SUMMARY
Studies were undertaken to assess the induction of defence response pathways in pearl millet (Pennisetum glaucum) in response to infection with the leaf rust fungus Puccinia substriata. Pretreatment of pearl millet with salicylic acid (SA) conferred resistance to a virulent isolate of the rust fungus, whereas methyl jasmonate (MeJA) did not significantly reduce infection levels. These results suggest that the SA defence pathway is involved in rust resistance. In order to identify pearl millet genes that are specifically regulated in response to SA and not MeJA, and thus could play a role in resistance to P. substriata, gene expression profiling was performed. Substantial overlap in gene expression responses between the treatments was observed, with MeJA and SA treatments exhibiting 17% co‐regulated transcripts. However, 34% of transcripts were differentially expressed in response to SA treatment, but not in response to MeJA treatment. SA‐responsive transcripts represented genes involved in SA metabolism, defence response, signal transduction, protection from oxidative stress and photosynthesis. The expression profiles of pearl millet plants after treatment with SA or MeJA were more similar to one another than to the response during a compatible infection with P. substriata. However, some SA‐responsive genes were repressed during P. substriata infection, indicating possible manipulation of host responses by the pathogen.
INTRODUCTION
Pearl millet (Pennisetum glaucum) is an important staple crop in the semi‐arid tropics of Africa and Asia, and is also extensively grown as a summer annual grazing crop in the southern USA (Goldman et al., 2003). Of all the major cereals, it is the most able to tolerate both extremes of heat and drought. It yields reliably in regions too hot and too dry to consistently support good yields of maize or even sorghum. Although pearl millet suffers less from disease and insect pests than do sorghum and maize (National Research Council, 1996), yields are still limited by disease. One such disease is rust caused by Puccinia substriata, which is fairly widespread throughout the Americas, Asia and Africa. Previous synonyms include Puccinia penniseti Zimm. and Puccinia substriata var. penicillariae (de Carvalho et al., 2006). Puccinia substriata is a macrocyclic rust that causes small reddish‐orange round uredinia mainly on pearl millet foliage. As the severity of the infection increases, leaf tissue wilts and becomes necrotic from the leaf apex to the base (Wilson, 2000). Although disease resistance has been introduced into agronomically acceptable forage and grain cultivars (Singh et al., 1990b), the diverse nature of P. substriata has hampered efforts to breed for stable resistance and biomass production (Wilson and Gates, 1999). Even low levels of rust infection lead to significant losses of digestible dry matter and, as a result, the disease has become an important limiting factor for grain and forage production.
Very few genetic and molecular studies have been performed in pearl millet. Genetic maps of pearl millet of some 300 loci spread over seven linkage groups are available (Liu et al., 1994). An extended map from multiple crosses incorporates not only molecular markers but also significant phenotypic traits (Devos et al., 2006). Recently, Mishra et al. (2007) published results on the isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of pearl millet seedlings that had been exposed to abiotic stresses (salinity, cold and drought). With regard to plant defence response in pearl millet, Van den Berg et al. (2004) constructed a pearl millet defence response cDNA library. A number of studies have also been published outlining the treatment of pearl millet seedlings with various elicitors and signalling molecules to improve resistance to downy mildew (Geetha and Shetty, 2002; Manjunatha et al., 2008; Sarosh et al., 2005; Shailasree et al., 2007; Sharathchandra et al., 2004).
Three major signalling molecules, salicylic acid (SA), jasmonic acid (JA) and ethylene (ET), are known to play key roles in various aspects of plant defence signal transduction, but others, such as abscisic acid (ABA), auxin, gibberellin (GA), cytokinin (CK) and brassinosteroid (BL), have recently been linked to plant defence and may affect SA and/or JA signalling (Robert‐Seilaniantz et al., 2007). SA, JA and ET are involved in two major defence signalling pathways: an SA‐dependent pathway and an SA‐independent pathway that involves JA and ET (Kunkel and Brooks, 2002). Evidence from model plants, such as Arabidopsis, suggests that SA‐mediated defence responses are effective against biotrophic fungi, bacteria and viruses, whereas JA and ET mediate plant defence against insects, and appear to mobilize antimicrobial defences that are predominantly effective against necrotrophic pathogens (Murray et al., 2002). Initial molecular studies have suggested that the SA and JA/ET defence signalling pathways are antagonistic, but these reports were restricted to the analysis of a relatively small number of genes or proteins (Kunkel and Brooks, 2002). However, recent defence signalling studies have applied DNA microarray global gene expression profiling (Glazebrook et al., 2003; Maleck et al., 2000; Salzman et al., 2005; Schenk et al., 2000; Zhu‐Salzman et al., 2004), and indicate the existence of a substantial network of regulatory interactions and coordination during plant defence signalling, notably between the SA and JA pathways.
Induced resistance in plants is activated either by a prior pathogen infection (biologically induced resistance) or by treatment with a chemical (chemically induced resistance). The best studied form of induced resistance is systemic acquired resistance (SAR) (Vlot et al., 2008), which is induced by pathogens, and results in broad‐spectrum disease resistance that involves the up‐regulation of a range of pathogenesis‐related (PR) genes. Significantly, SAR is dependent on SA for the activation of resistance at local and systemic sites, and the methylated form of SA is now thought to be an important mobile signal (Vlot et al., 2008). Although molecular information is available on chemically and biologically induced resistance mechanisms in the Dicotyledoneae (Vlot et al., 2008), this is largely missing for the Monocotyledoneae, including most of the important cereals used in agriculture. Biologically induced resistance has been demonstrated in barley, wheat, rice and maize (Cho and Smedegaard‐Petersen, 1986; Djonovic et al., 2007; Smith and Metraux, 1991), and the chemical benzo(1,2,3)thiadiazole‐7‐carbothioic acid S‐methyl ester (BTH) has been found to induce resistance against several cereal pathogens (Beßer et al., 2000; Gorlach et al., 1996; Morris et al., 1998; Shimono et al., 2007).
A number of gene expression studies have been performed to identify genes that are differentially regulated in interactions between cereals and rust fungi. Zhang et al. (2003) employed cDNA‐amplified fragment length polymorphism (AFLP) and isolated genes that were expressed during a compatible interaction between leaf rust (Puccinia triticina) and wheat. More recently, studies have been undertaken that have profiled the wheat–leaf rust interaction using cDNA microarrays. Fofana et al. (2007) constructed a cDNA array containing 7728 wheat ESTs, and used the array to identify host genes that were differentially expressed in a wheat line containing the leaf rust resistance gene Lr1, following inoculation with compatible and incompatible races of leaf rust fungus P. triticina. One hundred and ninety‐two genes were found to have significantly altered gene expression between compatible and incompatible interactions. Among these were genes involved in photosynthesis, the production of reactive oxygen species (ROS), ubiquitination, signal transduction and the shikimate/phenylpropanoid pathway. Coram et al. (2008) employed an Affymetrix GeneChip Wheat Genome Array to characterize the resistance of wheat to stripe rust (Puccinia striiformis) conferred by the Yr5 resistance (R) gene. The Yr5‐mediated incompatible interaction resulted in a rapid and amplified resistance response involving signalling pathways and defence‐related transcripts known to occur during R gene‐mediated responses. These included protein kinase signalling and the production of ROS, leading to the hypersensitive response (HR).
In this paper, we report the chemically induced resistance and expression of defence‐related genes in pearl millet. We hypothesized that part of the pathogen's strategy is to enhance antagonism between the SA and methyl jasmonate (MeJA) pathways; however, the comparison of expression profiles did not support this. Our results demonstrate, nevertheless, that the treatment of pearl millet with SA, but not MeJA, induces resistance to rust. Subsequent gene expression profiling of pearl millet leaves treated with SA or MeJA identified a number of genes specifically up‐regulated in response to SA, but not MeJA, which could play a role in resistance to rust in pearl millet. We also examined pearl millet's molecular response following infection with a compatible rust fungus, and identified SA‐inducible genes repressed by P. substriata.
RESULTS
Pearl millet exhibits chemically induced resistance to rust
The causal agent of rust collected from pearl millet plants in Kwazulu‐Natal province, South Africa, was identified as P. substriata by rDNA sequencing (Maier et al., 2007). Pearl millet (line ICML12=P7), although reported as being moderately resistant to P. pennesiti in India (Singh et al., 1990a), was susceptible to the South African isolate of P. substriata. Glasshouse inoculations of this pearl millet accession with P. substriata urediniospores resulted in the production of chlorotic lesions within 5 days post‐inoculation (dpi) and fully developed rust pustules within 8 dpi (Fig. 1A,B).
Figure 1.
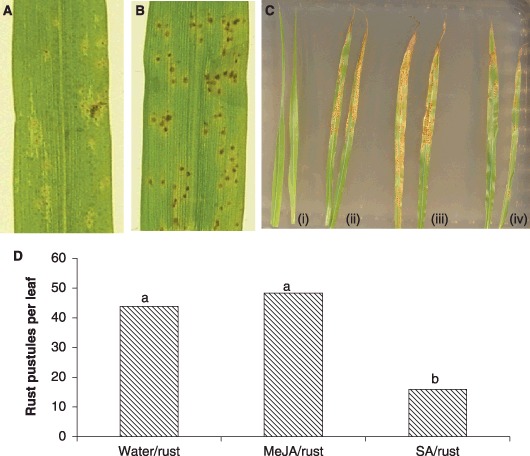
Development of rust symptoms on pearl millet (cv. ICML12=P7) leaves after inoculation with Puccinia substriata (A, B), and the reduction of symptoms after pretreatment with salicylic acid (SA), but not methyl jasmonate (MeJA) (C, D). (A) Typical symptoms 5 days post‐inoculation (dpi) – chlorotic spots with a small orange centre, indicative of pustule formation, are present on the leaf surface. (B) Typical symptoms at 8 dpi – rust pustules have developed and contain urediniospores. (C) Pretreatment of pearl millet plants with SA (iv) results in delayed and reduced rust symptoms, but typical symptoms develop on plants pretreated with water (ii) or MeJA (iii) (photograph taken at 10 dpi). Three‐week‐old pearl millet plants were sprayed with water (i, ii), MeJA (iii) or SA (iv), and the fourth leaf of each plant was inoculated with P. substriata urediniospores 24 h later (ii, iii, iv). (D) Mean number of rust pustules per pearl millet leaf at 10 dpi with P. substriata. Bars with identical letters are not significantly different from each other (Students t‐test at P < 0.05).
Experiments were performed to assess whether the treatment of pearl millet with the defence signalling molecules MeJA and SA elicited a defence response that would render pearl millet less susceptible to rust infection. Pearl millet plants (line ICML12=P7) were treated with water, MeJA or SA, and the fourth leaf of each pearl millet plant was inoculated 24 h later with freshly collected P. substriata urediniospores. Rust pustules began to develop on water‐ and MeJA‐treated leaves within 7 days of inoculation, whereas rust symptoms only became evident on SA‐treated leaves after 9 days (Fig. 1C; photograph taken at 10 dpi). Some of the SA‐treated leaves developed chlorotic lesions only and did not develop full rust‐like symptoms. The number of pustules per leaf after SA treatment was significantly less than that on water‐ and MeJA‐treated plants (Fig. 1D; Students t‐test; P < 0.05). It thus appears that the application of SA to pearl millet leaves 24 h prior to infection is able to protect the plant against subsequent attack by a compatible rust fungus.
Expression profiling reveals SA‐specific responses in contrast with MeJA and rust treatment
Gene expression profiling of pearl millet plants after treatment with SA, MeJA or during a compatible interaction with P. substriata was used to identify genes that are likely to contribute to chemically induced resistance based on the criterion that they are differentially expressed in response to SA and not to the other treatments. Furthermore, we aimed to identify similarities between the compatible response and MeJA treatment, which may indicate that part of the pathogen's strategy is to enhance antagonism between the SA and MeJA responses, which has been shown in Arabidopsis (Robert‐Seilaniantz et al., 2007) We exploited glass slide microarrays of pearl millet cDNA libraries that had previously been constructed using suppression subtractive hybridization (SSH) and that were enriched for transcripts either up‐ or down‐regulated in response to elicitors or wounding (Van den Berg et al., 2004).
Gene expression changes over time following treatment with either SA or MeJA were assessed using a direct‐sequential loop design with biological and technical replication (Fig. S1, see Supporting Information) (Naidoo et al., 2005). Expression profiles obtained with these designs derive from pairwise comparisons of adjacent time points, allowing direct comparison of expression differences between time points. The direct‐sequential loop design increases precision for some pairwise comparisons in the time course, which reduces the mean variance for data collected in this way (Alba et al., 2004). We chose this approach instead of a common reference design, which has the disadvantage that such comparisons can only be made indirectly, and which may have made subtle differences from one time point to another difficult to detect (Alba et al., 2004).
Samples were collected from MeJA‐ and SA‐treated plants at 0, 12, 24 and 48 h post‐inoculation (hpi), the same time points as used by Schenk et al. (2000), and from P. substriata‐inoculated plants at an early time point before symptoms appeared (20 h) and during symptom development at 5 and 8 dpi (Fig. 1A). High‐quality microarray hybridizations were obtained (Fig. S2, see Supporting Information), data were analysed using limmaGUI (Wettenhall and Smyth, 2004) (see Experimental procedures), and expression ratios for each cDNA were calculated for each time point relative to t = 0 of the same treatment. Differential expression was assessed using the moderated t‐test implemented in limmaGUI. Three hundred and fifty‐five of 1920 cDNAs were significantly regulated (more than twofold induced or repressed; P < 0.05) in at least one of the nine conditions analysed (three treatments with three time points relative to t = 0). The transcript abundances of 108 cDNAs for SA and 189 cDNAs for MeJA were specifically increased as a result of treatment with these signal molecules (Fig. 2). The transcript abundances of 42 and 16 cDNAs were reduced after treatment with SA and MeJA, respectively (Fig. 2). A number of transcripts (17%) were found to be co‐regulated by MeJA and SA (Fig. 2). Similar co‐regulation has been observed previously in Arabidopsis (Schenk et al., 2000) and sorghum (Salzman et al., 2005).
Figure 2.
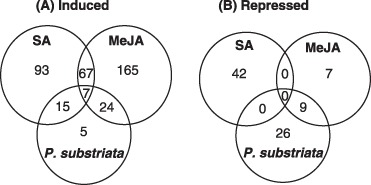
Venn diagrams depicting regulatory relationships of pearl millet transcripts significantly induced (A) or repressed (B) by more than twofold (P < 0.05) relative to untreated controls by methyl jasmonate (MeJA), salicylic acid (SA) and/or Puccinia substriata treatments.
A subset of 135 cDNAs that showed the most significant differential expression after SA or MeJA treatment were selected for sequencing. Putative functions were assigned by comparing them to public databases using the blastx program (Altschul et al., 1990) with an E value cut off of 10−5. Eighty five (63%) were found to have homology to previously known genes, and 50 (37%) represented genes with no matches in the database. In total, the selected cDNAs were found to represent 66 UniGenes (51% redundancy). Of the 66 UniGenes, 44 exhibited similarity to annotated genes, and 22 showed no similarity to sequences in the database (Table 1).
Table 1.
Microarray expression profiles of sequenced cDNAs that are more than twofold induced [expression ratio > 2, P < 0.05 (moderated t‐test, Bonferroni corrected, limmaGUI), shaded in grey] or repressed [expression ratio < 0.5, P < 0.05 (moderated t‐test, Bonferroni corrected, limmaGUI), underlined] by methyl jasmonate (MeJA), salicylic acid (SA) or Puccinia substriata treatments (relative to t = 0 h). Clone names in italics represent genes that are differentially expressed only in response to SA, but not MeJA, treatment.
| Clone | blastx similarity | Gene ID | E‐value | Species | MeJA | SA | P. substriata | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 h | 24 h | 48 h | 12 h | 24 h | 48 h | 20 h | 5 d | 8 d | |||||
| Defence | |||||||||||||
| 2‐H11 | HSP70 | gi|115440955 | 5.E‐38 | Oryza sativa | 1.1 | 0.9 | 1.0 | 3.3 | 1.1 | 0.9 | 1.2 | 0.5 | 0.2 |
| 3‐B6 | Serine carboxypeptidase | gi|115480844 | 4.E‐48 | Oryza sativa | 1.2 | 0.8 | 3.2 | 6.4 | 4.6 | 3.5 | 8.3 | 17.7 | 91.8 |
| 4‐A1 | Pathogenesis‐related protein 1 | gi|75994061 | 4.E‐23 | Zea mays | 1.1 | 2.9 | 2.3 | 1.1 | 2.2 | 2.2 | 16.9 | 67.4 | 49.5 |
| 5‐B12 | Calcium‐binding EF‐hand protein | gi|6900307 | 1.E‐53 | Hordeum vulgare | 2.2 | 0.7 | 1.7 | 4.9 | 3.0 | 3.2 | 0.5 | 0.0 | 0.5 |
| 6‐H2 | S‐Adenosylmethionine decarboxylase | gi|3913427 | 5.E‐57 | Zea mays | 1.3 | 1.3 | 1.3 | 2.1 | 1.4 | 1.1 | 1.1 | 1.3 | 1.4 |
| 8‐B2 | UDP‐glucose: salicylic acid glucosyltransferase | gi|115480183 | 1.E‐07 | Oryza sativa | 1.3 | 1.8 | 2.7 | 6.1 | 2.8 | 3.6 | 72.5 | 49.5 | 7.8 |
| 8‐D7 | MATE transporter protein | gi|34394668 | 9.E‐37 | Oryza sativa | 1.8 | 0.7 | 1.6 | 3.4 | 1.8 | 1.5 | 0.8 | 0.7 | 0.6 |
| 10‐C3 | Ethylene‐responsive element binding protein | gi|145390028 | 1.E‐07 | Oryza sativa | 1.6 | 2.5 | 1.6 | 1.2 | 1.0 | 0.9 | 1.8 | 1.6 | 1.4 |
| 11‐A10 | Brown plant hopper susceptibility protein | gi|33771375 | 2.E‐05 | Oryza sativa | 0.5 | 1.4 | 1.1 | 0.6 | 1.4 | 0.7 | 2.1 | 1.7 | 1.8 |
| 11‐F3 | β‐Glucosidase | gi|12746303 | 1.E‐43 | Musa acuminata | 1.5 | 2.7 | 1.5 | 2.4 | 2.1 | 1.5 | 2.4 | 3.4 | 3.8 |
| 15‐G7 | Thionin | gi|246215 | 1.E‐14 | Hordeum jubatum | 5.3 | 3.3 | 3.2 | 2.4 | 2.6 | 6.0 | 1.1 | 1.3 | 1.4 |
| 16‐E11 | Pore‐forming toxin‐like protein Hfr‐2 | gi|57233444 | 4.E‐14 | Triticum aestivum | 1.8 | 5.7 | 5.2 | 1.1 | 0.6 | 1.0 | 0.8 | 0.7 | 0.9 |
| 19‐H3 | Leucine‐rich repeat protein | gi|108862896 | 8.E‐82 | Oryza sativa | 1.7 | 1.9 | 2.1 | 1.4 | 1.0 | 0.9 | 3.2 | 2.6 | 1.7 |
| Metabolism | |||||||||||||
| 01‐H12 | C4 phosphoenolpyruvate carboxylase | gi|20257597 | 2.E‐22 | Setaria italica | 1.1 | 0.9 | 1.6 | 1.8 | 2.3 | 2.9 | 0.4 | 0.9 | 0.7 |
| 1‐D3 | Farnesyl‐pyrophosphate synthetase | gi|115439441 | 7.E‐62 | Oryza sativa | 2.2 | 2.5 | 2.6 | 1.2 | 2.3 | 1.6 | 1.1 | 2.4 | 0.4 |
| 5‐B6 | Phosphoglycerate kinase | gi|129916 | 9.E‐57 | Triticum aestivum | 1.1 | 1.0 | 1.1 | 2.2 | 1.6 | 1.2 | 0.8 | 1.3 | 1.5 |
| 6‐B6 | Alanine aminotransferase | gi|461498 | 3.E‐18 | Panicum miliaceum | 0.9 | 1.0 | 1.4 | 2.7 | 1.8 | 1.3 | 1.4 | 1.1 | 1.1 |
| 6‐F1 | 2‐Phosphoglycerate dehydrogenase (enolase) | gi|119355 | 3.E‐54 | Zea mays | 3.1 | 2.7 | 2.5 | 2.4 | 2.1 | 1.3 | 0.9 | 0.7 | 0.7 |
| 7‐E2 | Pyrophosphate‐energized vacuolar membrane proton pump | gi|18274925 | 8.E‐47 | Oryza sativa | 5.7 | 5.2 | 4.5 | 1.1 | 1.1 | 1.1 | 7.8 | 11.9 | 8.9 |
| 7‐G5 | Glyceraldehyde‐3‐phosphate dehydrogenase | gi|118498764 | 6.E‐63 | Urochloa decumbens | 14.8 | 15.1 | 11.7 | 4.2 | 2.2 | 0.7 | 1.8 | 0.8 | 0.5 |
| 10‐H1 | Tryptophan synthase alpha chain | gi|115470901 | 1.E‐30 | Oryza sativa | 1.5 | 2.4 | 1.2 | 1.4 | 1.2 | 1.0 | 3.0 | 2.1 | 1.0 |
| 14‐B12 | Glucose‐6‐phosphate isomerase | gi|1346073 | 5.E‐67 | Zea mays | 0.7 | 1.2 | 0.6 | 0.5 | 0.8 | 0.9 | 0.6 | 0.9 | 1.6 |
| 14‐C1 | Pyruvate dehydrogenase kinase isoform 1 | gi|3746431 | 4.E‐24 | Zea mays | 1.8 | 5.0 | 5.7 | 1.1 | 0.8 | 1.3 | 0.4 | 0.6 | 0.9 |
| Oxidative stress | |||||||||||||
| 1‐G9 | Glutaredoxin | gi|55584168 | 9.E‐50 | Oryza sativa | 1.3 | 1.1 | 1.3 | 2.5 | 1.7 | 1.0 | 1.1 | 2.0 | 3.4 |
| 12‐C6 | Peroxidase | gi|115480874 | 6.E‐53 | Oryza sativa | 1.1 | 1.9 | 0.9 | 0.5 | 0.4 | 0.5 | 1.2 | 1.5 | 0.5 |
| 18‐E3 | Catalase isoenzyme 3 | gi|1345683 | 1.E‐21 | Zea mays | 0.3 | 2.9 | 2.5 | 0.3 | 2.6 | 1.2 | 1.0 | 3.3 | 3.6 |
| Photosynthesis | |||||||||||||
| 12‐F9 | Chlorophyll a/b‐binding protein of LHCII type I | gi|115771 | 2.E‐14 | Zea mays | 0.3 | 1.0 | 1.1 | 0.5 | 1.0 | 1.1 | 0.6 | 1.4 | 1.6 |
| 14‐H11 | Chlorophyll a/b‐binding protein of LHCII type III | gi|115793 | 3.E‐34 | Hordeum vulgare | 0.4 | 2.8 | 2.6 | 0.4 | 2.9 | 1.4 | 1.1 | 2.8 | 3.1 |
| 16‐B10 | Photosystem II subunit PsbS precursor | gi|33867383 | 4.E‐47 | Zea mays | 1.7 | 0.6 | 0.9 | 1.9 | 1.0 | 1.2 | 0.3 | 0.5 | 0.5 |
| 18‐D6 | Photosystem I reaction centre subunit II | gi|115477831 | 1.E‐24 | Oryza sativa | 0.6 | 0.6 | 0.4 | 0.8 | 0.6 | 1.0 | 0.4 | 0.9 | 1.6 |
| Protein metabolism | |||||||||||||
| 1‐E5 | Ubiquitin‐associated protein | gi|115447045 | 2.E‐26 | Oryza sativa | 7.0 | 6.7 | 5.0 | 1.4 | 2.0 | 3.3 | 1.3 | 0.8 | 1.0 |
| 2‐E10 | Aspartic proteinase 1 | gi|15186732 | 1.E‐17 | Glycine max | 1.6 | 1.6 | 1.3 | 1.8 | 1.6 | 1.2 | 2.1 | 1.2 | 1.5 |
| 3‐F10 | 26S proteasome regulatory subunit 4 | gi|115474241 | 2.E‐56 | Oryza sativa | 3.1 | 3.7 | 3.5 | 1.5 | 1.4 | 1.0 | 2.6 | 1.1 | 1.5 |
| 7‐A8 | Elongation factor 1‐α | gi|93209512 | 3.E‐16 | Litchi chinensis | 1.1 | 1.2 | 1.1 | 2.0 | 1.8 | 1.3 | 1.4 | 0.9 | 0.8 |
| 7‐E6 | Protein translation factor Sui 1 | gi|115464699 | 4.E‐16 | Oryza sativa | 7.9 | 9.4 | 7.5 | 3.4 | 2.1 | 1.2 | 1.7 | 1.1 | 1.0 |
| Unknown | |||||||||||||
| 4‐B11 | Nodulin MtN3 family protein | gi|18400517 | 8.E‐25 | Arabidopsis thaliana | 3.2 | 1.1 | 1.9 | 4.1 | 1.6 | 1.4 | 0.5 | 0.5 | 0.2 |
| 6‐F5 | Expressed protein | gi|115455153 | 3.E‐10 | Oryza sativa | 2.0 | 0.7 | 1.4 | 1.9 | 3.8 | 6.4 | 1.0 | 4.2 | 14.1 |
| 7‐A10 | Hypothetical protein | gi|125528888 | 2.E‐06 | Oryza sativa | 2.6 | 2.7 | 2.1 | 1.0 | 0.9 | 0.8 | 1.4 | 1.6 | 1.2 |
| 13‐C4 | Retrotransposon protein, Ty1‐copia subclass | gi|77555220 | 4.E‐19 | Oryza sativa | 1.1 | 0.9 | 0.9 | 0.9 | 0.6 | 1.0 | 2.0 | 1.0 | 0.8 |
| 13‐D2 | Rhodanese‐like domain‐containing protein | gi|115447077 | 1.E‐42 | Oryza sativa | 0.9 | 1.1 | 0.7 | 0.5 | 0.5 | 0.7 | 1.1 | 0.8 | 0.7 |
| 13‐G1 | Dehydration‐responsive protein RD22 | gi|115477082 | 3.E‐17 | Oryza sativa | 1.4 | 9.5 | 4.9 | 1.9 | 4.0 | 1.4 | 1.9 | 1.6 | 1.0 |
| 15‐G10 | Expressed protein | gi|198345411 | 7.E‐30 | Panicum virgatum | 3.8 | 0.4 | 0.7 | 8.1 | 1.0 | 1.6 | 0.6 | 0.3 | 0.2 |
| 16‐B8 | ABA response protein | gi|584786 | 6.E‐37 | Solanum lycopersicon | 0.9 | 1.7 | 2.4 | 0.6 | 2.4 | 2.3 | 1.0 | 1.4 | 3.3 |
| 20‐G6 | bZIP transcription factor | gi|108862927 | 8.E‐05 | Oryza sativa | 0.4 | 0.8 | 0.7 | 0.2 | 0.8 | 0.7 | 0.8 | 1.1 | 1.6 |
| 1‐G12 | No significant similarity | 3.8 | 4.5 | 3.7 | 1.5 | 1.1 | 0.9 | 1.3 | 0.8 | 0.8 | |||
| 2‐D3 | No significant similarity | 2.5 | 1.6 | 1.4 | 1.9 | 1.3 | 1.1 | 1.2 | 1.3 | 0.5 | |||
| 3‐C6 | No significant similarity | 17.3 | 21.0 | 16.2 | 4.1 | 2.6 | 1.1 | 2.0 | 0.8 | 1.0 | |||
| 4‐A9 | No significant similarity | 2.0 | 2.2 | 1.9 | 1.2 | 1.3 | 0.8 | 1.6 | 0.9 | 1.5 | |||
| 5‐B1 | No significant similarity | 0.8 | 1.5 | 1.4 | 1.8 | 1.7 | 0.8 | 3.0 | 1.7 | 1.5 | |||
| 5‐B9 | No significant similarity | 2.0 | 2.0 | 1.8 | 0.9 | 1.0 | 0.8 | 1.0 | 1.2 | 1.4 | |||
| 5‐F7 | No significant similarity | 2.6 | 2.4 | 2.8 | 1.0 | 3.0 | 2.1 | 1.1 | 0.9 | 1.1 | |||
| 5‐H11 | No significant similarity | 0.6 | 0.3 | 0.4 | 0.9 | 1.3 | 0.9 | 0.1 | 0.2 | 0.5 | |||
| 6‐A1 | No significant similarity | 2.2 | 2.1 | 1.8 | 1.1 | 1.0 | 1.1 | 0.9 | 0.9 | 0.8 | |||
| 6‐A4 | No significant similarity | 0.4 | 0.3 | 0.3 | 0.7 | 0.5 | 0.9 | 0.4 | 0.7 | 1.3 | |||
| 6‐C3 | No significant similarity | 7.8 | 8.7 | 8.2 | 2.9 | 1.7 | 1.4 | 2.1 | 1.2 | 0.9 | |||
| 6‐E1 | No significant similarity | 5.3 | 6.0 | 4.6 | 1.9 | 1.5 | 1.3 | 2.1 | 1.0 | 0.5 | |||
| 6‐F5 | No significant similarity | 5.5 | 6.2 | 5.4 | 1.9 | 1.5 | 1.0 | 2.0 | 1.2 | 0.4 | |||
| 6‐G9 | No significant similarity | 4.0 | 4.5 | 3.8 | 1.4 | 0.9 | 0.9 | 2.0 | 1.7 | 0.7 | |||
| 6‐H12 | No significant similarity | 4.7 | 5.4 | 3.6 | 1.6 | 1.3 | 0.8 | 2.0 | 1.2 | 0.9 | |||
| 7‐A7 | No significant similarity | 1.0 | 0.7 | 1.5 | 1.9 | 2.5 | 3.2 | 0.3 | 0.9 | 0.8 | |||
| 8‐F10 | No significant similarity | 1.0 | 1.3 | 1.2 | 1.2 | 1.2 | 0.8 | 2.4 | 2.0 | 1.7 | |||
| 10‐C4 | No significant similarity | 1.4 | 2.5 | 1.1 | 1.0 | 0.6 | 0.9 | 2.2 | 2.0 | 1.3 | |||
| 11‐F11 | No significant similarity | 0.6 | 0.8 | 1.0 | 0.5 | 1.7 | 1.4 | 1.0 | 1.8 | 2.1 | |||
| 14‐B2 | No significant similarity | 1.8 | 0.8 | 1.9 | 0.6 | 0.7 | 1.1 | 1.1 | 0.4 | 0.5 | |||
| 15‐B6 | No significant similarity | 0.9 | 1.6 | 1.3 | 0.7 | 2.0 | 1.1 | 1.9 | 2.5 | 2.8 | |||
| 16‐B9 | No significant similarity | 0.6 | 1.5 | 1.3 | 0.6 | 2.1 | 1.1 | 1.5 | 2.2 | 2.7 | |||
In this study, a number of candidate genes were significantly induced by SA, but not up‐regulated to the same extent by MeJA (Table 1). Some highly SA‐responsive genes included well‐characterized defence response genes encoding a uridine diphosphate glucose:salicylic acid glucosyltransferase (UDP‐glucose:SA‐GTase), a glutaredoxin, a multidrug and toxin extrusion (MATE) transporter protein, heat shock protein 70 (HSP70), a serine carboxypeptidase, a calcium‐binding EF‐hand protein, S‐adenosylmethionine (SAM) decarboxylase and a peroxidase (Table 1). Several genes encoding components of photosynthesis were down‐regulated by SA (Table 1).
Interestingly, transcripts of thionin (a marker in dicots of the MeJA defence pathway), PR protein 1 (PR1, a marker in dicots of the SA defence pathway), a β‐glucosidase, a β‐ZIP transcription factor, a nodulin‐like protein, a dehydration‐responsive protein RD22 and an ABA response protein were up‐regulated by both MeJA and SA treatment (Table 1). Putative defence‐related genes that were significantly and specifically up‐regulated by MeJA treatment included those encoding an ET‐responsive element binding protein, a pore‐forming toxin‐like protein Hfr‐2, a leucine‐rich repeat protein and a ubiquitin‐associated protein (Table 1).
We compared the expression profiles after each chemical treatment with the profile during a compatible rust infection, as we hypothesized that part of the successful strategy of the virulent fungus may be to suppress SA responses and induce MeJA responses. Clustering of the expression data for the pearl millet UniGene set showed that ‘within‐treatment’ expression profiles were more similar to one another than were ‘between‐treatment’ profiles, as expected (Fig. 3). Interestingly, MeJA and SA responses were more similar to one another than to the response to a compatible P. substriata infection (Fig. 3). However, several genes that showed induction by SA and/or MeJA demonstrated strong up‐regulation in response to a compatible rust infection, namely those encoding PR1 (up to 50‐fold), SA‐GTase (up to 73‐fold) and serine carboxypeptidase (up to 92‐fold) (Table 1). In contrast, some genes that were induced by SA were repressed or not expressed during P. substriata infection, namely those encoding HSP70, MATE transporter protein, calcium‐binding EF‐hand protein, SAM decarboxylase, an expressed protein (clone 15‐G10) and thionin (Table 1). These may be targets for manipulation by effectors of the compatible pathogen, as has been shown in other systems (Truman et al., 2007).
Figure 3.
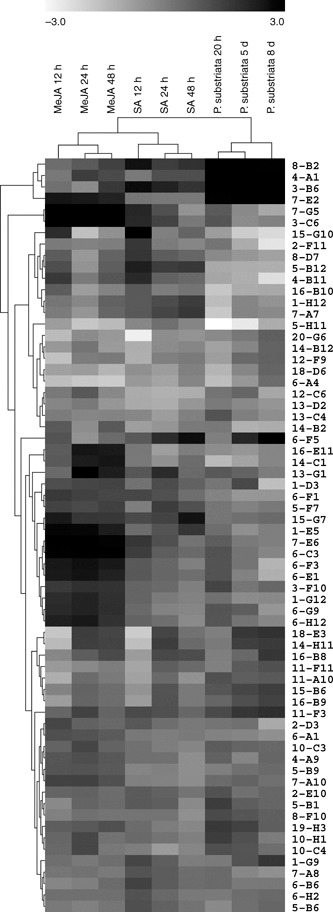
Hierarchical cluster of sequenced pearl millet cDNAs with twofold (P < 0.05) or more changes in transcript abundance in response to methyl jasmonate (MeJA), salicylic acid (SA) or Puccinia substriata treatment. Each gene is represented by a single row of boxes. Each column represents a time point following a particular treatment. Expression ratios relative to time = 0 for each treatment range from white (repressed) to black (induced) with a log2 fold‐change scale bar shown above the cluster.
To verify the differential expression levels of the genes observed in cDNA microarray analysis, quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR) was performed for selected SA‐responsive genes. Greater and/or earlier up‐regulation in response to SA, but not MeJA, was measured by qRT‐PCR for transcripts of seven of the tested genes (Fig. 4A–E,G,H); in particular, transcripts of HSP70, serine carboxypeptidase, MATE transporter protein and alanine aminotransferase were significantly up‐regulated at 12 and 24 h after treatment with SA, but not MeJA (Fig. 4A–D). The calcium‐binding EF‐hand protein showed up‐regulation at all time points after treatment with SA (Fig. 4E), and the expressed protein (clone 15‐G10) and SA‐GTase showed up‐regulation in response to SA at 12 hours post‐treatment (hpt) (Fig. 4G,H). Down‐regulation of peroxidase transcripts in response to SA, but not MeJA, was also observed (Fig. 4F). Expression trends observed over time for each of the treatments using microarray analysis were similar to qRT‐PCR expression trends, with the changes in expression levels observed using qRT‐PCR being similar to or greater than the levels obtained by microarray analysis (Fig. 4). This has been observed previously for comparison of microarray and PCR‐based estimates of gene expression changes (Salzman et al., 2005). The expression of selected genes in response to compatible P. substriata infection was also verified using qRT‐PCR, and the results mirrored the microarray data, namely SA‐GTase was up‐regulated in response to both SA and P. substriata, whereas calcium‐binding EF‐hand protein, thionin and the expressed protein (clone 15‐G10), which were induced by SA, were repressed during P. substriata infection, indicating possible manipulation by the pathogen (Table 1; qRT‐PCR data not shown).
Figure 4.
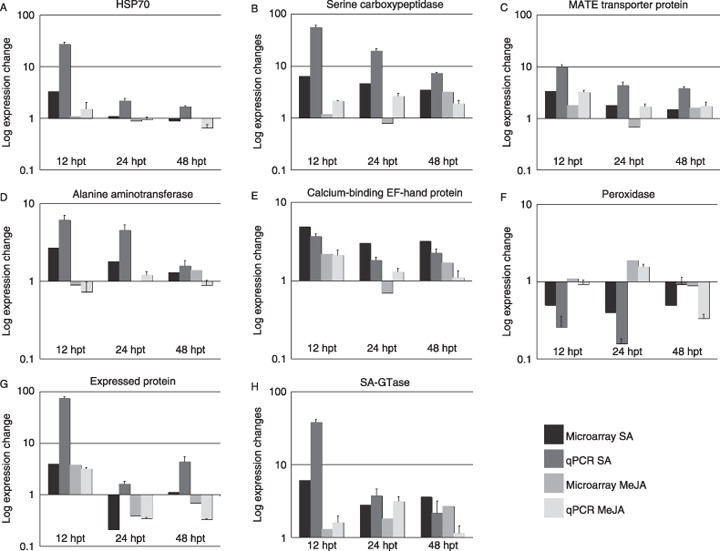
Differential expression of salicylic acid (SA)‐responsive genes in pearl millet measured by microarray analysis was confirmed by quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR). Relative abundance of mRNA at 12, 24 and 48 h after treatment of pearl millet (cultivar ICML12=P7) with SA (dark boxes) or methyl jasmonate (MeJA) (light boxes) was measured by qRT‐PCR using primers designed to genes encoding the following proteins: (A) heat shock protein 70 (HSP70); (B) serine carboxypeptidase; (C) multidrug and toxin extrusion (MATE) transporter protein; (D) alanine aminotransferase; (E) calcium‐binding EF‐hand protein; (F) peroxidase; (G) expressed protein (clone 15‐G10); and (H) salicylic acid glucosyltransferase (SA‐GTase). Expression data were normalized using the standard curves for each target gene and the endogenous control gene (pearl millet 18S rRNA). Expression data are given as log(expression change) for each time point relative to untreated samples (t = 0 h). Error bars represent standard deviations for three biological replicates. Microarray data for the same samples were calculated from Table 1.
DISCUSSION
SA mediates induced disease resistance in pearl millet and other cereals
Under our test conditions, SA is effective in reducing rust disease in pearl millet caused by P. substriata (Fig. 1). In contrast, prior application of MeJA to pearl millet plants, followed by subsequent P. substriata inoculation, did not reduce the number of rust pustules formed relative to the water control, but showed a small but not statistically significant increase in pustule formation (Fig. 1). These results suggest that the SA defence pathway is responsible for resistance to P. substriata in pearl millet plants. P. substriata is a biotrophic fungal pathogen, and thus our data further support evidence from dicotyledonous systems that resistance to biotrophic pathogens is commonly regulated by the SA‐dependent pathway.
Similar results have been obtained in other cereals, in which the application of BTH, an analogue of SA, has conferred resistance to the biotrophic fungal pathogens Blumeria graminis f.sp. hordei (Bgh) in wheat and barley, Puccinia recondita in wheat (Beßer et al., 2000; Gorlach et al., 1996), Peronosclerospora sorghi in maize (Morris et al., 1998) and Magnoporthe grisea in rice (Shimono et al., 2007). BTH, 2,6‐dichloroisonicotinic acid (DCINA) and 2,5‐dichlorosalicylic acid (DCSA) were more potent inducers than SA of both defence gene expression and resistance to Bgh in barley (Beßer et al., 2000; Kogel et al., 1995).
SA‐responsive genes in pearl millet have defence response homologues in other plants
Examination of the SA‐specific genes from pearl millet (Table 1) indicated that a number of homologues in other plant species have previously been shown to play an important role in the defence response. UDP‐glucose:SA‐GTase is induced in SA‐treated or tobacco mosaic virus‐inoculated tobacco leaves (Enyedi and Raskin, 1993). SA‐GTase enzymes convert SA to SA‐2‐O‐β‐d‐glucose (SAG, also known as SGA) or SA glucose ester (SGE) (Dean et al., 2005). Studies in tobacco cell suspensions have shown that most SAG is produced by a cytoplasmic SA‐GTase, and then transported to the vacuoles by an H+‐ATPase (Dean et al., 2005). Two SA‐GTase paralogues in Arabidopsis, UGT74F1 (encoded by At2g43840) and UGT74F2 (encoded by At2g43820) (also called ATSF1; Song, 2006), are induced by Pseudomonas syringae infection and SA (Dean and Delany, 2008; Song, 2006). Pearl millet SA‐GTase, identified in this study, shows greater sequence similarity to the main producer of SAG in Arabidopsis (UGT74F1) than to UGT74F2 (data not shown). In rice, sevenfold and 60‐fold inductions of SA‐GTase were observed after treatment with exogenous SA and BTH, respectively (Shimono et al., 2007; Silverman et al., 1995).
HSP70 and a MATE transporter protein were found to be up‐regulated when pearl millet was treated with SA, but not MeJA. Virus‐induced gene silencing (VIGS) of HSP70 has shown that this protein is an essential component of the plant defence signal transduction pathway (Kanzaki et al., 2003). MATE transporter proteins are putative secondary transporters, unique to plants and microbes, that remove toxins and secondary metabolites from the plant cell cytoplasm for storage in the vacuole (Diener et al. 2001). EDS5, an SA‐inducible component of SA‐dependent disease resistance in Arabidopsis, is a MATE transporter protein (Nawrath et al., 2002). MATE transporters have been shown to be induced in barley during two different incompatible interactions with Pyrenophora teres (Bogacki et al., 2008), and in maize in response to Cochliobolus heterostrophus and Cochliobolus carbonum (Simmons et al., 2003).
SA has been found to induce genes involved in basic and secondary metabolism that have links with the plant defence response. Serine carboxypeptidase is a wound‐inducible gene product (Moura et al., 2000) that functions in signal transduction (Li et al., 2001). A recent study by Liu et al. (2008) outlined the isolation of a serine carboxypeptidase‐like gene from rice that was significantly up‐regulated after treatments with BTH, SA, JA and 1‐amino cyclopropane‐1‐carboxylic acid (ACC), and also up‐regulated in incompatible interactions between rice and the blast fungus M. grisea. Transgenic plants overexpressing the rice serine carboxypeptidase‐like gene showed enhanced resistance to Pseudomonas syringae pv. tomato and Alternaria brassicicola, as well as increased resistance to oxidative stress and up‐regulated expression of oxidative stress‐related genes. SAM decarboxylase catalyses the conversion of SAM into decarboxylated SAM, which provides the aminopropyl moiety required for spermidine and spermine biosynthesis from putresine. Spermine has been hypothesized to act as an inducer of PR proteins, and as a trigger for caspase‐like activity and hence HR (Walters, 2003).
Although a calcium‐binding EF‐hand protein gene was found to be up‐regulated in pearl millet in response to both MeJA and SA treatments, microarray data indicated that this gene was significantly up‐regulated at all time points after SA treatment (Table 1), whereas it was only up‐regulated at 12 hpt with MeJA. These results were confirmed by qRT‐PCR (Fig. 4). Calcium‐binding EF‐hand proteins are one of four similar monomers which form a multiprotein calcium‐dependent protein kinase (CDPK). Calcium signalling is known to play a role in the response to pathogens (Harmon et al., 2000). The treatment of barley plants with SA or its analogues, and wheat with an incompatible rust pathogen, also resulted in the induction of a Ca‐binding EF‐hand protein (Beßer et al., 2000; Coram et al., 2008).
Studies in both rice (Yang et al., 2004) and Arabidopsis (Mateo et al., 2006) have shown that SA plays an important role in modulating the redox balance and protecting against oxidative stress. Our study also indicates that SA may play a role in protecting pearl millet from oxidative damage, as certain genes involved in oxidative stress tolerance, namely glutaredoxin, peroxidase and catalase (Table 1), are affected by SA treatment. Glutaredoxins protect plants from oxidative stress by catalysing dithiol‐disulphide exchange reactions or reducing protein‐mixed glutathione disulphides (Rouhier et al., 2006). Interestingly, some glutaredoxin targets include catalases and peroxidases, as well as alanine aminotransferase and HSP (Marchand et al., 2004; Rouhier et al., 2006).
Zhu‐Salzman et al. (2004) examined transcriptional regulation in sorghum in response to SA, MeJA and a phloem‐feeding aphid, and Salzman et al. (2005) profiled sorghum's response to SA, MeJA and ACC. Comparison of SA and MeJA treatments from these studies with our study of pearl millet indicates similarities in gene expression responses. Alanine aminotransferase and the calcium‐binding EF‐hand protein genes were induced in response to SA treatment in both species, and chlorophyll a/b‐binding protein gene expression was suppressed. In sorghum, catalase, HSP and serine carboxypeptidase genes were also induced in response to SA treatment, but, unlike pearl millet, these genes were also induced by MeJA treatment. Furthermore, a β‐glucosidase gene was up‐regulated in response to SA and MeJA in both species.
PR gene expression in pearl millet in response to SA and MeJA
Although the expression patterns of PR genes vary in different plant species (Ryals et al., 1996), the induction of PR2, PR 5 and, particularly, PR1 by pathogens and chemicals occurs in most dicots and, consequently, these genes have often been used as markers of SAR onset (Lawton et al., 1996). In this study, PR1 was found to be significantly up‐regulated in response to MeJA treatment, but not so significantly after SA treatment, although a twofold average induction was observed (Table 1). Microarray studies of pearl millet after inoculation with a compatible rust pathogen showed considerable induction of PR1 expression (Table 1). Sorghum did not show a significant increase in PR1 gene expression following MeJA or SA treatment (Salzman et al., 2005; Zhu‐Salzman et al., 2004). The expression of genes encoding basic PR1 from barley and acidic PR1 from maize has also been shown to be induced after plant treatment with DCINA and BTH, respectively (Kogel et al., 1994; Morris et al., 1998; Muradov et al., 1993). However, wheat PR1.1 and PR1.2 genes were induced on infection with either compatible or incompatible isolates of the fungal pathogen Bgh, but these genes did not respond to activators of SAR, such as SA, BTH or DCINA (Molina et al., 1999). Treatment of barley seedlings with DCINA correlated with the accumulation of barley defence‐related genes encoding PR1, peroxidase (PR9) and chitinase, but not β‐1,3‐glucanase (PR2) (Kogel et al., 1994). In contrast with these results, a transcript for peroxidase (PR9) was down‐regulated in pearl millet following SA treatment (Table 1, Fig. 4). A pearl millet β‐1,3‐glucanase gene (GenBank accession number AF488414) showed no significant induction by SA or MeJA when tested by qRT‐PCR (results not shown). β‐1,3‐glucanase was neither induced nor repressed in response to MeJA and SA treatment in sorghum; however, chitinase was expressed in sorghum in response to SA treatment, but not MeJA treatment (Salzman et al., 2005; Zhu‐Salzman et al., 2004).
The onset of chemically induced resistance in barley by DCINA, DCSA, SA and BTH correlated with the accumulation of mRNA encoding thionin (Beßer et al., 2000; Kogel et al., 1995). These authors also showed that the thionin polypeptide exhibited antifungal activity against the biotrophic cereal pathogens Bgh and Puccinia graminis f.sp. tritici. In pearl millet, thionin was also up‐regulated following SA treatment (Table 1), suggesting potential involvement in resistance to the biotrophic pathogen P. substriata. However, the thionin transcript was also up‐regulated following MeJA treatment, which did not effectively reduce P. substriata infection. On the whole, monocotyledonous plants appear to be less uniform than dicots in the expression of particular PR marker genes in response to pathogen infection and SA treatment.
Gene expression in pearl millet during a compatible interaction with P. substriata
Successful virulence of a pathogen on a plant host has been documented as the ability of the pathogen to either evade or manipulate host responses (Jones and Dangl, 2006; Truman et al., 2007). We hypothesized that P. substriata may suppress endogenous SA responses during a compatible interaction and, if SA and MeJA pathways are antagonistic, as observed in dicots, the transcriptome response to P. substriata would be more similar to MeJA than to SA. However, the pearl millet microarray time course data did not reveal this pattern: SA and MeJA profiles were more similar to each other than to the P. substriata profile (Fig. 3). In our experiment, samples were taken at a later time point after P. substriata treatment than after SA or MeJA treatment; however, this was based on the appearance of symptoms during which time defence response signalling would be expected to occur. Profiling the whole transcriptome of pearl millet would provide a global view of expression patterns. Whole genome arrays for pearl millet are not available; however, this approach is now feasible with new transcriptome sequencing technologies on 454 or Solexa platforms. Nevertheless, we observed possible evidence of manipulation of host responses in the compatible interaction. Transcripts of SA‐induced defence genes, such as HSP70, MATE transporter protein, calcium‐binding EF‐hand protein, SAM decarboxylase, an expressed protein (clone 15‐G10) and thionin, were repressed or not induced in response to virulent P. substriata (Table 1).
To our knowledge, this is the first study to apply transcriptome analysis to biotic stress responses in pearl millet. We have identified a number of genes that were significantly differentially expressed in response to SA, but not MeJA, treatment. These are likely to play a role in conferring induced resistance to P. substriata in pearl millet, as well as during incompatible interactions. The latter could not be tested in this study as a pearl millet line with resistance to the South African isolate of P. substriata has not been identified. Functional characterization of these genes using RNA interference (RNAi), VIGS, targeting induced local lesions in genomes (TILLING) or overexpression approaches will confirm their role in resistance to rust disease. Methods are available for the transformation of pearl millet (O’Kennedy et al., 2004) to silence selected genes. VIGS has been demonstrated to work in cereal crops (Hein et al., 2005), and experiments are currently underway to establish this technology in pearl millet.
Future plant production needs to focus on durable resistance strategies that have many advantages over race‐specific resistance traits (Kogel and Langen, 2005). Bion®, the commercial formulation of the SA analogue BTH, has been shown to provide field‐level protection against cereal pathogens, such as powdery mildew, in wheat (Gorlach et al., 1996). Although BTH was not tested in our study, our results indicate that SA analogues, such as Bion® (Gorlach et al., 1996), could provide a solution for farmers to improve their pearl millet yields under rust disease pressure.
EXPERIMENTAL PROCEDURES
Plant and fungal material
Pearl millet breeding line ICML12=P7, obtained from the International Crops Research Institute for the Semi‐Arid Tropics, Patancheru, Andhra Pradesh, India, has been reported (Singh et al., 1990a) to be resistant to downy mildew, caused by the Straminipila Sclerospora graminicola, and a rust pathotype from India (P. pennesiti), now known as Puccinia substriata Ellis & Barth. var. indica Ramachar & Cummin. (Wilson, 2000). Pearl millet seed (line ICML12=P7) was sterilized by briefly rinsing with 70% ethanol, followed by a 20‐min incubation in 0.7% sodium hypochlorite. Following three washes with sterile distilled water, seeds were plated on half‐strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962), and incubated at 25 °C with a 16‐h light/8‐h dark photoperiod. After 1 week, seedlings were transferred to trays containing sterilized vermiculite that had been fertilized with Hoagland's solution (Hoagland and Arnon, 1950). Plants were grown under 16‐h light (140 µmol/m2/s) and 8‐h dark cycles at a constant temperature of 25 °C and 85% relative humidity. Puccinia substriata cultures, isolated from infected pearl millet plants grown in KwaZulu‐Natal, South Africa, were maintained on pearl millet ICML12=P7 plants.
Chemical treatment/pathogenicity trials
Three‐week‐old ICML12=P7 plants were treated with water, 5 mm sodium salicylate (Sigma Aldrich, Irvine, Ayrshire, UK) prepared in 0.1% Tween‐20 or 500 µm MeJA (Sigma) in 0.1% ethanol containing 0.1% Tween‐20 until run off. Plants were incubated for 24 h, after which 50 µL of freshly collected P. substriata urediniospores were applied to the fourth leaf of each plant. Seedlings were incubated in the dark for 2 days at 22 °C, and were then grown in the conditions described in the previous paragraph until rust pustules developed on the leaf surface. Each treatment consisted of two biological replicates, each containing seven plants. The results were analysed in Microsoft Excel using Student's t‐test, assuming unequal variances.
Defence signalling molecule treatment, sampling and RNA isolation
Leaf tissue was harvested from pearl millet plants at 0, 12, 24 and 48 h after treatment with SA or MeJA, and at 0 h, 20 h, 5 days and 8 days after inoculation with P. substriata. Treatment of 7‐week‐old plants with SA or MeJA, or inoculation with P. substriata, was performed as described above. Leaf tissue from two replications of nine plants each was immediately frozen in liquid nitrogen and stored at –80 °C until RNA purification. Total RNA was isolated from frozen leaf tissue using Qiazol™ Lysis Reagent, treated with RNAse‐free DNAseI (Qiagen, Hilden, Germany) and further purified using an RNeasy® Minelute™ Kit (Qiagen).
Microarray experiments
A pearl millet SSH library, which was enriched for genes either up‐ or down‐regulated in pearl millet leaves at various time points following wounding or treatment with the defence elicitors, has been constructed previously and screened using a high‐throughput DNA microarray method (Van den Berg et al., 2004). This quantitative approach allowed us to identify and exclude clones that were not derived from truly up‐ or down‐regulated transcripts (Berger et al., 2007). cDNA microarrays containing 1920 cDNA probes in duplicate were fabricated, processed and scanned at the ACGT Microarray Facility (Pretoria, South Africa) (http://www.microarray.up.ac.za), as described previously (Berger et al., 2007). Targets were prepared by indirect aminoallyl labelling of cDNA from total RNA and subsequent NHS‐Cyanine dye (Amersham BioSciences, Little Chalfont, Buckinghamshire, UK) coupling reactions. Labelled targets for each treatment (SA, MeJA or P. substriata) were hybridized using a direct‐sequential loop design with dye swap (Fig. S1, see Supporting Information) (Naidoo et al., 2005).
Microarray data analysis
Scanned images (tiff images) were imported into GenePix Pro 5.0 (Axon Instruments, Sunnyvale, CA, USA), and the fluorescence intensity for each spot was quantified in both red (Cyanine™‐5 dye) and green (Cyanine™‐3 dye) channels. Grids were predefined and manually adjusted to ensure optimal spot recognition, and spots with dust or locally high background were flagged as bad. Fluorescence data from a total of 12 slides per treatment (SA, MeJA or P. substriata) were imported into limmaGUI (linear models for microarray data Graphical User Interface) (Wettenhall and Smyth, 2004) in the R computing environment, where the data were normalized (‘within‐array’ global loess normalization and ‘between‐array’ quantile normalization), and linear models were fitted in order to contrast post‐treatment expression values with those of the non‐treated sample (time = 0 h). Differentially expressed genes for each treatment were defined as those with a fold‐change (expression ratio) greater than or equal to two between a particular time point and t = 0, and were significant at P ≤ 0.05 (moderated t‐test as implemented in limmaGUI with Bonferroni correction for multiple testing) (Wettenhall and Smyth, 2004). Finally, the data were filtered in Microsoft Excel to retain genes that fulfilled this definition of ‘differentially expressed’ for at least one time point in any of the treatments (SA, MeJA or P. substriata). Hierarchical clustering of expression ratios for the unigene set after treatment with SA, MeJA or P. substriata was carried out using TIGR Multi‐experiment Viewer (MeV) (Saeed et al., 2003) with Euclidean distance measures and average linkage clustering. Microarray data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (Edgar et al., 2002), and are accessible through GEO Series accession number GSE13481 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13481).
DNA sequence analysis
Nucleotide sequences of selected cDNA clones were determined on an ABI PRISM 377 DNA analyser (Perkin‐Elmer Applied Biosystems, Streetsville, Ont., Canada) using a BigDye Termination Cycle Sequencing Ready Reaction Kit (V3) (Perkin Elmer Applied Biosystems). Vector and SSH adaptor sequences were removed manually using Vector NTI® Suite V.6 (InforMax®, North Bethesda, MD, USA) Sequence homologies were determined using blast programs (Altschul et al., 1990) at the NCBI (http://www.ncbi.nih.gov/BLAST). Pearl millet cDNA sequences have been deposited in GenBank dbEST with the accession numbers GD180624–GD180688.
qRT‐PCR
DNAse I (Qiagen)‐treated total RNA from each of the four MeJA and SA post‐treatment time points was reverse transcribed using a Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, Mannheim, Germany). Thereafter, the expression profiles of selected genes were assessed using a LightCycler FastStart DNA MasterPLUS SYBR Green I Kit (Roche Diagnostics). The cycling conditions were as follows: denaturation cycle (95 °C for 10 min); amplification and quantification cycle repeated 40 times (95 °C for 10 s, 58 °C for 10 s, 72 °C for 6 s, with a single fluorescence measurement); melting curve cycle (65–95 °C with a heating rate of 0.1 °C/s and continuous fluorescence measurement); finally, a cooling step to 40 °C.
Primers were designed as balanced pairs of between 58 and 60 °C T m to amplify fragments of between 71 and 112 bp using Primer3 (Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA, USA) and NetPrimer (Premier Biosoft, Palo Alto, CA, USA). The primer sequences were as follows: pearl millet 18S rRNA (5′‐GCCATCGCTCTGGATACATT‐3′; 5′‐TCATTACTCCGATCCCGAAG‐3′); alanine aminotransferase (5′‐GAAGATGCTCGGTCAAAAGG‐3′; 5′‐TCACATTGGTTGTCCTCAGC‐3′); calcium‐binding EF‐hand protein (5′‐TAACATCCGCAGAGATCGAG‐3′; 5′‐ATTAGTCCCCATTCCCCTTC‐3′); HSP70 (5′‐ATCACCGTGTGCTTCGACAT‐3′; 5′‐GCCCTTATCGTTGGTAATCG‐3′); an expressed protein (clone 15‐G10) (5′‐TGTTCTGGTGCAACTCTGCT‐3′; 5′‐ATTGCGGAGGACTGAATCAC‐3′); a MATE transporter protein (5′‐GCTCAAGTTCTACGCCAAGG‐3′; 5′‐CTCCGTGATCTTGGACCATT‐3′); peroxidase (5′‐GGCAATATTAAGCCCGTCAC‐3′; 5′‐CCGCCACATCCATGTTTCTA‐3′); SA‐GTase (5′‐AAGGCAAAGAAGTCCATGAGC‐3′; 5′‐CGCTTCGAGCTATCACCAAT); serine carboxypeptidase (5′‐CTACGTTGGCACCCAAGAGT‐3′; 5′GTGAGGTTGTGGGCGTAAGT‐3′).
Expression data were normalized using the standard curve for the specific target gene and the endogenous control gene, pearl millet 18S rRNA, as described previously (Applied Biosystems, User Bulletin No. 2, 2001).
Supporting information
Fig. S1 Diagrammatic representation of a direct‐sequential loop design applied to analyse microarray gene expression changes in salicylic acid (SA)‐treated pearl millet plants over time. Each circle represents an RNA sample extracted from pearl millet leaves at a specified time post‐SA treatment. The head of the arrow indicates that the sample was labelled with Cyanine™‐5 dye (shown in red), whereas the tail represents a sample that was labelled with the Cyanine™‐3 dye (shown in green). Each arrow represents a single hybridization experiment. The same experimental design was applied to analyse pearl millet plants that had been treated with methyl jasmonate or Puccinia substriata, although samples were taken at 0 h, 20 h post‐inoculation, 5 days post‐inoculation (dpi) and 8 dpi for the latter experiment.
Fig. S2 Example of a pearl millet microarray image following hybridization with differentially labelled RNA samples, and scanning with a Genepix™ 4000B scanner (Axon Instruments). In this particular example, RNA extracted from pearl millet plants 0 h post‐methyl jasmonate (MeJA) treatment was labelled with Cyanine™‐5 dye, and RNA isolated from plants 48 h post‐MeJA treatment was labelled with Cyanine™‐3 dye.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
The authors wish to thank Walter de Miliano for providing P. substriata‐infected pearl millet material, and Daniel Theron, Wiesner Vos and Fourie Joubert for assistance with microarray data analyses. This study was made possible through funding from the African Centre for Gene Technologies (ACGT), CSIR (Council for Scientific and Industrial Research) and NRF (National Research Foundation), South Africa.
REFERENCES
- Alba, R. , Fei, Z. , Payton, P. , Liu, Y. , Moore, S.L. , Debbie, P. , Cohn, J. , D’Ascenzo, M. , Gordon, J.S. , Rose, J.K.C. , Martin, G. , Tanksley, S.D. , Bouzayen, M. , Jahn, M.M. and Giovannoni, J. (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J. 39, 697–714. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Berger, D.K. , Crampton, B.G. , Hein, I. and Vos, W. (2007) Screening cDNA libraries on glass slide microarrays In: Microarrays, Vol. II, Applications and Data Analysis, 2nd edn. (Brampal J., ed.), pp. 177–203. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Beßer, K. , Jarosch, B. , Langen, G. and Kogel, K.‐H. (2000) Expression analysis of genes induced in barley after chemical activation reveals distinct disease resistance pathways. Mol. Plant Pathol. 1, 277–286. [DOI] [PubMed] [Google Scholar]
- Bogacki, P. , Oldach, K.H. and Williams, K.J. (2008) Expression profiling and mapping of defence response genes associated with the barley–Pyrenophora teres incompatible interaction. Mol. Plant Pathol. 9, 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho, A.de.O. , Soares, D.J. , Do Carmo, M.G.F. , De Costa, A.C.T. and Pimentel, C. (2006) Description of the life‐cycle of the pearl millet rust fungus Puccinia substriata var. penicillariae with a proposal of reducing var. indica to a synonym. Mycopathologia, 161, 331–336. [DOI] [PubMed] [Google Scholar]
- Cho, B.H. and Smedegaard‐Petersen, V. (1986) Induction of resistance to Erysiphe graminis f.sp. hordei in near‐isogenic barley lines. Phytopathology, 76, 301–305. [Google Scholar]
- Coram, T.E. , Wang, M. and Chen, X. (2008) Transcriptome analysis of the wheat–Puccinia striiformis f.sp. tritici interaction. Mol. Plant Pathol. 9, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, J.V. and Delany, S.P. (2008) Metabolism of salicylic acid in wild‐type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana . Physiol. Plant 132, 417–425. [DOI] [PubMed] [Google Scholar]
- Dean, J.V. , Mohammed, L.A. and Fitzpatrick, T. (2005) The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta, 221, 287–296. [DOI] [PubMed] [Google Scholar]
- Devos, K.M. , Hanna, W.W. and Ozias‐Akins, P. (2006) Pearl millet In: The Genomes, Vol. 1: Cereals and Millets (Kole C., ed.), pp. 478–506. New Delhi: Indus International. [Google Scholar]
- Diener, A.C. , Gaxiola, R.A. and Fink, G.R. (2001) Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. The Plant Cell, 13, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonovic, S. , Vargas, W.A. , Kolomiets, M.V. , Horndeski, M. , Wiest, A. and Kenerley, C.M. (2007) A proteinaceous elicitor sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. and Lash, AE . (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids. Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi, A.J. and Raskin, I. (1993) Induction of UDP‐glucose:salicylic acid glucosyltransferase activity in tobacco mosaic virus‐inoculated tobacco (Nicotiana tabacum) leaves. Plant Physiol. 101, 1375–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofana, B. , Banks, T.W. , McCallum, B. , Strelkov, S.E. and Cloutier, S. (2007) Temporal gene expression profiling of the wheat leaf rust pathosystem using cDNA microarray reveals differences in compatible and incompatible defence pathways. Int. J. Plant Genomics, 2007, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha, H.M. and Shetty, H.S. (2002) Induction of resistance in pearl millet against downy mildew disease caused by Sclerospora graminicola using benzothiadiazole, calcium chloride and hydrogen peroxide—a comparative evaluation. Crop Prot. 21, 601–610. [Google Scholar]
- Glazebrook, J. , Chen, W. , Estes, B. , Chang, H.‐S. , Nawrath, C. , Métraux, J.‐P. , Zhu, T. and Katagiri, F. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 24, 217–228. [DOI] [PubMed] [Google Scholar]
- Goldman, J.J. , Hanna, W.W. , Fleming, G. and Ozias‐Akins, P. (2003) Fertile transgenic pearl millet [Pennisetum glaucum (L.) R. Br.] plants recovered through microprojectile bombardment and phosphinothricin selection of apical meristem‐, inflorescence‐, and immature embryo‐derived embryogenic tissues. Plant Cell Rep. 21, 999–1009. [DOI] [PubMed] [Google Scholar]
- Gorlach, J. , Volrath, S. , KnaufBeiter, G. , Hengy, G. , Beckhove, U. , Kogel, K.H. , Oostendorp, M. , Staub, T. , Ward, E. , Kessmann, H. and Ryals, J. (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell, 8, 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon, A.C. , Gribskov, M. and Harper, J.F. (2000) CDPKs—a kinase for every Ca2+ signal? Trends. Plant Sci. 5, 154–159. [DOI] [PubMed] [Google Scholar]
- Hein, I. , Barciszewska‐Pacak, M. , Hrubikova, K. , Williamson, S. , Dinesen, M. , Soenderby, I.E. , Sundar, S. , Jarmolowski, A. , Shirasu, K. and Lacomme, C. (2005) Virus‐induced gene silencing‐based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol. 138, 2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland, D.R. and Arnon, D.I. (1950) The water culture method of growing plants without soil. Calif. Agric. Exp. Station Circ. 347. The College of Agriculture, University of California, Berkeley, CA. [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kanzaki, H. , Saitoh, H. , Ito, A. , Fujisawa, S. , Kamoun, S. , Katou, S. , Yoshioka, H. and Terauchi, R. (2003) Cytosolic HSP90 and HSP70 are essential components of INF1‐mediated hypersensitive response and non‐host resistance to Pseudomonas cichorii in Nicotiana benthamiana . Mol. Plant Pathol. 4, 383–391. [DOI] [PubMed] [Google Scholar]
- Kogel, K.‐H. and Langen, G. (2005) Induced disease resistance and gene expression in cereals. Cell. Microbiol. 7, 1555–1564. [DOI] [PubMed] [Google Scholar]
- Kogel, K.H. , Beckhove, U. , Dreschers, J. , Munch, S. and Romme, Y. (1994) Acquired resistance in barley (the resistance mechanism induced by 2,6‐dichloroisonicotinic acid is a phenocopy of a genetically based mechanism governing race‐specific powdery mildew resistance). Plant Physiol. 106, 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogel, K.H. , Ortel, B. , Jarosch, B. , Atzorn, R. , Schiffer, R. and Wasternack, C. (1995) Resistance in barley against the powdery mildew fungus (Erysiphe graminis f.sp. hordei) is not associated with enhanced levels of endogenous jasmonates. Eur. J. Plant Pathol. 101, 319–332. [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Lawton, K.A. , Friedrich, L. , Hunt, M. , Weymann, K. , Delaney, T. , Kessmann, H. , Staub, T. and Ryals, J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82. [DOI] [PubMed] [Google Scholar]
- Li, J. , Lease, K.A. , Tax, F.E. and Walker, J.C. (2001) BRS1, a serine carboxypeptidase regulates BRI1 signaling in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA 98, 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.J. , Witcombe, J.R. , Hash, C.T. , Busso, C.S. , Pittaway, T.S. , Nash, M. and Gale, M.D. (1994) An RFLP‐based genetic map of pearl millet (Pennisetum glaucum ). Theor. Appl. Genet. 89, 481–487. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Wang, X. , Zhang, H. , Yang, Y. , Ge, X. and Song, F. (2008) A rice serine carboxypeptidase‐like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene, 420, 57–65. [DOI] [PubMed] [Google Scholar]
- Maier, W. , Wingfield, B.D. , Mennicken, M. and Wingfield, M.J. (2007) Polyphyly and two emerging lineages in the rust genera Puccinia and Uromyces . Mycol. Res. 111, 176–185. [DOI] [PubMed] [Google Scholar]
- Maleck, K. , Levine, A. , Eulgem, T. , Morgan, A. , Schmid, J. , Lawton, K.A. , Dangl, J.L. and Dietrich, R.A. (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Manjunatha, G. , Roopa, K. , Prashanth, G.N. and Shekar, S.H. (2008) Chitosan enhances disease resistance in pearl millet against downy mildew caused by Sclerospora graminicola and defence‐related enzyme activation. Pest. Manag. Sci., 64, 1250–1257. [DOI] [PubMed] [Google Scholar]
- Marchand, C. , Le Maréchal, P. , Meyer, Y. , Miginiac‐Maslow, M. , Issakidis‐Bourguet, E. and Decottignies, P. (2004) New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics, 4, 2696–2706. [DOI] [PubMed] [Google Scholar]
- Mateo, A. , Funck, D. , Muhlenbock, P. , Kular, B. , Mullineaux, P.M. and Karpinski, S. (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57, 1795–1807. [DOI] [PubMed] [Google Scholar]
- Mishra, R. , Reddy, P. , Nair, S. , Markandeya, G. , Reddy, A. , Sopory, S. and Reddy, M. (2007) Isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of Pennisetum glaucum seedlings. Plant Mol. Biol. 64, 713–732. [DOI] [PubMed] [Google Scholar]
- Molina, A. , Gorlach, J. , Volrath, S. and Ryals, J. (1999) Wheat genes encoding two types of pr‐1 proteins are pathogen inducible, but do not respond to activators of systemic acquired resistance. Mol. Plant–Microbe Interact. 12, 53–58. [DOI] [PubMed] [Google Scholar]
- Morris, S.W. , Vernooij, B. , Titatarn, S. , Starrett, M. , Thomas, S. , Wiltse, C.C. , Frederiksen, R.A. , Bhandhufalck, A. , Hulbert, S. and Uknes, S. (1998) Induced resistance responses in maize. Mol. Plant–Microbe Interact. 11, 643–658. [DOI] [PubMed] [Google Scholar]
- Moura, D.S. , Bergey, D.R. and Ryan, C.A. (2000) Characterization and localization of a wound‐inducible type I serine‐carboxypeptidase from leaves of tomato plants (Lycopersicon esculentum Mill.). Planta, 212, 222–230. [DOI] [PubMed] [Google Scholar]
- Muradov, A. , Petrasovits, L. , Davidson, A. and Scott, K.J. (1993) A cDNA clone for a pathogenesis‐related protein 1 from barley. Plant Mol. Biol. 23, 439–442. [DOI] [PubMed] [Google Scholar]
- Murashige, T. and Skoog, F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Murray, S.L. , Denby, K.J. , Berger, D.K. and Loake, G.J. (2002) Disease resistance signalling in Arabidopsis: applications in the study of plant pathology in South Africa. S. Afr. J. Sci. 98 161–165. [Google Scholar]
- Naidoo, S. , Denby, K.J. and Berger, D.K. (2005) Microarray experiments: considerations for experimental design. S. Afr. J. Sci. 101, 347–354. [Google Scholar]
- National Research Council (1996) Pearl millet In: Lost Crops of Africa: Vol. I: Grains (Board on Science and Technology for International Development , ed.), pp. 77–92. Washington DC: National Academy Press. [Google Scholar]
- Nawrath, C. , Heck, S. , Parinthawong, N. and Metraux, J.P. (2002) EDS5, an essential component of salicylic acid‐dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell, 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kennedy, M.M. , Burger, J.T. and Botha, F.C. (2004) Pearl millet transformation system using the positive selectable marker gene phosphomannose isomerase. Plant Cell Rep. 22, 684–690. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Navarro, L. , Bari, R. and Jones, J.D. (2007) Pathological hormone imbalances. Curr. Opin. Plant Biol. 10, 372–379. [DOI] [PubMed] [Google Scholar]
- Rouhier, N. , Couturier, J. and Jacquot, J.P. (2006) Genome‐wide analysis of plant glutaredoxin systems. J. Exp. Bot. 57, 1685–1696. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A. , Neuenschwander, U.H. , Willits, M.G. , Molina, A. , Steiner, H.Y. and Hunt, M.D. (1996) Systemic acquired resistance. Plant Cell, 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed, A.I. , Sharov, V. , White, J. , Li, J. , Liang, W. , Bhagabati, N. , Braisted, J. , Klapa, M. , Currier, T. , Thiagarajan, M. , Sturn, A. , Snuffin, M. , Rezantsev, A. , Popov, D. , Ryltsov, A. , Kostukovich, E. , Borisovsky, I. , Liu, Z. , Vinsavich, A. , Trush, V. and Quackenbush, J. (2003) TM4: a free, open‐source system for microarray data management and analysis. Biotechniques, 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Salzman, R.A. , Brady, J.A. , Finlayson, S.A. , Buchanan, C.D. , Summer, E.J. , Sung, F. , Klein, P.E. , Klein, R.R. , Pratt, L.H. , Cordonnier‐Pratt, M. and Mullet, J.E. (2005) Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol. 138, 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarosh, B.R. , Sivaramakrishnan, S. and Shetty, H.S. (2005) Elicitation of defense related enzymes and resistance by l‐methionine in pearl millet against downy mildew disease caused by Sclerospora graminicola . Plant Physiol. Biochem. 43, 808–815. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M. , Kazan, K. , Wilson, I. , Anderson, J.P. , Richmond, T. , Somerville, S.C. and Manners, J.M. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA, 97, 11 655–11 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shailasree, S. , Ramachandra, K.K. and Shetty, S.H. (2007) β‐Amino butyric acid‐induced resistance in pearl millet to downy mildew is associated with accumulation of defence‐related proteins. Australas. Plant Pathol. 36, 204–211. [Google Scholar]
- Sharathchandra, R.G. , Raj, S.N. , Shetty, N.P. , Amruthesh, K.N. and Shetty, H.S. (2004) A chitosan formulation Elexa(TM) induces downy mildew disease resistance and growth promotion in pearl millet. Crop. Prot. 23, 881–888. [Google Scholar]
- Shimono, M. , Sugano, S. , Nakayama, A. , Jiang, C.J. , Ono, K. , Toki, S. and Takatsuji, H. (2007) Rice WRKY45 plays a crucial role in benzothiadiazole‐inducible blast resistance. Plant Cell, 19, 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, P. , Seskar, M. , Kanter, D. , Schweizer, P. , Metraux, J.P. and Raskin, I. (1995) Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 108, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, C.R. , Fridlender, M. , Navarro, P.A. and Yalpani, N. (2003) A maize defense‐inducible gene is a major facilitator superfamily member related to bacterial multidrug resistance efflux antiporters. Plant Mol. Biol. 52, 433–446. [DOI] [PubMed] [Google Scholar]
- Singh, S.D. , King, S.B. and Malla Reddy, P. (1990a) Registration of five pearl millet germplasm sources with stable resistance to downy mildew. Crop Sci. 30, 1164. [Google Scholar]
- Singh, S.D. , King, S.B. and Malla Reddy, P. (1990b) Registration of five pearl millet germplasm sources with stable resistance to rust. Crop Sci. 30, 1165. [Google Scholar]
- Smith, J.A. and Metraux, J.P. (1991) Pseudomonas syringae pv. syringae induces systemic resistance to Pyricularia oryzae in rice. Physiol. Mol. Plant Pathol. 39, 451–461. [Google Scholar]
- Song, J.T. (2006) Induction of a Salicylic Acid Glucosyltransferase, AtSGT1, is an early disease response in Arabidopsis thaliana . Mol. Cells 22, 233–238. [PubMed] [Google Scholar]
- Truman, W. , Bennett, M.H. , Kubigsteltig, I. , Turnbull, C. and Grant, M. (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. 104, 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg, N. , Crampton, B.G. , Hein, I. , Birch, P.R.J. and Berger, D.K. (2004) High‐throughput screening of suppression subtractive hybridization cDNA libraries using DNA microarray analysis. Biotechniques, 37, 818–824. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Klessig, D.F. and Park, S.W. (2008) Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11, 436–442. [DOI] [PubMed] [Google Scholar]
- Walters, D. (2003) Resistance to plant pathogens: possible roles for free polyamines and polyamine catabolism. New Phytol. 159, 109–115. [DOI] [PubMed] [Google Scholar]
- Wettenhall, J.M. and Smyth, G.K. (2004) limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics, 20, 3705–3706. [DOI] [PubMed] [Google Scholar]
- Wilson, J.P. (2000) Pearl Millet Diseases: A Compilation of Information on the Known Pathogens of Pearl Millet, Pennisetum glaucum (L.) R. Br. Tifton: United States Department of Agriculture, Agricultural Research Service. [Google Scholar]
- Wilson, J.P. and Gates, R.N. (1999) Disease resistance and biomass stability of forage pearl millet hybrids with partial rust resistance. Plant Dis. 83, 733–738. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Qi, M. and Mei, C. (2004) Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40, 909–919. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Meakin, H. and Dickinson, M. (2003) Isolation of genes expressed during compatible interactions between leaf rust (Puccinia triticina) and wheat using cDNA‐AFLP. Mol. Plant Pathol. 4, 469–477. [DOI] [PubMed] [Google Scholar]
- Zhu‐Salzman, K. , Salzman, R.A. , Ahn, J.‐E. and Koiwa, H. (2004) Transcriptional regulation of sorghum defense determinants against a phloem‐feeding aphid. Plant Physiol. 134, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Diagrammatic representation of a direct‐sequential loop design applied to analyse microarray gene expression changes in salicylic acid (SA)‐treated pearl millet plants over time. Each circle represents an RNA sample extracted from pearl millet leaves at a specified time post‐SA treatment. The head of the arrow indicates that the sample was labelled with Cyanine™‐5 dye (shown in red), whereas the tail represents a sample that was labelled with the Cyanine™‐3 dye (shown in green). Each arrow represents a single hybridization experiment. The same experimental design was applied to analyse pearl millet plants that had been treated with methyl jasmonate or Puccinia substriata, although samples were taken at 0 h, 20 h post‐inoculation, 5 days post‐inoculation (dpi) and 8 dpi for the latter experiment.
Fig. S2 Example of a pearl millet microarray image following hybridization with differentially labelled RNA samples, and scanning with a Genepix™ 4000B scanner (Axon Instruments). In this particular example, RNA extracted from pearl millet plants 0 h post‐methyl jasmonate (MeJA) treatment was labelled with Cyanine™‐5 dye, and RNA isolated from plants 48 h post‐MeJA treatment was labelled with Cyanine™‐3 dye.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


