted open reading frame (ORF) | Distance (bp) | Chromosome | Predicted function |
|---|---|---|---|---|
| 6F9 | Loss of imp160::gfp | |||
| 6C3 | niaD (no excision) | |||
| 5A11 | FOXG_11479 | 235 (5′) | 10 | Uracil phosphoribosyltransferase Similar to chromosome condensation protein CrcB |
| FOXG_11478 | 438 (5′) | 10 | ||
| 7A1 | FOXG_00076 | 439 (5′) | 1 | Putative zinc finger transcription factor (cDNA clone sequenced) |
| 6G5 | FOXG_02277 | 239 (5′) | 5 | Similar to Aspergillus nidulans nitrogen response regulator MeaB |
| 6F7 | FOXG_08661 | 483 (5′) | 2 | Conserved hypothetical protein |
| 17A1 | FOXG_00016 | ORF | 1 | Similar to A. nidulans developmental regulator VeA |
Virulence defects in revertants 7A1, 6G5 and 6F7 are unlinked to the transposon insertion site
Of the revertants analysed, strain 7A1 exhibited the most dramatic virulence phenotype. Interestingly, this revertant shared a number of virulence‐related phenotypes with the previously characterized Δfmk1 mitogen‐activated protein kinase mutant (Di Pietro et al., 2001), including a lack of clear halo production on polygalacturonic acid (PGA) medium, inability to penetrate cellophane membranes and decreased colony hydrophobicity. The insertion site of the imp160::gfp element was located 439 bp upstream of a predicted ORF (gene transfer encompassing supercontig 1, 224 954–228 632) overlapping with FOXG_00076. The predicted ORF consists of five exons whose position was confirmed by sequencing of a cDNA clone (data not shown), and encodes a putative zinc finger transcription factor. Southern analysis with a genomic PCR fragment confirmed that the wild‐type 1.7‐kb hybridizing XhoI genomic fragment was replaced in strain 7A1 by a 2.6‐kb band, consistent with the insertion of the 0.9‐kb imp160::gfp element (Fig. 2A). To determine whether the reduced virulence phenotype of strain 7A1 was linked to the insertion of the imp160::gfp element, we replaced the FOXG_00076 locus in the wild‐type strain 4287 with an inactivated allele in which 2481 bp of the predicted coding region had been replaced with the hygromycin resistance cassette. The analysis of eight hygromycin‐resistant transformants by PCR with different combinations of gene‐specific primers identified six transformants with PCR amplification products indicative of homologous integration‐mediated gene replacement (data not shown). Southern blot analysis in one of these transformants confirmed that the 1.7‐kb XhoI hybridizing fragment of the wild‐type had been replaced by a 4.5‐kb fragment (Fig. 2A). All ΔFOXG_00076 strains were analysed for clear halo production on PGA medium, ability to cross cellophane membranes, colony hydrophobicity and invasive growth on apple cuttings. In contrast with revertant 7A1, the ΔFOXG_00076 strains displayed phenotypes identical to those of the wild‐type and the T12 reporter strain (Fig. 3), including virulence on tomato plants (data not shown). In addition, a complementation approach was performed by co‐transforming protoplasts of revertant 7A1 with a 5445‐bp PCR fragment of the wild‐type FOXG_00076 locus, together with the phleomycin resistance marker. Phleomycin‐resistant transformants were analysed for the presence of the FOXG_00076 wild‐type allele by PCR with gene‐specific primers, and several co‐transformants carrying the functional FOXG_00076 gene were subjected to virulence assays. These strains still displayed reduced virulence on tomato plants and apple slices, similar to the 7A1 revertant (data not shown). These results indicated that the virulence phenotype of strain 7A1 was not caused by a loss of function of the FOXG_00076 gene. We also considered the possibility that insertion of the imp160::gfp element upstream of FOXG_00076 could produce a genetic effect other than a loss of function (e.g. gain of function). To test this hypothesis, we generated a mutant allele in which the hygromycin resistance cassette was inserted precisely at the same genomic site as the imp160::gfp element in strain 7A1, 439 bp upstream of the predicted start codon of FOXG_00076. After PCR analysis of hygromycin‐resistant transformants with different combinations of gene‐specific primers, two transformants with homologous integration‐mediated gene replacement were identified (data not shown). Phenotypic characterization of these strains, denominated 7A1::hyg, revealed the same phenotype as in the wild‐type strain (Fig. 3). The next predicted ORF, FOXG_00077, was located 3795 bp from the insertion site. Taken together, these results strongly suggest that the virulence defect of revertant 7A1 is caused by genetic events unlinked to the insertion of the imp160::gfp element.
Figure 2.
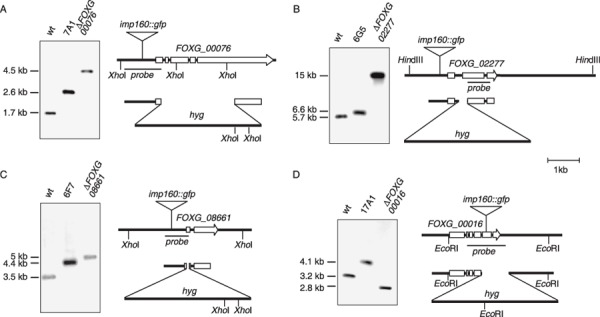
Physical map of transposon‐tagged genomic loci in Fusarium oxysporum revertants and the construction of targeted gene knockout mutants. Schematic representation of the insertion site of the imp160::gfp element in the F. oxysporum genome, as determined by inverse polymerase chain reaction (PCR) sequencing, and, below, the corresponding gene disruption constructs. For Southern blot analysis, 10 µg of genomic DNA from the wild‐type strain 4287, each revertant and knockout mutant was treated with the indicated restriction enzyme, separated on a 0.7% agarose gel, transferred to a nylon membrane and hybridized with the indicated DNA probe. The molecular sizes of the hybridizing fragments are indicated on the left. wt, wild‐type.
Figure 3.
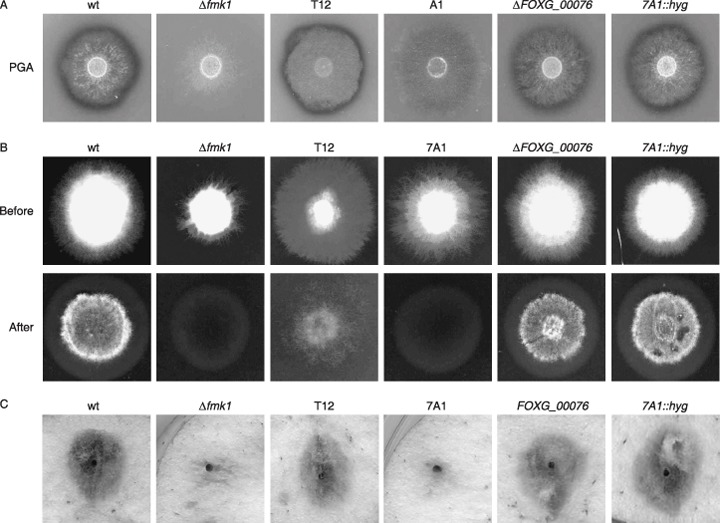
Shared virulence‐related phenotypes of revertant 7A1 with the Δfmk1 mitogen‐activated protein kinase mutant are unlinked to the insertion of the imp160::gfp element. Microconidial suspensions of the following strains were used for the different phenotypic assays: wild‐type (wt), Δfmk1 knockout mutant, co‐transformant T12, revertant 7A1, ΔFOXG_00076 knockout strain and strain 7A1::hyg carrying the hygromycin resistance cassette inserted at the identical genomic position as the imp160::gfp element in revertant 7A1. (A) Clear halo production on polygalacturonic acid (PGA) plates for visualization of extracellular polygalacturonase activity (visible as a dark halo underneath the fungal colony). (B) Penetration of cellophane sheets. Colonies were grown for 4 days on a plate with minimal medium covered with a cellophane sheet (before); the cellophane with the colony was removed and the plate was incubated for one additional day (after). (C) Infection assay on apple slices with the indicated strains. Suspensions containing 5 × 104 freshly obtained microconidia were applied to apple slices. Colonization and maceration of the surrounding fruit tissue were determined after 4 days of incubation at 28 °C.
In revertant 6G5, the imp160::gfp insertion was mapped 239 bp upstream of FOXG_02277 (Fig. 2B), whereas the next ORF, FOXG_02276, was located at 5031 bp. Southern blot analysis with the FOXG_02277 probe revealed that the 5.7‐kb wild‐type HindIII genomic fragment was replaced in strain 6G5 by a new band with a 0.9‐kb increase in size, consistent with insertion of the imp160::gfp element (Fig. 2B). FOXG_02277 encodes a putative homologue of the A. nidulans bZIP transcription factor MeaB, which mediates nitrogen metabolite repression by negatively regulating the transcriptional activator AREA (Polley and Caddick, 1996; Wong et al., 2007). MeaB mutants are derepressed for genes that are subject to nitrogen metabolite repression, including the nitrate reductase gene niaD, resulting in sensitivity to chlorate in the presence of ammonium (Polley and Caddick, 1996). Indeed, colony growth of revertant 6G5 on MM containing 10 mm ammonium and 100 mm chlorate was strongly impaired compared with the wild‐type, suggesting that FOXG_02277 encodes a functional orthologue of A. nidulans MeaB (Fig. 4A).
Figure 4.
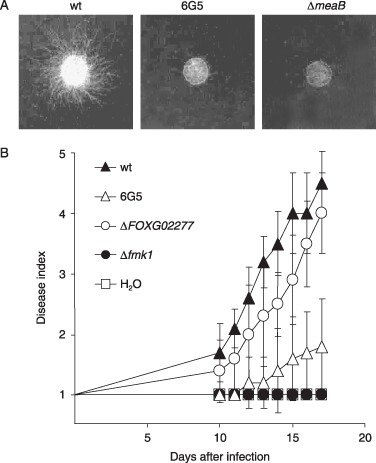
The meaB‐related nitrogen growth phenotype, but not the virulence phenotype of revertant 6G5, is linked to insertion of the imp160::gfp element. (A) Assay for repression of nitrate reductase activity by ammonium. The indicated strains were grown for 3 days on minimal medium containing 10 mm ammonium plus 100 mm chlorate. Wild‐type hyphal growth under these conditions requires meaB‐mediated repression of nitrate reductase activity, whereas derepression results in chlorate toxicity. (B) Infection assay on tomato plants. Groups of 10 plants were inoculated by immersing the roots into a suspension of 5 × 106 freshly obtained microconidia/mL of the indicated strains. The severity of disease symptoms was recorded at different times after inoculation, using an index ranging from 1 (healthy plant) to 5 (dead plant). wt, wild‐type.
Fusarium oxysporum mutants lacking a functional copy of the gene were obtained by homologous integration of a ΔFOXG_ 02277 allele, with the hygromycin resistance cassette replacing 178 bp of the second predicted exon (Fig. 2B). The ΔFOXG_02277 mutants showed slightly more impaired growth than revertant 6G5 on chlorate + ammonium, confirming that the nitrogen growth phenotype of this revertant is linked to the insertion of the imp160:: gfp element (Fig. 4A). However, in contrast with revertant 6G5, the ΔFOXG_02277 strain was as virulent as the wild‐type in a tomato plant infection assay (Fig. 4B). This suggests that, as in 7A1, the virulence defect in 6G5 is unlinked to the insertion of imp160::gfp.
In revertant 6F7, the insertion site of the modified TE was mapped 483 bp upstream of FOXG_08661, encoding a conserved hypothetical protein of unknown function (Fig. 2C), whereas the next ORF, FOXG08660, was located at 1305 bp. Southern blot analysis of strain 6F7 with the FOXG_08661 probe confirmed the replacement of the wild‐type hybridizing XhoI genomic fragment by a new band with a 0.9‐kb increase in size. Targeted mutants carrying an inactivated FOXG_08661 allele were generated by transforming the wild‐type strain with a construct in which 30 bp of the first predicted exon was replaced by the hygromycin resistance cassette, and homologous integration in the transformants was tested by Southern blot (Fig. 2C). As in the previous two cases, the ΔFOXG_08661 knockout strains were as virulent as the wild‐type on both apple cuttings and tomato plants (data not shown), suggesting that the virulence defect in 6F7 was not linked to the insertion of the imp160::gfp element.
The virulence defect in revertant 17A1 is caused by insertion of the imp160::gfp element into a velvet‐like gene
The insertion site of imp160::gfp in revertant 17A1 was located within the predicted ORF FOXG_00016. Southern blot analysis with the FOXG_00016 probe confirmed that the wild‐type 3.2‐kb hybridizing EcoRI genomic fragment was replaced in strain 17A1 by a 4.1‐kb band, consistent with the insertion of imp160::gfp (Fig. 2D). The predicted gene product has homology with members of the fungal Velvet protein family. Velvet was originally identified as a light‐dependent regulator of conidiation in A. nidulans (Mooney and Yager, 1990), and controls diverse cellular processes in fungi, including asexual and sexual development, as well as secondary metabolism (Calvo, 2008). We noted that colonies of revertant 17A1 had reduced aerial mycelium compared with those of the wild‐type strain (Fig. 5A). To determine whether the virulence and colony phenotypes of strain 17A1 were caused by the imp160::gfp insertion, targeted mutants carrying an inactivated FOXG_00016 allele were generated by transforming the wild‐type strain with a construct in which 791 bp encompassing the last two exons was replaced by the hygromycin resistance cassette. Transformants showing homologous integration were identified by Southern blot (Fig. 2D). The ΔFOXG_00016 knockout mutants displayed a similar colony phenotype to the 17A1 revertant (Fig. 5A), and significantly delayed virulence on tomato plants (Fig. 5B) and apple slices (not shown). For complementation experiments, a 4002‐bp PCR fragment carrying the wild‐type FOXG_00016 locus was used to co‐transform protoplasts of revertant 17A1 or knockout mutant ΔFOXG_0001, together with the phleomycin cassette. Phleomycin‐resistant transformants were analysed for the presence of the FOXG_00016 wild‐type allele by PCR with gene‐specific primers (data not shown). Co‐transformants carrying the functional FOXG_00016 allele had wild‐type colony morphology (Fig. 5A) and fully restored virulence on tomato plants (Fig. 5B). These results strongly suggest that the developmental and virulence phenotypes of revertant 17A1 are caused by insertion of the imp160::gfp element into the FOXG_00016 gene.
Figure 5.
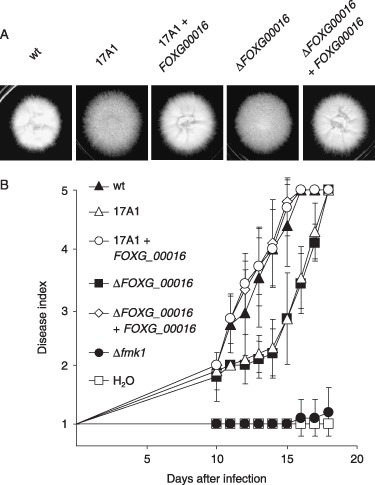
Mutation of the velvet‐like gene FOXG_00016 affects colony morphology and virulence. (A) Colonies of the indicated strains grown on yeast extract–peptone–glucose (YPG) for 3 days. (B) Infection assay on tomato plants. Groups of 10 plants were inoculated with the indicated strains as described in Fig. 4. wt, wild‐type.
Evidence for increased genetic instability in co‐transformant T12
The high proportion of revertants whose virulence phenotype was unlinked to the imp160::gfp insertion site (five of six revertants tested) indicates the presence of alternative sources of genetic variation in the strain T12. This co‐transformant was originally selected as reporter strain for transposon mutagenesis because it produced a very large number of Nia+ revertants in a manner dependent on the constitutively expressed imp160 transposase (see above). To test whether T12 has an increased level of genetic variability, we monitored the appearance of chlorate‐resistant mutants in five independent colonies from three different T12 revertant strains, in comparison with the wild‐type strain. In the three T12 revertants tested, chlorate‐resistant mutants appeared in all five colonies, compared with only one of five untreated wild‐type colonies (Table 2). The large variation in the number of mutants between individual colonies is most probably related to the different time points at which the mutations occurred during colony growth. To confirm that the number of chlorate‐resistant mutants allows an accurate estimation of genetic instability, five independent wild‐type colonies were subjected to UV treatment at a defined time point during growth (see ‘Experimental procedures’). As shown in Table 2, chlorate‐resistant mutants were detected in all five UV‐treated wild‐type colonies, suggesting that UV treatment dramatically increased the mutation rate. In contrast with the revertants, very similar numbers of chlorate‐resistant mutants were obtained from different UV‐treated colonies, as expected, as the same dose of UV radiation was applied to each colony at the same time point during growth. These results suggest an increased genetic instability in strain T12 compared with the F. oxysporum wild‐type strain.
Table 2.
Frequency of mutations arising in different fungal strains. Five independent cultures of each strain were grown for 7 days on potato dextrose agar (PDA), and 2.5 × 106 spores collected from each culture were spread onto PDA plates containing 0.15 m chlorate. Chlorate‐resistant colonies were counted after 3 days of incubation at 28 °C. Wild‐type (wt) ultraviolet (UV)‐treated colonies were irradiated with 600 J/m2 after 4 days of growth. Mean values and standard deviations were calculated from two replicate plates.
| Culture | Strain | ||||
|---|---|---|---|---|---|
| wt | wt UV‐treated | 7A1 | 21D7 | 23F7 | |
| 1 | 0 ± 0 | 39.5 ± 3.5 | 3.5 ± 0.7 | 4.5 ± 2.1 | 4 ± 1.1 |
| 2 | 0 ± 0 | 38.5 ± 2.1 | 4.5 ± 0.7 | 9.5 ± 2.1 | 3 ± 1.4 |
| 3 | 35.5 ± 0.71 | 72 ± 5.6 | 349 ± 29.7 | 1.5 ± 2.1 | 1 ± 1.4 |
| 4 | 0 ± 0 | 75.5 ± 4.9 | 5.5 ± 0.7 | 2.5 ± 2.1 | 11 ± 0 |
| 5 | 0 ± 0 | 50 ± 14.1 | 4.5 ± 2.1 | 1 ± 0 | 3 ± 2.8 |
DISCUSSION
In this study, we set out to explore the feasibility of transposon mutagenesis for the genome‐wide identification of virulence genes in F. oxysporum f. sp. lycopersici strain 4287, a trans‐kingdom pathogen model able to cause disease in both tomato plants and immunodepressed mice (Di Pietro et al., 2003; Ortoneda et al., 2004). For the large‐scale approach, we used a two‐component TE system with a novel modified version of impala160. The choice of impala was based on two criteria. First, this element has been shown to tolerate genetically engineered modifications, a prerequisite for use in a two‐component system (Firon et al., 2003; Hua‐Van et al., 2001; Villalba et al., 2001). Second, different versions of impala160 have been used successfully in previous transposon tagging studies, including a smaller screen of 350 revertants of M. grisea leading to the isolation of a virulence gene (Villalba et al., 2001), as well as a large‐scale screen with 2386 revertants to identify essential genes in A. fumigatus (Firon et al., 2003). The analysis of the revertants obtained in our study suggested that the characteristics of the transposition process of imp160::gfp in F. oxysporum f. sp. lycopersici 4287 are highly similar to those reported for other impala160 versions in different Fusarium spp. (Hua‐Van et al., 2001), M. grisea (Villalba et al., 2001), A. nidulans (Li Destri Nicosia et al., 2001) and A. fumigatus (Firon et al., 2003). This result is consistent with the concept that TEs from the Tc1‐mariner superfamily transpose through a conserved mechanism which is independent of host‐specific factors (Plasterk et al., 1999).
Efficiency of the imp160::gfp element for insertional mutagenesis
Ideally, a random insertional mutagenesis tool should combine: (i) ease of production of insertional mutants; (ii) absence of rearrangements associated with insertions; and (iii) random distribution of insertions along the genome. With regard to the first point, the structure of the imp160::gfp element allowed the rapid generation of a large collection of insertional mutants from a single parental strain by simple selection for wild‐type growth on nitrate. The estimated re‐insertion rate of imp160::gfp was around 75%, similar to that reported in previous studies with impala160 (Firon et al., 2003; Hua‐Van et al., 2001; Li Destri Nicosia et al., 2001). Concerning the second point, we found no evidence for rearrangements at the insertion site of imp160::gfp in the five revertants analysed, which is also in agreement with previous studies using different versions of impala (Firon et al., 2003; Hua‐Van et al., 2001; Li Destri Nicosia et al., 2001). The absence of DNA rearrangements represents a major advantage of transposon mutagenesis versus REMI and ATMT, because it facilitates the characterization of the transposon‐tagged loci (Daboussi and Capy, 2003). Based on the relatively low number of revertants analysed in this study, re‐insertion of imp160::gfp in the genome of F. oxysporum f. sp. lycopersici appeared to be random at both the nucleotide and chromosomal levels. However, in four of the five revertants, TE had re‐inserted outside a predicted coding region, suggesting a preference for 5′ non‐coding regions; this appears to be independent of the host species, because a similar trend has been reported in two previous studies in A. nidulans and A. fumigatus (Firon et al., 2003; Li Destri Nicosia et al., 2001). This preference for non‐coding regions will most probably result in a lower mutagenesis rate than expected.
Usefulness of the apple slice infection screening assay for the identification of virulence mutants
One of the hypotheses tested in this study was that an invasive growth assay on apple slices could be used as a rapid and efficient method to identify virulence mutants in the tomato vascular wilt pathogen F. oxysporum f. sp. lycopersici. This idea originated from previous studies, which showed that most gene knockout mutants of F. oxysporum f. sp. lycopersici strain 4287 with reduced virulence in tomato root infection assays were also affected in their ability to grow invasively on tomato fruits (Delgado‐Jarana et al., 2005; Di Pietro et al., 2001; Madrid et al., 2003; Martinez‐Rocha et al., 2008; Martin‐Urdiroz et al., 2008). We chose the apple slice infection assay because it has a number of characteristics that make it ideally suited for a high‐throughput virulence screening approach. First, it is at least five times faster and much less labour intensive than the tomato root infection assay (only 4 days vs. > 20 days until the final evaluation of disease symptoms). Second, it only requires a fraction of the space because the inoculated apple slices can be incubated in standard Petri dishes, obviating the need for large glasshouse or growth chamber facilities. The protocol used in this study allows for the processing and screening of a large collection of revertants by a single researcher within a few months under standard laboratory conditions.
After screening 2072 Nia+ revertants, we identified 14 that were less invasive on apple fruits. When these revertants were re‐tested using the standard tomato root infection assay, seven still showed significantly reduced virulence, a 50% correlation between virulence phenotypes on apple fruits and tomato plants. This rate of correlation validates the usefulness of the apple slice infection assay for large‐scale virulence screening in F. oxysporum, given the savings in time, labour costs and space. However, it is important to note that certain types of mutation affecting tissue‐ or host‐specific infection mechanisms, such as tomato root‐ or xylem‐related virulence factors, may remain undetected by the apple slice assay. Moreover, mutations in genes contributing only partly to virulence on tomato plants are difficult to detect with this assay (M. S. López‐Berges et al., unpublished data). Failure to detect these classes of mutants may partially explain the relatively low overall frequency of virulence mutants obtained in our revertant screening (seven of 2072, or 0.33%), although similar frequencies were obtained in infection screens with M. grisea using transposon mutagenesis (0.3%) (Villalba et al., 2001) or REMI (0.5%) (Balhadere and Talbot, 2001; Sweigard et al., 1998). The similar frequency obtained with impala160 in a standard infection assay of M. grisea (Villalba et al., 2001) suggests that the low rate of virulence mutants obtained in our study may be caused by factors intrinsic to impala, such as the preference for non‐coding regions, rather than to the characteristics of the apple slice infection assay used in our virulence screening.
Limitations of transposon tagging in F. oxysporum
Random insertional mutagenesis is an excellent approach for dissecting complex biological traits, such as pathogenicity, because it does not require prior information or assumptions on gene function. In previous studies, REMI has been used successfully for the identification of novel virulence factors in F. oxysporum (Duyvesteijn et al., 2005; Imazaki et al., 2007; Inoue et al., 2002; Madrid et al., 2003). ATMT has also been proposed as an efficient insertional mutagenesis tool for this species (Mullins et al., 2001), and a large‐scale ATMT approach is currently being carried out in the sequenced strain 4287 (Michielse et al., 2008). However, although transposon gene tagging with the impala160 TE has been applied successfully in different fungal species (Firon et al., 2003; Li Destri Nicosia et al., 2001; Villalba et al., 2001), not a single virulence gene has been identified in F. oxysporum by transposon mutagenesis. A previous attempt at transposon insertional mutagenesis in F. oxysporum f. sp. melonis yielded several virulence mutants, but no tagged genes (Migheli et al., 2000).
The present large‐scale transposon tagging approach has led to the isolation of a novel virulence gene: FOXG_00016. A detailed analysis of the role of this velvet‐like gene is currently underway. Our result shows that transposon insertional mutagenesis is feasible in F. oxysporum, although the extremely low success rate (only one tagged virulence mutant from 2072 revertants screened) hardly makes it the method of choice in this species. The low efficiency is most probably a result of a combination of different factors: preference of impala160 for non‐coding regions; inherent limitations of the apple slice assay; and, most importantly, the very low percentage of virulence mutations linked to the transposon insertion (only one of seven virulence mutants tested). Although it is still possible that the aberrant expression of additional ORFs located close to the insertion site might be responsible for the virulence phenotype in some of the revertants (e.g. 6F7, see ‘Results’ section), we suspect that at least part of the high genetic variability may be inherent to co‐transformant T12, the reporter strain used for revertant selection. To test this hypothesis, we followed the number of spontaneous chlorate‐resistant mutants as a measure of genetic variability, and found that all T12‐derived revertant colonies tested generated chlorate‐resistant mutants, compared with only one of five colonies in the wild‐type F. oxysporum strain. A probable explanation of the high level of genetic instability in T12 is the increased transposition of endogenous TEs. Inspection of the complete genome sequence of F. oxysporum f. sp. lycopersici strain 4287 revealed the presence of a large number of Tes, which can be mobilized by the constitutively expressed impala160 transposase, including nine non‐autonomous copies of impala (five impE and four impD) and 37 copies of the miniature inverted‐repeat TEs mimp1 and mimp2 (Dufresne et al., 2007; M. Dufresne et al., unpublished data). Mobilization in trans may even be possible for other mimp elements, such as mimp3, mimp5 and mimp6. Finally, as T12 was among the co‐transformants providing the highest frequency of revertants, it cannot be ruled out that other inherent mechanisms, related to the two previous transformation events, could also contribute to the high genetic variability of this strain.
In summary, a large‐scale transposon tagging approach has led to the isolation of a novel virulence gene, FOXG_00016, the first identified by transposon tagging in F. oxysporum. Our study demonstrates that two‐component transposon mutagenesis can be used for the identification of virulence genes in the sequenced strain of F. oxysporum f. sp. lycopersici, provided that background genetic variability caused by endogenous TEs can be reduced. Using a tightly regulable rather than a constitutive promoter to control the heterologous source of transposase may provide a means to address this issue in future studies.
EXPERIMENTAL PROCEDURES
Biological material and culture conditions
Fusarium oxysporum f. sp. lycopersici race 2 strain 4287 (FGSC 9935) and nit1− mutant CO6 derived thereof (Garcia‐Pedrajas and Roncero, 1996) were used. Fungal strains were stored as microconidial suspensions with 30% glycerol at –80 °C. For the extraction of DNA, production of microconidia, and fungal development and conidiation studies, cultures were grown for the indicated times in liquid potato dextrose broth (PDB; Difco, Detroit, MI, USA) at 28 °C with shaking at 170 r.p.m. (Di Pietro and Roncero, 1998). For phenotypic analysis of colony growth, drops of water containing 2 × 104 microconidia were spotted onto potato dextrose agar (PDA; Difco) or YPGA (1% yeast extract, 1% peptone, 7% glucose, 1.5% agar) plates and incubated at 28 °C for 3 days. Assays for polygalacturonase production on PGA plates were performed as described previously (Scott‐Craig et al., 1990). For nitrate reductase repression assays, MM plates containing 10 mm 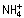 and 100 mm
and 100 mm 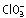 were spot inoculated with 5 × 103 microconidia and incubated at 28 °C for 3 days (Polley and Caddick, 1996). For cellophane invasion assays, autoclaved cellophane sheets were placed on MM plates, and the centre of each plate was spot inoculated with 5 × 104 microconidia. After 4 days at 28 °C, the cellophane sheet with the fungal colony was removed carefully. The presence or absence of fungal mycelium on the underlying medium was recorded after incubation of the plates for an additional 24 h at 28 °C (Prados Rosales and Di Pietro, 2008). For Nia+ reversion studies, microconidia of co‐transformants were obtained by washing and filtration from a single spore culture grown on solid medium, and spread at different dilutions on nitrate minimal agar medium supplemented with 0.06% Triton X‐100, as described previously (Hua‐Van et al., 2001).
were spot inoculated with 5 × 103 microconidia and incubated at 28 °C for 3 days (Polley and Caddick, 1996). For cellophane invasion assays, autoclaved cellophane sheets were placed on MM plates, and the centre of each plate was spot inoculated with 5 × 104 microconidia. After 4 days at 28 °C, the cellophane sheet with the fungal colony was removed carefully. The presence or absence of fungal mycelium on the underlying medium was recorded after incubation of the plates for an additional 24 h at 28 °C (Prados Rosales and Di Pietro, 2008). For Nia+ reversion studies, microconidia of co‐transformants were obtained by washing and filtration from a single spore culture grown on solid medium, and spread at different dilutions on nitrate minimal agar medium supplemented with 0.06% Triton X‐100, as described previously (Hua‐Van et al., 2001).
Plasmids
The vector pPtrap10B containing the imp160::gfp element was constructed by introducing an 862‐bp SmaI fragment containing the coding region of the egfp gene into the SmaI site of a transposase‐deleted impala160 version containing 61 bp and 31 bp of the 5′ and 3′ border sequences, respectively. The construct was then used to replace the impala element previously cloned into the promoter region of the A. nidulans niaD gene in the pNI160+ plasmid (Hua‐Van et al., 2001).
Plasmid pHEO62 contains the coding region of the impalaE transposase (Hua‐Van et al., 1998), cloned between the A. nidulans gpdA promoter and trpC terminator. The entire cassette was extracted from plasmid pEO62 (Li Destri Nicosia et al., 2001) as an EcoRI/HindIII fragment, and cloned into plasmid pBC1004 (Carroll et al., 1994) containing the hygromycin resistance gene.
Nucleic acid manipulations, PCRs and gene disruption
Genomic DNA was extracted from F. oxysporum mycelium as described by Raeder and Broda (1985). DNA was treated with appropriate restriction enzymes and subjected to Southern hybridization analysis following standard protocols (Sambrook, et al., 1989) using the non‐isotopic digoxigenin labelling kit (Roche Diagnostics SL, Barcelona, Spain), or 32P‐labelled using the Rediprime II DNA labeling kit (Amersham Biosciences, Freiburg, Germany), according to the manufacturers’ instructions. The 419‐bp niaD probe was generated with primers niaCG1 and niaCG2 (Dufresne et al., 2007). Genomic DNA of F. oxysporum strains was used for PCR amplification on a Perkin‐Elmer GeneAmp System (Madrid, Spain). PCRs were routinely performed with the Long Template PCR system (Roche Diagnostics SL), and products were precipitated with 4 m 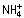 acetate, pH 5.4, and isopropanol, as described in standard protocols. PCR products from agarose gels were purified with the GENECLEAN® Turbo Kit (MP Biomedicals, LLC, Santa Ana, CA, USA). [Table S1 (‘Supporting Information’) provides a complete overview of the primers used in this study.]
acetate, pH 5.4, and isopropanol, as described in standard protocols. PCR products from agarose gels were purified with the GENECLEAN® Turbo Kit (MP Biomedicals, LLC, Santa Ana, CA, USA). [Table S1 (‘Supporting Information’) provides a complete overview of the primers used in this study.]
For the amplification of genomic sequences flanking the imp160::gfp element, inverse PCR was performed as described by Dufresne et al. (2007). Briefly, 100–500 ng of genomic DNA of Nia+ revertants was treated with the appropriate restriction enzyme, followed by heat inactivation of the restriction enzyme and self‐ligation with T4 DNA ligase, and 7.5–75 ng of self‐ligated DNA was used for inverse PCR with primers Gfp5b and Gfp3c. A second PCR round was performed, if needed, using 0.1–1 µL of the first round inverse PCR product and primers Gfp3c and Div5. Amplified DNA fragments were cloned into the pGEM‐T vector (Promega, Madison, WI, USA). Sequencing of both DNA strands of the obtained clones was performed at the Servicio de Secuenciación Automática de DNA, SCAI (University of Córdoba, Spain) using the Dyedeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an ABI Prism 377 Genetic Analyzer apparatus (Applied Biosystems). The Fusarium group genome database (http://www.broad.mit.edu/annotation/fgi/) and the National Center for Biotechnology Information (NCBI) database were searched using the blast algorithm (Altschul et al., 1990).
Gene knockout constructs were obtained by fusion PCR (Yang et al., 2004). The hygromycin B resistance gene, under the control of the A. nidulans gpdA promoter and trpC terminator (Punt et al., 1987), was amplified with primers M13‐For and M13‐Rev and used for the final fusion PCR with two fragments of the flanking sequences of each gene and external primers. [Details on the primers used are given in Table S1 (‘Supporting Information’).]
Fungal transformation
To obtain reporter strain T12, plasmids pPtrap10B and pHEO62 (see above) were used to co‐transform protoplasts of nit1−strain CO6 as described previously (Hua‐Van et al., 2001). Co‐transformants were identified from hygromycin‐resistant transformants by colony hybridization with the gfp probe.
For targeted gene knockout, PCR‐generated disruption cassettes were used to transform protoplasts of F. oxysporum strain 4287, and hygromycin‐resistant transformants were selected and purified by monoconidial isolation, as described by Di Pietro et al. (2001). Gene knockout was routinely confirmed by PCR and Southern blot analysis.
For complementation experiments, a PCR fragment encompassing the entire gene of interest was obtained using F. oxysporum genomic DNA as a template. The fragment obtained was introduced into protoplasts of the revertant or knockout strain by co‐transformation with the phleomycin resistance cassette amplified from plasmid pAN8‐1 (Mattern et al., 1988), and phleomycin‐resistant transformants were isolated as described by Di Pietro et al. (2001). The presence of the wild‐type allele in the complemented transformants was routinely confirmed by PCR with gene‐specific primers.
Isolation of revertants and storage of the revertant collection
Co‐transformant T12 was used to generate the entire mutant collection. Individual cultures originating from germinated single microconidia were grown in liquid PDB, and tested for the ability to produce revertants with wild‐type growth on plates with MM and sodium nitrate as the sole nitrogen source. Spores of these cultures were then spread at low concentration on to solid MM and, after 6–8 h at 26 °C, germinated single microconidia were selected under a stereomicroscope and transferred to individual microtitre wells with solid nitrate MM. After 1–3 weeks at 26 °C, revertant colonies were transferred to new microtitre plates with nitrate MM, and the collection was stored at –80 °C.
Infection assays
Tomato root infection assays were performed as described by Di Pietro et al. (2001) using the susceptible cultivar Monika (Syngenta Seeds, Almeria, Spain). All plant infection experiments were performed at least twice with similar results. For apple slice infection screening assays, individual revertants from the collection were re‐grown for 4 days on plates containing nitrate MM with 2% sorbose to restrict hyphal extension (Seiler and Plamann, 2003), transferred into 2‐mL Eppendorf tubes containing 1 mL of liquid PDB, and incubated at 28 °C and 170 r.p.m. for 4 days. Five microlitres of the culture containing approximately 107 microconidia/mL were spot inoculated on to slices of apple fruits (thickness, 0.8 cm; variety, Golden Delicious) placed in a Petri dish, and the slices were incubated at 28 °C and 100% humidity. Each strain was inoculated in duplicate. After 4 days, invasion and maceration of the surrounding tissue were evaluated macroscopically.
Assay for estimation of genetic instability of fungal strains
Five independent cultures of each strain originating from a single germinated microconidium were transferred to PDA plates and grown at 28 °C. After 7 days, microconidia were recovered from the colony by washing with sterile water and filtration, 2.5 × 106 microconidia were spread onto PDA plates containing 0.15 m chlorate (two plates per culture), and chlorate‐resistant colonies were counted after 3 days of incubation at 28 °C. As a positive control, five independent wild‐type cultures grown on PDA as described above were subjected to UV irradiation after 4 days of growth. The UV dose at 254 nm (Vilbert Lourtmat lamp, Paris, France) was 600 J/m at a dose rate of 4 J/m2/s. The incident fluence was measured with a VLX254 radiometer. UV‐treated cultures were incubated for an additional 3 days to complete the total of 7 days of growth, and the number of chlorate‐resistant colonies was determined as described above. The experiment was performed twice with similar results.
Supporting information
Fig. S1 Schematic illustration of the two‐component impala transposon tagging system in Fusarium oxysporum. (A) Physical maps of constructs pPtrap10B, with a modified version of the impala element (imp160::gfp) inserted upstream of the Aspergillus nidulans niaD gene, and pHEO62, carrying the imp160 transposase coding region fused to the constitutive A. nidulans gpdA promoter. (B, C) After co‐transformation of F. oxysporum nit− strain CO6 with pPtrap10B and pHEO62, transposition of the imp160::gfp element (B) is monitored by recovering vigorously growing Nia+ colonies on nitrate (C).
Fig. S2 Large‐scale infection assay on apple slices for the identification of virulence mutants. (A) Revertants stored in microtitre plates at –80 °C were grown on solid minimal medium with colony restrictor to confirm the Nia+ phenotype. (B) Suspensions containing 5 × 104 freshly obtained microconidia from liquid cultures grown in Eppendorf tubes with potato dextrose broth (PDB) were applied to apple slices. Colonization and maceration of the surrounding fruit tissue were determined after 4 days of incubation at 28 °C. Two experimental replicates are shown. Arrows point to a mutant with reduced virulence.
Fig. S3 Infection assay with different revertants on tomato plants. Groups of 10 plants were inoculated by immersing the roots into a suspension of 5 × 106 freshly obtained microconidia/mL of the indicated strains. The severity of disease symptoms was recorded at different times after inoculation, using an index ranging from 1 (healthy plant) to 5 (dead plant). Revertants 5A11, 6C3 and 17A1 show moderate virulence reduction, 6G5, 6F7 and 6F9 show strong virulence reduction, and 7A1 is non‐pathogenic. The non‐pathogenic Δfmk1 mutant was included as a negative control.
Table S1 Primers used in this study.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
The authors are grateful to Esther Martinez Aguilera, Universidad de Córdoba, for valuable technical assistance. This work was supported by grants BIO‐2004‐00276 and BIO‐2004‐01240 from the Spanish Ministerio de Ciencia y Tecnología (MCyT), by the Marie Curie RTN‐019277 (SIGNALPATH) and by the Centre National de la Recherche Scientifique (CNRS). M.S.L.B. has a PhD fellowship from MCyT.
REFERENCES
- Alonso, J.M. , Stepanova, A.N. , Leisse, T.J. , Kim, C.J. , Chen, H. , Shinn, P. , Stevenson, D.K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C.C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , Deen, H. , Geralt, M. , Hazari, N. , Hom, E. , Karnes, M. , Mulholland, C. , Ndubaku, R. , Schmidt, I. , Guzman, P. , Aguilar‐Henonin, L. , Schmid, M. , Weigel, D. , Carter, D.E. , Marchand, T. , Risseeuw, E. , Brogden, D. , Zeko, A. , Crosby, W.L. , Berry, C.C. and Ecker, J.R. (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Balhadere, P.V. and Talbot, N.J. (2001) PDE1 encodes a P‐type ATPase involved in appressorium‐mediated plant infection by the rice blast fungus Magnaporthe grisea . Plant Cell, 13, 1987–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts, M.F. , Tucker, S.L. , Galadima, N. , Meng, Y. , Patel, G. , Li, L. , Donofrio, N. , Floyd, A. , Nolin, S. , Brown, D. , Mandel, M.A. , Mitchell, T.K. , Xu, J.R. , Dean, R.A. , Farman, M.L. and Orbach, M.J. (2007) Development of a high throughput transformation system for insertional mutagenesis in Magnaporthe oryzae . Fungal Genet. Biol. 44, 1035–1049. [DOI] [PubMed] [Google Scholar]
- Blaise, F. , Remy, E. , Meyer, M. , Zhou, L. , Narcy, J.P. , Roux, J. , Balasdent, M.H. and Rouxel, T. (2007) A critical assessment of Agrobacterium tumefaciens‐mediated transformation as a tool for pathogenicity gene discovery in the phytopathogenic fungus Leptosphaeria maculans . Fungal Genet. Biol. 44, 123–138. [DOI] [PubMed] [Google Scholar]
- Brown, J.S. and Holden, D.W. (1998) Insertional mutagenesis of pathogenic fungi. Curr. Opin. Microbiol. 1, 390–394. [DOI] [PubMed] [Google Scholar]
- Calvo, A.M. (2008) The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 45, 1053–1061. [DOI] [PubMed] [Google Scholar]
- Carroll, A.M. , Sweigard, J.A. and Valent, B. (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41, 22. [Google Scholar]
- Choi, J. , Park, J. , Jeon, J. , Chi, M.H. , Goh, J. , Yoo, S.Y. , Jung, K. , Kim, H. , Park, S.Y. , Rho, H.S. , Kim, S. , Kim, B.R. , Han, S.S. , Kang, S. and Lee, Y.H. (2007) Genome‐wide analysis of T‐DNA integration into the chromosomes of Magnaporthe oryzae . Mol. Microbiol. 66, 826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clergeot, P.H. , Gourgues, M. , Cots, J. , Laurans, F. , Latorse, M.P. , Pepin, R. , Tharreau, D. , Notteghem, J.L. and Lebrun, M.H. (2001) PLS1, a gene encoding a tetraspanin‐like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea . Proc. Natl. Acad. Sci. USA, 98, 6963–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi, M. and Langin, T. (1992) Fot1, a new family of fungal transposable elements. Mol. Gen. Genet. 232, 12–16. [DOI] [PubMed] [Google Scholar]
- Daboussi, M.J. and Capy, P. (2003) Transposable elements in filamentous fungi. Annu. Rev. Microbiol. 57, 275–299. [DOI] [PubMed] [Google Scholar]
- Delgado‐Jarana, J. , Martinez‐Rocha, A.L. , Roldan‐Rodriguez, R. , Roncero, M.I. and Di Pietro, A. (2005) Fusarium oxysporum G‐protein beta subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways. Fungal Genet. Biol. 42, 61–72. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. and Roncero, M.I. (1998) Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum . Mol. Plant–Microbe Interact. 11, 91–98. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. , Garcia‐MacEira, F.I. , Meglecz, E. and Roncero, M.I. (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Di Pietro, A. , Madrid, M.P. , Caracuel, Z. , Delgado‐Jarana, J. and Roncero, M.I.G. (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–326. [DOI] [PubMed] [Google Scholar]
- Dufresne, M. , Perfect, S. , Pellier, A.L. , Bailey, J.A. and Langin, T. (2000) A GAL4‐like protein is involved in the switch between biotrophic and necrotrophic phases of the infection process of Colletotrichum lindemuthianum on common bean. Plant Cell, 12, 1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne, M. , Hua‐Van, A. , El Wahab, H.A. , Ben M’Barek, S. , Vasnier, C. , Teysset, L. , et al (2007) Transposition of a fungal miniature inverted‐repeat transposable element through the action of a Tc1‐like transposase. Genetics, 175, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyvesteijn, R.G. , Van Wijk, R. , Boer, Y. , Rep, M. , Cornelissen, B.J. and Haring, M.A. (2005) Frp1 is a Fusarium oxysporum F‐box protein required for pathogenicity on tomato. Mol. Microbiol. 57, 1051–1063. [DOI] [PubMed] [Google Scholar]
- Finnegan, D. (1992) Transposable elements. Curr. Opin. Genet. Dev. 2, 861–867. [DOI] [PubMed] [Google Scholar]
- Firon, A. , Villalba, F. , Beffa, R. and D’Enfert, C. (2003) Identification of essential genes in the human fungal pathogen Aspergillus fumigatus by transposon mutagenesis. Eukaryot. Cell, 2, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Pedrajas, M.D. and Roncero, M.I. (1996) A homologous and self‐replicating system for efficient transformation of Fusarium oxysporum . Curr. Genet. 29, 191–198. [DOI] [PubMed] [Google Scholar]
- Gordon, T. and Martyn, R. (1997) The evolutionary biology of Fusarium oxysporum . Annu. Rev. Phytopathol. 35, 111–128. [DOI] [PubMed] [Google Scholar]
- De Groot, M.J. , Bundock, P. , Hooykaas, P.J. and Beijersbergen, A.G. (1998) Agrobacterium tumefaciens‐mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842. [DOI] [PubMed] [Google Scholar]
- Hua‐Van, A. , Hericourt, F. , Capy, P. , Daboussi, M.J. and Langin, T. (1998) Three highly divergent subfamilies of the impala transposable element coexist in the genome of the fungus Fusarium oxysporum . Mol. Gen. Genet. 259, 354–362. [DOI] [PubMed] [Google Scholar]
- Hua‐Van, A. , Pamphile, J.A. , Langin, T. and Daboussi, M.J. (2001) Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol. Gen. Genet. 264, 724–731. [DOI] [PubMed] [Google Scholar]
- Idnurm, A. , Reedy, J.L. , Nussbaum, J.C. and Heitman, J. (2004) Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell, 3, 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imazaki, I. , Kurahashi, M. , Iida, Y. and Tsuge, T. (2007) Fow2, a Zn(II)2Cys6‐type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum . Mol. Microbiol. 63, 737–753. [DOI] [PubMed] [Google Scholar]
- Inoue, I. , Namiki, F. and Tsuge, T. (2002) Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell, 14, 1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J. , Park, S.Y. , Chi, M.H. , Choi, J. , Park, J. , Rho, H.S. , Kim, S. , Goh, J. , Yoo, S. , Park, J.Y. , Yi, M. , Yang, S. , Kwon, M.J. , Han, S.S. , Kim, B.R. , Khang, C.H. , Park, B. , Lim, S.E. , Jung, K. , Kong, S. , Karunakaran, M. , Oh, H.S. , Kim, H. , Kang, S. , Choi, W.B. and Lee, Y.H. (2007) Genome‐wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 39, 561–565. [DOI] [PubMed] [Google Scholar]
- Jeon, J.S. , Lee, S. , Jung, K.H. , Jun, S.H. , Jeong, D.H. , Lee, J. , Kim, C. , Jung, S. , Yang, K. , Nam, J. , An, K. , Han, M.J. , Sung, R.J. , Choi, H.S. , Yu, J.H. , Choi, J.H. , Cho, S.Y. , Cha, S.S. , Kim, S.I. and An, G. (2000) T‐DNA insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570. [DOI] [PubMed] [Google Scholar]
- Kempken, F. and Kuck, U. (2000) Tagging of a nitrogen pathway‐specific regulator gene in Tolypocladium inflatum by the transposon Restless. Mol Gen Genet. 263, 302–308. [DOI] [PubMed] [Google Scholar]
- Kidwell, M.G. and Lisch, D. (1997) Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA, 94, 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, A. , Takano, Y. , Furusawa, I. and Okuno, T. (2001) Peroxisomal metabolic function is required for appressorium‐mediated plant infection by Colletotrichum lagenarium . Plant Cell, 13, 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladendorf, O. , Brachmann, A. and Kamper, J. (2003) Heterologous transposition in Ustilago maydis . Mol. Genet. Genomics, 269, 395–405. [DOI] [PubMed] [Google Scholar]
- Langin, T. , Capy, P. and Daboussi, M.J. (1995) The transposable element impala, a fungal member of the Tc1‐mariner superfamily. Mol. Gen. Genet. 246, 19–28. [DOI] [PubMed] [Google Scholar]
- Li Destri Nicosia, M.G. , Brocard‐Masson, C. , Demais, S. , Hua Van, A. , Daboussi, M.J. and Scazzocchio, C. (2001) Heterologous transposition in Aspergillus nidulans . Mol. Microbiol. 39, 1330–1344. [PubMed] [Google Scholar]
- Madrid, M.P. , Di Pietro, A. and Roncero, M.I. (2003) Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47, 257–266. [DOI] [PubMed] [Google Scholar]
- Martinez‐Rocha, A.L. , Roncero, M.I. , Lopez‐Ramirez, A. , Marine, M. , Guarro, J. , Martinez‐Cadena, G. and Di Pietro, A. (2008) Rho1 has distinct functions in morphogenesis, cell wall biosynthesis and virulence of Fusarium oxysporum . Cell. Microbiol . [DOI] [PubMed]
- Martin‐Urdiroz, M. , Roncero, M.I. , Gonzalez‐Reyes, J.A. and Ruiz‐Roldan, C. (2008) ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum . Eukaryot. Cell, 7, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattern, I.E. , Punt, P.J. , Van Den Hondel, D.A. (1988) A vector of Aspergillus transformation conferring phleomycin resistance. Fungal Genet. Newsl. 35, 25. [Google Scholar]
- Meng, Y. , Patel, G. , Heist, M. , Betts, M.F. , Tucker, S.L. , Galadima, N. , Donofrio, N.M. , Brown, D. , Mitchell, T.K. , Li, L. , Xu, J.R. , Orbach, M. , Thon, M. , Dean, R.A. and Farman, M.L. (2007) A systematic analysis of T‐DNA insertion events in Magnaporthe oryzae . Fungal Genet. Biol. 44, 1050–1064. [DOI] [PubMed] [Google Scholar]
- Michielse, C.B. , Hooykaas, P.J. , Van Den Hondel, C.A. and Ram, A.F. (2005) Agrobacterium‐mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 48, 1–17. [DOI] [PubMed] [Google Scholar]
- Michielse, C. , Van Wijk, R. , Reijnen, L. , Cornelissen, B. and Rep, M. (2008) Insight into the molecular requirements for pathogenicity of Fusarium oxysporum f. sp. lycopersici through large‐scale insertional mutagenesis. ECFG Conference, Edinburgh, UK: Fungal Genetics Ltd., Nottingham, UK. [DOI] [PMC free article] [PubMed]
- Migheli, Q. , Steinberg, C. , Davière, J. , Olivain, C. , Gerlinger, C. , Gautheron, N. , Alabouvette, C. and Daboussi, M. (2000) Recovery of mutants impaired in pathogenicity after transposition of impala in Fusarium oxysporum f. sp. melonis . Phytopathology, 90, 1279–1284. [DOI] [PubMed] [Google Scholar]
- Mooney, J.L. and Yager, L.N. (1990) Light is required for conidiation in Aspergillus nidulans . Genes Dev. 4, 1473–1482. [DOI] [PubMed] [Google Scholar]
- Mullins, E. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D. and Kang, S. (2001) Agrobacterium‐mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Nucci, M. and Anaissie, E. (2007) Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortoneda, M. , Guarro, J. , Madrid, M.P. , Caracuel, Z. , Roncero, M.I. , Mayayo, E. and Di Pietro, A. (2004) Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72, 1760–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk, R.H. , Izsvak, Z. and Ivics, Z. (1999) Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15, 326–332. [DOI] [PubMed] [Google Scholar]
- Polley, S.D. and Caddick, M.X. (1996) Molecular characterisation of meaB, a novel gene affecting nitrogen metabolite repression in Aspergillus nidulans . FEBS Lett. 388, 200–205. [DOI] [PubMed] [Google Scholar]
- Prados Rosales, R.C. and Di Pietro, A. (2008) Vegetative hyphal fusion is not essential for plant infection by Fusarium oxysporum . Eukaryot Cell, 7, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt, P.J. , Oliver, R.P. , Dingemanse, M.A. , Pouwels, P.H. and Van Den Hondel, C.A. (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli . Gene, 56, 117–124. [DOI] [PubMed] [Google Scholar]
- Raeder, U. and Broda, P. (1985) Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1, 17–22. [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scott‐Craig, J.S. , Panaccione, D.G. , Cervone, F. and Walton, J.D. (1990) Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell, 2, 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S. and Plamann, M. (2003) The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa . Mol. Biol. Cell, 14, 4352–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard, J.A. , Carroll, A.M. , Farrall, L. , Chumley, F.G. and Valent, B. (1998) Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant–Microbe Interact. 11, 404–412. [DOI] [PubMed] [Google Scholar]
- Urban, M. , Bhargava, T. and Hamer, J.E. (1999) An ATP‐driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba, F. , Lebrun, M.H. , Hua‐Van, A. , Daboussi, M.J. and Grosjean‐Cournoyer, M.C. (2001) Transposon impala, a novel tool for gene tagging in the rice blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 14, 308–315. [DOI] [PubMed] [Google Scholar]
- Walton, F.J. , Idnurm, A. and Heitman, J. (2005) Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans . Mol. Microbiol. 57, 1381–1396. [DOI] [PubMed] [Google Scholar]
- Weil, C.F. and Kunze, R. (2000) Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae . Nat. Genet. 26, 187–190. [DOI] [PubMed] [Google Scholar]
- Wong, K.H. , Hynes, M.J. , Todd, R.B. and Davis, M.A. (2007) Transcriptional control of nmrA by the bZIP transcription factor MeaB reveals a new level of nitrogen regulation in Aspergillus nidulans . Mol. Microbiol. 66, 534–551. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Ukil, L. , Osmani, A. , Nahm, F. , Davies, J. , De Souza, C.P. , Dou, X. , Perez‐Balaguer, A. and Osmani, S.A. (2004) Rapid production of gene replacement constructs and generation of a green fluorescent protein‐tagged centromeric marker in Aspergillus nidulans . Eukaryot. Cell, 3, 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Schematic illustration of the two‐component impala transposon tagging system in Fusarium oxysporum. (A) Physical maps of constructs pPtrap10B, with a modified version of the impala element (imp160::gfp) inserted upstream of the Aspergillus nidulans niaD gene, and pHEO62, carrying the imp160 transposase coding region fused to the constitutive A. nidulans gpdA promoter. (B, C) After co‐transformation of F. oxysporum nit− strain CO6 with pPtrap10B and pHEO62, transposition of the imp160::gfp element (B) is monitored by recovering vigorously growing Nia+ colonies on nitrate (C).
Fig. S2 Large‐scale infection assay on apple slices for the identification of virulence mutants. (A) Revertants stored in microtitre plates at –80 °C were grown on solid minimal medium with colony restrictor to confirm the Nia+ phenotype. (B) Suspensions containing 5 × 104 freshly obtained microconidia from liquid cultures grown in Eppendorf tubes with potato dextrose broth (PDB) were applied to apple slices. Colonization and maceration of the surrounding fruit tissue were determined after 4 days of incubation at 28 °C. Two experimental replicates are shown. Arrows point to a mutant with reduced virulence.
Fig. S3 Infection assay with different revertants on tomato plants. Groups of 10 plants were inoculated by immersing the roots into a suspension of 5 × 106 freshly obtained microconidia/mL of the indicated strains. The severity of disease symptoms was recorded at different times after inoculation, using an index ranging from 1 (healthy plant) to 5 (dead plant). Revertants 5A11, 6C3 and 17A1 show moderate virulence reduction, 6G5, 6F7 and 6F9 show strong virulence reduction, and 7A1 is non‐pathogenic. The non‐pathogenic Δfmk1 mutant was included as a negative control.
Table S1 Primers used in this study.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


