SUMMARY
Phelipanche ramosa L. parasitizes major crops, acting as a competitive sink for host photoassimilates, especially sucrose. An understanding of the mechanisms of sucrose utilization in parasites is an important step in the development of new control methods. Therefore, in this study, we characterized the invertase gene family in P. ramosa and analysed its involvement in plant development. Invertase‐encoded cDNAs were isolated using degenerate primers corresponding to highly conserved regions of invertases. In addition to enzyme assays, gene expression was analysed using real‐time quantitative reverse transcriptase‐polymerase chain reaction during overall plant development. The dominant isoform was purified and sequenced using electrospray ionization‐liquid chromatography‐tandem mass spectrometry (ESI‐LC‐MS/MS). Five invertase‐encoded cDNAs were thus characterized, including PrSai1 which encodes a soluble acid invertase (SAI). Of the five invertases, PrSai1 transcripts and SAI activity were dominant in growing organs. The most active invertase corresponded to the PrSai1 gene product. The purified PrSAI1 displayed low pI and optimal pH values, specificity for β‐fructofuranosides and inhibition by metallic ions and competitive inhibition by fructose. PrSAI1 is a typical vacuolar SAI that is actively involved in growth following both germination and attachment to host roots. In addition, germinated seeds displayed enhanced cell wall invertase activity (PrCWI) in comparison with preconditioned seeds, suggesting the contribution of this activity in the sink strength of infected roots during the subsequent step of root penetration. Our results show that PrSAI1 and, possibly, PrCWI constitute good targets for the development of new transgenic resistance in host plants using proteinaceous inhibitors or silencing strategies.
INTRODUCTION
Broomrapes consist of almost 170 species devoid of chlorophyll and are thereby obligate parasitic plants. Seven species, including Phelipanche ramosa L. Pomel (syn. Orobanche ramosa L.), are serious parasitic weeds that cause great losses in many economically important crops around the Mediterranean Sea and in Asia Minor (Parker, 2009). Phelipanche ramosa attacks Solanaceae crops most severely and is well established in long winter crops (e.g. rapeseed) as well as in shorter spring and summer crops (hemp, melon, tobacco, tomato). Success in controlling broomrape is rare (Rispail et al., 2007) and there is an urgent need to continue to screen genetic resources and to characterize resistance mechanisms (Pérez‐de‐Luque et al., 2009). An understanding of the physiological and metabolic features of the parasite is particularly pertinent, because new control methods that target parasite physiology or metabolism are highly desirable (Aly et al., 2009).
Broomrape development occurs in five stages (Labrousse et al., 2001): germination (stage I), which is elicited by stimulants secreted by the roots of host plants; attachment to the host vascular system (stage II) through a haustorium containing both xylem and phloem that serves as both an attaching organ and a bridge for water and nutrient transfer from the host; establishment and growth of a tubercle (stage III), which gives rise to a subterranean shoot (stage IV) and then to a flowering spike after emergence from the soil (stage V). Via the haustorium, the parasite connects to the phloem sieve elements of the host plant (Hibberd and Jeschke, 2001), and thereby becomes a major competitive sink for photoassimilates. Sucrose has long been known to be the main host‐derived compound (Aber et al., 1983). However, there is evidence, notably in P. ramosa, that sucrose is not accumulated in the parasite, but is utilized for growth and for the accumulation of hexoses and, to a lesser extent, mannitol and starch (Abbes et al., 2009; Delavault et al., 2002; Singh et al., 1968). Broomrape displays both low transpiration rates and low water flux in xylem elements (Hibberd et al., 1999); osmotic adjustment through the strong accumulation of potassium and soluble sugars is thus considered to be an essential process in water movement from the host plant, and thus growth of the parasite (Abbes et al., 2009).
Considering the major role of soluble carbohydrates in osmotic adjustment and the sink strength of the parasite, there is a need to better understand the primary steps of sucrose allocation, which consist of the degradation of host‐derived sucrose. Sucrose degradation is catalysed by two types of enzyme in plants: invertases (EC 3.2.1.26) and sucrose synthases (SuSys) (EC 2.4.1.13). SuSys are glycosyl transferases which convert sucrose into fructose and UDP‐glucose in the presence of uridine diphosphate (UDP), whereas invertases hydrolyse sucrose into glucose and fructose. Sucrose‐degrading enzymes play multiple pivotal roles in plants: sugar transport and partitioning, cell turgor and expansion, response to insect wounding and pathogen infection, sugar signalling and plant development (Roitsch and Gonzalez, 2004; Sturm, 1999; Sturm and Tang, 1999). On the basis of their solubility, subcellular localization, pH optima and isoelectric point, three types of invertase isoenzyme have been identified: acid invertases (AIs), including soluble/vacuolar isoforms (soluble acid invertase, SAI) and insoluble/cell wall‐bound isoforms (cell wall invertase, CWI), and neutral/cytosolic invertases (soluble neutral invertase, SNI). Both vacuolar and cell wall isoenzymes are β‐fructofuranosidases which share a high degree of overall sequence homology. They correspond to acid and glycosylated invertases, showing an acidic pH optimum, and accept other fructofuranosides, such as stachyose and raffinose, as substrates. Vacuolar isoforms control sugar composition in the vacuole and the remobilization of sucrose for metabolic processes, and CWI isoforms are ionically bound to the cell wall and coordinate phloem unloading in sink cells with hexose transporters. Moreover, the sucrose gradient mediated by CWI at sites of phloem unloading is crucial for the normal development of sink organs (Zanor et al., 2009). SNI proteins are unglycosylated proteins and have neutral/alkaline pH optima and high specificity for sucrose. For a long time, SNI isoenzymes had only been described in the cytosol, where they play a housekeeping role and maintain hexose concentrations. Recent work has demonstrated that cytosolic SNIs are essential for normal growth and that mitochondrial and chloroplastic isoforms contribute to intracellular carbon trafficking (Barratt et al., 2009; Vargas et al., 2008). Similarly, SuSys are located in the cytosol and in mitochondria and possibly in plastids as well (Hennen‐Bierwagen et al., 2009). Since SuSy is commonly associated with glucan synthesis, it is a key regulator of starch accumulation and thickening of secondary cell walls in the xylem (Baroja‐Fernández et al., 2009; Coleman et al., 2009).
Information on the metabolic and molecular bases of parasitic plant–plant interactions is rare. Glucose and fructose are strongly accumulated in broomrape from host‐derived sucrose, suggesting that invertases are the dominant sucrose‐degrading enzyme in the parasite (Abbes et al., 2009; Delavault et al., 2002). The present study characterizes the invertase family of the parasite P. ramosa at both molecular and biochemical levels, and thereby its involvement in the development of the parasitic plant. Patterns of expression of the different invertase‐encoding genes and of SAI, SNI and CWI activities were analysed during germination and post‐attachment stages of broomrape development. In addition, the most active invertase isoform was purified and partially sequenced. We characterized the gene encoding this dominant invertase and its associated isoform, which are closely involved in broomrape development before and after attachment to the host plant. This work provides a better understanding of the interaction and opens up new perspectives in host resistance engineering.
RESULTS
Cloning and characterization of P. ramosa invertase cDNAs
Five partial P. ramosa cDNAs were obtained by reverse transcriptase‐polymerase chain reaction (RT‐PCR) using sets of primers designed from conserved regions of plant invertase sequences. Using rapid amplification of cDNA ends (RACE) strategies, four full‐length cDNAs, PrSai1, PrCwi, PrSni1 and PrSni2 (accession numbers: GU997130, GU99132, GU99133 and GU99134, respectively), and one partial cDNA, PrSai2 (accession number: GU997131), were isolated. The characteristics of P. ramosa invertase cDNAs and the corresponding protein are presented in Table 1.
Table 1.
Characteristics of Phelipanche ramosa invertase cDNAs and their corresponding proteins.
| Gene | cDNA (bp) | 5′‐UTR (bp) | ORF (bp) | 3′‐UTR (bp) | Protein (kDa) | pI (theor.) | Identity (%) | Accession number | |
|---|---|---|---|---|---|---|---|---|---|
| PrSai1 | 2160 | 62 | 1986 | 112 | 73.1 | 5.42 | 65.6 | Nicotiana tabacum | CAC83577 |
| 64.6 | Daucus carota | CAA77267 | |||||||
| PrSai2 * | 1711 | — | — | 74 | — | — | 71 | D. carota | CAA77266 |
| 65.4 | Camellia sinensis | BAF34362 | |||||||
| PrCwi | 2187 | 40 | 1761 | 386 | 66.2 | 8.68 | 66.6 | Lycopersicon esculentum | AAM28823 |
| 63.8 | D. carota | CAA55188 | |||||||
| PrSni1 | 2032 | 115 | 1710 | 207 | 64.8 | 6.72 | 80.9 | L. esculentum | ABQ28669 |
| 78.6 | Vitis vinifera | CAP59642 | |||||||
| PrSni2 | 2445 | 67 | 2001 | 377 | 76.4 | 7.16 | 69 | V. vinifera | ABS52644 |
| 67.9 | D. carota | CAA76145 |
The percentage of identity is that with the closest annotated homologue.
Not full‐length.
ORF, open reading frame; Pr, Phelipanche ramosa; UTR, untranslated region.
Three distinct cDNAs encoding AIs were identified. The amino acid sequence encoded by PrSai1 has 65.6% identity to a Nicotiana tabacum vacuolar invertase (CAC83577). The partial PrSai2‐encoded sequence shares 71% identity with a Daucus carota SAI (CAA77266) (Sturm, 1996). The PrCwi‐encoded sequence shares 66.6% identity with a Solanum lycopersicum CWI (AAM28823) (Fridman and Zamir, 2003). All three deduced proteins contain the conserved motifs of plant AIs, including a β‐fructosidase motif ‘DPNG’ and a putative catalytic domain ‘WEC(I/V/P)D’ (Roitsch et al., 1995; Sturm and Chrispeels, 1990). The plant fructan‐metabolizing fructosyltransferases share motifs that are conserved in AIs (Vijn and Smeekens, 1999). Nevertheless, in contrast with plant fructosyltransferases, AIs contain the amino acid triplet WIN or WMN (Gallagher et al., 2004). Thus, the presence of the WIN triplet in the PrSai1‐ and PrCwi‐encoded sequences (residues 139–141 and 57–59, respectively) and WMN in the PrSai2‐encoded sequence confirmed that the three genes encoded AIs. As shown in Fig. 1, the PrSai1‐encoded protein has a closer evolutionary relationship to vacuolar invertases (SAIs) than to apoplasmic isoforms, whereas the protein encoded by PrCwi belongs to the CWI family. The partial PrSai2‐encoded sequence was also closely related to the SAI family. Three features in the three P. ramosa invertase‐encoded sequences also supported this classification: (i) the fourth residues of the conserved catalytic domain WEC(V/I/P)D in PrSai1‐ and PrSai2‐encoded proteins were isoleucine and valine, respectively, present in vacuolar invertases; the corresponding amino acid residue in the PrCwi‐encoded protein was proline, a characteristic feature of CWI (Roitsch et al., 1995); (ii) the calculated pI values of the PrSai1‐ and PrCwi‐encoded proteins were 5.42 and 8.68, respectively (Table 1), in accordance with previous observations indicating that vacuolar invertases display acidic pI values and CWIs show basic pI values (Roitsch and Gonzalez, 2004; Sturm, 1999); (iii) the TargetP 1.1 program strongly predicted that the PrCwi‐encoded protein is exported to the apoplast, in accordance with the assumed apoplastic location of PrCwi.
Figure 1.
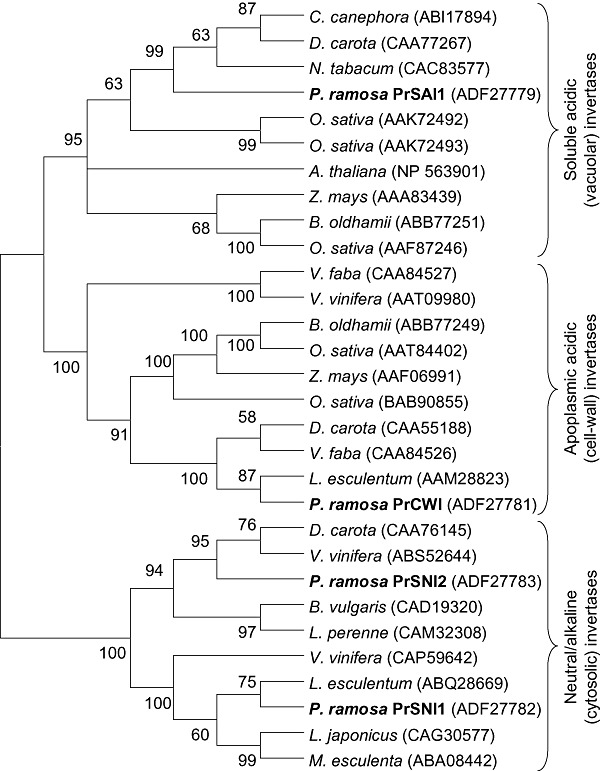
Dendrogram of invertase amino acid sequences from various plants including sequences deduced from Phelipanche ramosa full‐length invertase genes. The neighbour‐joining consensus tree was inferred from 500 bootstrap replicates. Branches occurring in less than 50% bootstrap replicates were collapsed.
In addition, two distinct cDNAs encoding putative neutral/alkaline invertase isoforms were isolated. PrSni1‐ and PrSni2‐encoded proteins can be classified as neutral invertases. The deduced amino acid sequences were indeed highly homologous to neutral/alkaline invertases from other plants (Table 1). The full‐length sequence encoded by PrSni1 shows 80.9% identity with a Solanum lycopersicum invertase (ABQ28669) and 78.6% identity with a Vitis vinifera putative neutral invertase (CAP59642). The full‐length sequence encoded by PrSni2 shares 69% identity with a V. vinifera neutral invertase (ABS52644) and 67.9% identity with a D. carota neutral invertase (CAA76145) (Sturm et al., 1999). As shown in Fig. 1, PrSni1‐ and PrSni2‐encoded proteins have a closer evolutionary relationship to neutral/alkaline (cytoplasmic) invertases than to acidic (apoplasmic or vacuolar) invertases. In addition, the pI values of PrSni1‐ and PrSni2‐encoded proteins were calculated to be 6.72 and 7.16, respectively (Table 1), in agreement with previous observations that neutral invertases have neutral pI values (Roitsch and Gonzalez, 2004).
Development‐related changes in the transcript levels of invertase‐encoding genes
The accumulation of transcripts encoding different putative invertases was expressed relative to the level of transcripts of the constitutive elongation factor 1‐α gene (EF1α) (Fig. 2a,c). Regardless of the invertase gene assayed, preconditioned seeds displayed low transcript accumulation (Fig. 2a). Transcript accumulation remained low in germinated seeds for PrSai2, PrSni1, PrSni2 and PrCwi genes. In contrast, large amounts of PrSai1 transcripts were found in germinating seeds developing a radicle. Similarly, whatever the organ or stage of development after attachment to host roots, transcript accumulation of PrSai2, PrSni1, PrSni2 and PrCwi was low in comparison with the high PrSai1 transcript levels (Fig. 2c). PrSni1 showed the lowest transcript levels. Moreover, the results showed that PrSai1 transcripts accumulated particularly in growing subterranean shoots and emerged flowering shoots (apices, FS.V). The base of flowering shoots (Bp.V) displayed significantly lower levels of PrSai1 transcripts. PrSai1 transcripts also accumulated substantially in fruits containing developing seeds (F.V).
Figure 2.
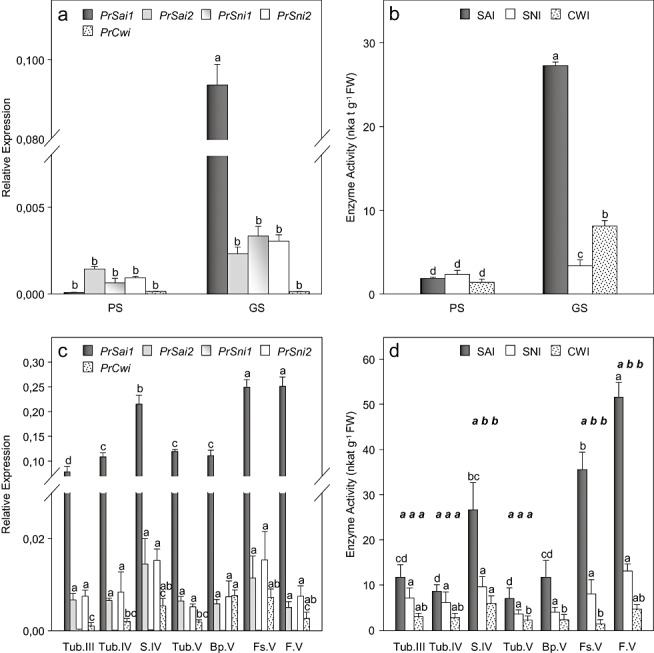
Development‐related changes in the levels of invertase‐encoded gene transcripts and invertase activities in Phelipanche ramosa: (a, b) independent development following seed germination; PS, preconditioned seeds; GS, germinated seeds; (c, d) parasitic stages following attachment to tomato roots. See Fig. 5 for abbreviations of the various plant organs. (a, c) Transcript accumulation expressed relative to elongation factor 1‐α gene (EF1α) transcript levels. Data are means ± standard error (n= 3). For each gene, values with the same letter are not significantly different (anova, SNK test, P < 0.05). (b, d) Invertase activities. Data are means ± standard error (n= 6). For each isoform, activities with the same letter (plain typeface) are not significantly different (anova, SNK test, P < 0.05). For each stage of development and organ, activities with the same letter (italic and bold typeface) are not significantly different (anova, SNK test, P < 0.05).
Development‐related changes in invertase activities and in situ localization of SAI activity
Activities were measured in preconditioned and germinated seeds and in different organs of the subterranean and emerged broomrape (Fig. 2b,d). SAI, SNI and CWI activities were low in preconditioned seeds, and increased following germination by factors of 14‐, 1.5‐ and five‐fold, respectively (Fig. 2b). Once the parasite had attached to host roots, levels of CWI, SNI and SAI activities in tubercles were similar and did not change significantly during overall plant development (Fig. 2d). In contrast, enzyme activity increased substantially in both subterranean shoots (S.IV) and at the apices of emerged flowering shoots (FS.V), primarily as a result of a large increase in SAI activity (Fig. 2d). Fruits containing developing seeds (F.V) displayed the highest SAI activity, whereas the base of emerged flowering shoots (Bp.V) showed low SAI activity. In situ AI activities were visualized in cross‐sections of basal and apical parts of emerged flowering shoots (Fig. 3). Activity was confined to the medullar parenchyma in basal parts, but extended to the cortical parenchyma in the apices (Fig. 3a2,b2). Staining on fixed sections was attributed to SAI (Sergeeva and Vreugdenhil, 2002), because no CWI activity was revealed on unfixed sections (data not shown). The higher SAI activity observed in the apices of emerged shoots may be related to the more specific localization of SAI activity in these young, growing tissues (2, 3). SAI staining from germinated seeds also showed high SAI activity in the radicle apex (Fig. 3d).
Figure 3.
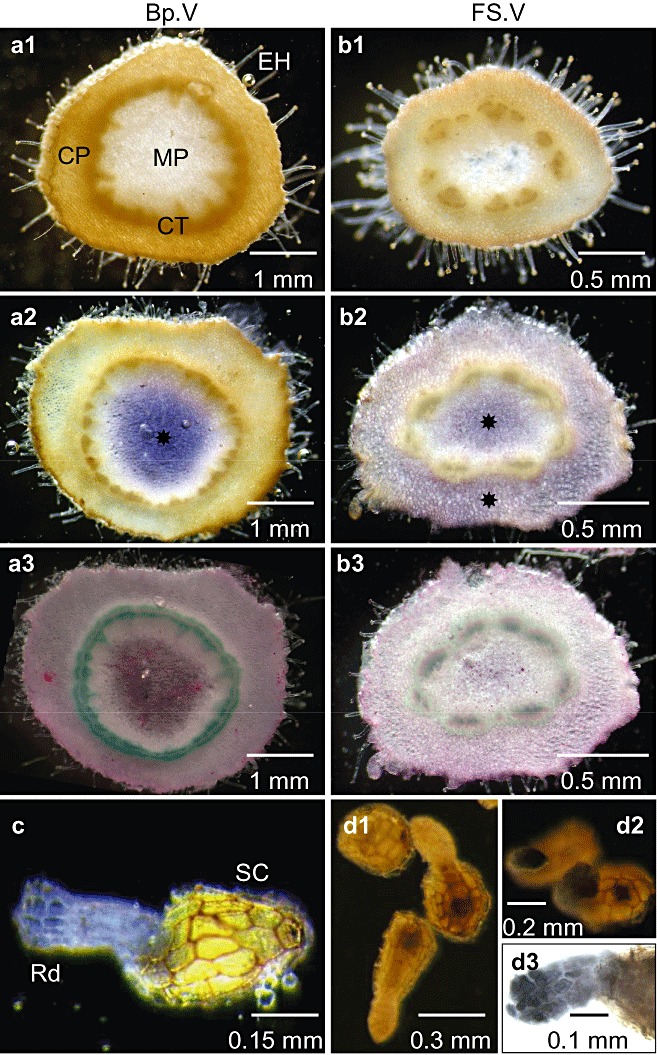
Tissue localization of soluble acid invertase (SAI) activity in seeds and flowering shoots of Phelipanche ramosa parasitizing tomato plants. (a1, b1) Control cross‐sections of basal (Fp.V) and apical (FS.V) shoots incubated after fixation in reaction medium without sucrose. (a2, b2) Assay sections incubated after fixation in a reaction mixture containing sucrose. SAI activity ( ) is shown in blue in medullar and cortical parenchyma. No cell wall invertase (CWI) activity was detected on unfixed sections (data not shown). Staining on fixed sections was thus attributed to SAI. (a3, b3) Post‐activity staining of cellulosic (purple) and lignified (green) tissues using Mirande's reagent. (c) Germinated seeds incubated in the reaction mixture containing sucrose. (d1) NTB staining of seeds without sucrose in reaction medium. (d2) NTB staining of seeds with sucrose in reaction medium. (d3) Zoom of the radicle region stained with NTB: accumulation of reducing sugars produced from SAI‐mediated hydrolysis of sucrose is shown in black in the apex of the radicle. CP, cortical parenchyma; CT, conductive tissues; EH, epidermal hairs; MP, medullar parenchyma; NTB, nitrotetrazolium blue; Rd, radicle; SC, seed coat.
) is shown in blue in medullar and cortical parenchyma. No cell wall invertase (CWI) activity was detected on unfixed sections (data not shown). Staining on fixed sections was thus attributed to SAI. (a3, b3) Post‐activity staining of cellulosic (purple) and lignified (green) tissues using Mirande's reagent. (c) Germinated seeds incubated in the reaction mixture containing sucrose. (d1) NTB staining of seeds without sucrose in reaction medium. (d2) NTB staining of seeds with sucrose in reaction medium. (d3) Zoom of the radicle region stained with NTB: accumulation of reducing sugars produced from SAI‐mediated hydrolysis of sucrose is shown in black in the apex of the radicle. CP, cortical parenchyma; CT, conductive tissues; EH, epidermal hairs; MP, medullar parenchyma; NTB, nitrotetrazolium blue; Rd, radicle; SC, seed coat.
Figure 2 highlights the strong similarity between the patterns of PrSai‐1 expression and SAI activity. Figure S1 (see Supporting Information) illustrates the strong relationship between SAI activity (X) and PrSai1 transcript levels (Y= 168.14X; R 2= 0.78).
SAI purification, characterization and sequencing
SAI proteins were purified from growing subterranean shoots (S.IV) and partially sequenced following trypsin digestion to verify that the PrSai1 gene encodes the most active SAI isoform. The latter was highly purified by a factor of 700‐fold with 2% of activity recovery (Table S3, see Supporting Information). The purified SAI displayed an apparent molecular mass of 84 kDa in native conditions and 35 kDa in denaturing conditions (Table 2). The identity of purified SAI was demonstrated on native gels by both activity staining and immunoblotting (Fig. S2, see Supporting Information). As shown by liquid isoelectric focusing experiments and enzyme assays in the pH range 2.0–8.0, purified SAI displayed the expected pI and optimal acidic pH (4.63 and 4.5, respectively) (Table 2). When assayed at pH 4.5 and at temperatures ranging from 20 to 80 °C, activity was optimal at 50 °C. Purified SAI was active on stachyose, raffinose and, especially, sucrose, where it showed very high activity and affinity. No activity was detected with any other carbohydrates tested as substrate. Fructose inhibited SAI activity in a linear competitive manner (K i= 1.63 mm), whereas glucose acted as a linear noncompetitive inhibitor (K i= 12 mm). SAI characterization was carried out by testing mineral ions as potential inhibitors (Table 2). Partial inhibition was shown with Ag+, Cu2+, K+, Na+, Mg2+ and Pb2+ ions, and SAI activity was affected strongly by Hg2+.
Table 2.
Characteristics of the soluble acid invertase PrSAI1 purified from growing subterranean shoots of Phelipanche ramosa parasitizing tomato plants.
| Kinetic properties | |
| pI | 4.6 ± 0.1 |
| Optimal pH | 4.5 ± 0.1 |
| Optimal temperature (°C) | 50 ± 3 |
| Molecular mass (kDa) | |
| Native form | 86 ± 4 |
| Denatured form | 35 ± 2 |
| Substrate specificity (mm) | |
| K m(sucrose) | 2.7 ± 0.3 |
| K m(stachyose) | 16.9 ± 2.4 |
| K m(raffinose) | 75.6 ± 5.5 |
| K m(cellobiose, lactose, maltose, melezitose, melibiose, methyl‐glucose, trehalose, verbascose) | — |
| Inhibition | |
| K i(fructose) (mm) | 1.6 ± 0.1 |
| K i(glucose) (mm) | 12.0 ± 0.4 |
| MnCl2, CoCl2 (1 mm, % control) | 110 ± 3.1 |
| CaCl2 (1 mm, % control) | 86 ± 6.1 |
| AgNO3, KCl, NaCl, MgCl2 (1 mm, % control) | 70.5 ± 2.4 |
| CuSO4, ZnSO4, PbNO3 (1 mm, % control) | 48.7 ± 7.3 |
| HgCl2 (1 mm, % control) | 9 ± 2.0 |
Values are means ± standard error (n= 3).
The purified SAI protein was extracted from the band showing both SAI staining and immunoblotting on native gels, and then sequenced using electrospray ionization‐liquid chromatography‐tandem mass spectrometry (ESI‐LC‐MS/MS). Trypsin digestion generated 14 peptides, which were aligned on the deduced peptide sequence of the PrSai1 gene (Fig. 4). These peptides cover 26% of the deduced sequence, attesting that PrSai1 encodes for the SAI purified in the present study.
Figure 4.

Amino acid sequence as deduced from the PrSai1 gene and amino acid sequences of the peptides generated by trypsin digestion of the soluble acid invertase (SAI) isoform purified from the subterranean shoots of Phelipanche ramosa parasitizing tomato plants. Peptides matching the putative P. ramosa acid invertase sequence are shaded, and tryptic cleavage sites are indicated by arrows. The β‐fructofuranosidase motif and a highly conserved peptide domain with a catalytic cysteine residue are underlined (Sturm, 1999).
DISCUSSION
The present study was designed to both identify and characterize the invertase genes and isoforms that determine sucrose utilization during broomrape development, using P. ramosa as a model. We covered the major steps of parasite development, starting from the independent germinating seedling to the ripening parasitic plant after attachment to host roots and emergence from the soil.
The invertase gene family in P. ramosa
Because information on the broomrape genome was scarce when the present study started, we set out to isolate cDNAs encoding invertases from P. ramosa using traditional degenerate primers corresponding to highly conserved regions of plant SAI, CWI and SNI genes (Tables S1 and S2, see Supporting Information). The first group of sequences characterized encoded AIs, including three cDNAs, PrSai1, PrSai2 and PrCwi. Based on their close evolutionary relationship to vacuolar invertases or CWIs (Fig. 1), the identity of the fourth residue of the conserved catalytic domain (WEC(V/I/P)D) of plant invertases (Roitsch et al., 1995), the pI value (Roitsch and Gonzalez, 2004; Sturm, 1999) and the predicted apoplastic location, the cDNAs PrSai1 and PrSai2 encode vacuolar isoforms, whereas the PrCwi‐encoded protein belongs to the CWI isoform. The second group of sequences isolated encoded soluble neutral/alkaline invertases, including two cDNAs, PrSni1 and PrSni2. In parallel with our study, the Parasitic Plant Genome Project (PPGP) sequenced cDNAs representing transcripts from key life stages of three members of the Orobanchaceae, including P. aegyptiaca (syn. Orobanche aegyptiaca), a close relative of P. ramosa (http://ppgp.huck.psu.edu). The PrSai1, PrCwi, PrSni1 and PrSni2 genes share 96.7%, 86.7%, 97.9% and 95.5% identity with four partial sequences encoding putative invertases in P. aegyptiaca: OrAeBC3‐2697, OrAEBC3‐21925, OrAEBC3‐35083 and OrAEBC3‐36350, respectively. Other CWI‐ and SNI‐encoding cDNAs exhibiting lower identity could be identified in P. aegyptiaca, suggesting the occurrence of other invertase genes in P. ramosa. Furthermore, the sequence OrAeBC3‐2783 encoding the key enzyme in mannitol production, mannose 6‐phosphate reductase (EC 1.1.1.224), shares 99.3% identity with the PrM6PR sequence from P. ramosa (Aly et al., 2009; Delavault et al., 2002). These data highlight the strong identity of nucleic acid sequences from both species, and the PPGP database will clearly facilitate future studies on P. ramosa and probably other broomrape species as well.
SAI isoform expression in P. ramosa is simple, with a typical dominant PrSai1‐encoded isoform
Multiple SAI isoforms have been reported in many species. For example, three of five SAI isoforms suspected in mature leaves of Arabidopsis thaliana have been characterized and their properties compared (Tang et al., 1996). They exhibited similar pI, pH and temperature optima and molecular mass values. Further, sucrose is their preferred substrate with a K m for sucrose varying from 5 to 12 mm, according to the isoform. To date, only two genes, Atβfruct3 and Atβfruct4, encoding vacuolar invertases have been identified in A. thaliana (Haouazine‐Takvorian et al., 1997). Heterogeneity in SAI isoforms is better understood in other species and involves the expression of different Sai genes (e.g. carrot; Unger et al., 1994) or post‐translational modification of a single gene product (e.g. radish, tomato; Elliott et al., 1993; Faye et al., 1986). As mentioned above, P. ramosa exhibits two genes, PrSai1 and PrSai2, that encode putative vacuolar isoforms (Table 1). Nevertheless, the pattern of expression of SAI isoforms is simple in growing shoots: only one SAI isoform was detected. This finding suggests that the second SAI isoform is not active in shoots, or only has a low basal activity that could not be detected on purification. In addition, patterns of PrSai1 and PrSai2 gene expression indicate that PrSai1 transcripts accumulate much more strongly than do PrSai2 transcripts (Fig. 2a,c). The purified SAI isoform shares high identity with the deduced peptide sequence of the highly expressed PrSai1 gene (Fig. 4). We thus assume that it corresponds to the PrSai1‐encoded protein, therefore called ‘PrSAI1’. Furthermore, PrSai1 transcript levels and SAI activity correlated significantly with overall parasite development (Fig. S1), indicating that SAI activity can be attributed to PrSAI1 in all organs.
PrSAI1 was visualized in native conditions by activity staining on polyacrylamide gels or by immunoblotting using an antibody raised against grape vacuolar invertase (Fig. S1). The molecular mass of the PrSai1‐deduced protein (73 kDa) is lower than that of the native PrSAI1 (86 kDa, Table 2), which is in agreement with the fact that the native form is glycosylated. This finding also demonstrates that PrSAI1 is monomeric. Accordingly, multiple SAI forms varying from 30 to 220 kDa have been detected in plants, but monomeric or dimeric forms of 50–70 kDa predominate (Sturm, 1999).
PrSAI1 also exhibited kinetic features typical of many other SAIs. Of all the carbohydrates tested as potential substrates, PrSAI1 was active only on β‐fructofuranosides, including sucrose, stachyose and raffinose (Table 2), and was competitively inhibited by fructose, confirming that PrSAI1 is a β‐fructofuranosidase, as generally reported for invertases (Sturm, 1999). PrSAI1 exhibited a K m value for sucrose in a lower millimolar range than for raffinose and stachyose, demonstrating that sucrose is the preferred substrate. The optimal temperature of PrSAI1 was high (50 °C), as demonstrated for other plant SAIs (Marouf and Zeki, 1982; Miller and Ranwala, 1994). PrSAI1 activity was inhibited more or less strongly by metallic ions (Table 2). As generally observed in plant SAIs, PrSAI1 exhibited high sensitivity to Hg2+, suggesting the presence of a sulphydryl group at the catalytic site of the enzyme (Sturm, 1999).
Sucrose utilization is mediated mainly by PrSAI1 during both host‐independent and post‐attachment development
In the work presented here, the expression of the five genes encoding invertases and the activities of SAI, SNI and CWI isoforms were followed in both preconditioned and germinated seeds of P. ramosa (Fig. 2a,b). Although the transcript levels and activities were low in preconditioned seeds, both PrSai1 transcript levels and putative PrSAI1 activities increased greatly in seedlings after germination. The gene PrSai1 was by far the most expressed invertase‐encoding gene in germinated seeds, where PrSAI1 activity also predominated, suggesting that sucrose degradation for radicle growth during the mobilization of seed reserves is controlled mainly by PrSAI1. This was confirmed by the high PrSAI1 activity detected at the radicle apex (Fig. 3d3). Considering that broomrape seeds store essentially lipids as reserves, starch being very uncommon (Bar‐Nun and Mayer, 2002; Velasco et al., 2000), additional investigations on the mobilization of the lipid reserves in broomrape seeds are recommended. Most of the germination stimulants identified so far are strigolactones (Yoneyama et al., 2010). Recent studies on arbuscular mycorrhizal fungi have shown that strigolactones stimulate mitochondrial metabolism and catabolic processes, such as β‐oxidation of fatty acid degradation (Besserer et al., 2008; Bücking et al., 2008). Accordingly, we speculate that lipids are converted into sucrose, followed by PrSAI1‐mediated sucrose utilization, in young P. ramosa seedlings following germination. However, this does not preclude PrSAI1 involvement in gentianose hydrolysis, which is stimulated in germinating broomrape seeds (e.g. A. Okazawa, Graduate School of Engineering, Osaka University, 2‐1 Yamadaoka, Suita, Osaka, Japan, unpublished data).
Voegele et al. (2006) have reported that, on penetration into the host faba bean leaf, the rust fungus Uromyces fabae establishes new sinks which compete with other occurring sink organs through strong expression of the Uf‐INV1 gene encoding an apoplastic invertase in haustoria. Thus, the fungal invertase activity might limit sucrose export from the infected leaf and might condition the tissue for a conversion from source to sink. Such alterations in translocation patterns are well documented (Whipps and Lewis, 1981). Interestingly, the PrCWI activity increased by five‐fold in broomrape germinated seeds in comparison with preconditioned seeds, suggesting that the parasite CWI could influence the sink strength of the infected roots during the subsequent step of host root penetration. Additional studies should be performed to assess the role of PrCWI in host root phloem unloading during broomrape attachment. In contrast, the expression of the studied PrCwi gene was low in both preconditioned and germinated seeds. At least three cDNAs encoding putative CWI proteins could be detected in the close relative P. aegyptiaca (OrAe41G2B1‐99085, OrAe3GB1‐12647 and OrAe3GB1‐11068 from PGPP), sharing 56%, 54% and 60% identity, respectively, with the PrCwi sequence. We can thus hypothesize that at least another cell wall‐encoding gene in addition to PrCwi exists in P. ramosa. Nevertheless, the existence of regulatory mechanisms other than transcriptional regulation cannot be excluded to explain the gain in CWI activity in germinated seeds.
Once the radicle reaches the host roots and establishes a functional haustorium, the attached broomrape first develops a tubercle and then an achlorophyllous shoot. Sucrose uptake from the host leads to hexose accumulation, mainly in the growing shoot, during both subterranean and aerial development (Abbes et al., 2009; Delavault et al., 2002), suggesting that invertases also play a central role during overall post‐attachment development. In the present study, we followed the expression of the five genes encoding invertases and the activities of SAI, SNI and CWI in tubercles, shoots and fruits containing developing seeds of P. ramosa parasitizing tomato plants (Fig. 2c,d). Of the five transcripts, PrSai1 transcript levels predominated in all organs regardless of the developmental stage. Nonetheless, the highest levels of PrSai1 transcripts and of putative PrSAI1 activities occurred in growing organs, including the subterranean shoot, the apex of emerged flowering shoots and fruits containing developing seeds. In old tissues, such as the basal parts of emerged flowering shoots, levels of PrSai1 transcripts and PrSAI1 activity were significantly lower. Moreover, in these older plant parts, PrSAI1 activity was confined to the medullar parenchyma. In contrast, PrSAI1 was located in both cortical and medullar parenchyma in apices of flowering shoots (Fig. 3b2), as well as in the whole subterranean shoot (data not shown). These findings indicate that PrSAI1 controls major processes in parasite development, including the sink strength for sucrose in these young tissues by reducing water potential through the massive accumulation of hexoses in the vacuole, cell expansion in elongating zones by maintaining turgor through hexose accumulation and adjustment of the hexose to sucrose ratio. As sucrose cleavage by vacuolar PrSAI1 should be the primary sucrose utilization pathway in these cells, PrSAI1 is assumed to actively drive phloem unloading of sucrose in the growing sinks. Sucrose hydrolysis mediated by an SAI isoform has also been demonstrated to be efficient in phloem unloading, hexose accumulation and the regulation of cell turgor and expansion in carrot roots, bean stems and tomato fruits (Klann et al., 1996; Morris and Arthur, 1985; Ohyama et al., 1995; Ricardo and Ap Rees, 1970).
In contrast, PrSni1‐ and PrSni2‐encoding isoforms display low activity (Fig. 2), as do SNI isoforms in carrot roots (Ricardo and Ap Rees, 1970) and sugar beet (Masuda et al., 1987). Nevertheless, a role of SNI in sucrose utilization cannot be excluded in P. ramosa. Indeed, it is commonly assumed that SNI substitutes for SuSy and provides starch and cell wall precursors when SuSy is deficient (Barratt et al., 2009). The study of these activities at the cellular or tissue level is needed to understand their respective roles in sucrose utilization in P. ramosa.
Control of steady‐state PrSAI1 activity
High sucrose levels and invertase can co‐occur in plant vacuoles. Post‐translational inhibition of vacuolar invertases can result from inhibition by hexose products, proteinaceous inhibitors, structural modification and hexose‐mediated modulation of expression of some invertase inhibition‐related genes (Privat et al., 2008; Rausch and Greiner, 2004). Searching the O. aegyptiaca sequences from PPGP with known proteinaceous invertase inhibitor sequences leads to hits for putative invertase inhibitors in the Orobanchaceae. Nevertheless, inhibition of PrSAI is unlikely in a parasite which contains only negligible amounts of sucrose (Delavault et al., 2002). Furthermore, the extractable PrSAI1 activity was shown to be typically modulated by glucose as a noncompetitive inhibitor and fructose as a competitive inhibitor (Table 2), indicating that PrSAI1 inhibition by hexoses was overcome in cells, resulting in massive accumulation of hexoses in the vacuole, as observed in growing shoots (Delavault et al., 2002).
Steady‐state levels of plant invertases are commonly controlled by a dominant transcriptional regulation mechanism. Likewise, patterns of PrSAI1 transcript levels and activity were highly correlated (Fig. S1). As suggested in germinating legume seeds, which show increased CWI activity in response to cytokinins, as well as the induction of d‐type cyclin‐encoding genes in response to hexose products (Roitsch and Gonzalez, 2004), cytokinins and hexoses may enhance, in a cooperative manner, cell proliferation and sink metabolism in growing organs of P. ramosa. Interestingly, high cytokinin levels have been reported in xylem‐feeding parasitic plants (Jiang et al., 2005; Lechowski and Bialczyk, 1996); nevertheless, no data are available for phloem‐feeding parasitic plants such as P. ramosa.
In addition, PrSAI1 may also be controlled by a post‐translational regulatory mechanism involving proteolytic cleavage. We suggest this mechanism as a result of differences in the molecular mass predicted from the Prsai1 gene (73‐kDa polypeptide) and that estimated from PrSAI1 (86 kDa and 35 kDa in native and denaturing conditions, respectively) (Table 2). PrSAI1 in subterranean shoots probably corresponds to two peptides of about 35 kDa produced by proteolytic cleavage. This type of post‐translational mechanism has been described for the monomeric 68‐kDa and 70‐kDa vacuolar forms in mung bean and carrot, respectively (Arai et al., 1991; Unger et al., 1994), in which the fragments are tightly associated under native conditions, and the complex is active. Disassociation appears to be under developmental control in both species, as the ratio between fragments and full‐length protein increases with plant age. Developmental control of PrSAI1 in P. ramosa was not studied here. In addition, the physiological function of fragmentation remains unclear in plants.
The findings presented in this work indicate that, among the studied invertases, vacuolar PrSAI1 dominates in conditioning the sink strength and growth during both host‐independent and post‐attachment phases. However, additional studies are needed to confirm that PrCWI acts on the sink strength of infected roots during root penetration. Consequently, inhibition of PrSAI1 and possibly PrCWI activities may be a good strategy for curbing parasite development. Given that the phloem mobility of proteins and nucleic acids between host and parasite has been demonstrated (Aly et al., 2006; Davis and Wurdack, 2004), our results call for additional studies to evaluate the efficiency of new transgenic resistance in host plants based on either the production of proteinaceous inhibitors specifically targeted against PrSAI1 (or PrCWI) or the PrSai1 (or PrCwi) silencing technology described by Aly et al. (2009) and Tomilov et al. (2008). The efficiency of the silencing strategy has already been demonstrated: a silencing mechanism targeted against the parasite's mannose 6‐phosphate reductase‐encoding gene causes P. aegyptiaca to lose accumulated solutes and, as a result, a significant increase in parasite mortality (Aly et al., 2009). Previous studies have shown that the mannose 6‐phosphate reductase protein is indeed a key enzyme in mannitol production in broomrape species (Delavault et al., 2002; Harloff and Wegmann, 1993). Moreover, because no transformation method is available for P. ramosa, the silencing strategy through host genetic modification could be useful to confirm the key role of PrSAI1 in parasite sink strength and growth, and to analyse the relative contribution of the slightly expressed genes PrCwi and PrSni. To obtain a complete picture of the sink strategy in broomrape species, more investigations are needed on the mechanisms of sucrose unloading and sucrose allocation between mannitol, hexoses and starch in sink cells, and the involvement of other classes of sucrose‐degrading enzymes, e.g. SuSy.
EXPERIMENTAL PROCEDURES
Plant materials, cultivation and sampling
Seeds were collected from flowering spikes of P. ramosa L. Pomel in France (at Saint Jean d'Angely, 2005) and stored in darkness at 25 °C until use. They were mixed homogeneously with a 1:1:1 peat–sand–clay mixture in 3‐L pots (10 mg/L of soil); the pots were watered and protected from light with a black plastic film for 1 week at 20–25 °C (day–night temperature). Then, one tomato seed (Solanum lycopersicum L., var. Robusta, Seminis) was sown directly into each pot. The plants were sprinkled weekly with a sterile solution of 50% Coïc nutrient solution (Labrousse et al., 2001) and grown under a photoperiod of 16 h (photosynthetic fluence rate of 300 µmol/m2/s over a waveband of 400–700 nm) at a temperature of 20–25 °C. Twelve weeks after sowing, tomato plants were gently uprooted from the soil and the different developmental stages of P. ramosa seedlings were harvested (Fig. 5). Fruits containing developing seeds were also harvested from flowering spikes (stage V).
Figure 5.
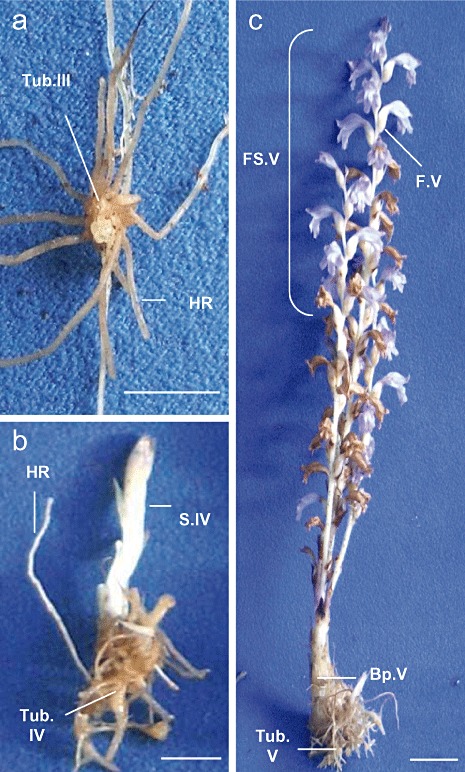
Developmental stages in Phelipanche ramosa parasitizing tomato plants: (a) stage III, growing tubercle (Tub.III); (b) stage IV, tubercle (Tub.IV) bearing the growing subterranean shoot (S.IV); (c) stage V, tubercle (Tub.V) bearing the emerged flowering shoot. Apical part of the flowering shoot is growing (FS.V) and bears fruits containing developing seeds (F.V). Basal part (Bp.V) does not bear flowers and is larger and more fibrous than the flowering shoot. HR, host roots (tomato). Bar, 1 cm.
Phelipanche ramosa seeds were surface sterilized as described previously by Labrousse et al. (2001). They were preconditioned on sterile, watered, glass‐fibre filters placed in 9‐cm‐diameter Petri dishes after a 6‐day period in darkness at 21 °C. Germination of a preconditioned seed set was induced following an additional 6‐day period in darkness in the presence of GR24 (1 µg/L; Johnson et al., 1976).
Seeds and plant organs were immediately frozen in liquid nitrogen and stored at −80 °C prior to RNA and enzyme extraction.
Total RNA extraction and cDNA preparation
Frozen tissues were ground in liquid nitrogen and total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. Extracts were treated with DNase I enzyme (0.02 U/µL; New England Biolabs, Ipswich, MA, USA) to eliminate residual genomic DNA. The integrity of total RNA was determined by electrophoresis on 1% (w/v) agarose gel. Using oligo(dT20) as a primer, cDNA was prepared from samples (0.5 µg) of total RNA using the Superscript II reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA).
Isolation of cDNAs from the invertase family in P. ramosa
The cDNAs isolated from an emerged parasite (stage V) were used for PCR amplification. Degenerate primers corresponding to highly conserved regions of AIs (SAI and CWI) and neutral invertases (cytosolic, SNI) were designed (Table S1). After denaturation at 94 °C for 5 min, the amplification consisted of 35 cycles of 45 s at 94 °C, 45 s at 52, 54 or 57 °C for PrSai (1 and 2), PrCwi and PrSni (1 and 2), respectively, and 1 min 30 s at 72 °C. An additional final step of elongation was performed at 72 °C for 5 min. The amplified DNA fragment was purified and cloned into a pGEM‐T Easy vector (Promega, Charbonnières‐Les‐Bains, France). Recombinant plasmid DNA was prepared and then sequenced by GATC Biotech (Konstanz, Germany). Based on the initial sequence information, new primers were generated for RACE of each fragment using the Generacer kit (Invitrogen, Eragny Sur Joise, France). RACE products corresponding to different invertase genes were cloned and sequenced. An incomplete 5′ RACE product was amplified for PrSai2. To amplify full‐length PrSai1, PrCwi, PrSni1 and PrSni2 cDNAs, specific primer pairs were designed. After denaturation at 94°C for 5 min, the amplification consisted of 35 cycles of 1 min at 94 °C, 1 min at 58, 60 or 54 °C for PrSai1, PrCwi and PrSni (1 and 2), respectively, and 2 min 30 s at 72 °C. An additional final step of elongation was performed at 72 °C for 7 min. The amplified DNA fragments were purified and cloned as mentioned above.
Real‐time quantitative PCR
The determined sequences were used to design gene‐specific primers for real‐time quantitative PCR. Six primer pairs were designed (maximum length, 150 bp; optimal melting temperature T m at 60 °C; GC percentage between 30% and 80%) with Primer Express 3.0 software (Applied Biosystems, Foster City, CA, USA). The selected primers underwent an extensive search using the blast tool to avoid any significant homology with other known nucleotide sequences.
Quantitative PCR was carried out on an Applied Biosystems 7300 real‐time PCR system using SYBR Green technology. Each reaction contained 300 nm of each primer, 5 µL of cDNA sample (∼5 ng of input RNA) and 2 × Power SYBR Green PCR master mix (Applied Biosystems) in a total volume of 25 µL. Thermal cycling was performed using the following cycling conditions: 95 °C for 10 min, 40 cycles at 95 °C for 15 s and 60 °C for 1 min. All quantitative PCR runs contained negative controls with no cDNA template, so as to exclude or detect possible contamination. The specificity of the PCR amplification was checked with a heat dissociation protocol (from 60 to 95 °C) following the final cycle of PCR. The efficiency of the primer sets was calculated by performing quantitative PCR on several dilutions of the strands generated in the first cycle. The efficiency of the different primer sets used was checked and found to be similar.
Results were standardized to PrEF1α1 (accession number: HM219554) gene expression levels; PrEF1α1 displays no development‐related changes. Experiments were carried out on three biological replicates.
Sequence analysis
Bioinformatics analyses were performed using Vector NTI 9.1.0 software (Invitrogen, Invitrogen Corporation, Carlsbad, CA, USA). Sequence homologies were verified against GenBank databases using blast programs (http://www.ncbi.nlm.nih.gov/blast). The subcellular location of the deduced proteins was predicted using the TargetP 1.1 Server (http://www.cbs.dtu.dk/services/TargetP). Phylogenetic analyses were conducted using mega4 software (Tamura et al., 2007). Amino acid sequences of invertase isoenzymes were aligned using Clustal W.
Invertase extraction and enzyme assays
Frozen tissue (1 g fresh weight) was homogenized at 4 °C (Ultra‐Turrax T25, IKA®‐Labortechnik, Staufen, Germany) in 6 mL of buffer A (Table 3) and 0.5 g polyvinylpolypyrrolidone (PVPP). Following centrifugation (10 000 g, 15 min, 4 °C), the supernatant underwent a soluble invertase assay. The residue was rinsed with 50 mL of buffer A and the supernatant was discarded after centrifugation. CWI was extracted from the washed pellet in 3 mL of buffer A containing 1 m NaCl with gentle shaking for 24 h at 4 °C. After centrifugation, the supernatant was used for the CWI assay. Proteins were measured according to Bradford (1976).
Table 3.
Chromatographic steps for PrSAI1 purification from growing subterranean shoots of Phelipanche ramosa parasitizing tomato plants.
| Buffer | Flow rate (mL/min) | Gradient | Collected fractions (mL) | |
|---|---|---|---|---|
| Extraction | A: 100 mm Tris‐HCl (pH 7.0) containing 5 mm EDTA, 10 mm ascorbic acid, 1 mmβ‐mercaptoethanol, 0.1 mm PMSF, 1 mm benzamidine | — | — | — |
| Concanavalin‐A Sepharose | B: 10 mm sodium phosphate (pH 6.5) containing 1 mm EDTA, 10 mm ascorbic acid, 1 mm DTT, 0.1 mm PMSF, 1 mm benzamidine, 1 mm CaCl2, 1 mm MgCl2, 1 mm MnCl2 and 0.5 mm NaCl | 3 | 0–25 mβ‐methyl‐mannoside (60 mL) | 3 |
| High Q anion exchanger | C: 50 mm Tris‐HCl (pH 8.0) containing 1 mm DTT, 0.1 mm PMSF and 1 mm benzamidine | 0.5 | 0–0.5 m NaCl (50 mL) | 2 |
| HR Sephacryl | D: 10 mm sodium phosphate (pH 6.5) containing 5 mm EDTA, 0.1 mm PMSF, 1 mm benzamidine, 0.05% (w/v) NaN3 | 0.5 | — | 1 |
| Liquid isoelectric focusing | 50‐mL solution containing an aliquot of HR‐Sephacryl fraction, 2.5% (v/v) Bio‐Lyte® pH 3/10 ampholyte or Bio‐Lyte® pH 3/5 ampholyte (Bio‐Rad, France) | — | — | 2.5 |
DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; PMSF, phenylmethylsulphonyl.
Prior to assay, the enzyme extract (2.5 mL) was desalted on a PD‐10 Sephadex column (GE Healthcare, Orsay, France), previously equilibrated with 10 mm sodium phosphate buffer (pH 5 for AIs and pH 7.5 for the neutral/alkaline invertase, SNI) containing 1 mm benzamidine and 0.1 mm phenylmethylsulphonylfluoride (PMSF). The enzymatic reaction (1 mL) was initiated by adding sucrose to a final concentration of 100 mm in the medium containing 100 mm sodium phosphate buffer (pH 5.0 for AIs and pH 7.5 for SNI) and 1–50 µg of proteins. The reaction mixture was incubated at 30 °C for 30 min and the reaction was stopped by the addition of 0.5 mL of 100 mm sodium phosphate buffer (pH 7.5) and heating at 95 °C for 3 min. After cooling, the reaction mixture was centrifuged and 0.5 mL of reagent, consisting of 1.5 mm NADP, 1.5 mm ATP, 2 mm MgCl2 and 1 U of both hexokinase (EC 2.7.1.1) and glucose 6‐phosphate dehydrogenase (EC 1.1.1.49) (Sigma, Saint‐Quentin Fallavier, France), was added to the supernatant. Glucose conversion was complete after 30 min at 30 °C. Enzyme activity was determined spectrophotometrically (A 340) and calculated from the amount of NADPH (nmol) produced per second (nkat).
In situ staining of soluble AI activity
All procedures were carried out at 4 °C. SAI activity was determined in both seeds and flowering spikes using histochemical staining according to Sergeeva and Vreugdenhil (2002). The samples, including seeds and handmade transverse sections of basal and apical parts of flowering shoots, were fixed immediately in 50 mm sodium phosphate buffer (pH 5.0) containing 2% (v/v) paraformaldehyde, 2% (w/v) polyvinylpyrrolidone 40, 1 µm dithiothreitol (DTT), 5 mm ethylenediaminetetraacetic acid (EDTA), 2 mm ethyleneglycol‐bis(β‐aminoethylether)‐N,N′‐tetraacetic acid (EGTA), 1 mm benzamidine and 2 mm ascorbic acid for 1 h. The fixation step was omitted for control samples intended for CWI staining. The endogenous soluble carbohydrates were removed by washing the samples three times in fixation buffer without paraformaldehyde, and then three times in water over a period of 5 h. Then, the samples were stained in 50 mm sodium phosphate buffer (pH 5.0) containing 294 µm nitrotetrazolium blue chloride, 460 µm phenazine methosulphate, 25 U/mL glucose oxidase (EC 1.1.3.4) and 100 mm sucrose for 1 h at 37 °C. Controls corresponded to samples incubated in the staining medium without sucrose. In parallel, sections were immersed in sodium hypochlorite (26% active chlorine) for 30 min, rinsed twice in water, incubated for 10 min in 10% (v/v) acetic acid, and then immersed in 0.4% (w/v) Mirande's reagent for 30 min for better visualization of cellulosic and lignified tissues (Mondolot et al., 2001). After three washes in water, photographs were taken with a Olympus SZX10 stereo microscope using an E330 camera (Olympus Europa GMBH, Hamburg, Germany). Fixed samples revealed both CWI and SAI activity, and unfixed samples revealed only CWI activity.
Purification of the SAI isoform
All procedures were carried out at 4 °C. Subterranean shoots (100 g fresh weight, stage IV) were homogenized in 0.6 L of buffer A (Table 3) in the presence of 50 g PVPP (Ultra‐Turrax T25, IKA®‐Labortechnik) and centrifuged (10 000 g, 20 min). Solid ammonium sulphate was added to 80% saturation of the supernatant (516 g/L). The precipitate formed after 2 h of gentle stirring and was collected following centrifugation, and dissolved in a minimal volume of buffer B (Table 3). Following centrifugation, the supernatant was desalted on PD‐10 Sephadex columns (Amersham Pharmacia), previously equilibrated with buffer B.
SAI was purified sequentially on a concanavalin‐A Sepharose affinity gel (Amersham Pharmacia, 10 cm × 1 cm), anion exchanger (Econo‐Pac High Q, 1 mL cartridge, Bio‐Rad, Marnes‐La‐Coquette, France), HR Sephacryl column (30 cm × 1 cm, size exclusion chromatography, GE Healthcare, Orsay, France) with liquid isoelectric focusing on a Rotofor preparative cell (Bio‐Rad, Hercules, CA, USA) (Table 3). Protein fractions were desalted on PD‐10 Sephadex columns prior to SAI assay on 96‐well microplates using the 3,5‐dinitrosalicylic acid method (Miller, 1959; iEMS 96 well Microplate Reader, MTX Lab Systems, Vienna, VA, USA). The active fractions were pooled and concentrated in Vivaspin 5‐kDa microcentrifuge tubes (Sartorius Stedim France SAS, Aubagne, France). The molecular mass of the purified SAI was determined using an HR Sephacryl column following calibration using molecular mass standards (dextran, 2000 kDa; thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa; ovalbumin, 43 kDa; ribonuclease A, 13.5 kDa; GE Healthcare).
Native and sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), SAI activity staining on native gels and Western blotting
Nondenaturing PAGE was performed on a 12% native gel in a Tris‐glycine buffer (pH 8.3) using the Mini Protean® II gel system (Bio‐Rad, USA). The gels were stained with Coomassie Brilliant Blue G250 (Neuhoff et al., 1988), or SAI activity was revealed using the method described by Gabriel and Wang (1969), or the proteins were transferred to a nitrocellulose membrane (Hybond‐ECL™ Nitrocellulose membrane, GE Healthcare) for 1 h at 200 mA using the Mini Trans‐Blot Transfer Cell (Bio‐Rad, USA). SAI immunolabelling was performed using polyclonal antibodies raised against grape (Vitis vinifera) vacuolar invertase (Dambrouck et al., 2005; 1:2500 dilution) and peroxidase‐labelled goat anti‐rabbit IgG antibodies (AffiniPure F(ab′)2 fragment, 1:12 500 dilution; Jackson Immuno‐Research Europe Ltd., Newmarket, Suffolk, UK), according to the manufacturer's instructions (ECL Plus Western Blotting Detection System, GE Healthcare). Immunolabelled SAI was imaged on Kodak BioMax XAR Film (Sigma). In addition, SDS‐PAGE of purified SAI was performed on a 12% gel using Precision Plus Protein™ unstained standards (Bio‐Rad, Marnes‐La‐Coquette, France) as molecular mass standards (Laemmli, 1970).
Analysis by ESI‐LC‐MS/MS
The band showing SAI activity and immunolabelling was excised from native Coomassie Brilliant Blue‐stained gels. In‐gel enzymatic digestion of SAI, MS analysis and protein identification‐databank searching and validation were performed according to Mary et al. (2010). Mass data collected during LC‐MS/MS analysis were processed and searched for protein identification against the Uni‐Prot/Swiss‐Prot, Uni‐Prot/TrEMBL and Institute of Genomic Research (TIGR) expressed sequence tag (EST) protein databanks.
Statistical analysis
An analysis of variance (anova) was performed on the results from enzyme assays and quantitative PCR analyses with the isoform (or gene) or the development stage as the tested factor using SigmaPlot® ver. 10.0. Means of three independent RNA isolations or six independent enzyme extractions were tested at P < 0.05 (SNK test).
Supporting information
Fig. S1 Correlation between levels of PrSAI1 transcripts and soluble acid invertase (SAI) activities in seeds and different organs of Phelipanche ramosa parasitizing tomato plants. GS, germinated seed; PS, preconditioned seed; Tub.III, growing tubercle; Tub.IV, tubercle bearing a growing subterranean shoot (S.IV); Tub.V, tubercle bearing an emerged flowering shoot. Apical part of the flowering shoot is growing (FS.V) and bears fruits containing developing seeds (F.V). Basal part (Bp.V) does not bear flowers and is larger and more fibrous than the flowering shoot. The accumulation of PrSai1 transcripts was expressed relative to elongation factor 1‐α gene (EF1α) transcript levels. Data are means ± standard error (n = 3 for quantitative polymerase chain reaction analysis, n = 6 for enzyme assay).
Fig. S2 Polyacrylamide gel electrophoresis (PAGE) of PrSAI1 purified from growing subterranean shoots of Phelipanche ramosa parasitizing tomato plants. (a) Colloidal blue staining. (b) Soluble acid invertase (SAI) staining. (c) Immunoblotting using polyclonal antibodies targeted against grape (Vitis vinifera) vacuolar invertase. CE, 50 μg crude extract; Con‐A, 10 μg concanavalin‐A fraction; MM, molecular markers; Mono Q, 5 μg Mono Q fraction; 80% SA, 25 μg 80% ammonium sulphate fraction.
Table S1 Degenerate primer pairs designed for cloning invertase polymerase chain reaction (PCR) fragments.
Table S2 Primer sequences for cloning full‐length invertase cDNAs.
Table S3 PrSAI1 purification from subterranean shoots of Phelipanche ramosa parasitizing tomato plants.
Supporting info item
ACKNOWLEDGEMENTS
We gratefully acknowledge Dominique Bozec and Johannes Schmidt for their help in the glasshouse. MS analyses were conducted at the platform ‘Biopolymers‐Interaction‐Structural Biology’ located at the INRA Centre of Angers‐Nantes (http://www.angers‐nantes.inra.fr/plateformes_et‐plateaux_techniques/plate‐forme_bibs). This research was supported by grants from the Syrian and French Ministries of Education and Research (grants to RD and TP, respectively).
REFERENCES
- Abbes, Z. , Kharrat, M. , Delavault, P. , Chaïbi, W. and Simier, P. (2009) Nitrogen and carbon relationships between the parasitic weed Orobanche foetida and susceptible and tolerant faba bean lines. Plant Physiol. Biochem. 47, 153–159. [DOI] [PubMed] [Google Scholar]
- Aber, M. , Fer, A. and Sallé, G. (1983) Etude des substances organiques de l'hôte (Vicia faba) vers le parasite (Orobanche crenata Forsk.). Z. Pflanzenphysiol. 112, 297–308. [Google Scholar]
- Aly, R. , Plakhin, D. and Achdari, G. (2006) Expression of sarcotoxin IA gene via a root‐specific tob promoter enhanced host resistance against parasitic weeds in tomato plants. Plant Cell Rep. 25, 297–303. [DOI] [PubMed] [Google Scholar]
- Aly, R. , Cholakh, H. , Joel, D.M. , Leibman, D. , Steinitz, B. , Zelcer, A. , Naglis, A. , Yarden, O. and Gal‐On, A. (2009) Gene silencing of mannose 6‐phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol. J. 7, 487–498. [DOI] [PubMed] [Google Scholar]
- Arai, M. , Mori, H. and Imaseki, H. (1991) Roles of sucrose‐metabolizing enzymes in growth of seedlings. Purification of acid invertase from growing hypocotyls of mung bean seedlings. Plant Cell Physiol. 32, 1291–1298. [Google Scholar]
- Bar‐Nun, N. and Mayer, A.M. (2002) Composition and changes in storage compounds in Orobanche aegyptiaca seeds during preconditioning. Isr. J. Plant Sci. 50, 277–279. [Google Scholar]
- Baroja‐Fernández, E. , Munoz, J.J. , Montero, M. , Etxeberria, E. , Sesma, M.T. , Ovecka, M. , Bahaji, I. , Ezquer, I. , Li, J. , Prat, S. and Pozueta‐Romero, J. (2009) Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.) tubers results in increased levels of starch, ADPglucose and UDPglucose and total yield. Plant Cell Physiol. 50, 1651–1662. [DOI] [PubMed] [Google Scholar]
- Barratt, D.H.P. , Derbyshire, P. , Findlay, K. , Pike, M. , Wellner, N. , Lunn, J. , Feil, R. , Simpson, C. , Maule, A.J. and Smith, A.M. (2009) Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA, 106, 13 124–13 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer, A. , Bécard, G. , Jauneau, A. , Roux, C. and Séjalon‐Delmas, N. (2008) GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 148, 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bücking, H. , Abubaker, J. , Govindarajulu, M. , Tala, M. , Pfeffer, P. , Nagahashi, G. , Lammers, P. and Shachar‐Hill, Y. (2008) Root exudates stimulate the uptake and metabolism of organic carbon in germinating spores of Glomus intraradices . New Phytol. 180, 684–695. [DOI] [PubMed] [Google Scholar]
- Coleman, H.D. , Yan, J.M. and Mansfield, S.D. (2009) Sucrose synthase affects carbon partitioning to increase cellulose production and alter cell wall ultrastructure. Proc. Natl. Acad. Sci. USA, 106, 13 118–13 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambrouck, T. , Marchal, R. , Cilindre, C. , Parmentier, M. and Jeandet, P. (2005) Determination of the grape invertase content (using PTA‐ELISA) following various fining treatments versus changes in the total protein content of wine. Relationships with wine foamability. J. Agric. Food Chem. 53, 8782–8789. [DOI] [PubMed] [Google Scholar]
- Davis, C.C. and Wurdack, K.J. (2004) Host‐to‐parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science, 305, 676–678. [DOI] [PubMed] [Google Scholar]
- Delavault, P. , Simier, P. , Thoiron, S. , Véronési, C. , Fer, A. and Thalouarn, P. (2002) Isolation of mannose 6‐phosphate reductase cDNA, changes in enzyme activity and mannitol content in broomrape (Orobanche ramosa) parasitic on tomato roots. Physiol. Plant. 115, 48–55. [DOI] [PubMed] [Google Scholar]
- Elliott, K.J. , Butler, W.O. , Dickinson, C.D. , Konno, Y. , Vedvick, T.S. , Fitzmaurice, L. and Mirkov, T.E. (1993) Isolation and characterization of fruit vacuolar invertase genes from two tomato species and temporal differences in mRNA levels during fruit ripening. Plant Mol. Biol. 21, 515–524. [DOI] [PubMed] [Google Scholar]
- Faye, L. , Mouatassim, B. and Ghorbel, A. (1986) Cell wall and cytoplasmic isozymes of radish (Raphanus sativus cultivar Longue rave saumonee) beta‐fructosidase have different N‐linked oligosaccharides. Plant Physiol. 80, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, E. and Zamir, D. (2003) Functional divergence of a syntenic invertase gene family in tomato, potato, and Arabidopsis. Plant Physiol. 131, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, O. and Wang, S.F. (1969) Determination of enzymatic activity in polyacrylamide gels: I. Enzymes catalyzing the conversion of nonreducing substrates to reducing products. Anal. Biochem. 27, 545–554. [DOI] [PubMed] [Google Scholar]
- Gallagher, J.A. , Cairns, A.J. and Pollock, C.J. (2004) Cloning and characterization of a putative fructosyltransferase and two putative invertase genes from the temperate grass Lolium temulentum L. J. Exp. Bot. 55, 557–569. [DOI] [PubMed] [Google Scholar]
- Haouazine‐Takvorian, N. , Tymowska‐Lalanne, Z. , Takvorian, A. , Tregear, J. , Lejeune, B. , Lecharny, A. and Kreis, M. (1997) Characterization of two members of the Arabidopsis thaliana gene family, Atbfruct3 and Atbfruct4, coding for vacuolar invertases. Gene, 197, 239–251. [DOI] [PubMed] [Google Scholar]
- Harloff, H.J. and Wegmann, K. (1993) Evidences for a mannitol cycle in Orobanche ramosa and O. crenata . J. Plant Physiol. 141, 513–520. [Google Scholar]
- Hennen‐Bierwagen, T.A. , Lin, Q. , Grimaud, F. , Planchot, V. , Keeling, P.L. , James, M.G. and Myers, A.M. (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol. 149, 1541–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd, J.M. and Jeschke, W.D. (2001) Solute flux into parasitic plants. J. Exp. Bot. 52, 2043–2049. [DOI] [PubMed] [Google Scholar]
- Hibberd, J.M. , Quick, W.P. , Press, M.C. , Scholes, J.D. and Jescke, W.D. (1999) Solute fluxes from tobacco to the parasitic angiosperm Orobanche cernua and the influence of infection on host carbon and nitrogen relations. Plant Cell Environ. 22, 937–947. [Google Scholar]
- Jiang, F. , Vselova, S. , Veselov, D. , Kudoyarova, G. , Jeschke, W.D. and Hartung, W. (2005) Cytokinin flows from Hordeum vulgare to the hemiparasite Rhinanthus minor and the influence of infection on host and parasite cytokinins relations. Funct. Plant Biol. 32, 619–629. [DOI] [PubMed] [Google Scholar]
- Johnson, A.W. , Rosebery, G. and Parker, C. (1976) A novel approach to Striga and Orobanche control using synthetic germination stimulants. Weed Res. 16, 223–227. [Google Scholar]
- Klann, E.M. , Hall, B. and Bennett, A.B. (1996) Antisense acid invertase (TIVI) gene alters soluble sugar composition and size in transgenic tomato fruit. Plant Physiol. 112, 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse, P. , Arnaud, M.‐C. , Berville, A. and Thalouarn, P. (2001) Several mechanisms are involved in resistance to O. cumana Wallr. Ann. Bot. 88, 859–868. [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of the structural proteins of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lechowski, Z. and Bialczyk, J. (1996) Cytokinins in the hemi‐parasite Melampyrum arvense L. before and after attachment to the host. Biol. Plant. 38, 481–488. [Google Scholar]
- Marouf, B.A. and Zeki, L. (1982) Isolation and characterization of invertase from Iraqi date fruit. J. Food Sci. 47, 678–679. [Google Scholar]
- Mary, J. , Rogniaux, H. , Ress, J.F. and Zal, F. (2010) Response of Alvinella pompejana to variable oxygen stress: a proteomic approach. Proteomics, 10, 2250–2258. [DOI] [PubMed] [Google Scholar]
- Masuda, H. , Takahashi, T. and Sugawara, S. (1987) Purification and properties of starch hydrolyzing enzymes in mature roots of sugar beets. Plant Physiol. 84, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G.L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428. [Google Scholar]
- Miller, W.B. and Ranwala, A.P. (1994) Characterization and localization of three soluble invertase forms from Lilium longiflorum flower buds. Physiol. Plant. 92, 247–253. [Google Scholar]
- Mondolot, L. , Roussel, J.L. and Andary, C. (2001) New applications for an old lignified element staining reagent. Histochem. J. 33, 379–385. [DOI] [PubMed] [Google Scholar]
- Morris, D.A. and Arthur, E.D. (1985) Invertase activity, carbohydrate metabolism and cell expansion in the stem of Phaseolus vulgaris L. J. Exp. Bot. 36, 623–633. [Google Scholar]
- Neuhoff, V. , Stamm, R. and Eibl, H. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background sensitivity using coomassie brilliant blue G‐250 and R‐250. Electrophoresis, 9, 255–262. [DOI] [PubMed] [Google Scholar]
- Ohyama, A. , Ito, H. , Sato, T. , Nishimura, S. , Imai, T. and Hirai, M. (1995) Suppression of acid invertase activity by antisense RNA modifies the sugar composition of tomato fruit. Plant Cell Physiol. 36, 369–376. [Google Scholar]
- Parker, C. (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag. 65, 453–459. [DOI] [PubMed] [Google Scholar]
- Pérez‐de‐Luque, A. , Fondevilla, S. , Perez‐Vich, B. , Aly, R. , Thoiron, S. , Simier, P. , Castillejo, M.A. , Fernandez, J.M. , Jorrin, J. , Rubiales, D. and Delavault, P. (2009) Understanding broomrape–host plant interaction and developing resistance. Weed Res. 49, 8–22. [Google Scholar]
- Privat, I. , Fourcrier, S. , Prins, A. , Epalle, T. , Eychenne, M. , Kandalaft, L. , Gaillet, V. , Lin, C. , Tanksley, S. , Foyer, C. and McCarthy, J. (2008) Differential regulation of grain sucrose accumulation and metabolism in Coffea arabica (Arabica) and Coffea canephora (Robusta) revealed through gene expression and enzyme activity analysis. New Phytol. 178, 781–797. [DOI] [PubMed] [Google Scholar]
- Rausch, T. and Greiner, S. (2004) Plant protein inhibitors of invertases. Biochim. Biophys. Acta, 1696, 253–261. [DOI] [PubMed] [Google Scholar]
- Ricardo, C.P. and Ap Rees, T. (1970) Invertase activity during the development of carrot roots. Phytochemistry, 9, 239–247. [Google Scholar]
- Rispail, N. , Dita, M.A. , Gonzales‐Verdejo, C. , Perez‐de‐Luque, A. , Castillejo, M.A. , Prats, E. , Roman, B. , Jorrin, J. and Rubiales, D. (2007) Plant resistance to parasitic plants: molecular approaches to an old foe. New Phytol. 173, 703–712. [DOI] [PubMed] [Google Scholar]
- Roitsch, T. and Gonzalez, M.C. (2004) Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 9, 606–613. [DOI] [PubMed] [Google Scholar]
- Roitsch, T. , Bittner, M. and Godt, D.E. (1995) Induction of apoplastic invertase of Chenopodium rubrum by d‐glucose and a glucose analog and tissue‐specific expression suggest a role in sink–source regulation. Plant Physiol. 108, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva, L.I. and Vreugdenhil, D. (2002) In situ staining of activities of enzymes involved in carbohydrate metabolism in plant tissues. J. Exp. Bot. 53, 361–370. [DOI] [PubMed] [Google Scholar]
- Singh, M. , Singh, D.V. , Misra, P.C. , Tewari, K.K. and Krishnan, P.S. (1968) Biochemical aspects of parasitism by angiosperm parasites: starch accumulation. Physiol. Plant. 21, 525–538. [Google Scholar]
- Sturm, A. (1996) Molecular characterisation and functional analysis of sucrose‐cleaving enzymes in carrot (Daucus carota L.). J. Exp. Bot. 47, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Sturm, A. (1999) Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 121, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm, A. and Chrispeels, M.J. (1990) cDNA cloning of carrot extracellular beta‐fructosidase and its expression in response to wounding and bacterial infection. Plant Cell, 2, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm, A. and Tang, G.Q. (1999) The sucrose‐cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 4, 401–407. [DOI] [PubMed] [Google Scholar]
- Sturm, A. , Hess, D. , Lee, H.S. and Lienhard, S. (1999) Neutral invertase is a novel type of sucrose‐cleaving enzyme. Physiol. Plant. 107, 159–165. [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Ruffner, H.P. , Scholes, J.D. and Rolfe, S.A. (1996) Purification and characterization of soluble invertases from leaves of Arabidopsis thaliana . Planta, 198, 17–23. [DOI] [PubMed] [Google Scholar]
- Tomilov, A.A. , Tomolova, N.B. , Wroblewski, T. , Michelmore, R. and Yoder, J.I. (2008) Trans‐specific gene silencing between host and parasitic plants. Plant J. 56, 389–397. [DOI] [PubMed] [Google Scholar]
- Unger, C. , Hardegger, M. , Lienhard, S. and Sturm, A. (1994) cDNA cloning of carrot (Daucus carota) soluble acid beta‐fructofuranosidases and comparison with the cell wall isoenzyme. Plant Physiol. 104, 1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, W.A. , Pontis, H.G. and Salerno, G.L. (2008) New insights on sucrose metabolism: evidence for an active A/N‐Inv in chloroplasts uncovers a novel component of the intracellular carbon trafficking. Planta, 227, 795–807. [DOI] [PubMed] [Google Scholar]
- Velasco, L. , Goffman, F.D. and Pujadas‐Salva, A.J. (2000) Fatty acids and tocochromanols in seeds of Orobanche . Phytochemistry, 54, 295–300. [DOI] [PubMed] [Google Scholar]
- Vijn, I. and Smeekens, S. (1999) Fructan: more than a reserve carbohydrate? Plant Physiol. 120, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele, R.T. , Wirsel, S. , Möll, U. , Lechner, M. and Mendgen, K. (2006) Cloning and characterization of a novel invertase from the obligate biotroph Uromyces fabae and analysis of expression patterns of host and pathogen invertases in the course of infection. Mol. Plant–Microbe Interact. 19, 625–634. [DOI] [PubMed] [Google Scholar]
- Whipps, J.M. and Lewis, D.H. (1981) Patterns of translocation, storage and interconversion of carbohydrates In: Effects of Disease on the Physiology of the Growing Plants (Ayers P.G., ed.), pp. 47–83. Cambridge: Cambridge University Press. [Google Scholar]
- Yoneyama, K. , Awad, A.A. , Xie, X. , Yoneyama, K. and Takeuchi, Y. (2010) Strigolactones as germination stimulants for root parasitic plants. Plant Cell Physiol. 51, 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanor, M.I. , Osorio, S. , Nunes‐Nesi, A. , Carrari, F. , Lohse, M. , Usadel, B. , Kühn, C. , Bleiss, W. , Giavalisco, P. , Millmitzer, L. , Sulpice, R. , Zhou, Y.H. and Fernie, A.R. (2009) RNA interference of LIN5 in tomato confirms its role in controlling Brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 150, 1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Correlation between levels of PrSAI1 transcripts and soluble acid invertase (SAI) activities in seeds and different organs of Phelipanche ramosa parasitizing tomato plants. GS, germinated seed; PS, preconditioned seed; Tub.III, growing tubercle; Tub.IV, tubercle bearing a growing subterranean shoot (S.IV); Tub.V, tubercle bearing an emerged flowering shoot. Apical part of the flowering shoot is growing (FS.V) and bears fruits containing developing seeds (F.V). Basal part (Bp.V) does not bear flowers and is larger and more fibrous than the flowering shoot. The accumulation of PrSai1 transcripts was expressed relative to elongation factor 1‐α gene (EF1α) transcript levels. Data are means ± standard error (n = 3 for quantitative polymerase chain reaction analysis, n = 6 for enzyme assay).
Fig. S2 Polyacrylamide gel electrophoresis (PAGE) of PrSAI1 purified from growing subterranean shoots of Phelipanche ramosa parasitizing tomato plants. (a) Colloidal blue staining. (b) Soluble acid invertase (SAI) staining. (c) Immunoblotting using polyclonal antibodies targeted against grape (Vitis vinifera) vacuolar invertase. CE, 50 μg crude extract; Con‐A, 10 μg concanavalin‐A fraction; MM, molecular markers; Mono Q, 5 μg Mono Q fraction; 80% SA, 25 μg 80% ammonium sulphate fraction.
Table S1 Degenerate primer pairs designed for cloning invertase polymerase chain reaction (PCR) fragments.
Table S2 Primer sequences for cloning full‐length invertase cDNAs.
Table S3 PrSAI1 purification from subterranean shoots of Phelipanche ramosa parasitizing tomato plants.
Supporting info item


