SUMMARY
NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1 (NPR1; also known as NIM1) is a master regulator of systemic acquired resistance (SAR). SAR is induced by salicylic acid (SA), leading to the expression of PATHOGENESIS‐RELATED (PR) genes. Current evidence suggests that NPR1 is part of a transcription complex tethered to activation sequence‐1 (as‐1)‐like cis‐acting elements in PR‐1 gene promoters through TGA transcription factors, and that SA‐dependent PR‐1 gene expression is regulated by NIM1‐INTERACTING (NIMIN) proteins. In Arabidopsis, NPR1 is active only after SA induction. Regulation of Arabidopsis NPR1 activity has been proposed to comprise cysteine‐156 (Cys‐156), mediating SA‐induced cytoplasmic oligomer–nuclear monomer exchange, and Cys‐521 and Cys‐529, mediating SA‐dependent transcriptional activation. Tobacco NPR1 does not harbour these residues. To understand the function of tobacco NPR1, we analysed its biochemical capabilities in a heterologous system: yeast. Tobacco NPR1 differs from Arabidopsis NPR1 in its subcellular localization and its transactivation potential. Yet, both tobacco and Arabidopsis NPR1, as well as tobacco NIM1‐like1, alter some of their biochemical activities in response to SA. Whereas the addition of SA to yeast growth medium induces transcriptional activity in tobacco NPR1, its interaction with NIMIN2‐type proteins is suppressed. The effects of SA are specific, sensitive and occur coordinately. They are abolished completely by mutation of the arginine residue within the invariable penta‐amino acid motif LENRV, as present in the nonfunctional Arabidopsis nim1‐4 allele. Furthermore, NPR1 proteins with the LENRV domain coincidently harbour a broad and strongly conserved NIMIN1/NIMIN2 binding site. Our data suggest that NPR1 and some NPR1‐like proteins are sensitive to the plant hormone SA, altering some of their biochemical capabilities to enable stimulus‐dependent gene expression. The sensitivity of NPR1 proteins to SA, together with their differential interaction with diverse NIMIN proteins, seems a plausible molecular basis for the timely and coordinated activation of PR genes during SAR.
INTRODUCTION
Systemic acquired resistance (SAR) is a long‐lasting and broad‐spectrum disease resistance state in plants, which is induced after infection with necrotizing pathogens (Ross, 1961). SAR is regulated by increasing levels of salicylic acid (SA; Delaney et al., 1994; Gaffney et al., 1993; Malamy et al., 1990; Métraux et al., 1990; Uknes et al., 1993), resulting in the coordinate induction of a heterogeneous group of SAR markers, termed PATHOGENESIS‐RELATED (PR) proteins (Uknes et al., 1992; Ward et al., 1991), that are supposed to confer general resistance against biotrophic pathogens, such as fungi, viruses and bacteria (van Loon and van Strien, 1999). Notably, PR gene induction and SAR can also be triggered by the exogenous application of SA or the functional SA analogues benzoic acid (BA; White, 1979), 2,6‐dichloroisonicotinic acid (INA; Vernooij et al., 1995) and benzo(1,2,3)thiadiazole‐7‐carbothioic acid S‐methyl ester (BTH; Friedrich et al., 1996; Lawton et al., 1996). The SA‐regulated induction of PR genes depends on cis‐acting sequences structurally related to activation sequence‐1 (as‐1). Such as‐1‐like elements occur, for example, in the tobacco (Nt) PR‐1a and Arabidopsis (At) PR‐1 promoters (Lebel et al., 1998; Strompen et al., 1998). Originally, SA‐responsive as‐1 was identified in the 35S RNA promoter from Cauliflower mosaic virus (Qin et al., 1994). as‐1‐like elements from the PR‐1 upstream regions, like as‐1, interact with basic leucine zipper (bZIP) TGA transcription factors (Després et al., 2000; Strompen et al., 1998; Zhang et al., 1999) containing a highly conserved DNA binding domain (BD). In Nt and At, TGA factors are organized into small gene families comprising several members, some of which mediate transcription activation (Niggeweg et al., 2000; Zhou et al., 2000).
The SA‐dependent expression of PR genes also relies on NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES1 (NPR1; also known as NIM1 or SAI1), which has been identified as a positive key regulator of SAR (Cao et al., 1997; Ryals et al., 1997; Shah et al., 1997). At plants with a defective NPR1 gene are no longer able to induce the expression of PR genes, and are compromised in disease resistance in response to SA (Cao et al., 1994; Delaney et al., 1995). Importantly, homologues of NPR1 are present in multiple other plant species, including Nt, rice and rape (Chern et al., 2005a; Liu et al., 2002; Potlakayala et al., 2007; Zwicker et al., 2007). These NPR1 homologues have been shown to confer resistance against various pathogens, suggesting that the induction of SAR via NPR1 is conserved among higher plants. However, although NPR1 overexpression experiments resulted in enhanced disease resistance of At plants against bacterial and oomycete pathogens, PR gene expression was not detected in the overexpressors before induction either by chemicals or by pathogen infection (Cao et al., 1998; Friedrich et al., 2001). These data indicate that At NPR1 requires some kind of activation in order to elicit the SAR response.
At NPR1 acts downstream of the SA signal. It contains two conserved protein–protein interaction motifs, an ankyrin repeat domain and a broad complex, tramtrack, and bric à brac/poxvirus and zinc finger (BTB/POZ) domain (Aravind and Koonin, 1999). These motifs occur also in NPR1 proteins from other plant species, e.g. Nt (Zwicker et al., 2007; Fig. S1, see Supporting Information). Indeed, use of NPR1 as a bait in yeast two‐hybrid (Y2H) screens has identified two classes of interacting proteins. Intriguingly, TGA transcription factors have been isolated as NPR1 partner proteins (Chern et al., 2001; Després et al., 2000; Zhang et al., 1999; Zhou et al., 2000; Zwicker et al., 2007). Furthermore, a chimeric TGA2:Gal4 DNA BD protein is able to activate a reporter gene in an INA‐ and NPR1‐dependent fashion in At plants (Fan and Dong, 2002), and a triple knockout mutant, At tga2tga5tga6, is blocked in the induction of PR genes and pathogen resistance (Zhang et al., 2003).
In addition to TGA factors, a group of small novel proteins, termed NIM1‐INTERACTING (NIMIN) proteins, were isolated with the NPR1 bait (Chern et al., 2005b; Weigel et al., 2001; Zwicker et al., 2007). At contains diverse NIMIN genes: NIMIN1, NIMIN2 and NIMIN3. The encoded proteins interact differentially with NPR1. NIMIN1 and NIMIN2 harbour structurally related NPR1 interaction motifs (Weigel et al., 2001). Both proteins bind to the C‐terminus of NPR1, whereas NIMIN3 interacts with the N‐terminal end (Weigel et al., 2001). Furthermore, At TGA factors and NIMIN proteins bind independently from each other to NPR1 (Weigel et al., 2001). Together, these proteins are able to form a ternary complex on the as‐1 sequence motif in yeast (Weigel et al., 2005). In contrast, only one type of NIMIN gene has been identified in Nt and rice. The encoded proteins have been classified as NIMIN2, as they all carry the At NIMIN2 NPR1 interaction motif (Zwicker et al., 2007). The Nt NIMIN2 genes are organized in a small gene family, comprising at least three members: NIMIN2a, NIMIN2b and NIMIN2c. NIMIN1 and NIMIN2 genes from At and Nt are strongly induced by SA, thus clearly linking them to the SAR response (Horvath et al., 1998; 2001, 2005; Zwicker et al., 2007). Notably, Nt NIMIN2 genes are activated prior to the PR‐1a gene (Horvath et al., 1998; Zwicker et al., 2007). Moreover, overexpression of At NIMIN1 and rice NRR, a NIMIN2‐type protein, suppressed the induction of PR genes and caused enhanced susceptibility to bacterial pathogens (2005b, 2008; Weigel et al., 2005). In Nt, overexpression of NIMIN2a delayed PR‐1 protein induction, whereas suppression of NIMIN2 transcripts enhanced the accumulation of PR‐1 proteins (Zwicker et al., 2007). Taken together, current evidence suggests that NPR1 proteins fulfil their function as part of a transcription complex which is tethered to as‐1‐like elements of PR gene promoters via TGA transcription factors, and that this complex may be regulated through the action of NIMIN proteins.
Further efforts to understand the activation of At NPR1 after pathogen infection have, however, produced conflicting results. On the one hand, it has been proposed that constitutively expressed NPR1 accumulating in unchallenged At plants is deposited in an oligomeric form in the cytoplasm (Kinkema et al., 2000; Mou et al., 2003). On induction, monomeric NPR1 is released from the cytoplasmic complex by the thioredoxin‐mediated reduction of cysteine‐156 (Cys‐156) and, via a C‐terminal bipartite nuclear localization signal (NLS), monomeric NPR1 is translocated to the nucleus where it initiates PR‐1 gene expression (Kinkema et al., 2000; Tada et al., 2008). To maintain protein homeostasis, monomeric NPR1 can be oligomerized by S‐nitrosylation of Cys‐156 (Tada et al., 2008). Nuclear monomer accumulation is also seen with two unrelated NPR1 mutant proteins, C82A and C216A, which both are able to induce PR‐1 gene expression in the absence of SA (Mou et al., 2003). In contrast, mere nuclear translocation of a fusion of At NPR1 with the hormone BD of rat glucocorticoid receptor did not prove to be sufficient to activate PR‐1 gene expression, occurring only after additional stimulation by INA (Kinkema et al., 2000). Furthermore, it has been reported recently that nuclear At NPR1 is continuously disposed via the proteasome in unchallenged plants in order to prevent untimely PR gene activation, whereas, after a stimulus, serine‐11 (Ser‐11) and Ser‐15 are phosphorylated, leading to NPR1 turnover as well, which is required for full induction of target genes and the establishment of SAR (Spoel et al., 2009). In other studies, constitutively expressed NPR1 was detected in both the cytoplasm and the nucleus, and NPR1 was even found to be associated with the PR‐1 promoter in the nuclei of unchallenged At plants (Després et al., 2000; Rochon et al., 2006). Moreover, on the basis of in vivo plant transcription assays, transcriptional activation of PR genes has been attributed to the core of the NPR1 BTB/POZ domain, relieving PR gene repression by oligomeric TGA2, and to the oxidation of Cys‐521 and Cys‐529, comprising a novel type of transactivation domain (Boyle et al., 2009; Rochon et al., 2006). The mechanism of switching NPR1 from an inactive to an active form has, however, remained elusive. Curiously, functionally relevant residues Cys‐156, Cys‐521 and Cys‐529, as described for At NPR1, are not present in Nt NPR1 (Fig. S1). Likewise, the bipartite NLS identified in At NPR1 is not fully conserved in Nt NPR1 (Fig. S1). These observations prompted us to analyse Nt NPR1 at the biochemical level. We chose a heterologous system, yeast, which does not contain interfering endogenous plant defence components. Here, we show that Nt NPR1, in some aspects, is biochemically distinct from At NPR1. Yet, both Nt and At NPR1, as well as Nt NIM1‐like1, are responsive to SA in yeast. Together, our data suggest that NPR1 is an SA‐sensitive protein which changes some of its biochemical capabilities to promote timely, coordinate and sudden PR gene expression in response to the SAR signal SA.
RESULTS
Amino acid sequences and interaction partners of Nt NPR1 proteins and expression of the NPR1 gene
In this study, we used two cDNA clones, Nt NPR1.AF and Nt NPR1.DQ, both isolated from Nt (Nicotiana tabacum L.; Liu et al., 2002; Zwicker et al., 2007). Silencing of the Nt NPR1.AF gene has been shown to lead to a loss of resistance to Tobacco mosaic virus (Liu et al., 2002). The Nt NPR1 proteins are highly similar to At NPR1 (52% identity, 68%–72% similarity) and nearly identical to each other (97% identity, 98% similarity; Fig. S1). Consistently, Nt NPR1 proteins, like At NPR1, interact strongly with NIMIN1 and NIMIN2 proteins and with Nt TGA8 (Zwicker et al., 2007). They do not, however, bind to At NIMIN3. Most experiments shown were performed independently with both Nt cDNA clones. In cases in which only one sequence was used in an experiment, the choice was based solely on technical feasibility.
The expression of Nt NPR1 was monitored by reverse transcriptase‐polymerase chain reaction (RT‐PCR) analyses. Nt NPR1 mRNA was likewise detected at rather low levels in seedlings grown on Murashige and Skoog (MS) medium and in leaves from untreated Nt plants (Fig. S2A,C, see Supporting Information), showing that the gene, like At NPR1, is expressed constitutively. The addition of SA to the seedling growth medium induced the SAR genes NIMIN2c and PR‐1a to high levels, yet had no comparable effects on Nt NPR1 expression (Fig. S2B).
Nt NPR1, unlike At NPR1, is targeted to the nucleus
To analyse the subcellular targeting of Nt NPR1, we expressed different NPR1–yeast‐enhanced green fluorescent protein (yEGFP) fusion proteins. In full agreement with previous observations on At NPR1–GFP and At NPR1–β‐glucuronidase (GUS) fusions expressed in plant tissue (Kinkema et al., 2000; Weigel et al., 2001), the chimeric At NPR1:yEGFP protein, like yEGFP alone, is detected in the cytoplasm of yeast cells (Fig. 1A). In contrast, Nt NPR1.DQ:yEGFP accumulates exclusively in the nuclei (Fig. 1A). Even though the Nt NPR1.DQ fusion protein is more strongly expressed than the At NPR1 fusion (Fig. 1C), we have never observed any fluorescence with Nt NPR1.DQ:yEGFP in the cytoplasm of yeast cells. These data suggest that Nt NPR1 harbours a strong nuclear targeting sequence. The addition of SA to the yeast growth medium did not alter the localization of fusion proteins (data not shown).
Figure 1.
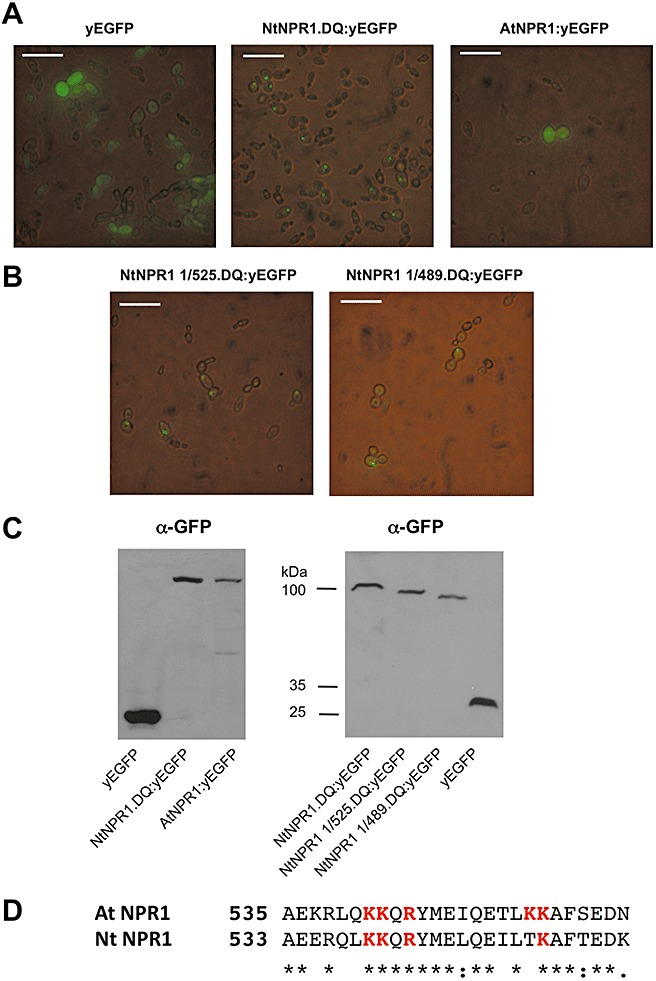
Tobacco NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1 (NPR1) contains a strong nuclear localization signal (NLS). (A) Localization of yeast‐enhanced green fluorescent protein (yEGFP) and tobacco (Nt) NPR1:yEGFP fusion protein and Arabidopsis (At) NPR1:yEGFP fusion protein in yeast. Yeast cells were examined by fluorescence microscopy. Scale bars represent 25 µm. (B) Localization of Nt NPR1:yEGFP fusion proteins truncated at the C‐terminus in yeast. The nuclear targeting sequence in Nt NPR1 is distinct from the NLS in At NPR1. (C) Expression of NPR1:yEGFP fusion proteins in yeast. For the immunodetection of fusion proteins, an antiserum directed against GFP was used. The size of fusion proteins, as estimated from marker proteins, is indicated. (D) Alignment of At and Nt NPR1 proteins in the region of the At NPR1 NLS. Amino acids comprising the At NPR1 NLS are marked in red. Identical amino acids are denoted by asterisks and conservative exchanges by dots.
To test whether the signal sequence identified in At NPR1 directs Nt NPR1 to the nucleus, two C‐terminal deletions, Nt NPR1 1/525.DQ:yEGFP (amino acids 1–525) and Nt NPR1 1/489.DQ:yEGFP (amino acids 1–489), were analysed which lack the proposed At NPR1 NLS (Fig. 1C,D). A schematic representation of all NPR1 constructs used in this study is depicted in Fig. S11 (see Supporting Information). Both deletions, like Nt NPR1:yEGFP, accumulate in the nuclei of yeast cells (Fig. 1B), demonstrating that the NLS of Nt NPR1 is different from that identified in At NPR1. This finding is consistent with the notion that the At NPR1 NLS sequence motif is not fully conserved in Nt NPR1 (Fig. 1D). Together, our results indicate that constitutively expressed Nt NPR1 may accumulate in the nuclei of unchallenged Nt cells, and that regulation of Nt NPR1 activity may comprise other means than cytoplasmic oligomer–nuclear monomer exchange, as described for At NPR1.
Nt NPR1, unlike At NPR1, harbours a strong transactivation domain in its N‐terminal moiety
Given the fact that Nt NPR1 is a nuclear localized protein, we sought a function of Nt NPR1 in the context of the NIMIN2–NPR1–TGA factor transcription complex. Intriguingly, yeast one‐hybrid (Y1H) assays, as shown in Fig. 2A, which we routinely performed as negative controls in Y2H analyses, seemed to indicate that Nt NPR1 itself may exert transcriptional activity. This conclusion appeared to be even more valid as comparable constructs, e.g. pGBT9/AtNPR1, did not yield any reporter gene activities above the background level (Fig. 2A). Notably, Gal4 BD:NPR1 fusion proteins from At and Nt are expressed equally in yeast cells (Fig. 2F). To explore the transcription activation potential of Nt NPR1, we generated several deletion constructs (Fig. S11C). Three truncated proteins derived from different regions, NtNPR1 1/228 (comprising amino acids 1–228), NtNPR1 1/315 (comprising amino acids 1–315) and NtNPR1 386/588 (comprising amino acids 386–588), displayed clear lacZ reporter gene activities (Fig. 2B). The transcription activation potential of Gal4 BD:NtNPR1 1/315 appeared to be particularly high. It clearly exceeds the activity of the plant bZIP transcription factors Nt TGA1a and Nt TGA8, and even that of the strong viral VP16 transactivation domain in Y1H assays (Fig. 2C). This is noteworthy, as SA‐induced transcriptional activity has been associated with two Cys residues, Cys‐521 and Cys‐529, near the C‐terminal end in At NPR1. In yeast, subclones from the At NPR1 C‐terminal half were inactive (data not shown), whereas the Gal4 BD:At NPR1 1/313 fusion (comprising amino acids 1–313) produced rather low, yet clear, transcriptional activity in the range of Gal4 BD:Nt NPR1 1/228 (Fig. 2D). This finding confirms previous reports on low‐level transcriptional activity associated with the At NPR1 N‐terminal moiety in yeast and plants (Rochon et al., 2006; Zhang et al., 1999). Together, both Nt and At NPR1 harbour autonomous transactivation domains in their N‐terminal moieties which are functional in yeast. The transcription activation potential of Nt NPR1 appears, however, to be clearly stronger, indicating that Nt NPR1 itself may be an efficient activator of PR genes in Nt plants exhibiting SAR.
Figure 2.
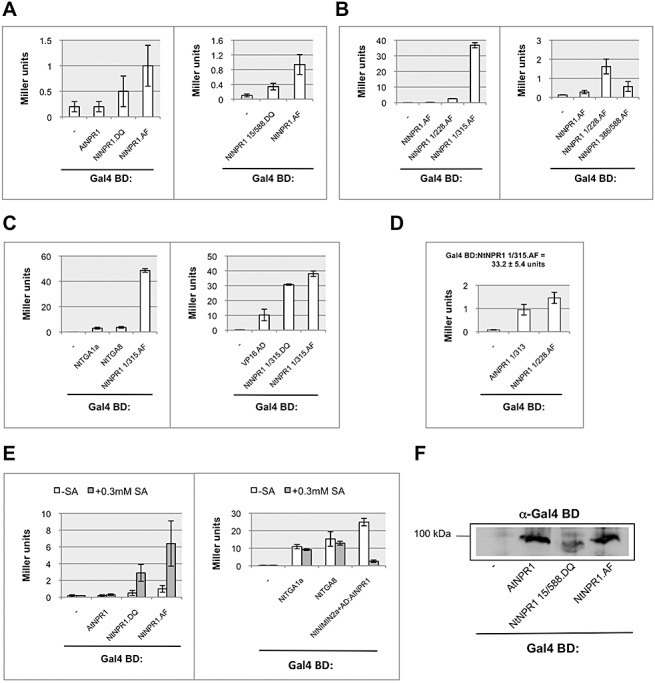
Tobacco NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1 (NPR1) harbours a strong transactivation domain in its N‐terminal half and can be activated by salicylic acid (SA) in yeast. (A) Transcriptional activity of tobacco (Nt) NPR1 and Arabidopsis (At) NPR1 Gal4 DNA binding domain (BD) fusion proteins. Quantitative yeast one‐hybrid assays were performed under standard conditions. β‐Galactosidase activities are the means of three independent colonies tested, each in duplicate, ±SD. (B) Transcriptional activity of truncated Nt NPR1:Gal4 BD fusion proteins in yeast one‐hybrid assays. (C) Transcriptional activity of Nt NPR1 1/315:Gal4 BD in yeast one‐hybrid assays in comparison with the transcriptional activators Nt TGA1a and Nt TGA8 and the VP16 transcription activation domain expressed as Gal4 BD fusion proteins. (D) Transcriptional activity of At NPR1 1/313:Gal4 BD in yeast one‐hybrid assays in comparison with Nt NPR1 1/228:Gal4 BD and Nt NPR1 1/315:Gal4 BD. (E) Transcriptional activity of Nt NPR1:Gal4 BD and At NPR1:Gal4 BD, and of transcriptional activators Nt TGA1a and Nt TGA8, expressed as Gal4:BD fusion proteins in yeast one‐hybrid assays in the absence and presence of SA. The transcription activation domain of Nt NPR1 can be partially uncovered by the addition of SA to the yeast growth medium. (F) Expression of Nt NPR1 and At NPR1:Gal4 BD fusion proteins in yeast. Immunodetection was performed using an antiserum directed against Gal4 BD.
The transcription activation domain of Nt NPR1 can be partially uncovered by SA
Differential transcriptional activities of full‐length Nt NPR1 and deletion mutant Nt NPR1 1/315 in yeast would suggest that the transcription activation domain in Nt NPR1 is masked, and that it may be uncovered only in SA‐challenged Nt expressing high levels of PR genes. Such a change in transcriptional activity could most easily be accomplished by a simple conformational switch in Nt NPR1 mediated by agents released during the defence response. In order to test whether the signalling compound SA itself could be such an agent acting on Nt NPR1, we added SA to the growth medium of yeast cells expressing the Gal4 BD:Nt NPR1 fusion protein. It has been shown previously that SA is taken up by yeast cells from the medium (Scharff et al., 1982). Furthermore, the addition of SA up to 0.3 mm in liquid culture medium does not interfere with the growth of yeast cells (data not shown) or with the expression of Gal4 fusion proteins (Fig. 3D). Quite surprisingly, the transcriptional activity of Nt NPR1 was indeed clearly higher in the presence of SA than in untreated cells (Fig. 2E), although it was not as high as with deletion Nt NPR1 1/315. SA did not affect At NPR1 in this assay, nor the transcriptional activity of the bZIP factors Nt TGA1a and Nt TGA8 (Fig. 2E).
Figure 3.
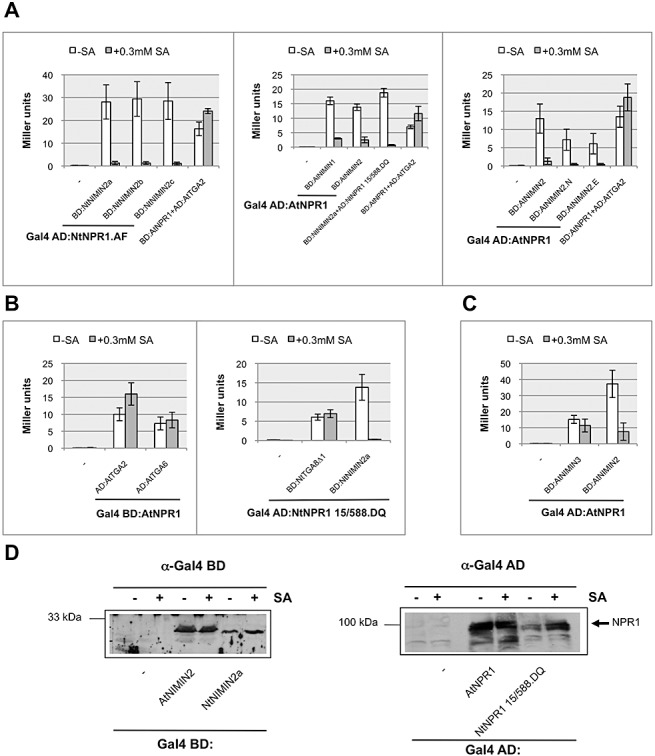
Salicylic acid (SA) suppresses the interaction of NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1 (NPR1) with NIM1‐INTERACTING (NIMIN) proteins NIMIN1 and NIMIN2, but not with NIMIN3 and TGA factors in yeast. (A) Interaction of tobacco (Nt) NPR1 and Arabidopsis (At) NPR1‐Gal4 activation domain (AD) fusion proteins with NIMIN1, NIMIN2 or truncated At NIMIN2:Gal4 binding domain (BD) fusions in yeast two‐hybrid assays in the absence and presence of SA. (B) Interaction of At NPR1:Gal4 BD and Nt NPR1:Gal4 AD fusion proteins with TGA factors expressed as Gal4 AD or Gal4 BD fusions in yeast two‐hybrid assays in the absence and presence of SA. (C) Interaction of the At NPR1:Gal4 AD fusion protein with At NIMIN3:Gal4 BD in yeast two‐hybrid assays in the absence and presence of SA. (D) Expression of NIMIN2:Gal4 BD and NPR1:Gal4 AD fusion proteins in yeast in the absence and presence (0.3 mm) of SA. Immunodetection was performed using antisera directed against the Gal4 BD or Gal4 AD as indicated.
SA suppresses the interaction of NPR1 with NIMIN1 and NIMIN2 proteins
To further explore whether SA could exert an influence on the biochemical properties of NPR1, we tested its ability to bind to the strongly interacting NIMIN proteins in the presence of SA. Y2H assays were routinely performed in parallel with cells grown in medium with or without the addition of SA. The interaction of Nt NPR1 with the Nt NIMIN proteins, NIMIN2a, NIMIN2b and NIMIN2c, was nearly abolished by SA (Figs 3A and S3A, see Supporting Information). Similarly, the binding of At NPR1 to At NIMIN1 or At NIMIN2 was reduced significantly by SA in the growth medium, albeit to a lesser extent than the interaction of Nt NIMIN2a with Nt NPR1 (Fig. 3A). Furthermore, interaction of At NPR1 with the truncated NIMIN2 proteins At NIMIN2.N and At NIMIN2.E, comprising only 56 and 37 amino acids, respectively, including the NPR1 interaction motif (Weigel et al., 2001), is still sensitive to SA (Fig. 3A), indicating that SA targets NPR1 to induce the loss of NIMIN2 binding. Notably, reduced binding of NIMIN2 proteins to NPR1 is not a result of altered protein expression levels in yeast cells exposed to SA (Fig. 3D).
SA does not impair the binding of TGA transcription factors or NIMIN3 to NPR1
Unlike the NIMIN1/NIMIN2–NPR1 interaction, binding of TGA factors to NPR1, Nt and At, as well as binding of At NIMIN3 to At NPR1, were not weakened significantly by SA (Figs 3B,C and S3B). In contrast, the interaction of TGA factors with NPR1 seemed to be moderately enhanced in the presence of SA, considering that reporter gene activities were generally slightly lower in assays from cells grown in SA‐containing medium (see, for example, 2, 3). A reinforcement of the At NPR1–TGA factor interaction in the presence of SA is in agreement with previous findings reporting enhanced At NPR1–At TGA2 binding in SA‐treated At plants (Fan and Dong, 2002). In summary, SA added to the yeast growth medium seems to target NPR1, mainly affecting its C‐terminal end, to disrupt the interaction of NPR1 with the SA‐inducible NIMIN1 and NIMIN2 proteins, whereas the binding of partner proteins to the N‐terminus and the central region of NPR1 appears rather undisturbed. Moreover, in Nt NPR1, SA may change the conformation of the protein, thus promoting transcription activation of target genes. The effects of SA on NPR1, as detected by His reporter gene activity in Y1H and Y2H analyses, are summarized in Fig. S3B.
The effects of SA on NPR1 are specific and sensitive, and occur coordinately
To further test the specificity of SA on the activity of NPR1, we added functional and nonfunctional SA analogues to the yeast growth medium. Like SA, these chemicals are taken up by yeast cells and are nonmetabolizable (Henriques et al., 1997; Macris, 1975; Scharff et al., 1982). BA is a strong inducer of PR‐1 genes and SAR in Nt, whereas 4‐hydroxy‐benzoic acid (4‐OH BA) is inactive (Fig. 4A; White, 1979; Xie et al., 1998). Similar patterns were observed when these compounds were added to the yeast growth medium. BA induced transcriptional activity in Nt NPR1 and also compromised the interaction of NPR1, Nt and At with NIMIN2 proteins, whereas binding to TGA factors was slightly enhanced in BA‐containing medium (Figs 4B and S4, see Supporting Information). In contrast, 4‐OH BA had no effect on NPR1 activity (Figs 4B and S4).
Figure 4.
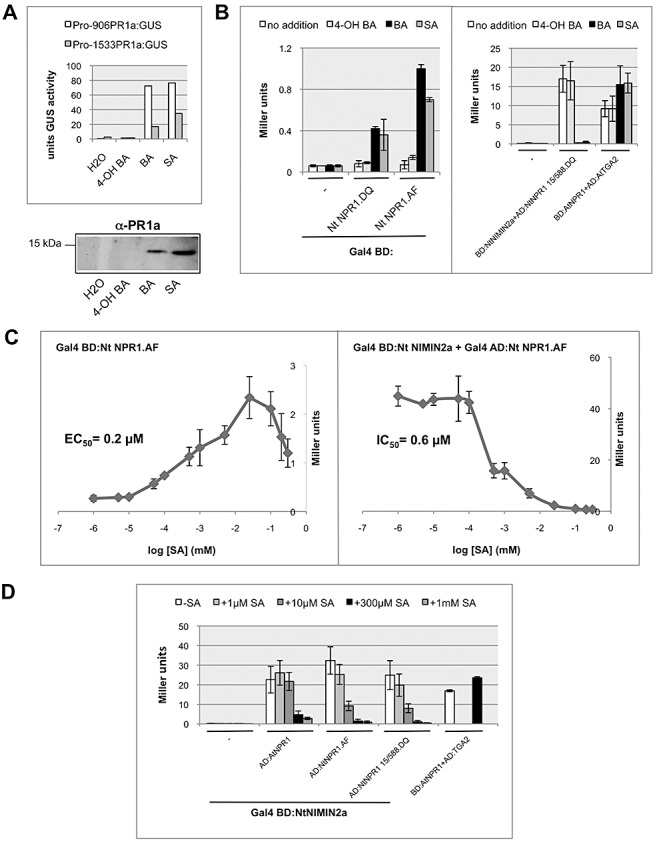
The effects of salicylic acid (SA) on NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1 (NPR1) in yeast are specific and sensitive and occur coordinately. (A) Effect of the SA analogues benzoic acid (BA) and 4‐hydroxy‐benzoic acid (4‐OH BA) on the expression of PR‐1a promoter‐β‐glucuronidase (GUS) fusion genes and the endogenous PR‐1 genes in tobacco. GUS reporter enzyme activities were determined in transgenic plants containing PR‐1a promoter constructs comprising a 906‐ or 1533‐bp upstream sequence. Immunodetection was performed using an antiserum directed against the PR‐1a protein. (B) Effect of the SA analogues BA and 4‐OH BA (0.3 mm) on the transcriptional activity of tobacco (Nt) NPR1:Gal4 binding domain (BD) fusion proteins in yeast one‐hybrid assays and on the interaction of NPR1 fusion proteins with Nt NIMIN2a or Arabidopsis (At) TGA2 co‐expressed as Gal4:BD and Gal4:AD fusions, respectively, in yeast two‐hybrid assays. The effects of SA on NPR1 in yeast are specific. (C) Effect of different concentrations of SA on the transcriptional activity of Nt NPR1:Gal4 BD in yeast one‐hybrid assays and on the interaction of Nt NPR1:Gal4 activation domain (AD) with Nt NIMIN2a:Gal4 BD in yeast two‐hybrid assays. The effects of SA on NPR1 in yeast are highly sensitive. The gain of transcriptional activity and the loss of NIMIN2 binding by SA occur coordinately in Nt NPR1. (D) Effect of different concentrations of SA on the interactions of Nt and At NPR1:Gal4 AD fusion proteins with Nt NIMIN2a:Gal4 BD in yeast two‐hybrid assays. At NPR1 is less sensitive to SA than Nt NPR1. NIMIN, NIM1‐INTERACTING.
The sensitivity of NPR1 to SA was monitored by dose–response experiments. Both the uncovering of the Nt NPR1 transactivation domain and impairment of the Nt NPR1–Nt NIMIN2 interaction are rather sensitive to SA. Half‐maximal transcription activation of Nt NPR1 occurred at a concentration of 0.2 µm SA in the yeast growth medium, and 50% inhibition of Nt NIMIN2a binding to Nt NPR1 was reached at 0.6 µm SA (Fig. 4C). A similar sensitivity to SA was monitored for the Nt NPR1–Nt NIMIN2c interaction (Fig. S5A, see Supporting Information). Thus, these responses of Nt NPR1 to SA are coordinated. Furthermore, transcription activation of Nt NPR1 was clearly inhibited at SA concentrations beyond 100 µm (Fig. 4C). At NPR1, on the other hand, seemed less responsive to SA. More than 50% impairment of the At NPR1–At NIMIN1/NIMIN2 interaction occurred only at concentrations of more than 10 µm SA (Fig. S5B,C). To compare directly the SA sensitivity of the NPR1 proteins from At and Nt, we performed dose–response experiments with Nt NIMIN2a as the common interactor. The NPR1 interaction motif of Nt NIMIN2a is 87% (93%) identical (similar) to that of At NIMIN2 (Zwicker et al., 2007). Consequently, being equally expressed in yeast (Fig. 3D; Zwicker et al., 2007), the At and Nt NPR1 fusion proteins interact to similar levels with Nt NIMIN2a (Fig. 4D; Zwicker et al., 2007). As depicted in Fig. 4D, the At NPR1–Nt NIMIN2a interaction indeed seems to be less sensitive than the Nt NPR1–Nt NIMIN2a interaction to SA. The high sensitivity of Nt NPR1 to SA supplemented in the yeast growth medium is consistent with PR‐1 gene induction in SA‐treated Nt cell cultures, whereas PR‐1 induction is less sensitive to exogenous SA in Nt plants (Glocova et al., 2005; Xie et al., 1998).
The nim1‐4 mutation abolishes both transcriptional activity and SA‐mediated suppression of NIMIN2 binding in Nt NPR1
Differential sensitivity of the interaction of At NPR1 with NIMIN3, TGA factors and NIMIN1/NIMIN2 towards SA suggested that SA may act on the C‐terminal end of NPR1. Furthermore, if SA sensitivity is of functional relevance to NPR1 activity in planta, mutants disrupting this feature should be affected when mounting the SAR response. Several defective npr1/nim1 alleles have been characterized in At. Of these, only nim1‐4 carries a single amino acid exchange, arginine‐432 (Arg‐432) to lysine (Lys), in the C‐terminal part of the gene (Ryals et al., 1997). The allele has been classified as severely compromised with respect to chemical PR‐1 gene induction and resistance to fungal infection. The mutant gene is expressed to normal levels, suggesting that the protein may be nonfunctional. The nim1‐4 mutation was introduced by recombinant DNA techniques in the At and Nt NPR1 proteins (cDNA sequences Nt NPR1.AF and Nt NPR1.DQ; Fig. S11). The mutant fusion proteins are expressed to the same levels as the respective wild‐type fusions in yeast (Fig. 5E). Furthermore, the mutation does not interfere with the binding of NIMIN proteins and TGA transcription factors to At or Nt nim1‐4 (Figs 5A and S6, see Supporting Information). Yet, binding of NIMIN1 and NIMIN2 proteins to At and Nt nim1‐4 was preserved in the presence of SA (Figs 5A and S6B). In addition, Nt nim1‐4 does not exhibit any transcriptional activity in Y1H assays in the absence or presence of SA (Fig. 5B), supporting the view that the suppression of NIMIN2 binding and transcriptional activation are coordinated in Nt NPR1.
Figure 5.
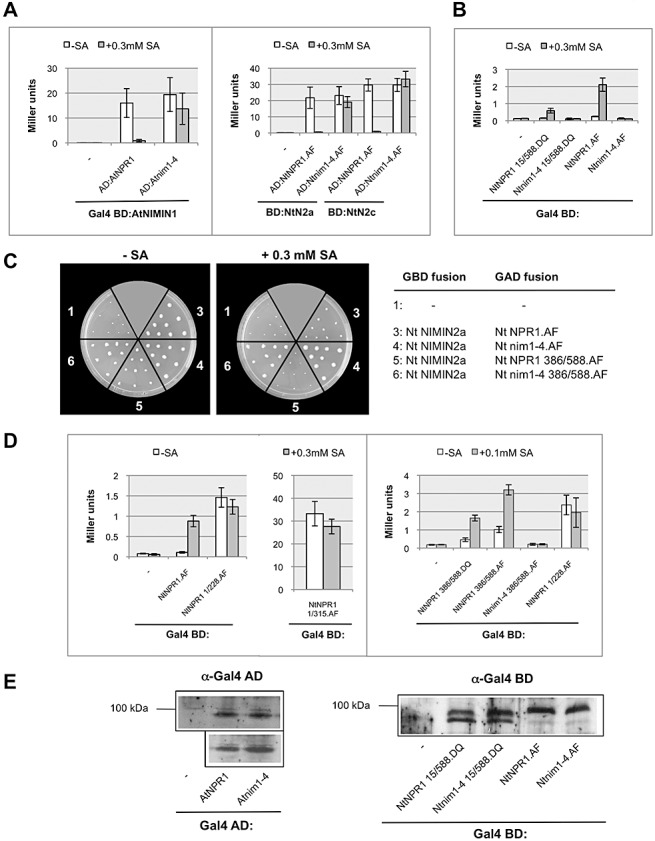
The nim1‐4 mutant protein is insensitive to salicylic acid (SA) in yeast. (A) Effect of SA on the interactions of Arabidopsis (At) and tobacco (Nt) nim1‐4:Gal4 activation domain (AD) fusion proteins with NIMIN1 and NIMIN2 co‐expressed as Gal4 binding domain (BD) fusions in yeast two‐hybrid assays. BD:NtN2a(c), Gal4 BD:NtNIMIN2a(c). (B) Transcriptional activity of Nt nim1‐4:Gal4 BD fusion proteins in yeast one‐hybrid assays. (C) Effect of SA on the interactions of the truncated proteins Nt NPR1 386/588:Gal4 AD and Nt nim1‐4 386/588:Gal4 AD with Nt NIMIN2a:Gal4 BD in yeast two‐hybrid assays. His reporter gene activity was monitored in yeast two‐hybrid assays with or without the addition of SA to the medium as indicated. (D) Effect of SA on the transcriptional activities of truncated Nt NPR1:Gal4 BD fusion proteins and on Nt nim1‐4 386/588:Gal4 BD in yeast one‐hybrid assays. The activation of Nt NPR1 by SA in yeast localizes to the protein's C‐terminal third (amino acids 386–588). The activation of Nt NPR1 by SA is abolished in the nim1‐4 mutant. (E) Expression of At nim1‐4:Gal4 AD and Nt nim1‐4:Gal4 BD fusion proteins in yeast. Immunodetection was performed using antisera directed against Gal4 AD or Gal4 BD as indicated. NIMIN, NIM1‐INTERACTING; NPR1, NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1.
To determine whether the impact of SA indeed localizes to the C‐terminal end of Nt NPR1, we tested the truncated protein Nt NPR1 386/588 and the corresponding mutant Nt nim1‐4 386/588.AF (Fig. S11). Both deletions are able to interact with Nt NIMIN2a, albeit to a lesser extent than their full‐length correspondents (Fig. 5C). In the wild‐type deletion, Nt NIMIN2a binding is sensitive to SA, whereas the mutation of Arg‐431 to Lys (corresponding to R432K in At NPR1) abolishes the SA‐sensitive loss of NIMIN2 interaction (Fig. 5C). Likewise, importantly, Nt NPR1 386/588, but not Nt nim1‐4 386/588, displays SA‐dependent transcription activation (Fig. 5D), whereas Nt NPR1 1/228.AF and Nt NPR1 1/315.AF, representing the N‐terminal half of Nt NPR1, are not affected in their transcriptional activity by SA (Fig. 5D). These results support the notion that SA indeed acts in an autonomous manner on the NPR1 C‐terminal third. Furthermore, the effects of SA on NPR1, as detected in the heterologous system yeast, seem to be specific and relevant to the function of NPR1 in planta.
The binding of NIMIN2 proteins to Nt NIM1‐like1, an NPR1‐related protein, is suppressed in the presence of SA
In addition to NPR1, both At and Nt possess NPR1‐related genes. The functional relevance of these NPR1‐like genes is not yet fully understood. It is, however, generally accepted that NPR1‐like genes from At are not positive regulators of SA‐induced SAR gene expression (Liu et al., 2005; Zhang et al., 2006). To analyse the effects of SA on an NPR1‐like protein, we isolated a cDNA clone for Nt NIM1‐like1. The encoded protein is identical to a sequence described in GenBank except for one amino acid exchange, proline‐534 (Pro‐534) to threonine (Thr). Notably, Nt NIM1‐like1 harbours Arg‐435 (corresponding to Arg‐431 in Nt NPR1 and Arg‐432 in At NPR1), which is embedded in a region of five identical amino acids shared by NPR1 proteins from different plant species (Fig. 7D). Yet, Nt NIM1‐like1 does not contain Cys residues at the positions corresponding to Cys‐156, Cys‐521 and Cys‐529 of At NPR1. The overall identity (similarity) of Nt NIM1‐like1 to Nt NPR1 is only 43% (61%), and 39% (58%) to At NPR1. Like Nt NPR1, Nt NIM1‐like1 is expressed constitutively (Fig. S7, see Supporting Information). The transcript levels are not elevated to high levels by SA.
Figure 7.
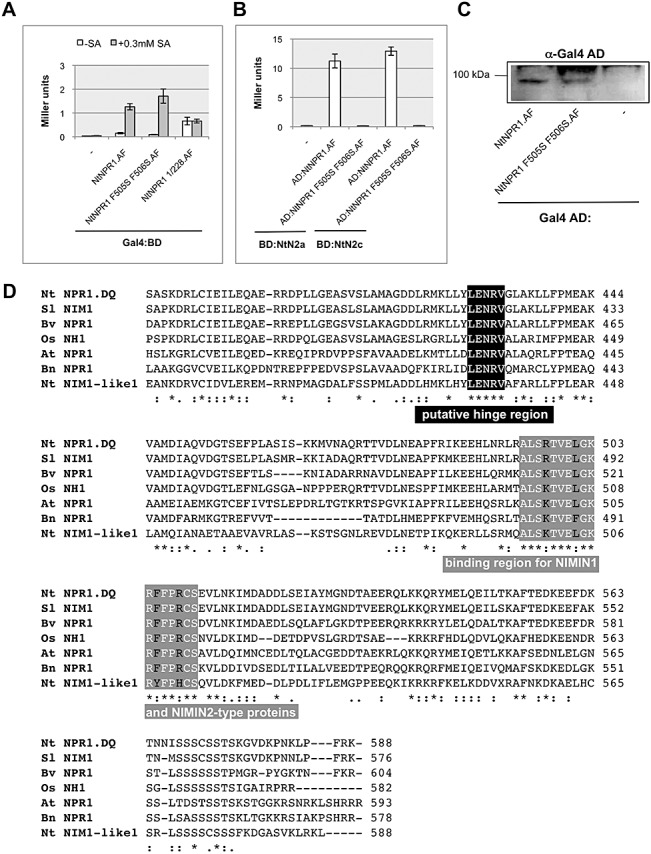
The occurrence of the conserved penta‐amino acid domain LENRV and a binding site for salicylic acid (SA)‐inducible NIM1‐INTERACTING (NIMIN) proteins is coincident in NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1 (NPR1) from different species. (A) Transcriptional activity of the mutant tobacco (Nt) NPR1 F505S F506S:Gal4 binding domain (BD) fusion protein in yeast one‐hybrid assays in the absence and presence of SA. The Nt NPR1 F505S F506S mutant protein exhibits wild‐type‐level transcriptional activity which is enhanced by SA in the yeast growth medium. (B) Co‐expression of the mutant Nt NPR1 F505S F506S:Gal4 activation domain (AD) and Nt NIMIN2:Gal4 BD fusion proteins in yeast. The Nt NPR1 F505S F506S mutant protein does not interact with NIMIN2 proteins. BD:NtN2a(c), Gal4 BD:NtNIMIN2a(c). (C) Expression of the mutant Nt NPR1 F505S F506S:Gal4 AD fusion protein in yeast. Immunodetection was performed using an antiserum directed against the Gal4 AD. (D) Alignment of NPR1 proteins from Nt, tomato (Sl), beet (Bv), rice (Os), Arabidopsis (At) and rape (Bn) and of Nt NIM1‐like1. The alignment encompassing the C‐terminal thirds of the proteins is shown. The conserved penta‐amino acid domain LENRV (black box), which may serve as a hinge region in NPR1, and the binding site for NIMIN1/NIMIN2 proteins (grey box) are indicated. Amino acids within these motifs which are identical in all proteins are shown in white. Dashes indicate gaps introduced to maximize alignments. Amino acids shared by all proteins are denoted by asterisks, and conservative exchanges are denoted by dots.
The effect of SA on Nt NIM1‐like1 was monitored in Y1H and Y2H assays. We were unable to detect transcriptional activity with the Gal4 BD:Nt NIM1‐like1 fusion protein (data not shown). Yet, Nt NIM1‐like1 interacts with TGA factors from Nt and At (Nt TGA8, At TGA2 and At TGA6), but not with Nt TGA1a (Fig. S8A, see Supporting Information). Similarly, it has been reported previously that two NPR1‐like proteins from At, NPR3 and NPR4, bind TGA2, TGA3, TGA5 and TGA6, but not TGA1 and TGA4 (Liu et al., 2005; Zhang et al., 2006). Nt NIM1‐like1 also interacts with the NIMIN1 and NIMIN2 proteins from At and Nt. Like Nt NPR1, Nt NIM1‐like1 does not, however, bind to At NIMIN3 (Fig. S8B). Thus, Nt NIM1‐like1 possesses binding properties in yeast as observed for Nt NPR1 (Zwicker et al., 2007). Surprisingly, the binding of Nt NIMIN2 proteins to Nt NIM1‐like1 is also sensitive to SA (Fig. 6).
Figure 6.
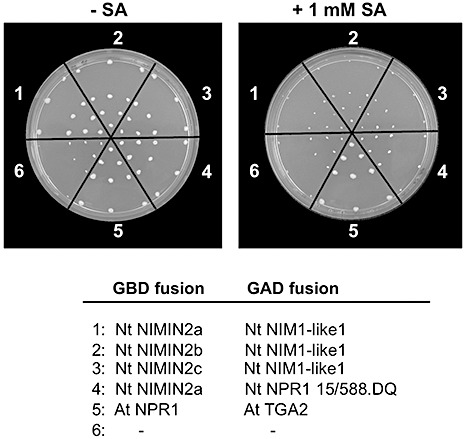
Tobacco (Nt) NIM1‐like1 is sensitive to salicylic acid (SA) in yeast. His reporter gene activity was monitored in yeast two‐hybrid assays co‐expressing Nt NIM1‐like1:Gal4 activation domain (AD) and Nt NIMIN2:Gal4 binding domain (BD) fusion proteins with and without the addition of SA to the yeast growth medium. NIMIN, NIM1‐INTERACTING.
The occurrence of the conserved penta‐amino acid domain LENRV and a binding site for SA‐inducible NIMIN proteins is coincident in NPR1 proteins from different species and in Nt NIM1‐like1
Our finding that Nt NPR1 is sensitive to SA, altering its transcription activation potential and its capacity for binding NIMIN2 proteins in yeast, might suggest that SA‐inducible NIMIN proteins regulate NPR1 activity in planta. In line with this, the occurrence of the conserved C‐terminal Arg‐containing penta‐amino acid motif, which mediates responsiveness to SA in Nt and At NPR1, should be correlated with the ability to bind NIMIN1 and NIMIN2 proteins. To test this idea, we set out to identify the NIMIN2 binding site in Nt NPR1.
Previous results have indicated that the NIMIN1/NIMIN2 interaction domain localizes to the C‐terminal end of At NPR1 (Weigel et al., 2001). Consistently, Nt NPR1 386/588 is able to interact with Nt NIMIN2a (Fig. 5C). In addition to the conserved penta‐amino acid motif, At and Nt NPR1 harbour a 20‐residue‐long stretch of near identity in their C‐terminal ends (At NPR1 from amino acids 493 to 512, Nt NPR1 from amino acids 491 to 510; Fig. S1). Two mutations, phenylalanine‐505 (Phe‐505) to Ser and Phe‐506 to Ser, were introduced in this region of Nt NPR1.AF (Fig. S11). The wild‐type and mutant fusion proteins are expressed to similar levels in yeast (Fig. 7C). Furthermore, Nt NPR1 F505S F506S is still able to interact with Nt TGA8 to normal levels (Fig. S9A, see Supporting Information) and also displays transcriptional activity which is enhanced in the presence of SA (Fig. 7A). However, the mutant protein is no longer able to interact with Nt NIMIN2a or Nt NIMIN2c (Figs 7B and S9B). These data clearly demonstrate that the Phe‐505 Phe‐506 mutation specifically destroys the NIMIN2 binding ability in Nt NPR1 without interfering with the protein's interaction with TGA factors, its transcription activation potential and its responsiveness to SA. Thus, sensitivity towards SA and binding of the SA‐inducible NIMIN2 proteins are mediated by distinct domains in Nt NPR1, which both lie in the C‐terminal third of the protein. Databank searches revealed that the Arg‐containing penta‐amino acid domain and a conserved binding site for NIMIN2‐type proteins, which are widely found throughout the plant kingdom (Zwicker et al., 2007), indeed co‐occur in NPR1 proteins from different species and in Nt NIM1‐like1 (Fig. 7D). Interestingly, NPR1‐related proteins from At, known as BLADE‐ON‐PETIOLE, BOP1 and BOP2 (Hepworth et al., 2005), harbour neither of these domains (Fig. S10, see Supporting Information).
DISCUSSION
In At, NPR1 is a master regulator of SAR which initiates PR gene expression in response to the SA signal. Current evidence suggests that At NPR1 itself is activated in challenged plants. Intensive search for cues mediating this activation event has produced results leading to diverse and highly complex models for NPR1‐mediated PR‐1 gene induction. To understand the action of NPR1 at the molecular level, we have dissected another NPR1 protein, Nt NPR1, which lacks critical Cys residues previously implied in At NPR1 activation. We have chosen a completely different approach, analysis of Nt NPR1 and its comparison with At NPR1 in yeast, a system allowing us to monitor the activities of NPR1 proteins in the absence of interfering plant defence compounds and signal cascades. This approach has led to novel insights into the biochemical capabilities of NPR1 and NPR1‐related proteins from Nt and At.
Nt NPR1 harbours a potent transcription activation interface that can be uncovered by SA
It has been reported previously that At NPR1 possesses transcriptional activation capability. In full agreement with our findings, low‐level transcriptional activity was observed with the At NPR1 N‐terminal half in yeast (Zhang et al., 1999), whereas, using in vivo assays, transactivation was detected in both the N‐terminal and C‐terminal moieties of At NPR1 (Rochon et al., 2006). Transcription activation in the C‐terminal half proved to be induced by SA and was attributed to the oxidation of Cys‐521 and Cys‐529. In contrast, the N‐terminal At NPR1 transactivation region was constitutively active and its nature has remained elusive. Similarly, transcriptional activity, as determined by us in yeast, is associated with different regions of the Nt NPR1 protein (2, 5). We monitored low‐level transcriptional activity with Nt NPR1 386/588, representing the protein's C‐terminal third. This activity is increased on SA treatment of yeast cells, and thus appears to resemble the SA‐induced transactivation of C‐terminal At NPR1, as detected by in planta transcription assays. The transcriptional activity cannot, however, be attributed to Cys residues occurring in Nt NPR1 at the same positions as in At NPR1. Indeed, Cys residues do not occur at these positions in any other NPR1 protein, including Brassica NPR1 (Fig. 7D). Furthermore, we detected transcriptional activity in yeast with two deletions comprising regions from the Nt NPR1 N‐terminal half. Neither is affected by SA, and thus these activities appear to resemble the constitutive transactivation of N‐terminal At NPR1, as detected by in planta transcription assays. Transcription activation with Nt NPR1 1/228 is moderate, whereas transactivation with Nt NPR1 1/315 is very strong, clearly exceeding transactivation from the viral VP16 activation domain (Fig. 2C). It appears unlikely that Nt NPR1 could contain three distinct transcription activation domains exhibiting different strengths and different specificities towards the SA signal. Rather, it seems conceivable that Nt NPR1 may harbour a broad and potent transactivation interface. This transactivation interface can be masked, rendering Nt NPR1 inactive. Yet, in the presence of SA, Nt NPR1 may undergo a conformational switch, thus exposing its transactivation interface and adopting an active state to promote induced transcription of its target genes.
The Nt NPR1‐type proteins are biochemically distinct from At NPR1 in some aspects
We have noted previously that Nt NPR1, albeit interacting strongly with the At NIMIN1 and NIMIN2 proteins, does not bind At NIMIN3. Likewise, Nt NIM1‐like1 does not interact with At NIMIN3 (Fig. S8B). These findings are consistent with our observation that At NIMIN3 homologous genes have not been identified to date outside the family of Brassicaceae, suggesting that At NPR1 may be biochemically distinct from Nt NPR1 proteins (Zwicker et al., 2007). Our conclusion is corroborated by further evidence. Albeit expressed to similar levels, At and Nt NPR1 differ in yeast in their subcellular localization preference (Fig. 1A–C), their transcription activation potential (Fig. 2) and their sensitivity towards SA (Fig. S5). Furthermore, At and Nt NPR1 seem to use different NLSs (Fig. 1B–D). On the other hand, At NPR1, Nt NPR1 and Nt NIM1‐like1 interact similarly with TGA transcription factors and with the SA‐inducible NIMIN proteins. Thus, SAR gene expression seems to rely on evolutionary conserved components, NPR1, TGA transcription factors, as‐1‐like cis‐acting elements and NIMIN proteins in both species, which nevertheless may have adapted to the different lifestyles and environmental needs encountered by At and Nt, respectively.
NPR1 and some NPR1‐related proteins are sensitive to the SAR signal molecule SA
The most important finding of our work is that NPR1 and some NPR1‐related proteins are sensitive to the low‐molecular‐weight signal molecule SA. The effects of SA are specific and highly sensitive (Fig. 4). We have shown that three distinct proteins, At NPR1, Nt NPR1 and Nt NIM1‐like1, diminish their binding capacities for NIMIN1 and NIMIN2 proteins, but not for At NIMIN3 or TGA factors, in the presence of SA in yeast (3, 6). Indeed, we cannot differentiate whether SA acts in a direct or a more indirect fashion resulting in the modification of the NPR1 protein. Yeast does not, however, harbour an NPR1 homologous gene, thus excluding the possibility that SA‐treated yeast could modify NPR1 specifically. With Nt NPR1, we monitored two distinct biochemical changes in response to SA: loss of NIMIN2 binding ability and enhancement of transcription activation. These two changes in Nt NPR1 activity occur coordinately (Fig. 4C), and thus are probably elicited by the same signal transduction event. Although a loss of function is highly compatible with a nonspecific modification of a protein, a gain of function is not. Therefore, it seems conceivable that the signal molecule SA may target directly the plant protein Nt NPR1 in yeast and, by inducing a conformational switch in Nt NPR1, alter its biochemical capabilities, thus activating the protein. Efforts to demonstrate the binding of SA to NPR1 have, however, not been successful to date.
SA likewise controls two distinct biochemical abilities of Nt NPR1: transcription activation and its interaction with NIMIN2‐type proteins. Changes in both activities are transmitted through the holoprotein as well as through the Nt NPR1 386/588 truncated protein, but not through deletions 1/218 and 1/315, indicating that SA can be sensed by Nt NPR1 386/588 (Fig. 5C,D). Indeed, SA seems to act through the C‐terminal end of the protein. In both Nt and At NPR1, SA sensitivity is mediated by the Arg residue within the strictly conserved penta‐amino acid motif LENRV, which occurs at similar positions in the C‐terminal regions of NPR1 proteins from multiple plant species and in Nt NIM1‐like1 (Fig. 7D). Mutation of the Arg residue within the penta‐amino acid motif abolishes SA‐mediated PR‐1 gene induction and pathogen resistance in At plants (Ryals et al., 1997), and also abolishes the loss of NIMIN1/NIMIN2 interaction in SA‐treated yeast (Figs 5A and S6B). In Nt NPR1 and Nt NPR1 386/588, mutation of the Arg residue abolishes transcriptional activity and loss of NIMIN2 binding in SA‐treated yeast (Fig. 5A–D). The sensitivity towards SA is not, however, transmitted through the binding site for the SA‐inducible NIMIN proteins (Fig. 7A), which also occurs in the NPR1 C‐terminal region not far from the conserved penta‐amino acid domain.
Although it is evident from our experiments that the conserved penta‐amino acid domain transduces the SA signal, it is not clear whether this motif itself is able to sense SA. Application of SA to yeast cells enhances the transcriptional activity of Nt NPR1 which is already detectable in untreated cells (Fig. 2E). This enhancement of basal transcriptional activity is consistent with the assumption that Nt NPR1 can adopt two different states in yeast—active and inactive—and that the protein can switch to the active state in the heterologous host in the presence of SA. Mutation of Arg‐431 completely abolishes any transcriptional activity of Nt NPR1, irrespective of whether expressed in SA‐treated or nontreated yeast cells (Fig. 5B,D), implying that the Nt NPR1 R431K mutant protein is caught in an inactive form, and that the Arg‐containing penta‐amino acid motif LENRV may comprise a hinge region, allowing Nt NPR1 to switch between two conformational states exhibiting differential activities. We do not exclude the possibility, however, that the putative hinge region may be involved in the perception of the SA signal. In this context, it is interesting to note that an NPR1‐related protein, Nt NIM1‐like1, also harbours the conserved penta‐amino acid domain LENRV (Fig. 7D) and is relieved from NIMIN2 proteins in the presence of SA (Fig. 6). These data suggest that SA may affect NPR1 and also some NPR1‐related proteins.
Relief from NIMIN2 binding and gain of transcriptional activity are coupled in Nt NPR1
Previous results have shown that the overexpression of NIMIN proteins suppresses the induction of defence‐related genes, whereas reduced NIMIN transcript levels enhance PR‐1 gene induction, strongly suggesting that the SA‐inducible NIMIN1‐ and NIMIN2‐type proteins negatively regulate NPR1 activity in At, rice and Nt (2005b, 2008; Weigel et al., 2005; Zwicker et al., 2007). Our data on the differential biochemical capabilities of NPR1 in the presence and absence of SA in yeast are highly compatible with these in planta observations. Relief of Nt NPR1 from NIMIN2 binding by SA in yeast cells is correlated with a gain in transcriptional activity (Fig. 4C), indicating that activation and interaction with NIMIN2 proteins may be opposite functions of NPR1. Consequently, NIMIN1‐ and NIMIN2‐type proteins, whilst bound to NPR1, could inhibit the activation of NPR1, whereas reduced NIMIN levels could lead to a premature activation of the protein and PR gene induction. It also seems conceivable that the binding of NIMIN proteins to NPR1 may delay the expression of some PR genes, such as the late induced PR‐1 genes, thus enabling the plant to control different stages of SAR. In this respect, it is of interest to note that the binding site for NIMIN1 and NIMIN2 proteins appears to be rather broad (17 amino acids), is highly conserved in NPR1 proteins from different plant species and occurs at a conserved distance with regard to the penta‐amino acid putative hinge region (Fig. 7D), possibly indicating that occupancy of the NIMIN binding pocket may control the accessibility of SA to NPR1.
CONCLUSION
Based on functional analyses of NPR1 and NIMIN proteins in yeast and plants, we suggest a simple model for the induction of SAR genes after pathogen attack (Fig. 8). Our model implies that the activation of NPR1 and some NPR1‐related proteins depends directly on the ambient concentration of the signal molecule SA. SA is sensed by and the signal is transduced through the C‐terminal regions of NPR1 and some NPR1‐like proteins harbouring the penta‐amino acid motif LENRV and a strictly conserved binding site for NIMIN1/NIMIN2 proteins. NIMIN proteins are likely to control the activation of NPR1 through differential binding, allowing NPR1 activation only after distinct threshold levels of SA have been reached. This model does not necessarily contradict all aspects of previous models on the activation of At NPR1 (Boyle et al., 2009; Rochon et al., 2006; Spoel et al., 2009; Tada et al., 2008). Notably, we provide functional significance for two highly conserved, yet hitherto unrecognized, domains occurring in the C‐terminal regions of NPR1 proteins from multiple plant species. Furthermore, our work strongly supports the view that SA acts as a true hormone in plants, which is perceived by the key regulatory component of the SAR response, and thus adds NPR1 and some NPR1‐related proteins to an increasing number of plant proteins which are directly targeted and activated by low‐molecular‐weight signal molecules, including TRANSPORT INHIBITOR RESPONSE1 (TIR1), which perceives auxin (Tan et al., 2007), CORONATINE INSENSITIVE1 (COI1), which functions as a receptor for jasmonic acid isoleucine (Yan et al., 2009), and REGULATORY COMPONENT OF ABA RECEPTOR (RCAR)/PYRABACTINE RESISTANCE (PYR), which binds abscisic acid (Ma et al., 2009; Park et al., 2009).
Figure 8.
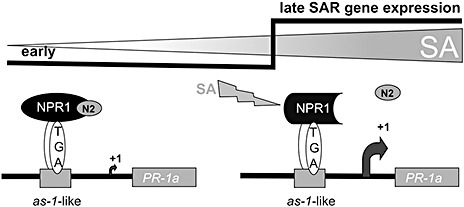
Model for the salicylic acid (SA)‐mediated activation of the tobacco (Nt) PR‐1a gene late in systemic acquired resistance (SAR). Low levels of SA elicited by pathogen attack induce the expression of NIMIN2 genes. At this stage, premature PR‐1a induction via the TGA–NPR1 complex is inhibited by binding of NIMIN2‐type proteins to the C‐terminal end of NPR1. Inability to confine the pathogen leads to a further increase in SA levels in the noninfected leaves of the plant, eventually resulting in the release of NIMIN2, the activation of NPR1 and induction of the PR‐1a gene. The model implies that PR genes are activated at distinct threshold levels of SA (indicated by the step), and that direct measurement of the ambient cellular SA concentration occurs through SA‐sensitive complexes formed between the NPR1 C‐terminal end and NIMIN2 proteins. NIMIN, NIM1‐INTERACTING; NPR1, NONEXPRESSOR OF PATHOGENESIS‐RELATED PROTEINS1; PR, PATHOGENESIS‐RELATED.
EXPERIMENTAL PROCEDURES
DNA constructs
For subcellular localization studies, NPR1 coding regions were fused in‐frame to the N‐terminal sequence of yEGFP in pEGFP C‐FUS, which was obtained from pGFP C‐FUS (Niedenthal et al., 1996) by the exchange of GFP for the codon‐optimized yEGFP (Cormack et al., 1997). Nt NPR1.DQ was excised from pGAD424/NtNPR1 (Zwicker et al., 2007) and ligated as a BamHI fragment to pEGFP C‐FUS. For fusions with Nt NPR1 proteins truncated at their C‐terminus, the 3′ end of Nt NPR1 was replaced by PCR‐generated fragments using an internal HindIII restriction endonuclease site occurring at position 1226 from the ATG start codon in the Nt NPR1 cDNA sequence. The primers were NtNPR1‐11 (5′‐AAGGATCCCAAGTGCTCCTCTTTAATCC) for Nt NPR1 1/489 and NtNPR1‐12 (5′‐TTGGATCCTATCTCAGACAAGTCATCAGC) for Nt NPR1 1/525. The resulting partial Nt NPR1 sequences were added as BamHI fragments to pEGFP C‐FUS. At NPR1 was amplified by PCR with primers NPR1 fwd (5′‐ACGGATCCCCATGGACACCACCATTGATGG) and NPR1 3′ Eco (5′‐GTGAATTCCCGACGACGATGAGAGAG), and cloned into pEGFP C‐FUS cut with BamHI and EcoRI.
Most plasmids used in Y1H and Y2H analyses have been described previously (Weigel et al., 2001; Zwicker et al., 2007). Nt NPR1 15/588 comprises a nearly full‐length protein encoded by a cDNA clone isolated from an Nt cDNA library in a Y2H screen (Zwicker et al., 2007). To map the transcriptional activation domain in Nt NPR1, deletions were generated using convenient restriction endonuclease sites. Nt NPR1 1/228 was created by inserting the 0.7‐kb EcoRI/EcoRV fragment from the Nt NPR1 5′ end in EcoRI/SmaI‐cleaved pGBT9. To generate Nt NPR1 1/315, pGBT9/NtNPR1 was cut with EcoRI and partially with BglII, and the 0.9‐kb fragment comprising the N‐terminal half of Nt NPR1 was ligated to EcoRI/BamHI‐cleaved pGBT9. A similar strategy was employed for At NPR1 1/313. The strong transactivation domain of VP16 was obtained as a 0.3‐kb EcoRI/BamHI fragment from the Y2H bait vector pBT3‐N (Dualsystems Biotech AG, Schlieren, Switzerland), and fused to the sequence encoding the Gal4 DNA BD in pGBT9.
The nim1‐4 mutation was introduced into At and Nt NPR1 by recombinant DNA technology. The 3′ end of At NPR1 was amplified using primers NPR1Mut2 (5′‐GGCGGCCGATGAATTGAAGATGACGCTGCTCGATCTTGAAAATAAAGTTGCAC) and NPR1bck (5′‐GTGTCGACCGACGACGATGAGAGA). NPR1Mut2 contains the sequence for an XmaIII restriction endonuclease site and changes the codon at position 1294 in the At NPR1 open reading frame from AGA (Arg‐432) to AAA (Lys‐432). The wild‐type At NPR1 3′ end was replaced by the PCR fragment cleaved with XmaIII and SalI, and the At nim1‐4 sequence was cloned as a BamHI/SalI fragment into pGBT9 and pGAD424. To generate Nt nim1‐4.DQ 15/588, a 0.2‐kb internal fragment was amplified from Nt NPR1.DQ with primers NtNPR1‐9 (5′‐TTGGTACCGGAGCTAGACCTTCTG) and NtNPR1‐10 (5′‐AAGGATCCAGGCCTACTTTATTTTCAAGATATAACAGCTTCATACGCAAATCATCGCCCGCC). NtNPR1‐10 introduces the nim1‐4 mutation in the PCR fragment (AGA to AAA, Arg‐431 to Lys) and a StuI restriction endonuclease site 3′ to the mutation (GTTGGCCTG to GTAGGCCTG). The Nt NPR1.DQ 3′ end was added as a StuI/BamHI fragment, which was amplified by PCR using primers NtNPR1‐8 (5′‐TTGGTACCAGGCCTGGCTAAACTCC) and NtNPR1‐6 (5′‐CCGGATCCTTTCCTAAAAGGGAGCTTATTGGG). The 5′ end was obtained by cleaving pUC19ΔHindIII/NtNPR1Δ1 (Zwicker et al., 2007) with EcoRI and partially with HindIII. The Nt nim1‐4.DQ 15/588 sequence was ligated to EcoRI/BamHI‐cut pGBT9 and pGAD424. To clone Nt nim1‐4.AF, the 0.2‐kb NsiI/NheI fragment in Nt NPR1.AF was replaced by the fragment from Nt nim1‐4.DQ 15/588 comprising the nim1‐4 mutation. Both restriction enzyme sites, NsiI and NheI, occur at the same positions in Nt NPR1.AF and Nt NPR1.DQ, and the amino acid sequences in this region are identical in both genes. Nt nim1‐4.AF was cloned as a BamHI/SalI fragment in pGBT9 and pGAD424. The C‐terminal fragment encompassing amino acids 386–588 was amplified from Nt NPR1.AF and Nt nim1‐4.AF with primers NtNPR1‐10.AF (5′‐AAGGATCCGTTCTGCTTCGAATGATCGG) and NtNPR1‐2.AF (5′‐TTGTCGACCTATTTCCTAAAAGGGAGC), respectively, and ligated to BamHI/SalI‐cut pGAD424.
To map the NIMIN2 binding site in Nt NPR1, Phe‐505 and Phe‐506 were mutated to Ser using overlap extension PCR (Ho et al., 1989). The primers for mutagenesis were NtNPR1‐5.AF (5′‐TGGAAAACGCAGCAGTCCACGTTGTTC) and NtNPR1‐6.AF (5′‐GAACAACGTGGACTGCTGCGTTTTCCA). The mutations were integrated into the Nt NPR1.AF sequence on a 0.3‐kb XbaI/SalI fragment, and the Nt NPR1 F505S F506S.AF sequence was cloned into BamHI/SalI‐cleaved pGBT9 and pGAD424.
A cDNA clone comprising the entire coding region of an NPR1‐like gene was amplified from DNA isolated from an Nt (N. tabacum L. cv. Samsun NN) Y2H cDNA library (Börnke, 2005). The primers were NtNIMlike‐1 (5′‐AAGAATTCATGGCTTGTTCTGCTGAACC) and NtNIMlike‐2 (5′‐GGGTCGACTCATAGTTTCCTAAGTTTGACACTTGC). The sequence is identical to the previously described Nt NIM1‐like1 cDNA, except for three nucleotide exchanges and one amino acid exchange (Pro‐534 to Thr). The Nt NIM1‐like1 sequence was inserted as an EcoRI/SalI fragment in pGBT9 and pGAD424.
All clones were analysed for the correct orientation of the inserted fragments, and the clones generated by PCR amplification were verified by DNA sequence analysis.
One‐hybrid and two‐hybrid assays and expression of yEGFP fusion proteins in yeast
For Y1H analyses, cDNA sequences comprising truncated or full‐length proteins were fused in frame to the Gal4 DNA BD sequence in pGBT9. For Y2H analyses, cDNAs were fused in frame to the Gal4 BD in pGBT9 and to the Gal4 activation domain sequence in pGAD424. Plasmids were transformed into Saccharomyces cerevisiae HF7c. Yeast cells were grown in the absence or presence of different concentrations of SA or SA analogues as indicated. Cells cultivated in liquid medium were harvested after 24 h for the determination of reporter gene activities. Qualitative protein interaction assays, quantitative determination of lacZ reporter gene activities and immunodetection of Gal4 fusion proteins were carried out as described previously (Weigel et al., 2001). β‐Galactosidase activities are the means of at least three independent colonies tested, each in duplicate, plus or minus the standard deviation (SD).
Plasmids encoding yEGFP fusion proteins were transformed into S. cerevisiae CEN.PK. Yeast cells were mounted in water and viewed with a Nikon Eclipse TS100 microscope (Nikon GmbH, Düsseldorf, Germany). yEGFP was visualized with a filter block limiting fluorescence excitation in the range 450–490 nm and allowing low‐pass emission detection beyond 515 nm. Fluorescence and bright field images were captured at ×200 magnification with an Olympus C7070 camera (Olympus Imaging Europa GmbH, Hamburg, Germany). The images were merged and processed in Adobe Photoshop. Protein expression was monitored by immunodetection using a rabbit polyclonal antiserum against GFP (Santa Cruz Biotechnology, Heidelberg, Germany).
RT‐PCR analyses, GUS reporter gene assays and immunodetection of PR‐1 proteins
Total RNA was isolated from soil‐grown Nt plants (N. tabacum L. cv. Samsun NN) and from seedlings cultivated on MS medium or MS medium with the addition of 1 mm SA. RNA isolation and analysis by RT‐PCR were performed as described previously (Zwicker et al., 2007). In the case of Nt NPR1 and Nt NIM1‐like1, RT‐PCR products were hardly visible by ethidium bromide staining on agarose gels. Therefore, Nt NPR1 and Nt NIM1‐like1 RT‐PCR products were diluted as indicated, and the diluted cDNAs were amplified by another round of PCR. The primer combinations used for the detection of the different mRNAs are listed in Table S1 (see Supporting Information). When available, 1 ng of plasmid DNA harbouring the respective cDNA was subjected to PCR amplification in parallel with the RNA samples (lanes c).
PR‐1 gene induction in Nt leaf discs by 1 mm solutions of SA, BA or 4‐OH BA, extraction of proteins, determination of GUS reporter enzyme activity and immunodetection of PR‐1 proteins were performed as described previously (Glocova et al., 2005).
Accession numbers
Sequence data from this article can be found in the GenBank /EMBL or Arabidopsis Genome Initiative (AGI) databases under accession numbers AF480488 and DQ837218 (Nicotiana tabacum NPR1), AY640382 (Nicotiana tabacum NIM1‐like1), AY640378 (Solanum lycopersicum NIM1), ABM55236 (Beta vulgaris NPR1), AY923983 (Oryza sativa NH1), At1g64280 (Arabidopsis thaliana NPR1), At3g57130 (Arabidopsis thaliana BOP1), At2g41370 (Arabidopsis thaliana BOP2) and AF527176 (Brassica napus NPR1).
Supporting information
Fig. S1 Alignment of tobacco (Nt) and Arabidopsis (At) NPR1 proteins.
Fig. S2 The tobacco (Nt) NPR1 gene is expressed constitutively.
Fig. S3 The addition of salicylic acid to the growth medium alters the biochemical capabilities of NPR1 proteins in yeast.
Fig. S4 The structural analogue 4‐hydroxy‐benzoic acid does not suppress the interaction of tobacco (Nt) NPR1 with NIMIN2 proteins in yeast.
Fig. S5 Sensitivity of tobacco (Nt) and Arabidopsis (At) NPR1 proteins towards salicylic acid in yeast.
Fig. S6 The nim1‐4 mutant protein is insensitive to salicylic acid in yeast.
Fig. S7 Tobacco (Nt) NIM1‐like1 is expressed constitutively.
Fig. S8 Tobacco (Nt) NIM1‐like1 interacts with TGA transcription factors and NIMIN1 and NIMIN2 proteins in yeast.
Fig. S9 Protein interaction of the tobacco (Nt) NPR1 F505S F506S mutant in yeast.
Fig. S10 Alignment of NPR1 proteins from tobacco (Nt) and Arabidopsis (At) with Arabidopsis NPR1‐related proteins BLADE‐ON‐PETIOLE, BOP1 and BOP2.
Fig. S11 Schematic representation of the NPR1 constructs used in this study. Structural domains as well as serine and cysteine residues with relevance to the function of At NPR1 are indicated (see also Fig. S1). The diagrams are drawn approximately to scale. (A) Constructs used in subcellular localization studies in yeast. (B) At NPR1 constructs used in Y1H and Y2H analyses. (C) Nt NPR1 deletion constructs used in Y1H and Y2H analyses. (D) Nt NPR1 mutant constructs used in Y1H and Y2H analyses. (E) Schematic representation of Nt NPR1. The diagram includes the LENRV domain mediating transduction of the SA signal, and the NIMIN1/NIMIN2 binding domain (N2 BD) which were identified in this work. Both domains are also present in NPR1 proteins from different plant species including Arabidopsis (see also Fig. 7D).
Table S1 Primers and control plasmids used in reverse transcriptase‐polymerase chain reaction (RT‐PCR) analyses.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We are grateful to Xinnian Dong (Duke University, Durham, NC, USA) for the generous gift of a cDNA clone encoding Nt NPR1.AF, and to Ralf Weigel and Vlatka Stos for providing DNA constructs.
REFERENCES
- Aravind, L. and Koonin, E.V. (1999) Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. [DOI] [PubMed] [Google Scholar]
- Börnke, F. (2005) The variable C‐terminus of 14‐3‐3 proteins mediates isoform‐specific interaction with sucrose‐phosphate synthase in the yeast two‐hybrid system. J. Plant Physiol. 162, 161–168. [DOI] [PubMed] [Google Scholar]
- Boyle, P. , Le Su, E. , Rochon, A. , Shearer, H.L. , Murmu, J. , Yan Chu, J. , Fobert, P.R. and Després, C. (2009) The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell, 21, 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Bowling, S.A. , Gordon, A.S. and Dong, X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell, 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Glazebrook, J. , Clarke, J.D. , Volko, S. and Dong, X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell, 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Cao, H. , Li, X. and Dong, X. (1998) Generation of broad‐spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA, 95, 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern, M. , Fitzgerald, H.A. , Canlas, P.E. , Navarre, D.A. and Ronald, P.C. (2005a) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant–Microbe Interact. 18, 511–520. [DOI] [PubMed] [Google Scholar]
- Chern, M. , Canlas, P.E. , Fitzgerald, H.A. and Ronald, P.C. (2005b) Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 43, 623–635. [DOI] [PubMed] [Google Scholar]
- Chern, M. , Canlas, P.E. and Ronald, P.C. (2008) Strong suppression of systemic acquired resistance in Arabidopsis by NRR is dependent on its ability to interact with NPR1 and its putative repression domain. Mol. Plant, 1, 552–559. [DOI] [PubMed] [Google Scholar]
- Chern, M.‐S. , Fitzgerald, H.A. , Yadav, R.C. , Canlas, P.E. , Dong, X. and Ronald, P.C. (2001) Evidence for a disease‐resistance pathway in rice similar to the NPR1‐mediated signaling pathway in Arabidopsis . Plant J. 27, 101–113. [DOI] [PubMed] [Google Scholar]
- Cormack, B.P. , Bertram, G. , Egerton, M. , Gow, N.A.R. , Falkow, S. and Brown, A.J.P. (1997) Yeast‐enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans . Microbiology, 143, 303–311. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , Gaffney, T. , Gut‐Rella, M. , Kessmann, H. , Ward, E. and Ryals, J. (1994) A central role of salicylic acid in plant disease resistance. Science, 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P. , Friedrich, L. and Ryals, J.A. (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA, 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C. , DeLong, C. , Glaze, S. , Liu, E. and Fobert, P.R. (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell, 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Fan, W. and Dong, X. (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid‐mediated gene activation in Arabidopsis. Plant Cell, 14, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, L. , Lawton, K. , Ruess, W. , Masner, P. , Specker, N. , Gut Rella, M. , Meier, B. , Dincher, S. , Staub, T. , Uknes, S. , Métraux, J.‐P. , Kessmann, H. and Ryals, J. (1996) A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 10, 61–70. [Google Scholar]
- Friedrich, L. , Lawton, K. , Dietrich, R. , Willits, M. , Cade, R. and Ryals, J. (2001) NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol. Plant–Microbe Interact. 14, 1114–1124. [DOI] [PubMed] [Google Scholar]
- Gaffney, T. , Friedrich, L. , Vernooij, B. , Negrotto, D. , Nye, G. , Uknes, S. , Ward, E. , Kessmann, H. and Ryals, J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science, 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glocova, I. , Thor, K. , Roth, B. , Babbick, M. , Pfitzner, A.J.P. and Pfitzner, U.M. (2005) Salicylic acid (SA)‐dependent gene activation can be uncoupled from cell death‐mediated gene activation: the SA‐inducible NIMIN‐1 and NIMIN‐2 promoters, unlike the PR‐1a promoter, do not respond to cell death signals in tobacco. Mol. Plant Pathol. 6, 299–314. [DOI] [PubMed] [Google Scholar]
- Henriques, M. , Quintas, C. and Loureiro‐Dias, M.C. (1997) Extrusion of benzoic acid in Saccharomyces cerevisiae by an energy‐dependent mechanism. Microbiology, 143, 1877–1883. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R. , Zhang, Y. , McKim, S. , Li, X. and Haughn, G.W. (2005) BLADE‐ON‐PETIOLE‐dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell, 17, 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N. , Hunt, H.D. , Horton, R.M. , Pullen, J.K. and Pease, L.R. (1989) Site‐directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Horvath, D.M. , Huang, D.J. and Chua, N.‐H. (1998) Four classes of salicylate‐induced tobacco genes. Mol. Plant–Microbe Interact. 11, 895–905. [DOI] [PubMed] [Google Scholar]
- Kinkema, M. , Fan, W. and Dong, X. (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell, 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton, K.A. , Friedrich, L. , Hunt, M. , Weymann, K. , Delaney, T. , Kessmann, H. , Staub, T. and Ryals, J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82. [DOI] [PubMed] [Google Scholar]
- Lebel, E. , Heifetz, P. , Thorne, L. , Uknes, S. , Ryals, J. and Ward, E. (1998) Functional analysis of regulatory sequences controlling PR‐1 gene expression in Arabidopsis. Plant J. 16, 223–233. [DOI] [PubMed] [Google Scholar]
- Liu, G. , Holub, E.B. , Alonso, J.M. , Ecker, J.R. and Fobert, P.R. (2005) An Arabidopsis NPR1‐like gene, NPR4, is required for disease resistance. Plant J. 41, 304–318. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Marathe, R. and Dinesh‐Kumar, S.P. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N‐mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. and Van Strien, E.A. (1999) The families of pathogenesis‐related proteins, their activities, and comparative analysis of PR‐1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Ma, Y. , Szostkiewicz, I. , Korte, A. , Moes, D. , Yang, Y. , Christmann, A. and Grill, E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Macris, B.J. (1975) Mechanism of benzoic acid uptake by Saccharomyces cerevisiae . Appl. Microbiol. 30, 503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J. , Carr, J.P. , Klessig, D.F. and Raskin, I. (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science, 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Métraux, J.P. , Signer, H. , Ryals, J. , Ward, E. , Wyss‐Benz, M. , Gaudin, J. , Raschdorf, K. , Schmid, E. , Blum, W. and Inverardi, B. (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science, 250, 1004–1006. [DOI] [PubMed] [Google Scholar]
- Mou, Z. , Fan, W. and Dong, X. (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell, 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Niedenthal, R.K. , Riles, L. , Johnston, M. and Hegemann, J.H. (1996) Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast, 12, 773–786. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R. , Thurow, C. , Weigel, R. , Pfitzner, U. and Gatz, C. (2000) Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA‐binding properties. Plant Mol. Biol. 42, 775–788. [DOI] [PubMed] [Google Scholar]
- Park, S.‐Y. , Fung, P. , Nishimura, N. , Jensen, D.R. , Fujii, H. , Zhao, Y. , Lumba, S. , Santiago, J. , Rodrigues, A. , Chow, T.F. , Alfred, S.E. , Bonetta, D. , Finkelstein, R. , Provart, N.J. , Desveaux, D. , Rodriguez, P.L. , McCourt, P. , Zhu, J.‐K. , Schroeder, J.I. , Volkman, B.F. and Cutler, S.R. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potlakayala, S.D. , DeLong, C. , Sharpe, A. and Fobert, P.R. (2007) Conservation of NON‐EXPRESSOR OF PATHOGENESIS‐RELATED GENES1 function between Arabidopsis thaliana and Brassica napus . Physiol. Mol. Plant Pathol. 71, 174–183. [Google Scholar]
- Qin, X.F. , Holuigue, L. , Horvath, D.M. and Chua, N.H. (1994) Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as‐1 element. Plant Cell, 6, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon, A. , Boyle, P. , Wignes, T. , Fobert, P.R. and Després, C. (2006) The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C‐terminal cysteines. Plant Cell, 18, 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A.F. (1961) Systemic acquired resistance induced by localized virus infections in plants. Virology, 14, 340–358. [DOI] [PubMed] [Google Scholar]
- Ryals, J. , Weymann, K. , Lawton, K. , Friedrich, L. , Ellis, D. , Steiner, H.Y. , Johnson, J. , Delaney, T.P. , Jesse, T. , Vos, P. and Uknes, S. (1997) The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell, 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff, T.G. , Badr, M.Z. and Doyle, R.J. (1982) The nature of salicylate inhibition of sugar transport in yeast. Fundam. Appl. Toxicol. 2, 168–172. [DOI] [PubMed] [Google Scholar]
- Shah, J. , Tsui, F. and Klessig, D.F. (1997) Characterization of a salicylic acid‐insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA‐inducible expression of the tms2 gene. Mol. Plant–Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. , Mou, Z. , Tada, Y. , Spivey, N. , Genschik, P. and Dong, X. (2009) Proteasome‐mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell, 137, 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen, G. , Grüner, R. and Pfitzner, U.M. (1998) An as‐1‐like motif controls the level of expression of the gene for the pathogenesis‐related protein 1a from tobacco. Plant Mol. Biol. 37, 871–883. [DOI] [PubMed] [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. , Zuo, J. and Dong, X. (2008) Plant immunity requires conformational changes of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, X. , Calderon‐Villalobos, L.I.A. , Sharon, M. , Zheng, C. , Robinson, C.V. , Estelle, M. and Zheng, N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature, 446, 640–645. [DOI] [PubMed] [Google Scholar]
- Uknes, S. , Mauch‐Mani, B. , Moyer, M. , Potter, S. , Williams, S. , Dincher, S. , Chandler, D. , Slusarenko, A. , Ward, E. and Ryals, J. (1992) Acquired resistance in Arabidopsis. Plant Cell, 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes, S. , Winter, A.M. , Delaney, T. , Vernooij, B. , Morse, A. , Friedrich, L. , Nye, G. , Potter, S. , Ward, E. and Ryals, J. (1993) Biological induction of systemic acquired resistance in Arabidopsis . Mol. Plant–Microbe Interact. 6, 692–698. [Google Scholar]
- Vernooij, B. , Friedrich, L. , Ahl Goy, P. , Staub, T. , Kessmann, H. and Ryals, J. (1995) 2,6‐Dichloroisonicotinic acid‐induced resistance to pathogens without the accumulation of salicylic acid. Mol. Plant–Microbe Interact. 8, 228–234. [Google Scholar]
- Ward, E.R. , Uknes, S.J. , Williams, S.C. , Dincher, S.S. , Wiederhold, D.L. , Alexander, D.C. , Ahl‐Goy, P. , Métraux, J.P. and Ryals, J.A. (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell, 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, R.R. , Bäuscher, C. , Pfitzner, A.J.P. and Pfitzner, U.M. (2001) NIMIN‐1, NIMIN‐2 and NIMIN‐3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46, 143–160. [DOI] [PubMed] [Google Scholar]
- Weigel, R.R. , Pfitzner, U.M. and Gatz, C. (2005) Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell, 17, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R.F. (1979) Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology, 99, 410–412. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Fan, B. and Chen, Z. (1998) Induction of PR‐1 proteins and potentiation of pathogen signals by salicylic acid exhibit the same dose–response and structural specificity in plant cell cultures. Mol. Plant–Microbe Interact. 11, 568–571. [Google Scholar]
- Yan, J. , Zhang, C. , Gu, M. , Bai, Z. , Zhang, W. , Qi, T. , Cheng, Z. , Peng, W. , Luo, H. , Nan, F. , Wang, Z. and Xie, D. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell, 21, 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Fan, W. , Kinkema, M. , Li, X. and Dong, X. (1999) Interaction of NPR1 with basic leucine zipper transcription factors that bind sequences required for salicylic acid induction of the PR‐1 gene. Proc. Natl. Acad. Sci. USA, 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Tessaro, M.J. , Lassner, M. and Li, X. (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell, 15, 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Cheng, Y.T. , Qu, N. , Zhao, Q. , Bi, D. and Li, X. (2006) Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. [DOI] [PubMed] [Google Scholar]
- Zhou, J.‐M. , Trifa, Y. , Silva, H. , Pontier, D. , Lam, E. , Shah, J. and Klessig, D.F. (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR‐1 gene required for induction by salicylic acid. Mol. Plant–Microbe Interact. 13, 191–202. [DOI] [PubMed] [Google Scholar]
- Zwicker, S. , Mast, S. , Stos, V. , Pfitzner, A.J.P. and Pfitzner, U.M. (2007) Tobacco NIMIN2 proteins control PR gene induction through transient repression early in systemic acquired resistance. Mol. Plant Pathol. 8, 385–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment of tobacco (Nt) and Arabidopsis (At) NPR1 proteins.
Fig. S2 The tobacco (Nt) NPR1 gene is expressed constitutively.
Fig. S3 The addition of salicylic acid to the growth medium alters the biochemical capabilities of NPR1 proteins in yeast.
Fig. S4 The structural analogue 4‐hydroxy‐benzoic acid does not suppress the interaction of tobacco (Nt) NPR1 with NIMIN2 proteins in yeast.
Fig. S5 Sensitivity of tobacco (Nt) and Arabidopsis (At) NPR1 proteins towards salicylic acid in yeast.
Fig. S6 The nim1‐4 mutant protein is insensitive to salicylic acid in yeast.
Fig. S7 Tobacco (Nt) NIM1‐like1 is expressed constitutively.
Fig. S8 Tobacco (Nt) NIM1‐like1 interacts with TGA transcription factors and NIMIN1 and NIMIN2 proteins in yeast.
Fig. S9 Protein interaction of the tobacco (Nt) NPR1 F505S F506S mutant in yeast.
Fig. S10 Alignment of NPR1 proteins from tobacco (Nt) and Arabidopsis (At) with Arabidopsis NPR1‐related proteins BLADE‐ON‐PETIOLE, BOP1 and BOP2.
Fig. S11 Schematic representation of the NPR1 constructs used in this study. Structural domains as well as serine and cysteine residues with relevance to the function of At NPR1 are indicated (see also Fig. S1). The diagrams are drawn approximately to scale. (A) Constructs used in subcellular localization studies in yeast. (B) At NPR1 constructs used in Y1H and Y2H analyses. (C) Nt NPR1 deletion constructs used in Y1H and Y2H analyses. (D) Nt NPR1 mutant constructs used in Y1H and Y2H analyses. (E) Schematic representation of Nt NPR1. The diagram includes the LENRV domain mediating transduction of the SA signal, and the NIMIN1/NIMIN2 binding domain (N2 BD) which were identified in this work. Both domains are also present in NPR1 proteins from different plant species including Arabidopsis (see also Fig. 7D).
Table S1 Primers and control plasmids used in reverse transcriptase‐polymerase chain reaction (RT‐PCR) analyses.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


