SUMMARY
Uromyces fabae on Vicia faba is a model system for obligate biotrophic interactions. Searching for potential effector proteins we investigated the haustorial secretome of U. fabae (biotrophic stage) and compared it with the secretome of in vitro grown infection structures, which represent the pre‐biotrophic stage. Using the yeast signal sequence trap method we identified 62 genes encoding proteins secreted from haustoria and 42 genes encoding proteins secreted from in vitro grown infection structures. Four of these genes were identical in both libraries, giving a total of 100 genes coding for secreted proteins. This finding indicates a strong stage‐specific regulation of protein secretion. Similarity with previously identified proteins was found for 39 of the sequences analysed, 28 of which showed similarity to proteins identified among members of the order Uredinales only. This might be taken as an indication for possible roles in virulence and host specificity unique to the Uredinales.
The biotrophic lifestyle is defined by a dependence of the fungal parasite on living host tissue for propagation. Rust fungi and powdery mildew fungi share a common structure, the haustorium. The haustorium provides an intimate contact with the host cell, as its cytoplasm is only separated from the host cytoplasm by the haustorial membrane, the extrahaustorial matrix and the extrahaustorial membrane (Voegele and Mendgen, 2003). This makes the haustorium the ideal structure for nutrient uptake from the plant and also for signalling between host and parasite.
The role of haustoria in nutrient uptake surmised since the discovery and naming of these structures [haurire (lat.) = to drink] by de Bary (1863) was clarified recently (Voegele et al., 2001), whereas a role of these structures in signalling between fungus and plant remains enigmatic. However, there is some evidence that signals are released by the pathogen, not only in order to influence host metabolism to satisfy its nutritional demands, but also to suppress plant defence responses (Heath, 1997a). Possible signals of the fungal partner may include metabolites such as sugars or sugar alcohols. It has been shown that mannitol and d‐arabitol are secreted by Uromyces fabae and have the potential to suppress plant defence responses (Link et al., 2005; Voegele et al., 2005). Effector proteins were first identified in bacteria (Alfano and Collmer, 1996). These parasites are able to inject effector proteins directly into host cells by means of type‐three secretion systems (Mota et al., 2005). However, while a comparable transfer machinery has not been identified in eukaryotes thus far, there are also proteins transferred into the host cell in the eukaryotic host–parasite interactions that occur in malaria (Hiller et al., 2004) and oomycete plant diseases (Kamoun, 2006). Recently, it was shown that proteins are transferred from the haustorium into the plant cytoplasm for Melampsora lini (Dodds et al., 2004) and U. fabae (Kemen et al., 2005). In order to identify new effector proteins we set out to investigate the secretome of U. fabae.
To identify secreted proteins from U. fabae haustoria we used the yeast signal sequence trap method (Jacobs et al., 1997, 1999), which allows selection of signal sequences through complementation of invertase deficiency. Plasmid pSuc2tM13ori and Saccharomyces cerevisiae strain YTK12 (Jacobs et al., 1999) were obtained from Wyeth Research (Cambridge, MA).
RNA was prepared from isolated haustoria as described (Hahn and Mendgen, 1992, 1997) except that pellets enriched with haustoria were resuspended in peqGOLD RNAPure (PeqLab, Erlangen, Germany) immediately after centrifugation and ground in a mortar for 20 min at 4 °C. From this point onward the RNA preparation method followed the recommendations of the manufacturer. mRNA was prepared using the Oligotex Mini Kit (Qiagen, Hilden, Germany) and reverse transcribed (Jacobs et al., 1999). Following addition of an EcoRI cleavage site to the cDNA using an EcoRI–NotI adapter (Stratagene, La Jolla, CA; Table 1), size fractionation was performed using cDNA Size Fractionation Columns (Invitrogen, Carlsbad, CA). cDNA fragment size was determined by ligating aliquots of the fractions into plasmid pSuc2tM13ori (Jacobs et al., 1999) and subsequent colony PCR. Numbers of transformants from this test were also used to determine optimal insert/vector ratios for the main ligation. cDNA fragments with a length no less than 200 bp were cloned into plasmid pSuc2tM13ori. Plasmids were amplified in Escherichia coli (ElectroMAX™ DH10B™, Invitrogen), prepared from 250‐mL cultures by alkali lysis (Heilig et al., 1998) and directly transformed into yeast strain YTK12 (Jacobs et al., 1999) using the lithium acetate method (Becker and Lundblad, 1991) without the addition of carrier DNA. Invertase selection was performed as described (Jacobs et al., 1999; Treco and Lundblad, 1993). We obtained 260 000 clones in E. coli and 650 000 transformants in yeast, of which 1650 tested positive for invertase secretion.
Table 1.
Oligonucleotides used in this study.
| Name | Sequence | Used for |
|---|---|---|
| Random nonamers | 5′‐CGATTGAATTCTAGACCTGCCTCGAGNNNNNNNNA‐3′ | Reverse transcription |
| 5′‐CGATTGAATTCTAGACCTGCCTCGAGNNNNNNNNC‐3′ | ||
| 5′‐CGATTGAATTCTAGACCTGCCTCGAGNNNNNNNNG‐3′ | ||
| 5′‐CGATTGAATTCTAGACCTGCCTCGAGNNNNNNNNT‐3′ | ||
| pSuc5′‐2 | 5′‐CTCGTCATTGTTCTCGTTCCC‐3′ | PCR |
| pSuc5′ | 5′‐CAAGCATACAATCAACTCCAAG‐3′ | PCR and sequencing |
| pSuc3′‐2 | 5′‐CTTCTCTTCTAAATACTCCTCTG‐3′ | PCR and sequencing |
| pSuc3′ | 5′‐GTCAAATCATCGGAAGTAGC‐3′ | PCR |
| EcoRI–NotI | 5′‐AATTCGCGGCCGC‐3′ | Adaptors |
| 3′‐GCGCCGGCG‐5′ |
Positive clones were isolated and inserts were sequenced. For PCR, yeast cells were incubated in 20 µL lysis buffer (1% Triton ×100, 2 mm EDTA, 20 mm Tris, pH 8.9) at 99 °C for 10 min. Three microlitres were transferred to 20 µL PCR‐Mix [75 mM Tris‐HCl (pH 8.8 at 25 °C), 20 mM (NH4)2SO2, 2.5 mm MgCl2, 0.1% BSA, 0.2 mm dNTPs, 0.5 µm primer pSuc5′‐2, 0.5 µm, primer pSuc3′, 0.05 u/µL Taq (Fermentas, St. Leon‐Rot, Germany)]. PCR cycling was done on a Mastercycler Gradient (Eppendorf, Hamburg, Germany). Sequencing was done by GATC (Konstanz, Germany), or by MWG‐Biotech (Ebersberg, Germany).
From the number of clones in E. coli, the total number of transformants in yeast and the number of yeast transformants tested positive for invertase secretion, we calculated that 0.25% of all clones code for secreted proteins and that our library should comprise about 660 different clones. In total, 315 yeast clones were sequenced (this was when only one out of ten clones yielded a new sequence) and sequences were grouped into 82 contigs and unisequences. Trace files were inspected using Chromas2.13 (Technylesium Pty. Ltd, Queensland, Australia). Alignment of DNA and protein sequences was done using the SeqMan and MegAlign modules of the Lasergene software packet (DNASTAR, Madison, WI). Prediction of open reading frames (ORFs) was performed using Gene Runner 3.05 (Hastings Software, Inc., Hudson, NY). Prediction of signal sequences was done using SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP) (Bendtsen et al., 2004) and re‐checked with PsortII (http://psort.ims.u‐tokyo.ac.jp), SigCleave (http://bioweb.pasteur.fr/seqanal/interfaces/sigcleave.html) and Phobius (http://phobius.cgb.ki.se). For all analyses, default settings and cutoffs were used.
Of the initial 82 contigs and unisequences seven had to be discarded because the sequences were so short that Gene Runner was not able to identify an ORF, eight did not have an in silico detectable signal sequence, and four had high similarities to plant (chloroplast) proteins and one to ribosomal RNA. The rate of false positives was well within the range reported by other authors working with the yeast signal sequence trap method (Belanger et al., 2003; Hugot et al., 2004; Jacobs et al., 1997; Klein et al., 1996). Kaiser et al. (1987) showed that about 20% of random sequences can mimic a signal sequence and this again shows that our values were in the expected range.
The remaining 62 sequences supposedly coding for proteins secreted from the haustorium were called Uf‐HSPs (haustorially secreted proteins of U. fabae). BLAST searches (Altschul et al., 1990) on these sequences were performed on the NCBI databases including the genome sequence of Puccinia graminis (http://www.ncbi.nlm.nih.gov/BLAST/), the databases of the Broad Institute (http://www.broad.mit.edu/annotation/fgi/) and PIR (http://pir.georgetown.edu/pirwww/). We found similarities to previously identified proteins for 27 genes other than those already tested for U. fabae. Only for eight Uf‐HSPs were similarities outside the Uredinales noted (Table 2).
Table 2.
Haustorial secreted proteins of U. fabae.
| No* | R† | P‡ | BLAST hits Uredinales | E‐value | BLAST hits others | E‐value |
|---|---|---|---|---|---|---|
| 4 | 3 | A2 | Pgt § G768P6997FB8.T0 | 5E‐16 | ||
| 5 | 3 | A3 | Pgt supercont2.7 1183850‐1183978 | 1.5E‐25 | U. maydis UM06290.1 | 3E‐14 |
| M. lini, HESP‐379 Q2MV38 | 7E‐22 | |||||
| 8 | 1 | A5 | Pgt supercont2.11 934678‐934770 | 2.1E‐18 | F. graminearum glucose regulated protein homolog FG09471.1 | 3E‐12 |
| C. ribicola, Cro r II Q9C1C1 | 3E‐17 | |||||
| 16 | 4 | A6 | Pgt supercont2.41 478877‐479068 | 0.0 | ||
| M. lini, HESP‐735 Q2MV34 | 6E‐8 | |||||
| 21 | 3 | A10 | Pgt supercont2.84 260975‐261352 | 1.5E‐6 | ||
| 36 | 1 | B7 | M. lini, HESP‐C49 Q2MV26 | 1E‐7 | ||
| 38 | 3 | B8 | F. graminearum FG10084.1 | 1E‐9 | ||
| 39 | 1 | B9 | Pgt supercont2.17 571823‐571930 | 1.1E‐10 | ||
| 40 | 2 | B10 | Pgt supercont2.36 736872‐737096 | 9.0E‐19 | ||
| 41 | 2 | B11 | Pgt supercont2.36 562961‐563092 | 6.1E‐13 | ||
| 42a | 10 | B12 | Pgt supercont2.25 856646‐857233 | 4.4E‐27 | ||
| 42c | 7 | C1 | Pgt supercont2.25 856646‐857233 | 3.5E‐25 | ||
| 44 | 3 | C2 | Pgt supercont2.88 168566‐168661 | 8.4E‐22 | ||
| 45 | 1 | C3 | Pgt supercont2.84 260939‐261352 | 1.4E‐8 | ||
| 48 | 1 | C4 | Pgt supercont2.41 283205‐283390 | 9.1E‐7 | ||
| 51 | 2 | C7 | Pgt supercont2.2 1602184‐1602438 | 9.2E‐6 | ||
| 52 | 14 | C8 | Pgt supercont2.25 590077‐590817 | 6.9E‐15 | ||
| 56 | 4 | C10 | Pgt supercont2.51 349176‐349712 | 7.3E‐18 | ||
| 66 | 1 | D4 | Pgt supercont2.19 744692‐744802 | 1.2E‐8 | ||
| 69 | 1 | D7 | P. triticina chitinase Q7ZA28 | 2E‐34 | F. graminearum FG03212.1 | 3E‐12 |
| 74 | 1 | D9 | Pgt supercont2.8 969421‐969621 | 4.8E‐9 | S. pombe trehalose‐6p phosphatase gi|19115342|ref|NP_594430.1 | 3E‐6 |
| 83 | 1 | E4 | Pgt supercont2.3 2565537‐2565830 | 1.5E‐33 | U. maydis UM00567.1 | 8E‐34 |
| F. neoformans B2‐aldehyde forming enzyme Q5KCB0 | 2E‐18 | |||||
| 84 | 1 | E5 | Pgt supercont2.19 1098198‐1098407 | 3.3E‐31 | M. sympodialis, Allergen O93972 | 4E‐20 |
| 97 | 1 | E9 | Pgt supercont2.4 531225‐531458 | 5.1E‐21 | ||
| 98 | 1 | E10 | Pgt supercont2.22 467324‐467842 | 1.4E‐6 | S. cerevisiae gi|6322237 | 9E‐9 |
| 116 | 1 | — | Pgt supercont2.8 911042‐911119 | 4.5E‐6 | ||
| 117 | 1 | — | Pgt supercont2.21 821694‐821816 | 1.1E‐4 |
Proteins are numbered in the order of their identification. Homologies are divided in homologues among the Uredinales and outside the Uredinales.
Uf‐HSP number.
Redundancy of sequences of a given protein.
Position on macroarray.
Puccinia graminis f. sp. tritici.
From the redundancy of clones per contig the relative expression level of Uf‐HSPs can be estimated (Hahn and Mendgen, 1997). We found few proteins to be very strongly expressed, whereas most could be identified only once. We also compared our results with the haustorial expressed sequence tag (EST) library of U. fabae (Jakupovic et al., 2006). Here we found 25 matches. Supplementary Table S1 shows the full set of Uf‐HSPs. A comparison of these data sets showed good correlation with respect to expression level.
In order to complement the haustorial secretome, which is equivalent to the biotrophic stage, and to be able to compare it with a different developmental stage we analysed the secretome of U. fabae in vitro grown infection structures representing the pre‐biotrophic stage of the dicaryotic phase.
We used in vitro grown infection structures which had grown on polyethylene membranes for 22 h as described (Deising et al., 1991). These represent pre‐biotrophic infection structures up to the haustorial mother cell. Preparation of RNA from urediospores and in vitro infection structures followed US patent 5,973,137 (Purescript®, Gentra‐Systems Inc., Minneapolis, MN).
Again using the signal sequence trap we obtained 150 000 clones in E. coli and 250 000 clones in yeast. In total, 2800 yeast clones were positive for secreted invertase.
Based on these numbers we estimated that 1.1% of all mRNAs code for secreted proteins and that our library should comprise about 1800 different clones of secreted proteins. Two hundred and fifty yeast clones were sequenced (this was when only one out of ten clones yielded a new sequence) and sequences were grouped into 51 contigs and unisequences. Nine sequences had to be discarded (upstream ORFs, ribosomal genes or ubiquitin), so that 42 genes encoding in vitro infection structure secreted proteins (Uf‐ISSPs) remained. The results of database searches for these and redundancy of clones are given in Table 3. We found similarities to 15 proteins. For six proteins similarities outside the Uredinales were found.
Table 3.
Secreted proteins of U. fabae in vitro infection structures.
| No.* | R† | BLAST hits Uredinales | E‐value | BLAST hits others | E‐value |
|---|---|---|---|---|---|
| 4 | 4 | M. lini, HESP‐379 Q2MV38 | 5E‐22 | U. maydis UM01555.1 | 9E‐10 |
| Pgt ‡ supercont2.7 1181504‐1181632 | 9.4E‐20 | ||||
| 7 | 1 | Pgt supercont2.36 736872‐737087 | 4.8E‐14 | ||
| 10 | 3 | Pgt supercont2.3 702991‐703086 | 1.5E‐12 | ||
| 11 | 3 | Pgt supercont2.14 296966‐297481 | 1.2E‐42 | P. nodorum gb|EAT83001.1| | 2E‐19 |
| 12 | 2 | Pgt supercont2.161 59636‐59884 | 2.2E‐17 | ||
| 18 | 1 | Pgt supercont2.7 1184076‐1184180 | 9.5E‐19 | U. maydis UM06290.1 | 2E‐8 |
| M. lini, HESP‐379 Q2MV38 | 2E‐11 | ||||
| 24 | 6 | Pgt supercont2.36 562961‐563092 | 5.6E‐13 | ||
| 34 | 2 | Pgt supercont2.27 414813‐415091 | 1.6E‐11 | F. neoformans, Chitin deacetylase, putative Q5KFG8 | 5E‐7 |
| 42 | 2 | Pgt supercont2.17 571808‐571984 | 1E‐15 | ||
| 43 | 1 | Pgt supercont2.36 42792‐442911 | 1.4E‐6 | A. nidulans AN3310.2 | 3E‐13 |
| 44 | 7 | Pgt supercont2.7 1130413‐1130688 | 1.1E‐12 | ||
| 48 | 2 | Pgt supercont2.30 821627‐821830 | 5.3E‐23 | ||
| 62 | 1 | P. triticina chitinase Q7ZA28 | 8E‐10 | ||
| 66 | 1 | M. grisea MG08593.4 | 7E‐06 | ||
| 68 | 1 | Pgt supercont2.16 164395‐164553 | 1.6E‐8 |
Proteins are numbered in the order of their identification. Homologies are divided in homologues among the Uredinales and outside the Uredinales.
Uf‐ISSP number.
Redundancy of sequences.
Puccinia graminis f. sp. tritici.
Three matches were obtained with the haustorial EST library (Jakupovic et al., 2006). A comparison of the two secretome libraries gave four identities: Uf‐ISSP7 = Uf‐HSP40, Uf‐ISSP18 = Uf‐HSP5, Uf‐ISSP24 = Uf‐HSP41 and Uf‐ISSP54 = Uf‐HSP20. The full set of Uf‐ISSPs is presented in supplementary Table S2.
Summarizing the results obtained with our two libraries, we found 100 genes encoding proteins secreted by U. fabae. Thirty‐nine of these genes had similarities in the public databases and 28 of these had similar sequences among the Uredinales only.
Yeast signal sequence trap secretome libraries can be affected by cloning bias (Lee et al., 2006) and specific genes may be left out because cDNA fusions might be deleterious to translation or to functionality of the invertase, i.e. through steric hindrance or folding defects (Jacobs et al., 1997). It is also possible that some signal sequences defy the theory that signal peptides are functional among all organisms and are not functional in yeast (Jacobs et al., 1997). Yet, the major reason why genes may have not been picked up in our screen is a weak level of expression as sequencing of the libraries was not done to complete saturation.
Before the start of this study screening of a haustorium‐specific EST library had yielded nine genes that are expressed in haustoria and possess a signal peptide: PIG5, PIG9, PIG12, PIG14, PIG15, PIG23, BGL1, INV1 and RTP1 (Haerter and Voegele, 2004; Kemen et al., 2005; Voegele et al., 2006; M. Hahn, unpublished data). Of these, PIG5, PIG9, PIG12, PIG15 and BGL1 were found again in this study. Given this fact and knowing that we had sequenced about half of all independent clones of the library, we suggest that our library represents about half of the haustorial secretome. The library covering in vitro grown infection structures was sequenced to similar saturation. No proteins secreted from in vitro infection structures had been identified thus far, although activities of several plant cell‐wall‐degrading enzymes have been detected (Deising et al., 1995; Hahn et al., 1997). The fact that no plant cell‐wall‐degrading enzymes could be identified in this study can be explained by a low expression level of these enzymes compared with other secreted proteins, which takes them below the detection limit of our library.
Based on the assumption that our libraries represent about half of the complete secretome of U. fabae, this can be estimated to consist of about 200 proteins. Compared with the secretomes of the necrotrophic fungal pathogen Magnaporthe grisea, which was shown to comprise 739 proteins (Dean et al., 2005), or the saprotroph Neurospora crassa with about half that number (Dean et al., 2005) our number seems to be very low. On the other hand, there are also reports indicating much lower numbers. The secretome of Candida albicans, for example, was predicted to comprise fewer than 300 proteins (Lee et al., 2003), and Catanzariti et al. (2006) found only 20 haustorially expressed secreted proteins (HESPs) in M. lini. Both values are much closer to the numbers found in our study. The number of secreted proteins may vary strongly between different fungi. Our results also corroborate the assumption that secretory activity among obligate biotrophs is limited and strongly regulated (Mendgen and Hahn, 2002). Yet the total number of secreted proteins of U. fabae could only be inferred from genomic studies, or by additionally investigating other developmental stages, i.e. parts of the haploid phase.
The results of our database searches giving only few similar sequences and most of them among the Uredinales are similar to those of Catanzariti et al. (2006) who found only four homologues for their HESPs (note: the genome sequence of P. graminis was not then finished). Most of the proteins secreted from the haustorium therefore seem to be specific to U. fabae or at least to rust fungi. Of the nine similar sequences outside the Uredinales five stem from the plant pathogenic fungi Ustilago maydis and Fusarium graminearum. Taken together, these results strongly indicate that large portions of the haustorial secretome of U. fabae may have virulence functions. The fact that more than half of the Uf‐HSPs do not have similarity among other rust fungi could be due to host specificity. Plant responses against pathogen attack depend on the recognition of the pathogen. This can be effected either by Pathogen Associated Molecular Pattern (PAMP) receptors, which constitute plant innate immunity, or R‐proteins, which specifically recognize avirulence proteins (Avr proteins) in a gene for gene manner (Jones and Dangl, 2006). This exerts a strong selective pressure on effectors. Consistent with this hypothesis, Tyler et al. (2006) found accelerated evolution among secreted proteins comparing the genome sequences of Phytophthora ramorum and P. sojae. The fact that no similarities were found to more than half of the Uf‐HSPs even among other rust fungi could be due to this selective pressure and to host specificity. BLAST results for Uf‐ISSPs are very similar to those for Uf‐HSPs. Again, the small number of similar sequences might indicate accelerated evolution due to strong selective pressure. The fact that the expression of enzymes already known to be present in in vitro grown infection structures of U. fabae seems to be so low that they could not be identified in the present study can also be interpreted as part of the pathogen's strategy to avoid detection (Heath, 1997a).
Only four of the Uf‐HSPs were identical to Uf‐ISSPs. To verify the strong stage specificity of protein secretion suggested by this low number of identities between the two secretomes, we analysed the expression pattern of the secreted proteins. Two approaches were used. On the one hand, a macroarray for Uf‐HSPs was performed. The array was hybridized with cDNA prepared from haustorial RNA and RNA of in vitro grown infection structures by reverse transcription using oligo dT primers and simultaneous labelling with digoxygenin (Fig. 1). Although many genes showed only weak hybridization, a clear majority showed stronger or exclusive hybridization with haustorial cDNA. On the other hand, the expression pattern of selected genes was analysed by Northern blotting. For Northern blots, RNA was separated on 0.5% agarose gels with 20 mm MOPS, 5 mm sodium acetate, 1 mm EDTA and 6.6% formaldehyde as running buffer. Blotting and hybridization were performed according to Engler‐Blum et al. (1993). Gels were blotted onto Hybond‐N+‐membranes (GE Healthcare, Chalfont St. Giles, Herts., UK). Hybridization was done with homologous probes at 68 °C. Detection was carried out using alkaline‐phosphatase‐conjugated anti‐digoxigenin Fab fragments, CSPD (Roche Diagnostics GmbH, Mannheim, Germany) as substrate, and autoradiography. Alternatively, the AlkPhos Direct Labelling and Detection System (GE Healthcare) was used. Preparation of RNA from the different in vitro stages was carried out as described above, preparation of RNA from infected and non infected Vicia faba leaves followed US patent 5,973,137 (Purescript®, Gentra‐Systems Inc.).
Figure 1.
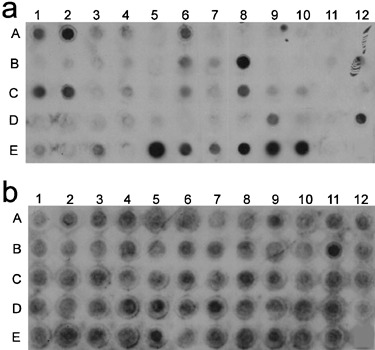
Macroarray of Uf‐HSPs: Uf‐HSP DNA is spotted in numerical order. For reference to individual spots see supplementary Table S1, spot E12 is empty. (a) Membrane hybridized with cDNA from haustorial RNA; (b) membrane hybridized with cDNA from RNA from in vitro infection structures.
Figure 2 shows Northern blots for genes that were represented in the haustorial library only (Uf‐HSP42), in both libraries (Uf‐HSP40 = Uf‐ISSP7) and in the library of in vitro infection structures only (Uf‐ISSP2). These data clearly confirm the results gained by comparison of the libraries. Sixteen additional Northern blots gave similar results (data not shown).
Figure 2.
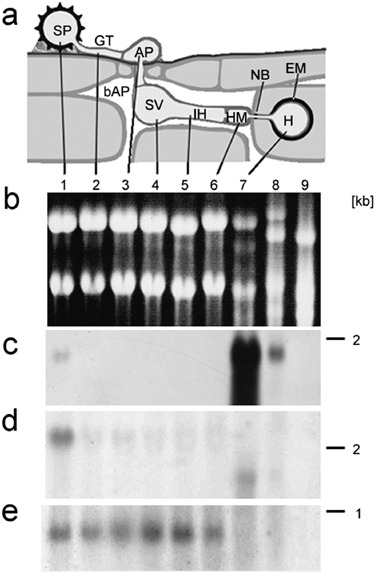
Northern blots resolving the expression pattern of individual genes. (a) Schematic representation of rust infection structures. (b) Ethidium‐bromide‐stained denaturing agarose gel (loading control). (c–e) Northern blots for Uf‐HSP42 (c), Uf‐HSP40 = Uf‐ISSP7 (d) and Uf‐ISSP2 (e). Probes were selected to show one gene that was represented in the haustorial library, one that was represented in both libraries, and one that was represented in the library of in vitro infection structures only. 1, uredospore (SP); 2, germ tube (GT) after 4 h germination; 3–6, in vitro infection structures harvested after 6 h (3, AP, appressorium), 12 h (4, SV, substomatal vesicle), 18 h (5, IH, infection hypha), 22 h (6, HM, haustorial mother cell, corresponds to the in vitro infection structures library); 7, isolated haustoria (HA), corresponds to the haustorial library; 8, infected leaves; 9, non‐infected leaves. bAP, bulk apoplast; NB, neckband; EM, extrahaustorial matrix. Numbers on the right give the size estimate in kb.
The small subgroup of four proteins, which are constitutively expressed in the pre‐biotrophic and biotrophic stage of the dicaryotic phase, also differs from the remaining secretome in the fact that a majority of three out of four of these proteins do have similar database entries. This might indicate that these proteins have other functions than in virulence. All other data concerning the expression pattern of Uf‐HSPs indicate strong stage specificity. Therefore, we speculate that a majority of Uf‐HSPs might have effector function.
Evidence that fungal proteins can be transferred from the haustorium into the plant cytoplasm is very recent (Dodds et al., 2004; Kemen et al., 2005), and knowledge about these proteins is therefore very limited. The transfer mechanism(s) and effector functions of these proteins are still unknown. However, the majority of transferred proteins thus far have been identified as Avr proteins, i.e. elicitors of plant defences (Catanzariti et al., 2006). These proteins have several allelic sequences and also homologues in other strains but ‘show no significant sequence similarity to other known proteins or peptide sequence motifs’ (Catanzariti et al., 2006). The only non‐avirulence transferred protein thus far identified is RTP1p from U. fabae (Kemen et al., 2005). Therefore, it seems promising to look among Uf‐HSPs without sequence similarity outside the Uredinales to find proteins that are transferred into the plant cytoplasm and that might have functions other than avirulence.
Supporting information
Table S1 Haustorical secreted proteins of U. fabae.
Table S2 Secreted proteins of U. fabae in vitro infection structures.
This material is available as part of the online article from:
http://www.blackwell‐synergy.com/doi/abs/10.1111/j.1574.6968.2007.00943.x
(This link will take you to the article abstract.)
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We are grateful to Matthias Hahn for sharing his sequence data, to Felix ten Brink, Martina Schönherr, Martina Gerst, Karin Lautenschlager, Florian Holland and Martina Strittmatter who did undergraduate work on this project and to Kurt Mendgen for support and critical reading of the manuscript. This work was supported by a grant provided by the Deutsche Forschungsgemeinschaft to R.T.V. (VO595/3‐1).
REFERENCES
- Alfano, J.R. and Collmer, A. (1996) Bacterial pathogens in plants: life up against the wall. Plant Cell, 8, 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- De Bary, A. (1863) Recherches sur le developpement de quelques champignons parasites. Ann. Sci. Nat. Part Bot. 20, 5–148. [Google Scholar]
- Becker, D.M. and Lundblad, V. (1991) Introduction of DNA into yeast cells In: Current Protocols in Molecular Biology (Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A. and Struhl K., eds), pp. 137.1–13.7.10. New York: John Wiley & Sons. [DOI] [PubMed] [Google Scholar]
- Belanger, K.D. , Wyman, A.J. , Sudol, M.N. , Singla‐Pareek, S.L. and Quatrano, R.S. (2003) A signal peptide secretion screen in Fucus distichus embryos reveals expression of glucanase, EGF domain‐containing, and LRR receptor kinase‐like polypeptides during asymmetric cell growth. Planta, 217, 931–950. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , Nielsen, H. , Von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. , Thon, M. , Kulkarni, R. , Xu, J.R. , Pan, H. , Read, N.D. , Lee, Y.H. , Carbone, I. , Brown, D. , Oh, Y.Y. , Donofrio, N. , Jeong, J.S. , Soanes, D.M. , Djonovic, S. , Kolomiets, E. , Rehmeyer, C. , Li, W. , Harding, M. , Kim, S. , Lebrun, M.H. , Bohnert, H. , Coughlan, S. , Butler, J. , Calvo, S. , Ma, L.J. , Nicol, R. , Purcell, S. , Nusbaum, C. , Galagan, J.E. and Birren, B.W. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- Deising, H. , Jungblut, P.R. and Mendgen, K. (1991) Differentiation‐related proteins of the broad bean rust fungus Uromyces viciae‐fabae, as revealed by high resolution two‐dimensional polyacrylamide gel electrophoresis. Arch. Microbiol. 155, 191–198. [Google Scholar]
- Deising, H. , Rauscher, M. , Haug, M. and Heiler, S. (1995) Differentiation and cell wall degrading enzymes in the obligately biotrophic rust fungus Uromyces viciae‐fabae . Can. J. Bot. 73, 624–631. [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Ayliffe, M.A. and Ellis, J.G. (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler‐Blum, G. , Meier, M. , Frank, J. and Müller, G.A. (1993) Reduction of background problems in nonradioactive northern and southern blot analyses enables higher sensitivity than 32P‐based hybridizations. Anal. Biochem. 210, 235–244. [DOI] [PubMed] [Google Scholar]
- Haerter, A.C. and Voegele, R.T. (2004) A novel β‐glucosidase in Uromyces fabae: feast or fight? Curr Genet. 45, 96–103. [DOI] [PubMed] [Google Scholar]
- Hahn, M. and Mendgen, K. (1992) Isolation by ConA binding of haustoria from different rust fungi and comparison of their surface qualities. Protoplasma, 170, 95–102. [Google Scholar]
- Hahn, M. and Mendgen, K. (1997) Characterization of in planta‐induced rust genes isolated from a haustorium‐specific cDNA library. Mol. Plant–Microbe Interact. 10, 427–437. [DOI] [PubMed] [Google Scholar]
- Hahn, M. , Deising, H. , Struck, C. and Mendgen, K. (1997) Fungal morphogenesis and enzyme secretion during pathogenesis In: Resistance of Crop Plants Against Fungi (Hartleb H., Heitefuss R. and Hoppe H.H., eds), pp. 33–57. Jena: G. Fischer. [Google Scholar]
- Heath, M.C. (1997a) Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Bot. 80, 713–720. [Google Scholar]
- Heath, M.C. (1997b) Evolution of plant resistance and susceptibility to fungal species In: The Mycota, Vol. V Part B, Plant Relationships ( Carroll, G.C. and Tudzynski, P. eds), pp. 257–276. Berlin: Springer‐Verlag. [Google Scholar]
- Heilig, J.S. , Elbing, K.L. and Brent, R. (1998) Large‐scale preparation of plasmid DNA In: Current Protocols in Molecular Biology (Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A. and Struhl K. eds), pp. 17.1–1.7.16. New York: John Wiley & Sons. [DOI] [PubMed] [Google Scholar]
- Hiller, N.L. , Bhattacharjee, S. , Van Ooij, C. , Liolios, K. , Harrison, T. , Lopez‐Estrano, C. and Haldar, K. (2004) A host‐targeting signal in virulence proteins reveals a secretome in malarial infection. Science, 306, 1934–1937. [DOI] [PubMed] [Google Scholar]
- Hugot, K. , Riviere, M.P. , Moreilhon, C. , Dayem, M.A. , Cozzitorto, J. , Arbiol, G. , Barbry, P. , Weiss, C. and Galiana, E. (2004) Coordinated regulation of genes for secretion in tobacco at late developmental stages: association with resistance against oomycetes. Plant Physiol. 134, 858–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, K.A. , Collins‐Racie, L.A. , Colbert, M. , Duckett, M. , Evans, C. , Golden‐Fleet, M. , Kelleher, K. , Kriz, R. , La Vallie, E.R. , Merberg, D. , Spaulding, V. , Stover, J. , Williamson, M.J. and McCoy, J.M. (1999) A genetic selection for isolating cDNA clones that encode signal peptides. Methods Enzymol. 303, 468–479. [DOI] [PubMed] [Google Scholar]
- Jacobs, K.A. , Collins‐Racie, L.A. , Colbert, M. , Duckett, M. , Golden‐Fleet, M. , Kelleher, K. , Kriz, R. , LaVallie, E.R. , Merberg, D. , Spaulding, V. , Stover, J. , Williamson, M.J. and McCoy, J.M. (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene, 198, 289–296. [DOI] [PubMed] [Google Scholar]
- Jakupovic, M. , Heintz, M. , Reichmann, P. , Mendgen, K. and Hahn, M. (2006) Microarray analysis of expressed sequence tags from haustoria of the rust fungus Uromyces fabae . Fungal. Genet. Biol. 43, 8–19. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaiser, C.A. , Preuss, D. , Grisafi, P. and Botstein, D. (1987) Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science, 235, 312–317. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 21–2.20. [DOI] [PubMed] [Google Scholar]
- Kemen, E. , Kemen, A.C. , Rafiqi, M. , Hempel, U. , Mendgen, K. , Hahn, M. and Voegele, R.T. (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant–Microbe. Interact. 18, 1130–1139. [DOI] [PubMed] [Google Scholar]
- Klein, R.D. , Gu, Q. , Goddard, A. and Rosenthal, A. (1996) Selection for genes encoding secreted proteins and receptors. Proc. Natl Acad. Sci. USA, 93, 7108–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.A. , Wormsley, S. , Kamoun, S. , Lee, A.F. , Joiner, K. and Wong, B. (2003) An analysis of the Candida albicans genome database for soluble secreted proteins using computer‐based prediction algorithms. Yeast, 20, 595–610. [DOI] [PubMed] [Google Scholar]
- Lee, S.J. , Kelley, B.S. , Damasceno, C.M. , St John, B. , Kim, B.S. , Kim, B.D. and Rose, J.K. (2006) A functional screen to characterize the secretomes of eukaryotic pathogens and their hosts in planta. Mol. Plant–Microbe Interact. 19, 1368–1377. [DOI] [PubMed] [Google Scholar]
- Link, T. , Lohaus, G. , Heiser, I. , Mendgen, K. , Hahn, M. and Voegele, R.T. (2005) Characterization of a novel NADP+‐dependent D‐arabitol dehydrogenase from the plant pathogen Uromyces fabae . Biochem. J. 389, 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendgen, K. and Hahn, M. (2002) Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 7, 352–356. [DOI] [PubMed] [Google Scholar]
- Mota, L.J. , Sorg, I. and Cornelis, G.R. (2005) Type III secretion: the bacteria‐eukaryotic cell express. FEMS Microbiol. Lett. 252, 1–10. [DOI] [PubMed] [Google Scholar]
- Treco, D.A. and Lundblad, V. (1993) Preparation of yeast media In: Current Protocols in Molecular Biology (Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A. and Struhl K. eds), pp. 131.1–13.1.7. New York: John Wiley & Sons. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H. , Aerts, A. , Arredondo, F.D. , Baxter, L. , Bensasson, D. , Beynon, J.L. , Chapman, J. , Damasceno, C.M. , Dorrance, A.E. , Dou, D. , Dickerman, A.W. , Dubchak, I.L. , Garbelotto, M. , Gijzen, M. , Gordon, S.G. , Govers, F. , Grunwald, N.J. , Huang, W. , Ivors, K.L. , Jones, R.W. , Kamoun, S. , Krampis, K. , Lamour, K.H. , Lee, M.K. , McDonald, W.H. , Medina, M. , Meijer, H.J. , Nordberg, E.K. , Maclean, D.J. , Ospina‐Giraldo, M.D. , Morris, P.F. , Phuntumart, V. , Putnam, N.H. , Rash, S. , Rose, J.K. , Sakihama, Y. , Salamov, A.A. , Savidor, A. , Scheuring, C.F. , Smith, B.M. , Sobral, B.W. , Terry, A. , Torto‐Alalibo, T.A. , Win, J. , Xu, Z. , Zhang, H. , Grigoriev, I.V. , Rokhsar, D.S. and Boore, J.L. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Voegele, R.T. and Mendgen, K. (2003) Rust haustoria: nutrient uptake and beyond. New Phytol. 159, 93–100. [DOI] [PubMed] [Google Scholar]
- Voegele, R.T. , Hahn, M. , Lohaus, G. , Link, T. , Heiser, I. and Mendgen, K. (2005) Possible roles for mannitol and mannitol dehydrogenase in the biotrophic plant pathogen Uromyces fabae . Plant Physiol. 137, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. , Struck, C. , Hahn, M. and Mendgen, K. (2001) The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae . Proc. Natl Acad. Sci. USA, 98, 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. , Wirsel, S. , Moll, U. , Lechner, M. and Mendgen, K. (2006) Cloning and characterization of a novel invertase from the obligate biotroph Uromyces fabae and analysis of expression patterns of host and pathogen invertases in the course of infection. Mol. Plant–Microbe Interact. 19, 625–634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Haustorical secreted proteins of U. fabae.
Table S2 Secreted proteins of U. fabae in vitro infection structures.
This material is available as part of the online article from:
http://www.blackwell‐synergy.com/doi/abs/10.1111/j.1574.6968.2007.00943.x
(This link will take you to the article abstract.)
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item


