SUMMARY
Eukaryotic translation initiation factors (eIFs) play a central role in potyviral infection. Accordingly, mutations in the gene encoding eIF4E have been identified as a source of recessive resistance in several plant species. In common bean, Phaseolus vulgaris, four recessive genes, bc‐1, bc‐2, bc‐3 and bc‐u, have been proposed to control resistance to the potyviruses Bean common mosaic virus (BCMV) and Bean common mosaic necrosis virus. In order to identify molecular entities for these genes, we cloned and sequenced P. vulgaris homologues of genes encoding the eIF proteins eIF4E, eIF(iso)4E and nCBP. Bean genotypes reported to carry bc‐3 resistance were found specifically to carry non‐silent mutations at codons 53, 65, 76 and 111 in eIF4E. This set of mutations closely resembled a pattern of eIF4E mutations determining potyvirus resistance in other plant species. The segregation of BCMV resistance and eIF4E genotype was subsequently analysed in an F2 population derived from the P. vulgaris all‐susceptible genotype and a genotype carrying bc‐3. F2 plants homozygous for the eIF4E mutant allele were found to display at least the same level of resistance to BCMV as the parental resistant genotype. At 6 weeks after inoculation, all F2 plants found to be BCMV negative by enzyme‐linked immunosorbent assay were found to be homozygous for the mutant eIF4E allele. In F3 plants homozygous for the mutated allele, virus resistance was subsequently found to be stably maintained. In conclusion, allelic eIF4E appears to be associated with a major component of potyvirus resistance present in bc‐3 genotypes of bean.
INTRODUCTION
The susceptibility of plants to virus infection relies on factors provided by the plant (Ahlquist et al., 2003) and, in particular, host translation initiation factors play a central role in infection by plant RNA viruses (Robaglia and Caranta, 2006). For several viral species in the family Potyviridae, mutated forms of translation initiation factor 4E (eIF4E) and/or its isoform eIF(iso)4E have been identified as host factors by their genetic appearance as recessive loci of resistance, phenotypically expressed as a lack of susceptibility (Lellis et al., 2002; Nicaise et al., 2003; Ruffel et al., 2002; Stein et al., 2005). eIF4E and its isoform are proteins that bind to the 5′ cap structure (m7GTP) of mRNA in plants (Carberry et al., 1991). The genome‐linked protein, VPg, of several potyviruses has been demonstrated to interact with one or more of these cap‐binding proteins (Beauchemin et al., 2007; Leonard et al., 2004; Schaad et al., 2000; Wittmann et al., 1997). The disruption of this interaction has been suggested as a mechanism to explain the loss of susceptibility observed in plants carrying mutant forms of the cap‐binding eIFs (Kang et al., 2005; Yeam et al., 2007). Accordingly, it has been found for several potyviruses that the ability to overcome eIF4E‐mediated resistance is associated with mutations in the coding region of VPg (Bruun‐Rasmussen et al., 2007; Keller et al., 1998; Moury et al., 2004; Yeam et al., 2007). In the model plant Arabidopsis thaliana, five different genes encode cap‐binding proteins (http://www.arabidopsis.org/browse/genefamily/eIF.jsp). Three of these genes, At4G18040, At1G29590 and At1G29550, encode the eIF4E proteins AteIF4E‐1, AteIF4E‐2 and AteIF4E‐3, respectively. A fourth gene, At5G35620, encodes AteIF(iso)4E, and a fifth gene, At5G18110, encodes the novel cap‐binding protein (nCBP) (Ruud et al., 1998). In A. thaliana, two of these five cap‐binding proteins described above, AteIF4E‐1 and AteIF(iso)4E, have been associated with potyviral infection (Lellis et al., 2002; Sato et al., 2005), and homologues of these two proteins in particular have been linked to potyviral resistance in other plant species (Hwang et al., 2009; Robaglia and Caranta, 2006; Ruffel et al., 2006; Rusholme et al., 2007).
Resistance to the potyviruses Bean common mosaic virus (BCMV) and Bean common mosaic necrosis virus (BCMNV) in Phaseolus vulgaris is affected by four different loci: bc‐1, bc‐2, bc‐3 and bc‐u (Drijfhout, 1978). Resistance controlled by alleles at these loci is inherited as recessive characters. In addition to the recessive bc genes, the dominant I gene in P. vulgaris confers resistance to BCMV and other potyviruses through a hypersensitive response (Collmer et al., 2000). Neither the bc genes nor the I gene have been identified at the molecular level, but some molecular markers have been developed for certain genetic backgrounds of bean (Johnson et al., 1997; Melotto et al., 1996; Miklas et al., 2000; Mukeshimana et al., 2005). Based on the importance of the eIF4E genes in potyvirus resistance, we decided to undertake a candidate gene approach and specifically investigate genes encoding cap‐binding proteins in P. vulgaris. Three such genes were cloned and their sequences were compared between genotypes carrying different combinations of bc genes and I gene. In order to test the possible linkage between the thereby observed allelic polymorphism and BCMV resistance, we developed a molecular marker and analysed the segregation of the DNA polymorphism and BCMV resistance using F2 and F3 populations generated from a cross between a resistant and a susceptible genotype.
RESULTS
Sequencing of cDNAs encoding cap‐binding proteins from P. vulgaris
cDNAs of P. vulgaris genes encoding homologues of AteIF4E‐1, AteIF(iso)4E and nCBP were targeted for amplification by the application of the PCR primer pairs FWa/RVa, FWb/RVb and FWd/RVd, respectively, on reverse transcribed RNA from the common bean cultivar Dubbele Witte (DW) susceptible to all strains of BCMV and BCMNV. Amplified fragments of the expected size were cloned and sequenced at least twice for each primer pair. A fourth primer pair, FWc/RVc, was subsequently used to amplify eIF(iso)4E full‐length cDNA, including the 5′ and 3′ untranslated regions. Amplified sequences were compared with the genome of A. thaliana using TAIR blast analysis (http://www.arabidopsis.org/Blast/index.jsp), and the cloning of three P. vulgaris cDNAs encoding homologues of AteIF4E‐1, AteIF(iso)4E and nCBP was thereby confirmed [PveIF4E (EF571267), PveIF(iso)4E (EF571278) and PvnCBP (EF558667)].
Genotype‐related differences in cDNAs encoding cap‐binding proteins
Nine P. vulgaris genotypes representing eight different gene combinations with regard to the bc genes and I gene (Table 1) were chosen in order to search for differences in the cDNA's encoding cap‐binding proteins. For each cultivar, RNA was extracted from two plants and used as template for oligo‐dT‐primed cDNA synthesis.
Table 1.
Sequence differences in eIF4E cDNAs of nine Phaseolus vulgaris genotypes representing eight gene combinations.
| Cultivar | Genotype | Nucleotide (nt)/amino acid (aa) positions | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 159/53 | 194/65 | 227/76 | 332/111 | |||||||
| nt | aa | nt | aa | nt | aa | nt | aa | |||
| DW* | ii | C | Asn | T | Phe | C | Ala | A | Asp | |
| SGR* | ii | bc‐u/bc‐u | — | — | — | — | — | — | — | — |
| Sanilac* | ii | bc‐u/bc‐u, bc‐2/bc‐2 | — | — | — | — | — | — | — | — |
| USCR8† | ii | bc‐3/bc‐3 | A | Lys | A | Tyr | A | Glu | G | Gly |
| Widusa* | II | — | — | — | — | — | — | — | — | |
| Topcrop* | II | bc‐u/bc‐u, bc‐1/bc‐1 | — | — | — | — | — | — | — | — |
| USCR7† | II | bc‐3/bc‐3 | A | Lys | A | Tyr | A | Glu | G | Gly |
| USLK1† | II | bc‐3/bc‐3 | A | Lys | A | Tyr | A | Glu | G | Gly |
| Raven‡ | II | bc‐u/bc‐u, bc‐3/bc‐3 | A | Lys | A | Tyr | A | Glu | G | Gly |
Genotype proposed by Drijfhout (1978). DW, Dubbele Witte; SGR, Stringless Green Refugee.
Genotype proposed by Miklas and Hang (1998a, 1998b) and Miklas and Kelly (2002).
Genotype proposed by Kelly et al. (1994).
Using the primer pair FWa/RVa, cDNA fragments of PveIF4E were amplified and sequenced from all nine genotypes. The alignment of the sequences revealed the presence of two eIF4E variants, denoted PveIF4E1 and PveIF4E2. The cDNA sequence corresponding to PveIF4E2 was found exclusively in four genotypes reported to carry the bc‐3 gene: Raven (EF571273), USCR7 (EF571274), USCR8 (EF571275) and USLK1 (EF571276). The PveIF4E1 variant identical to EF571267 from DW, which is susceptible to all strains of BCMV and BCMNV, was found in cultivars without the bc‐3 gene: Stringless Green Refugee (SGR) (EF571268), Widusa (EF571269), Topcrop (EF571270) and Sanilac (EF571271) (Table 1). There were four codon differences between PveIF4E1 and PveIF4E2 predicted to affect the following amino acid positions: 53 (Asn/Lys), 65 (Phe/Tyr), 76 (Ala/Glu) and 111 (Asp/Gly) (Table 1). When PveIF4E1 and PveIF4E2 of P. vulgaris were aligned with the eIF4E variants from Capsicum annuum (Kang et al., 2005; Ruffel et al., 2002), Lactuca sativa (Nicaise et al., 2003) and Pisum sativum (Bruun‐Rasmussen et al., 2007; Gao et al., 2004), it became evident that the polymorphism in PveIF4E resembled the polymorphisms previously shown to determine potyvirus resistance in C. annuum, L. sativa and P. sativum (Fig. 1). In particular, the differences at positions 65, 76 and 111 were closely aligned with codons near the predicted cap‐binding pocket mutated in potyvirus‐resistant lines of P. sativum (Gao et al., 2004).
Figure 1.
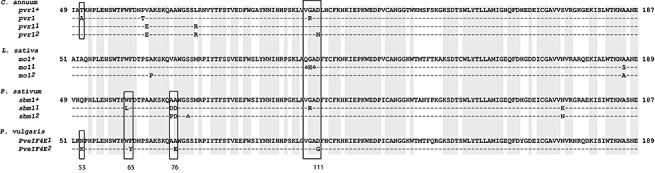
Comparison of amino acid polymorphisms in eukaryotic translation initiation factor 4E (eIF4E) found between susceptible and resistant/tolerant genotypes of Capsicum annuum (pvr1+, AY485127; pvr1, AY485129; pvr11, AY485130; pvr12, AY485131), Lactuca sativa (mo1+, AAP86602; mo11; mol2) and Pisum sativum (sbm1+, AAR04332; sbm1, AAT44121; sbm11, AAT44122) with eIF4E encoded by Phaseolus vulgaris alleles PveIF4E1 (EF571267) and PveIF4E2 (EF571275). PveIF4E2 found in the genotypes carrying the bc‐3 gene. The amino acids conserved in susceptible alleles of all four species are shaded in grey. Codons mutated in PveIF4E2 are indicated by numbers and similarly located mutations in eIF4E genes linked to potyvirus resistance in the other species are framed by rectangles. The symbol Δ indicates the amino acids deleted.
Comparison of the genes encoding the other cap‐binding proteins, eIF(iso)4E and nCBP, between cultivars of P. vulgaris only revealed silent mutations, and no correlation with resistance profiles could be identified (data not shown). Sequences have been posted in GenBank as described in the ‘Accession numbers’ section.
Generation of a cleaved amplified polymorphic sequence (CAPS) marker discriminating between PveIF4E1 and PveIF4E2 alleles
Sequence analysis revealed an RsaI site in PveIF4E2 at codon 65, which is not present in PveIF4E1. The primer pair ENM‐FWe/RVe was selected in order to amplify a fragment of PveIF4E genomic DNA (gDNA) containing the polymorphic RsaI site and spanning exon 1 and exon 2 as predicted from the A. thaliana genomic sequence. PCR using the primer pair ENM‐FWe/RVe on the P. vulgaris gDNA template generated a 541‐bp fragment from all genotypes tested. Digestion with RsaI resulted in cleavage into 381‐bp and 160‐bp fragments of PCR products derived from genotypes Raven, USCR7, USCR8 and USLK1, all predicted to carry the bc‐3 gene. PCR fragments derived from PveIF4E1 cultivars, including DW, SGR, Widusa, Topcrop and Sanilac, were not cleaved by RsaI (results shown for DW and USCR8, Fig. 2, lanes 2 and 3).
Figure 2.
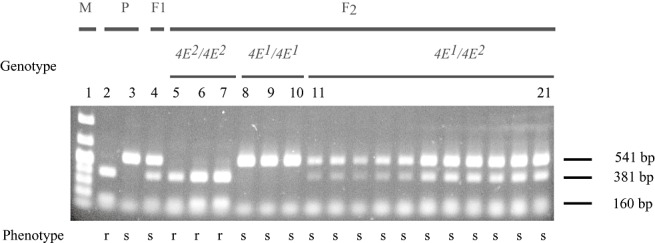
Segregation of Bean common mosaic virus (BCMV) resistance and a cleaved amplified polymorphic sequence (CAPS) marker differentiating eIF4E alleles PveIF4E2 (4E2) and PveIF4E1 (4E1). A Phaseolus vulgaris F2 population was generated from a cross between parents (P) USCR8 (4E2/4E2, lane 2) and Dubbele Witte (DW) (4E1/4E1, lane 3). Successful crossing was verified on F1 individuals (lane 4) and results from 17 F2 individuals are shown (lanes 5–21).The response to BCMV was assayed by enzyme‐linked immunosorbent assay (ELISA). Plants with an ELISA A405 (absorbance at 405 nm) reading of more than 2.5 times that of the mock‐inoculated control were rated as susceptible (s); plants with an ELISA A405 reading of less than 2.5 times that of the mock‐inoculated control were rated as resistant (r). The genotype was determined by restriction enzyme RsaI digestion of a 541‐bp polymerase chain reaction (PCR) product amplified with primers ENM‐FWe and ENM‐RVe from genomic DNA. DNA fragments were analysed by agarose gel electrophoresis with a 100‐bp ladder as marker (lane 1).
Generation of F2 plants segregating for bc‐3
Genotype USCR8 is reported to carry the Bc‐u, bc‐3 resistance gene and shows partial resistance towards some BCMV strains, including NL1 (Miklas and Hang 1998a). Bean genotypes USCR8 and all‐susceptible DW were therefore selected for the generation of F2 and F3 populations segregating for bc‐3 and the eIF4E allele identified (Fig. 2, lanes 2 and 3).
Crossings of USCR8 and DW were made and verified by testing candidate F1 plants for eIF4E heterozygosity with respect to PveIF4E1 and PveIF4E2 using the RsaI CAPS marker (Fig. 2, lane 4). F2 and F3 seeds were harvested from F1 plants heterozygous for eIF4E alleles and F2 plants homozygous for eIF4E2, respectively.
BCMV susceptibility and eIF4E genotype of F2 plants
In order to test the association of resistance to BCMV and the PveIF4E2 genotype, a total of 96 F2 plants (DW × USCR8) and a number of plants of the parental lines were inoculated with BCMV‐NL1 (Drijfhout 1978; Drijfhout et al., 1978). In two independent experiments, all plants were tested by enzyme‐linked immunosorbent assay (ELISA) for BCMV antigen at 6 weeks post‐inoculation (wpi). The PveIF4E genotype of each plant was determined by RsaI CAPS marker analysis (Fig. 2, lanes 5–21). In both experiments, plants homozygous for PveIF4E2 (denoted eIF4E2/eIF4E2) were found to display similar or less antigen on average than observed for the parental bc‐3 genotype, USCR8 (Fig. 3). At the level of individual plants, a marked under‐representation of ELISA‐positive plants was observed in the group of plants homozygous for PveIF4E2 (23% compared with 100% for the other genotypes eIF4E1/eIF4E1 and eIF4E1/eIF4E2; Table 2). The H0 hypothesis, stating that resistant and susceptible individuals were distributed equally between the three genotypes, was rejected on the basis of a χ 2 test (P < 0.001) (Table 2). All plants testing negative in BCMV ELISA (less than 2.5‐fold reading values compared with non‐inoculated USCR8) were found to be of genotype eIF4E2/eIF4E2. The eIF4E2/eIF4E2 group was not completely resistant, as 23% of individual plants were found to be ELISA positive. However, for parental line USCR8, carrying bc‐3, an even higher fraction of plants was found to be BCMV ELISA positive (55%, Table 2).
Figure 3.

Average Bean common mosaic virus (BCMV) antigen levels in parental lines and F2 plants grouped by eIF4E genotype. In two experiments, A and B, plants were inoculated with BCMV strain NL1 and analysed by BCMV enzyme‐linked immunosorbent assay (ELISA) of the top leaves at 6 weeks post‐inoculation (wpi). Dark columns show the average BCMV antigen levels and the bars on top indicate the standard deviation within each group. The ELISA control (CON, white column) is the average of two non‐inoculated plants of USCR8. The PveIF4E genotype was determined by application of the RsaI cleaved amplified polymorphic sequence (CAPS) marker, and plants were grouped into homozygous PveIF4E1 (4E1/4E1), homozygous PveIF4E2 (4E2/4E2) and heterozygous (4E1/4E2) plants. The number of plants in each group is indicated.
Table 2.
Enzyme‐linked immunosorbent assay (ELISA) assessment of Bean common mosaic virus (BCMV) infection in 96 Phaseolus vulgaris F2 plants grouped according to eIF4E genotype: eIF4E 1/eIF4E 1 (4E1/4E 1), eIF4E 1/eIF4E 2 (4E1/4E 2) or eIF4E 2/eIF4E2 (4E2/4E 2).
| Total plants | ELISA* 6 wpi | Genotype | χ 2 test† | |||
|---|---|---|---|---|---|---|
| 4E1/4E1 | 4E1/4E2 | 4E2/4E2 | ||||
| F2 | 96 | ELISA negative | 0 | 0 | 13 | P < 0.1% |
| ELISA positive | 33 | 46 | 4 | |||
| ELISA positive (%) | 100 | 100 | 23 | |||
| Parental line Dubbele Witte (DW) | 12 | ELISA positive (%) | 100 | — | — | |
| Parental bc‐3 line, USCR8 | 20 | ELISA positive (%) | — | — | 55 | |
Infection was assessed by ELISA and samples with A 405 > 2.5 times the mock‐inoculated controls were rated ELISA positive.
H0, equal distribution of infected and uninfected individuals between genotypes 4E1/4E1, 4E1/4E2 and 4E2/4E2.
BCMV resistance in parental, F2 and F3 plants homozygous for eIF4E2
BCMV accumulated in some USCR8 and F2 individuals homozygous for eIF4E2 to levels rated as ELISA positive (Table 2). To determine whether this difference in susceptibility was based on a genetic difference, resistance to BCMV in F3 plants derived from eIF4E2 homozygous F2 individuals was determined. Fifty F3 seeds derived from four ELISA‐positive and four ELISA‐negative F2 plants were germinated. To make sure that the seedlings were not infected by seed‐transmitted BCMV, seedlings were tested by ELISA prior to inoculation with BCMV‐NL1 (data not shown). At 6 wpi, the plants were tested by ELISA for the presence of BCMV antigen, and the relative absorbance ratio was compared with the results obtained for USCR8 and F2 plants homozygous for eIF4E2 (Fig. 4).
Figure 4.
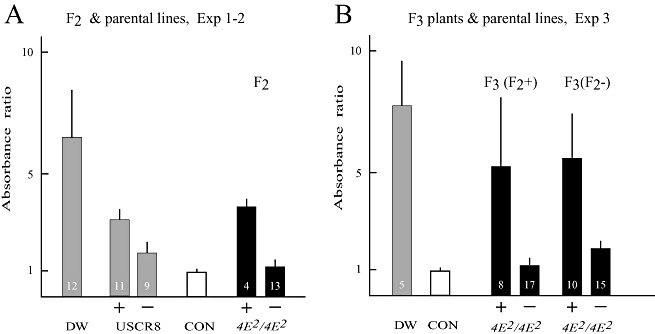
Average Bean common mosaic virus (BCMV) absorbance ratios in parental genotypes, F2 plants homozygous for PveIF4E2 (4E2/4E2) and F3 plants derived from 4E2/4E2 F2 plants. (A) 4E2/4E2 F2 individuals (dark columns) and parental USCR8 plants (USCR8, grey columns) from experiments 1 and 2 are divided into enzyme‐linked immunosorbent assay (ELISA)‐positive (+) and ELISA‐negative (–) groups and compared with parental cultivar Dubbele Witte (DW) (all‐susceptible, grey column). (B) F3 plants (dark columns) derived from ELISA‐positive (+) and ELISA‐negative (‐) 4E2/4E2 F2 (F2+ and F2–, respectively) plants were inoculated with BCMV strain NL1 and analysed by BCMV ELISA of the top leaves at 6 weeks post‐inoculation (wpi). F3 plants within each group were subdivided into ELISA positive (+) and ELISA negative (–) on the basis of the absorbance ratio. Columns show the absorbance ratios and the bars on top indicate the standard deviation within each group. The ELISA control (CON, white column) is the average of two non‐inoculated plants of USCR8. The number of plants in each group is indicated.
On average, both F3 plants derived from ELISA‐positive and ELISA‐negative F2 plants displayed similar antigen levels as observed for the parental bc‐3 genotype, USCR8 (data not shown). When F3 plants were divided into ELISA‐positive and ELISA‐negative plants, there was no difference between plants derived from ELISA‐positive and ELISA‐negative F2 plants. Plants that were rated positive in ELISA constituted 32% (8 of 25) of the F3 plants derived from ELISA‐positive F2 plants and 40% (10 of 25) of the plants derived from ELISA‐negative F2 plants (Fig. 4). This suggests that susceptibility, scored as ELISA‐positive plants, within the population of eIF4E2/eIF4E2 plants is not genetically based.
DISCUSSION
We report the cloning of P. vulgaris cDNA corresponding to homologues of eIF4E, eIF(iso)4E and nCBP. For eIF4E, two types of cDNA were identified, corresponding to two alleles, PveIF4E1 and PveIF4E2. The two alleles differed at four codons corresponding to predicted amino acid positions 53, 65, 76 and 111. The observed distribution of polymorphic codons resembled the polymorphism of eIF4E, reported previously to determine resistance or to be associated with resistance to potyviruses in three other species: C. annuum, L. sativa and P. sativum (reviewed in Robaglia and Caranta, 2006). Among nine P. vulgaris differentials of BCMV and BCMNV, the PveIF4E2 allele was found only in the four genotypes reported to carry bc‐3 resistance. A molecular CAPS marker was developed and, by subsequent analysis of the segregating F2 population, we observed that only plants homozygous for the PveIF4E2 allele resisted virus infection as determined by ELISA at 6 wpi. The resistance in USCR8 and in F2 plants homozygous for PveIF4E2 was not complete, because a fraction of the plants displayed ELISA readings that were rated as positive. When F3 plants derived from ELISA‐positive and ELISA‐negative F2 individuals homozygous for eIF4E2 were analysed, there was no difference between the two groups with respect to resistance. This suggests that the occurrence of ELISA‐positive plants after inoculation with BCMV is not a result of an inherited difference between the two groups. It may be speculated that resistance in the absence of bc‐u is not complete and that ELISA‐positive plants are a result of the occurrence of resistance‐breaking variants of BCMV‐NL1. Markers linked to bc‐3 in specific genotypes have been reported (Johnson et al., 1997; Mukeshimana et al., 2005), but these markers cannot discriminate between parental USCR8 and DW plants (data not shown). Taken together, the findings described above place PveIF4E2 as a strong candidate gene for bc‐3. However, complementation of BCMV infection in bc‐3 plants by co‐expression of the PveIF4E1 allele, or a similar direct test, is needed before a firm conclusion implying identity can be drawn.
Our findings represent new ground for potyviral research in bean, providing, for the first time, specific evidence for a conceptual alignment with a common mechanism of recessive potyviral resistance: eIF4E‐mediated. The molecular marker developed here for the observed eIF4E polymorphism in bean may serve as a useful tool to investigate further potyviral resistance in this species. In addition to the question of eIF4E complementation mentioned above, other questions relating to resistance, such as the effect of pyramiding P. vulgaris resistance genes and the influence of different genetic backgrounds on resistance, are likely to be more accessible to future investigation using this tool.
EXPERIMENTAL PROCEDURES
P. vulgaris germplasms and crossing
Seeds of P. vulgaris cultivars DW, Widusa, SGR, Topcrop, Sanilac (Drijfhout 1978) and Raven (Kelly et al., 1994) were obtained from CIAT (International Center for Tropical Agriculture, Cali, Colombia). Three other genotypes carrying the bc‐3 resistance gene, USCR7, USCR8 and USLK1 (Larsen et al., 2005; Miklas and Hang, 1998a, 1998b; Miklas and Kelly, 2002), were obtained from the United States Department of Agriculture, Agricultural Research Service (USDA‐ARS), Prosser, WA, USA. The proposed resistance genes of these genotypes are shown in Table 1. All plants were grown and multiplied under glasshouse conditions (16–18 °C, 16 h light).
An F2 population was generated from a cross between P. vulgaris genotypes DW and USCR8. DW is susceptible to BCMV and BCMNV with the proposed genotype ii/BcUBcU/Bc3Bc3; USCR8 has the proposed genotype ii/BcUBcU/bc3bc3 and displays resistance (absence of symptoms) towards a number of BCMV and BCMNV isolates (Larsen et al., 2005; Miklas and Hang 1998a). Crossings were made in the following way. Flower buds on recipient plants were opened and emasculated when they showed increased corolla size and reduced green colour compared with younger buds. Stamens with profuse pollen were removed from donor plants and hooked over the stigmas of emasculated recipient buds. Manually pollinated buds were finally closed to reduce the possibility of unintended crossing and to preserve humidity around the stigmas. Successful crossing was verified in F1 seedlings using a CAPS marker (Fig. 2), and the heterozygous plants were kept under glasshouse conditions for self‐pollination and the generation of F2 and F3 seeds.
Virus inoculation and detection
BCMV‐NL1 strain (Drijfhout et al., 1978) was received in infected seeds from USDA‐ARS. Inoculum of NL1 was propagated in DW, and test plants were mechanically inoculated at the primary leaf stage with infected DW leaf tissue homogenized in 100 mm cold phosphate buffer, pH 7.0, containing 2% polyvinylpyrrolidone and 0.2% Na2SO3. Carborundum (silicon carbide F400; Dragon, Maribo, Denmark) was added to infected sap before inoculation. Plants were recorded for disease symptoms (mosaic, leaf distortion and rugose) from 1 wpi, and plants without symptoms were re‐inoculated. BCMV antigen was measured in all plants at 6 wpi by indirect antigen‐first (AgF) ELISA (Albrechtsen 2006) with the following modifications: Maxisorb microplates (Nunc, Roskilde, Denmark) were used for coating with antigen. BCMV antisera AS‐0241 and AS‐0242 (DSMZ, Braunschweig, Germany) were mixed 1 : 1 and pre‐absorbed at a 1 : 1000 dilution in bean leaf homogenate before being applied to the plates. Anti‐rabbit IgG D‐306 (DAKO, Glostrup, Denmark), conjugated to alkaline phosphatase, was used as secondary antibody. The absorbance at 405 nm was recorded after 3 h of incubation with p‐nitrophenyl phosphate (Sigma, St. Louis, MO, USA) at room temperature on ELISA reader Thermo 354 (Electron Corporation, Waltham, MA, USA).
Design of primers to amplify cDNA and gDNA
Sequences from leguminous plants encoding cap‐binding proteins were retrieved using the Arabidopsis gene accessions At4G18040 (eIF4E‐1), At5G35620 (eIF(iso)4E) and At5G18110 (nCBP) as probes in National Center for Biotechnology Information (NCBI) blast nucleotide search. The primer pair (FWa, 5′‐TAACAATGGTTGTRGAAGAYRCCC‐3′; RVa, 5′‐TCAYACAACRTATTTRTTTTTAGCA‐3′) used to clone the P. vulgaris eIF4E homologue was designed to target conserved sequences of leguminous eIF4E cDNA identified by alignment of sequences from P. sativum (DQ641470), G. max eIF4E‐1 homologue 1 (CD412413, BU550435, BE661014) and G. max eIF4E‐1 homologue 2 (BM528551, AW348885). The software ClustalX was used for the alignment. For P. vulgaris eIF(iso)4E, two sets of primer pairs (FWb, 5′‐ATGGCAACAGACGAAGAAGT‐3′; RVb, 5′‐TTAYACRGTGTAYCGASCCT‐3′; and FWc, 5′‐GCCGAGAGAGAAAGAGAGAGA‐3′; RVc, 5′‐GCAYAAGTGCAAGAAGATGCTC‐3′) were similarly designed based on the coding sequence from P. sativum (DQ778076) and expressed sequence tags (ESTs) from P. coccineus (CA905562, CA905558), P. vulgaris (CV534941), G. max eIF(iso)4E homologue 1 (AI748763, BU926993, CD406294) and G. max eIF(iso)4E homologue 2 (CX710048). Primer pair FWc/RVc was designed to amplify P. vulgaris eIF(iso)4E cDNA, including the 5′ and 3′ untranslated regions. A P. vulgaris nCBP primer pair (FWd, 5′‐ATGGAATTCACAGTGGAGAAGGA‐3′; RVd, 5′‐CATCTAGCCTCTCAACCAAGTGTT‐3′) was designed using P. vulgaris EST sequences (CB540214, CV541716).
Nucleotide sequences of P. vulgaris eIF4E cDNA (EF571267 and EF571273) were used to design primers ENM‐FWe (5′‐ACCGATGAGCAAAACCCTA‐3′) and ENM‐RVe (5′‐CAACCAACTGGTATCGGATT‐3′) for a CAPS marker. The primers for the CAPS marker amplified a 541‐bp genomic fragment spanning exon 1 and exon 2 of P. vulgaris eIF4E and containing an RsaI restriction polymorphism spanning codon 65 (GTTC/GTAC).
RNA extraction, reverse transcriptase‐polymerase chain reaction (RT‐PCR), cloning and sequence analysis
Total RNA was extracted from 0.1 g of leaf tissue using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and eluted in 40 µL diethylpyrocarbonate (DEPC)‐treated H2O. Oligo‐dT‐primed cDNA synthesis by high‐fidelity reverse transcriptase (Roche, Mannheim, Germany) was performed using 1 µL of RNA template in a total volume of 20 µL according to the manufacturer's instructions. PCR amplification was performed using 5 µL of cDNA in 50 µL reactions containing 0.6 pm of oligonucleotide primers and 4 units of expand DNA polymerase (Roche). cDNA of eIF4E, eIF(iso)4E and nCBP was amplified in 40 cycles: 20 s of denaturation at 94 °C, 20 s of annealing at 57, 60 and 65 °C, respectively, and 35 s of elongation at 72 °C in a thermal cycler (GeneAmp PCR System 9700; Applied Biosystems, Naerum, Denmark) after an initial denaturation at 95 °C for 3 min.
PCR fragments for sequence analysis were obtained from RT reactions from two plants of each genotype. PCR products were purified using GFX™ PCR DNA and a Gel Band Purification Kit (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK), and sequenced directly or cloned into pCR 2.1 TOPO Vector (Invitrogen, Taastrup, Denmark). Sequencing was performed at MWG‐Biotech, Ebersberg, Germany. Sequence analysis and alignment were performed using the 4 Peaks sequence analysing program (http://mekentosj.com/4peaks/), ClustalW (http://www.ebi.ac.uk/clustalw/) and T_coffee (http://tcoffee.vital‐it.ch/cgi‐bin/Tcoffee/tcoffee_cgi/index.cgi).
gDNA extraction and CAPS marker eIF4E2‐RsaI analysis
gDNA was extracted from 0.1 g of leaf tissue that was homogenized in liquid nitrogen and dispersed in 500 µL of 2 × cetyltrimethylammonium bromide (CTAB) buffer [1.4 m NaCl, 20 mm ethylenediaminetetraacetic acid (EDTA), 2% CTAB, 2% polyvinylpyrrolidone and 100 mm TRIS, pH 8]. After incubation at 65 °C for 30 min, the homogenate was centrifuged for 10 min at 6300 g and the supernatant was extracted with 1 vol of chloroform–isoamyl alcohol (24 : 1) for 10 min. The phases were separated by centrifugation for 5 min at 15 000 g and DNA in the supernatant was precipitated using 0.8 vol of ice‐cold isopropanol and 0.1 vol of 3 m NaOAc, pH 4.6, followed by centrifugation for 10 min at 8000 g. The pellet was resuspended in 40 µL of dH2O.
For CAPS analysis, a fragment of 541 bp of eIF4E was amplified by PCR, using 0.3 µL of gDNA extract in a 25 µL reaction volume containing Taq‐buffer (New England Biolabs, Ipswich, MA, USA), 1.5 mm MgCl2, 6 pm of each of the oligonucleotide primers ENM‐FWe and ENM‐RVe and 5 units of Taq DNA polymerase (New England Biolabs). gDNA was amplified in 40 cycles: 20 s of denaturation at 94 °C, 20 s of annealing at 69 °C and 20 s of elongation at 72 °C in a thermal cycler (GeneAmp PCR System 9700) after an initial denaturation at 95 °C for 3 min. 8 µL of PCR product were subsequently subjected to RsaI (Roche) digestion and analysed by agarose gel electrophoresis.
ACCESSION NUMBERS
The nucleotide data appear in the GenBank database under accession numbers eIF4E: EF571267 (DW), EF571268 (SGR), EF571269 (Widusa), EF571270 (Topcrop), EF571271 (Sanilac), EF571273 (Raven), EF571274 (USCR7), EF571275 (USCR8) and EF571276 (USLK1); eIF(iso)4E: EF571278 (DW), EF571279 (Topcrop), EF571280 (Sanilac), EF571282 (USCR7), EF571283 (USCR8), EF571284 (USLK1), EF571285 (SGR), EF571286 (Widusa), EF571287 (Raven); and nCBP: EF558667 (DW), EF571261 (SGR), EF571262 (Topcrop), EF571263 (Sanilac), EF571265 (Raven), EF571266 (USCR7).
ACKNOWLEDGEMENTS
We are grateful to CIAT for the seeds of the bean genotypes. This project was supported financially by a PhD grant to MN from the Iranian Ministry of Science, Research and Technology and Ministry of Agriculture, and by the Danish Agricultural and Veterinary Research Council grant no. 53‐00‐0330 to OSL.
REFERENCES
- Ahlquist, P. , Noueiry, A.O. , Lee, W.M. , Kushner, D.B. and Dye, B.T. (2003) Host factors in positive‐strand RNA virus genome replication. J. Virol. 77, 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen, S.E. (2006) Testing Methods for Seed‐Transmitted Viruses: Principles and Protocols. Wallingford, Oxfordshire: CABI Publishing, pp. 111–115. [Google Scholar]
- Beauchemin, C. , Boutet, N. and Laliberte, J.F. (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta . J. Virol. 81, 775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun‐Rasmussen, M. , Møller, I.S. , Tulinius, G. , Hansen, J.K. , Lund, O.S. and Johansen, I.E. (2007) The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum . Mol. Plant–Microbe Interact. 20, 1075–1082. [DOI] [PubMed] [Google Scholar]
- Carberry, S.E. , Darzynkiewics, E. and Goss, D.J. (1991) A comparison of the binding of methylated cap analogues to wheat germ protein synthesis initiation factors 4F and (iso)4F. Biochemistry, 30, 1624–1624. [DOI] [PubMed] [Google Scholar]
- Collmer, C.W. , Marston, M.F. , Taylor, J.C. and Jahn, M. (2000) The I gene of bean: a dosage‐dependent allele conferring extreme resistance, hypersensitive resistance, or spreading vascular necrosis in response to the potyvirus Bean common mosaic virus . Mol. Plant–Microbe Interact. 13, 1266–1270. [DOI] [PubMed] [Google Scholar]
- Drijfhout, E. (1978) Genetic interaction between Phaseolus vulgaris and Bean common mosaic virus with implications for strain identification and breeding for resistance. Agricultural Research Report, Center for Agricultural Publishing and Documentation, Wageningen, The Netherlands, 98 pp.
- Drijfhout, E. , Silbernagel, M.J. and Burke, D.W. (1978) Differentiation of strains of Bean common mosaic virus . Neth. J. Plant Pathol. 84, 13–26. [Google Scholar]
- Gao, Z. , Johansen, E. , Eyers, S. , Thomas, C.L. , Ellis, T.H.N. and Maule, A.J. (2004) The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell‐to‐cell trafficking. Plant J. 40, 376–385. [DOI] [PubMed] [Google Scholar]
- Hwang, J.N. , Li, J. , Liu, W.‐Y. , An, S.‐J. , Cho, H. , Her, N.H. , Yeam, I. , Kim, D. and Kang, B.‐C. (2009) Double mutations in eIF4E and eIF(iso)4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells, 27, 329–336. [DOI] [PubMed] [Google Scholar]
- Johnson, W.C. , Guzman, P. , Mandala, D. , Mkandawire, A.B.C. , Temple, S. , Gilbertson, R.L. and Gepts, P. (1997) Molecular tagging of the bc‐3 gene for introgression into Andean common bean. Crop. Sci. 37, 248–254. [Google Scholar]
- Kang, B.‐C. , Yeam, I. , Frantz, J.D. , Murphy, J.F. and Jahn, M.M. (2005) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42, 392–405. [DOI] [PubMed] [Google Scholar]
- Keller, K.E. , Johansen, I.E. , Martin, R.R. and Hampton, R.O. (1998) Potyvirus genome‐linked protein (VPg) determines Pea seed‐borne mosaic virus pathotype‐specific virulence in Pisum sativum . Mol. Plant–Microbe Interact. 11, 124–130. [DOI] [PubMed] [Google Scholar]
- Kelly, J.D. , Hosfield, S.D. , Varner, G.V. , Uebersax, M.A. , Haley, S.D. and Taylor, J. (1994) Registration of ‘Raven’ black bean. Crop. Sci. 34, 1406–1407. [Google Scholar]
- Larsen, R.C. , Miklas, P.N. , Druffel, K.L. and Wyatt, S.D. (2005) NL3‐K is a stable and naturally occurring interspecific recombinant derived from Bean common mosaic necrosis virus and Bean common mosaic virus . Virology, 95, 1037–1042. [DOI] [PubMed] [Google Scholar]
- Lellis, A.D. , Kasschau, K.D. , Whitham, S.A. and Carrington, J.C. (2002) Loss‐of‐susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Leonard, S. , Viel, C. , Beauchemin, C. , Daigneault, N. , Fortin, M.G. and Laliberté, J.F. (2004) Interaction of VPg‐Pro of Turnip mosaic virus with the translation initiation factor 4E and the poly(A)‐binding protein in planta . J. Gen. Virol. 85, 1055–1063. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Afanador, L. and Kelly, J.D. (1996) Development of a SCAR marker linked to the I gene in common bean. Genome, 39, 1216–1219. [DOI] [PubMed] [Google Scholar]
- Miklas, P.N. and Hang, A.N. (1998a) Release of cranberry dry bean germ plasm lines USCR‐7 and USCR‐8 with resistance to Bean common mosaic and necrosis viruses . Annu. Rep. Bean. Improv. Coop. 41, 227–228. [Google Scholar]
- Miklas, P.N. and Hang, A.N. (1998b) Release of light red kidney dry bean germ plasm lines USLK‐1, ‐2, and ‐3 with resistance to Bean common mosaic and necrosis viruses . Annu. Rep. Bean. Improv. Coop. 41, 231–232. [Google Scholar]
- Miklas, P.N. and Kelly, J.D. (2002) Registration of two cranberry bean germplasm lines resistant to Bean common mosaic and necrosis potyviruses: USCR7 and USCR9. Crop. Sci. 42, 673–674. [Google Scholar]
- Miklas, P.N. , Larsen, R.C. , Riley, R. and Kelly, J.D. (2000) Potential marker‐assisted selection for bc‐12 resistance to Bean common mosaic potyvirus in common bean. Euphytica, 116, 11–219. [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. , Caranta, C. , Palloix, A. and Jacquemond, M. (2004) Mutations in Potato virus Y genome‐linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant–Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Mukeshimana, G. , Paneda, A. , Rodriguez‐Suarez, C. , Ferreira, J.J. , Giraldez, R. and Kelly, J.D. (2005) Markers linked to the bc‐3 gene conditioning resistance to Bean common mosaic potyviruses in common bean. Euphytica, 144, 291–299. [Google Scholar]
- Nicaise, V. , German‐Retana, S. , Sanjuan, R. , Dubrana, M.‐P. , Mazier, M. , Maisonneuve, B. , Candresse, T. , Caranta, C. and LeGall, O. (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus . Plant Physiol. 132, 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia, C. and Caranta, C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. [DOI] [PubMed] [Google Scholar]
- Ruffel, S. , Dussault, M.‐H. , Palloix, A. , Moury, B. and Bendahman, A. (2002) A natural recessive resistance gene against Potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32, 1067–1075. [DOI] [PubMed] [Google Scholar]
- Ruffel, S. , Gallois, J.‐L. , Moury, B. , Robaglia, C. , Palloix, A. and Caranta, C. (2006) Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent Pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87, 2089–2098. [DOI] [PubMed] [Google Scholar]
- Rusholme, R.L. , Higgins, E.E. , Walsh, J.A. and Lydiate, D.J. (2007) Genetic control of broad‐spectrum resistance to Turnip mosaic virus in Brassica rapa (Chinese cabbage). J. Gen. Virol. 88, 3177–3186. [DOI] [PubMed] [Google Scholar]
- Ruud, K.A. , Kuhlow, C. , Goss, D.J. and Browing, K.S. (1998) Identification and characterization of a novel cap‐binding protein from Arabidopsis thaliana . J. Biol. Chem. 273, 10325–10330. [DOI] [PubMed] [Google Scholar]
- Sato, M. , Nakahara, K. , Yoshii, M. , Ishikawa, M. and Uyeda, I. (2005) Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 579, 1167–1171. [DOI] [PubMed] [Google Scholar]
- Schaad, M.C. , Anderberg, R.J. and Carrington, J.C. (2000) Strain‐specific interaction of the Tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two‐hybrid system. Virology, 273, 300–306. [DOI] [PubMed] [Google Scholar]
- Stein, N. , Perovic, D. , Kumlehn, J. , Pellio, B. , Stracke, S. , Streng, S. , Ordon, F. and Graner, A. (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 42, 912–922. [DOI] [PubMed] [Google Scholar]
- Wittmann, S. , Chatel, H. , Fortin, M.G. and Laliberet, J.F. (1997) Interaction of the viral protein genome linked of Turnip mosaic potyvirus with the translation eukaryotic initiation factor (iso)4E of Arabidopsis thaliana using the yeast two hybrid system. Virology, 234, 84–92. [DOI] [PubMed] [Google Scholar]
- Yeam, I. , Cavatorta, J.R. , Ripoll, D.R. , Kang, B.‐C. and Jahn, M.M. (2007) Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell, 19, 2913–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]


