SUMMARY
White blister rust caused by Albugo candida (Pers.) Kuntze is a common and often devastating disease of oilseed and vegetable brassica crops worldwide. Physiological races of the parasite have been described, including races 2, 7 and 9 from Brassica juncea, B. rapa and B. oleracea, respectively, and race 4 from Capsella bursa‐pastoris (the type host). A gene named WRR4 has been characterized recently from polygenic resistance in the wild brassica relative Arabidopsis thaliana (accession Columbia) that confers broad‐spectrum white rust resistance (WRR) to all four of the above Al. candida races. This gene encodes a TIR‐NB‐LRR (Toll‐like/interleukin‐1 receptor‐nucleotide binding‐leucine‐rich repeat) protein which, as with other known functional members in this subclass of intracellular receptor‐like proteins, requires the expression of the lipase‐like defence regulator, enhanced disease susceptibility 1 (EDS1). Thus, we used RNA interference‐mediated suppression of EDS1 in a white rust‐resistant breeding line of B. napus (transformed with a construct designed from the A. thaliana EDS1 gene) to determine whether defence signalling via EDS1 is functionally intact in this oilseed brassica. The eds1‐suppressed lines were fully susceptible following inoculation with either race 2 or 7 isolates of Al. candida. We then transformed white rust‐susceptible cultivars of B. juncea (susceptible to race 2) and B. napus (susceptible to race 7) with the WRR4 gene from A. thaliana. The WRR4‐transformed lines were resistant to the corresponding Al. candida race for each host species. The combined data indicate that WRR4 could potentially provide a novel source of white rust resistance in oilseed and vegetable brassica crops.
INTRODUCTION
Arabidopsis thaliana has been an important genetic resource for the investigation of the molecular basis of genes that specify natural variation in disease resistance to bacterial, fungal, viral and oomycete pathogens of plants (Eulgem, 2005; Holub, 2001, 2008; Ryan et al., 2007). As in most plant species, a majority of these so‐called resistance (R) genes in A. thaliana encode intracellular receptor‐like proteins that are characterized by a central nucleotide‐binding (NB) domain and a C‐terminal leucine‐rich repeat (LRR) domain. They can be further grouped into two subclasses on the basis of either a TIR (similar to animal Toll‐like/interleukin‐1 receptors) or coiled‐coil (CC) domain at the N‐terminus (Jones and Jones, 1997).
Most R genes in A. thaliana confer disease resistance to a narrow spectrum of variants within a corresponding pathogen species, and therefore may never be cost‐effective for commercial investment leading to the development of transgenic crops. However, important exceptions of broad‐spectrum disease resistance have been reported across a wide taxonomic range of plants, including A. thaliana, in which a single, dominantly expressed R protein confers resistance to all known races of a pathogen. In an early report, a membrane‐bound receptor‐like kinase, designated Xa21, was described in rice, which is effective against 29 diverse isolates of the bacterium Xanthomonas oryzae pv. oryzae (Wang et al., 1996). Two genes in A. thaliana, designated RFO1 and RPW8, encode other non‐NB‐LRR proteins that confer resistance against diverse collections of Fusarium or powdery mildew fungi, respectively (Diener and Ausubel, 2005; Xiao et al., 2001). In the Solanaceae, two broad‐spectrum CC‐NB‐LRR proteins have been discovered, including Bs2 in pepper, which confers black spot resistance to X. campestris pv. vesicatoria, and RB (also named Rpi‐blb1) from the wild species Solanum bulbocastanum, which confers late blight resistance to current known races of the oomycete pathogen Phytophthora infestans in the USA and Europe (Song et al., 2003; van der Vossen et al., 2003). Similarly, TIR‐NB‐LRR genes have been reported, including WRR4 from A. thaliana, which confers resistance to four races of the oomycete Albugo candida (white blister rust) that occur naturally on other wild and domesticated host species, including Capsella bursa‐pastoris (the type host), Brassica rapa, B. juncea and B. oleracea (Borhan et al., 2008); RLM3 from A. thaliana, which confers resistance to several necrotrophic fungi (Staal et al., 2008); and RCT1 from Medicago truncatula which confers resistance to races of the anthracnose fungi Colletotrichum trifolii and C. destructivum (Yang et al., 2008).
WRR4 provides an important example to test the transgenic use in crops of an R gene that was originally derived from A. thaliana. Albugo candida (Pers.) Kuntze is an economically destructive crop pathogen. This biotrophic oomycete causes white blister rust in all vegetable and oilseed brassica crops, such as B. rapa (Chinese cabbage and turnip rape; diploid A genome), B. oleracea (cabbage, kale, broccoli and cauliflower; diploid C genome), B. juncea (oilseed mustard; allotetraploid A and B genomes) and B. napus (oilseed rape; allotetraploid A and C genomes) (Fan et al., 1983; Harper and Pittman, 1974; Kumari et al., 1970; Petrie, 1988; Pound and Williams, 1963). The parasite emerges from infected tissue as white rust pustules to release asexual zoosporangia. These pustules can emerge on all aerial parts of the host, and are often associated with abnormal growth in surrounding tissue, stimulated by a hormonal imbalance, such as a common severe symptom called a ‘staghead’ that occurs when the pathogen invades a floral stem. Susceptible oilseed crops (B. napus and B. juncea) are particularly vulnerable to floral infections, with devastating losses (30%–60% yield reduction) commonly occurring in North America, Australia, India and China (Bernier, 1972; 2007, 1996; Petrie, 1973). Oilseed B. juncea has better drought tolerance and more durable stem canker resistance (Leptosphaeria maculans) than oilseed B. napus (Marcroft et al., 2002), and therefore canola‐quality B. juncea varieties are being developed to extend production in low‐rainfall areas of Australia and North America (Li et al., 2007). Unfortunately, white rust looms as a major threat to B. juncea production in these areas.
Physiological races of Al. candida have been described which each have narrow host ranges, including races 2, 7 and 9 from B. juncea, B. rapa and B. oleracea, respectively (Hill et al., 1988; Pound and Williams, 1963). On the basis of the responses of different cultivars of B. rapa and B. juncea, races 2 and 7 have been further divided into pathotypes Ac2a, Ac2v, Ac7a and Ac7v (Rimmer et al., 2000). For example, Ac2a and Ac2v could be differentiated on the basis of being virulent on B. juncea cv. Burgonde and Cutlass, respectively. Similarly, A7a and Ac7v are virulent on B. rapa cv. Torch and Reward (Rimmer et al., 2000). The type specimen of Al. candida was collected from the invasive species Capsella‐bursa pastoris, and was later designated as race 4. These races are highly specialized; however, they are not necessarily restricted to the host species from which they were originally collected. For example, isolates of race 7 collected from B. rapa can cause disease in many cultivars of B. napus under field conditions (Bernier, 1972; Fan et al., 1983). Similarly, a low frequency (less than 10%) of A. thaliana accessions are susceptible to standard isolates of Al. candida races 2, 4 and 7 in a conducive laboratory environment (Borhan et al., 2008; E. B. Holub, unpublished work). In contrast, A. thaliana is universally a nonhost of Al. candida race 9. It is important to note here that a molecular taxonomic distinction has been made between Al. candida and a species now called Albugo laibachii which commonly causes white rust in A. thaliana under natural field conditions (Holub et al., 1995; Rehmany et al., 2000; Thines et al., 2009; Voglmayr and Riethmüller, 2006). We have previously referred to this common parasite of wild A. thaliana as Al. candida ssp. arabidopsis (Borhan et al., 2008).
WRR4 was identified as a gene in A. thaliana Columbia (Col‐0), which confers full immunity to Al. candida races 2, 4 and 7 when stably transformed into the accession Wassilewskija (Ws‐3); this accession is susceptible in the laboratory to all three races (Borhan et al., 2008). WRR4 also improved the partial resistance of Ws‐3 to race 9, conferring full immunity in transgenic lines. RNA interference (RNAi) suppression of the defence regulator protein enhanced disease susceptibility 1 (EDS1) conferred full susceptibility to race 2, indicating that WRR4 is fully dependent on the expression of this lipase‐like protein (Borhan et al., 2008; Parker et al., 1996). However, the same eds1‐suppressed lines exhibited enhanced colonization to varying degrees by races 4, 7 and 9, but still exhibited residual resistance that restricted the formation of rust pustules (least restricted with race 7, most restricted with race 9). This indicates that Col‐0 contains additional WRR genes which are independent of EDS1.
The purpose of the research described in this article was to determine whether WRR4 from A. thaliana could confer white rust resistance in a brassica species. We began transgenic experiments using RNAi‐mediated suppression of EDS1 in a white rust‐resistant breeding line of B. napus (transformed with a construct designed from the A. thaliana EDS1 gene) to confirm beforehand whether defence signalling via EDS1 was functionally intact in this crop species. We then transformed white rust‐susceptible cultivars of B. napus (susceptible to race 7) and B. juncea (susceptible to race 2) with the WRR4 gene from A. thaliana.
RESULTS
Resistance to Al. candida races 2 and 7 in B. napus is dependent on EDS1
We searched a B. napus expressed sequence tag (EST) database containing nearly 150 000 ESTs (generated at Saskatoon Research Centre, Agriculture and Agri‐Food Canada) for sequences that shared homology with A. thaliana EDS1 (referred to as At.EDS1). Eight B. napus ESTs were identified, and sequence assembly resulted in a single full‐length open reading frame (ORF) of 1800 bp that encodes a lipase‐like protein (referred to as Bn.EDS1). The DNA sequences of At.EDS1 and Bn.EDS1 share 71% identity when aligned, whereas the protein sequences share 61% identity (Figs S1 and S2). We also identified an EST of 634 bp from the B. juncea cv. Cutlass EST database (35 738 ESTs) (M. H. Borhan et al., unpublished work) with homology to the Arabidopsis EDS1 ORF (blastn: e‐value 0). The B. juncea EDS1‐like EST was most similar to a B. oleracea EDS1‐like gene (AJ620884) in the National Center for Biotechnology Information (NCBI) database (blastn: e‐value 1e−129), and also a B. napus EDS1‐like gene (EU6941108) (blastn: e‐value 9e−72). The B. juncea EST showed 81.8% identity with the B. napus EDS1 gene and 73.5% identity with the Arabidopsis EDS1 gene.
Given the close homology between these two genes, we used the At.EDS1‐RNAi construct described by Borhan et al. (2008) to suppress EDS1 in a white rust‐resistant, doubled haploid breeding line of B. napus (DH12075). Three independent RNAi‐transformed lines were identified using the herbicide selection marker phosphinotricin and confirmed by polymerase chain reaction (PCR). Seven‐day‐old seedlings of each T1 line were inoculated with Al. candida isolate Ac7a, and each family segregated in a ratio of approximately 3 : 1 of resistant (no pustules) to susceptible (large, profuse pustules) phenotypes (Table 1; Fig. 1). We then tested T2 lines of the susceptible T1 generation from each independent transformant. These lines were uniformly susceptible to Al. candida isolate Ac2v (Table 1). The combined data from the RNAi suppression lines indicate that the natural white rust resistance in DH12075 to Al. candida races 2 and 7 is dependent on EDS1 expression.
Table 1.
RNA interference suppression of enhanced disease susceptibility 1 (EDS1) and white rust resistance in Brassica napus.
| Host generation | Albugo isolate | Line no. | Total | R | S |
|---|---|---|---|---|---|
| T1 | Ac7a | 211 | 30 | 19 | 11 |
| 415 | 37 | 27 | 10 | ||
| 416 | 40 | 32 | 8 | ||
| T2 | Ac2v | 211–305 | 37 | 0 | 37 |
| 416–240 | 47 | 0 | 47 |
EDS1 is a lipase‐like protein that is commonly required for disease resistance conferred by TIR‐NB‐LRR (Toll‐like/interleukin‐1 receptor‐nucleotide binding‐leucine‐rich repeat) R genes in Arabidopsis thaliana. Suppression of EDS1 was achieved using a construct designed from the A. thaliana EDS1 gene. Transgenic lines were tested for susceptibility to Albugo candida races 2 (isolate Ac2v) and 7 (Ac7a) as follows: T1 seedlings of three independent lines were assessed for resistance (R) or susceptibility (S) to Ac7a at 9 days after inoculation (dai), and T2 seedlings derived from homozygous Ac7a‐susceptible T1 lines were assessed for response to Ac2v at 9 dai.
Figure 1.
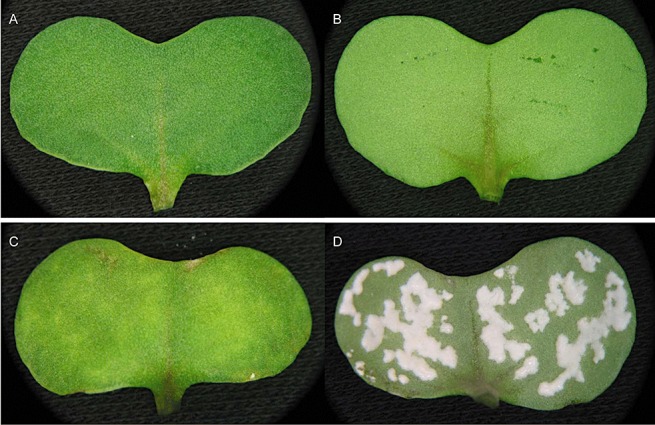
White rust resistance to two avirulent isolates of Albugo candida (Ac2V and Ac7A) is suppressed in Brassica napus line DH12075 with RNA interference (RNAi) suppression of the lipase‐like defence gene enhanced disease susceptibility 1 (EDS1). One‐week‐old seedlings were infected with either isolate, and interaction phenotypes (for Ac7A, shown here) were photographed 10 days later. (A, B) Top and bottom, respectively, of a fully resistant wild‐type cotyledon. (C, D) Top and bottom of a cotyledon from a fully susceptible EDS1‐RNAi‐suppressed line.
WRR4 from A. thaliana confers full resistance in transgenic B. napus and B. juncea
To test whether the WRR4 gene from A. thaliana can confer white rust resistance in B. napus and B. juncea, we transformed a susceptible accession of each oilseed crop, including the B. juncea cv. Cutlass, which is susceptible to Ac2v, and the B. napus breeding line ACS‐N32, which is susceptible to Ac7a. Arabidopsis thaliana WRR4 was expressed under its native promoter as described by Borhan et al. (2008).
Seven independent transgenic lines of B. juncea cv. Cutlass were identified containing a single insertion of WRR4 from A. thaliana using the selectable herbicide marker phosphinotricin. We confirmed the number of insertions by PCR and Southern blots (Table 2). T2 seeds were harvested from each transgenic line. Seven‐day‐old T2 seedlings were inoculated with Ac2v; some segregated in a resistant to susceptible ratio of c. 3 : 1 (Table 2), confirming the single insertion of WRR4 in each line. Growth of the parasite in a resistant transformant was usually localized to one to three penetrated cells at the site of infection (Fig. 2). However, a necrotic patch of cells was occasionally visible macroscopically which, under the microscope, was associated with more extensive growth of hyphae. A minute pustule rarely developed from these confined, necrosis‐inducing colonies of Al. candida. The necrotic patch phenotype may be indicative of a heterozygous genotype; however, this was not confirmed.
Table 2.
Segregation of white rust resistance amongst progeny of Brassica juncea cv. Cutlass and B. napus line ACS‐N32 following transformation with the WRR4 TIR‐NB‐LRR (Toll‐like/interleukin‐1 receptor‐nucleotide binding‐leucine‐rich repeat) gene from Arabidopsis thaliana.
| Brassica species | Line no. | No. of T1 insertions | Total T2 | R | S | χ2 | P |
|---|---|---|---|---|---|---|---|
| B. juncea | 1 | 1 | 42 | 30 | 12 | 0.28 | 0.59 |
| 2 | 2 | 41 | 32 | 9 | 0.20 | 0.65 | |
| 3 | 2 | 35 | 23 | 12 | 1.60 | 0.20 | |
| 4 | 1 | 41 | 37 | 4 | 5.08 | 0.02 | |
| 5 | 4 | 9 | 9 | 0 | 3.00 | 0.08 | |
| 6 | 1 | 40 | 27 | 13 | 1.20 | 0.27 | |
| 7 | 1 | 40 | 31 | 9 | 0.13 | 0.71 | |
| 8 | 1 | 41 | 27 | 14 | 1.82 | 0.17 | |
| B. napus | 1 | 4 | 41 | 37 | 4 | 5.08 | 0.02 |
| 2 | 1 | 39 | 22 | 17 | 7.18 | 0.007 | |
| 3 | 1 | 25 | 18 | 7 | 0.12 | 0.72 |
The number of transgene insertions in the T1 generation was determined by Southern blotting. The numbers of resistant (R; green cotyledon and no rust pustules) and susceptible (S; large pustules formed profusely on the underside of cotyledons) T2 seedlings were recorded at 10 days after inoculation.
Figure 2.
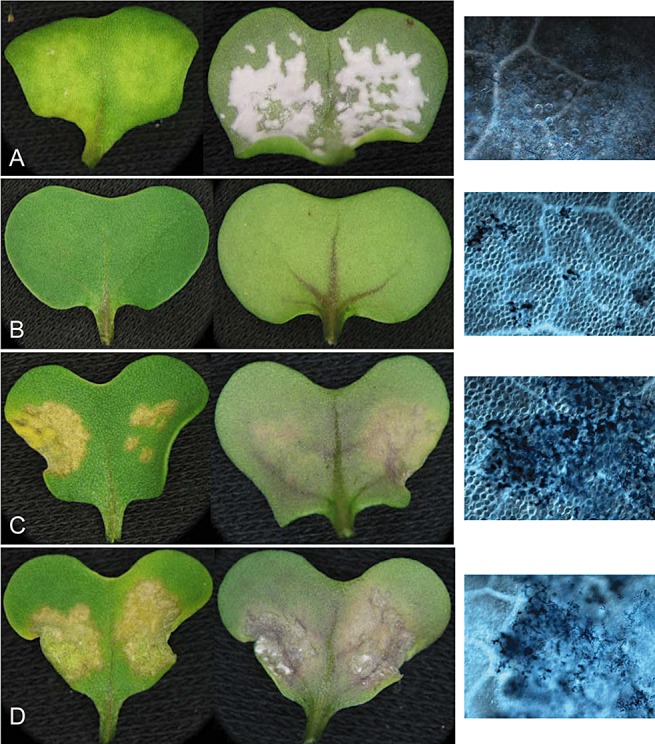
Arabidopsis thaliana WRR4 (WRR, white rust resistance) confers full resistance in the susceptible Brassica juncea cv. Cutlass. Photographs of cotyledons inoculated with the virulent isolate Ac2v at 10 days after inoculation. (A) Wild‐type Cutlass exhibiting full susceptibility with profuse development of white pustules on the lower surface and no host cell necrosis visible microscopically (far right). (B–D) Cotyledons of WRR4 transgenic Cutlass showing resistance phenotypes. (B) Full resistance with no formation of pustules on the upper or lower surfaces of the cotyledon; minute patches of necrotic cells are visible microscopically at the site of infection (far right). (C) Necrotic patches with no formation of pustules on the upper or lower surfaces of the cotyledon, and restriction of hyphae to cells surrounding the site of infection as shown by microscopy (far right). (D) Necrotic patch with a few minute sporadic pustules, and restriction of hyphae to cells surrounding the site of infection as shown by microscopy (far right).
Similarly, three transgenic lines from B. napus line ACS‐N32 were obtained containing the WRR4 gene from A. thaliana. T2 progeny from these lines were inoculated with Al. candida isolate Ac7a and segregated for resistance (Table 2). As in B. juncea, WRR4‐mediated resistance was typically associated with the restriction of the parasite to the site of infection (Fig. 3).
Figure 3.
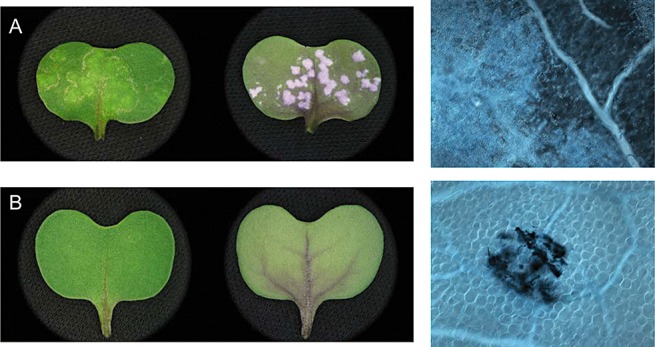
The white rust resistance gene WRR4 from Arabidopsis thaliana confers full resistance in a susceptible cultivar of Brassica napus (ACS‐N32). (A) Upper and lower surfaces of a wild‐type cotyledon 10 days after inoculation with the virulent isolate Ac7a, exhibiting full susceptibility with the profuse development of white pustules on the lower surface and no host cell necrosis visible microscopically (far right). (B) Upper and lower surfaces of an ACS‐N32 cotyledon stably transformed with WRR4, indicating full resistance with no formation of pustules on the upper or lower surfaces of the cotyledon, and minute patches of necrotic cells visible microscopically at the site of infection (far right).
DISCUSSION
Two decades of A. thaliana molecular biology have been central in shaping our current understanding of innate defence in plants and have also had an impact on the understanding of infectious disease in animals (Chisholm et al., 2006; Dangl and Jones, 2001; Holub, 2007; Jones et al., 2008). Early contributions to crop improvement came from a precedent that a virulent bacterial pathogen could be genetically modified to deliver an avirulence effector from a crop pathogen, and then used as a physiological probe to identify the matching receptor‐like R gene from a nonhost of the crop pathogen (Gassmann et al., 1999; Holub, 2001; Warren et al., 1998). This method holds great promise for the transient delivery of avirulence effectors from filamentous pathogens (Rentel et al., 2008; Sohn et al., 2007; Vleeshouwers et al., 2008). More importantly, the use of molecular markers derived from conserved domains in the most common NB‐LRR, R and other essential defence response genes enables the marker‐assisted selection of homologues in crops (Aarts et al., 1998; Botella et al., 1997; Leister et al., 1996; McHale et al., 2009; Shen et al., 1998). Lettuce provides a superb illustration of how even minor crops will benefit from this combined ‘model‐to‐crop’ knowledge transfer (Caldwell and Michelmore, 2009; McHale et al., 2009; Wroblewski et al., 2009).
Surprisingly, the direct use of A. thaliana R genes in transgenic crops has not been demonstrated, despite several examples of genes that could potentially confer broad‐spectrum resistance to brassica crop pathogens (Cooley et al., 2000; Grant et al., 1995; 2006, 2008; Xiao et al., 2001). Thus, the observation that WRR4 from A. thaliana confers white rust resistance in two oilseed brassica crops provides another important precedent for the utility of A. thaliana research. Albugo candida can cause severe yield losses in oilseed and vegetable crops of B. juncea, B. oleracea and B. rapa, with losses in oilseed mustard (B. juncea) often reaching 60% for small‐holding farmers in India (Bernier, 1972; Li et al., 1996; Li et al., 2007; Petrie, 1973). Brassica juncea has better drought tolerance and more durable stem canker resistance (L. maculans) than B. napus oilseed rape (Marcroft et al., 2002), and canola‐quality B. juncea varieties have therefore been developed to extend oilseed production in low‐rainfall areas of Australia and Canada (Li et al., 2007). Unfortunately, white rust looms as a major threat to production in these areas.
The durability of a single broad‐spectrum R gene for disease control in crops is not guaranteed, but will instead depend on how readily the pathogen can evolve variants to overcome host resistance (Leach et al., 2001). Pathogen effector molecules have been identified that correspond as the avirulence effector detected by broad‐spectrum R proteins, including Bs2 from pepper and Rb from potato (Kearney and Staskawicz, 1990; Vleeshouwers et al., 2008). The corresponding avirulence effectors in these examples occur species wide in each respective pathogen and appear to be significantly constrained from evolution as a result of a high penalty of mutation. Thus, slow or nonevolving effectors represent a plausible expectation that warrants investigation in pathogens from the other examples of broad‐spectrum R proteins, such as the predicted avirulence effector corresponding with WRR4 that may be shared amongst physiological races of Al. candida (Borhan et al., 2008).
NB‐LRR R genes may have evolved and expanded extensively as a gene family in plants because they typically induce defence only when it is likely to be beneficial after detection of an avirulent pathogen, and are unlikely to confer susceptibility to nontarget microorganisms. R genes, such as Bs2, Rb, RLM3 and WRR4, provide the additional advantage of broad‐spectrum disease control, and illustrate the potential for expanding the use of NB‐LRR genes from wild species in crop improvement. Interestingly, a trade‐off in enhanced susceptibility to nontarget pathogens has been observed for broad‐spectrum resistance genes that do not encode NB‐LRR proteins (Jarosch et al., 2003; Wang et al., 2007).
Genetic improvement of multiple agronomic traits (e.g. drought tolerance, nutrient‐use efficiency, yield performance and disease resistance) in crops that have large and complex genomes will continue to benefit from underpinning investment in model plants (Bevan and Waugh, 2007). In particular, A. thaliana is an excellent tool for crop scientists working with brassica crops, considering the significant synteny between the two genomes (Schranz et al., 2007). The results from the transgenic suppression of EDS1 and gain‐of‐function in resistance to Al. candida with WRR4 indicate that brassica species contain the genes essential for the direct use of NB‐LRR encoding other R genes from A. thaliana in brassica crops, such as the broad‐spectrum stem canker resistance genes RLM1 and RLM3 (2006, 2008). The advantage in both the white rust and stem canker examples is that economically devastating crop pathogens were strategically used from inception in the molecular genetics research of A. thaliana.
EXPERIMENTAL PROCEDURES
Pathogen handling and inoculation
Albugo candida races were propagated on the appropriate susceptible host (Rimmer et al., 2000). Pustules were harvested at 10–14 days after inoculation (dai) and stored at −20 °C, or used fresh to prepare the inoculum by suspending the spores in dH2O at a concentration of 2 × 104/mL sporangia. Inoculum was incubated at 14–16 °C for 2–4 h to ensure the release of motile zoospores. This inoculum was kept on ice during the inoculation of 5–7‐day‐old brassica seedlings. A repeater pipette was used to place a 10‐µL drop of the inoculum on each half of a cotyledon. Inoculated plants were kept at 14–16 °C under a micropropagator to maintain humidity. After 24 h, plants were transferred to a growth chamber with 12 h light, at temperatures of 18 °C during the night and 20 °C during the daytime. Cotyledon responses to Al. candida were scored at 7–14 dai.
Microscopy of infected tissues
To prepare inoculated tissues for microscopic observation of pathogen growth and plant response, cotyledons were excised from the seedlings at 7–14 dai, placed in a 50‐mL Falcon tube submerged in trypan blue (Parker et al., 1996) and transferred to a boiling water bath for 20 min. After this, stain was replaced with chloral hydrate. Tissues were left in chloral hydrate for 24 h. Cleared tissues were placed on a glass slide in 50% glycerol, covered with a coverslip and observed using differential interference contrast (DIC) with a Zeiss (Oberkochen, Germany) Axio Imager Z1 microscope.
Brassica transformation
Hypocotyl explants of B. napus or B. juncea were used for Agrobacterium tumefaciens (GV3101‐MP90)‐mediated transformation (De Block et al., 1989). Plants were grown in tissue culture growth room conditions (22 ± 1 °C under 16 h light, 100 µE/m2/s). Transgenic plants were selected on the basis of resistance to the herbicide phosphinotricin, transferred to soil and grown in the glasshouse (16 h light/8 h dark, 20 °C/17 °C). Transgenic plants were further confirmed by PCR. The WRR4 construct used for transformation has been described previously by Borhan et al. (2008).
Generation of B. napus EDS1‐RNAi lines
The EDS1‐RNAi construct described by Borhan et al. (2008) was transferred to the B. napus double haploid line DH12075, which is naturally resistant to Al. candida races 2 and 7. The response of seedlings of EDS1‐suppressed lines was monitored at 7–14 dai with Ac2 or Ac7.
Supporting information
Fig. S1 Alignment of Brassica napus enhanced disease susceptibility 1 (EDS1) homologue and Arabidopsis thaliana EDS1 open reading frames (ORFs). blastn search of B. napus sequence database identified eight expressed sequence tags (ESTs) with homology to the Arabidopsis EDS1. Assembly of these ESTs resulted in a cDNA of 2043 bp and an ORF of 1803 bp encoding a lipase‐like protein. Underlined sequences show the interval of Arabidopsis EDS1 used for the suppression of Brassica EDS1 by RNA interference.
Fig. S2 Alignment of Brassica napus protein with homology to the lipase‐like protein enhanced disease susceptibility 1 (EDS1) in Arabidopsis thaliana, which is required for several examples of TIR‐NB‐LRR (Toll‐like/interleukin‐1 receptor‐nucleotide binding‐leucine‐rich repeat)‐mediated disease resistance. The B. napus EDS1 open reading frame encodes a 600‐amino‐acid protein of 68.6 kDa with 70% similarity and 61% identity to A. thaliana EDS1.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Elena Beynon for technical assistance and Delwin Epp for generating the Brassica transgenic lines.
REFERENCES
- Aarts, M.G. , Te Lintel Hekkert, B. , Holub, E.B. , Beynon, J.L. , Stiekema, W.J. and Pereira, A. (1998) Identification of R‐gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana . Mol. Plant–Microbe Interact. 11, 251–258. [DOI] [PubMed] [Google Scholar]
- Bernier, C.C. (1972) Disease of rapeseed in Manitoba in 1971. Can. Plant Dis. Surv. 52, 108. [Google Scholar]
- Bevan, M. and Waugh, R. (2007) Applying plant genomics to crop improvement. Genome Biol. 8, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhan, M.H. , Gunn, N. , Cooper, A. , Gulden, S. , Tör, M. , Rimmer, S.R. and Holub, E.B. (2008) WRR4 encodes a TIR‐NB‐LRR protein that confers broad‐spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida . Mol. Plant–Microbe Interact. 21, 757–768. [DOI] [PubMed] [Google Scholar]
- Botella, M.A. , Coleman, M.J. , Hughes, D.E. , Nishimura, M.T. , Jones, J.D. and Somerville, S.C. (1997) Map positions of 47 Arabidopsis sequences with sequence similarity to disease resistance genes. Plant J. 1197–1211. [DOI] [PubMed] [Google Scholar]
- Caldwell, K.S. and Michelmore, R.W. (2009) Arabidopsis thaliana genes encoding defense signalling and recognition proteins exhibit contrasting evolutionary dynamics. Genetics, 181, 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cooley, M.B. , Pathirana, S. , Wu, H.J. , Kachroo, P. and Klessig, D.F. (2000) Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell, 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- De Block, M. , De Brouwer, D. and Tenning, P. (1989) Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the bar and neo genes in the transgenic plants. Plant Physiol. 91, 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, A.C. and Ausubel, F.M. (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease‐resistance gene, is not race specific. Genetics, 171, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78. [DOI] [PubMed] [Google Scholar]
- Fan, Z. , Rimmer, S.R. and Stefansson, B.R. (1983) Inheritance of resistance to Albugo candida in rape Brassica napus. L. Can. J. Genet. Cytol. 25, 420–424. [Google Scholar]
- Gassmann, W. , Hinsch, M.E. and Staskawicz, B.J. (1999) The Arabidopsis RPS4 bacterial‐resistance gene is a member of the TIR‐NBS‐LRR family of disease‐resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Grant, M.R. , Godiard, L. , Straube, E. , Ashfield, T. , Lewald, J. , Sattler, A. , Innes, R.W. and Dangl, J.L. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science, 11, 843–846. [DOI] [PubMed] [Google Scholar]
- Harper, F.R. and Pittman, U.J. (1974) Yield loss by Brassica campestris and Brassica napus from systemic stem infection by Albugo cruciferarum . Phytopathology, 64, 408–410. [Google Scholar]
- Hill, C.B. , Crute, I.R. , Sheriff, C. and Williams, P.H. (1988) Specificity of Albugo candida and Peronospora parasitica pathotypes towards rapid‐cycling crucifers. Cruciferae Newsl. 13, 112–113. [Google Scholar]
- Holub, E.B. (2001) The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. [DOI] [PubMed] [Google Scholar]
- Holub, E.B. (2007) Natural variation in innate immunity of a pioneer species. Curr. Opin. Plant Biol. 10, 415–424. [DOI] [PubMed] [Google Scholar]
- Holub, E.B. (2008) Natural history of Arabidopsis thaliana and oomycete symbioses. Eur. J. Plant. Pathol. 22, 99–109. [Google Scholar]
- Holub, E.B. , Brose, E. , Tor, M. , Clay, C. , Crute, I.R. and Beynon, J.L. (1995) Phenotypic and genotypic variation in the interaction between Arabidopsis thaliana and Albugo candida . Mol. Plant–Microbe Interact. 8, 916–928. [DOI] [PubMed] [Google Scholar]
- Jarosch, B. , Jansen, M. and Schaffrath, U. (2003) Acquired resistance functions in mlo barley, which is hypersusceptible to Magaporthe grisea . Mol. Plant–Microbe Interact. 16, 107–114. [DOI] [PubMed] [Google Scholar]
- Jones, D.A. and Jones, J.D.G. (1997) The role of leucine‐rich repeat proteins in plant defences. Adv. Bot. Res. 24, 89–167. [Google Scholar]
- Jones, A.M. , Chory, J. , Dangl, J.L. , Estelle, M. , Jacobsen, S.E. , Meyerowitz, E.M. , Nordborg, M. and Weigel, D. (2008) The impact of Arabidopsis on human health: diversifying our portfolio. Cell, 133, 939–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, B. and Staskawicz, B.J. (1990) Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2 . Nature, 346, 385–386. [DOI] [PubMed] [Google Scholar]
- Kumari, K. , Varghese, T.M. and Suryanarayana, D. (1970) Qualitative changes in the amino acid contents of hypertrophied organs in mustard due to Albugo candida . Curr. Sci. 39, 240–241. [Google Scholar]
- Leach, J.E. , Vera Cruz, C.M. , Bai, J. and Leung, H. (2001) Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 39, 187–224. [DOI] [PubMed] [Google Scholar]
- Leister, D. , Balivora, A. , Salamini, F. and Gebhardt, C. (1996) A PCR‐based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat. Genet. 14, 421–429. [DOI] [PubMed] [Google Scholar]
- Li, C.X. , Sivasithamparam, K. , Walton, G. , Salisbury, P. , Burton, W. , Banga Surinder, S. , Banga, S. , Chattopadhyay, C. , Kumar, A. , Singh, R. , Singh, D. , Agnohotri, A. , Liu, S.Y. , Liu, Y.C. , Fu, T.D. , Wang, Y.F. and Barbetti, M.J. (2007) Expression and relationships of resistance to white rust (Albugo candida) at cotyledonary, seedling and flowering stages in Brassica juncea germplasm from Australia, China and India. Aust. J. Agric. Res. 58, 259–264. [Google Scholar]
- Li, Q.J. , Parks, P. and Rimmer, S.R. (1996) Development of monogenic lines for resistance to Albugo candida from a Canadian Brassica napus cultivar. Phytopathology, 86, 1000–1004. [Google Scholar]
- Marcroft, S.J. , Purwantara, A. , Wratten, N. , Salisbury, P.A. , Potter, T.D. , Barbetti, M.J. , Khangura, R. and Howlett, B.J. (2002) Reaction of a range of Brassica species under Austalian conditions to the fungus, Leptosphaeria maculans, the causal agent of blackleg. Aust. J. Exp. Agric. 42, 587–594. [Google Scholar]
- McHale, L.K. , Truco, M.J. , Kozik, A. , Wroblewski, T. , Ochoa, O.E. , Lahre, K.A. , Knapp, S.J. and Michelmore, R.W. (2009) The genomic architecture of disease resistance in lettuce. Theor. Appl. Genet. 118, 565–580. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie, G.A. (1973) Disease of brassica species in Saskatchewan 1970–1972. I. Staghead and aster yellows. Can. Plant Dis. Surv. 53, 19–25. [Google Scholar]
- Petrie, G.A. (1988) Races of Albugo candida (white rust and stag head) on cultivated cruciferae in Saskatchewan. Can. J. Plant Pathol. 10, 142–150. [Google Scholar]
- Pound, G.A. and Williams, P.H. (1963) Biological races of Albugo candida . Phytopathology, 53, 1146–1149. [Google Scholar]
- Rehmany, A.P. , Lynn, J.R. , Tör, M. , Holub, E.B. and Beynon, J.L. (2000) A comparison of Peronospora parasitica (downy mildew) isolates from Arabidopsis thaliana and Brassica oleracea using amplified fragment length polymorphism and internal transcribed spacer 1 sequence analyses. Fungal Genet. Biol. 30, 95–103. [DOI] [PubMed] [Google Scholar]
- Rentel, M.C. , Leonelli, L. , Dahlbeck, D. , Zhao, B. and Staskawicz, B.J. (2008) Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial and viral pathogens. Proc. Natl. Acad. Sci. USA, 105, 1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer, S.R. , Mathur, S. and Wu, C.R. (2000) Virulence of isolates of Albugo candida from western Canada to brassica species. Can. J. Plant Pathol. 22, 229–235. [Google Scholar]
- Ryan, C.A. , Huffaker, A. and Yamaguchi, Y. (2007) New insights into innate immunity in Arabidopsis. Cellul. Microbiol. 9, 1902–1908. [DOI] [PubMed] [Google Scholar]
- Schranz, M.E. , Song, B.H. , Windsor, A.J. and Mitchell‐Olds, T. (2007) Comparative genomics in the Brassicaceae: a family‐wide perspective. Curr. Opin. Plant Biol. 10, 168–175. [DOI] [PubMed] [Google Scholar]
- Shen, K.A. , Meyers, B.C. , Islam‐Faridi, M.N. , Chin, D.B. , Stelly, D.M. and Michelmore, R.W. (1998) Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol. Plant–Microbe Interact. 11, 815–823. [DOI] [PubMed] [Google Scholar]
- Sohn, K.H. , Lei, R. , Nemri, A. and Jones, J.D. (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana . Plant Cell, 19, 4077–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. , Bradeen, J.M. , Naess, S.K. , Raasch, J.A. , Wielgus, S.M. , Haberlach, G.T. , Liu, J. , Kuang, H. , Austin‐Phillips, S. , Buell, C.R. , Helgeson, J.P. and Jiang, J. (2003) Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. USA, 100, 9128–9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal, J. , Kaliff, M. , Bohman, S. and Dixelius, C. (2006) Transgressive segregation reveals two Arabidopsis TIR‐NB‐LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J. 46, 218–230. [DOI] [PubMed] [Google Scholar]
- Staal, J. , Kaliff, M. , Dewaele, E. , Persson, M. and Dixelius, C. (2008) RLM3, a TIR domain encoding gene involved in broad‐range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 55, 188–200. [DOI] [PubMed] [Google Scholar]
- Thines, M. , Choi, Y.‐J. , Kemen, E. , Ploch, S. , Holub, E.B. , Shin, H.‐D. and Jones, J.D.G. (2009) A new species of Albugo parasitic to Arabidopsis thaliana reveals new evolutionary patterns in white blister rusts (Albuginaceae). Persoonia, 22, 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.K. , Wang, M. , Bouwmeester, K. , Vosman, B. , Visser, R.G. , Jacobsen, E. , Govers, F. , Kamoun, S. and Van der Vossen, E.A. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. Plos. ONE. 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr, M. and Riethmüller, A. (2006) Phylogenetic relationships of Albugo candida (white blister rusts) based on LSU rDNA sequence and oospore data. Mycol. Res. 110, 75–85. [DOI] [PubMed] [Google Scholar]
- Van Der Vossen, E. , Sikkema, A. , Hekkert, B.T.L. , Gros, J. , Stevens, P. , Muskens, M. , Wouters, D. , Pereira, A. , Stiekema, W. and Allefs, S. (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad‐spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882. [DOI] [PubMed] [Google Scholar]
- Wang, G.L. , Song, W.Y. , Wu, R.L. , Sideris, S. and Ronald, P.C. (1996) The cloned gene, Xa27, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol. Plant–Microbe Interact. 9, 850–855. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Devoto, A. , Turner, J.G. and Xiao, S. (2007) Expression of the membrane‐associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol. Plant–Microbe Interact. 20, 966–976. [DOI] [PubMed] [Google Scholar]
- Warren, R.F. , Henk, A. , Mowery, P. , Holub, E. and Innes, R.W. (1998) A mutation within the leucine‐rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell, 10, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski, T. , Caldwell, K.S. , Piskurewicz, U. , Cavanaugh, K.A. , Xu, H. , Kozik, A. , Ochoa, O. , McHale, L.K. , Lahre, K. , Jelenska, J. , Castillo, J.A. , Blumenthal, D. , Vinatzer, B.A. , Greenberg, J.T. and Michelmore, R.W. (2009) Comparative large‐scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia . Plant Physiol. 150, 1733–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S.Y. , Ellwood, S. , Calis, O. , Patrick, E. , Li, T.X. , Coleman, M. and Turner, J.G. (2001) Broad‐spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8 . Science, 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Yang, S. , Chenwu Xu, M.G. , Gao, J. , Deshpande, S. , Lin, S. , Roe, B.A. and Zhu, H. (2008) Alfalfa benefits from Medicago truncatula: the RCT1 gene from M. truncatula confers broad‐spectrum resistance to anthracnose in alfalfa. Proc. Natl. Acad. Sci. USA, 105, 12164–12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment of Brassica napus enhanced disease susceptibility 1 (EDS1) homologue and Arabidopsis thaliana EDS1 open reading frames (ORFs). blastn search of B. napus sequence database identified eight expressed sequence tags (ESTs) with homology to the Arabidopsis EDS1. Assembly of these ESTs resulted in a cDNA of 2043 bp and an ORF of 1803 bp encoding a lipase‐like protein. Underlined sequences show the interval of Arabidopsis EDS1 used for the suppression of Brassica EDS1 by RNA interference.
Fig. S2 Alignment of Brassica napus protein with homology to the lipase‐like protein enhanced disease susceptibility 1 (EDS1) in Arabidopsis thaliana, which is required for several examples of TIR‐NB‐LRR (Toll‐like/interleukin‐1 receptor‐nucleotide binding‐leucine‐rich repeat)‐mediated disease resistance. The B. napus EDS1 open reading frame encodes a 600‐amino‐acid protein of 68.6 kDa with 70% similarity and 61% identity to A. thaliana EDS1.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


