SUMMARY
Phenolics are aromatic benzene ring compounds with one or more hydroxyl groups produced by plants mainly for protection against stress. The functions of phenolic compounds in plant physiology and interactions with biotic and abiotic environments are difficult to overestimate. Phenolics play important roles in plant development, particularly in lignin and pigment biosynthesis. They also provide structural integrity and scaffolding support to plants. Importantly, phenolic phytoalexins, secreted by wounded or otherwise perturbed plants, repel or kill many microorganisms, and some pathogens can counteract or nullify these defences or even subvert them to their own advantage. In this review, we discuss the roles of phenolics in the interactions of plants with Agrobacterium and Rhizobium.
INTRODUCTION
Phenolics are one of the most ubiquitous groups of secondary metabolites found throughout the plant kingdom (Boudet, 2007; Harborne, 1980). They encompass a very large and diverse group of aromatic compounds characterized by a benzene ring (C6) and one or more hydroxyl groups. Generally, the classification of phenolics is based on the number of carbon atoms present in the molecule (Harborne and Simmonds, 1964).
Phenolics are formed by three different biosynthetic pathways: (i) the shikimate/chorizmate or succinylbenzoate pathway, which produces the phenyl propanoid derivatives (C6–C3); (ii) the acetate/malonate or polyketide pathway, which produces the side‐chain‐elongated phenyl propanoids, including the large group of flavonoids (C6–C3–C6) and some quinones; and (iii) the acetate/mevalonate pathway, which produces the aromatic terpenoids, mostly monoterpenes, by dehydrogenation reactions (for more details on these metabolic pathways, see http://www.plantcyc.org). Here, we focus on the classes of phenolics that are involved in interactions of plants with microbes, Agrobacterium and Rhizobium, belonging to the Rhizobiaceae family.
PLANTS SYNTHESIZE PHENOLICS IN RESPONSE TO BIOTIC AND ABIOTIC STRESS
Phenolics are often produced and accumulated in the subepidermal layers of plant tissues exposed to stress and pathogen attack (Cléet al., 2008; Schmitz‐Hoerner and Weissenbock, 2003). The concentration of a particular phenolic compound within a plant tissue is dependent on season and may also vary at different stages of growth and development (Lynn and Chang, 1990; Ozyigit et al., 2007; Thomas and Ravindra, 1999). Several internal and external factors, including trauma, wounding, drought and pathogen attack, affect the synthesis and accumulation of phenolics (Kefeli et al., 2003; Zapprometov, 1989). Furthermore, the biosynthesis of phenolics in chloroplasts and their accumulation in vacuoles are enhanced on exposure to light (Kefeli et al., 2003). Photoinhibition, as well as nutrient stresses, such as deficiencies in nitrogen, phosphate, potassium, sulphur, magnesium, boron and iron, also trigger the synthesis of phenylpropanoid compounds in some plant species (Dixon and Paiva, 1995). These may include members of the flavonoid biosynthetic pathway (Balasundram et al., 2006; Hollman and Katan, 1999).
PHENOLICS IN PLANT DEFENCE
Phenolics serve a dual function of both repelling and attracting different organisms in the plant's surroundings (Table 1). They act as protective agents, inhibitors, natural animal toxicants and pesticides against invading organisms, i.e. herbivores, nematodes, phytophagous insects, and fungal and bacterial pathogens (Dakora and Phillips, 1996; Lattanzio et al., 2006; Ravin et al., 1989). Simple phenolic acids, complex tannins and phenolic resins on the plant surface deter birds by interacting with the gut microflora and diminishing their digestive ability. The scent and pigmentation conferred by low‐molecular‐weight phenylpropanol derivatives attract symbiotic microbes, pollinators and animals that disperse fruits (Ndakidemi and Dakora, 2003; Vit et al., 1997).
Table 1.
Examples of different types of phenolics and diverse functions that they perform for plants and bacteria.
| Examples | Functions | |
|---|---|---|
| Phenolics | Coniferyl alcohol, sinapinic acid, cinnamic acid | vir gene inducers, determinants of scent and attractants of pollinators and symbiotic microbes in plants, etc. |
| Hydroxybenzoate, hydroxycinnamates, 5‐hydroxyanthraquinones | Allelochemicals for plant competition | |
| Umbelliferone, p‐hydroxybenzoic acid, vanillyl alcohol, isoflavones | Chemoattractants in Rhizobium | |
| Sinapinic acid, syringic acid, ethylsyringamide, propylsyringamide, carbethoxyethylensyringamide, parahydroxybenzoate, ferulic acid | vir gene inducers in Agrobacterium | |
| Vanillyl alcohol, bromo acetosyringone | Inhibitors of vir gene induction in Agrobacterium | |
| Acetosyringone, α‐hydroxyacetosyringone, p‐hydroxybenzoate | Chemoattractants in Agrobacterium and Rhizobium, and vir gene inducers in Agrobacterium | |
| Salicylic acid | Quorum quencher in Agrobacterium | |
| Hydroquinones | Allelochemical for plant competition | |
| Coumarins, xanthones, anthocyanidins | Determinants of colour and attractants of pollinators in plants | |
| Caffeic acid | vir gene inducer in Agrobacterium | |
| 3,4‐Dihydroxybenzoic acid | Chemoattractant in Agrobacterium and Rhizobium | |
| Protocatechuic acid, β‐resorcylic acid, protocatechuate, p‐resorcylate, catechol | vir gene inducer in Agrobacterium | |
| Chlorogenic acid | Precursor for lignin and suberin synthesis in plants | |
| Lignin, tannins and suberins | Structural components of plant cells | |
| Catechins | Plant defence | |
| Flavonoids, flavonols, flavones, genistein, daidzein, O‐acetyldaidzein, 6‐O‐malonylgenistin, 6‐O‐malonyl daidzin, glycitin, 6‐O‐malonylglycitin | nod gene inducers in Rhizobium | |
| Apigenin, naringenin, luteolin | Chemoattractants in Agrobacterium and Rhizobium, and nod gene inducers in Rhizobium | |
| Gallate, gallic acid, pyrogallic acid, syringic acid, kaempferol | vir gene inducers in Agrobacterium | |
| Flavanones, quercetin | nod gene inducers in Rhizobium | |
| Isoflavonoids | Chemoattractants and nod gene inducers in Rhizobium | |
| Cajanin, medicarpin, glyceoline, rotenone, coumestrol, phaseolin, phaseolinin, limonoids, tannins, flavonoids | Phytoalexins, phytoanticipins and nematicides in plant defence |
Phenolics have long been recognized as phyto‐estrogens in animals (Adams, 1989) and as allelochemicals for competitive plants and weeds (Weir et al., 2004; Xuan et al., 2005). Mainly, the volatile terpenoids, the toxic water‐soluble hydroquinones, hydroxybenzoates, hydroxycinnamates and the 5‐hydroxynapthoquinones are widely effective allelochemicals.
A number of simple and complex phenolics accumulate in plant tissues and act as phytoalexins, phytoanticipins and nematicides against soil‐borne pathogens and phytophagous insects (Akhtar and Malik, 2000; Lattanzio et al., 2006). Therefore, phenolic compounds have been proposed for some time to serve as useful alternatives to the chemical control of pathogens of agricultural crops (Langcake et al., 1981).
Most known effects of polyphenols on microbes are negative (Cushnie and Lamb, 2005; Ferrazzano et al., 2009; Taguri et al., 2006). Plants respond to pathogen attack by accumulating phytoalexins, such as hydroxycoumarins and hydroxycinnamate conjugates (Karou et al., 2005; Mert‐Türk, 2002). The synthesis, release and accumulation of phenolics—in particular, salicylic acid (Boller and He, 2009; Koornneef and Pieterse, 2008; Lu, 2009; Tsuda et al., 2008)—are central to many defence strategies employed by plants against microbial invaders.
Phenolics are synthesized when plant pattern recognition receptors recognize potential pathogens (Newman et al., 2007; Ongena et al., 2007; Schuhegger et al., 2006; Tran et al., 2007) by conserved pathogen‐associated molecular patterns (PAMPs), leading to PAMP‐triggered immunity (Zipfel, 2008). As a result, the progress of the infection is restricted long before the pathogen gains complete hold of the plant (Bittel and Robatzek, 2007; Nicaise et al., 2009).
PHENOLICS—MOLECULES FOR CROSS‐TALK IN THE RHIZOSPHERE
The plant rhizosphere is a dynamic ecosystem of different species, representing flora, fauna and microbes that interact with each other in a variety of complex reactions (Whipps, 2001). These interactions are mainly governed by a diverse array of phenolics exuded by the growing roots of plants, together with a host of other chemicals (Bais et al., 2006; Bertin et al., 2003; Dakora and Phillips, 2002). The functions performed by phenolics in the plant rhizosphere (Dakora, 2003) have been aptly termed the ‘rhizosphere effect’ (Hiltner, 1904). The root exudates generally include ions, free oxygen, water, enzymes, mucilage, a number of carbon‐containing primary and secondary metabolites and, most importantly, plant phenolics. The released phenolics differ from species to species and also with time, space and location. Their concentrations in the soil range from 2.1 to 4.4% in monocotyledonous plants and 0.1 to 0.6% in dicotyledonous plants (Hartley and Harris, 1981). Phenolics trigger redox reactions in soils and selectively influence the growth of soil microorganisms that colonize the rhizosphere. These then influence the hormonal balance, enzymatic activity, availability of phytonutrients and competition between neighbouring plants (Hättenschwiler and Vitousek, 2000; Kraus et al., 2003; Northup et al., 1998). As a result of this dynamic and ever‐changing interaction, the structure and chemistry of the soil are altered significantly depending on the quantity and identity of phenolics released by different plant species. The composition of the microorganism species in different root locations is also persistently modified and shaped. Moreover, as phenolics move through the rhizosphere, they are bound by soil organic matter and metabolized by the bacterial flora of the soil (Kefeli et al., 2003).
Microorganisms break down phenolics into elements that contribute towards the mineralization of soil nitrogen and the formation of humus (Halvorson et al., 2009). The phenolics chelate metals and improve soil porosity, providing active absorption sites and increasing the mobility and bioavailability of elements, such as potassium, calcium, magnesium, copper, zinc, manganese, molybdenum, iron and boron, for plant roots (Seneviratne and Jayasinghearachchi, 2003). Some phenolic metabolites, such as trans‐cinnamic acid, salicylic acid, coumarin, benzoic acid, parahydroxybenzoic acid and syringic acid, are phytotoxic. For example, the accumulation of phenolics in the soil can inhibit seed germination and seedling growth (Baleroni et al., 2000). This effect may be caused by interference with cell division and the normal functioning of cellular enzymes. Indeed, phenolics have been shown to inhibit phosphatase and prolyl aminopeptidase involved in seed germination (Madhan et al., 2009). In addition, phenolics affect the process of mineral uptake by the plants (Lodhi et al., 1987).
Many of the phenolic root exudates serve as chemotactic signals for a number of soil microorganisms that recognize them and move towards plant roots in the carbon‐rich environment of the rhizosphere (Perret et al., 2000; Taylor and Grotewold, 2005). Based on the nature and type of root‐derived chemicals, both positive and negative cross‐talk pathways are initiated between roots, roots and insects, and roots and microbes. Different organisms are repelled from or attracted to the same chemical signal, which then elicits different responses in different recipients. One specific example is the isoflavones from soybean roots which serve as a chemoattractant for both the symbiotic Bradyrhizobium japonicum and the pathogenic Phytopthora sojae (Morris et al., 1998). The number and activity of soil microorganisms around the root increase as a result of root–microbe cross‐talk (Bais et al., 2004). This subsequently leads to the colonization of roots. Generally, the zone of root elongation, just behind the tip, supports the growth of primary root colonizers that utilize the easily degradable sugars and organic acids. On the other hand, fungi and bacteria that are adapted to crowded, oligotrophic conditions inhabit the older root zones, comprising sloughed cells with lignified cellulose, hemicellulose and carbon deposits. Mature communities of fungi colonize relatively nutrient‐rich environments provided by the newly emerging lateral roots and the secondary non‐growing root tips. Colonization of vesicular–arbuscular mycorrhizal fungi in response to isoflavonoids from soybean roots leads to greater phosphorus acquisition for plant nutrition, improved water relations and, consequently, better plant growth (Bagayoko et al., 2000; Siqueira et al., 1991). During phosphate deficiency, plant roots also exude strigolactones. These apocarotenoid molecules are detected as host‐derived signalling compounds at the presymbiotic stage of beneficial fungal symbionts (Akiyama, 2007; Akiyama et al., 2005). Unlike various flavonoids that induce hyphal branching only in a limited number of hosts, strigolactones represent primary signalling factors for hyphal branching and growth of arbuscular mycorrhizal fungi (Steinkellner et al., 2007).
Other positive effects of root colonizers include the symbiotic associations with epiphytes and mycorrhizal fungi, the fixation of atmospheric nitrogen by different classes of proteobacteria (Moulin et al., 2001), increased biotic and abiotic stress tolerance imparted by the presence of endophytic microbes (Schardl et al., 2004), and several direct and indirect advantages caused by various plant growth‐promoting rhizobacteria (Gray and Smith, 2005). Some colonizing bacteria also interact with plants to produce protective biofilms or antibiotics, functioning as effective biocontrol against potential pathogens (Bais et al., 2004).
Although some types of colonization may lead to associations with symbiotic microorganisms, others result in plant infection by soil‐borne pathogens. The symbiotic Rhizobium and the pathogenic Agrobacterium spp. (except A. radiobacter) of the Rhizobiaceae family are examples of plant colonizers with positive and negative effects on their hosts, respectively. Specifically, rhizobia are important for their nitrogen‐fixing ability and endosymbiotic associations with leguminous plants, whereas most Agrobacterium species are phytopathogens. As members of the same family, these two bacteria exhibit similarities and differences in their basic mechanisms of symbiosis or infection, each enjoying a special ecological niche.
HOST PHENOLICS IN THE INFECTION CYCLES OF AGROBACTERIUM AND RHIZOBIUM
The type and concentration of phenolics in the surroundings govern the interactions of Agrobacterium and Rhizobium with their host plants (Table 2). These interactions may range from strong to weak to transient (Bais et al., 2006). Although plants exude different phenolic compounds that are toxic to most microorganisms, Agrobacterium and Rhizobium have evolved mechanisms to counteract, nullify and even utilize these defences for their own advantage (1, 2) (Hartmann et al., 2009; Matilla et al., 2007). The most significant of these mechanisms that involve phenolics are: (i) chemotaxis; (ii) activation of the bacterial nodulation (nod) and virulence (vir) gene networks; (iii) xenobiotic detoxification; and (iv) quorum signalling (Fig. 2).
Table 2.
Summary of events in host–bacteria (Rhizobiaceae) interactions affected by phenolics.
| Processes | Phenolics | Bacterial species | Bacterial factors |
|---|---|---|---|
| Chemotaxis | Acetosyringone, hydroxyacetosyringone, parahydroxybenzoic acid, 3,4‐dihydroxybenzoic acid, vanillyl alcohol, etc. | Agrobacterium tumefaciens, A. rhizogenes, A. vitis | VirA |
| Isoflavonoids, flavonoids, apigenin, luteolin, vanillyl alcohol, parahydroxybenzoic acid, 3,4‐dihydroxybenzoic acid, acetosyringone, umbelliferone, naringenin, etc. | Rhizobium meliloti, R. leguminosarum bv. phaseoli, viciae, trifolii | NodD | |
| Gene inducers | Acetosyringone, catechol, gallate, p‐resorcylate, protocatechuate, parahydroxybenzoate, vanillin, α‐hydroxyacetosyringone, ferulic acid, gallic acid, pyrogallic acid, protocatechuic acid, β‐resorcylic acid, syringic acid, kaempferol, ethylsyringamide, propyl syringamide, carbethoxyethylensyringamide, sinapinic acid | Agrobacterium spp. | VirA and VirG |
| Flavonoids, flavones, flavonols, vanillin, genistein, daidzein, O‐acetyl daidzin, 6‐O‐malonyl genistein, 6‐O‐malonyl daidzin, glycitin, 6‐O‐malonyl glycitin, genistin, apigenin, naringenin, luteolin | Rhizobium melitloti, R. leguminosarum bv. phaseoli, viciae, trifolii, Bradyrhzobium japonicum, Sinorhizobium meliloti | NodD | |
| Detoxification/mineralization | Phenolic vir gene inducers (high concentrations) | Agrobacterium spp. | VirH2 |
| Flavonoids, quercetin, daidzein, genistein, etc. | Rhizobium and Bradyrhzobium spp. | Enzymes that cleave C‐ring | |
| Quorum signalling | Negative influence of flavonoids | Rhizobium spp. | CinR/CinI, RhiI, RhiA, RhiB, RhiC, RhiR, RaiI, RaiR, TraI, TraB‐I, BisR and TraR |
| Indirect influence of phenolic vir gene inducers (via opines) | Agrobacterium spp. | TraI/R, products of tra and repABC operons | |
| Quorum quenching | Flavonoids | Rhizobium | BisR |
| Salicylic acid, phenolic vir gene inducers | Agrobacterium spp. | TraM, AttM, AttL, AttK |
Figure 1.
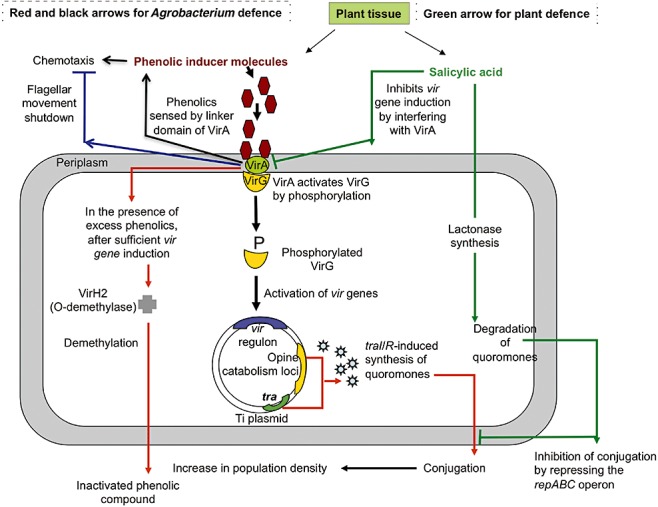
Phenolics in the Agrobacterium–plant interaction. Black arrows indicate how Agrobacterium uses phenolics to initiate pathogenesis in host plants for opine synthesis and nutrition, and red arrows indicate how it inactivates the excess amounts of the same phenolics using its own specific O‐demethylase system. Blue arrow indicates the involvement of the phenolic‐sensing bacterial VirA protein in negative chemotaxis. Green arrows show how plants synthesize and use some phenolics, such as salicylic acid, to interfere with VirA and hence pathogenesis, and also to degrade the quoromones that give ecological advantage to Agrobacterium over other bacteria in competition for the opines synthesized by the infected plant.
Figure 2.
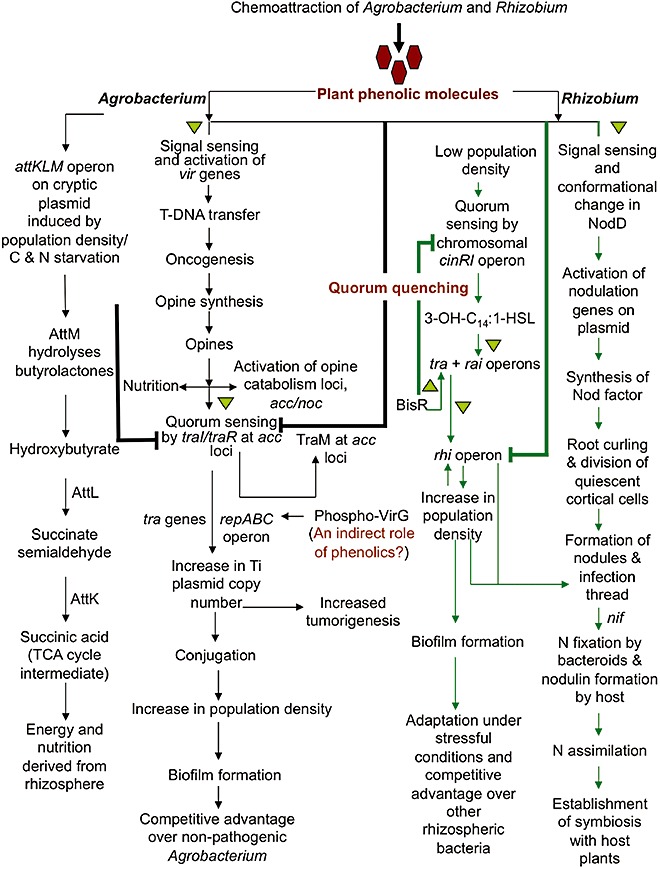
The use of phenolics by Agrobacterium and Rhizobium for survival and infection of the host plant. Black arrows indicate how Agrobacterium uses phenolics to initiate a complex process of pathogenesis, culminating with opine synthesis. In addition to their nutritional value, opines help Agrobacterium's competition with nonpathogenic bacteria, such as A. radiobacter, by increasing its population density and biofilm formation through quorum sensing. Agrobacterium also uses the attKLM operon to regulate its population density during times of nutritional starvation and to synthesize alternative sources of nutrients and energy by degrading γ‐butyrolactones produced by other rhizospheric bacteria. Green arrows indicate the use of phenolics by Rhizobium leguminosarum bv. viciae for the induction of nod genes followed by the process of symbiosis. Under stress conditions, phenolics also regulate the increase in population density, biofilm formation and effective nodulation by repressing the quorum‐sensing rhi operon. An increase in population density as a result of quorum sensing provides a competitive edge to rhizobia over other rhizospheric bacteria. Bold lines indicate the quorum‐quenching mechanisms, whereas triangles represent the steps of activation. TCA, tricarboxylic acid.
Chemotaxis
As in several other soil bacteria, phenolics play a pivotal role in the chemotactic responses of Agrobacterium and Rhizobium in their search for growth substrates and hosts. These serve as excellent models for signal transduction and plant–microbe interactions (Palmer et al., 2004; Samac and Graham, 2007). Diverse plant phenolic compounds with varying substitution patterns determine the chemotactic movement of Agrobacterium or Rhizobium across chemical gradients towards higher levels of potential nutrients and lower levels of inhibitors. For example, umbelliferone, vanillyl alcohol, p‐hydroxybenzoic acid, 3,4‐dihydroxybenzoic acid and acetosyringone evoke a strong chemotactic response in Rhizobium leguminosarum bv. phaseoli; R. leguminosarum bv. trifolii and Sinorhizobium meliloti are also chemoattracted by apigenin and luteolin (Brencic and Winans, 2005). In contrast, naringenin evokes only a weak to no chemotactic response in R. leguminosarum bv. viciae and trifolii (Zaat et al., 1987). It also suppresses the strong chemotaxis of S. meliloti by luteolin (Caetano‐Anolles et al., 1988). Although acetosyringone and umbelliferone are inhibitors of nod gene inducers, R. leguminosarum exhibits an exaggerated chemotactic response to high concentrations of these compounds (Aguilar et al., 1988). Probably, the complex nature of the root exudates of different plants in the rhizosphere (Djordjevic et al., 1987) necessitates such negative regulation by some phenolics. For example, it might be required to prevent competing rhizobia from targeting the same host plant and also for creating a favourable ecological niche for each of the species. Preventing nodule initiation in the vicinity of clover root tips by umbelliferone is a good example of such negative rhizospheric interactions (Djordjevic et al., 1987).
A variety of phenolic compounds affect directly the virA/G genes on the Ti plasmid of different Agrobacterium species (Lee et al., 1996; McCullen and Binns, 2006; Sheng and Citovsky, 1996). These, in turn, influence chromosomal genes, such as the 8‐kb chemotaxis operon, beginning with orf1 (Harighi, 2009; Wright et al., 1998) and the 7205‐bp putative operon involved in the formation of flagellar rods and associated proteins (Deakin et al., 1999; Shaw et al., 1991). Acetosyringone and hydroxyacetosyringone exuded from plant wounds are potent chemoattractants at very low concentrations, and they also act to induce the vir genes of Agrobacterium (Escobar and Dandekar, 2003). In addition to acetosyringone, a number of other phenolics and sugars are effective as chemoattractants (Brencic and Winans, 2005; Palmer et al., 2004; Peng et al., 1998). Even the non‐vir gene‐inducing vanillyl alcohol is a strong chemoattractant (Ashby et al., 1988). On the other hand, high concentrations of some polyphenols may have bacteriostatic or even bactericidal effects, and may block Agrobacterium's access to plant wound sites by inhibiting its chemotactic movement.
Activation of the bacterial nod and vir gene networks
Microbial gene expression following chemotaxis is influenced by a diverse array of substituted plant phenols. These plant‐derived signals are termed ‘host recognition factors’ or ‘xenognosins’ (Campbell et al., 2000). In both the symbiotic Rhizobium and pathogenic Agrobacterium, the same phenolics that act as chemoattractants may also regulate the expression of nod and vir genes, respectively (Djordjevic et al., 1987). For several decades, flavonoids were presumed to be the sole chemoattractants and inducers of nod gene expression in rhizobia (Cohen et al., 2001; Stougaard, 2000). Until the late 1980s, practically almost every study conducted on this subject revolved around the isoflavonoids from soybean (D'Arcy‐Lameta and Jay, 1987). It was only in the late 1990s that the nod gene‐inducing ability of other compounds, such as flavones and flavonols from broad beans, gained importance (Bekkara et al., 1998). With time, however, other organic molecules, mainly phenolics, were identified as potent inducers of nodABC genes, and the effect was dependent on the presence of a functional nodD gene (Perret et al., 2000; Subramanian et al., 2006). The nodD genes from different Rhizobium species are optimally responsive to specific phenolics (Gagnon and Ibrahim, 1998; Peck et al., 2006). Although some phenolics influence nod genes positively, others may have a negative effect. For example, the nod genes of some strains of Bradyrhizobium japonicum are induced by daidzein, genistein and isoflavonoids from soybean, whereas the nod genes of S. meliloti are inhibited by the same phenolics and, instead, are induced by luteolin (Begum et al., 2001; Kosslak et al., 1987). Some phenolics may also perform a dual function of serving as both nod inducers and chemoattractants, whereas others may perform only one of the two functions. One such example is isoliquiritigenin (2′,4′,4‐trihydroxychalcone), which is a strong nod gene inducer, but not a chemoattractant (Kape et al., 1992).
In addition to inducing the biosynthesis of Nod signals, flavonoids inhibit the transport of auxins at the site of rhizobial infection, once the rhizobia have entered the host cells (Brown et al., 2001). In Arabidopsis, two flavonoid‐binding protein complexes, AtAPM and AtMDR, are believed to regulate the polar transport of auxins through the PIN gene homologues (Subramanian et al., 2007). In legumes, the inhibition of flavonoid‐regulated auxin transport is critical for the formation of indeterminate nodules (Wasson et al., 2006). In contrast, the inhibition of auxin transport is not required for the formation of determinate nodules, which can develop even in the presence of very low levels of isoflavones (Grunewald et al., 2009; Subramanian et al., 2007).
In the case of Agrobacterium, a variety of phenolic compounds, one of the most potent of which is acetosyringone, are known to induce vir gene expression (Table 2) (Gelvin, 2009). These phenolics are detected by the VirA/VirG two‐component sensor–transducer system, which then induces all vir loci that encode most components of the protein machinery for T‐DNA transfer (Zupan et al., 2000). In addition to phenolics, VirA, with the help of a chromosomally encoded glucose/galactose‐binding protein ChvE, senses sugar components of the cell wall released from wound sites of susceptible hosts (Ankenbauer and Nester, 1990; He et al., 2009). A conformational change that ensues as a result of the binding of ChvE to VirA allows the latter to interact even with poor vir gene inducers, such as 4‐hydroxyacetophenone, p‐coumaric acid and phenol (Peng et al., 1998).
A specific structure of phenolic molecules is essential for vir gene induction. The aromatic hydroxyl group, together with several other structural features, is absolutely essential. Particularly, the phenolics bearing an unsaturated lateral chain have comparatively higher vir gene‐inducing ability (Joubert et al., 2002). The monomethoxy derivatives are more active than those that lack these methoxy substitutions on the phenol ring. The dimethoxy derivatives are invariably the most active of these three classes of phenolics. In addition, a chiral carbon at the centre of the phenolic molecule is critical for vir gene‐inducing activity. The polarity or acidity created by the para position of the aromatic hydroxyl group bound to the hydrogen bond is associated with a higher induction potential (McCullen and Binns, 2006).
As the introduction of an amide group in syringic acid enhances its vir gene‐inducing activity quite strongly, several new types of phenolic compounds, including three phenol amides, have been synthesized on the basis of syringic acid (Dye et al., 1997). Of these, the highest vir gene‐inducing activity was observed with ethylsyringamide, followed by propylsyringamide, carbethoxyethylensyringamide and syringic acid itself. Recently, benzene rings with a hydroxyl group at position 4, methoxy group at position 3 and another methoxy group at position 5 in a phenolic molecule have been shown to increase the induction of the vir gene significantly (Brencic and Winans, 2005). In addition to phenolics, other compounds, such as d‐glucose, d‐galactose and d‐xylose, are known to enhance vir gene induction (Wise et al., 2005).
Detoxification and biotransformation of xenobiotics into inactive and/or utilizable forms
Phenolics act as antimicrobial compounds because of their ability to disrupt nonspecifically the structural integrity of bacterial membranes and to inhibit specifically bacterial enzymes involved in electron transport (Hirsch et al., 2003). It is natural that bacteria would counteract or even nullify these toxic compounds. Many bacteria are endowed with the ability to degrade and utilize toxic phenols as a source of carbon (Dua et al., 2002; Lovely, 2003; Wackett, 2000; Wackett et al., 1987; Watanabe, 2001). Often the mechanisms for phenol detoxification are closely associated with those involved in establishing plant–bacteria interactions. For example, in the case of Agrobacterium, the virH (formerly pinF) locus that encodes factors for detoxification of harmful phenolics is itself located in the phenolic‐inducible vir regulon of the Ti plasmid (Fig. 1). This regulation ensures that the metabolism or inactivation of phenolics does not commence until the vir regulon is induced (Kalogeraki and Winans, 1998; Sheng and Citovsky, 1996). One of these factors, VirH2, bears strong resemblance to the xenobiotic detoxification enzymes, i.e. the cytochrome P450‐dependent mixed‐function oxidases (Brencic et al., 2004). VirH2 quenches or detoxifies wound‐released plant phenolics by catabolism, mineralization or conversion into sources of carbon, energy or nutrients (Fig. 1). The detoxification of xenobiotics generally involves the modification of xenobiotic compounds by transfer of polar or reactive groups (Guengerich, 2001). The modified compound is then conjugated with a charged species, such as glutathione sulphate (GSH), and metabolized by glutathione S‐transferases (GSTs), γ‐glutamyl transpeptidases and dipeptidases into easily removable acetylcysteine conjugates or mercapturic acids (Boyland and Chasseaud, 1969). These are finally removed from cells by a family of ATP‐binding cassette transporters (Konig et al., 1999). Both cytochrome P450‐dependent oxidases, such as VirH2, and a new class of GSTs exemplified by AtGST1 are present in Agrobacterium species (Kosloff et al., 2006).
What are the specific detoxification reactions catalysed by VirH2? O‐Demethylase encoded by virH2 (Brencic et al., 2004; Kalogeraki et al., 2000) is a specific detoxification enzyme that demethylates ferulic acid into caffeic acid (Fig. 3). VirH2‐dependent mineralization and O‐demethylation of 16 other vir‐inducing methoxyl group‐containing phenolics have also been detected (Brencic et al., 2004). Furthermore, VirH2 mediates the oxidation of vanillyl alcohol and vanillin into vanillate, which is then mineralized into protocatechuate via the β‐ketoadipate pathway (Shaw et al., 2006).
Figure 3.
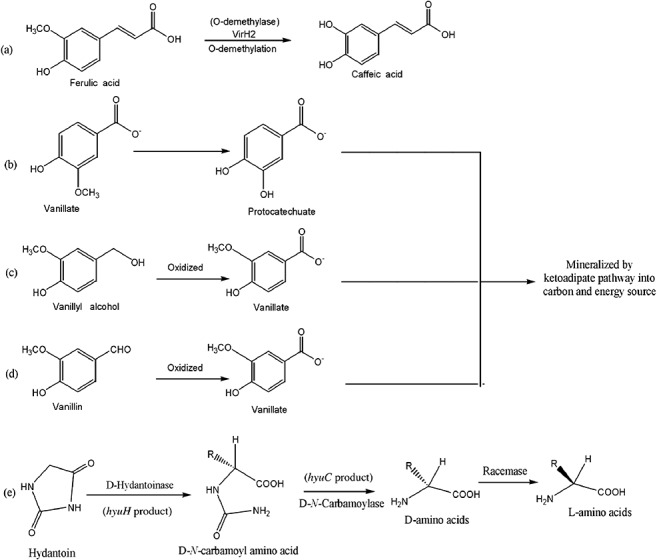
The xenobiotic detoxification reactions by Agrobacterium. (a) Conversion of the excess and toxic amounts of the vir‐inducing ferulic acid into its relatively less toxic caffeic acid by the VirH2 demethylase. (b–d) Oxidation and mineralization of vanillate, vanillyl alcohol and vanillin into sources of carbon and energy. (e) Biotransformation of hydantoin by hydantoinase into d‐N‐carbamoyl amino acid and further into d‐amino acids by d‐N‐carbamoylase. The d‐amino acid is finally converted into its easily utilizable l‐form of amino acid by racemase.
The detoxification of diverse xenobiotics is common in different species of pathogenic as well as nonpathogenic strains of Agrobacterium. Detoxification of hydantoins or glycolylurea is an important example in which the phenolic‐inducible hyuH gene encodes a hydantoinase that cleaves the amide bond at the second position of the hydantoin ring to produce d‐N‐carbamoylamino acid. The N‐carbamoylase enzyme encoded by the hyuC gene then converts the d‐N‐carbamoylamino acid into its corresponding d‐amino acid (Fig. 3) (Burton and Dorrington, 2004). The racemase enzyme present in some Agrobacterium strains can also convert the 5‐monosubstituted hydantoin from the l‐ to its d‐form (Martinez‐Rodriguez et al., 2004). Although both hyuH and hyuC genes have been cloned and characterized from most strains of Agrobacterium (Chao et al., 2000; Jiang et al., 2007; Jiwaji et al., 2009; Martinez‐Gomez et al., 2007), some strains contain only one of these genes (Hils et al., 2001). The activities of these detoxifying enzymes are tightly regulated by growth conditions and are particularly sensitive to nitrogen catabolism (Hartley et al., 2001). Although the molecular basis of hyu gene regulation is unknown, cellular glutamine levels regulate the reversible post‐translational modification of the hydantoinase, representing the pathway by which many Gram‐negative bacteria assimilate nitrogen under high‐nitrogen conditions (Jiwaji and Dorrington, 2009).
Both Agrobacterium and Rhizobium also possess hydrophobic efflux pumps for actively excluding toxic levels of flavonoids (Burse et al., 2004; González‐Pasayo and Martínez‐Romero, 2000; Palumbo et al., 1998; Parniske et al., 1991). Such flavonoid‐inducible efflux pumps confer enhanced resistance to phaseolin in R. etli (González‐Pasayo and Martínez‐Romero, 2000) and pump out isoflavonoids, such as medicarpin, coumestrol and formononetin in A. tumefaciens (Palumbo et al., 1998). It has been proposed that these pumps function by capturing hydrophobic or amphiphilic substrate molecules from the cytoplasmic membrane using a transporter protein (Bolhuis et al., 1997).
Although not threatened by plant‐derived toxic phenolics, rhizobia possess mechanisms for detoxifying various xenobiotics, mainly nonphenolic compounds, present in their rhizosphere. For example, four different Rhizobium species rapidly degrade glyphosate from their environment (Parker et al., 1999). Interestingly, Agrobacterium can also degrade glyphosate and other broad‐spectrum phosphonates and utilize them as a source of phosphorus (Liu et al., 1991). In most species of rhizobia and bradyrhizobia, phenolics are converted into forms that can be used as sources of carbon, nitrogen or energy (Vela et al., 2002). This confers selective advantage on the bacteria for their saprophytic and symbiotic survival in soil and host. The mineralization of catechins and substituted chloro‐aromatics by catechol‐1, 2‐dioxygenase into phloroglucinolcarboxylic acid and protocatechuate are good examples of such relationships (Fig. 4) (Latha and Mahadevan, 1997). Other phenolics, such as the flavonoids quercetin, daidzen and genistein, are also degraded by different Rhizobium and Bradyrhizobium species via the initial C‐ring cleavage (Fig. 4) (Brencic and Winans, 2005; Rao and Cooper, 1994). Even xenobiotic degradation of an industrial chemical, polychlorinated biphenyl (PCB), can occur during the catabolism of the flavonoids naringin and apigenin (Dzantor, 2007; Fletcher and Hedge, 1995). Such biodegradation of xenobiotic compounds presents attractive opportunities for rhizo‐engineering or rhizosphere manipulations for soil bioremediation.
Figure 4.
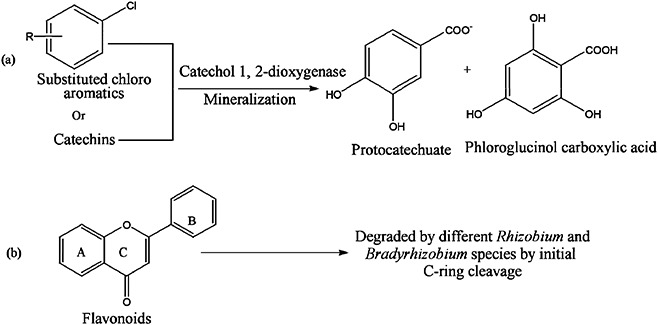
Reactions showing how rhizospheric phenolics are utilized as sources of carbon and energy by Rhizobium spp. for saprophytic and symbiotic survival in soil and host. (a) Mineralization of catechins and substituted chloro‐aromatics by catechol‐1,2‐dioxygenase into phloroglucinolcarboxylic acid and protocatechuate. (b) Degradation of flavonoids by C‐ring cleavage into utilizable forms.
Quorum signalling for attaining infection
An important mechanism by which members of Rhizobiaceae monitor their environment is quorum sensing (Bjarnsholt and Givskov, 2007; Gonzalez and Marketon, 2003). The production, release and sensing of homoserine lactones (HSLs) or their acylated forms (AHL) are important for quorum sensing (Parsek and Greenberg, 2000; Steidle et al., 2001). Quorum sensing allows bacterial cell–cell communication and promotes an advantageous lifestyle for both the survival and maintenance of pathogenic or symbiotic relationships within a range of environmental niches (Joint et al., 2002). In quorum sensing, cell density‐dependent regulation of gene expression enables bacteria to coordinate certain adaptive processes that cannot be performed by an individual microbe.
Quorum sensing helps rhizobia to synchronize themselves to phenolic signals on a population‐wide scale and to function as multicellular organisms for successful symbiosis (Fig. 2). The Rhizobium quorum sensing enhances nodulation efficiency, symbiosome development, exopolysaccharide production, nitrogen fixation and adaptation to stress (Danino et al., 2003; Gonzalez and Marketon, 2003). The extensively studied R. leguminosarum bv. viciae has four quorum sensing operons. Of these, only the cin operon, comprising cinr/i, is located on the chromosome (Lithgow et al., 2000) and regulates the synthesis of the long‐chain (C14.1) quoromones, i.e. AHL. Of the four operons involved in quorum sensing, these AHLs induce only the rai and tra operons located on two different plasmids. The plasmid pIJ9001 contains the raii/rair operon, which encodes the synthesis of short‐chain AHLs and 3‐OH‐C8‐HSL, whose functions are unknown. The raii/rair operon itself is regulated by AHLs produced by the cin operon and by the tra operon of the pRL1J1 plasmid (Wisniewski‐Dye and Downie, 2002). The tra operon encodes the synthesis of 3‐oxo‐C8‐HSL and influences the expression of rai, rhi and tra operons (Wilkinson et al., 2002). It also transcribes the repressor of the cin operon, i.e. the BisR regulator. The rhii genes of the rhii/r/abc operon located on the symbiotic plasmid pRL1J1 encode short‐chain (C6–C8) AHLs and induce their own rhi operon (Sanchez‐Contreras et al., 2007). The expression of the rhii/r/abc operons is repressed by flavonoids in R. leguminosarum (Economou et al., 1989), but, beyond this observation, little is known about the role of phenolics in the regulation of quorum sensing in rhizobia.
Quorum sensing in A. tumefaciens is regulated by the specific acc and divergent arc operon, closely linked to opine catabolism loci, and also by the tra (conjugal transfer genes) and repABC operons of the Ti plasmid (White and Winans, 2007). Mainly, TraI/R and TraM, in conjunction with the diffusible quoromones, N‐3‐(oxooctanoyl)‐l‐homoserine lactone (AAI) and 3‐oxo‐C8 HSL, regulate quorum sensing (Pappas et al., 2004; Zhu and Winans, 1999). In this mechanism, the N‐acylhomoserine lactone synthase or TraI, encoded by the first gene of a cluster located on the Ti plasmid (Hwang et al., 1994), synthesizes AAI, which then binds to the signal receptor and transcription regulator TraR to form a stable dimer (White and Winans, 2007). This, in turn, binds to the tra boxes (18 bp sequences) and activates the transcription of the Ti plasmid tra genes and repABC operon for conjugal transfer (White and Winans, 2007). The traR genes are expressed only in the presence of specific conjugal opines, indicating the indirect role of phenolics (Oger and Farrand, 2002; Pappas, 2008). The products of the repABC operon, in turn, increase the copy number of the Ti plasmid to lessen the metabolic burden during saprophytic growth (Cho and Winans, 2005). TraM is another important regulator of quorum sensing, and the gene encoding it is just adjacent to traR. Although AAI induces TraR at low cell density, the phenolic‐inducible TraM sequesters and inhibits TraR, resulting in quorum quenching (Chen et al., 2004; White and Winans, 2007). In addition to TraM, the products of the attKLM operon on the cryptic plasmid of Agrobacterium also inhibit quorum sensing specifically, either when the population density is low or under conditions of carbon and nitrogen starvation at high population density. AttM, AttL and AttK are also important for the utilization of other rhizospheric quoromones, such as the γ‐butyrolactones produced by other soil bacteria, as alternative nutrition and energy sources in times of food scarcity (Fig. 2).
Quorum sensing in Agrobacterium is also inhibited by salicylic acid (SA) which upregulates the attKLM operon, the products of which, in turn, degrade the bacterial quormone N‐acylhomoserine lactone (Yuan et al., 2007). Transcriptome analysis of A. tumefaciens revealed that, in this quorum‐quenching effect, SA functions additively with indole‐3‐acetic acid (IAA) and γ‐aminobutyric acid (GABA) (Yuan et al., 2008). In a complementary approach, a recent analysis of the Arabidopsis transcriptome also indicated the roles of SA and IAA, as well as of ethylene, in defence against Agrobacterium infection (Lee et al., 2009). The protective effect of SA against Agrobacterium is not limited to quorum quenching. SA can also shut down the expression of the vir genes (Anand et al., 2008; Yuan et al., 2007), mainly by attenuating the function of the VirA kinase domain and shutting down the virA/G regulatory system (Yuan et al., 2007). Interestingly, these inhibitory effects of SA on plant genetic transformation by Agrobacterium do not appear to be related to the classical role of SA in plant–pathogen interactions, i.e. the SA‐induced expression of pathogenesis‐related (PR) genes (Lee et al., 2009).
ACKNOWLEDGEMENTS
We apologise to colleagues whose works have not been cited because of the lack of space; because of these considerations, the reader is often referred to review articles rather than to the primary citations. We thank Miss Nandini Sharma, Division of Natural Plant Products, Institute of Himalayan Bioresource Technology (Council of Scientific and Industrial Research) for drawing the molecular structures for the chemical reactions. The work in VC's laboratory is supported by grants from National CFIDS Foundation/Chronic Fatigue Immune Dysfunction (CFIDS), National Institutes of Health, National Science Foundation, United States Department of Agriculture‐National Institute of Food and Agriculture, US‐Israel Binational Agricultural Research and Development Fund and US‐Israel Binational Science Foundation to VC.
REFERENCES
- Adams, N.R. (1989) Phytoestrogens In: Toxicants of Plant Origin (Cheeke P.R., ed.), pp. 23–51. Boca Raton: CRC Press. [Google Scholar]
- Aguilar, J.M.M. , Ashby, A.M. , Richards, A.J.M. , Loake, G.J. , Watson, M.D. and Shaw, C.H. (1988) Chemotaxis of Rhizobium leguminosarum biovar phaseoli towards flavonoid inducers of the symbiotic nodulation genes. J. Gen. Microbiol. 134, 2741–2746. [Google Scholar]
- Akhtar, M. and Malik, A. (2000) Roles of organic soil amendments and soil organisms in the biological control of plant‐parasitic nematodes: a review. Bioresour. Technol. 74, 35–47. [Google Scholar]
- Akiyama, K. (2007) Chemical identification and functional analysis of apocarotenoids involved in the development of arbuscular mycorrhizal symbiosis. Biosci. Biotechnol. Biochem. 71, 1405–1414. [DOI] [PubMed] [Google Scholar]
- Akiyama, K. , Matsuzaki, K. and Hayashi, H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature, 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Anand, A. , Uppalapati, S.R. , Ryu, C.M. , Allen, S.N. , Kang, L. , Tang, Y. and Mysore, K.S. (2008) Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens . Plant Physiol. 146, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbauer, R.G. and Nester, E.W. (1990) Sugar‐mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J. Bacteriol. 172, 6442–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby, A.M. , Watson, M.D. , Loake, G.J. and Shaw, C.H. (1988) Ti plasmid specified chemotaxis of Agrobacterium tumefaciens C58C toward vir‐inducing phenolic compounds and soluble factors from monocotyledonous and dicotyledonous plants. J. Bacteriol. 170, 4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagayoko, M. , George, E. , Romheld, V. and Buerkert, A. (2000) Effects of mycorrhizae and phosphorus on growth and nutrient uptake of millet, cowpea and sorghum on a West African soil. J. Agric. Sci. 135, 399–407. [Google Scholar]
- Bais, H.P. , Park, S.W. , Weir, T.L. , Callaway, R.M. and Vivanco, J.M. (2004) How plants communicate using the underground information superhighway. Trends Plant Sci. 9, 1360–1385. [DOI] [PubMed] [Google Scholar]
- Bais, H.P. , Weir, T.L. , Perry, L.G. , Gilroy, S. and Jorge, M. (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. [DOI] [PubMed] [Google Scholar]
- Balasundram, N. , Sundram, K. and Samman, S. (2006) Phenolic compounds in plants and agri‐industrial by‐products: antioxidant activity, occurrence, and potential uses. Food Chem. 99, 191–203. [Google Scholar]
- Baleroni, C.R.S. , Ferrarese, M.L.L. , Souza, N.E. and Ferrarese‐Filho, O. (2000) Lipid accumulation during canola seed germination in response to cinnamic acid derivatives. Biol. Plant. 43, 313–316. [Google Scholar]
- Begum, A.A. , Leibovitch, S. , Migner, P. and Zhang, F. (2001) Specific flavonoids induced nod gene expression and pre‐activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J. Exp. Bot. 52, 1537–1543. [DOI] [PubMed] [Google Scholar]
- Bekkara, F. , Jay, M. , Viricel, M.R. and Rome, S. (1998) Distribution of phenolic compounds within seed and seedlings of two Vicia faba cvs differing in their seed tannin content, and study of their seed and root phenolic exudations. Plant Soil, 203, 27–36. [Google Scholar]
- Bertin, C. , Yang, X.H. and Weston, L.A. (2003) The role of root exudates and allelochemicals in the rhizosphere. Plant Soil, 256, 67–83. [Google Scholar]
- Bittel, P. and Robatzek, S. (2007) Microbe‐associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10, 335–341. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt, T. and Givskov, M. (2007) Quorum‐sensing blockade as a strategy for enhancing host defenses against bacterial pathogens. Philos. Trans. R. Soc. B, 362, 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis, H. , Van Veen, H.W. , Poolman, B. , Driessen, A.J. and Konings, W.N. (1997) Mechanisms of multidrug transporters. FEMS Microbiol. Rev. 21, 55–84. [DOI] [PubMed] [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet, A. (2007) Evolution and current status of research in phenolic compounds. Phytochemistry, 68, 2722–2735. [DOI] [PubMed] [Google Scholar]
- Boyland, E. and Chasseaud, L.F. (1969) The role of glutathione and glutathione‐S‐transferase in mercapturic acid biosynthesis. Adv. Enzymol. 32, 173–219. [DOI] [PubMed] [Google Scholar]
- Brencic, A. and Winans, S.C. (2005) Detection of and response to signals involved in host–microbe interactions by plant‐associated bacteria. Microbiol. Mol. Biol. Rev. 69, 155–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brencic, A. , Ebehard, A. and Winans, S.C. (2004) Signal quenching, detoxification and mineralization of vir gene‐inducing phenolics by the VirH2 protein of Agrobacterium tumefaciens . Mol. Microbiol. 51, 1103–1115. [DOI] [PubMed] [Google Scholar]
- Brown, D.E. , Rashotte, A.M. , Murphy, A.S. , Normanly, J. , Tague, B.W. , Peer, W.A. , Taiz, L. and Muday, G.K. (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis . Plant Physiol. 126, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burse, A. , Weingart, H. and Ullrich, M.S. (2004) The phytoalexin‐inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora . Mol. Plant–Microbe Interact. 17, 43–54. [DOI] [PubMed] [Google Scholar]
- Burton, S.G. and Dorrington, R.A. (2004) Hydantoin‐hydrolyzing enzymes for the enantioselective production of amino acids: new insights and applications. Tetrahedron Asymmetry, 15, 2737–2741. [Google Scholar]
- Caetano‐Anolles, G.D. , Crist‐Estes, K. and Bauer, W.D. (1988) Chemotaxis of Rhizobium meliloti to the plant flavone luteolin requires functional nodulation genes. J. Bacteriol. 170, 3164–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, A.M. , Tok, J.B. , Zhang, J. , Wang, Y. , Stein, M. , Lynn, D.G. and Binns, A.N. (2000) Xenognosin sensing in virulence: is there a phenol receptor in Agrobacterium tumefaciens? Chem. Biol. 7, 65–76. [DOI] [PubMed] [Google Scholar]
- Chao, Y. , Chiang, C. , Lo, T. and Fu, H. (2000) Overproduction of d‐hydantoinase and cabamoylase in a soluble form in Escherichia coli . Appl. Microbiol. Biotechnol. 54, 348–353. [DOI] [PubMed] [Google Scholar]
- Chen, G. , Malenkos, J.W. , Cha, M.R. , Fuqua, C. and Chen, L. (2004) Quorum‐sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 52, 1641–1651. [DOI] [PubMed] [Google Scholar]
- Cho, H. and Winans, S.C. (2005) VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound‐released chemical signals. Proc. Natl. Acad. Sci. USA, 102, 14 843–14 848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clé, C. , Hill, L.M. , Niggeweg, R. , Martin, C.R. , Guisez, Y. , Prinsen, E. and Jansen, M.A.K. (2008) Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV‐tolerance. Phytochemistry, 69, 2149–2156. [DOI] [PubMed] [Google Scholar]
- Cohen, M.F. , Sakihama, Y. and Yamasaki, H. (2001) Roles of plant flavonoids in interactions with microbes: from protection against pathogens to the mediation of mutualism. Recent Res. Devel. Plant Physiol. 2, 157–173. [Google Scholar]
- Cushnie, T.T.P. and Lamb, A.J. (2005) Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents, 26, 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy‐Lameta, A. and Jay, M. (1987) Study of soybean and lentil root exudates. III. Influence of soybean isoflavonoids on the growth of rhizobia and some rhizospheric microorganisms. Plant Soil, 101, 267–272. [Google Scholar]
- Dakora, F.D. (2003) Defining new roles for plant and rhizobial molecules in sole and mixed plant cultures involving symbiotic legumes. New Phytol. 158, 39–49. [Google Scholar]
- Dakora, F.D. and Phillips, D.A. (1996) Diverse functions of isoflavonoids in legumes transcend anti‐microbial definitions of phytoalexins. Physiol. Mol. Plant Pathol. 49, 1–20. [Google Scholar]
- Dakora, F.D. and Phillips, D.A. (2002) Root exudates as mediators of mineral acquisition in low‐nutrient environments. Plant Soil, 245, 35–47. [Google Scholar]
- Danino, V.E. , Wilkinson, A. , Edwards, A. and Downie, J.A. (2003) Recipient‐induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum‐sensing relay. Mol. Microbiol. 50, 511–525. [DOI] [PubMed] [Google Scholar]
- Deakin, W.J. , Parker, V.E. , Wright, E.L. , Ashcroft, K.J. , Loake, G.J. and Shaw, C.H. (1999) Agrobacterium tumefaciens possesses a fourth flagellin gene located in a large gene cluster concerned with flagellar structure, assembly and motility. Microbiology, 145, 1397–1407. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A. and Paiva, N.L. (1995) Stress‐induced phenylpropanoid metabolism. Plant Cell, 7, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic, M.A. , Gabriel, D.W. and Rolfe, B.G. (1987) Rhizobium – the refined parasite of legumes. Annu. Rev. Phytopathol. 25, 145–168. [Google Scholar]
- Dua, M. , Singh, A. , Sethunathan, N. and Johri, A.K. (2002) Biotechnology and bioremediation: successes and limitations. Appl. Microbiol. Biotechnol. 59, 143–152. [DOI] [PubMed] [Google Scholar]
- Dye, F. , Berthelot, K. , Griffon, B. , Delay, D. and Delmotte, F.M. (1997) Alkylsyringamides, new inducers of Agrobacterium tumefaciens virulence genes. Biochimie, 79, 3–6. [DOI] [PubMed] [Google Scholar]
- Dzantor, E.K. (2007) Phytoremediation: the state of rhizosphere engineering for accelerated rhizodegradation of xenobiotic contaminants. J. Chem. Technol. Biotechnol. 82, 228–232. [Google Scholar]
- Economou, A. , Hawkins, F.K. , Downie, J.A. and Johnston, A.W. (1989) Transcription of rhiA, a gene on a Rhizobium leguminosarum bv. viciae Sym plasmid, requires rhiR and is repressed by flavanoids that induce nod genes. Mol. Microbiol. 3, 87–93. [DOI] [PubMed] [Google Scholar]
- Escobar, M.A. and Dandekar, A.M. (2003) Agrobacterium tumefaciens as an agent of disease. Trends Plant Sci. 8, 380–386. [DOI] [PubMed] [Google Scholar]
- Ferrazzano, G.F. , Amato, I. , Ingenito, A. , Natale De, A. and Pollio, A. (2009) Anti‐cariogenic effects of polyphenols from plant stimulant beverages (cocoa, coffee, tea). Fitoterapia, 80, 255–262. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.S. and Hedge, R.S. (1995) Release of phenols by perennial plant roots and their potential importance in bioremediation. Chemosphere, 31, 3009–3016. [Google Scholar]
- Gagnon, H. and Ibrahim, R.K. (1998) Aldonic acids: a novel family of nod gene inducers of Mesorhizobium loti, Rhizobium lupini, and Sinorhizobium meliloti . Mol. Plant–Microbe Interact. 11, 988–998. [Google Scholar]
- Gelvin, S.B. (2009) Agrobacterium in the genomics age. Plant Physiol. 150, 1665–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, J.E. and Marketon, M.M. (2003) Quorum sensing in nitrogen‐fixing rhizobia. Microbiol. Mol. Biol. Rev. 67, 574–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Pasayo, R. and Martínez‐Romero, E. (2000) Multiresistance genes of Rhizobium etli CFN42. Mol. Plant–Microbe Interact. 13, 572–577. Erratum in: Mol. Plant–Microbe Interact 13, 796. [DOI] [PubMed] [Google Scholar]
- Gray, E.J. and Smith, D.L. (2005) Intracellular and extracellular PGPR: commonalities and distinctions in the plant–bacterium signaling process. Soil Biol. Biochem. 37, 395–412. [Google Scholar]
- Grunewald, W. , Noorden van, G. , Isterdael, G.V. , Beeckman, T. , Gheysen, G. and Mathesius, U. (2009) Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell, 21, 2553–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich, F.P. (2001) Metabolism of chemical carcinogens. Carcinogenesis, 21, 345–351. [DOI] [PubMed] [Google Scholar]
- Halvorson, J.J. , Gonzalez, J.M. , Hagerman, A.E. and Smith, J.L. (2009) Sorption of tannin and related phenolic compounds and effects on soluble‐N in soil. Soil Biol. Biochem. 41, 2002–2010. [Google Scholar]
- Harborne, J.B. (1980) Plant phenolics In: Encyclopedia of Plant Physiology (Bell E.A. and Charlwood B.V., eds), pp. 329–395. Berlin Heidelberg, New York: Springer‐Verlag. [Google Scholar]
- Harborne, J.B. and Simmonds, N.W. (1964) Natural distribution of the phenolic aglycones In: Biochemistry of Phenolic Compounds (Harborne J.B., ed.), pp. 77–128. London: Academic Press. [Google Scholar]
- Harighi, B. (2009) Genetic evidence for CheB‐ and CheR‐dependent chemotaxis system in A. tumefaciens toward acetosyringone. Microbiol. Res. 164, 634–641. [DOI] [PubMed] [Google Scholar]
- Hartley, R.D. and Harris, P.J. (1981) Phenolic constituents of the cell walls of dicotyledons. Biochem. Syst. Ecol. 9, 189–203. [Google Scholar]
- Hartley, C.J. , Manford, F. , Burton, S.G. and Dorrington, R.A. (2001) Over‐production of hydantoinase and N‐carbamoylamino acid amido hydrolase enzymes by regulatory mutants of Agrobacterium tumefaciens . Appl. Microbiol. Biotechnol. 57, 43–49. [DOI] [PubMed] [Google Scholar]
- Hartmann, A. , Schmid, M. , Tuinen van, D. and Berg, G. (2009) Plant‐driven selection of microbes. Plant Soil, 321, 235–257. [Google Scholar]
- Hättenschwiler, S. and Vitousek, P.M. (2000) The role of polyphenols in terrestrial ecosystem nutrient cycling. Trends Ecol. Evol. 15, 238–242. [DOI] [PubMed] [Google Scholar]
- He, F. , Nair, G.R. , Soto, C.S. , Chang, Y. , Hsu, L. , Ronzone, E. , DeGrado, W.F. and Binns, A.N. (2009) Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens . J. Bacteriol. 191, 5802–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hils, M. , Munch, P. , Altenbucher, J. , Syldatk, C. and Mattes, R. (2001) Cloning and characterization of genes from Agrobacterium sp. IP I‐671 involved in hydantoin degradation. Appl. Microbiol. Biotechnol. 57, 680–688. [DOI] [PubMed] [Google Scholar]
- Hiltner, L. (1904) Uber neuere Erfahrungen und probleme auf dem Gebiete der Bodenbakteriologie unter besonderer Berucksichtigung der Grundungung und brache. Arbeilten der Deutschen Landwirtschaftlichen Gesellschaft, 98, 59–78. [Google Scholar]
- Hirsch, A.M. , Bauer, W.D. , Bird, D.M. , Cullimore, J. , Tyler, B. and Yoder, J.I. (2003) Molecular signals and receptors: controlling rhizosphere interactions between plants and other organisms. Ecology, 84, 858–868. [Google Scholar]
- Hollman, P.C.H. and Katan, M.B. (1999) Dietary flavonoids: intake, health effects and bioavailability. Food Chem. Toxicol. 37, 937–942. [DOI] [PubMed] [Google Scholar]
- Hwang, I. , Li, P.L. , Zhang, L. , Piper, K.R. , Cook, D.M. , Tate, M.E. and Farrand, S.K. (1994) TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N‐acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. USA, 91, 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Li, C. , Zhang, W. , Cai, Y. , Yang, S. and Jiang, W. (2007) Directed evolution and structural analysis of N‐carbamoyl‐D‐amino acid amidohydrolase provides insights into recombinant protein solubility in Escherichia coli . Biochem. J. 402, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiwaji, M. and Dorrington, R.A. (2009) Regulation of hydantoin‐hydrolyzing enzyme expression in Agrobacterium tumefaciens strain RU‐AE01. Appl. Microbiol. Biotechnol. 84, 1169–1179. [DOI] [PubMed] [Google Scholar]
- Jiwaji, M. , Hartley, C.J. , Clark, S.A. , Burton, S.G. and Dorrington, R.A. (2009) Enhanced hydantoin‐hydrolyzing enzyme activity in an Agrobacterium tumefaciens strain with two distinct N‐carbamoylases. Enzyme Microb. Technol. 44, 203–209. [Google Scholar]
- Joint, I. , Trait, K. , Callow, M.E. , Callow, J.A. , Milton, D. , Williams, P. and Cámara, M. (2002) Cell‐to‐cell communication across the prokaryote–eukaryote boundary. Science, 298, 1207. [DOI] [PubMed] [Google Scholar]
- Joubert, P. , Beaupère, D. , Lelièvre, P. , Wadouachi, A. , Sangwan, R.S. and Sangwan‐Norreel, B.S. (2002) Effects of phenolic compounds on Agrobacterium vir genes and gene transfer induction a plausible molecular mechanism of phenol binding protein activation. Plant Sci. 162, 733–743. [Google Scholar]
- Kalogeraki, V.S. and Winans, S.C. (1998) Wound‐released chemical signals may elicit multiple responses from an Agrobacterium tumefaciens strain containing an octopine‐type Ti plasmid. J. Bacteriol. 180, 5660–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeraki, V.S. , Zhu, J. , Stryker, J.L. and Winans, S.C. (2000) The right end of the vir region of an octopine‐type Ti plasmid contains four new members of the vir regulon that are not essential for pathogenesis. J. Bacteriol. 182, 1774–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kape, R. , Parniske, M. , Brandt, S. and Werner, D. (1992) Isoliquiritigenin, a strong nod gene and glyceollin resistance‐inducing flavonoid from soybean root exudate. Appl. Environ. Microbiol. 58, 1705–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karou, D. , Dicko, M.H. , Simpore, J. and Traore, A.S. (2005) Antioxidant and antibacterial activities of polyphenols from ethnomedicinal plants of Burkina Faso. Afr. J. Biotechnol. 4, 823–828. [Google Scholar]
- Kefeli, V.I. , Kalevitch, M.V. and Borsari, B. (2003) Phenolic cycle in plants and environment. J. Cell Mol. Biol. 2, 13–18. [Google Scholar]
- Konig, J. , Nies, A.T. , Cui, Y. , Leier, I. and Keppler, D. (1999) Conjugate export pumps of the multidrug resistance protein (MRP) family: localization, substrate specificity, and MRP2‐mediated drug resistance. Biochim. Biophys. Acta, 1461, 377–394. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. and Pieterse, C.M.J. (2008) Cross talk in defense signaling. Plant Physiol. 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosloff, M. , Han, G.W. , Sri Krishna, S. , Schwarzenbacher, R. , Fasnacht, M. , Elsliger, M.A. , Abdubek, P. , Agarwalla, S. , Ambing, E. , Astakhova, T. , Axelrod, H.L. , Canaves, J.M. , Carlton, D. , Chiu, H.J. , Clayton, T. , DiDonato, M. , Duan, L. , Feuerhelm, J. , Grittini, C. , Grzechnik, S.K. , Hale, J. , Hampton, E. , Haugen, J. , Jaroszewski, L. , Jin, K.K. , Johnson, H. , Klock, H.E. , Knuth, M.W. , Koesema, E. , Kreusch, A. , Kuhn, P. , Levin, I. , McMullan, D. , Miller, M.D. , Morse, A.T. , Moy, K. , Nigoghossian, E. , Okach, L. , Oommachen, S. , Page, R. , Paulsen, J. , Quijano, K. , Reyes, R. , Rife, C.L. , Sims, E. , Spraggon, G. , Sridhar, V. , Stevens, R.C. , Van Den Bedem, H. , Velasquez, J. , White, A. , Wolf, G. , Xu, Q. , Hodgson, K.O. , Wooley, J. , Deacon, A.M. , Godzik, A. , Lesley, S.A. and Wilson, I.A. (2006) Comparative structural analysis of a novel glutathione S‐transferase (Atu5508) from Agrobacterium tumefaciens at 2.0 Å resolution. Proteins Struct. Funct. Bioinform. 65, 527–537. [DOI] [PubMed] [Google Scholar]
- Kosslak, R.M. , Bookland, R. , Barkei, J. , Paaren, H.E. , Edward, R. and Appelbaum, E.R. (1987) Induction of Bradyrhizobium japonicum common nod genes by isoflavones isolated from Glycine max . Proc. Natl. Acad. Sci. USA, 84, 7428–7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, T.E.C. , Dahlgren, R.A. and Zasoski R.J. (2003) Tannins in nutrient dynamics of forest ecosystems – a review. Plant Soil, 256, 41–66. [Google Scholar]
- Langcake, P. , Irvine, J.A. and Jeger, M.J. (1981) Alternative chemical agents for controlling plant disease. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 295, 83–101. [Google Scholar]
- Latha, S. and Mahadevan, A. (1997) Role of rhizobia in the degradation of aromatic substances. World J. Microbiol. Biotechnol. 13, 601–607. [Google Scholar]
- Lattanzio, V. , Lattanzio, V.M.T. and Cardinali, A. (2006) Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. In: Phytochemistry Advances in Research (Imperato F. ed.), pp. 23–67. India: Research Signpost. [Google Scholar]
- Lee, C.W. , Efetova, M. , Engelmann, J.C. , Kramell, R. , Wasternack, C. , Ludwig‐Muller, J. , Hedrich, R. and Deeken, R. (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana . Plant Cell, 21, 2948–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.W. , Jin, S. , Sim, W.S. and Nester, E.W. (1996) The sensing of plant signal molecules by Agrobacterium: genetic evidence for the direct recognition of phenolic inducers by the VirA protein. Gene, 179, 83–88. [DOI] [PubMed] [Google Scholar]
- Lithgow, J.K. , Wilkinson, A. , Hardman, A. , Rodelas, B. , Wisniewski‐Dye, F. , Williams, P. and Downie, J.A. (2000) The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum‐sensing loci. Mol. Microbiol. 37, 81–97. [DOI] [PubMed] [Google Scholar]
- Liu, C.M. , McLean, P.A. , Sookdeo, C.C. and Cannon, F.C. (1991) Degradation of the herbicide glyphosate by members of the family Rhizobiaceae. Appl. Environ. Microbiol. 57, 1799–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi, M.A.K. , Bilal, R. and Malik, K.A. (1987) Allelopathy in agroecosystems: wheat phytotoxicity and its possible roles in crop rotation. J. Chem. Ecol. 13, 1881–1891. [DOI] [PubMed] [Google Scholar]
- Lovely, D.R. (2003) Cleaning up with genomics: applying molecular biology of self‐bioremediation. Nat. Rev. Microbiol. 1, 35–44. [DOI] [PubMed] [Google Scholar]
- Lu, H. (2009) Dissection of salicylic acid‐mediated defense signaling networks. Plant Signal. Behav. 4, 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, D.G. and Chang, M. (1990) Phenolic signals in cohabitation: implications for plant development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 497–526. [Google Scholar]
- Madhan, S.S.R. , Girish, R. , Karthik, N. , Rajendran, R. and Mahendran, V.S. (2009) Allelopathic effects of phenolics and terpenoids extracted from Gmelina arborea on germination of Black gram (Vigna mungo) and Green gram (Vigna radiata). Allelopathy J. 23, 323–332. [Google Scholar]
- Martinez‐Gomez, A.I. , Martinez‐Rodriguez, S. , Clemente‐Jimenez, J.M. , Pozo‐Dengra, J. , Rodriguez‐Vico, F. and Las Heras‐Vazquez, F.J. (2007) Recombinant polycistronic structure of hydantoinase process genes in Escherichia coli for the production of optically pure d‐amino acids. Appl. Environ. Microbiol. 73, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Rodriguez, S. , Las Heras‐Vazquez, F. , Clemente‐Jimenez, J. and Rodriguez‐Vico, F. (2004) Biochemical characterization of a novel hydantoin racemase from Agrobacterium tumefaciens C58. Biochimie, 86, 77–81. [DOI] [PubMed] [Google Scholar]
- Matilla, M. , Espinosa‐Urgel, M. , Rodriguez‐Herva, J. , Ramos, J. and Ramos‐Gonzalez, M. (2007) Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 8, R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen, C.A. and Binns, A.N. (2006) Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu. Rev. Cell. Dev. Biol. 22, 101–127. [DOI] [PubMed] [Google Scholar]
- Mert‐Türk, F. (2002) Phytoalexins: defense or just a response to stress? J. Cell Mol. Biol. 1, 1–6. [Google Scholar]
- Morris, S.W. , Vernooij, B. , Titatarn, S. , Starrett, M. , Thomas, S. , Wiltse, C.C. , Frederiksen, R.A. , Bhandhufalck, A. , Hulbert, S. and Uknes, S. (1998) Induced resistance responses in maize. Mol. Plant–Microbe Interact. 11, 643–658. [DOI] [PubMed] [Google Scholar]
- Moulin, L. , Munive, A. , Dreyfus, B. and Boivin‐Masson, C. (2001) Nodulation of legumes by members of the β‐subclass of proteobacteria. Nature, 411, 948–950. [DOI] [PubMed] [Google Scholar]
- Ndakidemi, P.A. and Dakora, F.D. (2003) Review: legume seed flavonoids and nitrogenous metabolites as signals and protectants in early seedling development. Funct. Plant Biol. 30, 729–745. [DOI] [PubMed] [Google Scholar]
- Newman, M.A. , Dow, J.M. , Molinaro, A. and Parrilli, M. (2007) Priming, induction and modulation of plant defense responses by bacteria lipopolysaccharides. J. Endotoxin Res. 13, 69–84. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , Roux, M. and Zipfel, C. (2009) Recent advances in PAMP‐triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150, 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northup, R.R. , Dahlgren, R.A. and McColl, J.G. (1998) Polyphenols as regulators of plant–litter–soil interactions in northern California's pygmy forest: a positive feedback? Biogeochemistry, 42, 189–220. [Google Scholar]
- Oger, P. and Farrand, S.K. (2002) Two opines control conjugal transfer of an Agrobacterium plasmid by regulating expression of separate copies of the quorum‐sensing activator gene traR . J. Bacteriol. 184, 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena, M. , Jourdan, E. , Adam, A. , Paquot, M. , Brans, A. , Joris, B. , Arpigny, J.L. and Thonart, P. (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Ozyigit, I.I. , Kahraman, M.V. and Ercan, O. (2007) Relation between explant age, total phenols and regeneration response of tissue cultured cotton (Gossypium hirsutum L). Afr. J. Biotechnol. 6, 3–8. [Google Scholar]
- Palmer, M.A. , Bernhardt, E. , Chornesky, E. , Collins, S. , Dobson, A. , Duke, C. , Gold, B. , Jacobson, R. , Kingsland, S. , Kranz, R. , Mappin, M. , Martinez, M.L. , Micheli, F. , Morse, J. , Pace, M. , Pascual, M. , Palumbi, S. , Reichman, O.J. , Simons, A. , Townsend, A. and Turner, M. (2004) Ecology for a crowded planet. Science, 304, 1251–1252. [DOI] [PubMed] [Google Scholar]
- Palumbo, J.D. , Kado, C.I. and Phillips, D.A. (1998) An isoflavonoid‐inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J. Bacteriol. 180, 3107–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas, K.M. (2008) Cell–cell signaling and the Agrobacterium tumefaciens Ti plasmid copy number fluctuations. Plasmid, 60, 89–107. [DOI] [PubMed] [Google Scholar]
- Pappas, K.M. , Weingart, C.L. and Winans, S.C. (2004) Chemical communication in proteobacteria: biochemical and structural studies of signal synthases and receptors required for intercellular signaling. Mol. Microbiol. 53, 755–769. [DOI] [PubMed] [Google Scholar]
- Parker, G.F. , Higgins, T.P. , Hawkes, T. and Robson, R.L. (1999) Rhizobium (Sinorhizobium) meliloti phn genes: characterization and identification of their protein products. J. Bacteriol. 181, 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M. , Ahlborn, B. and Werner, D. (1991) Isoflavonoid‐inducible resistance to the phytoalexin glyceollin in soybean rhizobia. J. Bacteriol. 173, 3432–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek, M.R. and Greenberg, E.P. (2000) Acyl‐homoserine lactone quorum sensing in Gram‐negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA, 97, 8789–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, M.C. , Fisher, R.F. and Long, S.R. (2006) Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti . J. Bacteriol. 188, 5417–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, W.T. , Lee, Y.W. and Nester, E.W. (1998) The phenolic recognition profiles of the Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar binding protein ChvE. J. Bacteriol. 180, 5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret, X. , Staehelin, C. and Broughton, W.J. (2000) Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64, 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, J.R. and Cooper, J.E. (1994) Rhizobia catabolize nod gene‐inducing flavonoids via C‐ring fission mechanisms. J. Bacteriol. 176, 5409–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin, H. , Andary, C. , Kovacs, G. and Molgaard, P. (1989) Caffeic acid esters as in vitro inhibitors of plant pathogenic bacteria and fungi. Biochem. Syst. Ecol. 17, 175–184. [Google Scholar]
- Samac, D.A. and Graham, M.A. (2007) Recent advances in legume–microbe interactions: recognition, defense response, and symbiosis from a genomic perspective. Plant Physiol. 144, 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Contreras, M. , Bauer, W.D. , Gao, M. , Robinson, J.B. and Downie, J.A. (2007) Quorum‐sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos. Trans. R. Soc. B, 362, 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl, C.L. , Leuchtmann, A. and Spiering, M.J. (2004) Symbiosis of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 55, 315–340. [DOI] [PubMed] [Google Scholar]
- Schmitz‐Hoerner, R. and Weissenbock, G. (2003) Contribution of phenolic compounds to the UV‐B screening capacity of developing barley primary leaves in relation to DNA damage and repair under elevated UV‐B levels. Phytochemistry, 64, 243–255. [DOI] [PubMed] [Google Scholar]
- Schuhegger, R. , Ihring, A. , Gantner, S. , Bahnweg, G. , Knappe, C. , Vogg, G. , Hutzler, P. , Schmid, M. , Van Breusegem, F. , Eberl, L. , Hartmann, A. and Langebartels, C. (2006) Induction of systemic resistance in tomato by N‐acyl‐L‐homoserine lactone‐producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918. [DOI] [PubMed] [Google Scholar]
- Seneviratne, G. and Jayasinghearachchi, H.S. (2003) Mycelial colonization by bradyrhizobia and azorhizobia. J. Biosci. 28, 243–247. [DOI] [PubMed] [Google Scholar]
- Shaw, C.H. , Loake, G.J. and Brown, A.P. (1991) Isolation and characterization of behavioral mutants and genes of Agrobacterium tumefaciens . J. Gen. Microbiol. 137, 1939–1953. [Google Scholar]
- Shaw, L.J. , Morris, P. and Hooker, J.E. (2006) Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ. Microbiol. 8, 1867–1880. [DOI] [PubMed] [Google Scholar]
- Sheng, J. and Citovsky, V. (1996) Agrobacterium–plant cell interaction: have virulence proteins, will travel. Plant Cell, 81, 699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira, J.O. , Safir, G.R. and Nair, M.G. (1991) Stimulation of vesicular arbuscular mycorrhiza formation and growth of white clover by flavonoid compounds. New Phytol. 118, 87–93. [Google Scholar]
- Steidle, A. , Sigl, K. , Schuhegger, R. , Ihring, A. , Schmid, M. , Gantner, S. , Stoffels, M. , Riedel, K. , Givskov M., Hartmann, A. , Langebartels, A. and Eberl, L. (2001) Visualization of N‐acylhomoserine lactone‐mediated cell–cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67, 5761–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkellner, S. , Lendzemo, V. , Langer, I. , Schweiger, P. , Khaosaad, T. , Toussaint, J.P. and Vierheilig, H. (2007) Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant–fungus interactions. Molecules, 12, 1290–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard, J. (2000) Regulators and regulation of legume root nodule development. Plant Physiol. 124, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S. , Stacey, G. and Yu, O. (2006) Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum . J. Biotechnol. 126, 69–77. [DOI] [PubMed] [Google Scholar]
- Subramanian, S. , Stacey, G. and Yu, O. (2007) Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci. 12, 282–285. [DOI] [PubMed] [Google Scholar]
- Taguri, T. , Tanaka, T. and Kouno, I. (2006) Antibacterial spectrum of plant polyphenols and extracts depending upon hydroxyphenyl structure. Biol. Pharm. Bull. 29, 2226–2235. [DOI] [PubMed] [Google Scholar]
- Taylor, L.P. and Grotewold, E. (2005) Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8, 317–323. [DOI] [PubMed] [Google Scholar]
- Thomas, P. and Ravindra, M.B. (1999) Shoot tip culture in mango: influence of medium, genotype, explant factors, season and decontamination treatments on phenolic exudation, explant survival and axenic culture establishment. J. Hortic. Sci. 72, 713–722. [Google Scholar]
- Tran, H. , Ficke, A. , Aslimwe, T. , Hofte, M. and Raaijmakers, J.M. (2007) Role of the cyclic lipopolypeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens . New Phytol. 175, 731–742. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Glazebrook, J. and Katagiri, F. (2008) The interplay between MAMP and SA signaling. Plant Signal. Behav. 3, 359–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela, S. , Häggblom, M.M. and Young, L.Y. (2002) Biodegradation of aromatic and aliphatic compounds by rhizobial species. Soil Sci. 167, 802–810. [Google Scholar]
- Vit, P. , Soler, C. and Tomás‐Barberán, F.A. (1997) Profiles of phenolic compounds of Apis mellifera and Melipona spp. honeys from Venezuela. Z. Lebensm. Unters. Forsch. A, 204, 43–47. [Google Scholar]
- Wackett, L.P. (2000) Environmental biotechnology. Trends Biotechnol. 18, 19–21. [DOI] [PubMed] [Google Scholar]
- Wackett, L.P. , Shames, S.L. , Venditti, C.P. and Walsh, C.T. (1987) Bacterial carbon phosphorus lyase: products, rates and regulation of phosphonic and phosphinic acid metabolism. J. Bacteriol. 169, 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson, A.P. , Pellerone, F.I. and Mathesius, U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell, 18, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, M.E. (2001) Can bioremediation bounce back? Nat. Biotechnol. 19, 1111–1115. [DOI] [PubMed] [Google Scholar]
- Weir, T.L. , Park, S.W. and Vivanco, J.M. (2004) Biochemical and physiological mechanisms mediated by allelochemicals. Curr. Opin. Plant Biol. 7, 472–479. [DOI] [PubMed] [Google Scholar]
- Whipps, J.M. (2001) Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511. [DOI] [PubMed] [Google Scholar]
- White, C.E. and Winans, S.C. (2007) Cell–cell communication in the plant pathogen Agrobacterium tumefaciens . Philos. Trans. R. Soc. B, 362, 1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, A. , Danino, V. , Wisniewski‐Dye, F. , Lithgow, J.K. and Downie, J.A. (2002) N‐acyl‐homoserine lactone inhibition of rhizobial growth is mediated by two quorum‐sensing genes that regulate plasmid transfer. J. Bacteriol. 184, 4510–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, A.A. , Voinov, L. and Binns, A.N. (2005) Intersubunit complementation of sugar signal transduction in VirA heterodimers and posttranslational regulation of VirA activity in Agrobacterium tumefaciens . J. Bacteriol. 187, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski‐Dye, F. and Downie, J.A. (2002) Quorum‐sensing in Rhizobium . Antonie Van Leeuwenhoek, 81, 397–407. [DOI] [PubMed] [Google Scholar]
- Wright, E.L. , Deakin, W.J. and Shaw, C.H. (1998) A chemotaxis cluster from Agrobacterium tumefaciens . Gene, 220, 83–89. [DOI] [PubMed] [Google Scholar]
- Xuan, T.D. , Shinkichi, T. , Khanh, T.D. and Chung, I.M. (2005) Biological control of weeds and plant pathogens in paddy rice by exploiting plant allelopathy: an overview. Crop Prot. 24, 197–206. [Google Scholar]
- Yuan, Z.C. , Edlind, M.P. , Liu, P. , Saenkham, P. , Banta, L.M. , Wise, A.A. , Ronzone, E. , Binns, A.N. , Kerr, K. and Nester, E.W. (2007) The plant signal salicylic acid shuts down expression of the vir regulon and activates quormone‐quenching genes in Agrobacterium . Proc. Natl. Acad. Sci. USA, 104, 11790–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Z.C. , Haudecoeur, E. , Faure, D. , Kerr, K.F. and Nester, E.W. (2008) Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole‐3‐acetic acid and gamma‐amino butyric acid reveals signaling cross‐talk and Agrobacterium–plant co‐evolution. Cell. Microbiol. 10, 2339–2354. [DOI] [PubMed] [Google Scholar]
- Zaat, S.A.J. , Wijffelman, C.A. , Spaink, H.P. , Van Brussel, A.A.N. , Okker, R.J.H. and Lugtenberg, B.J.J. (1987) Induction of the nodA promoter of Rhizobium leguminosarum sym plasmid pRLl JI by plant flavanones and flavones. J. Bacteriol. 169, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapprometov, M. (1989) The formation of phenolic compounds in plant cell and tissue cultures and possibility of its regulation. Adv. Cell Cult. 7, 240–245. [Google Scholar]
- Zhu, J. and Winans, S.C. (1999) Autoinducer binding by the quorum‐sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA, 96, 4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2008) Pattern‐recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. [DOI] [PubMed] [Google Scholar]
- Zupan, J. , Muth, T.R. , Draper, O. and Zambryski, P.C. (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 23, 11–28. [DOI] [PubMed] [Google Scholar]


