SUMMARY
Ribosome inactivating proteins are glycosidases synthesized by many plants and have been hypothesized to serve in defence against pathogens. These enzymes catalytically remove a conserved purine from the sarcin/ricin loop of the large ribosomal RNA, which has been shown in vitro to limit protein synthesis. The resulting toxicity suggests that plants may possess a mechanism to protect their ribosomes from depurination during the synthesis of these enzymes. For example, pokeweed antiviral protein (PAP) is cotranslationally inserted into the lumen of the endoplasmic reticulum and travels via the endomembrane system to be stored in the cell wall. However, some PAP may retrotranslocate across the endoplasmic reticulum membrane to be released back into the cytosol, thereby exposing ribosomes to depurination. In this work, we isolated and characterized a complexed form of the enzyme that exhibits substantially reduced activity. We showed that this complex is a homodimer of PAP and that dimerization involves a peptide that contains a conserved aromatic amino acid, tyrosine 123, located in the active site of the enzyme. Bimolecular fluorescence complementation demonstrated that the homodimer may form in vivo and that dimerization is prevented by the substitution of tyrosine 123 for alanine. The homodimer is a minor form of PAP, observed only in the cytosol of cells and not in the apoplast. Taken together, these data support a novel mechanism for the limitation of depurination of autologous ribosomes by molecules of the protein that escape transport to the cell wall by the endomembrane system.
INTRODUCTION
Ribosome inactivating proteins are N‐glycosidases that remove an adenine from the conserved sarcin/ricin loop of the large ribosomal RNA (rRNA) (Endo et al., 1988; Stirpe et al., 1988). Ribosomes depurinated in this manner do not efficiently bind elongation factor 2, which slows the rate of protein translation (Gessner and Irvin, 1980; Nilsson and Nygård, 1986). The role of these enzymes as defence against pathogens has traditionally been attributed to the toxicity caused by the inhibition of translation. The ribosome inactivating protein would come into contact with and depurinate ribosomes of infected cells, resulting in host cell death and a lack of pathogen proliferation. The heterologous expression of some ribosome inactivating proteins has generated plants with enhanced resistance to fungal or viral infection (Corrado et al., 2005; Zoubenko et al., 1997). Moreover, purified enzymes have been shown to inhibit directly the growth of fungal hyphae and to depurinate the genomes of some viruses, indicating that substrate selection is not limited to plant cell rRNA (Karran and Hudak, 2008; Nielsen et al., 2001). However, ribosomes isolated from plants that produce ribosome inactivating proteins are often depurinated and are unable to perform translation in vitro (Prestle et al., 1992; Taylor and Irvin, 1990).
Given the sensitivity of ribosomes to depurination, plants may avoid the toxicity of these enzymes by sequestering them in subcellular organelles. Most ribosome inactivating proteins contain an N‐terminal sequence that targets them for cotranslational insertion into the endoplasmic reticulum (Hartley and Lord, 1993). Once in the endomembrane system, the proteins are transported to the vacuole or the apoplast, thereby spatially separating the enzymes from autologous ribosomes (Carzaniga et al., 1994; Lord, 1985).
Pokeweed antiviral protein (PAP) is a ribosome inactivating protein produced in the leaves of the pokeweed plant Phytolacca americana (Irvin, 1975). PAP is synthesized by membrane‐bound ribosomes and travels by the endomembrane system to the cell wall (Ready et al., 1986). Its antiviral activity is hypothesized to result from either ribosome or virus genome depurination, after entry of the protein with viral particles into the cytosol of damaged cells (Picard et al., 2005; Taylor et al., 1994). Recently, it has been shown that PAP may pass from the endomembrane system to the cytosol, and that the C‐terminus of the protein, which is required for this activity, bears sequence homology to other ribosome inactivating proteins shown to retrotranslocate across the endoplasmic reticulum membrane (Baykal and Tumer, 2007). Given that pokeweed ribosomes are sensitive to depurination (Bonness et al., 1994), a mechanism may exist whereby the plant protects its ribosomes from this enzyme prior to its localization in the apoplast.
A complexed form of PAP has been identified previously from pokeweed leaves that exhibits reduced inhibition of translation in a cell‐free system (Desvoyes et al., 1997). The complexed form has an apparent mass of 57 kDa and cross‐reacts with antibody specific to PAP; however, its components were not identified. We expand on this initial observation to show that this complex is a homodimer of PAP that forms in planta, and is less toxic than the monomeric form because its ability to depurinate ribosomes is substantially reduced. The dimer was observed only in the cytosol of cells and was absent from the apoplast, substantiating our view that homodimerization may be a mechanism by which pokeweed avoids the depurination of its ribosomes during the processing of this protein.
RESULTS AND DISCUSSION
Expression of monomeric and complexed PAP in pokeweed
Immunoblot analysis of total cell lysates from pokeweed leaves indicated the presence of monomeric PAP at the expected 29 kDa, plus a protein migrating to a corresponding molecular weight of approximately 58 kDa. This complexed form was visible only when lysate samples were not boiled prior to sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE). Boiling in the presence of 2% SDS disassociated the complexed PAP; however, the anionic detergent alone did not disrupt the complex (Fig. 1A). Dissociation with detergent and boiling indicates that the complex is noncovalently formed, and the mass of 58 kDa is consistent with the possibility that it may be a homodimer of PAP.
Figure 1.
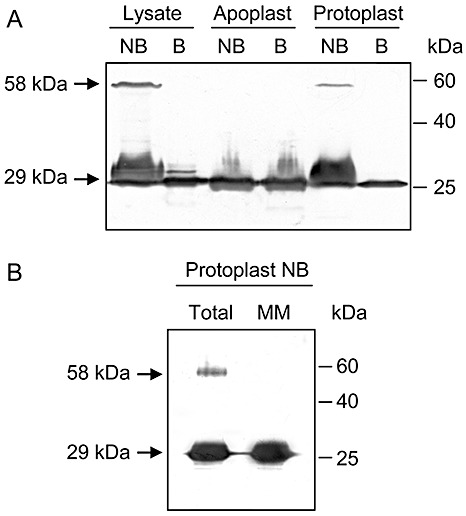
Expression of monomeric and complexed pokeweed antiviral protein (PAP) in pokeweed leaves. (A) Proteins (4 µg) of total cell lysate, apoplastic fluid or protoplasts were separated by 12% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred to nitrocellulose and probed with a polyclonal antibody specific to PAP (1:5000). (B) Total protoplast protein (8 µg) or proteins from the microsomal membrane (MM) fraction of protoplasts (8 µg) were also probed for the presence of PAP as described in (A). Samples that were boiled prior to separation are labelled B, and NB indicates samples that were not boiled. Molecular weights of monomeric (29 kDa) and complexed (58 kDa) PAP are indicated.
Given that PAP is synthesized by endoplasmically bound ribosomes and the mature form of the protein is stored in the cell wall (Ready et al., 1986), we analysed the intracellular and extracellular fractions for the presence of the complexed form. Apoplastic fluid, containing extracellular protein, and protoplasts, containing the plasma membrane and cellular proteins, were used to represent the extracellular and intracellular fractions, respectively. Complexed PAP was visualized only in protoplasts and not in the apoplastic fluid (Fig. 1A); therefore, the complexed form is not stored in the cell wall of pokeweed but is localized inside cells. A substantial amount of free PAP was also present inside protoplasts, with the complexed form representing a minor amount of total PAP. To determine the cellular localization of the monomeric form, microsomes of protoplasts were separated from the cytosolic fraction and immunoblotted for the presence of PAP (Fig. 1B). The majority of free PAP within cells was associated with microsomes, which are primarily composed of endoplasmic reticulum, and agrees with the proposed processing of PAP via the endomembrane system. The lack of the complexed form in microsomes suggests that it is present in the cytosol and may represent the small proportion of PAP that has re‐entered the cytosol from the lumen of the endoplasmic reticulum. The ability to sequester even small amounts of PAP into a complexed form may be important to cells, as nanomolar concentrations of the enzyme in the cytosol will disable pokeweed ribosomes (IC50= 2.9 nm; Bonness et al., 1994). The absence of the dimer from the cell wall and microsomal membrane fractions supports our contention that dimerization is a mechanism to trap PAP molecules that have re‐entered the cytoplasm, rather than a component of normal PAP processing within the endoplasmic reticulum.
Isolation and identification of the complexed form of PAP
Monomeric PAP and the 58‐kDa complexed form were purified by ion exchange chromatography. Both forms of PAP were eluted from an anionic resin with 350 mm NaCl, with the 58‐kDa complex eluting just prior to the free protein, suggesting a lower pI for complexed PAP compared with the monomeric form (Fig. 2). Complexed PAP sometimes appeared as two bands, which has been observed previously (Desvoyes et al., 1997). The doublet may represent two conformations with different folding, as the proteins were not boiled prior to separation through SDS‐PAGE.
Figure 2.
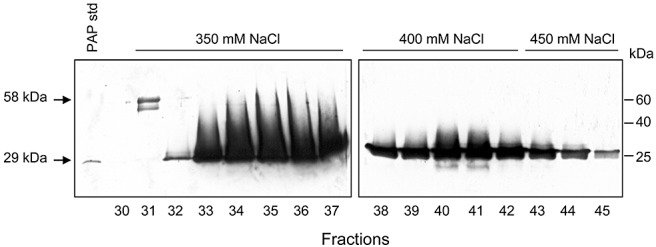
Separation of monomer from complexed pokeweed antiviral protein (PAP) by ion exchange chromatography. PAP was purified from the total cell lysate of pokeweed leaves by ion exchange. Positively charged proteins were eluted from the anionic column using an NaCl step gradient. Aliquots (10 µL) from each fraction were separated by 12% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) without prior boiling, transferred to nitrocellulose and probed with a polyclonal antibody specific to PAP (1:5000). Purified monomeric PAP was loaded (20 ng) as a standard. Molecular weights of monomeric (29 kDa) and complexed (58 kDa) PAP are indicated.
The complexed and free forms of PAP were subjected to in‐gel trypsin digestion and the peptide fragments were identified by mass spectrometry. All major peaks observed matched the expected masses of peptides from PAP, indicating that the complexed form was probably a PAP homodimer (Fig. 3A). To identify the regions of PAP that may be involved in dimerization, peptide fragments that were observed in the free form, but were undetected in the complexed form, were examined. In particular, the mass spectrum showed that the peak at 837.4, which matches the expected m/z ratio of the protonated peptide YPTLESK, was present in monomeric PAP, but not in the complexed form of PAP (Fig. 3A, compare spectrum i with iii), suggesting that the tryptic cleavage site in the latter instance was inaccessible as a result of dimerization. In addition, the peak at 865.4, which corresponds to the tryptic peptide NINFDSR immediately adjacent to the protected heptad in the PAP sequence, is reduced in intensity in the complexed PAP fraction relative to the free PAP sample. The peak at 842.5 corresponds to a peptide from trypsin and therefore serves as a reference, as the same amount of trypsin was added to the two samples. Taken together, these data suggest that dimerization inhibits the generation of the peptide YPTLESK, and reduces the efficiency of trypsin cleavage of the adjacent peptide NINFDSR, possibly as a result of steric hindrance. The peaks shown in Fig. 3A, ii and iv, illustrate the higher end of the same spectra as shown in Fig. 3A, i and iii. Two peaks are observed at 1112.5 and 1451.7, which correspond to peptides YATFLNDLR and the protonated YHIFNDISGTER, respectively. The prominence of both of these peaks with similar relative intensities serves as an internal control and suggests that they are not involved in the dimerization of PAP. Figure 3B is a molecular model of PAP illustrating the position of the protected peptide YPTLESK (coloured blue) and the location of residues required for enzyme activity (coloured pink). This three‐dimensional modelling of the protein, based on a 2.5‐Å crystal structure of PAP (Monzingo et al., 1993), shows that the protected heptad forms a surface‐exposed α‐helix. The dimerization of proteins often involves interaction between α‐helices (Mason and Arndt, 2004), and the surface exposure of the heptad YPTLESK supports our hypothesis that two PAP molecules may dimerize via this peptide.
Figure 3.
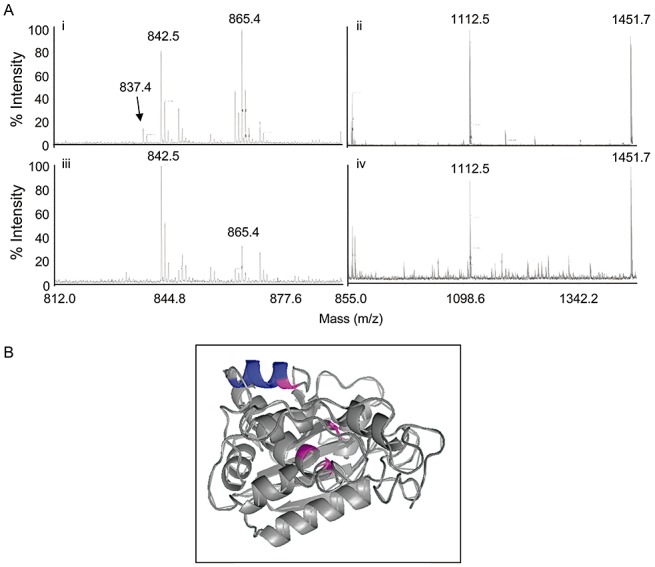
Mass spectrometric analysis of complexed pokeweed antiviral protein (PAP). (A) Partial peptide spectra of monomeric PAP (i, ii) and complexed PAP (iii, iv) following trypsin digestion. Peak 837.4 corresponds to peptide YPTLESK observed from the digestion of monomeric PAP, but not observed in the spectra of complexed PAP. (B) Molecular model of PAP; β‐strands and α‐helices are indicated, together with the helix containing the heptad YPTLESK (coloured blue). Amino acids essential for enzyme activity are coloured pink.
Bimolecular fluorescence complementation and in vitro cross‐linking
To investigate whether PAP dimerization occurs in vivo, pokeweed protoplasts were transfected with two plasmids, one encoding PAP fused to the N‐terminus of green fluorescent protein (GFP) and the other encoding PAP fused to the C‐terminus of GFP. Protoplasts transfected with these plasmids showed GFP fluorescence; therefore, the ability of PAP to dimerize was not an artefact of isolation or in vitro treatment, but rather demonstrates that dimerization occurs in planta (Fig. 4A). As a positive control, protoplasts were also transfected with a single plasmid encoding the full‐length GFP. As expected, a greater percentage of these cells were fluorescent relative to those transfected with the PAP constructs (65% compared with 15%), as fluorescence in cells containing PAP constructs relied on the interaction of individual PAP fusion molecules. No fluorescence was observed in cells transfected with plasmids containing only the N‐ and C‐termini of GFP, indicating that intact GFP did not form in the absence of PAP dimerization. The reconstitution of GFP through the interaction of PAP fusion proteins suggests that the dimerization of endogenous PAP may occur naturally in the cytosol of pokeweed cells.
Figure 4.
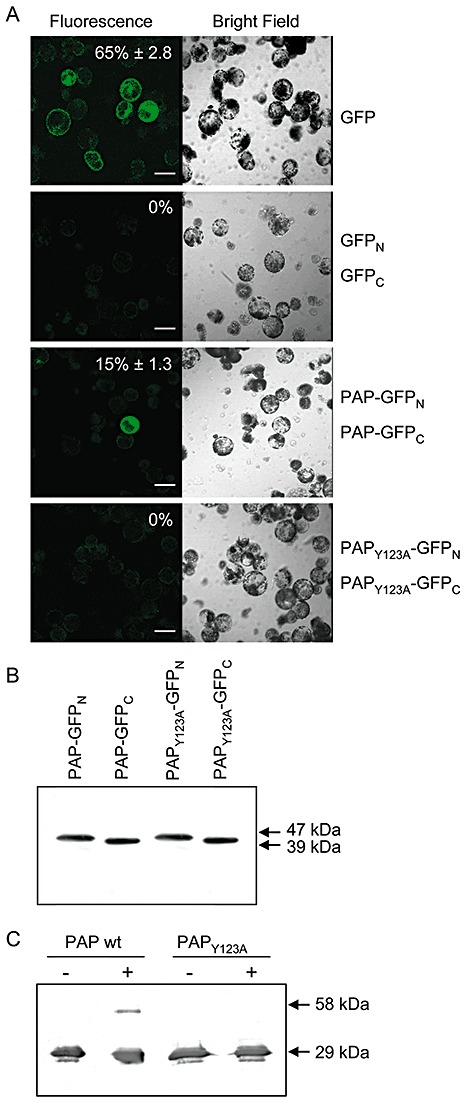
Bimolecular fluorescence complementation of pokeweed antiviral protein (PAP) dimerization in pokeweed protoplasts. Protoplasts were transfected with plasmids encoding the N‐ or C‐terminus of green fluorescent protein (GFP) fused to PAP or PAPY123A. As a negative control for bimolecular interaction, cells were transfected with plasmids encoding only the N‐ or C‐terminus of GFP. As a positive control for fluorescence, protoplasts were transfected with a plasmid encoding intact GFP. Transfected protoplasts were examined by fluorescence or bright field laser scanning microscopy. Percentage values indicate the mean numbers of fluorescent protoplasts relative to the total number of cells ± standard error from five fields of view. The scale bar indicates 50 µm. (B) Expression of individual constructs, PAP–GFPN, PAP–GFPC, PAPY123A–GFPN and PAPY123A–GFPC, following transfection into pokeweed protoplasts. Lysates of protoplasts (10 µg) were separated by 12% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred to nitrocellulose and probed with a polyclonal antibody specific to GFP (1:5000). The molecular weights of fusion proteins containing the N‐terminus of GFP (47 kDa) and the C‐terminus of GFP (39 kDa) are indicated. (C) In vitro chemical cross‐linking. Recombinant wild‐type PAP and PAPY123A (125 ng) were incubated with or without (+, −) 0.05% glutaraldehyde and separated by 12% SDS‐PAGE with prior boiling. Proteins were transferred to nitrocellulose and probed with a polyclonal antibody specific to PAP (1:5000). Molecular weights of monomeric (29 kDa) and complexed (58 kDa) PAP are indicated.
Given our mass spectrometry analysis indicating the protection of peptide YPTLESK from trypsin digestion, we repeated the transfection of pokeweed protoplasts with plasmids encoding a mutant form of PAP, PAPY123A, in place of wild‐type PAP. The mutant substitutes an alanine for tyrosine, the first amino acid of the heptad YPTLESK. We could not detect GFP fluorescence in these protoplasts, thus substantiating our mass spectrometry results indicating that tyrosine 123 is involved in dimerization (Fig. 4A). We hypothesize that the α‐helix is the critical component for dimerization. Tyrosine 123 may stabilize this helix, given that immediately adjacent to the helix, and N‐terminal to tyrosine 123, is an unstructured peptide NINFDSR. Tyrosine 123 may act as a border between the unstructured peptide and the α‐helix. Its mutation to alanine may destabilize this border and allosterically alter the shape of the critical α‐helix. To ensure that the fusion proteins were expressed in vivo, protoplasts were transfected with the individual fusion constructs and analysed by immunoblot for the presence of GFP. The molecular weight of proteins synthesized from the constructs should be the sum of PAP (29 kDa) and a portion of GFP (18 kDa for the N‐terminus and 10 kDa for the C‐terminus). The immunoblot illustrated proteins at 47 kDa for N‐terminal GFP–PAP and 39 kDa for C‐terminal GFP–PAP. Both wild‐type PAP and PAPY123A fusion constructs with GFP were expressed from the individual plasmids, and a lack of detectable catabolites suggests that neither fusion protein was unstable or susceptible to excessive proteolysis (Fig. 4B).
To test the ability of these proteins to dimerize in vitro, we purified recombinant wild‐type PAP and the point mutant PAPY123A from Escherichia coli, and incubated each with the chemical cross‐linker glutaraldehyde. The samples were separated by SDS‐PAGE, and immunoblots probed for PAP indicated that the wild‐type could be chemically cross‐linked in vitro, whereas cross‐linked PAPY123A was not detected (Fig. 4C). It is unlikely that wild‐type PAP cross‐linked randomly, because no trimeric forms of the protein were visible by immunoblot. Chemical cross‐linking forms a covalent bond and may not necessarily mimic the noncovalent in vivo interaction between PAP molecules. Dimerization of the purified monomeric form of PAP was not observed in the absence of cross‐linker, suggesting that isolated PAP monomers exhibit low binding affinity for each other. Perhaps a cytosolic factor may assist in the stabilization of the dimer in vivo. However, the inability of PAPY123A to form a complex under either in vivo or in vitro conditions supports the requirement of tyrosine 123 for PAP dimerization. Although this was a single amino acid substitution, we suggest that the change to alanine weakened the border of the α‐helical structure, which was essential for complex formation.
Tyrosine 123 is one of two aromatic amino acids that coordinate the enzyme's substrate, an adenine base, at the active site (Monzingo et al., 1993). Structural studies illustrate that the aromatic amino acids tyrosine 72 and tyrosine 123 sandwich the adenine base in an energetically favourable stacking conformation, whereas arginine 179 stabilizes an anionic charge on adenine and glutamic acid 176 stabilizes a cationic charge on the ribose (Ago et al., 1994). Together, these four amino acids (coloured pink in Fig. 3B) are directly involved in adenine binding and the hydrolysis of its glycoside bond. We have shown previously that this same point mutant of PAP (Y123A) expressed in yeast is not toxic and does not reduce significantly the rate of translation (Hudak et al., 2004), supporting the importance of this amino acid in the enzyme activity of the protein. Therefore, the dimerization of PAP may obscure tyrosine 123 and limit its accessibility to the adenine of rRNA, resulting in a complex with less ability for depurination.
Depurination of rRNA by PAP and complexed PAP
We compared the enzyme activity of the PAP dimer formed in vivo with wild‐type, monomeric PAP by incubating each protein with isolated pokeweed ribosomes and measuring the level of rRNA depurination by primer extension. Levels of depurination were measured by densitometry of band intensities of depurinated (Dep’n) cDNA products relative to 28S ribosomal cDNAs for each sample. As expected, rRNA of pokeweed was depurinated by endogenous PAP during its isolation, without the addition of purified PAP (Fig. 5; –PAP). However, the incubation of ribosomes with additional wild‐type monomeric PAP increased the level of rRNA depurination, whereas incubation with the same molar amount of dimeric PAP did not increase significantly rRNA depurination (9.2% compared with 2.3%). Therefore, the complexed form was less enzymatically active on pokeweed ribosomes, supporting our contention that dimerization of PAP is a mechanism to protect these ribosomes from depurination. To better visualize the level of enzyme activity of complexed PAP on ribosomes not already depurinated, we repeated the incubation of purified proteins with rabbit reticulocyte lysate. Ribosomes of lysate incubated with PAP dimer were depurinated significantly less relative to the total amount of 28S rRNA (1.6%), compared with depurination of ribosomes incubated with monomeric PAP (17.2%; Fig. 5), indicating that the dimer appeared to be consistently less active than the monomeric form of the protein.
Figure 5.
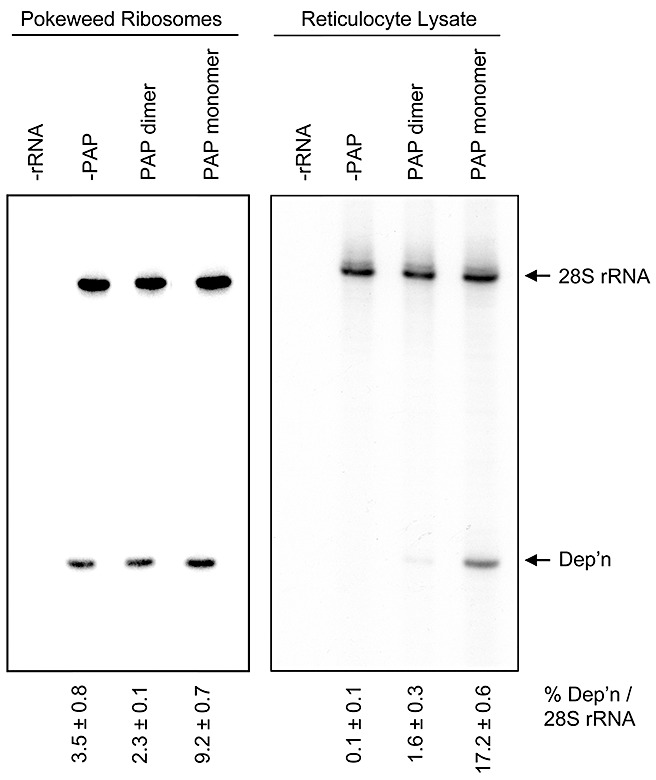
Depurination of rRNA by monomeric and complexed pokeweed antiviral protein (PAP). Isolated ribosomes of pokeweed and lysate of rabbit reticulocytes were incubated with monomeric or complexed PAP (5 nm) or buffer alone (–PAP). rRNA was isolated and primer extension analysis was performed using two end‐labelled primers, one annealing downstream of the expected depurination site (Dep’n) and the other annealing downstream of the 5′ end of 28S rRNA. cDNA products were separated on a 7 m urea/6% acrylamide gel, and extension patterns were visualized with a phosphorimager. –rRNA indicates extension reactions without rRNA template. The percentage depurination was estimated by the densitometry of band intensities of the Dep’n cDNA product relative to the 28S ribosomal cDNA product. Values represent means ± standard error for three independent experiments.
In vitro translation inhibition by PAP and complexed PAP
A decreased level of rRNA depurination suggested that the PAP dimer may be less toxic than the monomeric form. Given its comparably low enzyme activity, we tested the effect of the PAP dimer on the in vitro translation of poly(U) RNA in rabbit reticulocyte lysate. A synthetic poly(U) transcript was chosen as template for translation because we have shown previously that PAP can depurinate some mRNAs (Hudak et al., 2000), and we wished to examine the effect of dimeric relative to monomeric PAP on translation independent of RNA template. The level of [3H]‐phenylalanine incorporation was quantified by trichloroacetic acid (TCA) precipitation of total protein in each sample. As expected, the translation of poly(U) RNA was reduced with increasing concentration of monomeric PAP; however, the dimeric form of PAP did not substantially inhibit translation at these concentrations (Fig. 6). The level of rRNA depurination observed for reticulocyte ribosomes incubated with dimeric PAP was not sufficient to inhibit in vitro translation; therefore, the dimer represents a less toxic form of the protein.
Figure 6.
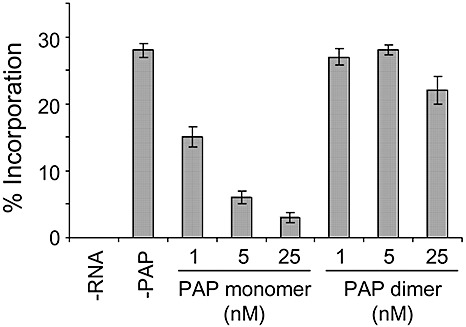
Translation inhibition by monomeric and complexed pokeweed antiviral protein (PAP). Poly(U) RNA was translated in rabbit reticulocyte lysate in the presence of increasing molar amounts of monomeric or complexed PAP and [3H]‐phenylalanine. –RNA indicates a translation reaction without mRNA and –PAP indicates a reaction without PAP. The protein products were precipitated in trichloroacetic acid (TCA) and quantified by liquid scintillation counting. Percentage incorporation refers to the amount of [3H]‐phenylalanine incorporated into protein relative to the total amount in the translation mix. Values represent means ± standard error for three independent experiments.
The control of enzyme activity by dimerization has been demonstrated for the ribotoxin α‐sarcin, an enzyme that hydrolyses the phosphodiester bond of the rRNA one nucleotide 3′ of the adenine that is removed by PAP (Endo and Wool, 1982). Although not a ribosome inactivating protein (i.e. N‐glycosidase), α‐sarcin forms a homodimer in solution that is stable during SDS‐PAGE (Sacco et al., 1983), similar to the PAP dimer reported here. The α‐sarcin homodimer is present as a minor form, in a 1:7 ratio relative to the monomeric form, and is enzymatically inactive (Cheung et al., 1996), characteristics shared with the PAP homodimer. Although the two proteins bear no sequence similarity to explain a common mechanism for dimerization, dimerization itself may represent a means by which cells protect their ribosomes from the toxic effects of these proteins during their synthesis.
Given that PAP has been hypothesized to function as an antiviral agent in pokeweed, the protein must be present in an active monomeric form during virus invasion. As much as 0.5% of the total soluble protein of the pokeweed plant consists of this antiviral protein, with the monomeric form stored after synthesis in the cell wall (1984, 1986). The dense concentration of the enzyme outside the cytosol is consistent with its proposed antiviral mechanism. The extracellular location prevents contact between PAP and ribosomes of healthy cells, yet provides a ready source of the enzyme in the event of viral infection. Unlike viruses that infect animals and gain access to cells by receptor‐mediated endocytosis, plant viruses rely on a physical breach of the cell wall and plasma membrane for introduction into cells. This is achieved either by a vector associated with transmission of the virus or simple mechanical damage to cells. We contend that this form of virus entry would also allow a substantial flow of PAP into the cytosol that would overwhelm the ability of the cell to dimerize each molecule. Given that PAP attacks ribosomes with a K cat value of 400 mol/mol/min (Ready et al., 1983), it is likely that protein synthesis would be inhibited in those cells invaded by virus.
The high affinity of ribosome inactivating proteins for their substrate rRNA, in combination with the substantial amount generally synthesized, suggests that protective mechanisms exist to deal with the small proportion of proteins that escape to the cytoplasm during processing. To our knowledge, this work with PAP is the first to describe, for any ribosome inactivating protein, that homodimerization occurs within the cytoplasm of living cells. The diminished enzyme activity, together with localization only to the cytosol, supports the biological role of homodimerization as a mechanism to limit toxicity to cells synthesizing PAP.
EXPERIMENTAL PROCEDURES
Pokeweed culture
Prior to sowing, pokeweed seeds were soaked in 98% sulphuric acid for 5 min, rinsed and imbibed with water for 6 h. Seeds were sown in Pro‐Mix BX medium and plants were fertilized once every 2 weeks with 0.5 g/L N20–P20–K20. Plants were raised in growth chambers (AC20; Biochambers, Winnipeg, Manitoba, Canada) at 25 °C/20 °C day/night temperature and 16 h day length with 180 µE/m2/s light intensity.
Cellular lysate preparation
Three leaf discs (1 cm in diameter) from pokeweed were homogenized with 200 µL of Buffer A [25 mm Tris‐HCl, pH 7.5, 1 mm ethyleneglycol‐bis(β‐aminoethylether)‐N,N′‐tetraacetic acid (EGTA), 1 mm dithiothreitol (DTT), 1 mm phenylmethylsulphonylfluoride (PMSF), 5% glycerol] in 1.5‐mL microtubes, and centrifuged at 16 110 g for 5 min. The supernatants were removed to new 1.5‐mL microtubes and the amount of total protein was quantified by the Bradford assay (Bradford, 1976).
Apoplast extraction
This protocol was adapted from the method of Hon et al. (1994). Briefly, pieces of leaves (1 cm2) from pokeweed were rinsed with deionized water and vacuum infiltrated in Buffer P (20 mm ascorbic acid, 20 mm CaCl2, pH 3.0) for 45 min. The leaf pieces were blotted dry and placed in small Miracloth (Calbiochem, San Diego, CA, USA) bags suspended in 1.5‐mL microtubes. The tubes were centrifuged at 134 g for 5 min and apoplastic fluid was collected from the bottom of each tube. Total protein was quantified by the Bradford assay (Bradford, 1976).
Protoplast isolation and microsome preparation
Protoplasts were isolated as described previously (Kroner et al., 1989). Briefly, the epidermis was removed from the abaxial surface of leaves. The peeled leaves were incubated in enzyme solution (10% cellulase, 0.5% macerozyme, 0.1% bovine serum albumin in 0.55 m mannitol) for 3 h in the dark at 25 °C with occasional agitation. Protoplasts were passed through one layer of Miracloth into Corex tubes, suspended in 4 mL of 0.55 m mannitol and centrifuged at 50 g for 5 min. The protoplast pellets were resuspended in 12 mL of 0.55 m mannitol, underlain with 4 mL of 20% sucrose, and centrifuged at 50 g for 5 min. Protoplasts were removed from the interphase and counted using a haemocytometer. To isolate proteins from the microsomal membrane fraction, protoplasts (2 × 105) were pelleted at 50 g for 5 min and resuspended in 3.0 mL of extraction buffer [250 mm sorbitol, 50 mm N‐2‐hydroxyethylpiperazine‐N′‐2‐ethanesulphonic acid (HEPES), pH 7.8, 10 mm ethylenediaminetetraacetic acid (EDTA), 2 mm PMSF] and vortexed vigorously to lyse the cells. Lysed cells were centrifuged at 250 000 g at 4 °C for 1 h, and the pellet was resuspended in 0.5 mL of extraction buffer followed by phenol–chloroform extraction. Proteins were recovered by precipitation with acetone, resuspended in protein storage buffer (100 mm NH4Cl, 50 mm HEPES, pH 7.8, 1 mm EDTA) and quantified by the Bradford assay (Bradford, 1976).
Purification of monomeric and complexed PAP from pokeweed
Leaves from pokeweed plants, harvested and stored at −80 °C, were homogenized without thawing in a blender with grinding buffer [50 mm Tris‐HCl, pH 7.5, 5 mmβ‐mercaptoethanol, 0.2 mm EDTA, 1% polyvinylpyrrolidone (PVP)] in a ratio of 100 g of leaves to 150 mL of grinding buffer. The leaf slurry was filtered twice through four layers of cheesecloth and then centrifuged at 8000 g for 10 min at 4 °C. The supernatant was brought to 60% saturation with ammonium sulphate and stirred for 3 h at 4 °C. This slurry was centrifuged at 15 000 g for 15 min at 4 °C, and the supernatant was brought to 100% saturation with ammonium sulphate and stirred overnight at 4 °C. The slurry was centrifuged at 20 000 g for 20 min at 4 °C, the pellet was resuspended in Q buffer (20 mm Tris‐HCl, pH 7.5, 1 mmβ‐mercaptoethanol, 0.2 mm EDTA) and dialysed against a total of 4 L of Q buffer (buffer was changed every 6–12 h). The dialysed sample was passed through a HiTrap Q FF column (GE Healthcare, Baie d'Urfe, Quebec, Canada) to remove negatively charged proteins. The collected flow through was loaded onto a HiTrap SP FF column (GE Healthcare) to bind all positively charged proteins. The column was washed with S buffer [10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.2, 0.2 mm EDTA, 0.1 mmβ‐mercaptoethanol] and a stepwise gradient of 100–500 mm NaCl was used to elute proteins in 2‐mL fractions. Fractions exhibiting peaks in UV280 reading were analysed for the presence of monomeric PAP or complexed PAP by 12% SDS‐PAGE, and concentrated with Centricon 30 centrifugation filters (Amicon, Billerica, MA, USA).
Mass spectrometry
Free and complexed PAP were separated by 12% SDS‐PAGE without prior boiling and stained with Coomassie brilliant blue. The protein bands at 29 and 58 kDa were excised from the gel, subjected to trypsin digestion and the peptide fragments were analysed by mass spectrometry by the Centre for Research in Mass Spectrometry, York University, Toronto, ON, Canada.
Bimolecular fluorescence complementation assay
cDNAs corresponding to mature PAP or PAPY123A were kindly provided by Nilgun Tumer, Rutgers University, New Brunswick, NJ, USA. Mature PAP is the processed form following cleavage of 22 and 29 amino acids from the N‐ and C‐termini, respectively (Hur et al., 1995). PAPY123A is a single amino acid mutant (Y123A) that exhibits reduced enzymatic activity (Hudak et al., 2004). These cDNAs were cloned upstream of the gfp gene in plasmids pGreen/ntGFP and pGreen/ctGFP encoding the N‐terminal half and C‐terminal half of GFP, respectively (kindly provided by Jean‐Francois Laliberté, INRS—Institut Armand‐Frappier, Laval, QC, Canada). pGreen constructs were transformed into pokeweed protoplasts according to Sheen (2002) with modifications. Briefly, plasmid DNA (20 µg) was gently mixed with 100 µL (∼4 × 104) protoplasts and 110 µL polyethyleneglycol (PEG)/Ca solution (200 mm mannitol, 100 mm CaCl2, 40% v/v PEG 4000; Fluka, Oakville, Ontario, Canada), and incubated for 20 min at room temperature. The mixture was diluted by the addition of 0.44 mL of W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 2 mm MES, pH 5.7) and centrifuged at 50 g for 1 min. Protoplasts were resuspended in 100 µL of W5 solution and transferred to 4‐cm Petri plates with 1.5 mL of incubation medium (0.55 m mannitol, 58.4 mm sucrose, 10 mm CaCl2, 1.0 mm KNO3, 1.0 mm MgSO4, 0.2 mm KH2PO4, 1.0 µm KI, 0.1 µm CuSO4, 0.01 mg/mL gentamicin sulphate). Protoplasts were incubated for 18 h under low light (25 µE/m2/s) at 23 °C and viewed on a confocal scanning laser microscope (Olympus Fluoview FV300, Olympus, Tokyo, Japan). An argon blue laser (488 nm) was used to observe GFP complementation. Images were generated using the Fluoview software package (Olympus).
Purification of recombinant proteins and cross‐linking in vitro
Escherichia coli (BL21) transformed with plasmids encoding histidine (His)‐tagged wild‐type PAP or PAPY123A was grown in Luria–Bertani (LB) medium with 100 µg/mL kanamycin at 37 °C. At an optical density at 600 nm (OD600) of 0.5, cells were induced by the addition of 400 µm isopropyl‐β‐d‐thiogalactopyranoside (IPTG) for 3 h. The cells were then centrifuged at 6000 g for 30 min, and the pellets were resuspended in buffer A (300 mm NaCl, 50 mm Tris‐HCl, pH 8.0, 5% glycerol), followed by lysis using a French press. Lysed cells were pelleted at 14 000 g for 30 min at 4 °C. The supernatant was loaded onto an Ni2+‐nitrilotriacetate (Ni‐NTA) resin and washed with buffer B (300 mm NaCl, 50 mm Tris‐HCl, pH 8.0, 10 mm imidazole, 5% glycerol), and samples were eluted with buffer C (300 mm NaCl, 250 mm imidazole, 50 mm Tris‐HCl, pH 8.0, 5% glycerol). Fractions containing PAP were pooled and concentrated using Centriplus 30 centrifugation filters (Amicon). For in vitro cross‐linking, His‐tagged wild‐type PAP or PAPY123A (0.25 µg) was incubated with either 0.05% glutaraldehyde or phosphate‐buffered saline (PBS) for 10 min at 4 °C. Following incubation, samples were separated through 12% SDS‐PAGE, transferred to nitrocellulose and probed for the presence of PAP using an anti‐PAP polyclonal antibody (1:5000).
Immunoblot analysis
Protein samples of equal amounts were separated by 12% SDS‐PAGE and transferred to nitrocellulose. The nitrocellulose was then blocked with 5% skimmed milk in phosphate‐buffered saline with 0.1% Tween‐20 (PBS‐T) for 2 h, and incubated overnight with anti‐PAP or anti‐GFP polyclonal antibody (1:5000) in 5% milk–PBS‐T. The blot was washed with PBS‐T and incubated with horseradish peroxidase‐conjugated goat anti‐rabbit immunoglobulin G (IgG) (1:10 000) in 5% milk–PBS‐T for 2 h. The blot was washed again, and PAP or GFP was visualized by chemiluminescence.
rRNA isolation and primer extension
Ribosomes of pokeweed were isolated as described previously (Tumer et al., 1997). Ribosomes (50 µg) or rabbit reticulocyte lysate (4 µL of 200 µg/µL total protein; Promega, Madison, WI, USA) was incubated with wild‐type PAP monomer or complexed PAP (5 nm), and incubated at 30 °C for 30 min in RIP buffer (60 mm KCl, 10 mm Tris‐HCl, pH 7.4, 10 mm MgCl2) in a final volume of 100 µL. Following incubation, an equal volume of 2 × extraction buffer (240 mm NaCl, 50 mm Tris‐HCl, pH 8.8, 20 mm EDTA, 2% SDS) was added. rRNA was extracted with phenol–chloroform and precipitated in 0.3 m NaOAc and ethanol. Depurination of rRNA (500 ng) was assessed by primer extension, as described previously (Hudak et al., 2000), with a tobacco 28S specific primer (5′‐CACGGTAGCTTCGCGCC‐3′) for the pokeweed rRNA samples and a human 28S specific primer (5′‐AGTCATAATCCCACAGATGGT‐3′) for the rabbit rRNA samples. A second primer, which annealed approximately 100 nucleotides 3′ of the 5′ end of each 28S rRNA (5′‐TTCACTCGCCGTTACTAAGG‐3′), was also extended and served as an internal control for RNA loading (Parikh et al., 2002).
In vitro translation with PAP or complexed PAP
In vitro translation of a synthetic transcript, poly(U) RNA (500 ng; Calbiochem), was performed in rabbit reticulocyte lysate (Promega) in the presence of wild‐type PAP monomer or complexed PAP at increasing concentrations (1, 5 or 25 nm). Samples were incubated at 30 °C for 1 h and the incorporation of [3H]‐phenylalanine was quantified by liquid scintillation counting of TCA‐precipitated protein.
ACKNOWLEDGEMENTS
The authors thank Dr Nilgun Tumer of Rutgers University, New Brunswick, NJ, USA for polyclonal antiserum to PAP and cDNAs of PAP and PAPY123A, and Dr Jean‐Francois Laliberté of INRS—Institut Armand‐Frappier, Laval, QC, Canada for plasmids pGreen/ntGFP and pGreen/ctGFP. Infrastructure support from the Canada Foundation for Innovation is gratefully acknowledged. This work was supported by the Natural Sciences and Engineering Research Council of Canada in Discovery Grants to KAH and KWMS, and a Premier's Research Excellence Award to KAH.
REFERENCES
- Ago, H. , Kataoka, J. , Tsuge, H. , Habuka, N. , Inagaki, E. , Noma, M. and Miyano, M. (1994) X‐ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding. Eur. J. Biochem. 225, 369–374. [DOI] [PubMed] [Google Scholar]
- Baykal, U. and Tumer, N.E. (2007) The C‐terminus of pokeweed antiviral protein has distinct roles in transport to the cytosol, ribosome depurination and cytotoxicity. Plant J. 49, 995–1007. [DOI] [PubMed] [Google Scholar]
- Bonness, M.S. , Ready, M.P. , Irvin, J.D. and Mabry, T.J. (1994) Pokeweed antiviral protein inactivates pokeweed ribosomes; implications for the antiviral mechanism. Plant J. 5, 173–183. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Carzaniga, R. , Sinclair, L. , Fordham‐Skelton, A.P. , Harris, N. and Croy, R.D.R. (1994) Cellular and subcellular distribution of saporins, type‐1 ribosome‐inactivating proteins, in soapwort (Saponaria officinalis L.). Planta, 194, 461–470. [Google Scholar]
- Cheung, J.I. , Wang, Y.R. and Lin, A. (1996) Substrate specificity of monomeric and dimeric alpha‐sarcin. FEBS Lett. 386, 60–64. [DOI] [PubMed] [Google Scholar]
- Corrado, G. , Delli Bovi, P. , Ciliento, R. , Gaudio, L. , Di Maro, A. , Aceto, S. , Lorito, M. and Rao, R. (2005) Inducible expression of a Phytolacca heterotepala ribosome‐inactivating protein leads to enhanced resistance against major fungal pathogens in tobacco. Phytopathology, 95, 206–215. [DOI] [PubMed] [Google Scholar]
- Desvoyes, B. , Poyet, J.L. , Schlick, J.L. , Adami, P. , Jouvenot, M. and Dulieu, P. (1997) Identification of a biological inactive complex form of pokeweed antiviral protein. FEBS Lett. 410, 303–308. [DOI] [PubMed] [Google Scholar]
- Endo, Y. and Wool, I.G. (1982) The site of action of alpha‐sarcin on eukaryotic ribosomes. The sequence at the alpha‐sarcin cleavage site in 28S ribosomal ribonucleic acid. J. Biol. Chem. 257, 9054–9060. [PubMed] [Google Scholar]
- Endo, Y. , Tsurugi, K. and Lambert, J.M. (1988) The site of action of six different ribosome‐inactivating proteins from plants on eukaryotic ribosomes: the RNA N‐glycosidase activity of the proteins. Biochem. Biophys. Res. Commun. 150, 1032–1036. [DOI] [PubMed] [Google Scholar]
- Gessner, S.L. and Irvin, J.D. (1980) Inhibition of elongation factor 2‐dependent translocation by the pokeweed antiviral protein and ricin. J. Biol. Chem. 255, 3251–3253. [PubMed] [Google Scholar]
- Hartley, M.R. and Lord, J.M. (1993) Structure, function and applications of ricin and related cytotoxic proteins In: Biosynthesis and Manipulation of Plant Products (Griesson D., ed.), pp. 210–239. New York: Chapman & Hall. [Google Scholar]
- Hon, W.C. , Griffith, M. , Chong, P. and Yang, D. (1994) Extraction and isolation of antifreeze proteins from winter rye (Secale cereale L.) leaves. Plant Physiol. 104, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak, K.A. , Wang, P. and Tumer, N.E. (2000) A novel mechanism for inhibition of translation by pokeweed antiviral protein: depurination of the capped RNA template. RNA, 6, 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak, K.A. , Parikh, B.A. , Di, R. , Baricevic, M. , Santana, M. , Seskar, M. and Tumer, N.E. (2004) Generation of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: evidence that ribosome depurination is not sufficient for cytotoxicity. Nucleic Acids Res. 32, 4244–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur, Y. , Hwang, D.J. , Zoubenko, O. , Coetzer, C. , Uckun, F.M. and Tumer, N.E. (1995) Isolation and characterization of pokeweed antiviral protein mutations in Saccharomyces cerevisiae: identification of residues important for toxicity. Proc. Natl. Acad. Sci. USA, 92, 8448–8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin, J.D. (1975) Purification and partial characterization of the antiviral protein from Phytolacca americana which inhibits eukaryotic protein synthesis. Arch. Biochem. Biophys. 169, 522–528. [DOI] [PubMed] [Google Scholar]
- Karran, R.A. and Hudak, K.A. (2008) Depurination within the intergenic region of Brome mosaic virus RNA3 inhibits viral replication in vitro and in vivo . Nucleic Acids Res. 36, 7230–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroner, P. , Richards, D. , Traynor, P. and Ahlquist, P. (1989) Defined mutations in a small region of the Brome Mosaic Virus 2α gene cause diverse temperature‐sensitive RNA replication phenotypes. J. Virol. 63, 5302–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, J.M. (1985) Precursors of ricin and Ricinus communis agglutinin. Glycosylation and processing during synthesis and intracellular transport. Eur. J. Biochem. 146, 411–416. [DOI] [PubMed] [Google Scholar]
- Mason, J.M. and Arndt, K.M. (2004) Coiled coil domains: stability, specificity, and biological implications. Chembiochem, 5, 170–176. [DOI] [PubMed] [Google Scholar]
- Monzingo, A.F. , Collins, E.J. , Ernst, S.R. , Irvin, J.D. and Robertus, J.D. (1993) The 2.5 Å structure of pokeweed antiviral protein. J. Mol. Biol. 233, 705–715. [DOI] [PubMed] [Google Scholar]
- Nielsen, K. , Payne, G.A. and Boston, R.S. (2001) Maize ribosome‐inactivating protein 1 has antifungal activity against Aspergillus flavus and Aspergillus nidulans . Mol. Plant–Microbe Interact. 14, 164–172. [DOI] [PubMed] [Google Scholar]
- Nilsson, L. and Nygård, O. (1986) The mechanism of the protein‐synthesis elongation cycle in eukaryotes. Effect of ricin on the ribosomal interaction with elongation factors. Eur. J. Biochem. 161, 111–117. [DOI] [PubMed] [Google Scholar]
- Parikh, B.A. , Coetzer, C. and Tumer, N.E. (2002) Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the alpha‐sarcin/ricin loop of rRNA. J. Biol. Chem. 277, 41 428–41 437. [DOI] [PubMed] [Google Scholar]
- Picard, D. , Kao, C.C. and Hudak, K.A. (2005) Pokeweed antiviral protein inhibits brome mosaic virus replication in plant cells. J. Biol. Chem. 280, 20 069–20 075. [DOI] [PubMed] [Google Scholar]
- Prestle, J. , Schonfelder, M. , Adam, G. and Mundry, K.‐W. (1992) Type 1 ribosome‐inactivating proteins depurinate plant 25S rRNA without species specificity. Nucleic Acids Res. 20, 3179–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready, M.P. , Bird, S. , Rothe, G. and Robertus, J.D. (1983) Requirements for antiribosomal activity of pokeweed antiviral protein. Biochim. Biophys. Acta, 740, 19–28. [DOI] [PubMed] [Google Scholar]
- Ready, M.P. , Adams, R.P. and Robertus, J.D. (1984) Dodecandrin, a new ribosome‐inactivating protein from Phytolacca dodecandra . Biochim. Biophys. Acta, 791, 314–319. [DOI] [PubMed] [Google Scholar]
- Ready, M.P. , Brown, D.T. and Robertus, J.D. (1986) Extracellular localization of pokeweed antiviral protein. Proc. Natl. Acad. Sci. USA, 83, 5053–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco, G. , Drickamer, K. and Wool, I.G. (1983) The primary structure of the cytotoxin α‐sarcin. J. Biol. Chem. 258, 5811–5818. [PubMed] [Google Scholar]
- Sheen, J. (2002) A transient expression assay using Arabidopsis mesophyll protoplasts. Available from URL: http://genetics.mgh.harvard.edu/sheenweb/ (accessed on Jun 14, 2010).
- Stirpe, F. , Bailey, S. , Miller, S.P. and Bodley, J.W. (1988) Modification of ribosomal RNA by ribosome‐inactivating proteins from plants. Nucleic Acids Res. 16, 1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B.E. and Irvin, J.D. (1990) Depurination of plant ribosomes by pokeweed antiviral protein. FEBS Lett. 273, 144–146. [DOI] [PubMed] [Google Scholar]
- Taylor, S. , Massiah, A. , Lomonossoff, G. , Roberts, L.M. , Lord, J.M. and Hartley, M. (1994) Correlation between the activities of five ribosome‐inactivating proteins in depurination of tobacco ribosomes and inhibition of tobacco mosaic virus infection. Plant J. 5, 827–835. [DOI] [PubMed] [Google Scholar]
- Tumer, N.E. , Hwang, D.‐J. and Bonness, M. (1997) C‐terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci. USA, 94, 3866–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubenko, O. , Uckun, F. , Hur, Y. , Chet, I. and Tumer, N. (1997) Plant resistance to fungal infection induced by nontoxic pokeweed antiviral protein mutants. Nat. Biotechnol. 15, 992–996. [DOI] [PubMed] [Google Scholar]


