SUMMARY
Host gene expression changes in the early response to potato virus YNTN interaction were compared in two differently sensitive potato cultivars: the resistant cultivar Santé and the sensitive cultivar Igor. Hybridization of potato TIGR cDNA microarrays allowed us to monitor the expression of approximately 10 000 genes simultaneously at 0.5 and 12 h post‐inoculation (hpi). Microarray data, analysed by statistics and data mining, were complemented by subtraction library construction and sequence analysis to validate the findings. The expression profiles of the two cultivars were similar and faint at 0.5 hpi, but they differed substantially at 12 hpi. Although, at 0.5 hpi, cv. Santé responded by the differential expression of a greater number of genes, at 12 hpi the number was higher in cv. Igor. The majority of genes in this cultivar were down‐regulated at 12 hpi, indicating a host gene shut‐off. Suites of genes that exhibited altered transcript abundance in response to the virus were identified, and included genes involved in the processes of photosynthesis, perception, signalling and defence responses. The expression of the considerable number of genes associated with photosynthesis was surprisingly up‐regulated as early as 0.5 hpi and down‐regulated at 12 hpi in both cultivars. The expression of genes involved in perception and signalling was increased in the sensitive cultivar at 12 hpi. By contrast, a simultaneous strong defence response at the transcriptional level was evident in the resistant cultivar, as shown by the up‐regulation of genes involved in brassinosteroid, polyamine and secondary metabolite biosynthesis, and of genes coding for pathogenesis‐related proteins.
INTRODUCTION
Plants respond to pathogens by activating a variety of active and passive defence mechanisms, which can be detected as a broad spectrum of physiological and histological changes (Whitham et al., 2003). Plant viruses enter the cells by mechanical damage of the cell wall and membrane. In the absence of an active defence response, the virus spreads to neighbouring cells through plasmodesmata and, finally, causes disease symptoms manifesting as structural and physiological changes. It has been shown that diverse susceptible hosts are not passive against a pathogen. However, even though they can set up a defence response, it is neither intensive nor rapid enough to stop viral replication and spread. Comparisons of induced defence reactions in compatible (susceptible) and incompatible (resistant) interactions indicate that effective resistance is reliant on the speed with which they occur (O'Donnell et al., 2003).
Generally, plant–virus interactions are the least studied pathogen interactions. Moreover, the very early response on the whole transcriptome level in these interactions is even less well understood. The majority of recently published transcriptome studies of plant–virus interactions were conducted just before or at the time of symptom development (Golem and Culver, 2003; Senthil et al., 2005; Smith et al., 2004; Whitham et al., 2003; Yang et al., 2007). Therefore, it was mainly the consequences of the infection, rather than plant active defence, that were assessed. Despite the fact that the true defence response is limited in time (Scheideler et al., 2002), it is very difficult to detect the early gene expression changes in the small proportion of affected cells of the inoculated leaves.
Potato virus Y (PVY), a member of the Potyviridae family, is classified into different strains. PVYNTN, a PVYN strain variety (Singh et al., 2008), is the most aggressive. Because of the importance of potato as a crop and the epidemic spread of PVYNTN in Europe and other continents from the 1980s onwards, many physiological and morphological parameters have been studied in PVYNTN‐infected plants. Most potato cultivars are susceptible to PVYNTN infection and develop diverse symptoms, including severe necrotic ringspots on tubers and necrotic spots (local lesions) or wrinkles and mosaic chloroses on leaves. The cultivar Igor is one of the most susceptible and sensitive cultivars, whereas cv. Santé is extremely resistant and not affected by PVYNTN infection. No hypersensitive reaction lesions are visible in this cultivar (Ravnikar, 2005). The reason for its resistance is the Rysto gene, which was bred into this cultivar and confers extreme resistance to the virus (Flis et al., 2005).
Several studies of individual physiological and morphological parameters in early potato–PVYNTN interactions (for example, Krečič‐Stres et al., 2005; Milavec et al., 2001b, 2008) have shown a link between plant sensitivity and response of the selected molecules, but have not tracked the overall changes arising from the potato–virus interaction. Recently, a study has been published focusing on the disease response of cv. Igor at the transcriptome level, several days after PVYNTN infection, using self‐made microarrays (Pompe‐Novak et al., 2006). Among others, changes in the expression of genes involved in photosynthesis and defence responses (β‐1,3‐glucanase, heat shock proteins, wounding‐induced protein) were reported.
Our goal was to investigate gene expression changes in the very early response [0.5 and 12 h post‐inoculation (hpi)] of two differently sensitive agronomically important potato cultivars of different origin: cv. Santé resistant to PVYNTN and cv. Igor sensitive to PVYNTN. The experiment was designed to control as many factors of variability as possible. Microarray data were analysed by rigorous statistics and data mining, and were complemented by subtraction library sequence analysis to validate the findings. Pathway analysis enabled us to conclude that the suites of co‐expressed genes involved in the same process, which were changed considerably after virus inoculation, were associated with photosynthesis, signalling and defence responses. Understanding the differences in and the time frame of the gene expression response in two differently sensitive potato cultivars enables a detailed insight to be made into the resistance mechanisms of potato–virus interaction. Furthermore, investigations in the very early stages following virus inoculation have enabled us to identify the earliest steps that could lead to the differential sensitivity of plants to the virus.
RESULTS
Expression profiles in the early response to virus inoculation
Gene expression levels in PVYNTN‐inoculated leaves of two differently sensitive potato cultivars (resistant cv. Santé and sensitive cv. Igor) were monitored 0.5 and 12 hpi, immediately on pathogen entrance into the plant.
Each cDNA microarray, representing approximately 10 000 potato genomic pool genes, was hybridized with a virus‐inoculated and a mock‐inoculated sample of cv. Santé or cv. Igor. After data pre‐processing, statistical analysis and data mining were carried out to obtain lists of differentially expressed genes and a list of rules determining resistance or sensitivity. Subtraction libraries, containing expressed sequence tags (ESTs) specific to the resistant cultivar, were constructed from the same biological material to complement the microarray data. Finally, all the results were compiled using the same functional annotation.
Microarray analysis resulted in lists of differentially expressed genes (Fig. 1; Table S1, see ‘Supporting Information’). More genes were differentially expressed at 0.5 hpi in cv. Santé than in cv. Igor. In contrast, at 12 hpi, the number of differentially expressed genes in cv. Igor was higher than that in cv. Santé. The difference was more pronounced in the number of down‐regulated genes, which suggests a massive host gene shut‐off, characteristic of compatible virus–plant interactions (Maule et al., 2002).
Figure 1.
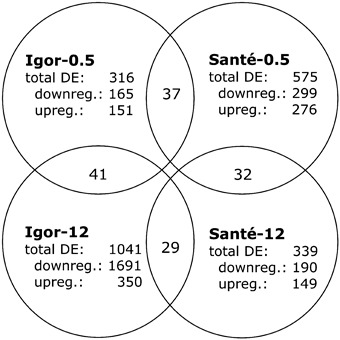
Numbers of differentially expressed (DE) genes (total, down‐regulated and up‐regulated) in virus‐ vs. mock‐inoculated plants in cv. Igor and Santé at 0.5 and 12 h following potato virus YNTN (PVYNTN) inoculation, and the numbers of genes in the intersections of the lists.
The pattern of gene expression was analysed further by studying the intersection of lists of the differentially expressed genes (Fig. 1). Each intersection consists of the genes that overlap in the two lists of differentially expressed genes. Approximately 10% of the genes overlap in the two lists. The majority of the genes present in the intersection between cv. Igor 0.5 hpi and cv. Santé 0.5 hpi, i.e. those that responded to virus infection at 0.5 hpi, were regulated in the two cultivars in the same manner, either up or down (Fig. 2).
Figure 2.
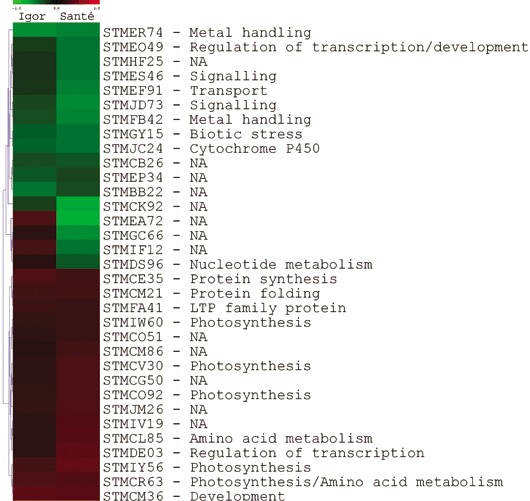
Comparison of the expression profiles of the cultivars at 0.5 h post‐inoculation (hpi). Genes (clone name, process) that were differentially expressed in both cultivars (Igor and Santé) at 0.5 hpi are shown. Log2 ratios of expression in virus‐ vs. mock‐inoculated plants (red, up‐regulated; green, down‐regulated) are clustered hierarchically (Euclidean distance, average linkage).
Pathway analysis
The classification of the differentially expressed genes using MapMan ontology enabled the co‐expression of suites of genes involved in the same processes to be identified (Table 1).
Table 1.
Significantly altered processes or protein families with corresponding P values* indicated by changes in gene expression level in cv. Igor and Santé at 0.5 h (0.5) and 12 h (12) after potato virus YNTN (PVYNTN) inoculation.
| Cultivar/time | Up‐regulated processes/protein families | P value | Down‐regulated processes/protein families | P value |
|---|---|---|---|---|
| Igor 0.5 | Photosynthesis light reactions | 0 | WRKY domain transcription factor family | 0 |
| Carbonic anhydrases | 0 | |||
| Santé 0.5 | Photosynthesis light reactions | 0 | Stress responses | 2 × 10−9 |
| Plastid ribosomal proteins | 4 × 10−16 | Secondary metabolism: simple phenols | 3 × 10−7 | |
| Tetrapyrrole synthesis | 7 × 10−15 | |||
| Igor 12 | Signalling | 2 × 10−6 | Photosynthesis reactions | 1 × 10−7 |
| WRKY domain transcription factor family | 1 × 10−7 | Invertase/pectin methylesterase inhibitor family | 9 × 10−5 | |
| Santé 12 | PR proteins | 0 | Photosynthesis | 8 × 10−7 |
| Biotic stress responses | 1 × 10−13 | |||
| Polyamine metabolism | 3 × 10−11 | |||
| Cell wall degradation: pectate lyases and polygalacturonases | 2 × 10−7 |
Wilcoxon rank sum test was carried out to predict BINs that exhibit different behaviour in terms of expression profile compared with all the other remaining BINs (Usadel et al., 2005).
Photosynthesis was changed significantly. It was up‐regulated at 0.5 hpi and down‐regulated at 12 hpi in both cultivars (Table 1; Fig. 3). However, at 0.5 hpi, the gene expression changes were more pronounced in the resistant cv. Santé. Moreover, in the resistant cv. Santé at 0.5 hpi, the increased expression of genes involved in photosynthesis was coupled with an increased expression of those involved in chlorophyll synthesis (magnesium protoporphyrin IX chelatase, hydroxymethylbilane synthase/porphobilinogen deaminase etc.; Fig. 3).
Figure 3.
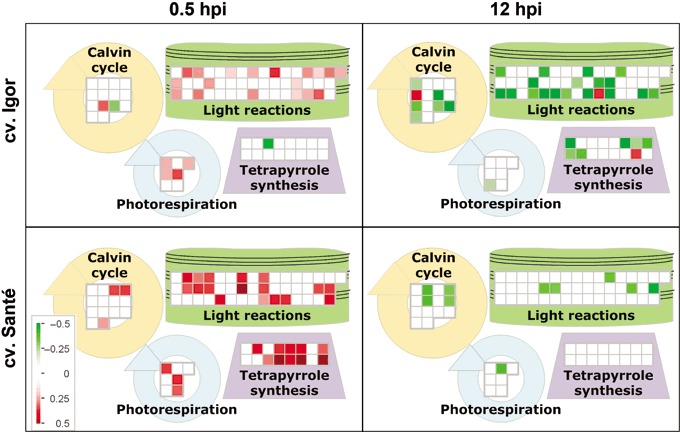
Expression of the genes involved in pathways connected to photosynthesis (MapMan bins: 1.1: PS.lightreaction; 1.2: PS.photorespiration; 1.3: PS.calvin cycle; 19: tetrapyrrole synthesis) in the sensitive cv. Igor and the resistant cv. Santé, 0.5 and 12 h following potato virus YNTN (PVYNTN) inoculation. Each square represents the log2 ratio of expression of one clone in virus‐ vs. mock‐inoculated plants (red, up‐regulated; green, down‐regulated). Detailed information about the differentially expressed genes shown in the figure can be found in Table S1.
Other major processes that were differentially regulated were general stress responses, which were down‐regulated in cv. Santé at 0.5 hpi, biotic stress responses, which were up‐regulated in cv. Santé at 12 hpi, and signalling, which was up‐regulated in cv. Igor at 12 hpi (Table 1).
Determination of resistance and sensitivity by data mining
In order to identify genes whose expression was a determinant of plant resistance or sensitivity, data mining was carried out as an alternative microarray data analysis approach.
The output of the data mining algorithm is expressed as a formula:
‘IF (gene_a:2=A AND gene_b:2 = A AND gene_c:1 = A AND gene_d:D = A AND gene_e:2 = P ...) THEN resistant’.
The result can be read as: ‘IF gene a AND gene b are down‐regulated at time 2 after the infection AND gene c is down‐regulated at time 1 after the infection AND gene d is down‐regulated at the difference of times post‐infection AND gene e is up‐regulated at time 2 post‐infection AND ... THEN the plant is resistant’.
In our study, there were 130 and 27 rules for cultivar resistance and sensitivity, respectively (Table S2, see ‘Supporting Information’). A lot of genes that were involved in the plant's resistance response were present in the list of rules for resistance (e.g. genes coding for proteinase inhibitors, secondary metabolites etc.), and the biological significance of this is discussed in the following sections. Twenty‐seven clones whose expression was determinant for resistance at 12 hpi were also found in the list of differentially expressed genes at 12 hpi for cv. Santé.
Complementation of microarray with subtraction library and quantitative polymerase chain reaction (qPCR) results
As an independent method to validate microarray results, two subtraction libraries were constructed from virus‐inoculated plant material of cv. Santé at 12 hpi: one enriched for genes that were up‐regulated (SO) and one for those that were down‐regulated (SS). cDNA inserts from 383 and 574 transformants from the SO and SS libraries, respectively, were sequenced. The sequences were trimmed and aligned into contigs. The SO library yielded 268 ESTs, 26 contigs (nine of which coded for viral proteins) and 242 singletons. The SS library contained 475 ESTs, 71 contigs and 404 singletons.
After removing viral sequences (derived from virus inoculum) and sequences that appeared in both libraries, there were 222 SO‐specific and 433 SS‐specific ESTs (Table S3, see ‘Supporting Information’). Different numbers of ESTs in the libraries are partly the consequence of different numbers of sequenced inserts from both libraries, and partly the consequence of virus sequences that were present in the SO library.
ESTs from the SO and SS libraries matched 209 and 368 different TIGR Potato Gene Index (StGI) accessions, respectively. There were nine (4%) and 37 (9%) ESTs in the SO and SS libraries without a significant match to the sequences in the StGI database. These ‘unknown’ ESTs [the majority had no match in the National Center for Biotechnology Information (NCBI) database either] could represent specific genes that code for previously unknown proteins with potentially crucial roles in the potato–virus interaction. Unfortunately, the sequences were too short to identify possible conserved protein domains which could help in the elucidation of the function of these proteins.
Complementation of microarray data analysed by statistical methods and data mining with subtraction library data greatly improves the significance of the results. Although both methods are based on hybridization, they are different in such a way that microarrays are limited to the sequences that are present on the microarray, whereas subtraction library data can yield previously unknown sequences. When comparing clones that were expressed in the subtraction libraries and differentially expressed following microarray analysis, the expression of 19 genes was confirmed in cv. Santé at 12 hpi. However, when comparing different genes having similar functions or involved in the same biological process, the agreement between the methods was much higher. As an example of an independent confirmation of the microarray results, the expression data for several clones representing 2‐oxoglutarate‐dependent dioxygenase (2‐ODD), an enzyme involved in secondary metabolite biosynthesis, were compared (Table 2). The expression of most of the clones was up‐regulated at 12 hpi and was found to be a determinant of resistance. In addition, two ESTs with high homology to the same gene were found in the SO library, confirming the microarray data. Moreover, the down‐regulation of 2‐ODD (clone STMCN85) at 12 hpi was found to be a determinant of cultivar sensitivity (Table S2).
Table 2.
Complementation of microarray and subtraction library data for the expression of 2‐oxoglutarate‐dependent dioxygenase. For each clone, the corresponding M value (log2 ratio of expression in virus‐ vs. mock‐inoculated plants), data mining resistance determining rules [12, 12 h post‐inoculation (hpi); D, difference between 0.5 and 12 hpi; P, up‐regulated) and expressed sequence tag (EST) sequences in the SO subtraction library (enriched for genes that were up‐regulated in cv. Santé at 12 hpi) showing high similarity (E < 10−10) to the clones are given.
| Clone | Statistics (M) | Data mining (resistance rules) | SSH (ESTs in the SO library) |
|---|---|---|---|
| STMCL06 | 0.84 | D = P; 12 = P | SO5T7snr004, SO5T7snr062 |
| STMGI29 | 0.78 | D = P; 12 = P | SO5T7snr004, SO5T7snr062 |
| STMGO18 | 0.59 | 12 = P | SO5T7snr004, SO5T7snr062 |
| STMCF39 | 0.73 | SO5T7snr004 | |
| STMCN85 | NA | D = P | SO5T7snr004, SO5T7snr062 |
Moreover, the expression of three selected genes was validated using qPCR (Table 3): a gene involved in secondary metabolism that was detected as up‐regulated by microarray analysis and subtractive libraries in cv. Santé at 12 hpi (2‐ODD), and two genes involved in photosynthesis [photosystem II 22‐kDa protein (PS II) and glyceraldehyde‐3‐phosphate dehydrogenase (GAP3DH)]. Changes in gene expression obtained by qPCR correlate with those obtained with microarrays, with the correlation coefficients between the data being 86%.
Table 3.
Validation of microarray results by quantitative polymerase chain reaction (qPCR): average M values (log2 ratio) of expression of 2‐oxoglutarate‐dependent dioxygenase (2‐ODD), photosystem II protein (PS II) and glyceraldehyde‐3‐phosphate dehydrogenase (GA3PDH) genes in virus‐ vs. mock‐inoculated plants, obtained by microarrays and qPCR. Asterisks (*) mark statistically significant (P < 0.05) results.
Differential responses of the two potato genotypes to PVYNTN at 12 hpi
At 0.5 hpi, gene expression changes were less prominent than at 12 hpi. Differentially expressed processes or protein families (Table 1) included the up‐regulation of photosynthesis (in both cultivars), carbonic anhydrases (cv. Igor) and plastid ribosomal proteins (cv. Santé), and the down‐regulation of WRKY transcription factors (cv. Igor) and stress responses (cv. Santé). However, major differences between the two cultivars’ responses to virus inoculation, which could be associated with cultivar sensitivity, were observed at 12 hpi (Fig. 4).
Figure 4.
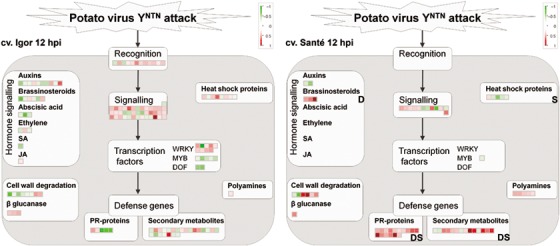
Differences in gene expression between the sensitive cv. Igor and the resistant cv. Santé 12 h following potato virus YNTN (PVYNTN) inoculation. Each square represents the log2 ratio of expression of one clone in virus‐ vs. mock‐inoculated plants (red, up‐regulated; green, down‐regulated). Bold letters mark the confirmation of results by data mining (D) or subtraction library (S). Detailed information about the differentially expressed genes shown in the figure can be found in Table S1 under the corresponding MapMan bins: recognition (20.1.2: stress.biotic.receptors); signalling (30: signalling; 20.1.3: stress.biotic.signalling); WRKY (27.3.32: RNA.regulation of transcription.WRKY domain transcription factor family); MYB (27.3.25: RNA.regulation of transcription.MYB domain transcription factor family); DOF (RNA.regulation of transcription.C2C2(Zn) DOF zinc finger family); PR‐proteins (20.1.7: stress.biotic.PR‐proteins); secondary metabolites (16: secondary metabolism; 20.1.8: stress.biotic.secondary metabolites); auxins (17.2: hormone metabolism.auxin); brassinosteroids (17.3: hormone metabolism.brassinosteroid); abscisic acid (17.1: hormone metabolism.abscisic acid); ethylene (17.5: hormone metabolism.ethylene); SA (17.8: hormone metabolism.salicylic acid); JA (17.7: hormone metabolism.jasmonate); cell wall degradation (10.6: cell wall.degradation); β‐glucanase (26.4: misc.beta 1,3 glucan hydrolases); heat shock proteins (20.2.1: stress.abiotic.heat); polyamines (22: polyamine metabolism).
In cv. Igor, several homologues of putative disease resistance genes harbouring leucine‐rich repeat (LRR), Toll‐interleukin‐1 receptor–nucleotide‐binding site–LRR (TIR‐NBS‐LRR) or NB‐ARC (nucleotide‐binding adaptor shared by APAF‐1, R proteins, and CED‐4) domains, and homologues of Cf‐ and Hcr‐like genes, involved in the perception of the signal, were up‐regulated (Fig. 4). Moreover, several genes involved in signalling were also up‐regulated: mediators of resistance‐mediated plant defence [suppressor of G2 allele of skp1 (SGT1), phytoalexin‐deficient 4 (PAD4) and enhanced disease susceptibility 1 (EDS1); reviewed in Muskett and Parker, 2003], leucine‐rich and thaumatin‐like receptor kinases, and several genes involved in calcium signalling (calnexin, calcineurin, calmodulin binding) and light signalling. However, in cv. Santé, the expression of genes involved in perception and signalling was not as prominent as in cv. Igor. Although some leucine‐rich receptor kinases were up‐regulated, there were no signs of up‐regulation of resistance gene homologues (Fig. 4). Differences in signalling between the cultivars were also evident in the expression of WRKY, MYB and DOF transcription factors, which were up‐ or down‐regulated in cv. Igor, whereas there were no transcriptional changes in cv. Santé (Fig. 4).
Genes involved in the metabolism and action of plant hormones were mostly down‐regulated in cv. Igor, whereas, in cv. Santé, we observed strong up‐regulation of genes involved in brassinosteroid (BR) biosynthesis (Fig. 4). Moreover, data mining showed this difference to be one of the features determining the resistance of the potato cultivars to infection by PVYNTN. In addition, genes involved in polyamine biosynthesis (S‐adenosyl methionine decarboxylase and agmatine deiminase) were up‐regulated in cv. Santé (Fig. 4).
Genes involved in cell wall degradation were mostly down‐regulated in cv. Igor, but up‐regulated in cv. Santé (Fig. 4).
Although genes involved in secondary metabolism were mostly up‐regulated in cv. Santé, their expression in cv. Igor was more diverse (Fig. 4). One of the genes showing the strongest up‐regulation in cv. Santé was that involved in several reactions in the biosynthesis of secondary metabolites, 2‐ODD. In addition, increased expression of this gene was found to be a determinant of cultivar resistance by data mining, and several clones showing high homology to this gene were found in the subtractive library (Table 2). Other important induced genes involved in secondary metabolism include phenylalanine ammonia lyase, cinnamyl‐alcohol dehydrogenase and cinnamic acid 4‐hydroxylase, all involved in lignin biosynthesis.
Although genes coding for some pathogenesis‐related (PR) proteins (e.g. chitinase) were induced in cv. Igor at 12 hpi, the expression of proteinase inhibitors was down‐regulated (Fig. 4). In contrast, PR proteins were strongly up‐regulated in cv. Santé. Moreover, the expression of some proteinase inhibitors was found to be ‘diagnostic’ of cultivar resistance. In addition, ESTs showing homology to PR‐10 (SO4T7snr023) and proteinase inhibitors (SO_Contig5, SO_Contig21, SO1T7snr069, SO3T7snr032, SO3T7snr039, SO7T7snr083) were found in the SO subtraction library, which validates our microarray analysis findings.
Another distinction between the profiles of the two cultivars was the expression of genes for heat shock proteins, which was up‐regulated at both time points in cv. Igor and down‐regulated in cv. Santé (Fig. 4). In addition, heat shock protein homologues (SS_Contig34, SS_Contig71, SS2T7snr046, SS4T7snr060, SS4T7snr074, SS5T7snr089, SS7T7snr078) were found in the SS subtraction library, confirming their down‐regulation following virus inoculation.
DISCUSSION
Transcriptome response in the early reaction to virus inoculation
Only a few studies have examined the early (minutes) plant response to virus inoculation in terms of gene expression levels (Ishihara et al., 2004; Marathe et al., 2004; Oh et al., 2006). The common finding of these studies is that the longer the time following inoculation, the more genes are differentially expressed. In the study of Oh et al. (2006), the majority of the examined genes showed considerable changes in their expression levels 48 h after Nicotiana tabacum inoculation with tobacco mosaic virus (TMV). Similarly, the study by Whitham et al. (2003) showed that the major differences in gene expression in Arabidopsis thaliana occurred 3–5 days rather than 1 or 2 days following inoculation with five different viruses. However, our experiment was based on previous studies, in which potato plants responded to PVYNTN at the biochemical level as early as 3 hpi (Milavec et al., 2008).
The study of the response of A. thaliana to cucumber mosaic virus (CMV) revealed that the number of differentially expressed genes increased from 3 to 18 hpi in both compatible and incompatible interactions (Ishihara et al., 2004). In contrast, in our experiment, the number of differentially expressed genes decreased with time in cv. Santé, whereas cv. Igor showed a greater response at the transcriptome level at 12 hpi (Fig. 1). It may be that, in cv. Santé, perception and signalling take place earlier to enable an effective defence response. The differential pattern of gene expression between the two potato genotypes at a very early stage of PVYNTN infection indicates that the cellular response for resistance differs from that resulting in sensitivity at the level detectable by the microarrays, as also found by Ishihara et al. (2004) in the cucumber–CMV interaction.
Shift in photosynthesis‐related gene expression
The microarray analysis revealed that photosynthesis was one of the most significantly changed processes in terms of gene expression levels following PVYNTN inoculation (Table 1; Fig. 3). Interestingly, in both cultivars, gene expression was up‐regulated as early as 0.5 hpi and down‐regulated at 12 hpi.
A decrease in photosynthetic activity (Hodgson et al., 1989) and chlorophyll content (Milavec et al., 2001a) and a down‐regulation of photosynthesis‐related genes (Pompe‐Novak et al., 2006) have been observed regularly in plants following virus attack. Nevertheless, the work presented here provides the first demonstration of a transient increase in photosynthesis‐related gene expression levels in both sensitive and resistant potato cultivars immediately after virus inoculation (Fig. 3; Table 1). The up‐regulation may be a consequence of a general stress response triggering an increase in energy consumption. It appears that the process of photosynthesis is not correlated with the sensitivity of the examined potato cultivars to PVYNTN. Moreover, at 0.5 hpi, when the expression of genes involved in photosynthesis in cv. Santé was up‐regulated, an increased expression of redox‐related genes was also detected. Photosynthesis metabolism produces intermediates with high positive or negative redox potential, and the redox system prevents the production of reactive oxygen species. Redox‐dependent regulatory mechanisms are active on the time scales of milliseconds to hours of immediate metabolic as well as transcriptomic control (Wormuth et al., 2007). Although a general decrease in photosynthetic activity and gene expression is usually associated with the appearance of the first symptoms, in our study, it is evident that the genes associated with photosynthesis decrease as early as 12 hpi. In both cultivars, the majority of regulated genes are involved in photosystem II (Fig. 3), which is consistent with the results of previous studies (Hodgson et al., 1989).
Induced perception and signalling in the compatible interaction
Effective perception and signalling are essential for an effective defence response. We observed the up‐regulation of disease resistance gene homologues that could be involved in the perception of the pathogen and genes involved in signalling in the sensitive cv. Igor at 12 hpi (Fig. 4). Moreover, the whole signalling group of genes was significantly up‐regulated in cv. Igor at 12 hpi (Table 1). Surprisingly, in cv. Santé, gene expression changes in perception and signalling were less obvious at this time point, when none of the disease resistance genes were differentially expressed (Fig. 4). However, the expression of putative resistance genes was down‐regulated at 0.5 hpi in cv. Santé, as observed in the A. thaliana resistome to CMV (Ishihara et al., 2004; Marathe et al., 2004). It is possible that the resistance genes that act mainly as receptors are constitutively expressed, or that their expression was low and therefore they were not detected, as proposed by Yoshimura et al. (1998). Alternatively, in our experiment, perception and signalling may have peaked between the two time points in the resistant cultivar. Thus, the sensitivity of cv. Igor may be a result of its delayed response. Interestingly, no resistance‐related genes were identified by data mining as being responsible for resistance. However, in the SO subtraction library, one EST (SO5T7snr018), with sequence similarity to the activated disease resistance 1 (ADR‐1) protein, was expressed. Further investigation of this EST is necessary to confirm its involvement in virus resistance.
Another indication of strong signalling in the sensitive cv. Igor is transcriptional activation at 12 hpi of several WRKY transcription factors known for their involvement in defence responses, whereas, at 0.5 hpi, they were repressed (Table 1). However, WRKY expression was unchanged in the resistant cv. Santé. This finding is not in accordance with that of Marathe et al. (2004), where WRKY transcription factors were identified as part of the A. thaliana resistome to CMV. However, some WRKY transcription factors may act as negative repressors of resistance responses (reviewed in Eulgem and Somssich, 2007). As suggested earlier, our results indicate a delayed host response to the virus by the sensitive cultivar, which could allow the virus to multiply and spread.
Signalling and defence responses specific to the resistant cultivar
In plants, hormone signalling plays a vital part in defence reactions. In our experiment, the main difference between the cultivars in hormone metabolism‐related gene expression involved BR metabolism, which was strongly up‐regulated in cv. Santé (Fig. 4) and a determinant of the cultivar's resistance. BRs are a group of naturally occurring plant steroid compounds with a wide range of biological activity, especially in the hormonal regulation of plant growth and development. However, their involvement in plant defence has been reported only recently (Nakashita et al., 2003). The mechanism by which BRs act against pathogens remains unknown. The induction of BR biosynthesis at the transcriptome level in our study suggests that BR‐mediated resistance is transcriptionally regulated.
When comparing the expression of genes involved in polyamine metabolism between the two differently sensitive potato cultivars, it was clear that, in the resistant cv. Santé bearing the Rysto gene, gene expression levels were increased at 12 hpi, whereas, in cv. Igor, there were minimal changes (Table 1; Fig. 4). Our results are consistent with those of Marini et al. (2001), where it was shown that, on inoculation with TMV, hypersensitive plants of tobacco (N. tabacum) reacted in a manner opposite to that of susceptible plants by up‐regulating polyamine synthesis and oxidation. The polyamines are implicated in many functions of the plant cell. Polyamine metabolism undergoes profound changes in response to infection by fungi and viruses (Walters, 2003). High levels of polyamine conjugates may be required for necrotic lesions to develop, thus limiting virus movement and preventing systemic infection (Torrigiani et al., 1997). Possible modes of action of polyamines could involve the direct inhibition of viruses (Bachrach, 2007), or they may be connected with nitric oxide signalling (Yamasaki and Cohen, 2006). The induction of polyamine metabolism at the transcriptional level, observed in cv. Santé at 12 hpi, may be associated with confining of the virus.
Another distinction between the plant responses of the two cultivars was the deployment of true defence responses, such as the expression of secondary metabolites and PR proteins in the resistant cultivar as early as 12 hpi. Not surprisingly, the transcription of a whole group of genes involved in biotic stress responses was increased (Table 1).
The induction of 2‐ODD expression in the resistant cv. Santé at 12 hpi, observed by microarray and subtraction library analysis (Table 2), indicates very early deployment of alkaloidal defences in the resistant cultivar, as was also the case in the black nightshade (Solanum nigrum) response to herbivore attack (Schmidt et al., 2005).
Phenylalanine ammonia lyase is the initial precursor of many secondary metabolites, which are induced as a reaction to plant pathogen infection; moreover, it catalyses the first step in lignin biosynthesis. In cv. Santé at 12 hpi, the lignin pathway was up‐regulated in several steps. Results from a similar study (Oh et al., 2006) showed the induction of the phenylalanine ammonia lyase gene at 9 hpi in the tobacco–TMV incompatible interaction.
The increased expression of genes coding for proteinase inhibitors was unique for the incompatible interaction at 12 hpi (Fig. 4). In addition to their role in constitutive processes, proteinase inhibitors are important in defence against pathogens and predators (Mosolov and Valueva, 2005). Although they have been shown to convey resistance of transgenic tobacco plants towards viruses (Gutierrez‐Campos et al., 1999; Gholizadeh et al., 2005), their induction was not reported in recent transcriptomic studies of compatible plant–virus interactions, which may be because these studies were conducted on Arabidopsis, where the expression of proteinase inhibitors may not be as diverse as in Solanaceae species.
Heat shock proteins play diverse roles in response to cellular stress, as well as in normal growth and development, and diverse DNA and RNA viruses induce their expression almost ubiquitously in eukaryotic hosts (reviewed in Whitham et al., 2006). However, it appears that this is not the case in incompatible interactions, as shown here (Fig. 4) and by the results of previous studies (Ishihara et al., 2004; Marathe et al., 2004). Most probably, the resistance‐related response interferes with the generic stress response.
CONCLUSIONS
The results suggest that the early response to PVYNTN in the sensitive potato cultivar Igor and in the resistant cultivar Santé involves changes in photosynthesis, perception and signalling, and plant defence. Although cv. Santé bears the Rysto gene, which is considered to confer extreme resistance to PVYNTN, based on the increased expression of secondary metabolites and PR protein genes, we can conclude that active defence is taking place in this cultivar. Nevertheless, the induced expression of genes involved in perception and signalling in cv. Igor indicates that, in this cultivar, a defence response is triggered, but it may not be sufficiently rapid or intense to stop the virus.
However, these predictions are based on changes in transcript abundance, which may not necessarily reflect protein abundance in these functional categories. In addition, our experimental design allowed us to monitor gene expression in a mixture of inoculated and non‐inoculated cells; therefore, gene expression changes in the inoculated cells could be masked. Nevertheless, the transcript abundance data raise several new and important testable hypotheses pertaining to how and when potato plants respond to virus invasion. Our study provides, for the first time, evidence indicating that differently sensitive cultivars react differently to virus inoculation at the transcriptional level, as early as 0.5 hpi, although the differences are more prominent at 12 hpi. Moreover, to date, only two studies have been published on the transcriptome changes of the early incompatible plant–virus interaction (Ishihara et al., 2004; Marathe et al., 2004). Both deal with the Arabidopsis–CMV interaction, whereas our experiment is the first transcriptomic study of the incompatible interaction of potato, an important crop species, with its economically important viral pathogen in the very early stages. The characterization of PVYNTN‐altered gene expression in the two genotypes will contribute to a better understanding of the molecular basis of compatible and incompatible interactions between virus and host plants. Host genes with altered expression profiles provide candidates for reverse genetic experiments to test for roles in viral pathogenesis, and provide an insight into the signalling pathways that are modulated by viral infection (Whitham et al., 2006).
EXPERIMENTAL PROCEDURES
Plant material: growth conditions, virus inoculation and experimental design
Healthy potato plants (Solanum tuberosum L.) of cultivars Igor and Santé were grown in stem node tissue culture. Two weeks after node segmentation, they were transferred to soil in a growth chamber and kept at 21 ± 2 °C in the light and 18 ± 1 °C in the dark, at a relative humidity of 75% ± 2%, with 70–90 µmol/m2/s2 radiation (L36W/77 lamp, Osram, Germany) and a 16‐h photoperiod.
After 4 weeks growth in soil, the potato plants of each cultivar were divided into two groups of 15–18 plants each: virus‐inoculated and mock‐inoculated groups. The PVYNTN (isolate NIB‐NTN, GenBank accession number AJ585342) inoculation procedure was carried out according to Milavec et al. (2008). The first group was virus inoculated by gently rubbing two or three bottom leaves with carborundum and then applying a buffered suspension of PVYNTN‐infected plant sap. Potato plants of the second group were mock inoculated with a buffered suspension of healthy potato plant sap and were used as controls. Samples of inoculated leaves from both groups of plants of each cultivar were collected at 0.5 and 12 hpi, using six to eight plants per time point, flash frozen in liquid nitrogen, ground to a fine powder and stored at −80 °C for analysis.
At least three experimental series of experiments were conducted independently at different times of the year. To control for environmental and circadian effects, all plants were maintained under identical conditions, with all inoculation and harvesting of leaf material carried out at the same time of the day. Data from all biological replicates were analysed and combined.
To confirm infection and health status of the inoculated plants, three plants from each group were grown for a further 13 days. After 5 and 12 days, local mosaic and systemic symptoms, respectively, were observed on each virus‐inoculated potato plant of sensitive cv. Igor; mock‐ and virus‐inoculated plants of resistant cv. Santé remained free of symptoms throughout the whole period. Enzyme‐linked immunosorbent assay (ELISA) testing confirmed the spread of the virus to non‐inoculated leaves in cv. Igor, whereas, in cv. Santé, the virus could not be detected in non‐inoculated leaves.
Microarray hybridization
Inoculated leaf tissue from at least six mock‐ or virus‐inoculated plants (for each cultivar, time point and biological replicate) was pooled for RNA isolation. Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen, Germany) from 300 mg of plant material. In order to remove residual genomic DNA, deoxyribonuclease I (DNase I, amplification grade, Invitrogen, CA, USA) digestion was carried out using 0.04 U/µg RNA in a reaction volume of 40 µL. The resulting RNA was further purified and concentrated using an RNeasy MinElute Cleanup Kit (Qiagen). RNA quality and quantity were assessed using agarose gel electrophoresis and an RNA 6000 Nano Lab Chip Kit in combination with a Bioanalyser (Agilent Technologies, CA, USA).
cDNA synthesis and tagging were carried out using a 3DNA Array 900 Kit (Genisphere, PA, USA) with Superscript II reverse transcriptase (Invitrogen); 100 pg of luciferase control RNA was added to the reverse transcription reaction as an external control. cDNA was concentrated using Microcon YM‐30 centrifugal filter devices (Millipore, MA, USA) to approximately 10 µL.
The microarrays used in all experiments were cDNA microarrays (TIGR 10K v2 or v3 potato arrays) containing approximately 10 000 potato cDNA clones (15 264 spots in duplicate; http://www.tigr.org/tdb/sol/sol_ma_microarrays.shtml).
Slide pre‐hybridization and subsequent washings were carried out as described by Hegde et al. (2000). Slides were dried by centrifugation (1000 g, 1 min). Dendrimer labelling technology was used for microarray hybridizations. A 3DNA Array 900 Kit with formamide‐based buffer was used for hybridization at 45 °C. LifterSlips (Erie Scientific Co., NH, USA) were used as coverslips. 3DNA‐tagged cDNAs were dendrimer labelled with cyanine‐3 (Cy3) and Cy5 dyes, according to the manufacturer's protocol and recommendations. Post‐hybridization washing and slide drying were carried out as described by Gruden et al. (2004). To account for inconsistencies in probe labelling and detection, each RNA pool was used to generate two reciprocally labelled sets of cDNA probes (Cy‐3 mock/Cy‐5 infected or Cy‐3 infected/Cy‐5 mock).
Microarray data analysis
Image acquisition and analysis
Microarrays were scanned using an LS200 scanner (TECAN, Switzerland) at 180 gain laser power and two times over‐sampling at both 543 and 633 nm, taking care not to saturate any spots. The TIFF images were quantified using ArrayPro® Analyzer 4.5 software (Media Cybernetics, MD, USA). For both 543‐ and 633‐nm channels, signal (trimmed mean of the whole variable spot area) and background (trimmed mean of the spot's local corners) intensities were calculated. Low‐quality spots were flagged and excluded from further analyses. These included: (i) non‐uniform spots [mean (signal)/standard deviation (signal) < 1]; (ii) spots with low signal to background ratio [mean (signal)/mean (background) < 1.5]; and (iii) spots with low signal to noise ratio (SNR < 3). In addition, empty or non‐validated spots were excluded from further analysis.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002), and are accessible through GEO Series accession number GSE12041 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12041).
Data pre‐processing, statistical analysis and data mining
Data analysis was performed in the R computing environment, limma package, as proposed previously by Rotter et al. (2008). Data quality was first assessed using various diagnostic plots. Background was subtracted using the option ‘half’. Two normalizations, ‘vsn’ and ‘loess’, were applied separately, thus producing two datasets. Duplicate within‐array replicate spot correlation was taken into account. A 2 × 2 factorial design analysis was applied with the factors being potato cultivar and time. The data were analysed using the linear model. Among other factors, information about the version of the microarray (versions 2 and 3) was taken into account. Statistical analysis was carried out for each dataset, giving lists of genes differentially expressed in virus‐ vs. mock‐inoculated plants for a given cultivar at a given time point. The intersection of both lists (‘vsn’ and ‘loess’ normalized datasets) was used as the final list of differentially expressed genes.
The significantly expressed microarray clones and their corresponding expression values (M) were exported to MapMan, using potato mapping (Rotter et al., 2007), where differential expression was visualized in the context of biological processes. Moreover, Wilcoxon rank sum test was carried out to predict BINs that exhibited different behaviour in terms of expression profile compared with all of the other remaining BINs (Usadel et al., 2005).
Data mining, using the closed itemset mining for class labelled data (RelSets) algorithm (Garriga et al., 2006), was carried out. In this way, two lists of rules defining class‐resistant or class‐sensitive cultivar in the early response to virus inoculation were obtained. The data mining results were annotated using MapMan ontology. Excel (Microsoft), MapMan (http://gabi.rzpd.de/projects/MapMan/), SmartDraw (SmartDraw.com) and MeV (TIGR) were used for the preparation of tables and figures.
qPCR
Changes in the expression of three selected genes, 2‐ODD, PS II and GA3PDH, were analysed by qPCR assay. Luciferase (Toplak et al., 2000) was used as an external quality control, and potato elongation factor 1‐α (EF‐1) and cytochrome oxidase (Cox; Weller et al., 2000) were used as endogenous controls for normalization. Publicly available sequences of transcripts from GenBank and StGI (Solanum tuberosum gene index, http://compbio.dfci.harvard.edu/tgi/cgi‐bin/tgi/gimain.pl?gudb=potato) were analysed to select the target sequences, and Primer Express (Applied Biosystems, CA, USA) was used for the design of primers and probes. The specificity of the designed amplicons was tested in silico with a blast search of public databases. Their sequences and concentrations used in qPCRs are shown in Table 4.
Table 4.
Target sequence (GenBank accession), primer (FP, RP) and probe (P) sequences and their optimized concentrations used in quantitative polymerase chain reactions (qPCRs) for elongation factor 1 (EF‐1), 2‐oxoglutarate‐dependent dioxygenase (2‐ODD), photosystem II protein (PS II) and glyceraldehyde‐3‐phosphate dehydrogenase (GA3PDH) genes.
| Amplicon | Target sequence | Sequence (5′–3′) | Reaction conc. (nm) | |
|---|---|---|---|---|
| EF‐1 | AB061263 | FP | GGAAGCTGCTGAGATGAACAAGA | 900 |
| RP | CTCACGTTCAGCCTTAAGTTTGTC | 900 | ||
| P | FAM‐TCATTCAAGTATGCCTGGGTGCT‐TAMRA | 250 | ||
| 2‐ODD | BQ113054 | FP | CGCATCATTGGAGCCCTTCAAAGTC | 900 |
| RP | AGGTAACAAAGGTAGGATACAAACTAGAAAGACATTG | 900 | ||
| P | FAM‐TTGTGTGCCAGAAGGATCAAGTCTATGCTG‐TAMRA | 250 | ||
| PS II | BQ111818 | FP | GACCCTGCCCCTGCTACTGG | 300 |
| RP | CTTTGTGAATCCAAAAAGTGGACCTCCTTC | 300 | ||
| GA3PDH | BQ113669 | FP | CCAGGCCGGAGCCAAGAAGG | 300 |
| RP | TCATGGTTGTAAAGTTCAGCATTGACACC | 300 | ||
| P | FAM‐CGCCCCCGGAAAAGGTGATATCCCTACTTA‐TAMRA | 150 | ||
DNAse‐treated RNA (1 µg), which was used for microarray hybridization, was spiked with 2 ng of luciferase mRNA (Promega, WI, USA) and reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCRs using SYBR Green (PS II) or TaqMan (all other amplicons) chemistry and C t value calculations were performed as described by Pompe‐Novak et al. (2006). Relative quantification was carried out as proposed by Pfaffl (2001). The expression in the individual samples was normalized to the expression of two endogenous control genes (EF‐1 and Cox), and luciferase was used for quality control only. The results were expressed as log2 of the ratio between gene expression in PVYNTN‐infected and mock‐inoculated plants (M). Average M values of the biological replicates were calculated, and Student's t‐test was used to check for the significance of the results.
Construction of subtraction libraries and sequence data analysis
Subtraction libraries were constructed from virus‐ and mock‐inoculated plant material of cv. Santé at 12 hpi, as described previously (Pompe‐Novak et al., 2006), using a PCR‐Select cDNA Subtraction Kit (Clontech, CA, USA). For the construction of the library enriched for up‐regulated genes (SO library), cDNA from virus‐inoculated plant material was used as a driver and cDNA from mock‐inoculated plant material as a tester. For the construction of the library enriched for down‐regulated genes (SS library), the driver and tester cDNAs were reversed. Unsubtracted cDNA molecules were ligated to pGEM‐T Easy plasmid vector (Promega) and introduced into Escherichia coli DH5α cells. Among the transformants obtained, 383 from the SO library and 574 from the SS library were picked individually and cDNA inserts were sequenced.
The sequences obtained were trimmed and assembled into contiguous sequences with the cap3 algorithm (Huang and Madan, 1999), using default values (overlap length cut‐off, 40; overlap percentage identity cut‐off, 80). From the resulting contigs and singletons, PVY‐related sequences and sequences that were present in both libraries were removed. The remaining ESTs were batch blasted towards NCBI's nr database (http://www.ncbi.nlm.nih.gov/; blastx) and StGI database (tblastx). In the case of high homology (E < 10−10), the corresponding StGI TC numbers were connected to the MapMan mapping file and the sequences were annotated using MapMan ontology.
The sequences were deposited at the NCBI dbEST database (http://www.ncbi.nlm.nih.gov/dbEST; GenBank accessions: FG550738–FG551325; GD190176–GD190491). Detailed information about the ESTs is given in Table S3.
Supporting information
Table S1 Lists of differentially expressed genes in virus‐ vs. mock‐inoculated plants in cv. Igor at 0.5 h post‐inoculation (hpi) (a) and 12 hpi (b) and in cv. Santé at 0.5 hpi (c) and 12 hpi (d).
Table S2 List of data mining rules determining cultivar resistance or sensitivity.
Table S3 List of expressed sequence tags (ESTs) (with GENBANK accession numbers and annotations) from subtraction libraries enriched for genes up‐regulated (SO, a) or down‐regulated (SS, b) in potato plants of cv. Santé at 12 hpi.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We are grateful to Mrs Lidija MatičIč for excellent technical support in plant material preparation and inoculation, and to Dr Roger Pain for language revision. This project was supported by the Slovenian Research Agency (grant numbers P4‐0165 and Z4‐9697).
REFERENCES
- Bachrach, U. (2007) Antiviral activity of oxidized polyamines. Amino Acids, 33, 267–272. [DOI] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. and Lash, A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T. and Somssich, I.E. (2007) Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Flis, B. , Hennig, J. , Strzelczyk‐Z.yta, D. , Gebhardt, C. and Marczewski, W. (2005) The Ry‐f(sto) gene from Solanum stoloniferum for extreme resistant to Potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122(718) in PVY resistant potato cultivars. Mol. Breed. 15, 95–101. [Google Scholar]
- Garriga, G. , Kralj, P. and Lavrač, N. (2006) Closed sets for labeled data In Knowledge Discovery in Databases: PKDD 2006 (Fürnkranz J., Scheffer T. and Spiliopoulou M., eds), pp. 163–174, Berlin, Heidelberg: Springer‐Verlag. [Google Scholar]
- Gholizadeh, A. , Santha, I.M. , Kohnehrouz, B.B. , Lodha, M.L. and Kapoor, H.C. (2005) Cystatins may confer viral resistance in plants by inhibition of a virus‐induced cell death phenomenon in which cysteine proteinases are active: cloning and molecular characterization of a cDNA encoding cysteine‐proteinase inhibitor (celostatin) from Celosia cristata (crested cock's comb). Biotechnol. Appl. Biochem. 42, 197–204. [DOI] [PubMed] [Google Scholar]
- Golem, S. and Culver, J.N. (2003) Tobacco mosaic virus induced alterations in the gene expression profile of Arabidopsis thaliana . Mol. Plant–Microbe Interact. 16, 681–688. [DOI] [PubMed] [Google Scholar]
- Gruden, K. , Kuipers, A.G. , Gunčar, G. , Slapar, N. , Strukelj, B. and Jongsma, M.A. (2004) Molecular basis of Colorado potato beetle adaptation to potato plant defence at the level of digestive cysteine proteinases. Insect Biochem. Mol. Biol. 34, 365–375. [DOI] [PubMed] [Google Scholar]
- Gutierrez‐Campos, R. , Torres‐Acosta, J.A. , Saucedo‐Arias, L.J. and Gomez‐Lim, M.A. (1999) The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 17, 1223–1226. [DOI] [PubMed] [Google Scholar]
- Hegde, P. , Qi, R. , Abernathy, K. , Gay, C. , Dharap, S. , Gaspard, R. , Hughes, J.E. , Snesrud, E. , Lee, N. and Quackenbush, J. (2000) A concise guide to cDNA microarray analysis. Biotechniques, 29, 548–556. [DOI] [PubMed] [Google Scholar]
- Hodgson, R.A. , Beachy, R.N. and Pakrasi, H.B. (1989) Selective inhibition of photosystem II in spinach by tobacco mosaic virus: an effect of the viral coat protein. FEBS Lett. 245, 267–270. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Madan, A. (1999) CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara, T. , Sakurai, N. , Sekine, K.T. , Hase, S. , Ikegami, M. , Shibata, D. and Takahashi, H. (2004) Comparative analysis of expressed sequence tags in resistant and susceptible ecotypes of Arabidopsis thaliana infected with cucumber mosaic virus. Plant Cell Physiol. 45, 470–480. [DOI] [PubMed] [Google Scholar]
- Krečič‐Stres, H. , Vučak, C. , Ravnikar, M. and Kovač, M. (2005) Systemic Potato virus YNTN infection and levels of salicylic and gentisic acids in different potato genotypes. Plant Pathol. 54, 441–447. [Google Scholar]
- Marathe, R. , Guan, Z. , Anandalakshmi, R. , Zhao, H. and Dinesh‐Kumar, S.P. (2004) Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol. Biol. 55, 501–520. [DOI] [PubMed] [Google Scholar]
- Marini, F. , Betti, L. , Scaramagli, S. , Biondi, S. and Torrigiani, P. (2001) Polyamine metabolism is upregulated in response to tobacco mosaic virus in hypersensitive, but not in susceptible, tobacco. New Phytol. 149, 301–309. [DOI] [PubMed] [Google Scholar]
- Maule, A. , Leh, V. and Lederer, C. (2002) The dialogue between viruses and hosts in compatible interactions. Curr. Opin. Plant Biol. 5, 279–284. [DOI] [PubMed] [Google Scholar]
- Milavec, M. , Ravnikar, M. and Kovač, M. (2001a) Peroxidases and photosynthetic pigments in susceptible potato infected with potato virus Yntn. Plant Physiol. Biochem. 39, 891–898. [Google Scholar]
- Milavec, M. , Ravnikar, M. and Kovač, M. (2001b) Peroxidases in the early response of potato (Solanum tuberosum L. cv. Igor) susceptible to potato virus YNTN. Acta Biol. Slov. 44, 3–11. [Google Scholar]
- Milavec, M. , Gruden, K. , Ravnikar, M. and Kovač, M. (2008) Peroxidases in the early responses of different potato cultivars to infection by Potato virus YNTN. Plant Pathol. 57, 861–869. [Google Scholar]
- Mosolov, V.V. and Valueva, T.A. (2005) Proteinase inhibitors and their function in plants: a review. Appl. Biochem. Microbiol. 41, 227–246. [PubMed] [Google Scholar]
- Muskett, P. and Parker, J. (2003) Role of SGT1 in the regulation of plant R gene signalling. Microb. Infect. 5, 969–976. [DOI] [PubMed] [Google Scholar]
- Nakashita, H. , Yasuda, M. , Nitta, T. , Asami, T. , Fujioka, S. , Arai, Y. , Sekimata, K. , Takatsuto, S. , Yamaguchi, I. and Yoshida, S. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J. , Schmelz, E.A. , Moussatche, P. , Lund, S.T. , Jones, J.B. and Klee, H.J. (2003) Susceptible to intolerance—a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33, 245–257. [DOI] [PubMed] [Google Scholar]
- Oh, S.K. , Lee, S. , Chung, E. , Park, J.M. , Yu, S.H. , Ryu, C.M. and Choi, D. (2006) Insight into Types I and II nonhost resistance using expression patterns of defense‐related genes in tobacco. Planta, 223, 1101–1107. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompe‐Novak, M. , Gruden, K. , Baebler, Š. , Krečič‐Stres, H. , Kovač, M. , Jongsma, M. and Ravnikar, M. (2006) Potato virus Y induced changes in the gene expression of potato (Solanum tuberosum L.). Physiol. Mol. Plant Pathol. 67, 237–247. [Google Scholar]
- Ravnikar, M. (2005) Potato virus Y and its interaction with potato In Plant Genomics and Bioinformatics Expression Micro Arrays and Beyond: A Course Book (Freitag J., ed.), pp. 66–71 Ljubljana, National Institute of Biology. [Google Scholar]
- Rotter, A. , Usadel, B. , Baebler, Š. , Stitt, M. and Gruden, K. (2007) Adaptation of the MapMan ontology to biotic stress responses: application in solanaceous species. Plant Methods, 3, http://www.plantmethods.com/ . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter, A. , Hren M., Baebler, Š. , Blejec, A. and Gruden, K. (2008) Finding differentially expressed genes in two‐channel DNA microarray datasets: how to increase reliability of data preprocessing. OMICS 12, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheideler, M. , Schlaich, N.L. , Fellenberg, K. , Beissbarth, T. , Hauser, N.C. , Vingron, M. , Slusarenko, A.J. and Hoheisel, J.D. (2002) Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem. 277, 10 555–10 561. [DOI] [PubMed] [Google Scholar]
- Schmidt, D.D. , Voelckel, C. , Hartl, M. , Schmidt, S. and Baldwin, I.T. (2005) Specificity in ecological interactions: attack from the same lepidopteran herbivore results in species‐specific transcriptional responses in two solanaceous host plants. Plant Physiol. 138, 1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil, G. , Liu, H. , Puram, V.G. , Clark, A. , Stromberg, A. and Goodin, M.M. (2005) Specific and common changes in Nicotiana benthamiana gene expression in response to infection by enveloped viruses. J. Gen. Virol. 86, 2615–2625. [DOI] [PubMed] [Google Scholar]
- Singh, R.P. , Valkonen, J.P.T. , Gray, S.M. , Boonham, N. , Jones, R.A.C. , Kerlan, C. and Schubert, J. (2008) Discussion paper: The naming of Potato virus Y strains infecting potato. Arch. Virol. 153, 1–13. [DOI] [PubMed] [Google Scholar]
- Smith, C.M. , Rodriguez‐Buey, M. , Karlsson, J. and Campbell, M.M. (2004) The response of the poplar transcriptome to wounding and subsequent infection by a viral pathogen. New Phytol. 164, 123–136. [DOI] [PubMed] [Google Scholar]
- Toplak, N. , Okršlar, V. , Stanič‐Racman, D. , Gruden, K. and Žel, J. (2000) A high throughput method for quantifying transgene expression in transformed plants using Real‐Time PCR analysis. Plant Mol. Biol. Rep. 22, 237–250. [Google Scholar]
- Torrigiani, P. , Rabiti, A.L. , Bortolotti, C. , Betti, L. , Marani, F. , Canova, A. and Bagni, N. (1997) Polyamine synthesis and accumulation in the hypersensitive response to TMV in Nicotiana tabacum . New Phytol. 135, 467–473. [Google Scholar]
- Usadel, B. , Nagel, A. , Thimm, O. , Redestig, H. , Blaesing, O.E. , Palacios‐Rojas, N. , Selbig, J. , Hannemann, J. , Piques, M.C. , Steinhauser, D. , Scheible, W.R. , Gibon, Y. , Morcuende, R. , Weicht, D. , Meyer, S. and Stitt, M. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, D. (2003) Resistance to plant pathogens: possible roles for free polyamines and polyamine catabolism. New Phytol. 159, 109–115. [DOI] [PubMed] [Google Scholar]
- Weller, S.A. , Elphinstone, J.G. , Smith, N.C. , Boonham, N. and Stead, D.E. (2000) Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real‐time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 66, 2853–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S.A. , Quan, S. , Chang, H.S. , Cooper, B. , Estes, B. , Zhu, T. , Wang, X. and Hou, Y.M. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Yang, C. and Goodin, M.M. (2006) Global impact: elucidating plant responses to viral infection. Mol. Plant–Microbe Interact. 19, 1207–1215. [DOI] [PubMed] [Google Scholar]
- Wormuth, D. , Heiber, I. , Shaikali, J. , Kandlbinder, A. , Baier, M. and Dietz, K.J. (2007) Redox regulation and antioxidative defence in Arabidopsis leaves viewed from a systems biology perspective. J Biotechnol. 129, 229–248. [DOI] [PubMed] [Google Scholar]
- Yamasaki, H. and Cohen, M.F. (2006) NO signal at the crossroads: polyamine‐induced nitric oxide synthesis in plants? Trends Plant Sci. 11, 522–524. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Guo, R. , Jie, F. , Nettleton, D. , Peng, J. , Carr, T. , Yeakley, J.M. , Fan, J.B. and Whitham, S.A. (2007) Spatial analysis of Arabidopsis thaliana gene expression in response to Turnip mosaic virus infection. Mol. Plant–Microbe Interact. 20, 358–370. [DOI] [PubMed] [Google Scholar]
- Yoshimura, S. , Yamanouchi, U. , Katayose, Y. , Toki, S. , Wang, Z.X. , Kono, I. , Kurata, N. , Yano, M. , Iwata, N. and Sasaki, T. (1998) Expression of Xa1, a bacterial blight‐resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA, 95, 1663–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Lists of differentially expressed genes in virus‐ vs. mock‐inoculated plants in cv. Igor at 0.5 h post‐inoculation (hpi) (a) and 12 hpi (b) and in cv. Santé at 0.5 hpi (c) and 12 hpi (d).
Table S2 List of data mining rules determining cultivar resistance or sensitivity.
Table S3 List of expressed sequence tags (ESTs) (with GENBANK accession numbers and annotations) from subtraction libraries enriched for genes up‐regulated (SO, a) or down‐regulated (SS, b) in potato plants of cv. Santé at 12 hpi.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


