SUMMARY
Xanthomonas citri ssp. citri (Xcc) causes citrus canker, one of the most economically damaging diseases affecting citrus worldwide. Biofilm formation is important for the pathogen to survive epiphytically in planta prior to the induction of canker symptoms. In this study, two EZ‐Tn5 transposon mutants of Xcc strain 306, affected in biofilm formation, were isolated; subsequent analyses led to the identification of a novel gene locus XAC3596 (designated as wxacO), encoding a putative transmembrane protein, and the rfbC gene, encoding a truncated O‐antigen biosynthesis protein. Sodium dodecylsulphate‐polyacrylamide gel electrophoresis revealed that lipopolysaccharide (LPS) biosynthesis was affected in both wxacO and rfbC mutants. The wxacO mutant was impaired in the formation of a structured biofilm on glass or host plant leaves, as shown in confocal laser scanning microscopy analysis of strains containing a plasmid expressing the green fluorescent protein. Both wxacO and rfbC mutants were more sensitive than the wild‐type strain to different environmental stresses, and more susceptible to the antimicrobial peptide polymyxin B. The two mutants were attenuated in swimming motility, but not in flagellar formation. The mutants also showed reduced virulence and decreased growth on host leaves when spray inoculated. The affected phenotypes of the wxacO and rfbC mutants were complemented to wild‐type levels by the intact wxacO and rfbC genes, respectively. This report identifies a new gene influencing LPS production by Xcc. In addition, our results suggest that a structurally intact LPS is critical for survival in the phyllosphere and for the virulence of Xcc.
INTRODUCTION
Citrus canker, caused by the bacterial pathogen Xanthomonas citri ssp. citri (Xcc) (synonyms: Xanthomonas citri, X. campestris pv. citri and X. axonopodis pv. citri) strain A (Cubero and Graham, 2002; Schaad et al., 2006; Vauterin et al., 1995), is an economically important disease that can cause extensive damage to most commercial citrus cultivars in subtropical citrus‐producing regions of the world (Gottwald et al., 2002; Graham et al., 2004). Xcc is spread by wind‐blown rain and invades the host directly through natural openings, such as stomata, and through wounds (Gottwald et al., 2002). Canker symptoms, characterized by raised necrotic lesions, appear in leaves, stems and fruits on severely affected trees, inducing defoliation, twig dieback, general tree decline, blemished fruit and premature fruit drop. Recurrent and severe attacks of the disease are responsible for serious economic losses in groves (Graham et al., 2004). Copper‐based bactericides are currently the most effective management approach to control citrus canker, especially in preventing infection of fruit (Cooksey, 1990).
Biofilms are known to protect bacteria from environmental stresses, host defence mechanisms and antimicrobial compounds (Karatan and Watnick, 2009). In addition, an association between biofilm formation and virulence has been reported for a variety of bacterial infections in plants (Danhorn and Fuqua, 2007). In the case of Xcc, the formation of biofilms on host leaves plays an important role in the epiphytic survival of this pathogen prior to the development of canker disease (Rigano et al., 2007). The characterization of the mechanisms involved in biofilm formation holds promise for the control of biofilm‐associated bacterial infections.
Lipopolysaccharide (LPS) is one of the major polysaccharide components on the cell surface of Gram‐negative bacteria (Vorholter et al., 2001). LPS generally contains three structural regions: lipid A that anchors LPS into the outer leaflet of the outer membrane; the core oligosaccharide, which is a short oligosaccharide attached to lipid A; and the O‐antigen, a distal polysaccharide (Raetz and Whitfield, 2002). It has been well documented that LPS is important for the interaction of bacteria with their surrounding environment and eukaryotic hosts (Raetz and Whitfield, 2002). LPS has been implicated in biofilm formation, and may serve as an important virulence factor in various animal and human pathogenic bacteria, such as Pseudomonas aeruginosa (Lau et al., 2009) and Salmonella enterica (Yethon et al., 2000). In plant–pathogen interactions, several roles have been proposed for LPS. It may act as a virulence factor and can be a protective barrier against plant antimicrobials (Newman et al., 2001). LPS is also being increasingly recognized as a major pathogen‐associated molecular pattern (PAMP) for plants, inducing plant defence responses including the production of phenolic compounds and the oxidative burst and the activation of nitric oxide synthase (NOS) (Newman et al., 2007). LPS has also been reported to suppress the hypersensitive reaction (HR) (Newman et al., 2007).
A role for LPS during biofilm formation and/or the development of different plant‐associated bacteria has been suggested by studies performed on Azospirillum brasilense, Pseudomonas fluorescens and Rhizobium leguminosarum. A mutant of A. brasilense SP7 defective in the synthesis of complete LPS showed impaired biofilm formation, and the same was reported for an R. leguminosarum mutant with an altered LPS structure (Lerner et al., 2009; Vanderlinde et al., 2009). However, the two mutants showed pleiotropic effects that could account for impaired biofilm formation. Although the Azospirillum mutant was affected in the monosaccharide composition of the exopolysaccharide (EPS), the R. leguminosarum mutant showed a lack of flagella and was nonmotile. Nevertheless, an LPS expression‐reduced mutant of P. fluorescens SBW25 did not show pleiotropic effects, but still exhibited a compromised ability to maintain the biofilm architecture, pointing to a direct role of LPS in biofilm development (Spiers and Rainey, 2005). For plant pathogenic bacteria of Xanthomonas spp., evidence is accumulating that LPS is involved in interactions with host and nonhost plants in X. campestris pv. campestris (Braun et al., 2005; Dow et al., 1995), X. campestris pv. citrumelo (Kingsley et al., 1993) and X. oryzae pv. oryzae (Wang et al., 2008a). However, it is unclear how LPS is involved in biofilm formation for the Xanthomonas genus.
The complete genome of Xcc strain 306 has been sequenced (da Silva et al., 2002) and has revealed a number of genes most probably involved in biofilm formation. Among them, researchers have so far characterized the role of the xanthan EPS biosynthetic genes gumB (Rigano et al., 2007) and gumD (Dunger et al., 2007), a haemagglutinin‐like protein encoding gene fhaB (Gottig et al., 2009) and the UTP‐glucose‐1‐phosphate uridylyltransferase (synonym: UDP‐glucose pyrophosphorylase) encoding gene galU (Guo et al., 2010) in biofilm formation. galU is required for the biosynthesis of EPS and capsular polysaccharide (CPS) (Guo et al., 2010). EPS may serve as a major component of the biofilm matrix (Rigano et al., 2007) to maintain the complex structure, and FhaB is necessary for surface attachment involved in biofilm formation (Gottig et al., 2009). However, the roles of genes putatively associated with LPS production in biofilm formation have not been investigated.
In this study, we characterized one novel gene locus XAC3596 (designated as wxacO) and the rfbC (XAC3598) gene of Xcc strain 306. The wxacO gene is predicted to encode a putative transmembrane protein and rfbC to encode a truncated O‐antigen biosynthesis protein. We presented evidence that both wxacO and rfbC are involved in the biosynthesis of LPS, and detailed the involvement of LPS in biofilm formation and bacterial ecological competence, as well as the virulence of the citrus canker pathogen.
RESULTS
Disruption of wxacO or rfbC affected biofilm formation
Two mutants, 247C8 (wxacO) and 236E9 (rfbC) (Table 1), which exhibited reduced ability to form biofilms in the polystyrene 96‐well plate assays with crystal violet (CV) staining (data not shown), were selected from the EZ‐Tn5 library of Xcc strain 306 (Guo et al., 2010). No difference was observed between the mutant and wild‐type strains in colony morphology on nutrient agar (NA) plates or growth dynamics in nutrient broth (NB) (data not shown). However, in the glass tube CV staining assays, approximately 4.4‐fold and 2.8‐fold reductions in biofilm formation (absorbance at 590 nm) were observed in 247C8 (wxacO) and 236E9 (rfbC) compared with the wild‐type strain 306 (Fig. 1).
Table 1.
Bacterial strains and plasmids.*
| Strains and plasmids | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F‐ recA1 endA1 hsdR17 supE44 thi‐1 gyrA96 relA1Δ (argE‐lacZYA)169ϕ80lazAΔ M15 | Hanahan (1983) |
| JM109 | recA1, endA1, gyrA96, thi, hsdR17 (rK‐,mK+), relA1, supE44, Δ(lac‐proAB), [F′, traD36, proAB, lacIqZΔM15] | Promega |
| HB101 | F‐ supE44, hsdS20(rB‐ mB‐), recA13, ara‐14, proA2, lacY1, galK2, rpsL20, xyl‐5, mtl‐1, leuB6, thi | Boyer and Roulland‐Dussoix (1969) |
| Xanthomonas citri ssp. citri | ||
| 306 | Rfr, cause citrus canker on citrus | Rybak et al. (2009) |
| 247C8 (wxacO) | wxacO (XAC3596): Tn5 derivative of strain 306, Rfr, Kmr | This study |
| 236E9 (rfbC) | rfbC (XAC3598): Tn5 derivative of strain 306, Rfr, Kmr | This study |
| 247C8V | 247C8 (wxacO) containing pUFR053, Kmr, Cmr, Gmr | This study |
| 236E9V | 236E9 (rfbC) containing pUFR053, Kmr, Cmr, Gmr | This study |
| C247C8 | 247C8 (wxacO) containing pJU3596, Kmr, Cmr, Gmr | This study |
| C236E9 | 236E9 (rfbC) containing pJU3598, Kmr, Cmr, Gmr | This study |
| Plasmids | ||
| pGEM®‐T Easy | PCR cloning and sequencing vector, lacZ', Apr | Promega |
| pRK2013 | ColE1 Kmr Tra+, conjugation helper plasmid | Figurski and Helinski (1979) |
| pUFR053 | IncW Mob+ mob(P) lacZα+ Par+, Cmr, Gmr, Kmr, shuttle vector | El Yacoubi et al. (2007) |
| pUFZ75 | Constitutive GFP expression vector; trp promoter cloned upstream of the GFP cassette; Kmr | Zhang et al. (2009) |
| pGE3596 | wxacO (XAC3596) gene from Xcc 306 cloned in pGEM®‐T‐Easy | This study |
| pGE3598 | rfbC (XAC3598) gene from Xcc 306 cloned in pGEM®‐T‐Easy | This study |
| pJU3596 | wxacO (XAC3596) gene on HindIII fragment from pGE3596 cloned in pUFR053 | This study |
| pJU3598 | rfbC (XAC3598) gene on HindIII fragment from pGE3598 cloned in pUFR053 | This study |
Apr, Cmr, Kmr, Gmr, Rfr and Tcr indicate resistance to ampicillin, chloromycetin, kanamycin, gentamicin, rifamycin and tetracycline, respectively.
GFP, green fluorescent protein; PCR, polymerase chain reaction.
Figure 1.
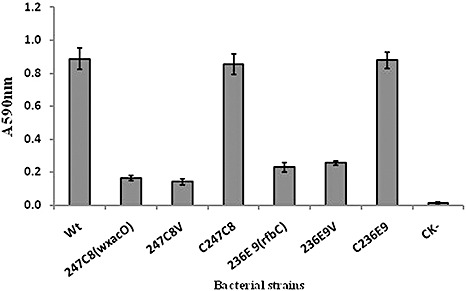
Ability of Xanthomonas citri ssp. citri strains to form biofilms in borosilicate glass tubes as determined using crystal violet staining. The experiment was repeated three times with five replicates each time. Averages and standard errors from one experiment of three with similar results are shown. Wt, X. citri ssp. citri wild‐type strain 306; 247C8(wxacO), wxacO mutant; 247C8V, 247C8 (wxacO) complemented with empty vector pUFR053 without the wxacO gene; C247C8, strain 247C8 (wxacO) complemented with pJU3596; 236E9(rfbC), rfbC mutant; 236E9V, 236E9 (rfbC) complemented with empty vector pUFR053 without the rfbC gene; C236E9, strain 236E9 (rfbC) complemented with pJU3598; CK‐, nutrient broth (NB) medium without inoculation of bacteria. A 590 nm, absorbance at 590 nm.
Random amplification of transposon ends‐polymerase chain reaction (RATE‐PCR) (Ducey and Dyer, 2002) was used to map transposon insertion sites in 247C8 (wxacO) and 236E9 (rfbC). A 0.5‐kb EZ‐Tn5‐flanking DNA fragment from 247C8 (wxacO) and a 0.6‐kb fragment from 236E9 (rfbC) were sequenced with the KAN‐2 FP‐1 primer (Table 2). A homology search of the GenBank database revealed that the 247C8 (wxacO) mutant had an insertion in the XAC3596 gene (166 bp downstream of the translation start site) of Xcc strain 306 (GenBank accession no. AE008923), and the transposon insertion site of the 236E9 (rfbC) mutant was mapped to 1117 bp downstream of the translation start site of the rfbC (XAC3598) gene (Fig. 2A). To verify the accuracy of transposon insertions in the sequence, PCR amplification was performed using primers (X96‐F/R or X98‐F/R) designed from the sequences flanking the EZ‐Tn5 sites in XAC3596 or the rfbC (XAC3598) gene (Table 2). The size of the PCR product from the 247C8 (wxacO) or 236E9 (rfbC) mutant was increased by insertion of a transposon (1.9 kb), indicating that the EZ‐Tn5 transposon was inserted in XAC3596 or the rfbC (XAC3598) gene of the Xcc strain 306 genome (data not shown). Analysis of the genome sequence showed that XAC3596 and the rfbC (XAC3598) gene were in a gene cluster of 15 genes putatively associated with the synthesis and transportation of LPS in Xcc strain 306 (da Silva et al., 2002). XAC3596 was the last gene of the operon containing XAC3593‐3596, whereas the adjacent downstream gene XAC3597 was transcribed separately from these four in reverse orientation (Caspi et al., 2010). The rfbC (XAC3598) gene consists of a single transcriptional unit (Fig. 2A). The affected biofilm formation by 247C8 (wxacO) or 236E9 (rfbC) could be restored fully to the wild‐type level by the introduction of the cloned wxacO and rfbC genes, respectively (Fig. 1). These results indicated that the transposon insertion in 247C8 (wxacO) or 236E9 (rfbC) was not a polar mutation, and the phenotypes of impaired biofilm formation in the two mutants resulted from the mutation of XAC3596 or the rfbC (XAC3598) gene, rather than from the malfunction of other genes in the cluster.
Table 2.
Primers used in this study.
| Primer | Sequence (5′ → 3′)* |
|---|---|
| Primers for cloning and sequencing Tn5‐flanking sequences | |
| Inv‐1 | ATGGCTCATAACACCCCTTGTATTA |
| Inv‐2 | GAACTTTTGCTGAGTTGAAGGATCA |
| KAN‐2 FP‐1 | ACCTACAACAAAGCTCTCATCAACC |
| KAN‐2 RP‐1 | CTACCCTGTGGAACACCTACATCT |
| Primers for amplification of DNA fragments of wxacO (XAC3596) and rfbC (XAC3598) | |
| X96‐F | GTGGGTGGAATTCTCACGTT |
| X96‐R | ACGATGCGATGTTCGTTGT |
| X98‐F | TATTGAGCGCAGACCAGTTG |
| X98‐R | TTGTCGATAACGGGTCCAAG |
| Primers for amplifying sequences used in complementation | |
| C96‐F | ACTGATaagcttGGTTCTACATGGCCGATACG |
| C96‐R | AACAGAaagcttTCACTGATGGGCTGTAGCTG |
| C98‐F | GCGACTaagcttTGCTGAGATTGACGGAACAG |
| C98‐R | GTCGGTaagattTGTGGATGCCACCAAAGTAA |
Lower case nucleotides are not exact matches to the sequence and were introduced to add the restriction enzyme site HindIII.
Figure 2.
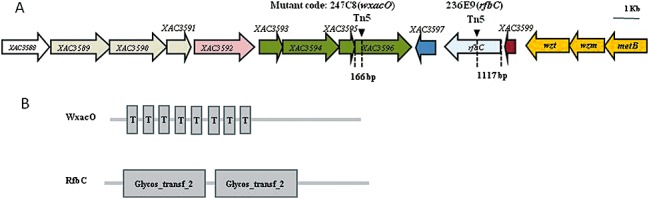
Sequence analysis of EZ‐Tn5 insertions in wxacO (XAC3596) and rfbC mutants. (A) Genetic organization of the putative lipopolysaccharide (LPS) gene cluster of Xanthomonas citri ssp. citri strain 306 and transposon insertion sites in the wxacO and rfbC mutants. The length of each arrow represents the relative open reading frame (ORF) size and indicates the direction of transcription. The triangles indicate the Tn5 insertion sites. Gene colour represents operon membership. The annotation information and sizes of the genes were obtained from the genome sequence database of Xcc strain 306 [National Center for Biotechnology Information (NCBI) Accession No: AE008923]. (B) Domain structure analyses of the putative WxacO and RfbC proteins. The domain structure prediction was performed using the smart program via the ExPASy Proteomics Server http://www.ca.expasy.org. Domain symbols: Glycos_transf_2, glycosyl transferase family 2 domain; T, transmembrane domain.
The protein encoded by XAC3596 was annotated as a 581‐amino‐acid hypothetical protein (da Silva et al., 2002), which has not been characterized previously. Domain structure analysis showed that XAC3596 contains a 21‐amino‐acid signal peptide and eight transmembrane helices located in the N‐terminal and central parts of the protein (Fig. 2B), and the prediction of protein localization sites suggested that the mature protein is located in the inner membrane based on PSORT analysis http://www.psort.org. A blastp search revealed that XAC3596 may be a transmembrane protein moderately conserved in other Xanthomonas species, including X. campestris pv. musacearum, X. oryzae pv. oryzae and X. oryzae pv. oryzicola, with over 30% amino acid identity (Table 3). The functions of XAC3596 homologues remain ambiguous, but XAC3596 was characterized to be involved in O‐antigen production in this study (see below and Fig. 3). Given this, and in accordance with the bacterial polysaccharide gene nomenclature (Reeves et al., 1996), XAC3596 was thus named as wxacO.
Table 3.
Comparison of the wxacO and rfbC genes of Xanthomonas citri ssp. citri strain 306 with genes of X. campestris pv. musacearum NCPPB4381*, X. oryzae pv. oryzae BXO8† and X. oryzae pv. oryzicola BLS256‡.
| Xcc 306 ORFs | Size of deduced product (aa) | Homologue | ||||
|---|---|---|---|---|---|---|
| Gene/locus_tag | Putative product | Size (aa) | Domain structure§ | Identity¶ (%) | ||
| wxacO | 581 | XcampmN_010100007915 | Hypothetical protein | 581 | Transmembrane segments (2); YhhN (1); Trep_Strep (1) | 97.4 |
| orf8 | Hypothetical protein | 521 | Transmembrane segments (10) | 44.5 | ||
| Xoryp_010100019000 | Transmembrane protein | 575 | Transmembrane segments (2); NnrS (1) | 30.1 | ||
| rfbC | 614 | XcampmN_010100007900 | Truncated O‐antigen biosynthesis protein | 566 | Glycos_transf_2 (2) | 98.0 |
| orf4 | O‐antigen biosynthesis protein | 611 | Glycos_transf_2 (2) | 66.5 | ||
According to sequence under accession number NZ_ACHT00000000.
According to sequence under accession number DQ907230.
According to sequence under accession number NZ_AAQN00000000.
Domains were predicted by the smart program via the ExPASy Proteomics Server http://ca.expasy.org. Domain symbols: Glycos_transf_2, glycosyl transferase family 2 domain; NnrS, haem‐ and copper‐containing membrane protein domain; Trep_Strep, hydrophobic protein domain; YhhN, possible transmembrane protein domain. The total number of domains is indicated in parentheses.
According to a blastp search.
aa, amino acid; ORF, open reading frame.
Figure 3.
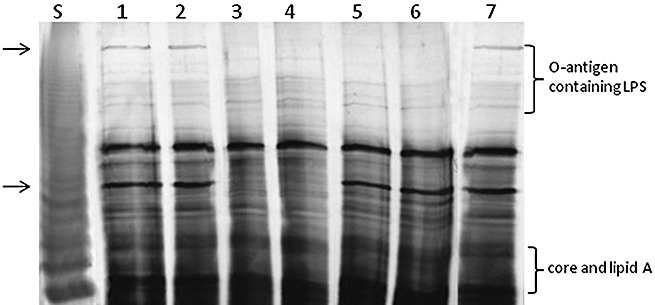
Sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of lipopolysaccharide (LPS) from proteinase K‐digested whole‐cell lysates of Xanthomonas citri ssp. citri strain 306 and its mutants carrying EZ‐Tn5 insertions in the wxacO and rfbC genes. Both wxacO and rfbC mutants were affected in the biosynthesis of O‐antigen. The wild‐type (Wt) LPS profile was restored in the complemented strains. S, LPS standard from Salmonella typhimurium (12.5 µg; Sigma). Lanes: 1, wild‐type strain 306; 2, C247C8 (wxacO mutant complemented with pJU3596); 3, 247C8 (wxacO) (wxacO mutant); 4: 247C8V (wxacO mutant complemented with empty vector pUFR053 without the wxacO gene); 5, 236E9 (rfbC) (rfbC mutant); 6, 236E9V (rfbC mutant complemented with empty vector pUFR053 without the rfbC gene); 7, C236E9 (rfbC mutant complemented with pJU3598). The gel was run in the SDS–glycine buffer system and silver stained using a silver stain kit (Bio‐Rad Laboratories, Inc.) following the manufacturer's instructions. The lost bands in the mutants are indicated by arrows.
The product of rfbC was annotated as a 614‐amino‐acid truncated O‐antigen biosynthesis protein that has a 24‐amino‐acid signal peptide and two separate glycosyl transferase family 2 (GT2) domains (Fig. 2B). The RfbC protein was highly conserved in the other two Xanthomonas spp. strains of X. campestris pv. musacearum NCPPB4381 and X. oryzae pv. oryzae BXO8, with 98% and 66% amino acid identity, respectively (Table 3). Their homologues with about 40%–48% identity were also present in Pseudomonas syringae, P. putida, Burkholderia sp., Rhizobium sp. and Acidovorax sp. (data not shown). The RfbC protein was similar to members of the subfamily of GT2 that is associated with O‐antigen biosynthesis in diverse bacteria (Wang et al., 2008b).
Detailed biofilm formation in vitro and in planta was studied in the wxacO mutant. The structural characteristics of the green fluorescent protein (GFP)‐labelled Xcc biofilm formed in vitro were analysed using confocal laser scanning microscopy (CLSM) over a 4‐day time course experiment in a culture dish with a glass bottom. At 3 days post‐inoculation (dpi), structured biofilm development with well‐established microcolonies and more complex bacterial aggregates was observed in the wild‐type strain (Fig. 4A). In contrast, a disorganized biofilm of uniform thickness composed of a confluent layer of cells and a few small microcolonies was observed in static cultures of the wxacO mutant over the course of the experiment (Fig. 4A). The results of in planta tests demonstrated that both the wild‐type strain 306 and the wxacO mutant were able to colonize grapefruit leaves and were distributed randomly on the leaves. The wild‐type strain formed large cell aggregates around the stomata at 2 dpi, which were visible as groups of fluorescent bacterial microcolonies (Fig. 4B). However, the wxacO mutant grew mainly as single cells and formed few small microcolonies on grapefruit leaves at the same time point (Fig. 4B).
Figure 4.
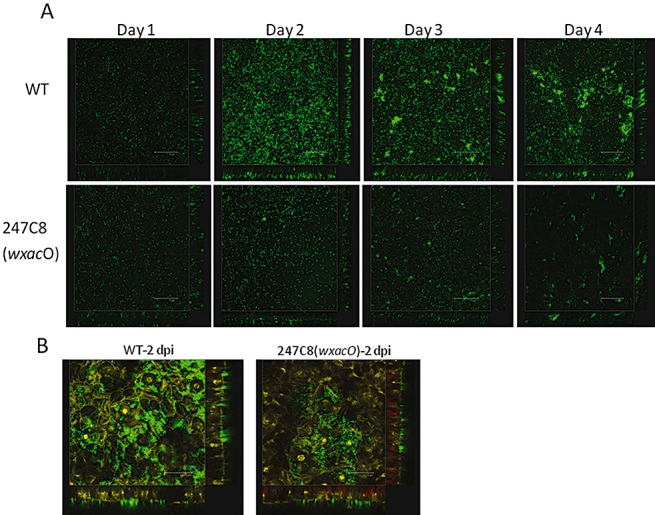
Confocal laser scanning microscopy (CLSM) of biofilms formed by wild‐type and wxacO mutant of Xanthomonas citri ssp. citri strain 306. (A) Green fluorescent protein (GFP)‐labelled cells grown on the glass bottoms of culture dishes were visualized at different stages of biofilm formation, 4 days post‐inoculation. The panels (left to right) show cell attachment to the glass surface. Scale bars, 75 µm. (B) Biofilm formation on grapefruit leaves. Bacterial suspensions were inoculated onto the abaxial leaf surface by spray. Scale bars, 34.78 µm. WT, wild‐type strain 306; 247C8 (wxacO), wxacO mutant.
LPSs of wxacO and rfbC mutants were altered
To determine whether wxacO or rfbC plays a role in LPS biosynthesis in Xcc strain 306, LPSs from the wild‐type, wxacO and rfbC mutants, and complemented wxacO and rfbC mutants, were separated by sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and compared with the well‐characterized LPS from Salmonella typhimurium (Fig. 3). The LPS of the wild‐type strain exhibited a ladder pattern that is characteristic of LPS with a different number of O‐repeat units in the O‐antigen, similar to that observed with Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia) (Huang et al., 2006). In the LPS profile of the wxacO mutant, one upper band, corresponding to the O‐antigen containing LPS, and another lower band, corresponding to an unknown component of LPS, were completely lost when compared with the wild‐type strain 306, whereas the rapidly migrating bands representing the core + lipid A structure were not affected, showing that the WxacO protein is involved in the biosynthesis and/or export of O‐antigen and other unknown components of LPS. In the LPS profile of the rfbC mutant, the upper band representing the O‐antigen containing LPS in the wild‐type was completely lost, but the other bands were unaffected, suggesting that this mutant contained LPS molecules having O‐antigens with reduced number of O‐repeat units or reduced chain length than that of the wild‐type. Therefore, it could be proposed that RfbC is involved in the biosynthesis of the LPS O‐antigen. The LPS phenotypes of the wxacO and rfbC mutants were restored to the level of the parental strains by in trans expression of wxacO and rfbC, respectively, driven by the lac promoter (Fig. 3). The empty vector pUFR053 did not complement LPS phenotypes in 247C8 (wxacO) and 236E9 (rfbC). No significant difference in EPS amount or capsule staining was observed between the two mutant strains and the wild‐type strain (data not shown).
Virulence and in planta growth of wxacO and rfbC mutants were reduced when spray inoculated
Plant inoculation by spray showed that both wxacO and rfbC mutants were reduced in virulence on grapefruit compared with the wild‐type strain 306 and the corresponding complemented mutants, as evidenced by decreased lesion numbers (Fig. 5A). Differences in lesion numbers were notable by 14 dpi and more distinctive over the remainder of the experiment. However, no detectable change in virulence compared with the wild‐type was observed in the two mutants in the pressure infiltration inoculation experiment. By day 10, in leaves subjected to pressure infiltration, the mutants showed typical canker lesions with a water‐soaking halo around, even at a lower inoculum concentration [106 colony‐forming units (cfu)/mL] (data not shown).
Figure 5.
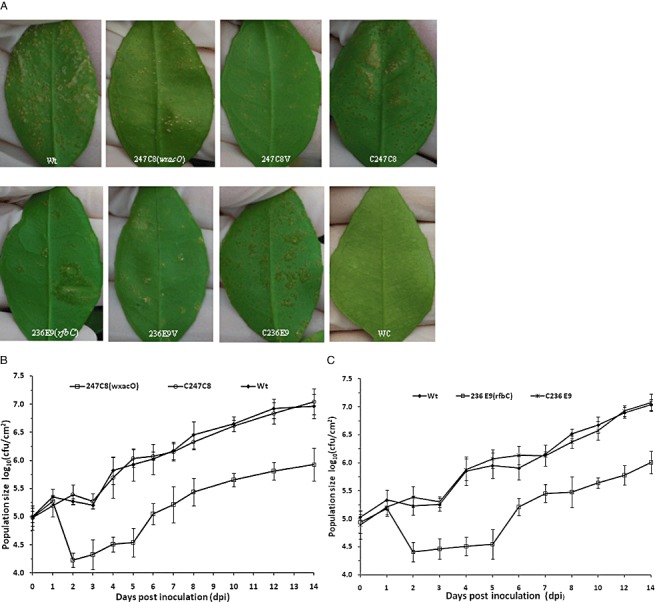
Virulence assays and growth of Xanthomonas citri ssp. citri strain 306 and derivative strains in planta. Bacterial strains were inoculated onto the abaxial leaf surfaces by spray. (A) Disease symptoms on grapefruit leaves inoculated with X. citri ssp. citri wild‐type strain 306 (Wt), wxacO mutant [247C8(wxacO)], 247C8 (wxacO) complemented with empty vector pUFR053 without the wxacO gene (247C8V), 247C8 (wxacO) complemented with pJU3596 (C247C8), rfbC mutant [236E9(rfbC)], 236E9 (rfbC) complemented with empty vector pUFR053 without the rfbC gene (236E9V), 236E9 (rfbC) complemented with pJU3598 (C236E9) and water control (WC). The inoculated leaves were photographed at 14 days after spraying. (B) Growth of X. citri ssp. citri wild‐type strain 306 (Wt), wxacO mutant [247C8 (wxacO)] and 247C8(wxacO) complemented with pJU3596 (C247C8) on grapefruit leaves. (C) Growth of X. citri ssp. citri wild‐type strain 306 (Wt), rfbC mutant [236E9 (rfbC)] and 236E9 (rfbC) complemented with pJU3598 (C236E9) on grapefruit leaves. Bacterial populations were determined by homogenizing leaf discs (0.8 cm in diameter) in 0.85% (w/v) NaCl, followed by dilution plating. The growth assays were repeated three times with similar results with three replicates each time. Data from one representative experiment are shown. Vertical bars represent the standard errors of the means.
The bacterial populations on the leaves of grapefruit were monitored for 14 days after spray. By 24 h after inoculation, the populations of the wild‐type strain and the wxacO and rfbC mutants had increased slightly, and no significant differences were observed between the wild‐type strain and the mutants (Fig. 5B,C). From that point onward, the population of the wild‐type strain remained approximately constant until 3 dpi, whereas the population of the wxacO or rfbC mutants declined significantly. At 3 dpi, the population of the wild‐type strain was nearly 10 times higher than that of the mutants (Fig. 5B,C). After 3 dpi, the populations of the wild‐type strain and the mutants began to increase slowly. Finally, the populations of the mutants were approximately 5–10‐fold lower than that of the wild‐type strain during 3–14 days after inoculation (Fig. 5B,C). The reduced growth of 247C8 (wxacO) and 236E9 (rfbC) was restored to wild‐type levels by complementation with the cloned wxacO and rfbC gene, respectively (Fig. 5B,C). No significant difference in growth between the wild‐type and mutant strains was observed after pressure infiltration (data not shown). The populations of the wild‐type and mutant strains continued to increase at a similar rate after inoculation, and increased by more than four orders of magnitude during the 10‐day monitoring period.
Mutation of wxacO or rfbC caused impairment in swimming motility, but no impact on flagellar formation
As alterations in LPS have been reported to affect flagellar formation and thus to result in impairment in swimming and/or swarming motility in a number of different bacteria (Huang et al., 2006), we aimed to determine whether the wxacO or rfbC gene mutation affected the flagellar formation and motility of Xcc. Transmission electron microscopy (TEM) analyses showed that both the wild‐type and the wxacO and rfbC mutant strains assembled a polar flagellum at the surface of the cell (data not shown), suggesting that the mutation of wxacO or rfbC had no impact on flagellar formation in Xcc. However, a significant reduction (P < 0.05) in swimming motility on 0.3% agar plates was observed in the wxacO and rfbC mutants compared with the wild‐type strain 306 (Table 4). The impaired swimming motility of 247C8 (wxacO) or 236E9 (rfbC) was restored to wild‐type levels following complementation with the cloned wxacO or rfbC gene, respectively,. In contrast, no significant difference in swarming motility was observed between the two mutants and the wild‐type strain 306 (Table 4).
Table 4.
Motility of wild‐type Xanthomonas citri ssp. citri strain 306 and derivative strains on nutrient agar (NA) medium.*
| Strain | Colony diameter (mm) | |
|---|---|---|
| Swimming motility | Swarming motility | |
| 306 | 38.5 ± 2.4 a | 12.6 ± 1.2 a |
| 247C8 (wxacO) | 22.1 ± 1.7 b | 10.8 ± 2.3 a |
| 236E9 (rfbC) | 24.3 ± 1.2 b | 11.5 ± 1.3 a |
| 247C8V | 21.6 ± 1.9 b | 10.7 ± 1.7 a |
| 236E9V | 23.2 ± 1.5 b | 10.2 ± 1.9 a |
| C247C8 | 37.8 ± 1.4 a | 12.4 ± 1.1 a |
| C236E9 | 40.7 ± 1.8 a | 11.6 ± 1.6 a |
Bacterial strains were inoculated to a central point on NA plates (0.3% agar in swimming assays and 0.7% agar for swarming assays), incubated at 28 °C for 72 h and the colony diameters were measured. The experiment was repeated three times with similar results. Means and standard errors of three replicates from one representative result are shown. Data with the same letters in the same column are not significantly different (P < 0.05).
Impact of wxacO or rfbC mutation on survival under environmental stress, and sensitivity to hydrogen peroxide and the antimicrobial peptide polymyxin B
The survival of the wxacO mutant 247C8 (wxacO) and the rfbC mutant 236E9 (rfbC) was tested under six environmental stresses, including UV radiation, heat shock, saline stress, high osmolarity, desiccation stress and SDS exposure. These experiments revealed that both 247C8 (wxacO) and 236E9 (rfbC) were more sensitive than the wild‐type strain to all stresses tested (Table 5). After 15 min of exposure to UV radiation, there were greater numbers of surviving cells of the wild‐type strain than of the two mutants. Following 5 min of exposure of bacteria to heat (54 °C), viable cells of the two mutants declined more rapidly than those of the wild‐type. When treated with 5% NaCl for 10 min, mutants showed significantly decreased survival compared with the wild‐type strain. After treatment with d‐sorbitol (40%) for 40 min, the survival rate of the wild‐type strain 306 was significantly higher than that of the two mutant strains. The two mutants also showed significantly lower survival rates than the wild‐type strain in response to desiccation (exposure to air and dried for 60 min). No viable cells of the two mutants were detected following treating with 0.1% SDS for 10 min. In addition, the two mutants exhibited higher sensitivity than the wild‐type strain to hydrogen peroxide (exposure to 0.03% H2O2 for 10 min).
Table 5.
Sensitivity of wild‐type Xanthomonas citri ssp. citri strain 306 and derivative strains to different stresses and the antimicrobial peptide polymyxin B.*
| Strain | Survival rate (%)† | |||||||
|---|---|---|---|---|---|---|---|---|
| UV radiation | Heat shock | Saline stress | Osmolarity stress | Desiccation tolerance | SDS exposure | H2O2 exposure | Polymyxin B | |
| 306 | 18.9 ± 2.2 a | 14.6 ± 1.7 a | 3.4 ± 1.4 a | 6.2 ± 2.5 a | 1.6 ± 0.7 a | 0.1 ± 0.1 a | 33.3 ± 5.5 a | 56.8 ± 6.4 a |
| 247C8 (wxacO) | 4.2 ± 0.6 b | 5.1 ± 2.1 b | 0.3 ± 0.1 b | 0.9 ± 0.4 b | 0.2 ± 0.1 b | 0.0 ± 0.0 b | 10.5 ± 4.3 b | 10.9 ± 3.4 b |
| 236E9 (rfbC) | 3.4 ± 1.4 b | 6.6 ± 1.6 b | 0.3 ± 0.2 b | 1.2 ± 0.5 b | 0.2 ± 0.1 b | 0.0 ± 0.0 b | 14.3 ± 2.6 b | 8.5 ± 3.2 b |
| 247C8V | 5.5 ± 1.8 b | 4.2 ± 2.2 b | 0.3 ± 0.1 b | 0.7 ± 0.2 b | 0.3 ± 0.1 b | 0.0 ± 0.0 b | 13.8 ± 3.5 b | 14.2 ± 4.5 b |
| 236E9V | 4.8 ± 1.6 b | 5.3 ± 1.4 b | 0.3 ± 0.1 b | 0.9 ± 0.3 b | 0.4 ± 0.2 b | 0.0 ± 0.0 b | 12.2 ± 2.9 b | 11.2 ± 2.6 b |
| C247C8 | 16.6 ± 1.9 a | 13.2 ± 3.2 a | 2.9 ± 1.3 a | 5.7 ± 2.1 a | 2.1 ± 0.4 a | 0.1 ± 0.1 a | 36.4 ± 4.7 a | 49.7 ± 5.3 a |
| C236E9 | 16.2 ± 2.0 a | 15.3 ± 2.5 a | 3.6 ± 0.8 a | 7.0 ± 2.4 a | 1.4 ± 0.5 a | 0.2 ± 0.1 a | 31.2 ± 2.7 a | 51.4 ± 7.0 a |
Bacterial cell viability was estimated by plating on nutrient agar (NA) at 28 °C before (T0) and after (T1) treatment. Percentage survival was calculated as the ratio of viable cell counts at T1 to those at T0. The treatments were applied as follows: for UV radiation, an overnight bacterial culture was exposed to short‐wave UV irradiation (254 nm) for 15 min; for heat‐shock, the bacterial culture was incubated at 54 °C for 5 min; for saline stress, the bacterial culture was treated with 5% NaCl for 10 min; for osmotic stress, the bacterial culture was treated with 40% d‐sorbitol for 40 min; for desiccation stress, the bacterial culture was placed on glass coverslips and air dried in a laminar flow apparatus for 60 min, and then resuspended in 0.85% NaCl and plated; for sodium dodecylsulphate (SDS) exposure, the bacterial culture was treated with 0.1% SDS for 10 min; for sensitivity to hydrogen peroxide, the bacterial culture was exposure to 0.03% H2O2 for 10 min; for sensitivity to polymyxin B, the bacterial culture was treated with 0.5 µg/mL polymyxin B for 2 h. Bacterial cells were serially diluted with nutrient broth (NB) medium and the colony‐forming units (cfu) were counted after being cultured on NA plates for 48 h. Each test, plated in triplicate, was repeated three times with similar results.
Means and standard errors of three replicates from one representative experiment are shown. Data with the same letters in the same column are not significantly different at P < 0.05 (Student's t‐test).
Previous studies have shown that an altered LPS is correlated with an increased sensitivity to antimicrobial peptides, including polymyxin B (Berry et al., 2009). The sensitivity of the wxacO and rfbC mutants to polymyxin B was assessed and compared with that of the wild‐type strain. The results showed that the mutant strains exhibited significantly higher sensitivity than the wild‐type strain to polymyxin B at 0.5 µg/mL for 2 h (Table 5). The wxacO and rfbC mutants were restored to the wild‐type levels of stress tolerance and hydrogen peroxide and polymyxin B sensitivity with the cloned wxacO and rfbC genes, respectively. No difference was observed in stress tolerance or hydrogen peroxide or polymyxin B sensitivity between the wild‐type strain 306 and the wxacO‐complemented strain C247C8 or the rfbC‐complemented strain C236E9 (Table 5). The empty vector pUFR053 did not complement stress tolerance or hydrogen peroxide or polymyxin B sensitivity in the wxacO or rfbC mutant.
DISCUSSION
In this article, we characterized one hypothetical gene wxacO and the rfbC gene, which are located in a gene cluster predicted to be involved in the biosynthesis of LPS in Xcc strain 306 (da Silva et al., 2002). Sequence analyses of the predicted wxacO product indicated that this gene is a member of a transmembrane protein family, which contains diverse enzymes or components involved in the assembly and/or export of surface polysaccharides (Popot and Engelman, 2000), and the rfbC gene belongs to the GT2 subfamily containing glycosyl transferase, which contributes to the biosynthesis of polysaccharides (Breton et al., 2006). In agreement with these findings, we demonstrated that mutation in wxacO or rfbC affected the LPS pattern of Xcc strain 306.
Homologous RfbCs are known to be involved in O‐antigen biosynthesis, but the functions of wxacO homologues remain ambiguous. In our study, the LPS profiles showed that both wxacO and rfbC mutants were affected in the production of O‐antigen and, in addition, the wxacO mutation caused a loss of another unknown LPS component (Fig. 3). These findings suggest that, similar to rfbC, wxacO contributes to O‐antigen biosynthesis, but it plays a role different from that of rfbC in LPS biosynthesis. The observation that wxacO was involved in the biosynthesis and/or export of the O‐antigen was consistent with that reported previously from mutants affected in the X. campestris pv. campestris wxcE gene, a gene coding for a transmembrane protein (Vorholter et al., 2001). WxacO was predicted to be located in the inner membrane and to contain eight transmembrane helices (Fig. 2B), but its role remains unclear.
Several lines of evidence indicate that rfbC encodes a glycosyltransferase involved in the biosynthesis of LPS O‐antigen. First, RfbC contains two GT2 domains (Fig. 2B). Second, disruption of rfbC results in the alteration of O‐antigen in the LPS profile (Fig. 3), consistent with the role of glycosyl transferases in LPS biosynthesis (Varki et al., 2009). Third, similar genes associated with the biosynthesis of the O‐antigen have been identified in other bacterial pathogens, including Escherichia coli (Kido et al., 1995), Myxococcus xanthus (Guo et al., 1996) and Yersinia enterocolitica (Zhang et al., 1993). However, the widely conserved GT2 is a superfamily of diverse functions, comprising cellulose synthase, chitin synthase, glucosyltransferase, mannosyltransferase, rhamnosyltransferase, etc. (Breton et al., 2006). RfbC in Xcc strain 306 may play a similar role to that in M. xanthus and Y. enterocolitica in LPS O‐antigen synthesis, but its mechanism of enzyme activity remains to be characterized.
LPS contributes to bacterial attachment to surfaces and to each other, and biofilm formation (Davey and O'Toole, 2000). Differential attachment has been reported for O‐antigen‐deficient mutants of P. fluorescens (Williams and Fletcher, 1996), S. maltophilia (Huang et al., 2006) and A. brasilense (Lerner et al., 2009) relative to the corresponding wild‐type strains. Our data showed that biofilm formation by the wxacO or rfbC mutant was decreased significantly on polystyrene plates and in glass tubes compared with the wild‐type strain (Fig. 1), and that GFP‐labelled wxacO mutant cells formed significantly less complex bacterial aggregates in vitro or in vivo compared with the wild‐type strain (Fig. 4). In addition, EPS or CPS production was not affected by mutation of wxacO or rfbC (data not shown). These findings, together with the demonstration that biofilm formation of the wxacO and rfbC mutants can be complemented by the cloned wxacO and rfbC genes, respectively, strongly suggest that a structurally intact LPS is critical to biofilm formation by Xcc. It has been suggested that biofilm development in Xcc is a three‐stage process, bacterial attachment, formation of aggregates or microcolonies and, finally, the formation of a structured biofilm (Rigano et al., 2007), and that EPS plays a role in biofilm development, particularly in the later stages (Rigano et al., 2007). Our data suggest that LPS, like EPS, plays a role in the biofilm development of Xcc, and may be one of the key elements that shape and provide structural support for bacterial biofilms.
The wxacO and rfbC mutants showed reduced swimming motility compared with the wild‐type strain (Table 4), consistent with previous reports for LPS mutants of other bacteria. For example, Huang et al. (2006) reported impaired swimming motility of an S. maltophilia mutant defective in xanB, which encodes an enzyme involved in the biosynthesis of GDP‐d‐mannose, which is necessary for O‐antigen biosynthesis in this bacterium. In addition, several research groups have recently described affected swimming motility of LPS mutants of A. brasilense (Lerner et al., 2009), P. aeruginosa (Lindhout et al., 2009) and R. leguminosarum (Vanderlinde et al., 2009). Alterations in LPS have been reported to interfere with the formation or function of flagella in numerous bacterial species, including E. coli (Genevaux et al., 1999), P. aeruginosa (Abeyrathne et al., 2005), S. maltophilia (Huang et al., 2006) and R. leguminosarum (Vanderlinde et al., 2009). Thus, it has been suggested that the defect in swimming motility of LPS mutants could be a result of a loss of formation or function of flagella. In contrast, the results of our study showed that flagellar formation in the wxacO and rfbC mutants was not affected (data not shown). Changes in LPS structure are not always accompanied by pronounced alterations in flagellar formation and function. Lindhout et al. (2009) reported that the deletion of genes involved in the biosynthesis of LPS by P. aeruginosa PAO1 caused no visible changes in flagellar assembly and function. The reduced motility of these mutants was suggested to be a result of changes in adhesion of bacterial cells to surfaces (Lindhout et al., 2009). The mechanism of the impaired swimming motility of the wxacO and rfbC mutants requires further characterization.
LPSs have been shown to contribute to the structural integrity of the cell envelope of Gram‐negative bacteria and are also involved in diverse interactions between bacterial cells and the environment (Raetz and Whitfield, 2002). As the wxacO and rfbC mutations affected LPS in Xcc strain 306, we examined whether the mutant strains exhibited a different response to diverse stresses relative to the wild‐type. As expected, these experiments revealed that both the wxacO and rfbC mutants were more sensitive than the wild‐type to several stresses that would be experienced in natural ecosystems, such as UV radiation, heat shock, desiccation, detergent, osmotic and oxidative stresses (Table 5). In addition, the two mutant strains were also found to be more susceptible than the wild‐type strain to polymyxin B (Table 5). These findings are consistent with previous reports that LPS‐defective mutants of a number of bacteria, including A. brasilense (Lerner et al., 2009), Erwinia amylovora (Berry et al., 2009), E. chrysanthemi (Touze et al., 2004), Salmonella enterica (Thomsen et al., 2003), S. maltophilia (Huang et al., 2006) and R. leguminosarum (Vanderlinde et al., 2009), exhibit compromised tolerance to stresses and antimicrobial agents. Our study, together with the others already mentioned, support the notion that LPS contributes to bacterial survival under diverse stress conditions. Alterations in the permeability of the outer membrane and the structural integrity of the cell envelope caused by defective LPSs may affect the tolerance of bacteria to stresses and antibiotics (Fralick and Burns‐Keliher, 1994; Garmiri et al., 2008; Lerner et al., 2009; Nikaido, 2003). Therefore, it is possible that the reduced stress adaptation and polymyxin B sensitivity of the wxacO and rfbC mutants may be attributed to alterations in the permeability of the outer membrane and the structural integrity of the cell envelope as a result of the altered LPS.
The disruption of wxacO or rfbC affected the virulence of Xcc strain 306 on host leaves when spray inoculated (Fig. 5A). This was expected because LPS‐defective mutants of many bacterial plant pathogens show reduced virulence compared with the corresponding wild‐type bacteria in numerous studies (Berry et al., 2009; Dow et al., 1995; Kingsley et al., 1993; Toth et al., 1999; Wang et al., 2008a). Interestingly, no significant change in virulence was observed in the two mutants following pressure infiltration inoculation (data not shown). Our observations are similar to those of studies in the gumB mutant of Xcc, which showed that a loss of pathogenicity of the mutant was more evident following spray infection rather than pressure infiltration (Rigano et al., 2007). It is known that the spray method mimics bacterial entry in nature, where the invasion of the host mesophyll is accomplished through natural openings, such as stomata and wounds, whereas pressure infiltration mechanically destroys the cuticle and cell wall that are the primary physical barriers in plant defence (Rigano et al., 2007). Therefore, we speculated that LPS plays an important role in the formation of biofilms that serve as a protective barrier against harsh environmental conditions and aid bacterial survival on the leaves, and subsequent entry into tissues at early stages of infection by Xcc. Consistent with this hypothesis, we showed that both wxacO and rfbC mutants are attenuated in growth on host leaves (Fig. 5B,C), but not in leaves following pressure infiltration inoculation (data not shown); this may be a result of impaired resistance against several stresses that may be experienced on the leaf surfaces (Table 5).
In conclusion, we characterized a hypothetical gene wxacO, as well as the rfbC gene, of Xcc strain 306. Our data provide new insights into the roles of LPS in Xcc–citrus interactions. We presented evidence that both wxacO and rfbC are associated with the biosynthesis of LPS, and emphasized that a structurally intact LPS is important for biofilm formation and bacterial ecological competence, as well as virulence, of the citrus canker pathogen.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids, media and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. The wild‐type Xcc strain 306 [rifamycin (Rif) resistant] (Rybak et al., 2009) and mutant strains were grown at 28 °C in NB or on NA (Daniels et al., 1984). Escherichia coli strains were grown in Luria–Bertani (LB) medium at 37 °C. When necessary, antibiotics were added at the following concentrations in the media: Rif, 50 µg/mL; kanamycin (Km), 50 µg/mL; ampicillin (Ap), 50 µg/mL; gentamicin (Gm), 5 µg/mL; chloramphenicol (Cm), 35 µg/mL.
DNA extraction and PCR
Bacterial genomic DNA and plasmid DNA were extracted using a Wizard genomic DNA purification kit and a Wizard miniprep DNA purification system following the manufacturer's instructions (Promega, Madison, WI, USA). The concentration and purity of DNA were determined using a Nanodrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). All conventional PCR amplications were performed with a Bio‐Rad DNAEngine Peltier thermal cycler (Bio‐Rad Laboratories, Inc., Hercules, CA, USA) using a Taq DNA polymerase (Promega) at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 52–58 °C (depending on the primers) for 45 s and 72 °C for 1–3 min (depending on the length of the amplicons). When necessary, PCR products were isolated from agarose gels using the Wizard® SV Gel and PCR Clean‐Up System (Promega). All of the primer sequences used for this study are listed in Table 2.
Transposon mutant library screen for biofilm formation‐impaired mutants
Initial screening for the biofilm formation‐affected mutants in a mutant library of Xcc 306, constructed using the EZ‐Tn5™ <R6Kγori/KAN‐2>Tnp Transposome™ Kit (Epicentre, Madison, WI, USA) (Guo et al., 2010), was performed using a flat‐bottomed polystyrene 96‐well plate (Nunclon surface, Nuncbrand, Roskilde, Denmark) assay system, as described by O'Toole and Kolter (1998) with modifications. Briefly, mutants were grown overnight in NB containing Km at 28 °C with shaking (200 rpm). The overnight cultures were grown for approximately 10 h to obtain a final optical density at 600 nm (OD600 nm) of 0.5. A 200‐µL portion of NB with 2% (w/v) d‐glucose was placed in each well of the 96‐well plates, and eight replicate wells were used for each analysis. A 2‐µL overnight culture was added to obtain a final OD600 nm of approximately 0.05, and the plates were incubated at 28 °C without agitation. After 72 h, the bacterial growth was checked and the culture was removed using a pipette. After repetitive washing of the plates, the adhered cells were stained with 0.1% (w/v) CV for 30 min at room temperature. The unbound stain was removed by washing under running tap water, and the CV stain was determined visually. Strains exhibiting significantly lower staining efficiencies (no or reduced CV binding) than the wild‐type were selected and subjected to further analyses. The mutants which passed the initial screening of the biofilm formation test were further confirmed using a quantitative biofilm assay in borosilicate glass tubes (Fischer Scientific, Pittsburgh, PA, USA), as described previously (Guo et al., 2010). Briefly, mid‐exponential phase cultures were diluted 1:10 in fresh NB containing Km. One millilitre of the diluted bacterial suspension was transferred into each glass tube and incubated at 28 °C without shaking for 48 h. Culture media were removed and bacterial cells attached to the tubes were gently washed three times with sterilized distilled water, incubated at 60 °C for 20 min and stained with 1.5 mL 0.1% CV for 45 min. The unbound CV was poured off, and the tubes were washed with water. The CV‐stained cells were solubilized in 1.5 mL of ethanol–acetone (80:20, v/v). The samples were measured at A 590 nm using an Agilent 8453 UV–visible spectrophotometer (Agilent Technologies, Santa Clara, CA, USA). Readings from five independent experiments were averaged. The test was repeated three times independently. Mutants that revealed reduced adherence were selected for further characterization.
Identification of transposon insertion sites
To identify the genes that were interrupted by the EZ‐Tn5 transposon, the RATE‐PCR method (Ducey and Dyer, 2002) was utilized with modifications. Briefly, the mutant genomic DNA was used as template in a standard PCR with the Inv‐1 or Inv‐2 primer (Table 2). The first 30 cycles of PCR were performed at 55 °C, with a 30‐s extension time. The second 30 cycles were carried out at 30 °C, with a 30‐s extension time. The last 30 cycles of the amplification were performed at 55 °C, with a 2‐min extension time. The RATE reactions were analysed on a 1.0% agarose TBE gel and the brightest band was purified with a PCR Purification Kit (Wizard® SV Gel and PCR Clean‐Up System, Promega); the resulting DNA was sequenced using the forward (KAN‐2 FP‐1) or reverse (KAN‐2 RP‐1) primer supplied with the EZ‐Tn5™ <R6Kγori/KAN‐2> Insertion Kit. After sequencing, the locations of the transposon inserts were determined by a blastn comparison with the genomic sequence of Xcc strain 306 on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/).
Complementation of the EZ‐Tn5 insertion mutants
To complement the wxacO mutant 247C8 (wxacO), a 2437‐bp DNA fragment containing an intact open reading frame (ORF) of wxacO, a 486‐bp sequence upstream of the translation start site and a 205‐bp sequence downstream from the translation stop site was amplified from the genomic DNA of Xcc strain 306 using the primers C96‐F and C96‐R, which contained a HindIII restriction site (Table 2). The PCR product was cloned into a pGEM®‐T Easy vector (Promega), following the manufacturer's instructions, resulting in pGE3596. The vector pJU3596 (Table 1) was constructed for complementation by excising the wxacO fragment from pGE3596 with HindIII and ligated into the HindIII site of the vector pUFR053 (El Yacoubi et al., 2007), which had been treated with calf intestinal alkaline phosphatase. Orientation of the fragment was confirmed by sequencing with M13 forward and reverse primers. The recombinant pJU3596 vector was then transformed into the mutant strain 247C8 (wxacO) to create a wxacO‐complemented strain C247C8 by triparental mating with E. coli helper strain HB101 containing pRK2013, as described elsewhere (Swarup et al., 1991). Transconjugants were selected on NA with Rif and Gm. The presence of pJU3596 was verified by PCR and sequencing. The plasmid pJU3598 (Table 1) was also constructed and transformed into the mutant strain 236E9 (rfbC) in this manner to create one rfbC‐complemented strain C236E9. The primers C98‐F and C98‐R (Table 2) were used to amplify and clone the full‐length rfbC coding sequence (also including about 500 bp upstream and 200 bp downstream) into pUFR053, resulting in the plasmid pJU3598.
Growth assays
Wild‐type and mutant strains were grown overnight in 5 mL NB supplemented with the antibiotics Rif and Km + Rif, respectively. Then, 0.3 mL of the cell suspensions (at about 108 cfu/mL) were transferred into 100‐mL flasks containing 30 mL of NB medium with appropriate antibiotics. Cultures were grown at 28 °C with agitation at 200 rpm, and OD600 nm was measured every 2–3 h over the course of 48 h using a spectrophotometer (WPA CO8000 Biowave Cell Density Meter, Biochrom Ltd, Cambridge, UK). Three replicates were included at each time point. The experiments were repeated twice.
LPS assays
LPS was extracted as described previously (Guo et al., 2010; Nesper et al., 2000) with modifications. Briefly, bacteria were grown overnight at 28 °C in NB to the exponential phase of growth. Cells from 5‐mL cultures were collected, washed in 1 mL of TNE [10 mm Tris (pH 8), 10 mm NaCl, 10 mM ethylenediaminetetraacetic acid (EDTA)] and resuspended in 540 µL of TNEX (TNE–1% Triton X‐100). Sixty microlitres of lysozyme (5 mg/mL; Sigma, St. Louis, MO, USA) were added, and the mixture was incubated for 20 min at 37 °C. The lysate was subsequently treated with proteinase K (30 µL; 20 µg/mL; Sigma) for 2 h at 65 °C and phenol for 5 min at room temperature. The aqueous phase was used for the analysis of the LPS pattern. LPS was separated on Precast Ready Gel Tris‐HCl polyacrylamide gels (containing 4% and 15% acrylamide in the stacking and separating gels, respectively) (Bio‐Rad Laboratories, Inc.) in buffer containing 2% SDS, and fixed overnight in buffer containing 40% methanol and 10% acetic acid. The gels were stained using a silver stain kit (Bio‐Rad Laboratories, Inc.) following the manufacturer's instructions. The experiment was repeated three times.
EPS assays
EPS in bacterial culture supernatants was determined quantitatively as described previously (Guo et al., 2010). Briefly, bacterial strains were grown in NB supplemented with 2% d‐glucose for 24 h at 28 °C with shaking at 200 rpm. A 10‐mL portion of the culture was collected, and the cells were removed by centrifugation (5000 g for 20 min). The supernatant was mixed with three volumes of 99% ethyl alcohol and the mixture was kept at 4 °C for 30 min. To determine the dry weights of EPS, the precipitated EPS was collected by centrifugation and dried at 55 °C overnight before measurement. Three replicates were used for each strain. The test was repeated three times.
Capsule assays
A bacterial capsule was stained using a capsule‐staining kit (Eng Scientific Inc., Clifton, NJ, USA) following the manufacturer's instructions. Briefly, bacteria were grown on NA with appropriate antibiotics for 72 h at 28 °C. A loop of culture was mixed and spread with one drop of 0.85% NaCl on a precleaned slide and air dried. The samples were then stained with 1% CV, washed with 20% copper sulphate supplied with the kit and photographed using a light microscope (Leica DMLB2; Leica Microsystems GmbH, Wetzlar, Germany) with a digital camera. The experiment was repeated three times.
Pathogenicity assays
Young (about 10‐week‐old) grapefruit (Citrus paradisi cv. Duncan grapefruit) plants were used as host plants. All plant inoculations involved a minimum of three immature leaves at a similar developmental stage from each plant, and five plants were inoculated for each bacterial strain. Bacterial suspensions of Xcc wild‐type and mutant strains (108 cfu/mL in sterile tap water) were inoculated by spraying the abaxial leaf surface and by pressure infiltration. Sterile tap water was used as control. After inoculation, the plants were kept in a quarantine glasshouse at the Citrus Research and Education Center, Lake Alfred, FL, USA, which was kept at approximately 28–35 °C and a relative humidity of more than 80%. Disease progression was monitored phenotypically in five separate biological assays. The test was repeated three times.
Bacterial growth assays in planta
For in planta growth assays, bacterial strains were inoculated onto leaves of grapefruit as described above. Bacterial cell counts were determined for at least three replicates at each sampling time point. Leaf discs (0.8 cm in diameter), randomly selected from the inoculated leaves, were ground in 1 mL of 0.85% (w/v) NaCl. Serial dilutions of the suspension were plated on NA plates containing appropriate antibiotics. Bacterial colonies were counted after 48 h and the number of cfu per square centimetre of leaf tissue was calculated. The experiment was repeated three times with similar results. Data from one representative experiment were presented.
Microscopic observation of biofilm formation in vitro and in planta
CLSM (Leica TCS SL; Leica Microsystems GmbH) was used to visualize biofilm formation by the bacterial cultures carrying the plasmid pUFZ75, which constitutively produces GFP (Zhang et al., 2009) both in vitro and in planta. For the in vitro study, biofilm formation assays were performed over a 4‐day time course experiment using glass‐bottomed culture dishes with a 0.085–0.13‐mm‐thick borosilicate glass base (no. p35G‐0‐20‐C; MatTek Corporation, Ashland, MA, USA). GFP‐labelled bacterial overnight cultures were diluted to OD600 nm= 0.05; 2 mL of the diluted cultures were inoculated into each dish, grown under static conditions at 28 °C for up to 4 days and imaged in the sequential mode with 40× objectives. Briefly, for the in planta assay, leaves of young grapefruit plants were infected by spray, as described above, with GFP‐labelled bacterial suspensions of Xcc wild‐type strain 306 and the wxacO mutant strain. Plants were kept in the same conditions as described above until canker formation. Areas of approximately 1 cm2 were cut from the leaves and placed on glass coverslips with the adaxial surface upwards, and imaged in the sequential mode with 63× objectives.
Motility assays
Swimming motility was examined on 0.3% (w/v) agar (Difco, Franklin Lakes, NJ, USA) medium containing NB as described elsewhere (Lindhout et al., 2009). Overnight cultures (2 µL) of bacteria grown in NB were spotted onto NA plates (diameter, 90 mm; containing 20 mL of NA) and incubated for 72 h at 28 °C. Swarming motility was determined on NA plates containing 0.7% (w/v) agar. Plates were inoculated with 2‐µL overnight cultures. Growth was analysed after 72 h of incubation at 28 °C. The experiment was repeated three times with three replicates each time.
Electron microscopy for flagella visualization
The presence of flagella on cells grown on NA plates for 48 h was examined by TEM on three separate occasions. Cells from agar cultures were resuspended in 1 mL of sterilized distilled water. A 400‐mesh Formvar carbon‐coated grid was placed on top of the cell suspension. The grid was rinsed with water, and excess water was removed by blotting onto Whatman filter paper no. 1 (Whatman Inc, Piscataway, NJ, USA). The grid was floated on 1 mL of 1% uranyl acetate solution, and excess solution was removed. Samples were examined using a Philips FEI Morgagni 268 transmission electron microscope (FEI Company, Eindhoven, the Netherlands) operating at 60 kV.
Stress tolerance assays
These experiments were performed as described previously (Lerner et al., 2009; Qian et al., 2008) with modifications. Xcc strains were cultured to early exponential stage (OD600 nm= 0.1) in NB, and experiments were conducted to test survival under six environmental stresses, including UV radiation, heat shock, saline stress, osmotic challenge, desiccation stress and SDS stress. In each stress treatment, Xcc cell viability was estimated by plating on NA at 28 °C before (T0) and after (T1) treatment. Survival was calculated as the ratio of viable cell counts at T1 to those at T0. The stress treatments were applied as follows: for UV radiation, the cells were exposed to short‐wave UV radiation (254 nm in a biological safety cabinet) at a distance of 60 cm for 15 min (T1); for heat‐shock stress, the culture was transferred to 54 °C for 5 min (T1); for sodium stress, NaCl (pH 7.5) was added to the bacterial culture at a final concentration of 5%, and survival was estimated after 10 min (T1); for osmotic challenge, d‐sorbitol (pH 7.0) was added to the bacterial culture at a final concentration of 40%, and survival was estimated after 40 min (T1); for desiccation stress, the bacterial culture was placed on glass coverslips (18 mm × 18 mm), air dried in a laminar flow apparatus for 60 min (T1) and then resuspended in 0.85% NaCl and plated; for SDS stress, SDS (pH 7.5) was added to the bacterial culture at a final concentration of 0.1%, and survival was estimated after 10 min (T1). Xcc cells were serially diluted with NB medium and the numbers of cfu were counted after being cultured on NA plates for 48 h. Each stress test, plated in triplicate, was repeated three times. Student's t‐test was used to test the significance of the differences.
Assessment of sensitivity to hydrogen peroxide and polymyxin B
These experiments were performed as described elsewhere (Berry et al., 2009) with modifications. Briefly, Xcc strains were cultured as described above. To assess bacterial sensitivity to hydrogen peroxide, hydrogen peroxide (30% solution) was added to the bacterial culture at a final concentration of 0.03%, and survival was estimated after 10 min. To assess the sensitivity to polymyxin B, polymyxin B (Sigma) was added to the bacterial culture at a final concentration of 0.5 µg/mL, and survival was determined after 2 h. Viable cells (cfu/mL) were determined by dilution plating as described above. The experiment was repeated three times with three replicates each time.
ACKNOWLEDGEMENTS
This work was supported by USDA‐CSREES Special Citrus Canker Grant Project 88811. We thank Yinping Guo for providing the GFP‐labelled Xcc wild‐type stain, Diann Achor for help with microscopy work and Lin Yang for assistance with glasshouse work.
REFERENCES
- Abeyrathne, P.D. , Daniels, C. , Poon, K.K. , Matewish, M.J. and Lam, J.S. (2005) Functional characterization of WaaL, a ligase associated with linking O‐antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187, 3002–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, M.C. , McGhee, G.C. , Zhao, Y. and Sundin, G.W. (2009) Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora . FEMS Microbiol. Lett. 291, 80–87. [DOI] [PubMed] [Google Scholar]
- Boyer, H. and Roulland‐Dussoix, D. (1969) A complementation analysis of the restriction and modification of DNA in Escherichia coli . J. Mol. Biol. 41, 459–472. [DOI] [PubMed] [Google Scholar]
- Braun, S.G. , Meyer, A. , Holst, O. , Pühler, A. and Niehaus, K. (2005) Characterization of the Xanthomonas campestris pv. campestris lipopolysaccharide substructures essential for elicitation of an oxidative burst in tobacco cells. Mol. Plant–Microbe Interact. 18, 674–681. [DOI] [PubMed] [Google Scholar]
- Breton, C. , Snajdrova, L. , Jeanneau, C. , Koca, J. and Imberty, A. (2006) Structures and mechanisms of glycosyltransferases. Glycobiology, 16, 29R–37R. [DOI] [PubMed] [Google Scholar]
- Caspi, R. , Altman, T. , Dale, J.M. , Dreher, K. , Fulcher, C.A. , Gilham, F. , Kaipa, P. , Karthikeyan, A.S. , Kothari, A. , Krummenacker, M. , Latendresse, M. , Mueller, L.A. , Paley, S. , Popescu, L. , Pujar, A. , Shearer, A.G. , Zhang, P. and Karp, P.D. (2010) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 38, D473–D479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey, D.A. (1990) Genetics of bactericide resistance in plant pathogenic bacteria. Annu. Rev. Phytopathol. 28, 201–219. [Google Scholar]
- Cubero, J. and Graham, J.H. (2002) Genetic relationship among worldwide strains of Xanthomonas causing canker in citrus species and design of new primers for their identification by PCR. Appl. Environ. Microbiol. 68, 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn, T. and Fuqua, C. (2007) Biofilm formation by plant‐associated bacteria. Annu. Rev. Microbiol. 61, 401–422. [DOI] [PubMed] [Google Scholar]
- Daniels, M. , Barber, C. , Turner, P. , Sawczyc, M. , Byrde, R. and Fielding, A. (1984) Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 3, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey, M.E. and O'Toole, G.A. (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, J.M. , Osbourn, A.E. , Wilson, T.J.G. and Daniels, M.J. (1995) A locus determining pathogenicity of Xanthomonas campestris is involved in lipopolysaccharide biosynthesis. Mol. Plant–Microbe Interact. 8, 768–777. [DOI] [PubMed] [Google Scholar]
- Ducey, T.F. and Dyer, D.W. (2002) Rapid identification of EZ:TN™ transposon insertion sites in the genome of Neisseria gonorrhoeae . EPICENTRE Forum, 9, 6–7. [Google Scholar]
- Dunger, G. , Relling, V.M. , Tondo, M.L. , Barreras, M. , Ielpi, L. , Orellano, E.G. and Ottado, J. (2007) Xanthan is not essential for pathogenicity in citrus canker but contributes to Xanthomonas epiphytic survival. Arch. Microbiol. 88, 127–135. [DOI] [PubMed] [Google Scholar]
- El Yacoubi, B. , Brunings, A. , Yuan, Q. , Shankar, S. and Gabriel, D. (2007) In planta horizontal transfer of a major pathogenicity effector gene. Appl. Environ. Microbiol. 73, 1612–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski, D.H. and Helinski, D.R. (1979) Replication of an origin‐containing derivative of plasmid RK2 dependent on a plasmid function provided in trans . Proc. Natl. Acad. Sci. USA, 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fralick, J.A. and Burns‐Keliher, L.L. (1994) Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coli K‐12. J. Bacteriol. 176, 6404–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmiri, P. , Coles, K. , Humphrey, T. and Cogan, T. (2008) Role of outer membrane lipopolysaccharides in the protection of Salmonella enterica serovar Typhimurium from desiccation damage. FEMS Microbiol. Lett. 281, 155–159. [DOI] [PubMed] [Google Scholar]
- Genevaux, P. , Bauda, P. , DuBow, M.S. and Oudega, B. (1999) Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172, 1–8. [DOI] [PubMed] [Google Scholar]
- Gottig, N. , Garavaglia, B.S. , Garofalo, C.G. , Orellano, E.G. and Ottado, J. (2009) A filamentous hemagglutinin‐like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS ONE, 4, e4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald, T.R. , Graham, J.H. and Schubert, T.S. (2002) Citrus canker: the pathogen and its impact. Plant Health Prog. Online. doi:10.1094/PHP‐2002‐0812‐01‐RV. [Google Scholar]
- Graham, J.H. , Gottwald, T.R. , Cubero, J. and Achor, D.S. (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Guo, D. , Bowden, M.G. , Pershad, R. and Kaplan, H.B. (1996) The Myxococcus xanthus rfbABC operon encodes an ABC transporter homolog required for O‐antigen biosynthesis and multicellular development. J. Bacteriol. 178, 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. , Sagaram, U.S. , Kim, J.S. and Wang, N. (2010) Requirement of the galU gene for polysaccharide production by and pathogenicity and growth in planta of Xanthomonas citri subsp. citri . Appl. Environ. Microbiol. 76, 2234–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, F. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Huang, T. , Somers, E.B. and Lee Wong, A.C. (2006) Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide‐coupled biosynthetic genes in Stenotrophomonas maltophilia . J. Bacteriol. 188, 3116–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan, E. and Watnick, P. (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido, N. , Torgov, V. , Sugiyama, T. , Uchiya, K. , Sugihara, H. , Komatsu, T. , Kato, K. and Jann, K. (1995) Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferases, and evidence for an ATP‐binding cassette transport system. J. Bacteriol. 177, 2178–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley, M.T. , Gabriel, D.W. , Marlow, G.C. and Roberts, P.D. (1993) The opsX locus of Xanthomonas campestris affects host range and biosynthesis of lipopolysaccharide and extracellular polysaccharide. J. Bacteriol. 175, 5839–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, P.C.Y. , Lindhout, T. , Beveridge, T.J. , Dutcher, J.R. and Lam, J.S. (2009) Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J. Bacteriol. 191, 6618–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, A. , Okon, Y. and Burdman, S. (2009) The wzm gene located on the pRhico plasmid of Azospirillum brasilense Sp7 is involved in lipopolysaccharide synthesis. Microbiology, 155, 791–804. [DOI] [PubMed] [Google Scholar]
- Lindhout, T. , Lau, C.Y.P. , Brewer, D. and Lam, J.S. (2009) Truncation in the core oligosaccharide of lipopolysaccharide affects flagella‐mediated motility in Pseudomonas aeruginosa PAO1 via modulation of cell surface attachment. Microbiology, 155, 3449–3460. [DOI] [PubMed] [Google Scholar]
- Nesper, J. , Kapfhammer, D. , Klose, K. , Merkert, H. and Reidl, J. (2000) Characterization of Vibrio cholerae O1 antigen as the bacteriophage K139 receptor and identification of IS1004 insertions aborting O1 antigen biosynthesis. J. Bacteriol. 182, 5097–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M.A. , Dow, J.M. and Daniels, M.J. (2001) Bacterial lipopolysaccharide and plant–pathogen interactions. Eur. J. Plant Pathol. 107, 95–102. [Google Scholar]
- Newman, M.A. , Dow, J.M. , Molinaro, A. and Parrilli, M. (2007) Priming, induction and modulation of plant defense responses by bacterial lipopolysaccharides. J. Endotoxin Res. 13, 69–84. [DOI] [PubMed] [Google Scholar]
- Nikaido, H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G.A. and Kolter, R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461. [DOI] [PubMed] [Google Scholar]
- Popot, J.‐L. and Engelman, D.M. (2000) Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69, 881–922. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Han, Z.J. , Tao, J. and He, C.Z. (2008) Genome‐scale mutagenesis and phenotypic characterization of two‐component signal transduction systems in Xanthomonas campestris pv. campestris ATCC 33913. Mol. Plant–Microbe Interact. 21, 1128–1138. [DOI] [PubMed] [Google Scholar]
- Raetz, C.R. and Whitfield, C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, P.R. , Hobbs, M. , Valvano, M.A. , Skurnik, M. , Whitfield, C. , Coplin, D. , Kido, N. , Klena, J. , Maskell, D. , Raez, C.R.H. and Rick, P.D. (1996) Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4, 495–503. [DOI] [PubMed] [Google Scholar]
- Rigano, L.A. , Siciliano, F. , Enrique, R. , Sendin, L. , Filippone, P. , Torres, P.S. , Questa, J. , Dow, J.M. , Castagnaro, A.P. , Vojnov, A.A. and Marano, M.R. (2007) Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri . Mol. Plant–Microbe Interact. 20, 1222–1230. [DOI] [PubMed] [Google Scholar]
- Rybak, M. , Minsavage, G.V. , Stall, E. and Jones, J.B. (2009) Identification of Xanthomonas citri subsp. citri host specificity genes in a heterologous expression host. Mol. Plant Pathol. 10, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, N. , Postnikova, E. , Lacy, G. , Sechler, A. , Agarkova, I. , Stromberg, P. , Stromberg, V. and Vidaver, A. (2006) Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29, 690–695. [DOI] [PubMed] [Google Scholar]
- da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Spiers, A.J. and Rainey, P.B. (2005) The Pseudomonas fluorescens SBW25 wrinkly spreader biofilm requires attachment factor, cellulose fibre and LPS interactions to maintain strength and integrity. Microbiology, 151, 2829–2839. [DOI] [PubMed] [Google Scholar]
- Swarup, S. , De Feyter, R. , Brlansky, R.H. and Gabriel, D.W. (1991) A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of Xanthomonas campestris to elicit canker‐like lesions on citrus. Phytopathology, 81, 802–809. [Google Scholar]
- Thomsen, L.E. , Chadfield, M.S. , Bispham, J. , Wallis, T.S. , Olsen, J.E. and Ingmer, H. (2003) Reduced amounts of LPS affect both stress tolerance and virulence of Salmonella enterica serovar Dublin. FEMS Microbiol. Lett. 228, 225–231. [DOI] [PubMed] [Google Scholar]
- Toth, I.K. , Thorpe, C.J. , Bentley, S.D. , Mulholland, V. , Hyman, L.J. , Perombelon, M.C.M. and Salmond, G.P.C. (1999) Mutation in a gene required for lipopolysaccharide and enterobacterial common antigen biosynthesis affects virulence in the plant pathogen Erwinia carotovora subsp. atroseptica . Mol. Plant–Microbe Interact. 12, 499–507. [DOI] [PubMed] [Google Scholar]
- Touze, T. , Goude, R. , Georgeault, S. , Blanco, C. and Bonnassie, S. (2004) Erwinia chrysanthemi O antigen is required for betaine osmoprotection in high‐salt media. J. Bacteriol. 186, 5547–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinde, E.M. , Muszynski, A. , Harrison, J.J. , Koval, S.F. , Foreman, D.L. , Ceri, H. , Kannenberg, E.L. , Carlson, R.W. and Yost, C.K. (2009) Rhizobium leguminosarum biovar viciae 3841, deficient in 27‐hydroxyoctacosanoate‐modified lipopolysaccharide, is impaired in desiccation tolerance, biofilm formation and motility. Microbiology, 155, 3055–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki, A. , Cummings, R.D. , Esko, J.D. , Freeze, H.H. , Stanley, P.C. , Bertozzi, R. , Hart, G.W. and Etzle, M.E. (2009) Essentials of Glycobiology, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories Press. [PubMed] [Google Scholar]
- Vauterin, L. , Hoste, B. , Kersters, K. and Swings, J. (1995) Reclassification of Xanthomonas . Int. J. Syst. Bacteriol. 45, 472–489. [Google Scholar]
- Vorholter, F.J. , Niehaus, K. and Puhler, A. (2001) Lipopolysaccharide biosynthesis in Xanthomonas campestris pv. campestris: a cluster of 15 genes is involved in the biosynthesis of the LPS O‐antigen and the LPS core. Mol. Genet. Genomics, 266, 79–95. [DOI] [PubMed] [Google Scholar]
- Wang, J.C. , So, B.H. , Kim, J.H. , Park, Y.J. , Lee, B.M. and Kang, H.W. (2008a) Genome‐wide identification of pathogenicity genes in Xanthomonas oryzae pv. oryzae by transposon mutagenesis. Plant Pathol. 57, 1136–1145. [Google Scholar]
- Wang, P. , Guo, H. , Yi, W. and Song, J.K. (2008b) Current understanding on biosynthesis of microbial polysaccharides. Curr. Top. Med. Chem. 8, 141–151. [DOI] [PubMed] [Google Scholar]
- Williams, V. and Fletcher, M. (1996) Pseudomonas fuorescens adhesion and transport through porous media are affected by lipopolysaccharide composition. Appl. Environ. Microbiol. 62, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yethon, J.A. , Gunn, J.S. , Ernst, R.K. , Miller, S.I. , Laroche, L. , Malo, D. and Whitfield, C. (2000) Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo . Infect. Immun. 68, 4485–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Al‐Hendy, A. , Toivan, P. and Skurnik, M. (1993) Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP‐L‐rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol. Microbiol. 9, 309–321. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.X. , Callaway, E.M. , Jones, J.B. and Wilson, M. (2009) Visualisation of hrp gene expression in Xanthomonas euvesicatoria in the tomato phyllosphere. Eur. J. Plant Pathol. 124, 379–390. [Google Scholar]


