SUMMARY
Evolutionary processes responsible for parasite adaptation to their hosts determine our capacity to manage sustainably resistant plant crops. Most plant–parasite interactions studied so far correspond to gene‐for‐gene models in which the nature of the alleles present at a plant resistance locus and at a pathogen pathogenicity locus determine entirely the outcome of their confrontation. The interaction between the pepper pvr2 resistance locus and Potato virus Y (PVY) genome‐linked protein VPg locus obeys this kind of model. Using synthetic chimeras between two parental PVY cDNA clones, we showed that the viral genetic background surrounding the VPg pathogenicity locus had a strong impact on the resistance breakdown capacity of the virus. Indeed, recombination of the cylindrical inclusion (CI) coding region between two PVY cDNA clones multiplied by six the virus capacity to break down the pvr23‐mediated resistance. High‐throughput sequencing allowed the exploration of the diversity of PVY populations in response to the selection pressure of the pvr23 resistance. The CI chimera, which possessed an increased resistance breakdown capacity, did not show an increased mutation accumulation rate. Instead, selection of the most frequent resistance‐breaking mutation seemed to be more efficient for the CI chimera than for the parental virus clone. These results echoed previous observations, which showed that the plant genetic background in which the pvr23 resistance gene was introduced modified strongly the efficiency of selection of resistance‐breaking mutations by PVY. In a broader context, the PVY CI coding region is one of the first identified genetic factors to determine the evolvability of a plant virus.
INTRODUCTION
The elucidation of the genetic bases of adaptation of populations to different environments is a goal which takes a central place in evolutionary biology. Viruses, which are characterized by rapid replication, large population size and high mutation rate (Drake and Holland, 1999; García‐Arenal et al., 2001), constitute particularly interesting models to address experimentally this type of question (Elena and Sanjuan, 2007). In addition, as obligate parasites, viruses undergo a continuous process of selection by their hosts and competition within the swarm of mutants generated during multiplication (Domingo et al., 2006).
Plant diseases are often combated with pesticides, but the limited efficiency of these control measures (Oerke and Dehne, 1997; Raccah, 1986) and the need to develop more sustainable production systems have fuelled a trend towards the substitution of pesticide applications by plant genetic resistances. The investigation of the evolutionary processes responsible for parasite adaptation to a new resistant host is therefore crucial for the development and sustainable use of plant resistances. The durability of host resistance is defined as the persistence of resistance efficiency when resistant cultivars are used over long periods, on large areas and in the presence of the target pathogen (Johnson, 1981, 1984). Therefore, it is dependent primarily on the rhythm of adaptive changes affecting pathogen populations in response to selection by the resistant host (Zhan et al., 2002). In contrast, resistance breakdown occurs when the parasite has gained the capacity to infect the resistant variety and impairs its efficiency and economic interest (Kang et al., 2005). It has been proposed that resistance durability to viruses may be partly a consequence of viral population dynamics (García‐Arenal and McDonald, 2003), the number of mutations in the virus genome required for shifts in pathogenicity (Harrison, 2002) and the constraints imposed on amino acid substitutions in the virus pathogenicity factors (Janzac et al., 2009).
Our case study involves the monogenic resistance to Potato virus Y (PVY; genus Potyvirus, family Potyviridae) conferred to pepper (Capsicum annuum) by the pvr23 gene. This resistance is recessive and thus results from the incompatibility between plant and virus factors. The outcome of the confrontation between pepper and PVY can be summed up, to a large extent, by the compatibility between two proteic partners: eIF4E (eukaryotic translation initiation factor 4E), encoded by the pepper pvr2 gene, and PVY VPg (genome‐linked viral protein). The ability of VPg to interact directly with eIF4E provides PVY with the capacity to multiply in its host and to infect it. Particular amino acid substitutions in eIF4E, such as those conferred by the pvr23 allele, render the pepper genotype resistant by impairing the interaction with VPg. In turn, amino acid substitutions in PVY VPg that restore the interaction confer resistance‐breaking properties (Charron et al., 2008). Five different amino acid substitutions in VPg of PVY have been shown to be independently responsible for pvr23 resistance breakdown in pepper (Ayme et al., 2006). This system is therefore a perfect example of a coevolutionary arms race between a parasite and its host.
Recent data have indicated that this plant–pathogen system cannot be entirely reduced to the eIF4E–VPg interaction. Palloix et al. (2009) have shown that the plant genetic background in which the major‐effect resistance gene pvr23 is introduced plays a critical role on the durability of pvr23. The frequency of resistance breakdown by PVY is high when pvr23 is introgressed into a susceptible genetic background, whereas no resistance breakdown occurs when pvr23 is combined with partial‐effect resistance quantitative trait loci (QTLs). The data of Palloix et al. (2009) strongly suggest that, after their appearance, the selection of PVY mutants possessing resistance‐breaking capacity is more efficient in susceptible than in partially resistant plant genetic backgrounds. This suggests that the same kind of effect may occur on the virus side of the interaction. In this study, we show that the viral genetic background, i.e. a region of the PVY genome outside the VPg major pathogenicity factor, also has a strong impact on the resistance breakdown capacity of the virus, and the evolutionary processes involved are characterized.
RESULTS
The viral genetic background modifies strongly the resistance‐breaking capacity
In a series of experiments, the doubled‐haploid line HD285 carrying the pvr23 resistance allele was challenged by the parental PVY isolates SON41p and LYE84.2 and by six synthetic chimeras between them (Fig. 1). The two parental isolates are genetically close: they share 92% genome identity at the nucleotide level and both belong to the C1 clade of PVY isolates (Moury, 2010). In HD285, SON41p was shown to infect about 29% of manually inoculated plants (Ayme et al., 2006), whereas LYE84.2 was not infectious at all (zero of >1000 inoculated plants; data not shown). Variable infection frequencies were observed at the systemic level, depending on the viruses and experiments (Table 1). Virus symptoms were obvious and consisted of vein necroses in leaves, often followed by necrosis of the main stem, and sometimes leading to leaf abscission and/or death of the plants. The presence of PVY in these plants and the absence of PVY in symptomless plants was confirmed by double‐antibody sandwich‐enzyme‐linked immunosorbent assay (DAS‐ELISA), and the presence of PVY in necrotic plants was confirmed by specific reverse transcription‐polymerase chain reaction (RT‐PCR) for experiment 3 (see below).
Figure 1.
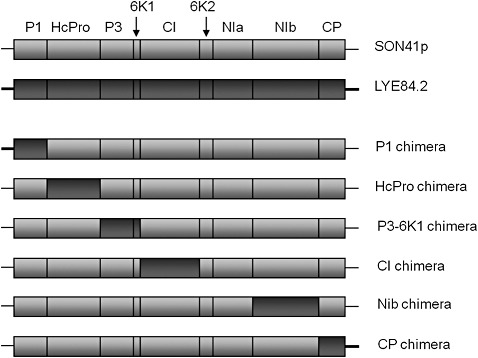
Schematic representation of the six chimera constructed by homologous recombination between the SON41p and LYE84.2 cDNA clones.
Table 1.
Frequency of breakdown of HD285 resistance (conferred by pvr23) by Potato virus Y (PVY) clone SON41p and six chimeras between SON41p and LYE84.2. Letters denote homogeneous groups in each column using Fisher's exact test at the P= 0.05 type‐I error threshold (corresponding to a 0.0024 type‐I error threshold after Bonferroni correction for multiple comparisons for experiments 1 and 2).
| Experiment 1 | Experiment 2 | Experiment 3 | Total | |
|---|---|---|---|---|
| P1 | 5/100b | 13/100b | ||
| HcPro | 0/100b | 3/100bc | ||
| P3‐6K1 | 0/100b | 0/100c | ||
| CI | 20/100a | 32/100a | 172/210a | 224/410 (54.6%) |
| Nib | 0/100b | 13/100b | ||
| CP | 1/100b | 2/100c | ||
| SON41p | 2/100b | 8/100bc | 30/225b | 40/425 (9.4%) |
Perfect correspondence between the systemic infection of HD285 after inoculation by SON41p and the occurrence of resistance‐breaking events has been established previously on a limited number of plants (Ayme et al., 2006). This correspondence was confirmed for both SON41p and the cylindrical inclusion (CI) chimera for 87% of infected plants if we considered only the mutations known to confer resistance‐breaking properties (Ayme et al., 2006), and for 99% of infected plants if we considered, in addition, the mutations that were candidates for resistance‐breaking properties (see below). Consequently, the capacity of SON41p and the six chimeras to break down the pvr23 resistance from HD285 can be estimated precisely by the frequency of systemically infected plants. Although the resistance breakdown frequency varied greatly between the three independent experiments (from 20% to more than 80% for the CI chimera; Table 1), the CI chimera showed consistently a higher resistance breakdown capacity than SON41p or any other chimera (Table 1). Depending on the experiment, resistance breakdowns were 4–10 times (six times on average) more frequent with the CI chimera than with SON41p (Table 1). In experiment 2, the P1 and Nib chimeras also showed a higher resistance breakdown capacity than the P3‐6K1 or CP chimeras (Table 1). With the exception of the CI chimera, there was no significant difference in resistance breakdown capacity between SON41p and the other chimeras (Table 1). As SON41p and the CI chimera shared the same sequence of the VPg cistron, which was shown to be directly involved in resistance breakdown (2006, 2007; Charron et al., 2008), these results showed that the viral genetic background (i.e. one or several genome parts outside the VPg cistron) was able to modify strongly, directly or indirectly, the resistance breakdown capacity of PVY.
Sequence of the CI coding region of resistance‐breaking PVY populations
A straightforward explanation for the higher resistance breakdown capacity of the CI chimera may be that the CI cistron constitutes another factor involved directly in the breakdown of pvr23 resistance. According to this assumption, the CI cistron would be an alternative pathogenicity factor to the VPg cistron, thus increasing the number of mutational pathways conferring resistance‐breaking properties to PVY, as described in the Lettuce mosaic virus–lettuce system (Abdul‐Razzak et al., 2009). This would imply that the resistance breakdown would be the result of mutations in the CI cistron. However, among sequences of the CI cistron from 10 randomly chosen HD285 plants infected by the CI chimera, no nucleotide substitutions were observed compared with the sequence of the CI cistron of the parental LYE84.2 cDNA clone.
Sequence of the VPg coding region of resistance‐breaking PVY populations
In contrast with the lack of variation in the CI cistron after resistance breakdown, mutations were observed in the VPg cistron of almost all (99%) infected HD285 plants (Table 2). Most of these mutations (190 of 196) were single nucleotide substitutions corresponding to a single amino acid change in the central part of the VPg encompassing amino acid positions 101–122 (Table 2). The large majority of these substitutions (166 of 190; 87.3%), corresponding to amino acid substitutions 101G, 115K, 119N and 120C, have been observed previously during breakdown of the resistance of HD285, and have been shown by a reverse genetics approach to be sufficient for resistance breakdown (Ayme et al., 2006). Five other substitutions (102K, 119G, 119E, 120I and 122K) were observed in this study. As they are located in close vicinity to (102K and 122K), or at the same amino acid position as (119G, 119E and 120I), the previous substitutions, it is highly probable that they are sufficient to confer resistance‐breaking properties. The distributions of the different VPg mutations were not significantly different between SON41p and the CI chimera as confirmed by Monte Carlo simulations (P < 0.05).
Table 2.
Number of infected HD285 plants (pvr23/pvr23) after inoculation by four Potato virus Y (PVY) inocula [SON‐1 and SON‐2, corresponding to SON41p, and CI‐1 and CI‐2, corresponding to the cylindrical inclusion (CI) chimera] and haplotype of the PVY populations. These haplotypes were determined by the consensus sequence of the genome‐linked viral protein (VPg) coding region and named after the observed amino acid substitutions compared with the VPg of SON41p, which is identical to that of the CI chimera. Letters denote homogeneous groups using Fisher's exact test at the 0.05 type‐I error threshold (corresponding to a 0.0085 type‐I error threshold after Bonferroni correction for multiple tests). Each type of VPg mutation was searched for through the single nucleotide polymorphism (SNP) report issued from high‐throughput sequencing of the corresponding inoculum. The presence or absence of the substitution within inocula is highlighted in light grey and dark grey, respectively.
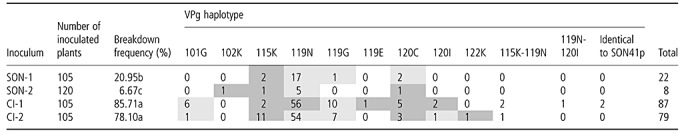
In four viral populations, two double‐picks were observed in the chromatogram of the VPg cistron sequence (Table 2). These cases could correspond to either a mixture of two molecules with a single nucleotide substitution each, or a mixture between a double mutant and a virus identical to SON41p. Because the resistance‐breaking substitutions 115K and 119N confer to PVY a huge competitive advantage against SON41p into the resistant plant genotype (Ayme et al., 2006), the former hypothesis is much more likely.
Interestingly, two populations issued from the CI chimera were identical to SON41p with regard to their VPg cistron sequence. The CI coding region of these populations was sequenced and did not reveal any nucleotide substitution when compared with the parental LYE84.2 cDNA clone. This suggests that SON41p was able to multiply and migrate, at least to some extent, into the resistant plant genotype.
A comparison between the single nucleotide polymorphism (SNP) reports of about 2000 sequences (from high‐throughput sequencing of the central part of the VPg cistron; see Experimental procedures section) obtained for each of the four inocula and the substitutions responsible (or candidates) for the corresponding resistance breakdowns showed that these substitutions could not be detected in inocula for 12 of 22 combinations between a resistance‐breaking substitution and an inoculum (Table 2). Moreover, there was no correlation between the frequency of resistance‐breaking variants into an inoculum and the frequency of resistance breakdowns in plants inoculated with this inoculum (Fig. 2). Among the different inocula, the most frequent resistance‐breaking substitution was 119N, which was present in 1% of sequences obtained from the SON‐1 inoculum. However, only 16% of HD285 plants inoculated with SON‐1 were infected by mutant 119N. Conversely, the highest breakdown frequency (53%) was caused by the resistance‐breaking variant 119N in HD285 plants inoculated with inoculum CI‐1. However, mutation 119N was not detected in inoculum CI‐1.
Figure 2.
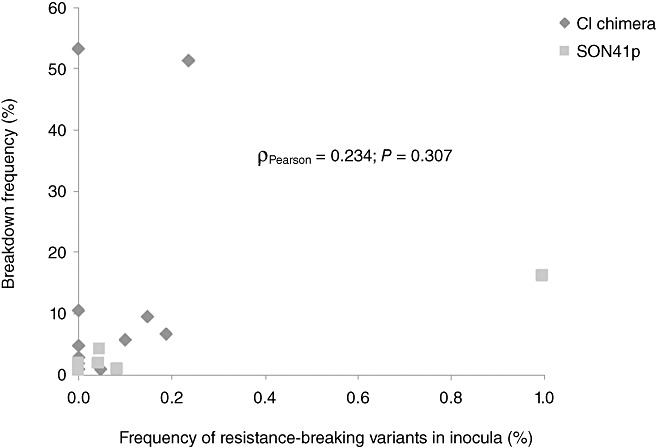
Relationship between the frequency of resistance‐breaking variants in inocula and the frequency of resistance breakdown in the corresponding inoculated HD285 plants. Pearson's product‐moment correlation and the corresponding significance are indicated.
Mutation frequencies, normalized Shannon indices and positive‐selection site frequencies did not reveal any significant differences in diversity between viral populations issued from SON41p and the CI chimera, whatever the host plant (Nicotiana spp., the resistant pepper HD285 and the susceptible pepper YW) (Table 3). Consequently, the hypotheses that the CI cistron of the parental LYE84.2 isolate recombined into the SON41p clone may have increased the global mutation rate or may have altered the mutant spectrum (as described in Lozano et al., 2009) of PVY were rejected. In PVY populations from HD285, the mutation frequency was even lower for the CI chimera than for SON41p, although only marginally significant (P= 0.066, Table 3). Haplotype frequencies and Shannon diversity indices showed that all sequenced PVY populations consisted of heterogeneous mutant distributions containing one major sequence surrounded by a mutant spectrum of closely related variants.
Table 3.
Comparison of mutation frequency (mutfr), normalized Shannon index (normH) and positive‐selection site frequency (PSfr) between viral populations issued from clone SON41p and from the cylindrical inclusion (CI) chimera into Nicotiana spp. (inocula), the resistant pepper HD285 and the susceptible pepper Yolo Wonder based on high‐throughput sequencing of the central part of the genome‐linked viral protein (VPg) cistron (nucleotide positions 5930–6168); 1577–2407 sequences were analysed for each sample. The Kruskal–Wallis chi‐squared test (KW) and corresponding significance (P) are indicated for each comparison between viruses.
| mutfr | normH | PSfr | |
|---|---|---|---|
| Inoculum | |||
| SON41p | 0.00048 | 0.265 | 0.100 |
| CI chimera | 0.00055 | 0.290 | 0.130 |
| (KW/P) | 2.40/0.1213 | 1.50/0.2207 | 0.17/0.6831 |
| YW | |||
| SON41p | 0.00049 | 0.235 | 0.100 |
| CI chimera | 0.00042 | 0.215 | 0.085 |
| (KW/P) | 2.40/0.1213 | 2.40/0.1213 | 0.00/1.0000 |
| HD285 | |||
| SON41p | 0.00054 | 0.280 | 0.120 |
| CI chimera | 0.00048 | 0.270 | 0.150 |
| (KW/P) | 3.37/0.0666 | 0.13/0.7213 | 1.21/0.2710 |
Competitiveness of SON41p‐119N and CI‐119N in HD285
The ability of CI‐119N to accumulate in competition with SON41p‐119N, both carrying the 119N mutation which was most frequently associated with HD285 resistance breakdown (Ayme et al., 2006; Table 2), was measured in HD285 plants inoculated with a mixture of these variants at 1:1 or 4:1 (conferring an advantage to SON41p‐119N) initial ratios (Fig. 3). For each initial ratio, CI‐119N showed a higher competitiveness than SON41p‐119N, and the majority [34 of 39 and 43 of 47 for the 1:1 ratio, and 35 of 45 and 34 of 46 for the 4:1 ratio, at 15 and 30 days post‐inoculation (dpi), respectively] of plants were infected by CI‐119N only, even as soon as 15 dpi. The proportion of plants in which CI‐119N had excluded SON41p‐119N was significantly higher than the initial proportion of that variant in the inoculum (Fisher's exact tests, P < 0.0001). However, this proportion was similar between 15 and 30 dpi (Fisher's exact tests, P= 0.306 and P= 0.185 for 1:1 and 4:1 initial ratios of the viruses, respectively).
Figure 3.
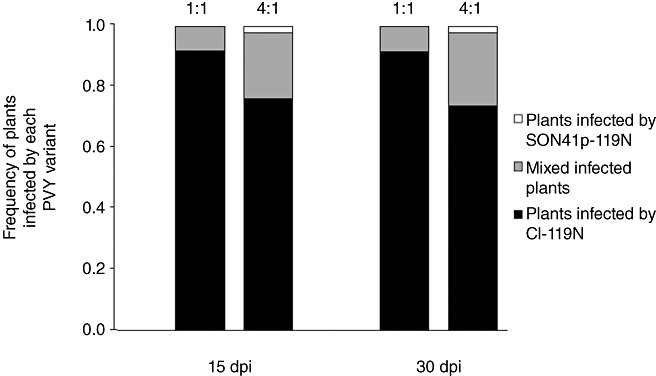
Relative competitiveness in HD285 of the pvr23 resistance‐breaking mutants SON41p‐119N and CI‐119N issued from clone SON41p and the cylindrical inclusion (CI) chimera, respectively, and carrying the 119N mutation in their genome‐linked viral protein (VPg). 1:1, SON41p‐119N and CI‐119N inocula were calibrated and mixed in the ratio of 1:1; 4:1, SON41p‐119N and CI‐119N inocula were calibrated and mixed in the ratio 4:1; dpi, days post‐inoculation.
DISCUSSION
In this study, we have shown that modification of the CI coding region in a PVY cDNA clone greatly increases the capacity of the virus to adapt to pvr23‐mediated resistance. Indeed, the resistance breakdown by the CI chimera is, on average, six times more frequent than by the parental SON41p clone (Table 1). The CI coding region has been shown previously to be responsible for resistance breakdown in other plant–potyvirus systems: rape–Turnip mosaic virus (2000, 2002), soybean–Soybean mosaic virus (Seo et al., 2009) and lettuce–Lettuce mosaic virus (Abdul‐Razzak et al., 2009). In all of these cases, reverse genetics approaches have shown that the CI coding region is directly involved in resistance breakdown. By contrast, the CI cistron of PVY isolate LYE84.2 is not a direct actor in resistance breakdown as: (i) it does not allow infection of 100% pvr23‐carrying pepper plants, either in the context of LYE84.2 or in the context of SON41p (i.e. in the CI chimera); and (ii) no nucleotide substitutions have been observed in the CI cistron of the CI chimera or in SON41p after resistance breakdown. For the same reasons, the CI cistron is not an alternative factor to the VPg cistron for resistance breakdown, as observed for the Lettuce mosaic virus, which can overcome mo11 resistance either by a mutation in the CI cistron or by undetermined mutation(s) in the VPg cistron (Abdul‐Razzak et al., 2009). The CI cistron is also not an additive factor to the VPg cistron for resistance breakdown, as it has been shown previously that VPg mutations observed in at least 87% of infected pvr23‐carrying pepper plants are sufficient for these breakdowns (Ayme et al., 2006). Consequently, we conclude that the CI cistron has an indirect effect on resistance breakdown, and confirm that the genetic background of the virus can have a strong influence on this previously defined gene‐for‐gene model (Charron et al., 2008).
In the same plant–virus system, Palloix et al. (2009) have shown that the plant genetic background in which the major‐effect pvr23 resistance gene is introduced also plays an important role in resistance breakdown. Indeed, when combined with three QTLs that, by themselves, did not impair the infection of the plants, but only delayed symptom appearance and decreased their intensity, the pvr23 gene was not broken down further. Consequently, from both the plant and virus sides of this biological system, small‐effect factors in the genetic background can drastically affect the outcome of the gene‐for‐gene interaction.
In this study, virus recombination was used as an artificial genetic tool for the measurement of the quantitative effects of the virus genetic background on resistance breakdowns. Recombination also affects significantly the evolutionary history of PVY in natural conditions (Glais et al., 2002; 2009a, 2009b; Moury et al., 2002; Revers et al., 1996), although the frequency of such events is largely ignored in plant viruses (but see Froissart et al., 2005) and is usually extremely low in plants coinfected by several potyvirus isolates. Consequently, this study allows the estimation of certain risks in terms of increased pathogenicity that can be expected from ‘accidental’ recombination events. Even if major‐effect genes remain untouched after recombination events, as does the VPg cistron in our case, major effects on such important traits as resistance breakdown can be expected from recombination. The observation of such an effect after the recombination of SON41p and LYE84.2 was unexpected because: (i) these two parental viruses show relatively low capacities of breakdown of pvr23 resistance (9.4% and 0%, respectively); (ii) they are genetically close (both belong to the C1 clade of PVY isolates that mainly affects pepper crops; Moury, 2010); and (iii) LYE84.2 is a tomato isolate that has a lower capacity than SON41p for infection and accumulation in pepper genotypes (data not shown). This illustrates that effects of recombination are largely unpredictable in viruses. It will be important to identify which mutations in the CI cistron are responsible for this increased resistance breakdown capacity to estimate whether this capacity can be acquired only by recombination or also by successive accumulation of a (limited) number of substitutions.
We examined two different evolutionary mechanisms that could be involved in the increased capacity of resistance breakdown of the CI chimera: mutation and selection. An increased mutation rate could indeed increase the probability of appearance of the VPg resistance‐breaking mutations in the PVY population. Alternatively, with an identical mutation rate, but an increased rate of multiplication of the virus, this probability would also be increased. These mechanisms were rejected because: (i) inoculum pressure was identical for SON41p and the CI chimera in our experiments; (ii) we observed no differences in diversity between viral populations issued from the CI chimera and SON41p, whatever the host plant considered (Table 3); and (iii) the CI chimera and SON41p accumulated to a similar level in seven susceptible (pvr2 +/pvr2 +) doubled‐haploid pepper lines issued from the same cross as HD285 and carrying different sets of resistance QTLs (Caranta et al., 1997, and data not shown). In contrast, we showed that the CI chimera carrying the 119N VPg mutation, which was most frequently (70% of cases) associated with resistance breakdown, was more competitive in pvr23‐carrying pepper plants than was the SON41p clone carrying the same VPg mutation (Fig. 3). This strongly suggests that the 119N mutation could be selected more rapidly by pvr23 resistance in a population issued from the CI chimera than in a population issued from SON41p. Therefore, the selection step that follows the appearance of the resistance‐breaking mutations is probably responsible for the difference in resistance breakdown observed between SON41p and the CI chimera.
Our results also indicate that the mutations involved in resistance breakdown are more likely in inoculated HD285 plants than in Nicotiana spp. plants that served as inoculum sources. This was suggested by: (i) the absence of approximately one‐half of these mutations in the inocula despite high‐throughput sequencing (i.e. from 2020–2411 sequences explored in the SNP reports of the four inocula); (ii) the lack of correlation between the frequency of these mutations in inocula and the frequency of resistance breakdowns (Fig. 2), which could be a result of the narrow bottlenecks and severe genetic drift that occur at the plant inoculation step (French and Stenger, 2003; Ohshima et al., 2010; Sacristan et al., 2003); and (iii) the fact that PVY carrying a wild‐type VPg (i.e. identical to that of SON41p) can, to some extent, multiply in and infect HD285 plants, as revealed in two cases (Table 2). Taken together, these results suggest an additional mechanism to explain the very low durability of pvr23. The residual multiplication of PVY in HD285 plants carrying pvr23 would favour the appearance of adaptive resistance‐breaking VPg mutations in these plants. Then, as soon as they appear, these mutants will be directly under the selective pressure of their host, avoiding the narrow bottlenecks that also occur during plant to plant transmission (Betancourt et al., 2008; Moury et al., 2007), and will outcompete the wild‐type virus population. This mutation fixation will be even more efficient and rapid as a large number of single nucleotide substitutions confer (or are strong candidates for) pvr23 resistance breakdown (at least nine in total; Table 2 and Ayme et al., 2006). Interestingly, the same evolutionary mechanism (i.e. selection) appears to be responsible for the effect of the plant genetic background on resistance breakdown (Palloix et al., 2009).
Beyond this evolutionary mechanism, it will be important to characterize more accurately the cellular and molecular mechanisms responsible for the resistance breakdown difference between the CI chimera and SON41p. The potyvirus CI protein possesses RNA‐binding domains (Fernandez et al., 1995) and helicase and ATPase domains (Fernandez et al., 1997; Gómez de Cedrón et al., 2006; Lain et al., 1990) that are essential for virus replication (Klein et al., 1994). CI is also involved in cell to cell movement of the virus through plasmodesmata (Carrington et al., 1998; Roberts et al., 1998; Wei et al., 2010). The CI self‐assembly is essential for interaction with the P3N‐PIPO potyvirus protein and for further building of cone‐shaped structures at plasmodesmata involved in the intercellular movement of potyviruses (Wei et al., 2010). The fact that the mechanism involved in increased resistance breakdown is more likely to be selection than mutation suggests that virus intercellular movement, rather than replication, could be the key mechanism here. The CI proteins of members of the Potyviridae accumulate as pinwheel‐shaped structures in the cytoplasm of infected cells. Electron microscopy observations of infected leaf tissues of HD285 plants did not reveal any difference between SON41p and the CI chimera (data not shown). Again, mapping of the region(s) and mutation(s) responsible for enhanced resistance breakdown will help to discriminate the causative function.
Recently, Pigliucci (2008) asked whether evolvability, the ability to facilitate phenotypic innovations in organisms, was an evolvable trait. In a stable environment, an organism that is well adapted and maximizes its fitness will find a need to reduce its adaptability in order to stay on this ‘fitness peak’. In contrast, in an unstable environment, this organism should increase its adaptability because it needs to react more rapidly to the changing peaks in the fitness landscape (as defined by Wright, 1932). The CI chimera clearly demonstrates that evolvability is indeed evolvable: recombination in the CI coding region multiplied by six the capacity of PVY to respond to the selective pressure of pvr23 resistance. Our results thus identify one of the first genetic factors that determine evolvability in a plant virus. However, it is not surprising that, in the notoriously unstable arms race world of host–parasite pathosystems, genetic factors increasing evolvability are selected for.
EXPERIMENTAL PROCEDURES
Plant and virus materials
Pepper (Capsicum annuum L.) genotypes used in this work were Yolo Wonder (YW) and HD285. YW is a bell pepper inbred line susceptible to all PVY isolates, and HD285 is a reference doubled‐haploid line carrying the pvr23 resistance allele in a susceptible genetic background (Palloix et al., 2009).
Two PVY isolates showing differences in their pathogenicity towards pepper were used to construct recombinant viruses: SON41p and LYE84.2 for which infectious cDNA clones have been constructed previously (Moury et al., 2004). SON41p was first isolated from Solanum nigrum and further passaged in pepper, whereas LYE84.2 is a tomato isolate. Six recombinant cDNA clones, in which a genome region of SON41p was substituted by the homologous region of LYE84.2, were constructed by homologous recombination in Saccharomyces cerevisiae (strain YPH501). For this, a vector corresponding to the SON41p cDNA clone deleted from the region of interest was recombined with a PCR product of the corresponding region of LYE84.2. In order to avoid potentially deleterious effects as a result of hybrid proteins, recombination points were chosen at the start and end of PVY proteins (Fig. 1). It should be noted that the segment spanning from 6K2 to the NIa‐proteinase coding regions was not used for recombination because it includes the VPg coding region that determines the breakdown of the resistance allele pvr23 (Ayme et al., 2006; Moury et al., 2004), and differs largely between SON41p and LYE84.2.
Measurement of the resistance breakdown capacity
Inoculations were carried out under insect‐proof glasshouse conditions. Primary inoculations with the cDNA clones SON41p, LYE84.2 and the six chimeric cDNAs were made by DNA‐coated tungsten particle bombardments of Nicotiana clevelandii or N. benthamiana juvenile plants (4 weeks old), as direct bombardment of pepper plants is not efficient (Moury et al., 2004). Crude extracts from the infected Nicotiana spp. plants were calibrated and adjusted by dilution using semiquantitative DAS‐ELISA, as shown by Ayme et al. (2006). For each virus, HD285 pepper plants were mechanically inoculated on their two cotyledons approximately 3 weeks after sowing (first‐leaf stage). YW plants were used as an inoculation control. This experiment was performed twice independently. For each test, 100 HD285 plants were inoculated by each virus. In a third experiment, the CI chimera and the parental clone SON41p were compared. Four viral populations [two independently prepared SON41p inocula (SON‐1 and SON‐2) and two independently prepared inocula of the CI chimera (CI‐1 and CI‐2) amplified in Nicotiana spp. plants] were manually inoculated to 435 HD285 pepper plants and to susceptible YW in a randomized experimental design. PVY infection in HD285, indicative of the breakdown of pvr23 resistance (Ayme et al., 2006 and the Results section), was checked by the presence of necrotic symptoms in apical noninoculated leaves, 5 weeks after inoculation, and by DAS‐ELISA (Moury et al., 2004). Fisher's exact test, with Bonferroni correction when necessary, was used to compare the breakdown frequencies between the different viruses.
Sequence analysis of resistance‐breaking PVY populations
For experiment 3, total RNAs of infected HD285 plants were purified from pools of three leaves per plant with the Tri Reagent Kit (Molecular Research Center Inc., Cincinnati, OH, USA) and used to amplify the entire VPg cistron by RT‐PCR with Avian myeloblastosis virus reverse transcriptase (Promega Corp., Madison, WI, USA), Taq DNA polymerase (Promega) and the same primers as described in Moury et al. (2004). The consensus sequence of the VPg cistron of the PVY populations corresponding to each infected plant was determined by classical Sanger sequencing (MWG, Ebersberg, Germany).
The entire CI cistron of 14 PVY populations, corresponding to the CI‐1 and CI‐2 inocula, 10 randomly chosen HD285 plants infected after inoculation with these two inocula, and two HD285 plants infected by PVY populations that did not show any mutation in the VPg cistron (see the Results section) was amplified by RT‐PCR with primers PVY‐CI (5′‐GATGCTGAAAGGAGTGATTG‐3′; nucleotide positions 3570–3589) and PVY‐CI‐REV (5′‐TTTCCCTTGGTGATGCACAG‐3′; nucleotide positions 5701–5720). The CI cistron of these 14 samples was sequenced using primers PVY‐CI, PVY‐CI‐REV and CI‐LYE (5′‐GCTAAAAGTTTCAGCCACTCCAGTGGGAAGGGAAGTTG‐3′; nucleotide positions 4253–4290). All sequences were aligned and compared with that of SON41p (accession number AJ439544) for the VPg cistron and of LYE84.2 (accession number AJ439545) for the CI cistron, using the software BioEdit (Hall, 1999) to identify substitutions potentially involved in resistance breakdown.
The distributions of pvr23 resistance‐breaking mutations observed with SON41p and the CI chimera were compared by Monte Carlo simulations as described in Ayme et al. (2006). The resistance‐breaking mutations observed with the CI chimera were chosen as the theoretical distribution and 10 000 Monte Carlo simulations of each of 29 random draws (i.e. the number of resistance breakdowns observed with SON41p; Table 2) were performed with R software version 2.9.2 (R Development Core Team, 2009) according to this theoretical distribution of mutants. The upper and lower frequencies of each mutant containing 95% of the simulations were then computed.
High‐throughput sequencing of the central part of the VPg cistron (nucleotide positions 5930–6168) was performed for 15 PVY populations corresponding to the four inocula SON‐1, SON‐2, CI‐1 and CI‐2, one PVY population from YW per inoculum and one (for CI‐2) or two (for SON‐1, SON‐2 and CI‐1) populations from HD285 per inoculum (Table 2), using the parallel‐tagged sequencing (PTS) method described by Meyer et al. (2008). PTS is a molecular barcoding method designed to adapt the recently developed high‐throughput 454 parallel sequencing technology for use with multiple samples, including pools of PCR products. PCR amplification of the VPg cistron was performed with primers PYRO‐FOR (5′‐ATTCATCCAATTCGTTGATCC‐3′; nucleotide positions 5930–5950) and PYRO‐REV (5′‐TGTCACAAACCTTAAGTGGG‐3′; nucleotide positions 6149–6168). PTS, emulsion PCR and sequencing were realized by GATC‐Biotech (Konstanz, Germany).
We received between 1756 and 2621 correctly assigned sequences per viral population, corresponding to a total of 33 254 sequences. Assembly was performed using default parameters of the software SeqMan (DNASTAR Lasergene, Madison, WI, USA), and an SNP report was built for each sample. SNP reports indicate, for each nucleotide position in the reference sequence (i.e. the sequence of the VPg cistron of SON41p), the frequency of each SNP, including insertions and deletions. Nucleotide substitutions are less likely in this technology than with traditional methods as the triphosphate nucleotides are flowed one at a time. However, indels are the most frequent 454 pyrosequencing errors (Huse et al., 2007). We then exported the aligned sequences and employed a program developed using R software to remove insertions and deletions and then to calculate haplotype frequencies. Because the program removed short sequences, the total number of cleaned sequences was reduced to 30 324, ranging from 1577 to 2407 per viral population.
For resistance breakdowns resulting from each inoculum, the nucleotide substitutions identified by classical sequencing in the VPg cistron, and either proven (Ayme et al., 2006) or suspected to be directly involved in resistance breakdown (Table 2), were searched for among mutations indicated by the SNP report of the inoculum. Subsequently, the relationship between the frequency of each of these nucleotide substitutions in each inoculum and the frequency of resistance breakdowns involving the same nucleotide substitution and corresponding to the same inoculum was tested with Pearson's product‐moment correlation using R software.
The mutation frequency and normalized Shannon index were used to characterize the mutant spectrum of each viral population (Lozano et al., 2009). Mutation frequencies were calculated by scoring the different types of mutation (i.e. repeated mutations were counted only once) relative to the reference sequence divided by the total number of nucleotides sequenced, and the normalized Shannon index was calculated as described by Montarry et al. (2006). The cleaned sequences were converted into codon format, using an R script developed by the authors, and analysed using the software quasi (Stewart et al., 2001). The codon‐based method used by quasi is advantageous over other methods with regard to the ratio between nonsynonymous and synonymous substitution rates because it is independent of the underlying phylogeny. This lower information level must be compensated for by a large number of sequences, making high‐throughput sequencing data ideal for this method. Positive selection (PS) sites were recorded along the consensus sequence. The frequencies of PS sites in each population were calculated as the percentage of the number of PS sites relative to the total number of amino acids in the consensus sequence. Statistical comparisons between SON41p and the CI chimera for the different variables were realized by a Kruskal–Wallis test using R software.
Estimation of virus competitiveness
The competitiveness of the mutant carrying the guanosine to adenosine substitution at position 6069 of SON41p (corresponding to the aspartic acid to asparagine substitution at position 119 of VPg, i.e. substitution 119N), named mutant SON41p‐119N, was compared with that of the mutant of the CI chimera carrying the same substitution, named CI‐119N, in HD285. The 119N substitution has been shown previously to confer resistance‐breaking properties towards the pvr23 allele present in HD285 (Ayme et al., 2006). SON41p‐119N and CI‐119N inocula were calibrated using semiquantitative DAS‐ELISA, as in Ayme et al. (2006), and mixed at two ratios (1:1 or 4:1) which were used to inoculate two independent sets of 50 HD285 plants per ratio, one set being analysed at 15 dpi and the other at 30 dpi. After inoculation, plants were grown in a growth chamber at 22–24 °C (night/day) and a 14‐h photoperiod. At 15 and 30 dpi, the presence of the two PVY variants in the viral populations was analysed using the cleaved amplified polymorphic sequence (CAPS) method (e.g. Montarry et al., 2009). For each HD285 plant, total RNAs were extracted from pools of three infected leaves and used to amplify the entire CI cistron as described above. In this region, two SNPs allowed SON41p to be distinguished from LYE84.2 and, consequently, SON41p‐119N from CI‐119N. A ClaI site (nucleotide positions 4961–4966) is present in CI‐119N, but not in SON41p‐119N, and an MscI site (nucleotide positions 4447–4452) is present in SON41p‐119N, but not in CI‐119N. For each infected plant, we determined whether one or the two viruses were present in the viral population after CAPS analysis and 1% agarose gel electrophoresis. Fisher's exact test, coupled with a Bonferroni correction, was used to compare the relative proportions of SON41p‐119N and CI‐119N in the inocula and the numbers of HD285 plants in which SON41p‐119N or CI‐119N only were detected by CAPS analysis.
ACKNOWLEDGEMENTS
This study was supported by an AIP Bio‐Resources from the Institut National de la Recherche Agronomique (INRA). We gratefully acknowledge Isabelle Bornard for electron microscopy observations, Mireille Jacquemond, Frédéric Fabre and Alain Palloix for useful discussions of the present work, and Mark Tepfer for raising our awareness of the high‐throughput sequencing technology. Josselin Montarry was the recipient of an INRA postdoctoral fellowship.
REFERENCES
- Abdul‐Razzak, A. , Guiraud, T. , Peypelut, M. , Walter, J. , Houvenaghel, M.C. , Candresse, T. , Le Gall, O. and German‐Retana, S. (2009) Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E‐mediated resistance against Lettuce mosaic potyvirus . Mol. Plant Pathol. 10, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadoeuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of Potato virus Y confer virulence on the pvr23 resistance in pepper. Mol. Plant–Microbe Interact. 19, 557–563. [DOI] [PubMed] [Google Scholar]
- Ayme, V. , Petit‐Pierre, J. , Souche, S. , Palloix, A. and Moury, B. (2007) Molecular dissection of the Potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. [DOI] [PubMed] [Google Scholar]
- Betancourt, M. , Fereres, A. , Fraile, A. and García‐Arenal, F. (2008) Estimation of the effective number of founders that initiate an infection after aphid transmission of a multipartite plant virus. J. Virol. 82, 12416–12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caranta, C. , Lefebvre, V. and Palloix, A. (1997) Polygenic resistance of pepper to potyviruses consists of a combination of isolate‐specific and broad‐spectrum quantitative trait loci. Mol. Plant–Microbe Interact. 10, 872–878. [Google Scholar]
- Carrington, J.C. , Jensen, P.E. and Schaad, M.C. (1998) Genetic evidence for an essential role for potyvirus CI protein in cell‐to‐cell movement. Plant J. 14, 393–400. [DOI] [PubMed] [Google Scholar]
- Charron, C. , Nicolaï, M. , Gallois, J.L. , Robaglia, C. , Moury, B. , Palloix, A. and Caranta, C. (2008) Natural variation and functional analyses provide evidence for co‐evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. [DOI] [PubMed] [Google Scholar]
- Domingo, E. , Martin, V. , Perales, C. , Grande‐Pérez, A. , Garcia‐Arriaza, J. and Arias, A. (2006) Viruses as quasispecies: biological implications. Curr. Top. Microbiol. Immunol. 299, 51–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J.W. and Holland, J.J. (1999) Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA, 96, 13910–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S.F. and Sanjuan, R. (2007) Virus evolution: insights from an experimental approach. Annu. Rev. Ecol. Syst. 38, 27–52. [Google Scholar]
- Fernandez, A. , Lain, S. and Garcia, J.A. (1995) RNA helicase activity of the plum pox potyvirus CI protein expressed in Escherichia‐coli—mapping of an RNA‐binding domain. Nucleic Acid Res. 23, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, A. , Guo, H.S. , Saenz, P. , SimonBuela, L. , deCedron, M.G. and Garcia, J.A. (1997) The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acid Res. 25, 4474–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, R. and Stenger, D.C. (2003) Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu. Rev. Phytopathol. 41, 199–214. [DOI] [PubMed] [Google Scholar]
- Froissart, R. , Roze, D. , Uzest, M. , Galibert, L. , Blanc, S. and Michalakis, Y. (2005) Recombination every day: abundant recombination in a virus during a single multi‐cellular host infection. PLoS Biol. 3, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Arenal, F. and McDonald, B.A. (2003) An analysis of the durability of resistance to plant viruses. Phytopathology, 93, 941–952. [DOI] [PubMed] [Google Scholar]
- García‐Arenal, F. , Fraile, A. and Malpica, J.M. (2001) Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39, 157–186. [DOI] [PubMed] [Google Scholar]
- Glais, L. , Tribodet, M. and Kerlan, C. (2002) Genomic variability in Potato potyvirus Y (PVY): evidence that (PVYW)‐W‐N and PVYNTN variants are single to multiple recombinants between PVYO and PVYN isolates. Arch. Virol. 147, 363–378. [DOI] [PubMed] [Google Scholar]
- Gómez de Cedrón, M. , Osaba, L. , López, L. and Garcia, J.A. (2006) Genetic analysis of the function of the plum pox virus CI RNA helicase in virus movement. Virus Res. 116, 136–145. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Harrison, B.D. (2002) Virus variation in relation to resistance breaking in plants. Euphytica, 124, 181–192. [Google Scholar]
- Hu, X.J. , Meacham, T. , Ewing, L. , Gray, S.M. and Karasev, A.V. (2009a) A novel recombinant strain of Potato virus Y suggests a new viral genetic determinant of vein necrosis in tobacco. Virus Res. 143, 68–76. [DOI] [PubMed] [Google Scholar]
- Hu, X.X. , He, C.Z. , Xiao, Y. , Xiong, X.G. and Nie, X.Z. (2009b) Molecular characterization and detection of recombinant isolates of Potato virus Y from China. Arch. Virol. 154, 1303–1312. [DOI] [PubMed] [Google Scholar]
- Huse, S.M. , Huber, J.A. , Morrison, H.G. , Sogin, M.L. and Mark Welch, D. (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8, R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzac, B. , Fabre, F. , Palloix, A. and Moury, B. (2009) Constraints on evolution of virus avirulence factors predict the durability of corresponding plant resistances. Mol. Plant Pathol. 10, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner, C.E. , Sanchez, F. , Nettleship, S.B. , Foster, G.D. , Ponz, F. and Walsh, J.A. (2000) The cylindrical inclusion gene of Turnip mosaic virus encodes a pathogenic determinant to the Brassica resistance gene TuRB01 . Mol. Plant–Microbe Interact. 13, 1102–1108. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Tomimura, K. , Ohshima, K. , Hughes, S.L. and Walsh, J.A. (2002) Mutations in Turnip mosaic virus P3 and cylindrical inclusion proteins are separately required to overcome two Brassica napus resistance genes. Virology, 300, 50–59. [DOI] [PubMed] [Google Scholar]
- Johnson, R. (1981) Durable resistance: definition of genetic control and attainment in plant breeding. Phytopathology, 71, 567–568. [Google Scholar]
- Johnson, R. (1984) A critical analysis of durable resistance. Annu. Rev. Phytopathol. 22, 309–330. [Google Scholar]
- Kang, B.C. , Yeam, I. and Jahn, M.M. (2005) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Klein, P.G. , Klein, R.R. , Rodriguez‐Cerezo, E. , Hunt, A.G. and Shaw, J.G. (1994) Mutational analysis of the Tobacco vein mottling virus genome. Virology, 204, 759–769. [DOI] [PubMed] [Google Scholar]
- Lain, S. , Riechmann, J.L. and Garcia, J.A. (1990) RNA helicase—a novel activity associated with a protein encoded by a positive strand RNA virus. Nucleic Acid Res. 18, 7003–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano, G. , Grande‐Perez, A. and Navas‐Castillo, J. (2009) Populations of genomic RNAs devoted to the replication or spread of a bipartite plant virus differ in genetic structure. J. Virol. 83, 12973–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, M. , Stenzel, U. and Hofreiter, M. (2008) Parallel tagged sequencing on the 454 platform. Nat. Protoc. 3, 267–278. [DOI] [PubMed] [Google Scholar]
- Montarry, J. , Corbière, R. , Lesueur, S. , Glais, I. and Andrivon, D. (2006) Does selection by resistant hosts trigger local adaptation in plant–pathogen systems? J. Evol. Biol. 19, 522–531. [DOI] [PubMed] [Google Scholar]
- Montarry, J. , Cartolaro, P. , Richard‐Cervera, S. and Delmotte, F. (2009) Spatio‐temporal distribution of Erysiphe necator genetic groups and their relationship with disease levels in vineyards. Eur. J. Plant Pathol. 123, 61–70. [Google Scholar]
- Moury, B. (2010) A new lineage sheds light on the evolutionary history of Potato virus Y . Mol. Plant Pathol. 11, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. and Jacquemond, M. (2002) Evidence for diversifying selection in Potato virus Y and in the coat protein of other potyviruses. J. Gen. Virol. 83, 2563–2573. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. , Caranta, C. , Palloix, A. and Jacquemond, M. (2004) Mutations in Potato virus Y genome‐linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant–Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Fabre, F. and Senoussi, R. (2007) Estimation of the number of virus particles transmitted by an insect vector. Proc. Natl. Acad. Sci. USA, 104, 17891–17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerke, E.C. and Dehne, H.W. (1997) Global crop production and the efficacy of crop protection—current situation and future trends. Eur. J. Plant Pathol. 103, 203–215. [Google Scholar]
- Ohshima, K. , Akaishi, S. , Kajiyama, H. , Koga, R. and Gibbs, A.J. (2010) The evolutionary trajectory of turnip mosaic virus populations adapting to a new host. J. Gen. Virol. 91, 788–801. [DOI] [PubMed] [Google Scholar]
- Palloix, A. , Ayme, V. and Moury, B. (2009) Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 183, 190–199. [DOI] [PubMed] [Google Scholar]
- Pigliucci, M. (2008) Opinion—is evolvability evolvable? Nat. Rev. Genet. 9, 75–82. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2009) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; ISBN 3‐900051‐07‐0. Available at http://www.R‐project.org/. [Google Scholar]
- Raccah, B. (1986) Nonpersistent viruses—epidemiology and control. Adv. Virus Res. 31, 387–429. [DOI] [PubMed] [Google Scholar]
- Revers, F. , Le Gall, O. , Candresse, T. , Le Romancer, M. and Dunez, J. (1996) Frequent occurrence of recombinant potyvirus isolates. J. Gen. Virol. 77, 1953–1965. [DOI] [PubMed] [Google Scholar]
- Roberts, I.M. , Wang, D. , Findlay, K. and Maule, A.J. (1998) Ultrastructural and temporal observations of the potyvirus cylindrical inclusions (CIs) show that the CI protein acts transiently in aiding virus movement. Virology, 245, 173–181. [DOI] [PubMed] [Google Scholar]
- Sacristan, S. , Malpica, J.M. , Fraile, A. and García‐Arenal, F. (2003) Estimation of population bottlenecks during movement of Tobacco mosaic virus in tobacco plants. J. Virol. 77, 9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.K. , Lee, S.H. and Kim, K.H. (2009) Strain‐specific cylindrical inclusion protein of Soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant–Microbe Interact. 22, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Stewart, J.J. , Watts, P. and Litwin, S. (2001) An algorithm for mapping positively selected members of quasispecies‐type viruses. BMC Bioinformatics, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Zhang, C. , Hong, J. , Xiong, R. , Kasschau, K.D. , Zhou, X. , Carrington, J.C. and Wang, A. (2010) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by viral protein P3N‐PIPO. PLoS Pathog. 6, e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. (1932) Evolution in Mendelian populations. Genetics, 16, 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, J. , Mundt, C.C. , Hoffer, M.E. and McDonald, B.A. (2002) Local adaptation and effect of host genotype on the rate of pathogen evolution: an experimental test in a plant pathosystem. J. Evol. Biol. 15, 634–647. [Google Scholar]


