SUMMARY
Taxonomy: Superkingdom Prokaryota; Kingdom Monera; Domain Bacteria; Phylum Firmicutes (low‐G+C, Gram‐positive eubacteria); Class Mollicutes; Candidatus (Ca.) genus Phytoplasma.
Host range: Ca. Phytoplasma comprises approximately 30 distinct clades based on 16S rRNA gene sequence analyses of ~200 phytoplasmas. Phytoplasmas are mostly dependent on insect transmission for their spread and survival. The phytoplasma life cycle involves replication in insects and plants. They infect the insect but are phloem‐limited in plants. Members of Ca. Phytoplasma asteris (16SrI group phytoplasmas) are found in 80 monocot and dicot plant species in most parts of the world. Experimentally, they can be transmitted by approximately 30, frequently polyphagous insect species, to 200 diverse plant species.
Disease symptoms: In plants, phytoplasmas induce symptoms that suggest interference with plant development. Typical symptoms include: witches’ broom (clustering of branches) of developing tissues; phyllody (retrograde metamorphosis of the floral organs to the condition of leaves); virescence (green coloration of non‐green flower parts); bolting (growth of elongated stalks); formation of bunchy fibrous secondary roots; reddening of leaves and stems; generalized yellowing, decline and stunting of plants; and phloem necrosis. Phytoplasmas can be pathogenic to some insect hosts, but generally do not negatively affect the fitness of their major insect vector(s). In fact, phytoplasmas can increase fecundity and survival of insect vectors, and may influence flight behaviour and plant host preference of their insect hosts.
Disease control: The most common practices are the spraying of various insecticides to control insect vectors, and removal of symptomatic plants. Phytoplasma‐resistant cultivars are not available for the vast majority of affected crops.
INTRODUCTION
Phytoplasmas are bacterial plant pathogens that can cause devastating yield losses in diverse low‐ and high‐value crops worldwide (Bertaccini, 2007; Lee et al., 2000). They are obligate symbionts of plants and insects, and in most cases need both hosts for dispersal in nature. In plants, they remain mainly restricted to the phloem tissue (Doi et al., 1967; Whitcomb and Tully, 1989), and spread throughout the plant by moving through the pores of the sieve plates that divide the phloem sieve tubes. Insect vectors of phytoplasmas are phloem feeders of the Order Hemiptera, mostly leafhoppers (Cicadellidae), planthoppers (Fulgoromorpha) and psyllids (Psyllidae) (Weintraub and Beanland, 2006). In insects, phytoplasmas invade the guts and salivary glands and many other tissues where they can accumulate at great numbers inside and outside cells (Ammar and Hogenhout, 2006). Phytoplasmas have to traverse the gut and salivary gland cells in order to reach the saliva for subsequent introduction into the phloem during insect feeding (Lefol et al., 1994; Lherminier et al., 1990; Nakashima and Hayashi, 1995). The latent period, i.e. the time between initial acquisition of the phytoplasmas by the insect vector from plants and the ability for the insect to introduce phytoplasmas back into plants, can vary between 7 and 80 days (Moya‐Raygoza and Nault, 1998; Murral et al., 1996). In plants, symptoms can develop at ~7 days after introduction of the phytoplasma by the insect vector, but can take much longer (6–24 months), depending on the phytoplasma and plant species.
Phytoplasmas have a broad plant host range, which depends on the plant feeding range of their insect vectors. With more than 100 isolates, the Aster Yellows Phytoplasma (AYP) subclade Candidatus (Ca.) Phytoplasma asteris (previously known as 16srI clade phytoplasmas) comprises the largest among the Ca. phytoplasma subclades (Marcone et al., 2000; Seemüller et al., 1998). AYPs are vectored by at least 30, often polyphagous, insect species and, as a consequence, are capable of infecting more than 80 plant species (Firrao et al., 2007), including many weeds that surround crop production fields (Marcone et al., 2000). For example, Aster Yellows phytoplasma strain Witches’ Broom (AY‐WB) can be transmitted by the polyphagous Macrosteles quadrilineatus (Forbes) to China aster (Fig. 1) and lettuce (Zhang et al., 2004), and Nicotiana benthamiana, tomato, Arabidopsis thaliana and maize (unpublished results). The broad plant and insect host ranges make phytoplasma outbreaks unpredictable. Furthermore, because of the long incubation periods of phytoplasmas in plants and insects, outbreaks are often detected too late, i.e. close to harvesting of the crops and after dispersion of the phytoplasmas by the insect vectors. This makes it harder to control or interrupt the disease cycle.
Figure 1.
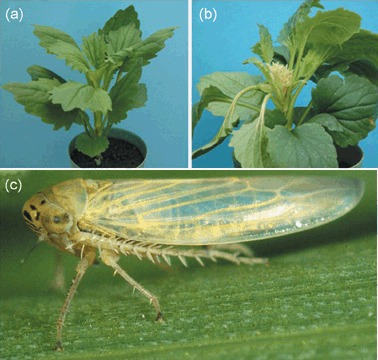
(a) Healthy China aster. (b) AY‐WB‐infected China aster. Note the witches’ broom symptoms. (c) Macrosteles quadrilineatus, the leafhopper‐vector of AY‐WB. (Photos in A and B are reprinted from Zhang J., Hogenhout S.A., Nault L.R., Hoy C.W. and Miller S.A. (2004). Molecular and symptom analyses of phytoplasma strains from lettuce reveal a diverse population. Phytopathology, 94, 842–849. with permission of the journal.)
Phytoplasmas are wall‐less pleiomorphic bacteria of ~500 nm in diameter. They have a single cell membrane and small genomes averaging ~750 kb (Bai et al., 2006; Gundersen et al., 1996; Marcone et al., 1999; Neimark and Kirkpatrick, 1993; Oshima et al., 2004). Phytoplasmas belong to the class Mollicutes, which encompasses small pleiomorphic bacteria with single membranes that have diverged from a Gram‐positive ancestor, most likely a Clostridium or Lactobacillus spp., through genome reductions and the loss of the outer cell wall (Weisburg et al., 1989; Woese, 1987). Other members of the class Mollicutes include mycoplasmas, ureaplasmas, spiroplasmas and acholeplasmas (Razin et al., 1998). Phytoplasmas comprise a monophyletic clade that arose from the Acholeplasma branch that is evolutionary distant from the mycoplasma/ureaplasma and spiroplasma branches (Fig. 2) (Gundersen et al., 1996; Lee et al., 2000; Lim and Sears, 1989, 1992). The phylogeny is consistent with the codon usage and metabolic requirements of the various mollicutes. Phytoplasmas and acholeplasmas have a standard codon usage with three stop codons, whereas mycoplasmas and spiroplasmas use one of these stop codons, the UGA codon, for tryptophane (Razin et al., 1998). Furthermore, acholeplasmas and phytoplasmas lack functional phosphotransferase transport systems (PTSs) for import of sugars (Bai et al., 2006; Cirillo, 1993; Oshima et al., 2004), whereas mycoplasmas and spiroplasmas have PTSs (Gaurivaud et al., 2000; Razin et al., 1998). All known phytoplasmas are insect‐transmitted plant pathogens, whereas mycoplasmas and ureaplasmas are human and livestock pathogens (Razin et al., 1998). The majority of spiroplasmas are associated with insects (Gasparich, 2002), but three Spiroplasma species are insect‐transmitted plant pathogens that can share host ranges with phytoplasmas. These are Spiroplasma citri, S. kunkelii and S. phoeniceum.
Figure 2.
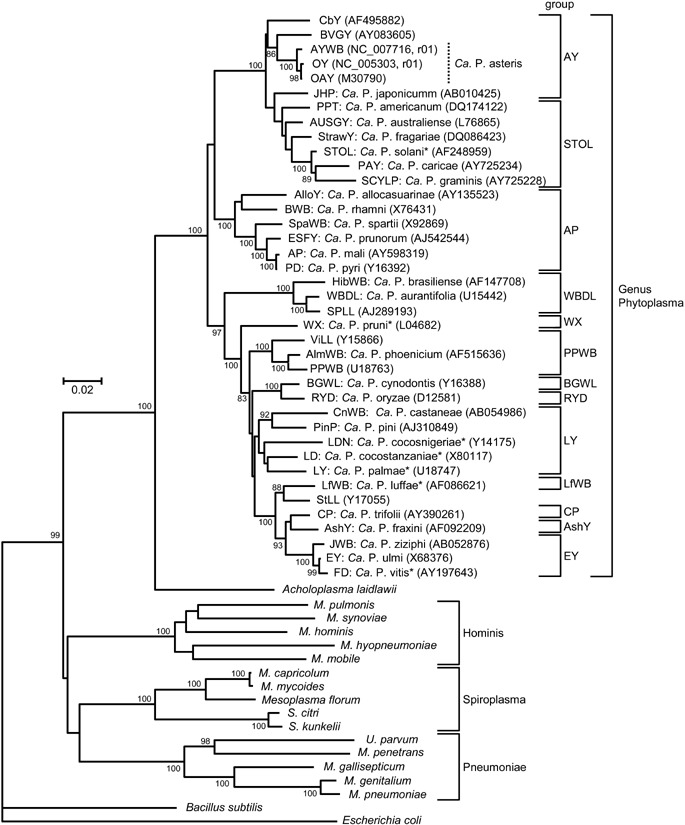
Phytoplasmas comprise a single clade that diverges from Acholeplasma spp. A phylogenetic tree was constructed by the neighbour‐joining method (Saitou and Nei, 1987) using 16S rRNA gene sequences from phytoplasmas, acholeplasma, mycoplasmas, spiroplasmas, Mesoplasma florum, Ureaplasma parvum, Bacillus subtilis and Escherichia coli (outgroup). Numbers on branches are the bootstrap values (only values > 80% are shown). GenBank accession numbers are given in parentheses. Asterisks indicate the proposed provisional names (IRPCM, 2004). The phylogeny presented here is consistent with IRPCM (2004) and the MolliGen database (http://cbi.labri.fr/outils/molligen/) (Barre et al., 2004). Abbreviations: AlloY, allocasuarina yellows; AlmWB, Almond witches’ broom; AP, apple proliferation; AshY, ash yellows; AUSGY, Australian grapevine yellows; AY, aster yellows; AY‐WB, aster yellows phytoplasma strain witches’ broom; BGWL, Bermuda grass white leaf; BVGY, Buckland valley grapevine yellows; BWB, buckthorn witches’ broom; Ca. P., Candidatus Phytoplasma; CbY, Chinaberry yellows; CnWB, chestnut witches’ broom; CP, clover proliferation; ESFY, European stone fruit yellows; EY, elm yellows; FD, flavescence dorée of grapevine; HibWB, Hibiscus witches’ broom; JHP, Japanese Hydrangea phyllody; JWB, jujube witches’ broom; LD, coconut lethal yellowing; LDN, coconut lethal yellowing; LY, coconut lethal yellowing; LfWB, loofah witches’ broom; M., Mycoplasma; OY, onion yellows; PAY, papaya; PD, pear decline; PPT, potato purple top wilt; PPWB, Caribbean pigeon pea witches’ broom; RYD, rice yellow dwarf; S., Spiroplasma; SCYLP, sugarcane yellow leaf syndrome; SpaWB, spartium witches’ broom; SPLL, sweet potato little leaf; STOL, stolbur; StrawY, strawberry yellows; ViLL, Vigna little leaf; WBDL, witches’ broom disease of lime; WX, western X‐disease.
Phytoplasmas and many mycoplasmas cannot be cultured outside their hosts in cell‐free artificial culture media. The phytoplasma genome reductions resulted in the loss of most metabolic pathways, including those for ATP synthesis by F0F1‐type ATP synthases, and amino acid and nucleotide synthesis (Bai et al., 2006; Oshima et al., 2004). Phytoplasmas have to obtain these essential metabolites from their hosts, and hence will require these metabolites in their culture media as well. Thus far, the genomes of many cultivable mycoplasmas have been sequenced to completion. However, in 2004 and 2006, the genomes of two phytoplasmas were published (Bai et al., 2006; Oshima et al., 2004). These genome sequences provided novel insights that might lead to the development of culture media and diagnostics assays for phytoplasmas. In addition, the genome sequences will help with the identification and characterization of phytoplasma virulence factors.
PHYTOPLASMA DISCOVERY AND CLASSIFICATION
Many different yellows, dwarf and witches’ broom diseases caused by phytoplasmas occur throughout the world. Mulberry dwarf disease was first observed in Japan during the Tokugawa Period (1603–1868), and spread widely causing severe damage to mulberry plants (Okuda, 1972). Cases of paulownia witches’ broom disease, rice yellow dwarf disease and yellows diseases have been reported since the early 1900s (Kunkel, 1926; Lee et al., 2000; Okuda, 1972). Before 1967, the causal agent of these diseases was thought to be a virus, because it could not be cultured in artificial media, was insect transmitted and the symptoms were often similar to those of viral diseases (Doi et al., 1967; Lee et al., 2000). However, no virus particles were definitively associated with these diseases.
Doi et al. (1967) discovered structures in ultrathin sections of the phloem of plants affected by these diseases, including mulberry dwarf, paulownia witches’ broom and aster yellows, that resembled animal and human mycoplasmas. The associated agents lacked rigid cell walls, were surrounded by a single membrane, were spherical or pleiomorphic in shape with size ranges similar to that of mycoplasmas (80–800 nm), and were sensitive to tetracycline antibiotics (Doi et al., 1967; Ishiie et al., 1967). This finding led to a drastic change in our understanding of the etiology of many yellows, dwarf and witches’ broom diseases. The structures that Doi et al. (1967) observed were similar to those observed in a veterinary laboratory of the same Faculty that had conducted electron microscope observations on mycoplasmas (Iida, 1972). Hence, since 1967, the term mycoplasma‐like organisms (MLOs) was used to refer to the causal agents of many yellows, dwarf and witches’ broom diseases, including those that turned out later to be phytoplasmas or spiroplasmas (Lee and Davis, 1992; McCoy et al., 1989).
Woese et al. (1980) suggested that the genera Mycoplasma, Spiroplasma and Acholeplasma originated from a single lineage derived from Gram‐positive bacteria (Woese, 1987), and this was confirmed by several other reports (Weisburg et al., 1989). In 1989, the 16S rRNA gene sequence from an MLO (Oenothera virescence phytoplasma) belonging to the aster yellows group was compared with those of Acholeplasma laidlawii, Spiroplasma citri and several mycoplasmas (Lim and Sears, 1989). Based on this analysis, it was suggested that the phytopathogenic MLO was a member of the class Mollicutes and that the MLO was more closely related to the Acholeplasma spp. than to the Spiroplasma spp. or animal mycoplasmas. These results were also confirmed by analysing several ribosomal protein sequences (Lim and Sears, 1991, 1992; Toth et al., 1994) and 16S rRNA gene sequences (Kuske and Kirkpatrick, 1992; Namba et al., 1993a,b; Seemüller et al., 1994; Toth et al., 1994). Phylogenetic analyses based on various conserved genes confirmed that MLOs represent a clearly distinct, monophyletic clade within the class Mollicutes (Fig. 2) (Gundersen et al., 1994; Jomantiene et al., 1998; Kuske and Kirkpatrick, 1992; Lee et al., 1998, , 2000; Lim and Sears, 1989; Namba et al., 1993, b; Seemüller et al., 1994; Toth et al., 1994). In 1994, the name ‘phytoplasma’ was adopted by the Phytoplasma Working Team at the 10th Congress of the International Organization of Mycoplasmology to collectively denote MLOs.
Since the establishment of PCR, sequencing and phylogenetic analyses of the 16S rRNA gene, sequences from many new phytoplasma strains have been determined, and have shown that thus far all phytoplasmas comprise a single clade diverged from Acholeplasma spp. within the class Mollicutes (Fig. 2) (Gundersen et al., 1994; Kuske and Kirkpatrick, 1992; Namba et al., 1993, b; Seemüller et al., 1994). At present, more than 900 sequences have been deposited in GenBank. Recently, it was proposed that phytoplasmas be placed within the novel genus ‘Candidatus (Ca.) Phytoplasma’ (IRPCM, 2004). Basically, a strain can be described as a novel ‘Ca. Phytoplasma’ species if its 16S rRNA gene sequence has < 97.5% similarity to that of any previously described ‘Ca. Phytoplasma’ species. Thus far, 26 Candidatus species have been described (Arocha et al., 2005; Davis et al., 1997; Griffiths et al., 1999; Hiruki and Wang, 2004; IRPCM, 2004; Jung et al., 2002, 2003a, 2003b; Lee et al., 2004a,2006a, 2004b; Marcone et al., 2004a,b; Montano et al., 2001; Sawayanagi et al., 1999; Schneider et al., 2005; Seemüller and Schneider, 2004; Valiunas et al., 2006; Verdin et al., 2003; Zreik et al., 1995).
THE PHYTOPLASMA LIFE CYCLE INVOLVES PLANT AND INSECT HOSTS
Phytoplasmas and the three insect‐transmitted plant pathogenic Spiroplasma spp. have a unique biology among plant pathogenic bacteria, because they require replication in diverse hosts, plants (Kingdom Plantae) and insects (Kingdom Animalia) for their survival and spread in nature (Fig. 3a). In plants, phytoplasmas are found mainly in phloem elements, including both mature sieve tubes devoid of nuclei and immature phloem cells that still have nuclei (Fig. 3b). In insects, phytoplasmas must traverse insect gut cells, replicate in various tissues of the insect (Fig. 3c,d) and traverse the salivary gland cells (Fig. 3e) in order to reach the saliva for subsequent introduction into plants (Fig. 3a). They can be found intra‐ and extracellularly in the insect tissues. Hence, phytoplasmas are intracellular as well as extracellular pathogens/symbionts of plants and insects.
Figure 3.

The phytoplasma life cycle involves replication in plants and insects. (a) Schematic illustration of the different stages of phytoplasma movement through the plant and leafhopper hosts. Phytoplasmas are indicated as dark red dots and the movement of the phytoplasmas is indicated with dark red arrows. Leafhoppers acquire phytoplasmas from the plant phloem (ph). Phytoplasmas that are ingested with plant sap move through the stylet's food canal and the intestinal tract, and invade epithelial and muscle cells of the oesophagus (Es), anterior midgut (Amg), mid midgut (Mmg), filter chamber (Fc), Malpighian tubules (Mt) and hindgut (Hg). Similarly to spiroplasmas (Özbek et al., 2003), phytoplasmas probably cross the basal lamina to enter the haemolymph (He), from where they can move to the salivary glands (Sg). They multiply in secretory salivary gland cells and then are transported along with the saliva to the salivary duct (Sd). Phytoplasmas are introduced back into the phloem tissue of host plants during feeding and salivation of leafhoppers. (b–e) Electron micrographs of phytoplasmas (arrowheads) in the plant phloem and in various leafhopper tissues as indicated in (a). (b) AY‐WB in adjacent sieve elements (se1) close to the nucleus (n) of an aster leaf; the arrow indicates a sieve pore between the sieve plates (sp), and asterisks indicate possibly dividing phytoplasma cells. (c) AY‐WB (arrowheads) in a midgut epithelial cell of the leafhopper vector M. quadrilineatus; asterisks indicate less dense areas of the cytoplasm. (d) Accumulations of maize bushy stunt phytoplasma (MBSP, arrowheads) in the cytoplasm of a cell in the muscle layer around the midgut of the leafhopper vector D. maidis; note that phytoplasmas are located close to the nucleus (n). (e) MBSP (arrowheads) in the cytoplasm of a salivary gland secretory cell in D. maidis; arrow indicates phytoplasma close to a secretory vesicle (sv). Other abbreviations: bl, basal lamina; Br, brain; Cng, compound nerve ganglion; mv, microvilli; pm, plasma membrane; sm, secretory material; Sp, salivary pump; St, stylet; Xy, xylem. Scale bars, 1 µm.
Several other bacterial plant pathogens, such as Xylella fastidiosa and Pantoea stewartii, require insects for dissemination in nature. However, the interactions of these pathogens with insects are fundamentally different from those of phytoplasmas. X. fastidiosa remains extracellularly in the lumen of the insect alimentary canal, where it forms a biofilm for attachment to the cuticular lining of the foregut (Newman et al., 2004; Redak et al., 2004). X. fastidiosa is introduced into plants by regurgitation or extravasation (egestion). The mechanism by which P. stewartii interacts with its flea beetle vector is still under investigation. Thus far, it is not clear whether P. stewartii can invade insect cells, or like X. fastidiosa, requires the formation of a biofilm for attachment to the cuticular lining of the alimentary canal of flea beetles. It is also not known whether flea beetles introduce P. stewartii into plants through regurgitation, salivation or defecation. However, preliminary studies suggest that P. stewartii can persist for several days in flea beetles, mainly at the lumen of the hindgut, indicating that defecation is a probable method of transmission (E.‐D. Ammar, V. Correa & S. A. Hogenhout, unpublished data). P. stewartii requires a specific secretion system, the type III secretion system (T3SS), for introducing virulence factors into plant cells and perhaps also insect host cells (Merighi et al., 2003). There is no evidence that phytoplasmas and the related Gram‐positive bacteria have T3SSs (Bai et al., 2006; Oshima et al., 2004; Rosch and Caparon, 2004); they apparently use the Sec‐dependent or other translocation pathways for introducing effector proteins into host cells (Kakizawa et al., 2001, 2004; Rosch and Caparon, 2004).
In plant hosts, the mature sieve tubes normally contain the highest concentration of phytoplasmas (Christensen et al., 2004). As phloem cells are live cells, this may be considered intracellular. In addition, intracellular phytoplasmas with various morphologies, some probably caused by budding or multiplying, were also found inside the cytoplasm of immature phloem elements (Fig. 3b). Furthermore, phytoplasmas apparently have been detected in the cytoplasm of phloem parenchyma cells adjacent to sieve elements (Sears and Klomparens, 1989), inside parenchyma cells in or close to the vascular system of Cascuta shoots (Siller et al., 1987), and in embryos of coconut (Cordova et al., 2003). However, some of the earlier observations of phytoplasmas (or mycoplasma‐like organisms) in parenchyma cells may have been due to misinterpretation of these structures or of early stages in the development of sieve elements (McCoy, 1979). Indeed, according to Esau (1977), ‘In many dicotyledons, sieve tube members and some parenchyma cells are derived from the same phloem initial.’ In contrast, X. fastidiosa and P. stewartii remain extracellular, mainly in the xylem vessels or between mesophyll cells (Braun, 1982; Wells et al., 1987), during their life cycles in plants.
When acquired by the insect vectors from plants, phytoplasmas will have to attach to the membranes of midgut epithelial cells, on or between microvilli, and initiate invasion of the midgut. Spiroplasmas and mycoplasmas have specific tip structures that are primarily responsible for attachment to cell membranes between microvilli (Ammar et al., 2004). Such tip structures have not yet been identified in phytoplasmas, but it is likely that phytoplasmas use a comparable mechanism for attachment. As will be further discussed below, at least one membrane protein, the antigenic membrane protein (AMP), important for insect invasion of Ca. Phytoplasma asteris strain OY, has been identified (Suzuki et al., 2006).
AY‐WB and Maize Bushy Stunt Phytoplasma (MBSP) traverse and apparently multiply in the midgut epithelial cells of their insect vectors (Fig. 3). Phytoplasmas can accumulate to high densities also outside the basal lamina of these epithelial cells in the hemocoel, haemocytes and particularly at muscle fibres and tracheae that form the outer layer of the midgut (Fig. 3d). S. citri and S. kunkelii accumulate at similar locations in their leafhopper vectors (Kwon et al., 1999; Özbek et al., 2003). There is evidence that S. kunkelii ruptures the extracellular matrix of the basal lamina of the midgut, possibly together with specific enzymes, for entering the hemocoel (Özbek et al., 2003). The haemolymph may provide a vehicle for these mollicutes to reach other tissues, including the salivary glands (Fig. 3).
Insect salivary glands also can become heavily infected with AY‐WB and MBSP. Individual phytoplasma cells appear to reside directly, and probably multiply, in the cytoplasm of salivary gland cells (Fig. 3e) sometimes close to the nucleus. Similarly to spiroplasmas (Kwon et al., 1999), phytoplasmas probably enter the canaliculi at the centre of secretory cells, before reaching the main salivary duct that leads to the stylet's salivary canal (Fig. 3a). They are then introduced into the plant phloem elements along with the insect salivary secretions during feeding.
AY‐WB can accumulate to high densities in individual phloem sieve elements (Fig. 3b), and is usually abundant in phloem tissue of sink areas, such as young shoots and roots, consistent with the development of typical symptoms (witches’ broom) in these young tissues (Zhang et al., 2004). Some phytoplasmas (e.g. MBSP) induce severe phloem necrosis in their host plants, indicating that these plants react to the phytoplasma infection. However, many phytoplasmas, including AY‐WB, do not seem to induce phloem necrosis but still accumulate to high densities in phloem elements. As has been shown for several other bacterial plant pathogens, it is likely that phytoplasmas produce a series of virulence proteins that suppress plant responses. Experiments demonstrating that phytoplasmas secrete virulence proteins that affect plant cells will be discussed further below.
Phytoplasmas clearly negatively impact the fitness of their plant hosts. Plants are frequently stunted and may not produce normal flowers, fruits or seeds. Phytoplasmas may or may not affect the fitness and survival of insect vectors. Some leafhopper species are negatively impacted by the phytoplasma infection as they die around the time they are capable of inoculating plants with the phytoplasmas, whereas other leafhopper species do not show obvious negative effects from the infection and sometimes can even benefit from the phytoplasma infection by living longer when deprived of a main food source and when exposed to lower suboptimal temperatures (Ebbert and Nault, 1994; Madden and Nault, 1983; Madden et al., 1984). The better adaptation of some leafhopper species to phytoplasma infection is, at least in the case of Dalbulus leafhopper spp., MBSP and S. kunkelii, a result of a long history of association between the organisms (Nault, 1990). Dalbulus leafhopper spp. that have been exposed to MBSP and S. kunkelii for longer times have developed tolerance to these bacteria and hence are well adapted to each other, whereas Dalbulus spp. that have had little or no history of exposure to these bacteria are maladapted and succumb quickly to infection (Nault, 1985, 1990). Phytoplasmas tend to accumulate at much higher levels in maladapted relative to tolerant leafhopper species (Ammar and Hogenhout, 2006). Thus, phytoplasmas can be pathogens or beneficial symbionts of hemipteran insects. However, even in the latter case, the cytoplasm in infected leafhopper midgut epithelial cells may appear less dense with fewer ribosomes and less endoplasmic reticulum compared with non‐infected leafhoppers (E.‐D. Ammar and S.A. Hogenhout, personal observations).
Beneficial symbiosis has also been observed for other leafhopper–phytoplasma interactions. The longevity and number of offspring of the aster leafhoppers (Macrosteles quadrilineatus) can significantly increase on AYP‐infected, as compared with healthy, China aster, lettuce, carrots (Daucus carota L.) and periwinkle (Vinca minor L.) (Beanland et al., 2000; Peterson, 1973; Schultz, 1973). Phytoplasmas can also manipulate plants to become new hosts for leafhoppers that normally do not use these plants as hosts. For example, the leafhopper Dalbulus maidis, which is a maize specialist, can feed and survive on AYP‐infected but not on healthy lettuce and China aster plants (Maramorosch, 1958; Purcell, 1988). In fact, the survival rates of D. maidis on AYP‐infected China aster plants and healthy maize (Zea mays L.) plants are comparable (Purcell, 1988). Similar findings were reported for nine leafhopper species on AYP‐infected celery (Apium graveolens L.) and China aster (Peterson, 1973; Schultz, 1973). Thus, phytoplasmas have a remarkable effect on the interactions between insects and plants, i.e. they may convert plants from being non‐hosts into hosts or better hosts for some phloem‐feeding insects.
The ways by which phytoplasmas affect their plant and insect hosts are consistent with their life cycle. Even though there is evidence that some phytoplasmas are transmitted at low rates to plant embryos or insect progeny (Botti and Bertaccini, 2006; Cordova et al., 2003; Khan et al., 2002; Nipah et al., 2007), they mainly depend on the interactions with both plants and insects for survival and dispersal. The mechanism by which phytoplasmas convert plants into more attractive hosts for insects is not yet known. However, leafhoppers prefer young green/yellow tissues for feeding as well as laying eggs, and the production of additional younger green/yellow tissues (witches’ broom, phyllody, virescence and yellowing) by phytoplasmas may provide an explanation for this conversion. It is also possible that phytoplasmas are involved in the down‐regulation of insect‐specific defence responses in plants to attract maladapted insects and improve insect fitness on these plants. The inability of a plant to serve as a host for an insect herbivore is often described as anti‐xenosis (non‐preference). It is clear that anti‐xenosis involves chemical responses in the plant. Plants deficient in the expression or recognition of jasmonates (JAs) are more susceptible to herbivore attack. For example, Nicotiana attenuata plants in which the JA cascade was silenced attracted more adapted herbivores, but also a novel (normally maladapted) leafhopper species, when planted in the native habitat (Kessler et al., 2004; Paschold et al., 2007). Phytoplasmas may be able to down‐regulate general defence responses to insects, such as the JA signalling pathway, in plants.
The above research findings suggest that phytoplasmas manipulate their plant and insect hosts to improve their own chances for survival and dispersal. Recent studies, discussed below, have also indicated that phytoplasmas produce a set of proteins involved in plant and insect infection. The completed genome sequences of two phytoplasmas, onion yellows strain M (OY‐M) and aster yellows strain witches broom (AY‐WB) that both belong to (a. Phytoplasma.asteris) provide unprecedented opportunities for the identification and characterization of phytoplasma virulence and other proteins.
LESSONS LEARNED FROM PHYTOPLASMA COMPLETE GENOME SEQUENCES
The complete genomic sequences of the Ca. Phytoplasma asteris OY‐M strain and the AY‐WB strain have been determined (Bai et al., 2006; Oshima et al., 2004). As phytoplasmas cannot be cultured in vitro, phytoplasma DNA was isolated from infected plants, purified by pulsed‐field gel electrophoresis, and used to construct large‐insert libraries and shotgun libraries. The genome of the Ca. P. asteris OY‐M strain consists of a single chromosome of 860 631 bp and two plasmids, while that of the Ca. P. asteris AY‐WB strain consists of a single chromosome of 706 569 bp and four plasmids. Phytoplasma chromosomes are larger than those of Mycoplasma genitalium (~580 kb) (Fraser et al., 1995), despite the loss of several metabolic genes, as discussed below. Phytoplasma chromosomes also have low G+C contents (approximately 28 mol%), similar to most analysed mollicutes (Glass et al., 2000) and endosymbiotic bacteria of insects (Tamas et al., 2002; Wernegreen, 2002).
Each phytoplasmal plasmid encodes a replication initiation protein (Rep) involved in rolling‐circle replication (Bai et al., 2006; Nishigawa et al., 2002, b), as well as several other unknown proteins. The phytoplasmal Reps are similar not only to the Reps encoded by bacterial plasmids, but also to the Rep of geminivirus (plant virus) (Nishigawa et al., 2001) and circovirus (animal virus) (Oshima et al., 2001a); therefore, it is of interest to analyse the evolutionary relationships between phytoplasmas and viruses. Although the functions of the other genes in the phytoplasma plasmids are unknown, the plasmids of spontaneous OY‐M mutants that were not insect‐transmissible (Oshima et al., 2001a,b) lacked several open reading frames (ORFs), suggesting a relationship between the plasmids and insect transmissibility (Nishigawa et al., 2002a).
As the small genomes of parasitic and/or symbiotic bacteria consist mostly of functional genes, comparisons of the metabolic pathways in these organisms often reveal fundamental divergences in the microbial way of life and their evolutionary origins (Moran, 2002). In general, small‐genome pathogenic bacteria have lost the genes for numerous biosynthetic pathways, most likely because many metabolites are available within the host cell environment, leading to a reduced selective constraint on genes for biosynthetic capabilities. In addition, selection favouring the loss of factors such as microbe/pathogen associated molecular patterns (MAMPs or PAMPs), which may trigger host responses (Jones and Dangle, 2006), is another likely explanation for gene loss events, especially for phytoplasmas that have to navigate between two diverse hosts. Mycoplasmas lack the genes for the tricarboxylic acid cycle, sterol biosynthesis, fatty acid biosynthesis, de novo nucleotide synthesis and the biosynthesis of most amino acids; thus, they must depend entirely on their host cells to supply them with the products of these pathways (Razin et al., 1998). Similarly, no genes for these biosynthetic pathways have been identified in phytoplasmas. However, the phytoplasmas have lost more metabolic pathway genes than the mycoplasmas (Bai et al., 2006; Oshima et al., 2004), including those of the pentose phosphate pathway. Instead, phytoplasmas harbour multiple copies of transporter‐related genes that are not found in mycoplasmas (Oshima et al., 2004). These genomic features imply that phytoplasmas are highly dependent on metabolic compounds from their host cells.
A unique feature of the phytoplasma genome is the absence of the gene encoding F0F1‐type ATP synthase. In general, bacteria use F0F1‐type ATP synthases to synthesize and hydrolyse ATP using ATP‐proton motive force interconversion. Like other eubacteria, mycoplasmas also possess an F0F1‐type ATP synthase (Razin et al., 1998). However, no genes for an F0F1‐type ATP synthase have been identified in the two sequenced Ca. P. asteris phytoplasmas (Bai et al., 2006; Oshima et al., 2004). Although mutagenesis of Bacillus subtilis (Kobayashi et al., 2003) demonstrated that the genes encoding ATP synthase are not essential for life, these genes have been found in all fully sequenced bacteria. Thus, Ca. P. asteris is the first example of a naturally occurring organism with no ATP synthase genes. Taking into account a previous report showing that glycolytic turnover was increased in B. subtilis strains in which the atp operon had been deleted (Santana et al., 1994), ATP synthesis in phytoplasma might be strongly dependent on the glycolytic pathway. Recently, it was reported that an approximately 30‐kb region that includes the glycolytic genes was tandemly duplicated in the genome of a phytoplasma that causes severe symptoms (Oshima et al., 2007). This also implies that the glycolytic pathway may play an important role in the life cycle of phytoplasmas.
Despite the absence of an ATP synthase gene, there is considerable membrane potential in phytoplasmas, as was shown using a potentiometric dye (Christensen et al., 2004). Phytoplasmas have five genes encoding P‐type ATPases, which are similar to animal Na+/K+ and H+/K+ pumps (Bai et al., 2006). These ATPases may generate electrochemical gradients across the membrane, and they may be involved in the energy‐yielding systems of phytoplasmas (Christensen et al., 2005).
The phytoplasma genome contains genes for basic cellular functions, such as DNA replication, transcription, translation and protein translocation; however, it contains fewer genes dedicated to the standard recombination pathway and SOS response (DNA repair system) than the mycoplasma genome (Bai et al., 2006). This suggests that phytoplasmas have deficient homologous recombination machinery, and genomic rearrangements via homologous recombination are likely to occur at a low frequency.
REPEATED GENE SEQUENCES OF PHYTOPLASMA GENOMES
Intriguingly, phytoplasma genomes contain clusters of repeated gene sequences, named putative mobile units (PMUs) (Bai et al., 2006) and sequence‐variable mosaics (SVMs) (Jomantiene and Davis, 2006; Jomantiene et al., 2007). PMUs and SVMs have similar compositions and contain similar genes; these gene clusters are referred to hereafter as PMUs. In the AY‐WB genome, PMUs are ~20 kb in size and consist of genes with similarities to sigF, hflB, dnaG, dnaB, tmk, ssb, himA and tra5, which is an IS3 family insertion sequence (Lee et al., 2005), and several genes with unknown functions organized in a conserved order (Fig. 4a) (Bai et al., 2006). These genes are also found in multiple copies, singly or in clusters, in other phytoplasma genomes (Arashida et al., 2008; Jomantiene and Davis, 2006; Jomantiene et al., 2007; Lee et al., 2005; Oshima et al., 2004). The repeated presence of PMUs, their gene contents, including genes for recombination (tra5, ssb, himA) and replication (dnaG, dnaB), and their conserved gene orders suggest that the PMUs are replicative composite transposons (Bai et al., 2006). The presence of multiple PMUs or apparently degenerated PMU‐like sequences, such as SVMs, on the one hand and the dramatic loss of basic metabolic pathways in phytoplasma genomes on the other hand (Bai et al., 2006; Oshima et al., 2004) suggest that PMUs are likely to be somehow important for phytoplasma fitness.
Figure 4.

Organization and localization of repeats in phytoplasma genomes. (a) Potential mobile units (PMUs) of the AY‐WB chromosome. (b) A PMU‐like region encoding the virulence protein SAP11. The chromosome is presented as a black line. ORFs are represented as block arrows in which paralogous genes in figures A and B have the same colours, with the exception of the grey‐coloured arrows that represent unique genes. Conserved 395‐bp regions upstream of tra5 genes are indicated as grey vertical transparent ovals (see Fig. 5a for alignment). The locations and directions of the ~330‐bp conserved repeats (rep) are indicated in closed black arrowheads (see Fig. 5b for alignment). The names of the ORFs with predicted functions are indicated above the arrows, with ORFs of predicted membrane‐targeted proteins indicated with an asterisk and predicted secreted proteins with ‘s’. The tra5 ORFs of PMU4 and the SAP11 PMU‐like region contain separate A and B ORFs that may produce a full‐length transposases upon single frameshifting events. The three transcripts predicted for PMU1 are indicated as thin grey arrows beneath the PMU1 region ((a) was modified from Bai et al., 2006, with permission from the American Society for Microbiology).
PMU1 of AY‐WB has been analysed in greater detail (T.Y. Toruño et al., unpublished data). The genes of PMU1 are organized in three transcription units (Fig. 4a). The first consists of an operon of 12 genes that starts with the specialized transcription factor sigF and contains several genes that are involved in recombination (ssb and himA) and that encode membrane‐targeted proteins, such as HflB. A promoter sequence and ribosome‐binding sites are predicted directly upstream of the sigF ORF, whereas the downstream 11 ORFs are preceded only by ribosome‐binding sites. The second predicted operon encompasses seven genes, including dnaG, dnaB and tmk, that function in DNA replication and synthesis. All seven ORFs have upstream ribosome‐binding sites. The tra5 gene, which encodes a full‐length ORFAB fusion transposase of the IS3 family insertion sequences, was predicted to be part of an independent transcription unit. The entire PMU1 is flanked by two ~330‐bp repeats in inverted orientations (Bai et al., 2006).
In most bacteria, the IS3 ORFs A and B are two separate ORFs in which the protein product of ORFA suppresses transposition activity, and the ORFAB fusion, which encodes the active transposase, is produced only in the event of a frameshift (Mahillon and Chandler, 1998). The separation of ORFs A and B is important to prevent too frequent transposition events and subsequent genome alterations. In contrast, the majority of PMU tra5 genes of AY‐WB encode full‐length ORFAB‐fused transposases (Fig. 4a) (Bai et al., 2006), suggesting that these transposons are continuously active. However, most tra5 ORFs are preceded by a conserved sequence of 395 bp (4, 5), indicating that tra5 expression is regulated. This conserved 395‐bp region coincides with the presence of conserved ~330‐bp repeats (Fig. 5b) that flank PMU1, and are present in PMU3, PMU4 and PMU‐like regions (Fig. 4). Indeed, PMU2 does not have the conserved ~330‐bp repeats and the 395‐bp region upstream of tra5 (Fig. 4a).
Figure 5.

Alignments of conserved repeats in PMUs and PMU‐like regions. (a) Alignment of the conserved 395‐bp upstream sequences of the tra5 genes of PMU1, PMU3, PMU4 and the SAP11 PMU‐like region (vertical transparent ovals in Fig. 4). (b) Alignment of the conserved ~330‐bp repeats that flank PMU1 and are present in PMU3, PMU4 and the SAP11 PMU‐like region (closed black arrowheads in Fig. 4). Conserved nucleotides are indicated with an asterisk above the alignments, and missing nucleotides are indicated with dashes. The positions of nucleotides within the repeat regions are indicated in the ruler line beneath the alignments.
It is likely that PMUs have had a major role in phytoplasma genome evolution. The AY‐WB genome is approximately 150 kb smaller than the OY‐M genome, primarily due to fewer PMU sequences (Bai et al., 2006). Furthermore, alignment of the AY‐WB and OY‐M genomes has revealed that regions containing PMUs in both genomes had undergone various rearrangements, whereas regions without PMUs had not (Bai et al., 2006). This confirms the observation that large repeated DNAs have a destabilizing effect on a genome, given that they can recombine, resulting in deletions, inversions and duplications of genome segments (Achaz et al., 2003). The PMUs themselves apparently undergo rapid degeneration, as evidenced by the presence of PMU‐like regions carrying truncated copies of PMU genes and lacking various segments of full‐length PMUs (Bai et al., 2006; Jomantiene and Davis, 2006; Jomantiene et al., 2007). Thus, it is not surprising that in AY‐WB and OY‐M the essential genes required for basic functions, such as replication and metabolism, are clustered together in ~250‐kb genome segments that do not contain PMUs (Bai et al., 2006; Oshima et al., 2004). The other ~500 kb of the phytoplasma genomes contain PMUs as single insertions and in clusters of two or more adjacent PMUs or PMU‐like regions in direct and inverted orientations (Bai et al., 2006; Jomantiene and Davis, 2006).
The presence of PMUs in many other phytoplasmas suggests that these clusters are exchanged among phytoplasmas. AY‐WB belongs to subgroup 16SrIA within Ca. Phytoplasma asteris (16SrI group phytoplasmas), but PMUs were also found in the genomes of subgroup 16SrIB phytoplasmas OY‐M (Bai et al., 2006; Oshima et al., 2004) and MBSP (Hogenhout et al., unpublished data), subgroup 16SrIC Clover phyllody (CPh) phytoplasma (Jomantiene and Davis, 2006), the 16SrVI phytoplasma BLTVA (Beet Leafhopper‐transmitted virescence phytoplasma) (Ca. Phytoplasma trifolii) (S. A. Hogenhout, unpublished data) and other phytoplasmas (Jomantiene et al., 2007). Evidence of PMU‐associated sequences undergoing horizontal gene transfer among mollicutes was provided by the mgs1 gene. This gene is part of PMU3 of AY‐WB and is flanked by two tra5 genes (Fig. 4a). A highly similar mgs1 gene is present in the genome of the distantly related Spiroplasma kunkelii (Bai et al., 2004b). Comparison of sequence identity percentages and phylogenetic analyses with mgs1, 16S rRNA and conserved metabolic genes provides evidence for horizontal gene transfer of mgs1 between the plant‐associated mollicutes (Bai et al., 2004b) (Fig. 6), consistent with its location between two full‐length tra5 genes in the AY‐WB genome (Fig. 4a) and an IS30 sequence in the S. kunkelii genome (our personal observations). The mgs1 sequence is also present in the genome of BLTVA, but not in those of MBSP or OY‐M (S. A. Hogenhout et al., unpublished results) (Fig. 6).
Figure 6.
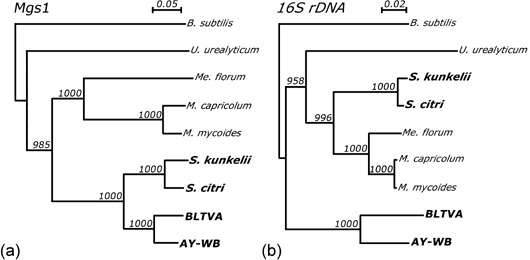
Evidence for horizontal gene transfer of mgs1 among the plant‐associated mollicutes. (a) The tree of the Mgs1 protein sequences shows clustering of the plant‐associated mollicutes. (b) The tree of 16S rRNA gene nucleotide sequences shows that the phytoplasmas (BLTVA and AY‐WB) form a separate cluster from the mycoplasmas, mesoplasmas, spiroplasmas and ureaplasmas, consistent with published phylogenetic analyses and differences in codon usage and metabolic activities among the mollicutes, see also Fig. 2 (Razin et al., 1998). Alignments were generated with ClustalX 1.83 (Thompson et al., 1997). Gaps were excluded from the trees that were generated by the neighbour‐joining method (Saitou and Nei, 1987) of ClustalX. Bootstrap values of 1000 replicates are indicated at the branches. The trees were rooted with the B. subtilis sequences. The insect‐transmitted plant‐associated mollicutes (AY‐WB, BLTVA, S. kunkelii and S. citri) are highlighted in bold font.
One PMU‐like region was shown to contain a gene encoding a putative effector (virulence) protein, SAP11, which contains a nuclear localization signal (NLS) (Fig. 4b) (X. Bai et al., unpublished data). SAP11 targets the nuclei of plant cells and changes plant gene transcription levels (X. Bai et al., unpublished data). In addition, the SAP11 region, PMU1 and other PMUs all carry genes encoding specialized transcription factors (sigF) and predicted membrane‐targeted proteins (Fig. 4). Overall these observations suggest that PMUs are probably involved in the adaptations of phytoplasmas to their plant and insect hosts. Indeed, repeated DNAs of mycoplasmas, which are also mollicutes, are also involved in host adaptations. The small genomes of mycoplasmas contain large repeats encoding major surface proteins and adhesins that take part in the antigenic variation strategies of mycoplasmas (Baseman et al., 1982; Rocha and Blanchard, 2002). In other bacteria, long repeats are involved in antigenic variation strategies as well (Rocha et al., 1999). SAP11 and other putative phytoplasma effector proteins will be further discussed below.
PHYTOPLASMA PROTEIN SECRETION MECHANISMS
In bacteria, there are at least five independent protein export systems (Economogu, 1999). These systems secrete various proteins such as toxins, adhesins and hydrolytic enzymes. Many Gram‐negative pathogens of plants and animals possess T3SSs that can inject bacterial virulence ‘effector’ proteins into host cells (Abramobitch et al., 2006 and Cornelis and van Gijsegem, 2000). The T3SSs are important for the pathogenicity of phytopathogens in the genera Pseudomonas, Xanthomonas, Ralstonia, Erwinia and Pantoea. T3SSs and flagella are evolutionarily related and share a remarkably similar basal structure. However, both T3SSs and flagella are restricted to Gram‐negative bacteria; thus, phytoplasmas, which belong phylogenetically to Gram‐positive bacteria, probably contain no T3SSs.
Bacterial type IV secretion systems (T4SSs) represent other important secretion mechanisms in plant and animal pathogens. T4SSs comprise a large family of translocation systems that use pilus‐like structures to transfer DNA or protein substrates to bacterial or eukaryotic cells (Grohmann et al., 2003; Tanaka and Sasakawa, 2005). Several component proteins of the conjugative transfer system in Gram‐positive and Gram‐negative bacteria have sequence similarities to the component proteins of T4SSs; therefore, conjugative transfer systems and T4SSs are thought to be ancestrally related (Grohmann et al., 2003). Thus, it has been suggested that T4SSs are widely distributed among Gram‐negative and Gram‐positive bacteria (Christie and Cascales, 2005). However, the OY‐M and AY‐WB genomes contain no genes with similarities to those encoding the component proteins of pili or T4SSs (Bai et al., 2006; Oshima et al., 2004), and electron microscopic analyses have not detected the existence of pilus‐like structures in phytoplasmas. In contrast, pilus‐like structures and four distinct traE genes that bear similarities to genes typically present in type IV secretory pathways have been reported in Spiroplasma kunkelii (Ammar et al., 2004; Bai et al., 2004a; Davis et al., 2005; Özbek et al., 2003). Similar traE genes are also present in S. citri (Joshi et al., 2005).
Of the five protein export systems in bacteria, only the Sec protein translocation system is essential for cell viability (Economogu, 1999; Tjalsma et al., 2000). The Escherichia coli Sec system, which is composed of at least 11 proteins and one RNA species, is the best characterized (Economogu, 1999). Among these proteins, SecA, SecY and SecE are essential for protein translocation and cell viability in E. coli (Economogu, 1999), and protein translocation activity can be reconstituted in vitro with only these three proteins (Akimaru et al., 1991). In OY phytoplasma, the genes encoding SecA, SecY and SecE have been identified (Kakizawa et al., 2001, 2004), and SecA expression has been confirmed in phytoplasma‐infected plants (Kakizawa et al., 2001; Wei et al., 2004). These three genes have also been identified in the AY‐WB genome (Bai et al., 2006), and secY genes have been cloned from several other phytoplasmas (Lee et al., 2006b). These results strongly suggest that a functional Sec system is common among phytoplasmas.
YidC is involved in the integration of newly synthesized membrane proteins (Dalbey and Kuhn, 2000). Initially, YidC was found to co‐purify with components of the Sec system (Scotti et al., 2000), and it was thought that YidC worked in conjunction with Sec translocase to insert the transmembrane regions of Sec‐dependent substrate proteins into the hydrophobic bilayer (Urbanus et al., 2001). However, it was recently demonstrated that YidC is sufficient to promote the membrane insertion of a membrane protein in vitro, suggesting that YidC can function independently of the Sec system (Serek et al., 2004). YidC is encoded in both the OY‐M and the AY‐WB genomes (Bai et al., 2006; Oshima et al., 2004); thus, phytoplasmas should have a YidC integration system. Because YidC is an essential protein in E. coli (Samuelson et al., 2000), it may also have an important role in phytoplasmas.
In summary, phytoplasmas have two secretion systems, the YidC system for the integration of membrane proteins, and the Sec system for the integration and secretion of proteins into the host cell cytoplasm. Antigenic membrane protein (Amp), a type of immunodominant membrane protein in phytoplasmas (Barbara et al., 2002), has been reported to be a substrate of the Sec system. Amp has a Sec signal sequence at its N‐terminus, which is cleaved in OY phytoplasma (Kakizawa et al., 2004), suggesting that the phytoplasma Sec system utilizes recognition and cleavage of a signal sequence, as in other bacterial Sec systems. This also suggests that signal sequence prediction programmes, such as SignalP (Nielsen et al., 1997) or PSORT (Nakai and Kanehisa, 1991), may be applicable to phytoplasmal proteins and could be used to identify secretory proteins (Kakizawa et al., 2004). As phytoplasmas lack a cell wall, their membrane proteins and secreted proteins function directly in the cytoplasm of the host plant or insect cell. Thus, the identification of secreted proteins encoded in the phytoplasma genome will be important for understanding host–phytoplasma interactions.
IDENTIFICATION AND FUNCTIONAL CHARACTERIZATION OF PHYTOPLASMA VIRULENCE FACTORS
Because membrane and secreted proteins are potential virulence factors, the complete AY‐WB genome was mined for the presence of membrane‐targeted proteins by determining whether deduced protein sequences carry N‐terminal signal peptide (SP) sequences (X. Bai et al., unpublished data) using the program SignalP (Bendtsen et al., 2004). This resulted in the identification of 76 AY‐WB proteins with SPs. Of these 76, 20 proteins contained an SP and additional transmembrane domains as predicted by TMHMM2.0 (Krogh et al., 2001; X. Bai et al., unpublished data). Thus, these 20 proteins are apparently initially secreted but remain associated with the membranes. Indeed, proteins AYWB_016, AYWB_395, AYWB_432, AYWB_599 (GenBank accession no. CP000061) and pIV04 (GenBank accession no. CP000065) have one additional transmembrane domain at their C‐termini and therefore are predicted to locate at the exterior of the phytoplasma cell. The accuracy of the SP and transmembrane domains predictions is validated by the presence of AYWB_599 among the list of 20 proteins. AY‐WB_599 has similarity to OY Amp that, as discussed above, has a cleavable SP (Barbara et al., 2002), is secreted by the Sec‐dependent translocation system (Kakizawa et al., 2004) and is an abundant cell surface protein (Milne et al., 1995). Amp is involved in phytoplasma virulence as it is required for uptake of phytoplasmas by the insect vector (Suzuki et al., 2006).
The remaining 56 proteins contain only SPs and no additional transmembrane domains, and are therefore probably secreted into the extracellular environment of AY‐WB (X. Bai et al., unpublished data). Most of the proteins have unknown functions. However, the list of 56 proteins contains the five solute‐binding proteins (SBPs) ArtI (AYWB_263), DdpA (AYWB_529), NlpA (AYWB_588), ZnuA (AYWB_624) and MalE (AYWB_667) (GenBank accession no. CP000061) (Bai et al., 2006; X. Bai et al., unpublished data). SBPs are secreted through the Sec‐dependent pathway in other bacteria (Higgins, 2001), thereby validating the SP predictions. SBPs are part of ATP‐binding cassette (ABC) transporters (Higgins 2001). As expected, the genes for the AY‐WB SBPs are located adjacent to genes of ABC transporter systems in the AY‐WB chromosome (Bai et al., 2006).
SBPs are frequently involved in bacterial virulence (Higgins, 2001). For example, the SBP Sc76 of another insect‐transmitted mollicute pathogen, Spiroplasma citri, is important for infection of the salivary glands and transmission by the leafhopper Circulifer haematoceps (Boutareaud et al., 2004). Many SBPs, including Sc76, are lipoproteins that remain attached to the bacterial membrane through a lipid anchor at the first conserved cysteine residue of the mature peptide (Taylor et al., 2006). None of the five AY‐WB SBPs and 51 other predicted secreted proteins has a conserved cysteine residue at the +1 position (X. Bai et al., unpublished data). Hence, the 56 SAPs are probably released in the extracellular environment of AY‐WB upon secretion, and therefore are putative effector proteins involved in the manipulation of plant and insect cells.
Additional independent evidence that the AY‐WB SAPs are effector proteins was obtained. Two phytoplasma effectors, SAP11 and SAP30, contain eukaryotic NLSs that are functional in plant cells (X. Bai et al., unpublished data). Yellow fluorescent protein (YFP) fusions of SAP11 and SAP30 solely accumulated in nuclei of N. benthamiana cells, whereas YFP fusions of SAP11 mutants without NLS domain were distributed throughout the plant cells (X. Bai et al., unpublished data). In N. benthamiana, nuclear localization of YFP‐SAP11 requires a host factor, importin α (X. Bai et al., unpublished data) that is required for import of proteins into plant nuclei (Adam et al., 1995; Gorlich et al., 1995), including virulence proteins of other plant pathogens (Kanneganti et al., 2007). Overexpression of SAP11 induced necrosis in N. benthamiana and alters gene transcription, notably of several transcription factors, in tomato (X. Bai et al., unpublished data). In immunofluorescence microscopy experiments with a specific antibody to SAP11, it was shown that SAP11 also located to nuclei of AY‐WB‐infected China aster cells (X. Bai et al., unpublished data). Thus, SAP11, and perhaps also SAP30, appears to be involved in the manipulation of plant processes during AY‐WB infection.
The immunofluorescence microscopy experiments with AY‐WB‐infected China aster plants detected SAP11 beyond the phloem tissue in nuclei of mesophyll cells and cells of other tissues (X. Bai et al., unpublished data), suggesting that SAP11 unloads from the phloem. In agreement with this finding, Imlau et al. (1999) have shown that GFP specifically produced in phloem cells under control of the AtSUC2 promoter in Arabidopsis can unload from the phloem into mesophyll cells and other cell types of developing (sink) tissues, including young rosette leaves, petals, anthers and roots, whereas this post‐phloem transport of GFP was not detected in fully developed (source) tissues. Indeed, the size exclusion limits (SELs) of plasmodesmata change during organ development. The SELs of sink tissue plasmodesmata range between 10 and 40 kDa, whereas those of source tissues are much smaller (Imlau et al., 1999). Interestingly, the majority of secreted AY‐WB proteins (51 of 56), including SAP11 and SAP30, are smaller than 40 kDa (X. Bai et al., unpublished data), indicating that most secreted protein of AY‐WB may move out of the phloem and target developing tissues of plants. This is consistent with the appearance of symptoms in predominantly developing tissues of phytoplasma‐infected plants, for instance witches’ broom symptoms of developing tissues (Fig. 1B). Our data are also consistent with the production of Sec transport pathway proteins by phytoplasmas in the plant phloem (Kakizawa et al., 2001). Thus, similarly to other pathogens, phytoplasmas secrete effector proteins that manipulate plant host components.
Membrane‐bound proteins that all have SPs and transmembrane domains were shown to determine insect specificity. A subset of membrane proteins, usually referred to as immunodominant membrane proteins (IDPs), constitute a major portion of the total cellular membrane proteins in most phytoplasmas (Shen and Lin, 1993). Immunogold‐labelling electron microscopy studies have demonstrated that IDP is located on the exterior surface of the cell membrane (Milne et al., 1995). As the mollicute membrane proteins probably play important roles in the attachment to the host cell, IDPs are candidates involved in host–phytoplasma interactions. Genes encoding IDPs have been isolated from several phytoplasmas (Barbara et al., 2002; Berg et al., 1999; Blomquist et al., 2001; Kakizawa et al., 2004, 2006a, 2006b; Morton et al., 2003) (Fig. 7). These IDPs were classified into three distinct types: (1) immunodominant membrane protein (Imp); (2) immunodominant membrane protein A (IdpA); and (3) antigenic membrane protein (Amp), and they are not orthologues of each other (Barbara et al., 2002; Kakizawa et al., 2006a,b). Namely, one of three non‐homologous types of IDP can have an identical role in different phytoplasmas, constituting the major portion of the plasma membrane proteins.
Figure 7.

Immunodominant membrane proteins (IDPs) of phytoplasma were classified into three distinct types. (a) Gene organizations around the genes encoding three IDPs. The gene organization of types 1, 2 and 3 were described using sequence from SPWB (U15224), WX (AF533231) and OY‐W (AB124806), respectively. (b) Schematic representation of the putative translocation products of three IDPs. Transmembrane regions are shown as blue regions and non‐transmembrane regions are shown as pink regions. The N‐terminal transmembrane region of Amp (type 3) is cleaved during protein localization (Kakizawa et al., 2004), and the cleavage site is indicated with a filled triangle. Abbreviations: C, C terminus; N, N terminus; aa, amino acids. (c) Schematic representation of the hypothetical transmembrane structures of three types of IDP. Abbreviations and colour lines are same as B.
Several investigators have reported that the IDPs within a type are highly variable (Barbara et al., 2002; Kakizawa et al., 2004; Morton et al., 2003). In general, coding regions rarely show lower identities than non‐coding regions because of functional constraints. However, the sequence similarity of IDP genes between phytoplasmas is lower than that of their upstream genes, downstream genes or non‐coding regions (Barbara et al., 2002; Kakizawa et al., 2004), suggesting that IDPs have been subjected to strong divergent selective pressures. In addition, the extracellular hydrophilic domains of IDPs are more divergent than the transmembrane domains or the export signal sequences, implying that the host–phytoplasma interactions promote the variability of IDPs (Barbara et al., 2002; Kakizawa et al., 2004). Moreover, it has been reported that the sequence identities of imp in several phytoplasmas were not correlated with that of the 16S rRNA gene, which suggests that the variability of IDPs reflects some factors other than evolutionary time (Morton et al., 2003). Recently, the Amp proteins were shown to be under positive selection, and the positively selected amino acids were in the central hydrophilic domain of the Amp (Kakizawa et al., 2006a,b), suggesting that Amp plays an important role in host–pathogen interactions, just like many positively selected proteins that were previously reported (Andrews and Gojobori, 2004; Bishop et al., 2000; Hughes and Nei, 1988; Jiggins et al., 2002; Nielsen and Yang, 1998; Urwin et al., 2002).
Recently, it has been reported that the Amp of OY phytoplasma interacts with an insect microfilament complex (Suzuki et al., 2006). OY phytoplasma was localized to the microfilaments of the insect's intestinal tract, and the Amp formed a complex with three insect proteins involved in a microfilament both in vitro and in vivo. In addition, the formation of Amp–microfilament complexes was correlated with the phytoplasma‐transmitting capability of leafhoppers, suggesting that the interaction between Amp and insect microfilament complexes plays a major role in determining the transmissibility of phytoplasmas (Fig. 8). The interactions between surface proteins of microbes and host microfilaments have often been reported (Cossart et al., 2003; Hayward and Koronakis, 2002). Taking into consideration the Amp–microfilament complexes, the interactions between a bacterial membrane protein and the microfilament of a host cell might be a general system, and are important for successful bacterial infection.
Figure 8.

The transmissibility of OY phytoplasma by leafhoppers (left column) was completely consistent with the formation of the Amp–microfilament complex (right column). + and –: OY‐Amp protein does or does not, respectively, form a complex with these insects’ microfilament (Suzuki et al., 2006).
The infection of insect hosts by phytoplasmas is composed of several different steps (Nielson, 1979). In these steps, there are two major barriers to pass through for successful infection of phytoplasma: the insect intestine and the salivary gland (Markham and Townsend, 1979; Purcell et al., 1981). Although the role of the Amp–microfilament complex remains unclear, it is likely that it may be necessary for the passage through these barriers, especially the insect intestine.
Also in the case of spiroplasma, the salivary gland is a specific barrier where spiroplasmas must pass for the infection with an insect (Markham and Townsend, 1979). S. citri and S. kunkelii are thought to invade from the gut epithelium of the insect host through a process of receptor‐mediated cell endocytosis (Ammar et al., 2004; Kwon et al., 1999; Özbek et al., 2003). Receptors on leafhopper gut epithelial cells probably recognize specific spiroplasma membrane proteins. Several candidate S. citri attachment protein genes have been isolated, including the immunodominant membrane protein (spiralin) (Duret et al., 2003), P58 (Ye et al., 1997), SARP1 (Berg et al., 2001) and P32 of pSci6 plasmid (Berho et al., 2006a,b; Killiny et al., 2006). It has been reported that the defective mutant of the spiralin was less effective in its transmissibility (Duret et al., 2003), and that the spiralin binds to glycoproteins of its insect vector (Killiny et al., 2005). Although details of the molecular function of spiralin are still unknown, spiralin is a candidate for the key component that determines insect vector specificity. Thus, although no similarity was detected between Amp and spiralin, the immunodominant membrane proteins of phytoplasmas and spiroplasmas would commonly play an important role in transmission by insect vectors.
The plasmid pSci6 confers insect transmissibility to a non‐transmissible strain of S. citri (Berho et al., 2006a,b). The pSci6 plasmid encodes protein P32, which was thought to be associated with insect transmissibility (Killiny et al., 2006). However, when only the p32 gene was introduced into the non‐transmissible strain of S. citri, its transmissibility was not restored (Killiny et al., 2006). Thus, pSci6‐encoded determinants other than P32 might be essential for insect transmission, and a more detailed analysis of pSci6 is expected. In phytoplasmas, the Amp–microfilament complex is probably essential for insect transmission, but this complex is probably not sufficient for the completion of all steps involved in insect transmission that requires passage of at least two barriers, the gut and salivary glands. Indeed, the plasmid‐encoded gene, ORF3, was suggested to take part in the insect transmission process as well (Nishigawa et al., 2002a). Hence, functional analysis of the protein encoded by ORF3 will probably reveal additional mechanisms that play a role in insect transmission.
FUTURE DIRECTIONS
Forty years have passed since phytoplasmas were first discovered. It is clear that much progress has since been made in understanding the molecular biology of phytoplasmas. It became clear that phytoplasmas are found all over the world and in the majority of plant species, are highly diverse (consisting of approximately 30 subclades), have highly flexible reduced genomes, and probably secrete a battery of virulence factors that interact with insect hosts (e.g. IDP of OY‐M) and plant hosts (e.g. SAP11 of AY‐WB). These new findings generate a series of new research questions.
The first question relates to the function of PMUs. PMUs seem to be involved in major phytoplasma genome rearrangements and size changes. In addition, the finding that PMUs contain genes for membrane‐targeted proteins suggests that they may mediate phytoplasma interactions with hosts. If so, phytoplasmas could adapt to different hosts by varying the number and types of PMUs. Alternatively, adaptation to hosts is regulated by a phase variation mechanism. PMUs may invert and/or form circular replisomes thereby changing the expression levels of PMU‐encoded genes. Finally, the possibility that PMUs are phage‐like parasitic DNAs that need the membrane proteins for escape and to enter other phytoplasmas should be considered. Several groups in the phytoplasma research community have started working on further characterization of PMU and PMU‐like sequences, and hence it is expected that more will be known about these intriguing structures in the near future.
Another question concerns the function of the genes that encode various membrane‐targeted and secreted proteins. It is clear that phytoplasma interactions with plants and insects are quite complex, and as reviewed herein requires the involvement of membrane‐associated proteins and secreted proteins (effector proteins). There are many other genes encoding putative effector proteins. The majority of these have unknown functions and are unique to phytoplasmas. Since phytoplasmas can efficiently infect insects and plants, it is possible that some of these effectors interact with proteins that are conserved among plants and animals. This could explain why phytoplasmas have broad host ranges.
Future research should reveal whether phytoplasma effectors are involved in the induction of developmental symptoms typical of phytoplasma‐infected plants and the enhanced susceptibility of plants to phloem‐feeding insects. Various pathogens produce effector proteins that target host pathways to enhance host susceptibility to the pathogen. Phytoplasmas may manipulate plants in a similar manner, but in addition, can apparently also affect the interaction of plants with their environment, including phloem‐feeding insects.
Phytoplasmas are obligate intracellular parasites and cannot be cultured in vitro. This has been the greatest barrier to characterizing these economically important, biologically interesting microorganisms at the molecular level. Determining the complete genomic sequence of individual species will enable the development of novel analytical techniques, including detection methods that could be used to gauge the prevalence of phytoplasmas in nature or to find the source of new outbreaks. Phytoplasma‐related diseases are expected to increase because of global warming/climate change, which is advantageous to the cold‐sensitive phytoplasma vectors, and the implementation of new and more restrictive regulations on the use of many pesticides to control phloem‐feeding insects, together with the increase in organic agriculture. Therefore, the detection of phytoplasmas will become more important in the future.
Real‐time PCR, nested‐PCR and microarray analysis for the detection of phytoplasmas have been developed, but most of these are based on 16S rRNA gene sequences (Hadidi et al., 2004; Hodgetts et al., 2007; Nicolaisen and Bertaccini, 2007; Torres et al., 2005). Genomic analyses have demonstrated that roughly 40% of the ORFs in phytoplasmas do not show significant similarity to sequences deposited in GenBank, and, among these, approximately 200 genes are conserved in the OY and AY‐WB genomes (Bai et al., 2006). These unique genes may be good targets for the detection of phytoplasmas.
In summary, phytoplasmas have a unique biology among the plant pathogens. Valuable insights into the pathogenicity and evolutionary relationships of phytoplasmas have been derived from the two complete genomic sequences obtained thus far. Several other genome sequencing projects are currently in progress, and the sequencing of additional phytoplasma genomes will further our understanding of these economically important and biologically attractive microorganisms at the molecular level.
REFERENCES
- Abramovitch, R.B. , Anderson, J.C. and Martin, G.B. (2006) Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achaz, G. , Coissac, E. , Netter, P. and Rocha, E.P. (2003) Associations between inverted repeats and the structural evolution of bacterial genomes. Genetics, 164, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, S.A. , Adam, E.J. , Chi, N.C. and Visser, G.D. (1995) Cytoplasmic factors in NLS‐mediated targeting to the nuclear pore complex. Cold Spring Harb. Symp. Quant. Biol. 60, 687–694. [DOI] [PubMed] [Google Scholar]
- Akimaru, J. , Matsuyama, S. , Tokuda, H. and Mizushima, S. (1991) Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli . Proc. Natl Acad. Sci. USA, 88, 6545–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar, E.‐D. and Hogenhout, S. A. (2006) Mollicutes associated with arthropods and plants In: Insect Symbiosis, Vol. 2 (Kostas B. and Miller T., eds), pp. 97–118. CRC Press, Taylor and Francis Group, Boca Raton, FL, USA. [Google Scholar]
- Ammar, E.‐D. , Fulton, D. , Bai, X. , Meulia, T. and Hogenhout, S.A. (2004) An attachment tip and pili‐like structures in insect‐and plant‐pathogenic spiroplasmas of the class Mollicutes . Arch. Microbiol. 181, 97–105. [DOI] [PubMed] [Google Scholar]
- Andrews, T.D. and Gojobori, T. (2004) Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis . Genetics, 166, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arashida, R. , Kakizawa, S. , Hoshi, A. , Ishii, Y. , Jung, H.Y. , Kagiwada, S. , Yamaji, Y. , Oshima, K. and Namba, S. (2008) Heterogeneic dynamics of the structures of multiple gene clusters in two pathogenetically different lines originating from the same phytoplasma. DNA Cell Biol. in press. [DOI] [PubMed] [Google Scholar]
- Arocha, Y. , Lopez, M. , Pinol, B. , Fernandez, M. , Picornell, B. , Almeida, R. , Palenzuela, I. , Wilson, M.R. and Jones, P. (2005) ‘Candidatus Phytoplasma graminis’ and ‘Candidatus Phytoplasma caricae’, two novel phytoplasmas associated with diseases of sugarcane, weeds and papaya in Cuba. Int. J. Syst. Evol. Microbiol. 55, 2451–2463. [DOI] [PubMed] [Google Scholar]
- Bai, X. , Fazzolari, T. and Hogenhout, S.A. (2004a) Identification and characterization of traE genes of Spiroplasma kunkelii . Gene, 336, 81–91. [DOI] [PubMed] [Google Scholar]
- Bai, X. , Zhang, J. , Ewing, A. , Miller, S.A. , Jancso Radek, A. , Shevchenko, D.V. , Tsukerman, K. , Walunas, T. , Lapidus, A. , Campbell, J.W. and Hogenhout, S.A. (2006) Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188, 3682–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X. , Zhang, J. , Holford, I.R. and Hogenhout, S.A. (2004b) Comparative genomics identifies genes shared by distantly related insect‐transmitted plant pathogenic mollicutes. FEMS Microbiol. 235, 249–258. [DOI] [PubMed] [Google Scholar]
- Barbara, D.J. , Morton, A. , Clark, M.F. and Davies, D.L. (2002) Immunodominant membrane proteins from two phytoplasmas in the aster yellows clade (chlorante aster yellows and clover phyllody) are highly divergent in the major hydrophilic region. Microbiology, 148, 157–167. [DOI] [PubMed] [Google Scholar]
- Barre, A.A. , De Daruvar, A. and Blanchard, A. (2004) MolliGen, a database dedicated to the comparative genomics of Mollicutes. Nucleic Acids Res. 32, D307–D310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseman, J.B. , Cole, R.M. , Krause, D.C. and Leith, D.K. (1982) Molecular basis for cytadsorption of Mycoplasma pneumoniae . J. Bacteriol. 151, 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanland, L. , Hoy, C.W. , Miller, S.A. and Nault, L.R. (2000) Influence of aster yellows phytoplasma on the fitness of aster leafhopper (Homoptera: Cicadellidae). Ann. Entomol. Soc. Am. 93, 271–276. [Google Scholar]
- Bendtsen, J.D. , Nielsen, H. , Von Heijne, G. and Brunak, S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Berg, M. , Davies, D.L. , Clark, M.F. , Vetten, H.J. , Maier, G. , Marcone, C. and Seemüller, E. (1999) Isolation of the gene encoding an immunodominant membrane protein of the apple proliferation phytoplasma, and expression and characterization of the gene product. Microbiology, 145, 1937–1943. [DOI] [PubMed] [Google Scholar]
- Berg, M. , Melcher, U. and Fletcher, J. (2001) Characterization of Spiroplasma citri adhesion related protein SARP1, which contains a domain of a novel family designated sarpin. Gene, 275, 57–64. [DOI] [PubMed] [Google Scholar]
- Berho, N. , Duret, S. and Renaudin, J. (2006a) Absence of plasmids encoding adhesion‐related proteins in non‐insect‐transmissible strains of Spiroplasma citri. Microbiology, 152, 873–886. [DOI] [PubMed] [Google Scholar]
- Berho, N. , Duret, S. , Danet, J.L. and Renaudin, J. (2006b) Plasmid pSci6 from Spiroplasma citri GII‐3 confers insect transmissibility to the non‐transmissible strain S. citri 44. Microbiology, 152, 2703–2716. [DOI] [PubMed] [Google Scholar]
- Bertaccini, A. (2007) Phytoplasmas: diversity, taxonomy, and epidemiology. Front. Biosci. 12, 673–689. [DOI] [PubMed] [Google Scholar]
- Bishop, J.G. , Dean, A.M. and Mitchell‐Olds, T. (2000) Rapid evolution in plant chitinases: molecular targets of selection in plant‐pathogen coevolution. Proc. Natl Acad. Sci. USA, 97, 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist, C.L. , Barbara, D.J. , Davies, D.L. , Clark, M.F. and Kirkpatrick, B.C. (2001) An immunodominant membrane protein gene from the Western X‐disease phytoplasma is distinct from those of other phytoplasmas. Microbiology, 147, 571–580. [DOI] [PubMed] [Google Scholar]
- Botti, S. and Bertaccini, A. (2006) Phytoplasma infection through seed transmission: further observations. In: Abstracts, 16th International Organization of Mycoplasmology Conference, Cambridge, UK, p. 76. [Google Scholar]
- Boutareaud, A. , Danet, J.L. , Garnier, M. and Saillard, C. (2004) Disruption of a gene predicted to encode a solute binding protein of an ABC transporter reduces transmission of Spiroplasma citri by the leafhopper Circulifer haematoceps . Appl. Environ. Microbiol. 70, 3960–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, E.J. (1982) Ultrastructural investigation of resistant and susceptible maize inbreds infected with Erwinia stewartii . Phytopathology, 72, 159–166. [Google Scholar]
- Christensen, N.M. , Axelsen, K.B. , Nicolaisen, M. and Schulz, A. (2005) Phytoplasmas and their interactions with hosts. Trends Plant Sci. 10, 526–535. [DOI] [PubMed] [Google Scholar]
- Christensen, N.M. , Nicolaisen, M. , Hansen, M. and Schulz, A. (2004) Distribution of phytoplasmas in infected plants as revealed by real‐time PCR and bioimaging. Mol. Plant–Microbe Interact. 17, 1175–1184. [DOI] [PubMed] [Google Scholar]
- Christie, P.J. and Cascales, E. (2005) Structural and dynamic properties of bacterial type IV secretion systems (review). Mol. Membr. Biol. 22, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo, V.P. (1993) Transport systems in mycoplasmas. Subcell. Biochem. 20, 293–310. [DOI] [PubMed] [Google Scholar]
- Cordova, I. , Jones, P. , Harrison, N.A. and Oropexa, C. (2003) In situ PCR detection of phytoplasma DNA in embryos from coconut palms with lethal yellowing disease. Mol. Plant Pathol. 4, 99–108. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R. and Van Gijsegem, F. (2000) Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54, 735–774. [DOI] [PubMed] [Google Scholar]
- Cossart, P. , Pizarro‐Cerda, J. and Lecuit, M. (2003) Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 13, 23–31. [DOI] [PubMed] [Google Scholar]
- Dalbey, R.E. and Kuhn, A. (2000) Evolutionarily related insertion pathways of bacterial, mitochondrial, and thylakoid membrane proteins. Annu. Rev. Cell Dev. Biol. 16, 51–87. [DOI] [PubMed] [Google Scholar]
- Davis, R.E. , Dally, E.L. , Gundersen, D.E. , Lee, I.M. and Habili, N. (1997) ‘Candidatus phytoplasma australiense,’ a new phytoplasma taxon associated with Australian grapevine yellows. Int. J. Syst. Bacteriol. 47, 262–269. [DOI] [PubMed] [Google Scholar]
- Davis, R.E. , Dally, E.L. , Jomantiene, R. , Zhao, Y. , Roe, B. , Lin, S. and Shao, J. (2005) Cryptic plasmid pSKU146 from the wall‐less plant pathogen Spiroplasma kunkelii encodes an adhesin and components of a type IV translocation‐related conjugation system. Plasmid, 53, 179–190. [DOI] [PubMed] [Google Scholar]
- Doi, Y. , Teranaka, M. , Yora, K. and Asuyama, H. (1967) Mycoplasma‐ or PLT group‐like microorganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches’ broom, aster yellows or paulownia witches’ broom. Ann. Phytopathol. Soc. Jpn, 33, 259–266. [Google Scholar]
- Duret, S. , Berho, N. , Danet, J.L. , Garnier, M. and Renaudin, J. (2003) Spiralin is not essential for helicity, motility, or pathogenicity but is required for efficient transmission of Spiroplasma citri by its leafhopper vector Circulifer haematoceps. Appl. Environ. Microbiol. 69, 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbert, M.A. and Nault, L.R. (1994) Improved overwintering ability in Dalbulus maidis (Homoptera: Cicadellidae) vectors infected with Spiroplasma kunkelii (Mycoplasmatales: Spiroplasmataceae). Environ. Entomol. 23, 634–644. [Google Scholar]
- Economogu, A. (1999) Following the leader: bacterial protein export through the Sec pathway. Trends Microbiol. 7, 315–320. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1977) Anatomy of Seed Plants. New York: Wiley. [Google Scholar]
- Firrao, G. , Garcia‐Chapa, M. and Marzachi, C. (2007) Phytoplasmas: Genetics, DIagnosis and Relationships with the Plant and Insect Host. Front. BioSci. 12, 1353–1375. [DOI] [PubMed] [Google Scholar]
- Fraser, C.M. , Gocayne, J.D. , White, O. , Adams, M.D. , Clayton, R.A. , Fleischmann, R.D. , Bult, C.J. , Kerlavage, A.R. , Sutton, G. , Kelley, J.M. , Fritchman, R.D. , Weidman, J.F. , Small, K.V. , Sandusky, M. , Fuhrmann, J. , Nguyen, D. , Utterback, T.R. , Saudek, D.M. , Phillips, C.A. , Merrick, J.M. , Tomb, J.F. , Dougherty, B.A. , Bott, K.F. , Hu, P.C. , Lucier, T.S. , Peterson, S.N. , Smith, H.O. , Hutchison, C.A. 3rd and Venter, J.C. (1995) The minimal gene complement of Mycoplasma genitalium. Science, 270, 397–403. [DOI] [PubMed] [Google Scholar]
- Gasparich, G.E. (2002) Spiroplasmas: evolution, adaptation and diversity. Front. Biosci. 7, d619–640. [DOI] [PubMed] [Google Scholar]
- Gaurivaud, P. , Laigret, F. , Garnier, M. and Bove, J. M. (2000) Fructose utilization and pathogenicity of Spiroplasma citri: characterization of the fructose operon. Gene, 252, 61–69. [DOI] [PubMed] [Google Scholar]
- Glass, J.I. , Lefkowitz, E.J. , Glass, J.S. , Heiner, C.R. , Chen, E.Y. and Cassell, G.H. (2000) The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature, 407, 757–762. [DOI] [PubMed] [Google Scholar]
- Gorlich, D. , Vogel, F. , Mills, A.D. , Hartmann, E. and Laskey, R.A. (1995) Distinct functions for the two importin subunits in nuclear protein import. Nature, 377, 246–248. [DOI] [PubMed] [Google Scholar]
- Griffiths, H.M. , Sinclair, W.A. , Smart, C.D. and Davis, R.E. (1999) The phytoplasma associated with ash yellows and lilac witches’‐broom: ‘Candidatus phytoplasma fraxini’. Int. J. Syst. Bacteriol. 49, 1605–1614. [DOI] [PubMed] [Google Scholar]
- Grohmann, E. , Muth, G. and Espinosa, M. (2003) Conjugative plasmid transfer in gram‐positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen, D.E. , Lee, I.M. , Rehner, S.A. , Davis, R.E. and Kingsbury, D.T. (1994) Phylogeny of mycoplasmalike organisms (phytoplasmas): a basis for their classification. J. Bacteriol. 176, 5244–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen, D.E. , Lee, I.M. , Schaff, D.A. , Harrison, N.A. , Chang, C.J. , Davis, R.E. and Kingsbury, D.T. (1996) Genomic diversity and differentiation among phytoplasma strains in 16S rRNA groups I (aster yellows and related phytoplasmas) and III (X‐disease and related phytoplasmas). Int. J. Syst. Bacteriol. 46, 64–75. [DOI] [PubMed] [Google Scholar]
- Hadidi, A. , Czosnek, H. and Barba, M. (2004) DNA microarrays and their potential applications for the detection of plant viruses, viroids, and phytoplasmas. J. Plant Pathol. 86, 97–104. [Google Scholar]
- Hayward, R.D. and Koronakis, V. (2002) Direct modulation of the host cell cytoskeleton by Salmonella actin‐binding proteins. Trends Cell Biol. 12, 15–20. [DOI] [PubMed] [Google Scholar]
- Higgins, C.F. (2001) ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152, 205–210. [DOI] [PubMed] [Google Scholar]
- Hiruki, C. and Wang, K. (2004) Clover proliferation phytoplasma: ‘Candidatus Phytoplasma trifolii’. Int. J. Syst. Evol. Microbiol. 54, 1349–1353. [DOI] [PubMed] [Google Scholar]
- Hodgetts, J. , Ball, T. , Boonham, N. , Mumford, R. and Dickinson, M. (2007) Use of terminol restriction fragment length polymorphism (T‐RFLP) for identification of phytoplasmas in plants. Plant Pathol. 56, 357–365. [Google Scholar]
- Hughes, A.L. and Nei, M. (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature, 335, 167–170. [DOI] [PubMed] [Google Scholar]
- Iida, T. (1972) Discovery of a plant pathogenic mycoplasma. Plant Prot. 26, 175–176. [Google Scholar]
- Imlau, A. , Truernit, E. and Sauer, N. (1999) Cell‐to‐cell and long‐distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell, 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IRPCM (2004) ‘Candidatus Phytoplasma’, a taxon for the wall‐less, non‐helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 54, 1243–1255. [DOI] [PubMed] [Google Scholar]
- Ishiie, T. , Doi, Y. , Yora, K. and Asuyama, H. (1967) Suppressive effects of antibiotics of tetracycline group on symptom development of mulberry dwarf disease. Ann. Phytopathol. Soc. Jpn, 33, 267–275. [Google Scholar]
- Jiggins, F.M. , Hurst, G.D. and Yang, Z. (2002) Host‐symbiont conflicts: positive selection on an outer membrane protein of parasitic but not mutualistic Rickettsiaceae . Mol. Biol. Evol. 19, 1341–1349. [DOI] [PubMed] [Google Scholar]
- Jomantiene, R. and Davis, R.E. (2006) Clusters of diverse genes existing as multiple, sequence‐variable mosaics in a phytoplasma genome. FEMS Microbiol. Lett. 255, 59–65. [DOI] [PubMed] [Google Scholar]
- Jomantiene, R. , Davis, R.E. , Maas, J. and Dally, E.L. (1998) Classification of new phytoplasmas associated with diseases of strawberry in Florida, based on analysis of 16S rRNA and ribosomal protein gene operon sequences. Int. J. Syst. Bacteriol. 48, 269–277. [DOI] [PubMed] [Google Scholar]
- Jomantiene, R. , Zhao, Y. and Davis, R.E. (2007) Sequence‐variable mosaics: composites of recurrent transposition characterizing the genomes of phylogenetically diverse phytoplasmas. DNA Cell. Biol. 26, 557–564. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Joshi, B.D. , Berg, M. , Rogers, J. , Fletcher, J. and Melcher, U. (2005) Sequence comparisons of plasmids pBJS‐O of Spiroplasma citri and pSKU146 of S. kunkelii: implications for plasmid evolution. BMC Genomics, 6, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, H.Y. , Sawayanagi, T. , Kakizawa, S. , Nishigawa, H. , Miyata, S. , Oshima, K. , Ugaki, M. , Lee, J.T. , Hibi, T. and Namba, S. (2002) ‘Candidatus Phytoplasma castaneae’, a novel phytoplasma taxon associated with chestnut witches’ broom disease. Int. J. Syst. Evol. Microbiol. 52, 1543–1549. [DOI] [PubMed] [Google Scholar]
- Jung, H.Y. , Sawayanagi, T. , Kakizawa, S. , Nishigawa, H. , Wei, W. , Oshima, K. , Miyata, S. , Ugaki, M. , Hibi, T. and Namba, S. (2003a) ‘Candidatus phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches’‐broom disease. Int. J. Syst. Evol. Microbiol. 53, 1037–1041. [DOI] [PubMed] [Google Scholar]
- Jung, H.Y. , Sawayanagi, T. , Wongkaew, P. , Kakizawa, S. , Nishigawa, H. , Wei, W. , Oshima, K. , Miyata, S. , Ugaki, M. , Hibi, T. and Namba, S. (2003b) ‘Candidatus Phytoplasma oryzae’, a novel phytoplasma taxon associated with rice yellow dwarf disease. Int. J. Syst. Evol. Microbiol. 53, 1925–1929. [DOI] [PubMed] [Google Scholar]
- Kakizawa, S. , Oshima, K. and Namba, S. (2006a) Diversity and functional importance of phytoplasma membrane proteins. Trends Microbiol. 14, 254–256. [DOI] [PubMed] [Google Scholar]
- Kakizawa, S. , Oshima, K. , Jung, H.Y. , Suzuki, S. , Nishigawa, H. , Arashida, R. , Miyata, S. , Ugaki, M. , Kishino, H. and Namba, S. (2006b) Positive selection acting on a surface membrane protein of the plant‐pathogenic phytoplasmas. J. Bacteriol. 188, 3424–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizawa, S. , Oshima, K. , Kuboyama, T. , Nishigawa, H. , Jung, H. , Sawayanagi, T. , Tsuchizaki, T. , Miyata, S. , Ugaki, M. and Namba, S. (2001) Cloning and expression analysis of Phytoplasma protein translocation genes. Mol. Plant–Microbe Interact. 14, 1043–1050. [DOI] [PubMed] [Google Scholar]
- Kakizawa, S. , Oshima, K. , Nishigawa, H. , Jung, H.Y. , Wei, W. , Suzuki, S. , Tanaka, M. , Miyata, S. , Ugaki, M. and Namba, S. (2004) Secretion of immunodominant membrane protein from onion yellows phytoplasma through the Sec protein‐translocation system in Escherichia coli . Microbiology, 150, 135–142. [DOI] [PubMed] [Google Scholar]
- Kanneganti, T.D. , Bai, X. , Tsai, C.W. , Win, J. , Meulia, T. , Goodin, M. , Kamoun, S. and Hogenhout, S.A. (2007) A functional genetic assay for nuclear trafficking in plants. Plant J. 50, 149–158. [DOI] [PubMed] [Google Scholar]
- Kessler, A. , Halitschke, R. and Baldwin, I.T. (2004) Silencing the jasmonate cascade: induced plant defenses and insect populations. Science, 305, 665–668. [DOI] [PubMed] [Google Scholar]
- Khan, A.J. , Botti, S. , Al‐Subhi, A.M. , Gundersen‐Rindal, D.E. and Bertaccini, A. (2002) Molecular identification of a new phytoplasma strain associated with alfafa witches’ broom in Oman. Phytopathology, 92, 1038–1047. [DOI] [PubMed] [Google Scholar]
- Killiny, N. , Batailler, B. , Foissac, X. and Saillard, C. (2006) Identification of a Spiroplasma citri hydrophilic protein associated with insect transmissibility. Microbiology, 152, 1221–1230. [DOI] [PubMed] [Google Scholar]
- Killiny, N. , Castroviejo, M. and Saillard, C. (2005) Spiroplasma citri spiralin acts in vitro as a lectin binding to glycoproteins from its insect vector Circulifer haematoceps . Phytopathology, 95, 541–548. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K. , Ehrlich, S.D. , Albertini, A. , Amati, G. , Andersen, K.K. , Arnaud, M. , Asai, K. , Ashikaga, S. , Aymerich, S. , Bessieres, P. , Boland, F. , Brignell, S.C. , Bron, S. , Bunai, K. , Chapuis, J. , Christiansen, L.C. , Danchin, A. , Debarbouille, M. , Dervyn, E. , Deuerling, E. , Devine, K. , Devine, S.K. , Dreesen, O. , Errington, J. , Fillinger, S. , Foster, S.J. , Fujita, Y. , Galizzi, A. , GardAn, R. , Eschevins, C. , Fukushima, T. , Haga, K. , Harwood, C.R. , Hecker, M. , Hosoya, D. , Hullo, M.F. , Kakeshita, H. , Karamata, D. , Kasahara, Y. , Kawamura, F. , Koga, K. , Koski, P. , Kuwana, R. , Imamura, D. , Ishimaru, M. , Ishikawa, S. , Ishio, I. , Le Coq, D. , Masson, A. , Mauel, C. , Meima, R. , Mellado, R.P. , Moir, A. , Moriya, S. , Nagakawa, E. , Nanamiya, H. , Nakai, S. , Nygaard, P. , Ogura, M. , Ohanan, T. , O’Reilly, M. , O’Rourke, M. , Pragai, Z. , Pooley, H.M. , Rapoport, G. , Rawlins, J.P. , Rivas, L.A. , Rivolta, C. , Sadaie, A. , Sadaie, Y. , Sarvas, M. , Sato, T. , Saxild, H.H. , Scanlan, E. , Schumann, W. , Seegers, J.F. , Sekiguchi, J. , Sekowska, A. , Seror, S.J. , Simon, M. , Stragier, P. , Studer, R. , Takamatsu, H. , Tanaka, T. , Takeuchi, M. , Thomaides, H.B. , Vagner, V. , Van Dijl, J.M. , Watabe, K. , Wipat, A. , Yamamoto, H. , Yamamoto, M. , Yamamoto, Y. , Yamane, K. , Yata, K. , Yoshida, K. , Yoshikawa, H. , Zuber, U. and Ogasawara, N. (2003) Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA, 100, 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , Von Heijne, G. and Sonnhammer, E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kunkel, L.O. (1926) Studies on aster yellows. Am. J. Bot. 23, 646–705. [Google Scholar]
- Kuske, C.R. and Kirkpatrick, B.C. (1992) Phylogenetic relationships between the western aster yellows mycoplasmalike organism and other prokaryotes established by 16S rRNA gene sequence. Int. J. Syst. Bacteriol. 42, 226–233. [DOI] [PubMed] [Google Scholar]
- Kwon, M. , Wayadande, A. and Fletcher, J. (1999) Spiroplasma citri movement into the intestines and salivary glands of its leafhopper vector, Circulifer tenellus . Phytopathology, 89, 1144–1151. [DOI] [PubMed] [Google Scholar]
- Lee, I.M. and Davis, R.E. (1992) Mycoplasmas which infect plant and insects. In: Mycoplasmas: Molecular Biology and Pathogenesis (Maniloff J., McElhansey R.N., Finch L.R. and Baseman J.B., eds), pp. 379–390. Washington, DC: American Society of Microbiology. [Google Scholar]
- Lee, I.M. , Bottner, K.D. , Secor, G. and Rivera‐Varas, V. (2006a) ‘Candidatus Phytoplasma americanum’, a phytoplasma associated with a potato purple top wilt disease complex. Int. J. Syst. Evol. Microbiol. 56, 1593–1597. [DOI] [PubMed] [Google Scholar]
- Lee, I.M. , Davis, R.E. and Gundersen‐Rindal, D.E. (2000) Phytoplasma: phytopathogenic mollicutes. Annu. Rev. Microbiol. 54, 221–255. [DOI] [PubMed] [Google Scholar]
- Lee, I.M. , Gundersen‐Rindal, D.E. , Davis, R.E. and Bartoszyk, I.M. (1998) Revised classification scheme of phytoplasmas based an RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Bacteriol. 48, 1153–1169. [DOI] [PubMed] [Google Scholar]
- Lee, I.M. , Gundersen‐Rindal, D.E. , Davis, R.E. , Bottner, K.D. , Marcone, C. and Seemüller, E. (2004a) ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 54, 1037–1048. [DOI] [PubMed] [Google Scholar]
- Lee, I.M. , Martini, M. , Marcone, C. and Zhu, S.F. (2004b) Classification of phytoplasma strains in the elm yellows group (16SrV) and proposal of ‘Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 54, 337–347. [DOI] [PubMed] [Google Scholar]
- Lee, I.M. , Zhao, Y. and Bottner, K.D. (2005) Novel insertion sequence‐like elements in phytoplasma strains of the aster yellows group are putative new members of the IS3 family. FEMS Microbiol. Lett. 242, 353–360. [DOI] [PubMed] [Google Scholar]
- Lee, I.M. , Zhao, Y. and Bottner, K.D. (2006b) SecY gene sequence analysis for finer differentiation of diverse strains in the aster yellows phytoplasma group. Mol. Cell Probes, 20, 87–91. [DOI] [PubMed] [Google Scholar]
- Lefol, C. , Lherminier, J. , Boudon‐Padieu, E. , Larrue, J. , Louis, C. and Caudwell, A. (1994) Propagation of flavescence doree MLO (Mycoplasma‐Like Organism) in the leafhopper vector Euscelidius variegatus . J. Invertebr. Pathol. 63, 285–293. [Google Scholar]
- Lherminier, J. , Prensier, G. , Boudon‐Padieu, E. and Caudwell, A. (1990) Immunolabeling of grapevine flavescence doree MLO in salivary glands of Euscelidius variegatus: a light and electron microscopy study. J. Histochem. Cytochem. 38, 79–85. [DOI] [PubMed] [Google Scholar]
- Lim, P.O. and Sears, B.B. (1989) 16S rRNA sequence indicates that plant‐pathogenic mycoplasmalike organisms are evolutionarily distinct from animal mycoplasmas. J. Bacteriol. 171, 5901–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, P.O. and Sears, B.B. (1991) The genome size of a plant‐pathogenic mycoplasmalike organism resembles those of animal mycoplasmas. J. Bacteriol. 173, 2128–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, P.O. and Sears, B.B. (1992) Evolutionary relationships of a plant‐pathogenic mycoplasmalike organism and Acholeplasma laidlawii deduced from two ribosomal protein gene sequences. J. Bacteriol. 174, 2606–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, L. and Nault, L.R. (1983) Differential pathogenicity of corn stunting mollicutes to leafhopper vectors in Dalbulus and Baldulus species. Phytopathology, 73, 1608–1614. [Google Scholar]
- Madden, L. , Nault, L.R. , Heady, S.E. and Styer, W.E. (1984) Effect of maize stunting mollicutes on survival and fecundity of Dalbulus leafhopper vectors. Ann. Appl. Biol. 105, 431–441. [Google Scholar]
- Mahillon, J. and Chandler, M. (1998) Insertion sequences. Microbiol. Mol. Biol. Rev. 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maramorosch, K. (1958) Beneficial effect of virus diseased plants on non‐vector insects. Tijdschr. Plantenziekten, 63, 383–391. [Google Scholar]
- Marcone, C. , Gibb, K.S. , Streten, C. and Schneider, B. (2004a) ‘Candidatus Phytoplasma spartii’, ‘Candidatus Phytoplasma rhamni’ and ‘Candidatus Phytoplasma allocasuarinae’, respectively associated with spartium witches’‐broom, buckthorn witches’‐broom and allocasuarina yellows diseases. Int. J. Syst. Evol. Microbiol. 54, 1025–1029. [DOI] [PubMed] [Google Scholar]
- Marcone, C. , Lee, I.M. , Davis, R.E. , Ragozzino, A. and Seemüller, E. (2000) Classification of aster yellows‐group phytoplasmas based on combined analyses of rRNA and tuf gene sequences. Int. J. Syst. Evol. Microbiol. 50, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Marcone, C. , Neimark, H. , Ragozzino, A. , Lauer, U. and Seemüller, E. (1999) Chromosome sizes of phytoplasmas composing major phylogeneic groups and subgroups. Phytopathology, 89, 805–810. [DOI] [PubMed] [Google Scholar]
- Marcone, C. , Schneider, B. and Seemüller, E. (2004b) ‘Candidatus Phytoplasma cynodontis’, the phytoplasma associated with Bermuda grass white leaf disease. Int. J. Syst. Evol. Microbiol. 54, 1077–1082. [DOI] [PubMed] [Google Scholar]
- Markham, P.J. and Townsend, R. (1979) Experimental vectors of spiroplasmas In: Leafhopper Vectors and Plant Disease Agents (Maramorosch K. and Harris K.F., eds), pp. 413–445. New York: Academic Press. [Google Scholar]
- McCoy, R.E. (1979) Mycoplasmas and yellows diseases In: The Mycoplasmas, Volume V, Spiroplasmas, Acholeplasmas, and Mycoplasmas of Plants and Arthropods (Whitcomb R.F. and Tully E.D., eds), pp. 545–640. San Diego: Academic Press. [Google Scholar]
- McCoy, R.E. , Caudwell, A. , Chang, C.J. , Chen, T.A. , Chiykowski, L.N. , Cousin, M.T. , Dale De Leeuw, G.T.N. , Golino, D.A. , Hackett, K.J. , Kirkpatrick, B.C. , Marwitz, R. , Petzold, H. , Sinha, R.C. , Suguira, M. , Whitcomb, R.F. , Yang, I.L. , Zhu, B.M. and Seemüller, E. (1989) Plant diseases associated with mycoplasma‐like organisms In: The Mycoplasmas (Whitcomb R.F. and Tully J.G., eds), pp. 546–640. New York: Academic Press. [Google Scholar]
- Merighi, M. , Majerczak, D.R. , Stover, E.H. and Coplin, D.L. (2003) The HrpX/HrpY two‐component system activates hrpS expression, the first step in the regulatory cascade controlling the Hrp regulon in Pantoea stewartii subsp. stewartii . Mol. Plant–Microbe Interact. 16, 238–248. [DOI] [PubMed] [Google Scholar]
- Milne, R.G. , Ramasso, E. , Lenzi, R. , Masenga, V. , Sarindu, N. and Clark, M.F. (1995) Preembedding and postembedding immunogold labeling and electron‐microscopy in plant host tissues of 3 antigenically unrelated MLOs‐primula yellows, tomato big bud and bermudagrass white leaf. Eur. J. Plant. Pathol. 101, 57–67. [Google Scholar]
- Montano, H.G. , Davis, R.E. , Dally, E.L. , Hogenhout, S. , Pimentel, J.P. and Brioso, P.S. (2001) ‘Candidatus Phytoplasma brasiliense’, a new phytoplasma taxon associated with hibiscus witches’ broom disease. Int. J. Syst. Evol. Microbiol. 51, 1109–1118. [DOI] [PubMed] [Google Scholar]
- Moran, N.A. (2002) Microbial minimalism: genome reduction in bacterial pathogens. Cell, 108, 583–586. [DOI] [PubMed] [Google Scholar]
- Morton, A. , Davies, D.L. , Blomquist, C.L. and Barbara, D.J. (2003) Characterization of homologues of the apple proliferation immunodominant membrane protein gene from three related phytoplasmas. Mol. Plant Pathol. 4, 109–114. [DOI] [PubMed] [Google Scholar]
- Moya‐Raygoza, G. and Nault, L.R. (1998) Transmission biology of maize bushy stunt Phytoplasma by the corn leafhopper (Homoptera: Cicadellidae). Ann. Entomol. Soc. Am. 91, 668–676. [Google Scholar]
- Murral, D.J. , Nault, L.R. , Hoy, C.W. , Madden, L. and Miller, S.A. (1996) Effects of temperature and vector age on transmission of two Ohio strains of aster yellows phytoplasma by the aster leafhopper (Homoptera: Cicadellidae). J. Econ. Entomol. 89, 1223–1232. [Google Scholar]
- Nakai, K. and Kanehisa, M. (1991) Expert system for predicting protein localization sites in gram‐negative bacteria. Proteins, 11, 95–110. [DOI] [PubMed] [Google Scholar]
- Nakashima, K. and Hayashi, T. (1995) Multiplication and distribution or rice yellow dwarf phytoplasma in infected tissues of rice and green rice leafhopper Nephytettix cincticeps . Ann. Phytopathol. Soc. Jpn, 61, 519–528. [Google Scholar]
- Namba, S. , Kato, S. , Iwanami, S. , Oyaizu, H. , Shiozawa, H. and Tsuchizaki, T. (1993a) Detection and differentiation of plant‐pathogenic mycoplasmalike organisms using polymerase chain‐reaction. Phytopathology, 83, 786–791. [Google Scholar]
- Namba, S. , Oyaizu, H. , Kato, S. , Iwanami, S. and Tsuchizaki, T. (1993b) Phylogenetic diversity of phytopathogenic mycoplasmalike organisms. Int. J. Syst. Bacteriol. 43, 461–467. [DOI] [PubMed] [Google Scholar]
- Nault, L.R. (1985) Evolutionary relationships between maize leafhoppers and their host plants In: The Leafhoppers and Planthoppers (Nault L.R. and Rodriguez J.G., eds), pp. 309–330. New York: John Wiley & Sons, Inc. [Google Scholar]
- Nault, L.R. (1990) Evolution of an insect pest: Maize and the corn leafhopper, a case study. Maydica, 35, 165–175. [Google Scholar]
- Neimark, H. and Kirkpatrick, B.C. (1993) Isolation and characterization of full‐length chromosomes from non‐culturable plant‐pathogenic Mycoplasma‐like organisms. Mol. Microbiol. 7, 21–28. [DOI] [PubMed] [Google Scholar]
- Newman, K.L. , Almeida, R.P. , Purcell, A.H. and Lindow, S.E. (2004) Cell‐cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl Acad. Sci. USA, 101, 1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaisen, M. and Bertaccini, A. (2007) An oligonucleotide microarray‐based assay for identification of phytoplasma 16S ribosomal groups. Plant Pathol. 56, 332–336. [Google Scholar]
- Nielsen, H. , Engelbrecht, J. , Brunak, S. and Von Heijne, G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Nielsen, R. and Yang, Z. (1998) Likelihood models for detecting positively selected amino acid sites and applications to the HIV‐1 envelope gene. Genetics, 148, 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson, M.W. (1979) Taxonomic relationships of leafhopper vectors of plant pathogens In: Leafhopper Vectors and Plant Disease Agents (Maramorosch K. and Harris K.F., eds), pp. 3–27. New York: Academic Press. [Google Scholar]
- Nishigawa, H. , Miyata, S. , Oshima, K. , Sawayanagi, T. , Komoto, A. , Kuboyama, T. , Matsuda, I. , Tsuchizaki, T. and Namba, S. (2001) In planta expression of a protein encoded by the extrachromosomal DNA of a phytoplasma and related to geminivirus replication proteins. Microbiology, 147, 507–513. [DOI] [PubMed] [Google Scholar]
- Nishigawa, H. , Oshima, K. , Kakizawa, S. , Jung, H.Y. , Kuboyama, T. , Miyata, S. , Ugaki, M. and Namba, S. (2002a) A plasmid from a non‐insect‐transmissible line of a phytoplasma lacks two open reading frames that exist in the plasmid from the wild‐type line. Gene, 298, 195–201. [DOI] [PubMed] [Google Scholar]
- Nishigawa, H. , Oshima, K. , Kakizawa, S. , Jung, H.Y. , Kuboyama, T. , Miyata, S. , Ugaki, M. and Namba, S. (2002b) Evidence of intermolecular recombination between extrachromosomal DNAs in phytoplasma: a trigger for the biological diversity of phytoplasma? Microbiology, 148, 1389–1396. [DOI] [PubMed] [Google Scholar]
- Nipah, J.O. , Jones, P. and Dickinson, M.J. (2007) Detection of lethal yellowing phytoplasma in embryos from coconut palms infected with Cape St Paul Wilt disease in Ghana. Plant Pathology, 56, 777–489. [Google Scholar]
- Okuda, S. (1972) Occurrence of diseases caused by mycoplasma‐like organisms in Japan. Plant Prot. 26, 180–183. [Google Scholar]
- Oshima, K. , Kakizawa, S. , Arashida, R. , Ishii, Y. , Hoshi, A. , Hayashi, Y. , Kagiwada, S. and Namba, S. (2007) Presence of two glycolytic gene clusters in a severe pathogenic line of Candidatus Phytoplasma asteris. Mol. Plant Pathol. 8, 481–489. [DOI] [PubMed] [Google Scholar]
- Oshima, K. , Kakizawa, S. , Nishigawa, H. , Jung, H.Y. , Wei, W. , Suzuki, S. , Arashida, R. , Nakata, D. , Miyata, S. , Ugaki, M. and Namba, S. (2004) Reductive evolution suggested from the complete genome sequence of a plant‐pathogenic phytoplasma. Nature Genet. 36, 27–29. [DOI] [PubMed] [Google Scholar]
- Oshima, K. , Kakizawa, S. , Nishigawa, H. , Kuboyama, T. , Miyata, S. , Ugaki, M. and Namba, S. (2001a) A plasmid of phytoplasma encodes a unique replication protein having both plasmid‐ and virus‐like domains: clue to viral ancestry or result of virus/plasmid recombination? Virology, 285, 270–277. [DOI] [PubMed] [Google Scholar]
- Oshima, K. , Shiomi, T. , Kuboyama, T. , Sawayanagi, T. , Nishigawa, H. , Kakizawa, S. , Miyata, S. , Ugaki, M. and Namba, S. (2001b) Isolation and characterization of derivative lines of the onion yellows phytoplasma that do not cause stunting or phloem hyperplasia. Phytopathology, 91, 1024–1029. [DOI] [PubMed] [Google Scholar]
- Özbek, E. , Miller, S.A. , Meulia, T. and Hogenhout, S.A. (2003) Infection and replication sites of Spiroplasma kunkelii (Class: Mollicutes) in midgut and Malpighian tubules of the leafhopper Dalbulus maidis . J. Invertebr. Pathol. 82, 167–175. [DOI] [PubMed] [Google Scholar]
- Paschold, A. , Halitschke, R. and Baldwin, I.T. (2007) Co(i)‐ordinating defenses: NaCOI1 mediates herbivore‐induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J. 51, 79–91. [DOI] [PubMed] [Google Scholar]
- Peterson, A.G. (1973) Host plant and aster leafhopper relationships. Proc. North Cent. Branch Entomol. Soc. Am. 28, 66–70. [Google Scholar]
- Purcell, A. , Richardson, J. and Finlay, A. (1981) Multiplication of the agent of X‐disease in a non‐vector leafhopper Macrosteles fascifrons . Ann. Appl. Biol. 99, 283. [Google Scholar]
- Purcell, A.H. (1988) Increased survival of Dalbulus maidis DeLong & Wolcott, a specialist on maize on nonhost plants infected with mollicute plant pathogens. Entomol. Exp. Appl. 46, 187–196. [Google Scholar]
- Razin, S. , Yogev, D. and Naot, Y. (1998) Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62, 1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redak, R.A. , Purcell, A.H. , Lopes, J.R. , Blua, M.J. , Mizell, R.F. 3rd and Andersen, P.C. (2004) The biology of xylem fluid‐feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu. Rev. Entomol. 49, 243–270. [DOI] [PubMed] [Google Scholar]
- Rocha, E.P. and Blanchard, A. (2002) Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30, 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, E.P. , Danchin, A. and Viari, A. (1999) Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol. Biol. Evol. 16, 1219–1230. [DOI] [PubMed] [Google Scholar]
- Rosch, J. and Caparon, M. (2004) A microdomain for protein secretion in Gram‐positive bacteria. Science, 304, 1513–1515. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. (1987) The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Samuelson, J.C. , Chen, M. , Jiang, F. , Moller, I. , Wiedmann, M. , Kuhn, A. , Phillips, G.J. and Dalbey, R.E. (2000) YidC mediates membrane protein insertion in bacteria. Nature, 406, 637–641. [DOI] [PubMed] [Google Scholar]
- Santana, M. , Ionescu, M.S. , Vertes, A. , Longin, R. , Kunst, F. , Danchin, A. and Glaser, P. (1994) Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J. Bacteriol. 176, 6802–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawayanagi, T. , Horikoshi, N. , Kanehira, T. , Shinohara, M. , Bertaccini, A. , Cousin, M.T. , Hiruki, C. and Namba, S. (1999) ‘Candidatus phytoplasma japonicum’, a new phytoplasma taxon associated with Japanese Hydrangea phyllody. Int. J. Syst. Bacteriol. 49, 1275–1285. [DOI] [PubMed] [Google Scholar]
- Schneider, B. , Torres, E. , Martin, M.P. , Schroder, M. , Behnke, H.D. and Seemüller, E. (2005) ‘Candidatus Phytoplasma pini’, a novel taxon from Pinus silvestris and Pinus halepensis . Int. J. Syst. Evol. Microbiol. 55, 303–307. [DOI] [PubMed] [Google Scholar]
- Schultz, G.A. (1973) Plant resistance to aster yellows. Proc. North Cent. Branch Entomol. Soc. Am. 28, 93–99. [Google Scholar]
- Scotti, P.A. , Urbanus, M.L. , Brunner, J. , De Gier, J.W. , Von Heijne, G. , Van Der Does, C. , Driessen, A.J. , Oudega, B. and Luirink, J. (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, B.B. and Klomparens, K.L. (1989) Leaf tip cultures of the evening primrose allow stable aseptic culture of mycoplasma‐like organisms. Can. J. Plant Pathol. 11, 343–348. [Google Scholar]
- Seemüller, E. and Schneider, B. (2004) ‘Candidatus Phytoplasma mali’, ‘Candidatus Phytoplasma pyri’ and ‘Candidatus Phytoplasma prunorum’, the causal agents of apple proliferation, pear decline and European stone fruit yellows, respectively. Int. J. Syst. Evol. Microbiol. 54, 1217–1226. [DOI] [PubMed] [Google Scholar]
- Seemüller, E. , Marcone, C. , Lauer, U. , Ragozzino, A. and Goschl, M. (1998) Current status of molecular classification of the phytoplasmas. J. Plant Pathol. 80, 3–26. [Google Scholar]
- Seemüller, E. , Schneider, B. , Maurer, R. , Ahrens, U. , Daire, X. , Kison, H. , Lorenz, K.H. , Firrao, G. , Avinent, L. and Sears, B.B. (1994) Phylogenetic classification of phytopathogenic mollicutes by sequence analysis of 16S ribosomal DNA. Int. J. Syst. Bacteriol. 44, 440–446. [DOI] [PubMed] [Google Scholar]
- Serek, J. , Bauer‐Manz, G. , Struhalla, G. , Van Den Berg, L. , Kiefer, D. , Dalbey, R. and Kuhn, A. (2004) Escherichia coli YidC is a membrane insertase for Sec‐independent proteins. EMBO J. 23, 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.C. and Lin, C.P. (1993) Production of monoclonal‐antibodies against a mycoplasmalike organism associated with sweet‐potato witches‐broom. Phytopathology, 83, 671–675. [Google Scholar]
- Siller, W. , Kuhbandner, B. , Marwitz, R. , Petzhold, H. and Seemüller, E. (1987) Occurence of mycoplasma like organisms in parenchyma cells of Cuscata odorata ruiz et pav . J. Phytopathol. 119, 147–159. [Google Scholar]
- Suzuki, S. , Oshima, K. , Kakizawa, S. , Arashida, R. , Jung, H.Y. , Yamaji, Y. , Nishigawa, H. , Ugaki, M. and Namba, S. (2006) Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect‐vector specificity. Proc. Natl Acad. Sci. USA, 103, 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas, I.L. , Klasson, B. , Canback, A.K. , Naslund, A.S. , Eriksson, J.J. , Wernegreen, J.J. , Sandstrom, J. P. , Moran, N.A. and Andersson, S.G. (2002) 50 million years of genomic statis in endosymbiotic bacteria. Science, 296, 2376–2379. [DOI] [PubMed] [Google Scholar]
- Tanaka, J. and Sasakawa, C. (2005) [Type IV secretion system in Helicobacter pylori: comparison with bacterial type III secretion apparatus]. Tanpakushitsu Kakusan Koso, 50, 36–43. [PubMed] [Google Scholar]
- Taylor, P.D. , Toseland, C.P. , Attwood, T.K. and Flower, D.R. (2006) LIPPRED: A web server for accurate prediction of lipoprotein signal sequences and cleavage sites. Bioinformation, 1, 176–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Gibson, T.J. , Plewniak, F. , Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL‐X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma, H. , Bolhuis, A. , Jongbloed, J.D. , Bron, S. and Van Dijl, J.M. (2000) Signal peptide‐dependent protein transport in Bacillus subtilis: a genome‐based survey of the secretome. Microbiol. Mol. Biol. Rev. 64, 515–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, E. , Bertolini, E. , Cambra, M. , Monton, C. and Martin, M. (2005) Real‐time PCR for simultaneous and quantitative detection of quarantine phytoplasmas from apple proliferation (16SrX) group. Mol. Cell. Probes, 19, 334–340. [DOI] [PubMed] [Google Scholar]
- Toth, K.F. , Harrison, N. and Sears, B.B. (1994) Phylogenetic relationships among members of the class Mollicutes deduced from rps3 gene sequences. Int. J. Syst. Bacteriol. 44, 119–124. [DOI] [PubMed] [Google Scholar]
- Urbanus, M.L. , Scotti, P.A. , Froderberg, L. , Saaf, A. , De Gier, J.W. , Brunner, J. , Samuelson, J.C. , Dalbey, R.E. , Oudega, B. and Luirink, J. (2001) Sec‐dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin, R. , Holmes, E.C. , Fox, A.J. , Derrick, J.P. and Maiden, M.C. (2002) Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol. Biol. Evol. 19, 1686–1694. [DOI] [PubMed] [Google Scholar]
- Valiunas, D. , Staniulis, J. and Davis, R.E. (2006) ‘Candidatus Phytoplasma fragariae’, a novel phytoplasma taxon discovered in yellows diseased strawberry, Fragaria ¥ ananassa . Int. J. Syst. Evol. Microbiol. 56, 277–281. [DOI] [PubMed] [Google Scholar]
- Verdin, E. , Salar, P. , Danet, J.L. , Choueiri, E. , Jreijiri, F. , El Zammar, S. , Gelie, B. , Bove, J.M. and Garnier, M. (2003) ‘Candidatus phytoplasma phoenicium’ sp. nov., a novel phytoplasma associated with an emerging lethal disease of almond trees in Lebanon and Iran. Int. J. Syst. Evol. Microbiol. 53, 833–838. [DOI] [PubMed] [Google Scholar]
- Wei, W. , Kakizawa, S. , Jung, H. , Suzuki, S. , Tanaka, M. , Nishigawa, H. , Miyata, S. , Oshima, K. , Ugaki, M. , Hibi, T. and Namba, S. (2004) An antibody against the SecA membrane protein of one phytoplasma reacts with those of phylogenetically different phytoplasmas. Phytopathology, 94, 683–686. [DOI] [PubMed] [Google Scholar]
- Weintraub, P.G. and Beanland, L. (2006) Insect vectors of phytoplasmas. Annu. Rev. Entomol. 51, 91–111. [DOI] [PubMed] [Google Scholar]
- Weisburg, W.G. , Tully, J.G. , Rose, D.L. , Petzel, J.P. , Oyaizu, H. , Yang, D. , Mandelco, L. , Sechrest, J. , Lawrence, T.G. , Van Etten, J. , Maniloff, J. and Woese, C.R. (1989) A phylogenetic analysis of the mycoplasmas: basis for their classification. J. Bacteriol. 171, 6455–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, J.M. , Raju, B.C. , Hung, H.Y. , Weisburg, W.G. , Mandelco‐Paul, L. and Brenner, D.J. (1987) Xylella fastidiosa gen. nov.: gram‐negative, xylem‐limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Bacteriol. 37, 136–143. [Google Scholar]
- Wernegreen, J.J. (2002) Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3, 850–861. [DOI] [PubMed] [Google Scholar]
- Whitcomb, R.F. and Tully, E.D. (1989) The Mycoplasmas, Vol. V. San Diego: Academic Press, Inc. [Google Scholar]
- Woese, C.R. (1987) Bacterial evolution. Microbiol. Rev. 51, 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R. , Maniloff, J. and Zablen, L.B. (1980) Phylogenetic analysis of the mycoplasmas. Proc. Natl Acad. Sci. USA, 77, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, F. , Melcher, U. and Fletcher, J. (1997) Molecular characterization of a gene encoding a membrane protein of Spiroplasma citri . Gene, 189, 95–100. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Hogenhout, S.A. , Nault, L.R. , Hoy, C.W. and Miller, S.A. (2004) Molecular and symptom analyses of phytoplasma strains from lettuce reveal a diverse population. Phytopathology, 94, 842–849. [DOI] [PubMed] [Google Scholar]
- Zreik, L. , Carle, P. , Bove, J.M. and Garnier, M. (1995) Characterization of the mycoplasmalike organism associated with witches’‐broom disease of lime and proposition of a Candidatus taxon for the organism, ‘Candidatus phytoplasma aurantifolia’. Int. J. Syst. Bacteriol. 45, 449–453. [DOI] [PubMed] [Google Scholar]


