SUMMARY
The capacity of Lettuce mosaic virus to overcome the lettuce resistance conferred by the mo11 and mo12 alleles of the gene for eukaryotic translation initiation factor 4E (eIF4E) was analysed using reverse genetics. Mutations in the virus genome‐linked protein (VPg) allowed mo11 only to be overcome, but mutations in the C‐terminal portion of the cylindrical inclusion (CI) protein allowed both alleles to be overcome. Site‐directed mutagenesis pinpointed a key role of the amino acid at position 621 in the virulence. This is the first example of the involvement of a potyviral CI protein in the breaking of an eIF4E‐mediated resistance.
INTRODUCTION
Of the plant viruses infecting lettuce (Lactuca sativa), the potyvirus Lettuce mosaic virus (LMV) is one of the most destructive (Le Gall, 2003). As for the other potyviruses, the viral genome consists of a single‐stranded, positive‐sense RNA molecule, about 10 kb in length, with a genome‐linked protein (VPg) covalently attached to its 5′ end and a poly‐A tail at its 3′ end. The viral RNA encodes a large polyprotein, processed by three virus‐encoded proteases (Revers et al., 1997). The only resistance genes currently used to protect lettuce crops worldwide are the recessive allelic genes mo11 and mo12, corresponding to mutant alleles of the gene coding for eukaryotic translation initiation factor 4E (eIF4E) (German‐Retana et al., 2008a; Nicaise et al., 2003).
LMV‐E is a resistance‐breaking isolate that induces symptoms in mo11and mo12 cultivars, whereas LMV‐0 is unable to induce symptoms in cultivars carrying the mo12 allele (tolerance) and does not systemically invade cultivars carrying the mo11 allele (resistance) (German‐Retana et al., 2008b; Revers et al., 1997). A reverse genetics analysis using full‐length infectious clones of LMV‐0 and LMV‐E has shown that the resistance‐breaking determinant(s) map to the 3′ half of the LMV‐E genome (Redondo et al., 2001), including the region encoding VPg. So far, VPg has been identified as the single potyvirus virulence determinant for the overcoming of eIF4E‐mediated recessive resistances (Ayme et al., 2006; Charron et al., 2008; Kang et al., 2005; Kuhne et al., 2003). In the present report, the LMV virulence region was narrowed down, and the use of site‐directed mutagenesis allowed the identification of a key amino acid for the ability of LMV isolates to overcome the mo1 eIF4E‐mediated resistance of lettuce.
RESULTS
Recombinants were constructed by exchanging unique restriction fragments between infectious cDNA clones of LMV‐0 (GenBank X97704) and LMV‐E (GenBank X97705) (Redondo et al., 2001; Yang et al., 1998). Following biolistics inoculation (German‐Retana et al., 2000) to susceptible lettuce cultivar Trocadéro, all isolates were transferred by mechanical inoculation to other cultivars carrying the mo11 or mo12allele (Mantilia and Salinas 88, respectively) or to susceptible cultivars (Trocadéro and Salinas) (Redondo et al., 2001). Salinas and Salinas 88 are nearly isogenic lettuce varieties differing at the mo1 locus (Irwin et al., 1999; Ryder, 1991).
Symptoms were monitored daily and leaves were harvested for the evaluation of viral accumulation 21 days post‐inoculation (dpi). Reverse transcriptase‐polymerase chain reaction (RT‐PCR) detection of the viral progenies was performed as described previously (Krause‐Sakate et al., 2002), and the identity and stability of each recombinant or mutant were assessed by restriction fragment length polymorphism (RFLP) analysis and/or sequence analysis. In all cases, genetic stability of the recombinants or mutants on inoculation of the resistant hosts was observed. The relative viral concentrations were estimated by semi‐quantitative double‐antibody sandwich enzyme‐linked immunosorbent assay (ELISA), as described previously (German‐Retana et al., 2000, 2003, 2008a), after tenfold dilution of the plant extracts, so that the relationship between A 405 and the antigen concentration was linear.
Figure 1 summarizes the behaviour of each recombinant with regard to viral accumulation and symptom induction at 21 dpi in three lettuce genotypes. All recombinants were infectious in susceptible cultivars Trocadéro and Salinas, and accumulated to levels comparable with those of the parental isolates (data not shown), indicating that their replication was not affected.
Figure 1.
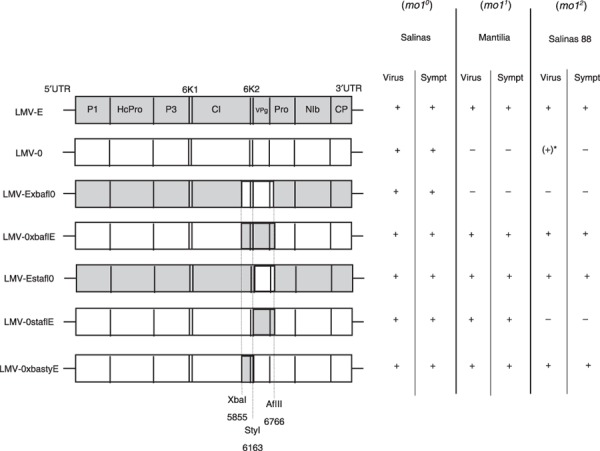
Structure and biological properties of susceptible and resistant lettuce plants of the Lettuce mosaic virus (LMV) recombinants used in this study. Left: schematic representations of the genomes of the recombinants constructed between LMV‐0 and LMV‐E. Coding sequences of LMV‐0 and LMV‐E are represented by white and grey boxes, respectively. Restriction sites used to construct the recombinants and their position along the LMV genome are indicated. Right: behaviour of the parental LMV isolates and of the recombinants in susceptible (mo10, cultivar Salinas) or resistant (mo11, cultivar Mantilia; mo1 2, cultivar Salinas 88) lettuce plants. Virus, viral accumulation in systemically infected leaves; Sympt, symptoms; –, no viral accumulation or no symptoms; +, viral accumulation or symptoms. *(+) denotes sporadic detection of low‐level (80% less than in susceptible plants), symptomless, systemic LMV‐0 accumulation in some Salinas 88 plants.
Recombinant LMV‐0xbaflE, containing a fragment from LMV‐E encoding the C‐terminus of the CI protein, 6K2, VPg and a small N‐terminal portion of the NIa proteinase, accumulated and induced symptoms in both mo11 and mo12 plants. The reciprocal recombinant (LMV‐Exbafl0) failed to overcome the mo11 and mo12 alleles (Fig. 1). Given that only two silent nucleotide changes separate the NIa proteinase coding region of the parental isolates, these results suggest that the region encoding the CI protein C‐terminus, 6K2 and VPg contains the mo1 resistance‐breaking determinant(s).
To refine this analysis, a second set of recombinants was constructed by exchanging only VPg and the NIa N‐terminal region (LMV‐0staflE and Estafl0). LMV‐0staflE multiplied and induced symptoms in Mantilia (mo11), showing that LMV‐E VPg is sufficient in the LMV‐0 background to enable the invasion of mo11 lettuce. However, this was not the case in Salinas 88 (mo12). Although it contains LMV‐0 VPg, the LMV‐Estafl0 reciprocal recombinant induced symptoms and accumulated in both resistant lettuce cultivars (Fig. 1), indicating that LMV‐E VPg is not necessary in the LMV‐E background to overcome mo1 resistance. Together, these results show that LMV‐E VPg is sufficient to enable LMV multiplication in mo11 plants, but does not confer mo12‐breaking properties, suggesting that another resistance‐breaking determinant must be present in the CI protein C‐terminus plus 6K2 region.
This hypothesis was evaluated using recombinant LMV‐0xbastyE, which contains the corresponding LMV‐E region in the LMV‐0 background. LMV‐0xbastyE invaded mo11 and mo12 plants, confirming that a virulence determinant conferring mo11‐ and mo12‐breaking ability is present in the C‐terminal region of the CI protein or 6K2. Despite extensive efforts, it has unfortunately not been possible to obtain the reciprocal recombinant (LMV‐Exbasty0) to further strengthen these results.
Sequence comparisons of the CI protein C‐terminus and 6K2 region from a collection of 16 LMV isolates representative of the biological and molecular diversity of LMV (seven isolates unable to multiply on mo11‐ or mo12‐carrying cultivars and nine mo1‐breaking isolates) showed that, of the five amino acids differing between LMV‐0 and LMV‐E in the part of the CI protein under study, only two (positions 602 and 621) differed between LMV‐0 and all mo1‐breaking isolates. These two positions were therefore selected for PCR‐based site‐directed mutagenesis to generate point mutants derived from LMV‐0 and LMV‐E: LMV‐0‐A602V, in which alanine 602 (codon GCA) is substituted by a valine (GTA); LMV‐0‐S621T, in which serine 621 (TCC) is substituted by a threonine (ACC); and the reciprocal mutant LMV‐E‐T621S, in which threonine 621 (ACC) is substituted by a serine (TCC). For each mutagenesis experiment, two fragments were amplified with primers A and B, and C and D, respectively (Table 1) (primers B and C include the mutated nucleotide indicated in bold). The fusion‐PCR step was further performed with primers A and D, by mixing the previous amplified fragments.
Table 1.
Oligonucleotides used for polymerase chain reaction (PCR)‐fusion‐based mutagenesis experiments.
| Mutant | Name | Position* | Polarity | Sequence† | |
|---|---|---|---|---|---|
| LMV‐E‐T621S | A | 5493 | Sense | 5′‐GCGCTCGAGAGTACAACTCCTTAGGAGCC‐3′ | LMV‐E |
| B | 5927 | Antisense | 5′‐CTCCATGTCGAAGGAACCCCCATCACT‐3′ | ||
| C | 5927 | Sense | 5′‐AGTGATGGGGGTTCCTTCGACATGGAG‐3′ | ||
| D | 6014 | Antisense | 5′‐TTTGAGACCGAGTCGTTTGCTAAGAGCTCC‐3′ | ||
| LMV‐0‐S621T | A | 5444 | Sense | 5′‐AAACTAAGCACGCTGGCAATAC‐3′ | LMV‐0 |
| B | 5927 | Antisense | 5′‐GAAGGTACCCCCGTCACC‐3′ (KpnI) | ||
| C | 5930 | Sense | 5′‐GACGGGGGTACCTTCGACAT‐3′ (KpnI) | ||
| D | 6146 | Antisense | 5′‐TTGGTGTCGAACTGCATCAGC‐3′ | ||
| LMV‐0‐A602V | A | 5444 | Sense | 5′‐AAACTAAGCACGCTGGCAATAC‐3′ | LMV‐0 |
| B | 5869 | Antisense | 5′‐CTGACTACCTGCAGTTTTTGC‐3′ (PstI) | ||
| C | 5871 | Sense | 5′‐AAAAACTGCAGGTAGTCAGAGACAC‐5′ (PstI) | ||
| D | 6146 | Antisense | 5′‐TTGGTGTCGAACTGCATCAGC‐3′ |
The position of the first nucleotide of the primer is indicated.
The mutated bases are indicated in bold in the nucleotide sequence of the primers. In some cases, oligonucleotides were designed to introduce recognition sites for restriction endonucleases used for screening purposes (recognition sites in italic, restriction endonucleases indicated in parentheses).
LMV‐0‐A602V, in which alanine 602 is substituted with valine as in mo1‐breaking isolates, accumulated and induced symptoms on the susceptible cultivar Salinas, but did not induce symptoms or accumulate in mo11 or mo12 cultivars (data not shown). The substitution at position 602 was therefore not sufficient to confer virulence.
LMV‐0‐S621T, in which serine 621 is substituted with threonine as in LMV‐E, induced symptoms similar to those caused by the wild‐type LMV‐0 on susceptible Salinas, and accumulated to a similar level (Fig. 2). However, unlike its parent LMV‐0, this mutant induced symptoms in both mo11 and mo12 cultivars (Mantilia and Salinas 88, respectively), and accumulated in both cultivars at 21 dpi. Variability in the virus accumulation levels was observed within and between our biological experiments. However, all mo1 plants inoculated with LMV‐0‐S621T became infected and displayed symptoms (gain of virulence), contrary to the parental LMV‐0 (Fig. 2). Nevertheless, the LMV‐0‐S621T efficiency of infection in Mantilia did not reach that of LMV‐E, LMV‐0xbastyE or LMV‐0xbaflE (symptoms less severe on Mantilia). The reciprocal mutant in the LMV‐E background, LMV‐E‐T621S, accumulated and induced symptoms in the susceptible cultivar Salinas, but not in the mo11and mo12 cultivars (Fig. 2). The stability of both point mutants was verified by sequencing following their propagation in the various hosts. No additional compensatory mutations were observed in the CI protein/6K2 and VPg coding regions (data not shown). The substitution of serine by threonine at position 621 is therefore sufficient to confer to LMV‐0 the ability to overcome the resistance afforded by the two mo1 alleles, whereas the reciprocal substitution results in a loss of virulence of LMV‐E, suggesting that amino acid 621 of the CI protein plays a critical role in LMV virulence in lettuce plants carrying mutations in eIF4E.
Figure 2.
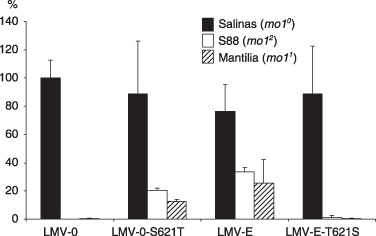
Effects of mutations of amino acid 621 on the accumulation of the various viruses in susceptible and resistant lettuce. Plants from the varieties Salinas (mo10, susceptible), Salinas 88 (mo12) and Mantilia (mo11) were assayed. The accumulation of each virus was determined by enzyme‐linked immunosorbent assay (ELISA) and expressed as a percentage of the average value measured for LMV‐0 on the susceptible lettuce cultivar Salinas. The samples (at least three plants for each virus) were collected from non‐inoculated leaves (level 3 from the top of the plant) at 21 days post‐inoculation (dpi) in at least four independent experiments. Average values are shown, together with standard deviations.
Overall, the results presented here show that the introduction of VPg from a virulent isolate into an avirulent one is sufficient to restore a full compatibility with lettuce varieties carrying the mo11 allele, but not the mo12allele. Simultaneously, the region coding for the C‐terminal portion of the CI protein and 6K2 allows both eIF4E alleles to be overcome. Site‐directed mutagenesis of the CI protein at position 621 was sufficient to affect, in a reciprocal manner, the infection phenotype of the parental viruses, in plants carrying both resistance alleles, further demonstrating that the C‐terminus of the CI protein is directly involved in the breaking of eIF4E‐mediated resistance in lettuce.
DISCUSSION
The interactions involved in the lettuce–LMV pathosystem appear to be more complex than those reported in other plant–potyvirus pathosystems, where VPg has been shown to be solely responsible for overcoming eIF4E‐mediated resistances (Ayme et al., 2006; Bruun‐Rasmussen et al., 2007; Charron et al., 2008; Kang et al., 2005), although the avirulence determinant corresponding to the sbm2 resistance gene in pea, shown to be tightly linked to the eIF(iso)4E gene, is the viral P3 of Pea seed‐borne mosaic virus (PSbMV) (Gao et al., 2004; Hjulsager et al., 2006).
It is possible that the LMV infection process in lettuce differs in essence from other host–potyvirus interactions, but the most parsimonious hypothesis is that the genetic diversity in LMV happens to reveal a CI protein role that has remained unnoticed so far in other pathosystems.
The potyvirus CI protein has RNA‐binding domains (Fernandez et al., 1995) and carries RNA helicase and ATPase activities in its N‐terminal two‐thirds, both essential for viral replication (Fernandez et al., 1997; Gomez de Cedron et al., 2006).
So far, no function has been assigned to the C‐terminal region of the CI protein, although it is a prominent feature of the potyviral CI protein, extending beyond the conserved domains. The identified activities located in the N‐terminal half of the protein (ATPase, RNA binding and RNA helicase) are not affected in vitro by removal of the last 103 amino acids of the CI protein (Fernandez et al., 1995). However, extensive mutagenesis experiments performed on Tobacco etch virus (TEV) and Potato virus A (PVA) identified mutations in the C‐terminal region able to abolish viral replication or systemic movement (Carrington et al., 1998; Kekarainen et al., 2002), suggesting that the CI protein C‐terminal region plays an important role(s) during viral infection. Furthermore, Choi et al. (2000) showed that the C‐terminus of the CI protein, helper component proteinase (HcPro), P1 and P3 of Wheat streak mosaic virus (WSMV), a Tritimovirus (family Potyviridae), interact with one another in yeast cells and in vitro. Recently, interactions between VPg and HcPro of LMV and between LMV VPg and lettuce eIF4E have been demonstrated in vitro (Roudet‐Tavert et al., 2007). The C‐terminus of the CI protein could therefore be involved in a network of viral and cellular protein interactions, involving VPg, the CI protein, eIF4E and, possibly, HcPro and the translation initiation factor eIF4G, shown to be required for LMV infection in Arabidopsis thaliana (Nicaise et al., 2007). Furthermore, as P3 is involved in the overcoming of sbm‐2 resistance (Hjulsager et al., 2006), the possibility exists that P3 could also participate in the proposed interaction of the CI protein and VPg with host translation initiation factors. Furthermore, as it has been shown that partially processed viral products of potyvirus can be accumulated in infected plants (Merits et al., 2002), including P3, the CI protein and VPg, it cannot be excluded that partially processed products could be the viral factor interacting with eIF4E. Mutations in eIF4E in resistant plants could destabilize this interaction network, resulting in resistance, whereas mutations in the CI protein could stabilize it, even in the presence of the mutated eIF4E, resulting in resistance breakdown. Further structural and biochemical studies should shed light on the role of the CI protein C‐terminal domain in the success of virus infection.
ACKNOWLEDGMENTS
This work was partially supported by EPR Aquitaine (ref. 20000307004) and by the French National Agency for Research (Poty4E, ref. ANR‐05‐BLAN‐0302‐01). AAR was supported by a fellowship from the Syrian Ministry of Higher Education, and TG by a joint fellowship between the Aquitaine region and the ‘Plant Health and the Environment’ division of Institut National de la Recherche Agronomique.
We thank Thierry Mauduit and Marylin Roncoroni for taking care of the plants, and all members of the Interactions Plantes Virus group for useful discussions.
REFERENCES
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadoeuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of potato virus Y confer virulence on the pvr2(3) resistance in pepper. Mol. Plant–Microbe Interact. 19, 557–563. [DOI] [PubMed] [Google Scholar]
- Bruun‐Rasmussen, M. , Moller, I.S. , Tulinius, G. , Hansen, J.K. , Lund, O.S. and Johansen, I.E. (2007) The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum . Mol. Plant–Microbe Interact. 20, 1075–1082. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. , Jensen, P.E. and Schaad, M.C. (1998) Genetic evidence for an essential role for potyvirus CI protein in cell‐to‐cell movement. Plant J. 14, 393–400. [DOI] [PubMed] [Google Scholar]
- Charron, C. , Nicolai, M. , Gallois, J.L. , Robaglia, C. , Moury, B. , Palloix, A. and Caranta, C. (2008) Natural variation and functional analyses provide evidence for co‐evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. [DOI] [PubMed] [Google Scholar]
- Choi, I.R. , Stenger, D.C. and French, R. (2000) Multiple interactions among proteins encoded by the mite‐transmitted wheat streak mosaic tritimovirus. Virology, 267, 185–198. [DOI] [PubMed] [Google Scholar]
- Fernandez, A. , Lain, S. and Garcia, J.A. (1995) RNA helicase activity of the plum pox potyvirus CI protein expressed in Escherichia coli. Mapping of an RNA binding domain. Nucleic Acids Res. 23, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, A. , Guo, H.S. , Saenz, P. , Simon‐Buela, L. , Gomez de Cedron, M. and Garcia, J.A. (1997) The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25, 4474–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z. , Eyers, S. , Thomas, C. , Ellis, N. and Maule, A. (2004) Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed‐borne mosaic virus. Theor. Appl. Genet. 109, 488–494. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Candresse, T. , Alias, E. , Delbos, R. and Le Gall, O. (2000) Effects of GFP or GUS tagging on the accumulation and pathogenicity of a resistance breaking LMV isolate in susceptible and resistant lettuce cultivars. Mol. Plant–Microbe Interact. 13, 316–324. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Redondo, E. , Tavert‐Roudet, G. , Le Gall, O. and Candresse, T. (2003) Introduction of a NIa proteinase cleavage site between the reporter gene and HC‐Pro only partially restores the biological properties of GUS‐ or GFP‐tagged LMV. Virus Res. 98, 151–162. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Walter, J. , Doublet, B. , Roudet‐Tavert, G. , Nicaise, V. , Lecampion, C. , Houvenaghel, M.C. , Robaglia, C. , Michon, T. and Le Gall, O. (2008a) Mutational analysis of plant cap‐binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J. Virol. 82, 7601–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German‐Retana, S. , Walter, J. and Le Gall, O. (2008b) Lettuce mosaic virus: from pathogen diversity to host interactors. Mol. Plant Pathol. 9, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez de Cedron, M. , Osaba, L. , Lopez, L. and Garcia, J.A. (2006) Genetic analysis of the function of the plum pox virus CI RNA helicase in virus movement. Virus Res. 116, 136–145. [DOI] [PubMed] [Google Scholar]
- Hjulsager, C.K. , Olsen, B.S. , Jensen, D.M. , Cordea, M.I. , Krath, B.N. , Johansen, I.E. and Lund, O.S. (2006) Multiple determinants in the coding region of Pea seed‐borne mosaic virus P3 are involved in virulence against sbm‐2 resistance. Virology, 355, 52–61. [DOI] [PubMed] [Google Scholar]
- Irwin, S.V. , Kesseli, R.V. , Waycott, W. , Ryder, E.J. , Cho, J.J. and Michelmore, R.W. (1999) Identification of PCR‐based markers flanking the recessive LMV resistance gene mo1 in an intraspecific cross in lettuce. Genome, 42, 982–986. [Google Scholar]
- Kang, B.C. , Yeam, I. , Frantz, J.D. , Murphy, J.F. and Jahn, M.M. (2005) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42, 392–405. [DOI] [PubMed] [Google Scholar]
- Kekarainen, T. , Savilahti, H. and Valkonen, J.P. (2002) Functional genomics on potato virus A: virus genome‐wide map of sites essential for virus propagation. Genome Res. 12, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause‐Sakate, R. , Le Gall, O. , Fakhfakh, H. , Peypelut, M. , Marrakchi, M. , Varveri, C. , Pavan, M.A. , Souche, S. , Lot, H. , Zerbini, F.M. and Candresse, T. (2002) Molecular characterization of Lettuce mosaic virus field isolates reveals a distinct and widespread type of resistance‐breaking isolate: LMV‐Most. Phytopathology, 92, 563–572. [DOI] [PubMed] [Google Scholar]
- Kuhne, T. , Shi, N. , Proeseler, G. , Adams, M.J. and Kanyuka, K. (2003) The ability of a bymovirus to overcome the rym4‐mediated resistance in barley correlates with a codon change in the VPg coding region on RNA1. J. Gen. Virol. 84, 2853–2859. [DOI] [PubMed] [Google Scholar]
- Le Gall, O. (2003) Lettuce mosaic virus In: CMI/AAB Description of Plant Viruses (Jones A.T., Robinson D.J., Boonham N. and Mumford R., eds), p. 399 Wellesbourne: Association of Applied Biologists; URL http://www.dpvweb.net/dpv/showdpv.php?dpvno=399 [accessed on 8 October 2008]. [Google Scholar]
- Merits, A. , Rajamaki, M.L. , Lindholm, P. , Runeberg‐Roos, P. , Kekarainen, T. , Puustinen, P. , Makelainen, K. , Valkonen, J.P. and Saarma, M. (2002) Proteolytic processing of potyviral proteins and polyprotein processing intermediates in insect and plant cells. J. Gen. Virol. 83, 1211–1221. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , German‐Retana, S. , Sanjuan, R. , Dubrana, M.P. , Mazier, M. , Maisonneuve, B. , Candresse, T. , Caranta, C. and LeGall, O. (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol. 132, 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. , Gallois, J.L. , Chafiai, F. , Allen, L.M. , Schurdi‐Levraud, V. , Browning, K.S. , Candresse, T. , Caranta, C. , Le Gall, O. and German‐Retana, S. (2007) Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana . FEBS Lett. 581, 1041–1046. [DOI] [PubMed] [Google Scholar]
- Redondo, E. , Krause‐Sakate, R. , Yang, S.J. , Lot, H. , Le Gall, O. and Candresse, T. (2001) Lettuce mosaic virus (LMV) pathogenicity determinants in susceptible and tolerant lettuce varieties map to different regions of the viral genome. Mol. Plant–Microbe Interact. 14, 804–810. [DOI] [PubMed] [Google Scholar]
- Revers, F. , Yang, S.J. , Walter, J. , Souche, S. , Lot, H. , Le Gall, O. , Candresse, T. and Dunez, J. (1997) Comparison of the complete nucleotide sequences of two isolates of Lettuce mosaic virus differing in their biological properties. Virus Res. 47, 167–177. [DOI] [PubMed] [Google Scholar]
- Roudet‐Tavert, G. , Michon, T. , Walter, J. , Delaunay, T. , Redondo, E. and Le Gall, O. (2007) Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HcPro. J. Gen. Virol. 88, 1029–1033. [DOI] [PubMed] [Google Scholar]
- Ryder, E.J. (1991) Salinas 88 lettuce. Hortscience, 26, 439–440. [Google Scholar]
- Yang, S.J. , Revers, F. , Souche, S. , Lot, H. , Le Gall, O. , Candresse, T. and Dunez, J. (1998) Construction of full‐length cDNA clones of Lettuce mosaic virus (LMV) and the effects of intron‐insertion on their viability in Escherichia coli and on their infectivity to plants. Arch. Virol. 143, 2443–2451. [DOI] [PubMed] [Google Scholar]


