SUMMARY
Maize streak virus (MSV; Genus Mastrevirus, Family Geminiviridae) occurs throughout Africa, where it causes what is probably the most serious viral crop disease on the continent. It is obligately transmitted by as many as six leafhopper species in the Genus Cicadulina, but mainly by C. mbila Naudé and C. storeyi. In addition to maize, it can infect over 80 other species in the Family Poaceae. Whereas 11 strains of MSV are currently known, only the MSV‐A strain is known to cause economically significant streak disease in maize. Severe maize streak disease (MSD) manifests as pronounced, continuous parallel chlorotic streaks on leaves, with severe stunting of the affected plant and, usuallly, a failure to produce complete cobs or seed. Natural resistance to MSV in maize, and/or maize infections caused by non‐maize‐adapted MSV strains, can result in narrow, interrupted streaks and no obvious yield losses. MSV epidemiology is primarily governed by environmental influences on its vector species, resulting in erratic epidemics every 3–10 years. Even in epidemic years, disease incidences can vary from a few infected plants per field, with little associated yield loss, to 100% infection rates and complete yield loss.
Taxonomy: The only virus species known to cause MSD is MSV, the type member of the Genus Mastrevirus in the Family Geminiviridae. In addition to the MSV‐A strain, which causes the most severe form of streak disease in maize, 10 other MSV strains (MSV‐B to MSV‐K) are known to infect barley, wheat, oats, rye, sugarcane, millet and many wild, mostly annual, grass species. Seven other mastrevirus species, many with host and geographical ranges partially overlapping those of MSV, appear to infect primarily perennial grasses.
Physical properties: MSV and all related grass mastreviruses have single‐component, circular, single‐stranded DNA genomes of approximately 2700 bases, encapsidated in 22 × 38‐nm geminate particles comprising two incomplete T = 1 icosahedra, with 22 pentameric capsomers composed of a single 32‐kDa capsid protein. Particles are generally stable in buffers of pH 4–8.
Disease symptoms: In infected maize plants, streak disease initially manifests as minute, pale, circular spots on the lowest exposed portion of the youngest leaves. The only leaves that develop symptoms are those formed after infection, with older leaves remaining healthy. As the disease progresses, newer leaves emerge containing streaks up to several millimetres in length along the leaf veins, with primary veins being less affected than secondary or tertiary veins. The streaks are often fused laterally, appearing as narrow, broken, chlorotic stripes, which may extend over the entire length of severely affected leaves. Lesion colour generally varies from white to yellow, with some virus strains causing red pigmentation on maize leaves and abnormal shoot and flower bunching in grasses. Reduced photosynthesis and increased respiration usually lead to a reduction in leaf length and plant height; thus, maize plants infected at an early stage become severely stunted, producing undersized, misshapen cobs or giving no yield at all. Yield loss in susceptible maize is directly related to the time of infection: infected seedlings produce no yield or are killed, whereas plants infected at later times are proportionately less affected.
Disease control: Disease avoidance can be practised by only planting maize during the early season when viral inoculum loads are lowest. Leafhopper vectors can also be controlled with insecticides such as carbofuran. However, the development and use of streak‐resistant cultivars is probably the most effective and economically viable means of preventing streak epidemics. Naturally occurring tolerance to MSV (meaning that, although plants become systemically infected, they do not suffer serious yield losses) has been found, which has primarily been attributed to a single gene, msv‐1. However, other MSV resistance genes also exist and improved resistance has been achieved by concentrating these within individual maize genotypes. Whereas true MSV immunity (meaning that plants cannot be symptomatically infected by the virus) has been achieved in lines that include multiple small‐effect resistance genes together with msv‐1, it has proven difficult to transfer this immunity into commercial maize genotypes. An alternative resistance strategy using genetic engineering is currently being investigated in South Africa.
Useful websites: 〈http://www.mcb.uct.ac.za/MSV/mastrevirus.htm〉; 〈http://www.danforthcenter.org/iltab/geminiviridae/geminiaccess/mastrevirus/Mastrevirus.htm〉.
MAIZE STREAK IN AFRICA: A CENTURY OF PATHOLOGY
Maize streak disease (MSD) was first recorded in South Africa by Claude Fuller (1901), the Government Entomologist of Natal. Fuller also quoted personal sources who noticed the disease of ‘mealie variegation’, as it was then described, as early as the 1870s. The disease was therefore not new at the time, and had probably been around as long as maize had been grown in the region. Fuller's investigation of MSD was motivated by an increase in incidence of the disease, marked by a serious outbreak in 1896. Although Fuller was ignorant as to its cause—he thought it was caused either by a soil nutrient deficiency or a ‘chemical enzyme’ acquired from the soil—he accurately described many features of the disease as it manifests today. Thus, he noted: (i) that the streaks were composed of ‘a series of elongated or almost circular spots’ (Fig. 1); (ii) that severely affected plants had fewer green leaves at the base than mildly diseased plants; (iii) that some plants with pronounced chlorosis grew normally and yielded cobs, whereas others were severely stunted and yielded nothing; and (iv) that severely diseased plants could be found next to perfectly healthy ones. His illustrations were also excellent, and he showed that the diseased sections of leaves or the ‘streaks’ contained few or no chloroplasts. Sadly, he was probably one of the first victims of motor vehicle accidents in Africa: he was killed by a car in Lourenço Marques (now Maputo) in Mozambique in 1905.
Figure 1.

Maize streak disease symptoms: chlorotic streaks on a maize leaf. Photo credit: Frederik Kloppers.
Over 100 years later, MSD remains the most significant viral disease of Africa's most important food crop (Bosque‐Pérez, 2000), costing between US$120M and US$480M per year according to one conservative estimate based on average annual yield losses of only 6%–10% (Martin and Shepherd, 2009). Despite considerable advances in control measures (see below), which could halve these monetary losses (Martin and Shepherd, 2009), poorer farmers continue to suffer serious crop losses to MSD.
DISEASE AETIOLOGY
The legendary H.H. Storey was the first to show that MSD was caused by a virus, and that this virus was obligately transmitted by a leafhopper (Storey, 1924, 1925). Storey and his colleagues subsequently elucidated the transmission cycle, latent periods in the vector, the fact that the vector's transmission ability was genetically determined, the existence of distinct strains of the virus and differential reactions of various host plants to the same viruses (Storey, 1928, 1931, 1932, 1938, 1939; Storey and McClean, 1930). McClean went on to describe streak virus infections in maize, sugarcane and wild grasses in South Africa (McClean, 1947)—work which reinforced the earlier finding that there were genetically distinct ‘streak viruses’ infecting maize, sugarcane and grasses.
Direct proof of the existence of maize streak virus (MSV) in infected tissue came with the visualization of virus particles in 1974. Bock et al. (1974) discovered that MSV virions have a novel, twinned, quasi‐icosahedral (geminate) shape, from which the name ‘geminivirus’ was born (Fig. 2). This was followed by the unexpected discovery in 1977 that geminivirus particles contain circular single‐stranded DNA (ssDNA), a genome type never before observed in plant viruses (Goodman, 1977a, 1977b; Harrison et al., 1977).
Figure 2.
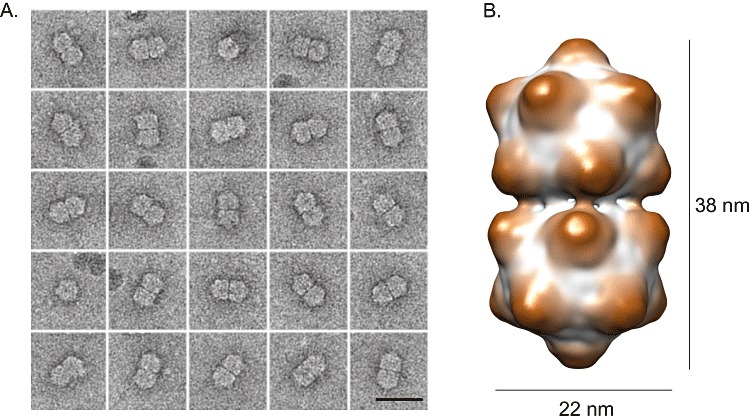
(A) Negatively stained geminate particles in different orientations. Size bar, 30 nm. (B) Three‐dimensional reconstruction of a geminate particle from cryoelectron microscopy data.
Despite being initially transmitted into phloem sieve tubes by the leafhopper, virus particles occur at high concentrations within the mesophyll cells of infected maize leaves (Lucy et al., 1996). Infection of chlorenchyma cells causes malformation of chloroplasts and reduced chlorophyll production (Pinner et al., 1993). The pattern of chlorotic lesions on infected maize leaves is directly correlated with the pattern of virus accumulation within the leaves (Lucy et al., 1996) and the virus can only be acquired by leafhoppers from these lesions (Peterschmitt et al., 1992; Storey, 1928).
MOLECULAR VIROLOGY
Each MSV virion contains a single, covalently closed, circular, ssDNA molecule (Francki et al., 1980) of approximately 2700 bases. It is generally accepted that geminiviruses replicate via double‐stranded DNA (dsDNA) intermediates using a rolling circle replication mechanism (1993a, 1993b; 1995a, 1995b; 1991, 1992). However, there is now good evidence that ‘recombination‐dependent replication’ mechanisms also play an important role in geminivirus replication (Jeske et al., 2001; Jovel et al., 2007; Preiss and Jeske, 2003; Saunders et al., 1991).
In replicative dsDNA molecules, genes are expressed from both strands, and diverge from an intergenic region which contains the virion‐sense origin of replication (Fig. 3). Transcription is thus bidirectional, with transcripts originating in the intergenic region and converging on the diametrically opposite side of the circular genome (Morris‐Krsinich et al., 1985). Rolling circle replication is initiated by binding of the virus replication‐associated protein (Rep) to the virion‐strand origin of replication, where the protein initiates and terminates virion‐strand DNA synthesis (Heyraud et al., 1993b; Heyraud‐Nitschke et al., 1995; 1995a, 1995b; Stanley, 1995; Stenger et al., 1991; Willment et al., 2007).
Figure 3.
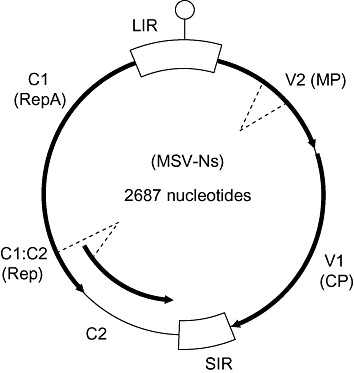
Genome organization of a representative maize streak virus isolate from Nigeria (MSV‐Ns). The virion‐sense origin of replication is represented by a stem‐loop symbol. Open reading frames (ORFs) are depicted by arrows in the direction of transcription, with broken lines indicating differential splice events. Non‐coding genomic regions are shown with open boxes. ORFs are named according to whether they are encoded in the virion or complementary sense (they are assigned either a V or C, respectively). Functional names of proteins (in parentheses below the ORF name) and the names of non‐coding genomic regions are abbreviated as follows: CP, coat protein; LIR, long intergenic region; MP, movement protein; Rep, replication‐associated protein; SIR, short intergenic region.
MSV Rep is the translation product of two complementary sense open reading frames (ORFs), C1 and C2. The C1:C2 transcript, which contains an intron, is translated to yield either Rep (spliced transcript) or RepA (unspliced transcript). Although Rep alone appears to be both necessary and sufficient for replication of mastreviruses in appropriate host cells (Liu et al., 1998; Schalk et al., 1989), RepA is thought to perform a variety of important additional functions during the mastrevirus life cycle. These include the modulation of host cell cycle regulation, and possibly other developmental pathways (Boulton, 2002; Gutierrez, 1999; Shepherd et al., 2005).
Apart from Rep and RepA, the MSV genome encodes only two additional proteins: the movement protein (MP) and the coat protein (CP). MP facilitates the movement of the virus out of the nucleus (the site of replication) and to adjacent cells (Boulton, 2002; Liu et al., 2001a). CP, as with most viruses, is responsible for the encapsidation of the viral nucleic acid, in this case ssDNA. However, mastrevirus CP performs a multitude of other functions: the CP of at least some leafhopper‐transmitted geminiviruses determines vector specificity, implying specific interactions with unknown vector factors (Boulton et al., 1989; Briddon et al., 1990); MSV CP is capable of binding non‐specifically to both ssDNA and dsDNA (Liu et al., 1997), and thereby facilitates the nuclear import of virus DNA (Liu et al., 1999); MSV CP is required for cell‐to‐cell and systemic spread of the virus in plants (1993, 1989; Lazarowitz et al., 1989; Liu et al., 2001b); moreover, MSV CP interacts specifically with MP to shuttle virus DNA out of the nucleus (Kotlizky et al., 2000; Liu et al., 2001a).
EPIDEMIOLOGY
Maize was first introduced to Africa via Ghana by Portuguese traders in the 16th century. The maize‐adapted MSV‐A strain can infect over 80 species in the Family Poaceae (Damsteegt, 1983), but it is probable that ancestral MSV‐A viruses that first came into contact with maize were specifically adapted to infecting species in the Genus Digitaria (Varsani et al., 2008a). MSV also infects other introduced grass species, such as wheat (Triticum aestivum), barley (Hordeum vulgare), rye (Secale cereal), oats (Avena sativa) and sugarcane (Saccharum officinarum), as well as cultivated indigenous species, such as finger millet (Eleusine coracana), pearl millet (Pennisetum americanum) and sorghum (Sorghum bicolor). Streak diseases in these crops are all probably of minor economic importance (Soto et al., 1982). One exception might be a sugarcane streak disease caused by the MSV‐A strain (van Antwerpen et al., 2008), which is increasing in prevalence in southern Africa and could have a serious impact on sugar production across this region.
Yield losses in MSV‐infected susceptible maize plants are directly related to times of infection: infected seedlings either die or produce no seed and, in one particular report, plants infected at the second, sixth and tenth leaf stages experienced approximately 55%, 40% and 25% losses in grain weight, respectively (Bock, 1982).
Rapid increases in virus populations and epidemic spread between crops are usually attributable to the convergence of factors, such as: (i) staggered growing seasons in which MSV‐A populations can bulk up in early planted maize and devastate seedlings that germinate in successive plantings (Dabrowski et al., 1991; Fajemisin and Shoyinka, 1976); (ii) the population density of wild grasses that are reservoirs of both MSV‐A and leafhopper vectors (Autrey and Ricaud, 1983); (iii) the presence within leafhopper populations of a high percentage of MSV transmitters; and (iv) environmental factors that drive the long‐distance movement of leafhoppers (Rose, 1978).
An important consideration for commercial maize farmers is the severe impact of MSD on ultra‐short season hybrids, which are becoming popular in southern Africa. The shorter growing time of these hybrids allows farmers to plant a second crop, such as wheat, during the winter, which also serves as a host for leafhoppers. Temporal overlap of these two crops provides a ‘green bridge’ (Kloppers, 2005), allowing survival of the leafhopper throughout the year. Although increased environmental virus titres towards the end of the growing season generally result in greater crop losses to MSD, ultra‐short season hybrids are especially vulnerable because of their sensitivity to MSV and the density with which they are planted. This provides a favourable microenvironment for the proliferation of leafhoppers that subsequently spread the virus. In addition, shortened growing seasons provide little chance for corrective action and recovery. This is because insecticidal control of leafhopper populations cannot usually be implemented in time to effectively control the spread of the disease (Kloppers, 2005).
Although maize is a favoured host for leafhopper feeding, leafhoppers preferentially breed on annual wild grass species (P. Markham, John Innes Centre, Norwich, East Anglia, UK). Approximately 70% of the more than 138 grass species on which leafhoppers feed are also MSV hosts (Konate and Traore, 1992), and the density and composition of grass populations in any region almost certainly has a major influence on MSD epidemiology in that region. For example, the maize‐adapted MSV‐A strain and the closely related grass‐adapted MSV‐B strain appear to be particularly well adapted to the infection of grasses in the Genus Digitaria (Varsani et al., 2008a).
Although outbreaks of MSD are governed by leafhopper acquisition and movement of severe MSV isolates from infected to non‐infected hosts, MSD epidemiology is complicated by the fact that different Cicadulina species have different proportions of individuals capable of transmitting the virus (ranging from 15% to 45%; Asanzi et al., 1995). In addition, not all of the 18 species of Cicadulina identified in Africa can transmit MSV (Bosque‐Pérez, 2000; Lett et al., 2002; Mesfin et al., 1995). Early studies (Markham et al., 1984; Storey, 1938), indicating that the insect gut wall acts as a barrier to MSV transmission in non‐vector species, were later confirmed using polymerase chain reaction (PCR) detection in individual insect organs (Lett et al., 2002): MSV was detected in the gut, haemolymph and head of a vector species (C. mbila), but restricted to the gut of a non‐vector species (C. chinai). In C. mbila, MSV crosses the gut in less than 3 h, indicating an active mechanism for transmembrane flow via a specific receptor (Lett et al., 2002). Although the main site for MSV accumulation is the alimentary canal (Ammar et al., 2009; Lett et al., 2002), amounts of viral DNA decrease considerably over time in both the gut and haemolymph. Interestingly, however, virus DNA copies remain stable over time in the head (presumably in the salivary glands, from which MSV is released into the phloem when the leafhopper feeds on a host plant), and it is probable that virus copies released by the salivary glands on feeding are continuously replaced from elsewhere, probably the haemolymph. As MSV is thought not to replicate in leafhoppers (Boulton and Markham, 1986; Reynaud and Peterschmitt, 1992), this flow towards the salivary glands, in addition to viral degradation, would explain the decrease in viral accumulation over time. Stable viral DNA levels in the salivary glands are also consistent with the observation that C. mbila can transmit the virus for 5 weeks after an acquisition access feeding period of only 3 h (Reynaud and Peterschmitt, 1992).
Although transmission to some hosts by C. mbila is remarkably efficient, studies of the feeding activities of this species by Mesfin et al. (1995) revealed vector preferences for certain hosts. On hosts from which the leafhoppers prefer not to feed, MSV transmission rates are decreased by reduced probing times (Bosque‐Pérez, 2000), indicating that feeding behaviour on a maize genotype influences its resistance to MSV infection. Although studies have shown that C. mbila is the species most often implicated in MSD outbreaks (Dabrowski, 1987; Magenya et al., 2008), Oluwafemi et al. (2007) found that C. storeyi is the better transmitter, indicating that transmission ability alone does not make an efficient vector. Other considerations are distribution (C. mbila is the most widely distributed species throughout Africa) and the fact that a larger proportion of C. mbila populations have the ability to transmit MSV compared with other Cicadulina species (Markham et al., 1984; Storey, 1928, 1933). This is partly a result of the proportion of C. mbila females (which are better transmitters), being two to three times higher than in other species (Wambugu and Wafula, 2000).
An additional factor contributing to transmission efficiency is the mobility of leafhoppers. In the warm wet season, C. mbila develops a longer body morph. This morph flies less than 10 m and, consequently, only isolated pockets of disease develop. However, with the onset of crop maturity or under drought conditions—both causing the food plants of leafhoppers to dry out—the stronger flying, short‐bodied C. mbila morph predominates. Extensive migration into irrigated crops occurs, spreading disease over great distances and resulting in widespread epidemics (Rose, 1978).
Environmental factors that have an influence on leafhopper population sizes also play an important role in MSD epidemiology. For example, MSD outbreaks are often associated with drought conditions, followed by irregular rains at the beginning of growing seasons (Efron et al., 1989), as in the savanna regions of West Africa in 1983 and 1984 (Rossel and Thottappilly, 1985), or in Kenya in 1988–89 (Njuguna et al., 1990). The relative abundance of various Cicadulina species with differing abilities to transmit the virus in different parts of Africa is influenced by altitude, temperature and rainfall (Dabrowski et al., 1987). In addition, late rainfall favours the development of leafhopper nymphs during the winter (Stanley et al., 1999). The interplay of all of these factors makes MSD epidemiology rather erratic, with the disease being devastating in some years and insignificant in others (Efron et al., 1989).
MSV DIVERSITY AND EVOLUTION
In addition to the demographics of grass and vector populations, the exact make‐up of MSV‐A populations in different parts of Africa probably also has a major influence on MSD epidemiology. Although there are fairly obvious differences in the genetic composition of MSV‐A populations in eastern, western and southern Africa (Briddon et al., 1994; Martin et al., 2001; Willment et al., 2001), it is not currently known whether these translate into differences in disease epidemiology. Of the four main lineages of MSV‐A currently circulating in Africa (MSV‐A1, MSV‐A2, MSV‐A3 and MSV‐A4; Fig. 4), MSV‐A1 and MSV‐A4 are apparently responsible for more than 95% of all MSD cases that have been analysed over the past 20 years. It is therefore safe to assume that these are the main lineages driving MSD epidemics throughout southern and East Africa where the bulk of virus sampling has been carried out (Martin et al., 2001; Owor et al., 2007; Varsani et al., 2008a; Willment et al., 2001).
Figure 4.
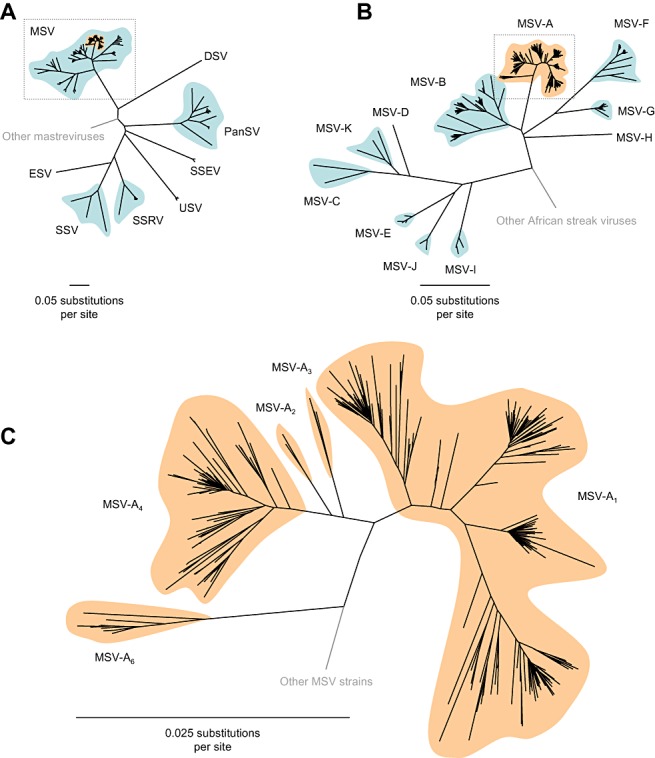
Phylogenetic relationships between viruses related to maize streak virus (MSV). (A) In addition to MSV, there are six other known African streak virus species, including Eragrostis streak virus (ESV), sugarcane streak virus (SSV), sugarcane streak Réunion virus (SSRV), Urochloa streak virus (USV), sugarcane streak Egypt virus (SSEV) and Panicum streak virus (PanSV). Digitaria streak virus (DSV), from the island of Vanuatu in the Pacific, is closely related to the African streak viruses and is included here for reference purposes. This tree has been rooted on the virus Chloris striate mosaic virus (not shown). The boxed area is expanded in (B). (B) There are 11 known MSV strains but only one of these, MSV‐A (in orange), is known to cause severe streak disease in maize. The boxed area is expanded in (C). (C) There are five major MSV‐A variants: MSV‐A6 has only been found on islands in the Indian Ocean; MSV‐A2 has only been found in West Africa; MSV‐A3 has only been found in East Africa; and MSV‐A4 has only been found in southern Africa. MSV‐A1 is found throughout mainland Africa.
Although MSV‐A4 is seemingly confined to southern Africa, MSV‐A1 has a geographical range that spans the whole of sub‐Saharan Africa (Martin et al., 2001; Owor et al., 2007; Varsani et al., 2008a; Willment et al., 2001). A characteristic of MSV‐A4, which may have some bearing on its more restricted geographical range, is that it is apparently less severe in maize than MSV‐A1 (Martin et al., 1999). The widespread distribution of MSV‐A1 is, however, somewhat unusual in that no other group of similar MSV variants (i.e. a MSV lineage displaying the same depth of genetic diversity as MSV‐A1) has ever been found to be spread between major regions of the continent, such as between East and West Africa or East and southern Africa.
Although MSV‐A is the only strain known to cause severe MSD (Martin et al., 2001; McClean, 1947; Storey and McClean, 1930), 10 non‐maize‐adapted MSV strains (MSV‐B to MSV‐K; Fig. 4) have also been identified (Martin et al., 2001; Schnippenkoetter et al., 2001a; Varsani et al., 2008a; Willment et al., 2002). Although they are normally found infecting wild grasses, some of these (MSV‐B, MSV‐C, MSV‐D and MSV‐E) are also known to produce mild infections in MSV‐susceptible maize genotypes (2001, 1999). Although they have no known direct impact on African agriculture, these MSV strains have probably had a large indirect influence on the evolution of the economically significant MSV‐A strain. For example, the main genetic feature differentiating MSV‐A4 from other MSV‐A lineages is that it is the product of a recombination event between MSV‐A and MSV‐B viruses found in southern Africa (Martin et al., 2001).
As with other geminiviruses, recombination has featured prominently in the evolution of MSV. In general, intra‐strain MSV recombination appears to have been far more prevalent during recent MSV‐A evolution than inter‐strain or inter‐species recombination (Owor et al., 2007; Varsani et al., 2008a). In all three recorded examples of recent natural inter‐strain recombination events involving MSV‐A viruses, fewer than 200 nucleotides have been exchanged, possibly indicating that inter‐strain recombination has little to offer in the way of substantial MSV‐A fitness improvements. This notion is backed up by the fact that laboratory constructed MSV‐A—MSV‐B and MSV‐A—MSV‐C chimaeras have invariably been severely defective (Martin and Rybicki, 2002; Martin et al., 2005; Schnippenkoetter et al., 2001b; van der Walt et al., 2008b). When maize plants are co‐infected with reciprocal MSV‐A—MSV‐B chimaeras (for example, laboratory constructed recombinants with mp and cp genes reciprocally swapped between MSV‐A and MSV‐B isolates), these viruses recombine very rapidly to produce MSV‐A‐like recombinants (van der Walt et al., 2009). Collectively, these observations indicate that the fitness of contemporary MSV‐A genotypes cannot be easily improved through inter‐strain recombination.
It is therefore somewhat surprising that the MSV‐A strain is thought to have arisen via a large recombination event that merged the mp and cp genes of an ancestral MSV‐B variant with the long intergenic region (LIR), short intergenic region (SIR) and rep genes of an ancestral virus resembling the progenitor of the MSV‐G and MSV‐F strains (Varsani et al., 2008a). It has been proposed that this recombination event may have triggered the emergence of MSV‐A as an agricultural pathogen. Given that, together with other geminiviruses (Duffy and Holmes, 2008; Ge et al., 2007), MSV probably has an evolution rate somewhere between 10−4 and 10−3 substitutions per site per year (Isnard et al., 1998; van der Walt et al., 2008a), this recombination event can be dated to any time between 100 years ago, when MSD was first described, and 500 years ago, when maize was first introduced to Africa—times that fit well with the hypothesis that this recombination event was pivotal in the adaptation of MSV‐A to maize.
Large inter‐strain recombination events and smaller inter‐species recombination events have also probably contributed substantially to the evolution of various MSV strains other than MSV‐A. Four strains (MSV‐F, MSV‐H, MSV‐J and MSV‐K) apparently arose via the exchange of large genomic regions (more than 1000 nucleotides) amongst two or more distinct MSV strains (Varsani et al., 2008a).
Other recombination events that are detectable in the genomes of grass‐adapted MSVs involve the introduction of short sequences (usually entirely within the SIR) from other streak virus species (Oluwafemi et al., 2008; Shepherd et al., 2008; Varsani et al., 2008b). These mostly perennial grass‐infecting viruses include Panicum streak virus, sugarcane streak virus, sugarcane streak Réunion virus, sugarcane streak Egypt virus, Eragrostis streak virus and Urochloa streak virus (Fig. 4). All of these viruses share largely overlapping geographical ranges, host species and leafhopper vectors with MSV, and it is perhaps surprising that, given the rampant inter‐species recombination observed in other geminiviruses (2007a, 2007b; Padidam et al., 1999), more inter‐species recombination is not found amongst these so‐called African streak viruses. As none of the African streak viruses other than MSV is considered to be a serious agricultural threat, and there is no evidence of genetic exchange between MSV‐A and these other viruses, it is currently doubtful whether they have any influence on MSV‐A epidemiology and evolution.
CONTROL
Suggested disease avoidance practices include barriers of bare ground between early‐ and late‐planted maize fields to reduce leafhopper movement and subsequent spread of MSV (Bosque‐Pérez, 2000), avoidance of maize plantings downwind from older cereal crops and the use of crop rotations that minimize invasion by viruliferous leafhoppers (Rose, 1978). The vector can be controlled by the application of systemic insecticides to the planting furrow during maize planting or, even more effectively, as seed treatments. However, expensive chemical seed treatments are generally not an option for poorer farmers—they provide only limited protection under severe pressure, and handling such treated seeds can be dangerous. The development and use of streak‐resistant cultivars is probably the most effective and economically viable means of preventing streak epidemics.
Thirty years after the first report of MSD, resistance in maize was discovered in the variety ‘Peruvian Yellow’ (Fielding, 1933), and several other maize genotypes have since been found to have varying degrees of resistance. Resistance usually manifests as reduced symptom severity combined with low virus titres, leading to low virus incidence in the field. Resistant varieties are therefore much poorer sources of inoculum during secondary disease spread (Rodier et al., 1995). Some resistant varieties produce good yields despite being infected (Bosque‐Pérez, 2000).
MSV‐resistant maize genotypes include Tzi4, a partially resistant inbred line from Nigeria originating from the TZ‐Y (Tropical Zea Yellow) population and developed at the International Institute of Tropical Agriculture (Kim et al., 1987), CML202, a sub‐tropical white inbred line from CIMMYT‐Zimbabwe (Welz et al., 1998), and D211 and CIRAD390, both from the Indian Ocean island of Réunion (Marchand et al., 1994; Pernet et al., 1999b; Rodier et al., 1995). The first resistant genotype to be mapped by molecular markers was Tzi4: a single, partially dominant gene was identified on the short arm of chromosome 1 and designated msv‐1 (Kyetere et al., 1995). As no other genomic region was associated with MSV resistance, the resistance was described as being monogenic. However, this resistant genotype would be better described as being MSV tolerant, with a rating in field tests of ‘3’ on a ‘1–5’ scale (‘1’ being completely immune; Kyetere et al., 1999).
Mapping of the other three resistant lines (CML202, D211 and CIRAD390) indicated that they all probably carry either msv‐1 or some allelic variation thereof. However, these three genotypes also seem to carry various additional small‐effect MSV resistance genes that are apparently not found in Tzi4. These genes may account for observable differences in the degrees of resistance between genotypes: CML202 was given a score in field tests of ‘2’ (Welz et al., 1998), whereas the two Réunion sources were rated as being completely immune to field infection by MSV (1999a, 1999b).
There are currently active MSV resistance breeding programmes in South Africa, Zimbabwe, Nigeria, Kenya, La Réunion, and elsewhere. However, despite the past success of these efforts, there are several difficulties in producing conventionally bred maize genotypes having high degrees of MSV resistance. The first is that all MSV resistance so far reported appears to rely quite heavily on the msv‐1 gene. If the enormous evolutionary potential of MSV (Isnard et al., 1998; van der Walt et al., 2008a) eventually yields virus genotypes capable of breaking this resistance, all current commercially available MSV‐resistant germplasm would be rendered largely ineffective. It is therefore imperative that alternative sources of MSV resistance be found.
A second problem is that natural resistance is not found in varieties with the best agronomic qualities. Current resistant sources are mostly tropical varieties with maturation and flowering characteristics that make them difficult to work with in the field. In addition, these resistant genotypes are not as high yielding as commercially favoured, albeit MSV‐sensitive, genotypes.
The third and probably largest obstacle to transferring very high degrees of MSV resistance to agronomically favourable genotypes is the coordinated transfer of multiple resistance genes scattered amongst different chromosomes. Transferring numerous resistance alleles, some of which may be recessive and/or have only small effects, during multiple breeding cycles is extremely difficult, particularly when these genes need to be separated from undesirable genetic backgrounds.
An alternative strategy to using conventional breeding would be to directly engineer MSV‐resistant maize genotypes using either natural maize MSV‐resistance genes or resistance genes from other sources. Great successes have been achieved in the genetic engineering of resistance to geminiviruses (see Shepherd et al., 2009 and Vanderschuren et al., 2007 for reviews) using the pathogen‐derived resistance concept (Sanford and Johnson, 1985), whereby pathogens themselves provide the genes for engineered resistance. This approach has already been successfully applied to the production of MSV‐resistant maize in South Africa (Shepherd et al., 2007).
Although direct genetic engineering provides better prospects than conventional breeding for the development of novel, varied and durable resistance strategies, this technology also has some significant drawbacks. These include: (i) maize genotypes with commercially appealing agronomic properties are not easy to transform or easily regenerated in tissue culture; (ii) it is technically difficult to genetically engineer maize because the commonly used transformation techniques (such as particle bombardment) generally introduce an unacceptably large number of gene copies at random locations in the maize genome, which can potentially disrupt important regulatory or coding sequences; (iii) a lengthy and costly risk assessment needs to be carried out to ensure that genetically engineered maize is both safe to eat and poses no harm to the environment and non‐target organisms; and (iv) public perception of genetically engineered foods can be unfavourable and they are banned in many African countries. This last obstacle may, however, become less formidable in the future. Since 2008, Burkino Faso, Egypt and Kenya have joined South Africa (which has permitted genetically modified crop farming since 1997) in allowing the production and use of genetically engineered crops. Meanwhile, several other African countries, such as Mali, Togo, Malawi, Zimbabwe and Cameroon, have the appropriate legal frameworks in place for the commercialization of genetically engineered crops, although they have yet to do so. For now, however, the use of conventionally bred resistant varieties, coupled with sound crop management practices, is still probably the best means of limiting the impact of MSD on maize yields.
ACKNOWLEDGEMENTS
DNS is supported by PANNAR (Pty) Ltd; DPM is supported by the Wellcome Trust.
REFERENCES
- Ammar, E.‐D. , Gargani, D. , Lett, J.M. and Peterschmitt, M. (2009) Large accumulations of maize streak virus in the filter chamber and midgut cells of the leafhopper vector Cicadulina mbila . Arch. Virol. 154, 255–262. [DOI] [PubMed] [Google Scholar]
- Van Antwerpen, T. , McFarlane, S.S. , Buchanan, G.F. , Shepherd, D.N. , Martin, D.P. , Rybicki, E.P. and Varsani, A. (2008) First report of maize streak virus infection of sugarcane in South Africa. Plant Dis. 92, 982. [DOI] [PubMed] [Google Scholar]
- Asanzi, M.C. , Bosque‐Pérez, N.A. , Nault, L.R. , Gordon, D.T. and Thottappilly, G. (1995) Biology of Cicadulina species (Homoptera: Cicadellidae) and transmission of maize streak virus. Afr. Entomol. 3, 173–179. [Google Scholar]
- Autrey, L.J.C. and Ricaud, C. (1983) The comparative epidemiology of two diseases of maize caused by leafhopper‐borne viruses in Mauritius In: Plant Virus Epidemiology (Plumb R.T. and Thresh J.M. eds), pp. 277–285. Oxford: Blackwell. [Google Scholar]
- Bock, K.R. (1982) Geminivirus diseases in tropical crops. Plant Dis. 66, 266–270. [Google Scholar]
- Bock, K.R. , Guthrie, E.J. and Woods, R.D. (1974) Purification of maize streak virus and its relationship to viruses associated with streak diseases of sugarcane and Panicum maximum . Ann. Appl. Biol. 77, 289–296. [Google Scholar]
- Bosque‐Pérez, N.A. (2000) Eight decades of maize streak virus research. Virus Res. 71, 107–121. [DOI] [PubMed] [Google Scholar]
- Boulton, M.I. (2002) Functions and interactions of mastrevirus gene products. Physiol. Mol. Plant Pathol. 60, 243–255. [Google Scholar]
- Boulton, M.I. and Markham, P.G. (1986) The use of squash‐blotting to detect plant pathogens in insect vectors In: Developments in Applied Biology 1: Developments and Applications in Virus Testing (Jones R.A.C. and Torrance L. eds), pp. 55–69. Wellesbourne, Warwicks., UK: Association of Applied Biologists. [Google Scholar]
- Boulton, M.I. , Pallaghy, C.K. , Chatani, M. , Macfarlane, S. and Davies, J.W. (1993) Replication of maize streak virus mutants in maize protoplasts—evidence for a movement protein. Virology, 192, 85–93. [DOI] [PubMed] [Google Scholar]
- Boulton, M.I. , Steinkellner, H. , Donson, J. , Markham, P.G. , King, D.I. and Davies, J.W. (1989) Mutational analysis of the virion‐sense genes of maize streak virus. J. Gen. Virol. 70, 2309–2323. [DOI] [PubMed] [Google Scholar]
- Briddon, R.W. , Lunness, P. , Chamberlin, L.C. and Markham, P.G. (1994) Analysis of the genetic variability of maize streak virus. Virus Genes, 9, 93–100. [DOI] [PubMed] [Google Scholar]
- Briddon, R.W. , Pinner, M.S. , Stanley, J. and Markham, P.G. (1990) Geminivirus coat protein gene replacement alters insect specificity. Virology, 177, 85–94. [DOI] [PubMed] [Google Scholar]
- Dabrowski, Z.T. (1987) Cicadulina ghaurii (Hem., Euscelidae): distribution, biology and maize streak virus (MSV) transmission. J. Appl. Entomol. 103, 489–496. [Google Scholar]
- Dabrowski, Z.T. , Nwilene, F. and Kumar, R. (1991) First regular observations on leafhoppers, Cicadulina spp., vectors of maize streak virus (MSV) in southeastern Nigeria. Insect Sci. Appl. 12, 249–261. [Google Scholar]
- Dabrowski, Z.T. , Wilson, M.R. and Nault, L.R. (1987). Comparative studies of Cicadulina leafhoppers in West Africa In: Proceedings of 2nd International Workshop on Leafhoppers and Planthoppers of Economic Importance, 28th July–1st August 1986 (Wilson M.R. ed.), pp. 35–39. Provo, UT: Brigham Young University. [Google Scholar]
- Damsteegt, V.D. (1983) Maize streak virus: I. Host range and vulnerability of maize germ plasm. Plant Dis. 67, 734–737. [Google Scholar]
- Duffy, S. and Holmes, E.C. (2008) Phylogenetic evidence for rapid rates of molecular evolution in the single‐stranded DNA begomovirus Tomato yellow leaf curl virus. J. Virol. 82, 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron, Y. , Kim, S.K. , Fajemisin, J.M. , Mareck, J.H. , Tang, C.Y. , Dabrowski, Z.T. , Rossel, H.W. , Thottappilly, G. and Buddenhagen, I.W. (1989) Breeding for resistance to maize streak virus: a multidisciplinary team approach. Plant Breed. 103, 1–36. [Google Scholar]
- Fajemisin, J.M. and Shoyinka, S.A. (1976) Maize streak and other maize virus disease in West Africa In: Proceedings of the International maize virus disease colloquium and workshop (Williams L.E., Gordon D.T., Nault L.R. eds), pp. 52–60. Wooster USA: Ohio Agricultural Research and Development Center. [Google Scholar]
- Fielding, W.L. (1933) Field experimental work on rotation crops. In: Empire Cotton Growing Association Progress Report 1931–1932, pp. 10–14.
- Francki, R.I.B. , Hatta, T. , Boccardo, G. and Randles, J.W. (1980) The composition of chloris striate mosaic virus, a geminivirus. Virology, 101, 233–241. [DOI] [PubMed] [Google Scholar]
- Fuller, C. (1901) Mealie variegation In: 1st Report of the Government Entomologist, Natal, 1899–1900. Pietermaritzburg, Natal, South Africa: P. Davis & Sons, Government Printers. [Google Scholar]
- Ge, L. , Zhang, J. , Zhou, X. and Li, H. (2007) Genetic structure and population variability of Tomato yellow leaf curl China virus. J. Virol. 81, 5902–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, R.M. (1977a) Infectious DNA from a whitefly transmitted virus of Phaseolus vulgaris . Nature, 266, 54. [Google Scholar]
- Goodman, R.M. (1977b) Single‐stranded DNA genome in a whitefly‐transmitted plant virus. Virology, 83, 171. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (1999) Geminivirus DNA replication. Cell Mol. Life Sci. 56, 313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, B.D. , Barker, H. , Bock, K.R. , Guthrie, E.J. , Meredith, G. and Atkinson, M. (1977) Plant viruses with circular single‐stranded DNA. Nature, 270, 760–762. [Google Scholar]
- Heyraud, F. , Matzeit, V. , Kammann, M. , Schaefer, S. , Schell, J. and Gronenborn, B. (1993a) Identification of the initiation sequence for viral‐strand DNA synthesis of wheat dwarf virus. EMBO J. 12, 4445–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyraud, F. , Matzeit, V. , Schaefer, S. , Schell, J. and Gronenborn, B. (1993b) The conserved nonanucleotide motif of the geminivirus stem‐loop sequence promotes replicational release of virus molecules from redundant copies. Biochimie, 75, 605–615. [DOI] [PubMed] [Google Scholar]
- Heyraud‐Nitschke, F. , Schumacher, S. , Laufs, J. , Schaefer, S. , Schell, J. and Gronenborn, B. (1995) Determination of the origin, cleavage and joining domain of geminivirus Rep proteins. Nucleic Acids Res. 23, 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnard, M. , Granter, M. , Frutos, R. , Reynaud, B. and Peterschmitt, M. (1998) Quasi‐species nature of the maize streak virus isolates obtained from a population used to assess maize cultivar response to infection. J. Gen. Virol. 79, 3091–3099. [DOI] [PubMed] [Google Scholar]
- Jeske, H. , Lutgemeier, M. and Preiss, W. (2001) DNA forms indicate rolling circle and recombination‐dependent replication of Abutilon mosaic virus. EMBO J. 20, 6158–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovel, J. , Preiss, W. and Jeske, H. (2007) Characterization of DNA intermediates of an arising geminivirus. Virus Res. 130, 63–70. [DOI] [PubMed] [Google Scholar]
- Kim, S.K. , Efron, Y. , Khadr, F.H. , Fajemisin, J.M. and Lee, M.H. (1987) Registration of 16 maize streak‐virus resistant tropical maize parental inbred lines. Crop Sci. 27, 824–825. [Google Scholar]
- Kloppers, F. (2005) Maize diseases: reflection on the 2004/2005 season. Available at: http://saspp.org/index2.php?option=com_content&do_pdf=1&id=2[accessed 6 July 2009.
- Konate, G. and Traore, O. (1992) Reservoir hosts of maize streak virus (MSV) in the Sudan–Sahel zone: identification and spatio‐temporal distribution. Phytoprotection, 73, 111–117. [Google Scholar]
- Kotlizky, G. , Boulton, M.I. , Pitaksutheepong, C. , Davies, J.W. and Epel, B.L. (2000) Intracellular and intercellular movement of maize streak geminivirus V1 and V2 proteins transiently expressed as green fluorescent protein fusions. Virology, 274, 32–38. [DOI] [PubMed] [Google Scholar]
- Kyetere, D. , Ming, R. , McMullen, M. , Pratt, R. , Brewbaker, J. , Musket, T. , Pixley, K. and Moon, H. (1995) Monogenic tolerance to maize streak virus maps to the short arm of chromosome 1. Maize Genet. Coop. News Lett. 69, 136–137. [Google Scholar]
- Kyetere, D.T. , Ming, R. , McMullen, M.D. , Pratt, R.C. , Brewbaker, J. and Musket, T. (1999) Genetic analysis of tolerance to maize streak virus in maize. Genome 42, 20–26. [Google Scholar]
- Laufs, J. , Schumacher, S. , Geisler, N. , Jupin, I. and Gronenborn, B. (1995a) Identification of the nicking tyrosine of geminivirus Rep protein. FEBS Lett. 377, 258–262. [DOI] [PubMed] [Google Scholar]
- Laufs, J. , Traut, W. , Heyraud, F. , Matzeit, V. , Rogers, S.G. , Schell, J. and Gronenborn, B. (1995b) In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA, 92, 3879–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz, S.G. , Pinder, A.J. , Damsteegt, V.D. and Rogers, S.G. (1989) Maize streak virus genes essential for systemic spread and symptom development. EMBO J. 8, 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre, P. , Lett, J.M. , Reynaud, B. and Martin, D.P. (2007a) Avoidance of protein fold disruption in natural virus recombinants. PLoS Pathog. 3, e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre, P. , Martin, D.P. , Hoareau, M. , Naze, F. , Delatte, H. , Thierry, M. , Varsani, A. Becker, N. , Reynaud, B. and Lett, J.M. (2007b) Begomovirus ‘melting pot’ in the south‐west Indian Ocean islands: molecular diversity and evolution through recombination. J. Gen. Virol. 88, 3458–3468. [DOI] [PubMed] [Google Scholar]
- Lett, J.‐M. , Granier, M. , Hippolyte, I. , Grondin, M. , Royer, M. , Blanc, S. , Reynaud, B. and Peterschmitt, M. (2002) Spatial and temporal distribution of geminiviruses in leafhoppers of the genus Cicadulina monitored by conventional and quantitative polymerase chain reaction. Phytopathology, 92, 65–74. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Boulton, M.I. and Davies, J.W. (1997) Maize streak virus coat protein binds single‐ and double‐stranded DNA in vitro. J. Gen. Virol. 78, 1265–1270. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Boulton, M.I. , Oparka, K.J. and Davies, J.W. (2001a) Interaction of the movement and coat proteins of Maize streak virus: implications for the transport of viral DNA. J. Gen. Virol. 82, 35–44. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Boulton, M.I. , Thomas, C.L. , Prior, D.A. , Oparka, K.J. and Davies, J.W. (1999) Maize streak virus coat protein is karyophyllic and facilitates nuclear transport of viral DNA. Mol. Plant–Microbe Interact. 12, 894–900. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Lucy, A.P. , Davies, J.W. and Boulton, M.I. (2001b) A single amino acid change in the coat protein of Maize streak virus abolishes systemic infection, but not interaction with viral DNA or movement protein. Mol. Plant Pathol. 2, 223–228. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Davies, J.W. and Stanley, J. (1998) Mutational analysis of bean yellow dwarf virus, a geminivirus of the genus Mastrevirus that is adapted to dicotyledonous plants. J. Gen. Virol. 79, 2265–2274. [DOI] [PubMed] [Google Scholar]
- Lucy, A.P. , Boulton, M.I. , Davies, J.W. and Maule, A.J. (1996) Tissue specificity of Zea mays infection by maize streak virus. Mol. Plant–Microbe Interact. 9, 22–31. [Google Scholar]
- Magenya, O.E.V. , Mueke, J. and Omwega, C. (2008) Significance and transmission of maize streak virus disease in Africa and options for management: a review. Afr. J. Biotechnol. 7, 4897–4910. [Google Scholar]
- Marchand, J.‐L. , Peterschmitt, M. and Reynaud, B. (1994) Les viroses de la striure, du stripe et de la mosaïque sur le maïs en région tropicale. Afrique et îles de l'Océan Indien. Agric. Dev. 4, 1–16. [Google Scholar]
- Markham, P.G. , Pinner, M.S. and Boulton, M.I. (1984) Dalbulus maidis and Cicadulina species as vectors of diseases in maize. Maize Virus Dis. Newsl. 1, 33–34. [Google Scholar]
- Martin, D.P. and Rybicki, E.P. (2002) Biological and genomic sequence characterization of Maize streak virus isolates from wheat. Phytopathology, 92, 81–86. [DOI] [PubMed] [Google Scholar]
- Martin, D.P. and Shepherd, D.N. (2009) The epidemiology, economic impact and control of maize streak disease. Food Secur. In Press.
- Martin, D.P. , Van Der Walt, E. , Posada, D. and Rybicki, E.P. (2005) The evolutionary value of recombination is constrained by genome modularity. PLoS Genet. 1, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.P. , Willment, J.A. , Billharz, R. , Velders, R. , Odhiambo, B. , Njuguna, J. , James, D. and Rybicki, E.P. (2001) Sequence diversity and virulence in Zea mays of Maize streak virus isolates. Virology, 288, 247–255. [DOI] [PubMed] [Google Scholar]
- Martin, D.P. , Willment, J.A. and Rybicki, E.P. (1999) Evaluation of maize streak virus pathogenicity in differentially resistant Zea mays genotypes. Phytopathology, 89, 695–700. [DOI] [PubMed] [Google Scholar]
- McClean, A.P.D. (1947) Some forms of streak virus occurring in maize, sugarcane and wild grasses. South African Department of Agriculture Bulletin, 265, 33pp. [Google Scholar]
- Mesfin, T. , Den Hollander, J. and Markham, P.G. (1995) Feeding activities of Cicadulina mbila (Hemiptera: Cicadellidae) on different host‐plants. Bull. Entomol. Res. 85, 387–396. [Google Scholar]
- Morris‐Krsinich, B.A. , Mullineaux, P.M. , Donson, J. , Boulton, M.I. , Markham, P.G. , Short, M.N. and Davies, J.W. (1985) Bidirectional transcription of maize streak virus DNA and identification of the coat protein gene. Nucleic Acids Res. 13, 7237–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njuguna, J.A.M. , Kendera, J.G. , Muriithi, L.M.M. , Songa, S. and Othiambo, R.B. (1990) Overview of maize diseases in Kenya In: Maize Review Workshop in Kenya, Kakamega, Kenya, pp 45–51. [Google Scholar]
- Oluwafemi, S. , Jackai, L.E.N. and Alegbejo, M.D. (2007) Comparison of transmission abilities of four Cicadulina species vectors of maize streak virus from Nigeria. Entomol. Exp. Appl. 124, 235–239. [Google Scholar]
- Oluwafemi, S. , Varsani, A. , Monjane, A.L. , Shepherd, D.N. , Owor, B. , Rybicki, E.P. and Martin, D.P. (2008) A new African streak virus species from Nigeria. Arch. Virol. 153, 1407–1410. [DOI] [PubMed] [Google Scholar]
- Owor, B. , Martin, D.P. , Shepherd, D.N. , Edema, R. , Monjane, A.L. , Rybicki, E.P. , Thomson, J.A. and Varsani, A. (2007) Genetic analysis of maize streak virus isolates from Uganda reveals widespread distribution of a recombinant variant. J. Gen. Virol. 88, 3154–3165. [DOI] [PubMed] [Google Scholar]
- Padidam, M. , Sawyer, S. and Fauquet, C.M. (1999) Possible emergence of new geminiviruses by frequent recombination. Virology, 265, 218–225. [DOI] [PubMed] [Google Scholar]
- Pernet, A. , Hoisington, D.A. , Dintinger, J. , Jewell, D.C. , Jiang, C. , Khairallah, M.M. , Letourmy, P. , Marchand, J.‐L. , Glaszmann, J.‐C. and González de Leon, D. (1999a) Genetic mapping of maize streak virus resistance from the Mascarene source. II. Resistance in line CIRAD390 and stability across germplasm. Theor. Appl. Genet. 99, 540–553. [DOI] [PubMed] [Google Scholar]
- Pernet, A. , Hoisington, D.A. , Franco, J. , Isnard, M. , Jewell, D.C. , Jiang, C. , Marchand, J.‐L. , Reynaud, B. , Glaszmann, J.‐C. and González de Leon, D. (1999b) Genetic mapping of maize streak virus resistance from the Mascarene source. I. Resistance in line D211 and stability against different virus clones. Theor. Appl. Genet. 99, 524–539. [DOI] [PubMed] [Google Scholar]
- Peterschmitt, M. , Quiot, J.B. and Reynaud, B. (1992) Detection of maize streak virus antigens over time in different parts of maize plants of a sensitive and a so‐called tolerant cultivar by ELISA. Ann. Appl. Biol. 121, 641–653. [Google Scholar]
- Pinner, M.S. , Medina, V. , Plaskitt, K.A. and Markham, P.G. (1993) Viral inclusions in monocotyledons infected by maize streak and related geminiviruses. Plant Pathol. 42, 75–87. [Google Scholar]
- Preiss, W. and Jeske, H. (2003) Multitasking in replication is common among geminiviruses. J. Virol. 77, 2972–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud, B. and Peterschmitt, M. (1992) A study of the mode of transmission of MSV by Cicadulina mbila using an enzyme‐linked immunosorbent assay. Ann. Appl. Biol. 121, 85–94. [Google Scholar]
- Rodier, A. , Assié, J. , Marchand, J.‐L. and Hervé, Y. (1995) Breeding maize lines for complete and partial resistance to maize streak virus (MSV). Euphytica, 81, 57–70. [Google Scholar]
- Rose, D.J.W. (1978) Epidemiology of maize streak disease. Ann. Rev. Entomol. 23, 250–282. [Google Scholar]
- Rossel, H.W. and Thottappilly, G. (1985) In: Virus Diseases of Important Food Crops in Tropical Africa, pp. 61 Ibadan: International Institute of Tropical Agriculture (IITA). [Google Scholar]
- Sanford, J.C. and Johnson, S.A. (1985) The concept of parasite‐derived resistance: deriving resistance genes from the parasite's own genome. J. Theor. Biol. 115, 395–405. [Google Scholar]
- Saunders, K. , Lucy, A. and Stanley, J. (1991) DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 19, 2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, K. , Lucy, A. and Stanley, J. (1992) RNA‐primed complementary‐sense DNA synthesis of the geminivirus African cassava mosaic virus. Nucleic Acids Res. 20, 6311–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk, H.J. , Matzeit, V. , Schiller, B. , Schell, J. and Gronenborn, B. (1989) Wheat dwarf virus, a geminivirus of graminaceous plants needs splicing for replication. EMBO J. 8, 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnippenkoetter, W.H. , Martin, D.P. , Hughes, F.L. , Fyvie, M. , Willment, J.A. , James, D. , Von Wechmar, M.B. and Rybicki, E.P. (2001a) The relative infectivities and genomic characterisation of three distinct mastreviruses from South Africa. Arch. Virol. 146, 1075–1088. [DOI] [PubMed] [Google Scholar]
- Schnippenkoetter, W.H. , Martin, D.P. , Willment, J.A. and Rybicki, E.P. (2001b) Forced recombination between distinct strains of Maize streak virus. J. Gen. Virol. 82, 3081–3090. [DOI] [PubMed] [Google Scholar]
- Shepherd, D.N. , Mangwende, T. , Martin, D.P. , Bezuidenhout, M. , Kloppers, F.J. , Carolissen, C.H. , Monjane, A.L. , Rybicki, E.P. and Thomson, J.A. (2007) Maize streak virus‐resistant transgenic maize: a first for Africa. Plant Biotechnol. J. 5, 759–767. [DOI] [PubMed] [Google Scholar]
- Shepherd, D.N. , Martin, D.P. , McGivern, D.R. , Boulton, M.I. , Thomson, J.A. and Rybicki, E.P. (2005) A three‐nucleotide mutation altering the Maize streak virus Rep pRBR‐interaction motif reduces symptom severity in maize and partially reverts at high frequency without restoring pRBR‐Rep binding. J. Gen. Virol. 86, 803–813. [DOI] [PubMed] [Google Scholar]
- Shepherd, D.N. , Martin, D.P. and Thomson, J.A. (2009) Strategies for developing crops resistant to geminiviruses. Plant Sci. 176, 1–11. [Google Scholar]
- Shepherd, D.N. , Varsani, A. , Windram, O. , Lefeuvre, P. , Monjane, A.L. , Owor, B. and Martin, D.P. (2008) Novel sugarcane streak virus and sugarcane streak Reunion virus‐like mastrevirus isolates from Southern Africa and La Reunion. Arch. Virol. 153, 605–609. [DOI] [PubMed] [Google Scholar]
- Soto, P.E. , Buddenhagen, I.W. and Asnani, V.L. (1982) Development of streak virus‐resistant maize populations through improved challenge and selection methods. Ann. Appl. Biol. 100, 539–546. [Google Scholar]
- Stanley, J. (1995) Analysis of African cassava mosaic virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology, 206, 707–712. [DOI] [PubMed] [Google Scholar]
- Stanley, J. , Boulton, M.I. and Davies, J.W. (1999) Geminiviridae in: Embryonic encyclopedia of Life Sciences. Nature Publishing group, London. http://www.els.net/elsonline/html/ [Google Scholar]
- Stenger, D.C. , Revington, G.N. , Stevenson, M.C. and Bisaro, D.M. (1991) Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling‐circle replication of a plant viral DNA. Proc. Natl. Acad. Sci. USA, 88, 8029–8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, H.H. (1924) The transmission of a new plant virus disease by insects. Nature, 144, 245. [Google Scholar]
- Storey, H.H. (1925) The transmission of streak disease of maize by the leafhopper Balclutha mbila Naudé. Ann. Appl. Biol. 12, 422–439. [Google Scholar]
- Storey, H.H. (1928) Transmission of maize streak disease. Ann. Appl. Biol. 15, 1–25. [Google Scholar]
- Storey, H.H. (1931) The inheritance by a leafhopper of the ability to transmit a plant virus. Nature, 127, 928. [Google Scholar]
- Storey, H.H. (1932) The inheritance by an insect vector of the ability to transmit a plant virus. Proc. R. Soc. Lond., B, 112, 46–60. [Google Scholar]
- Storey, H.H. (1933) Investigations of the mechanism of the transmission of plant viruses by insect vectors. I. Proc. R. Soc. Lond., B, 113, 463–485. [Google Scholar]
- Storey, H.H. (1938) Investigation of the mechanism of the transmission of plant viruses by insect vectors. II. The part played by puncture in transmission. Proc. R. Soc. Lond., B, 125, 455–477. [Google Scholar]
- Storey, H.H. (1939) Investigation of the mechanism of the transmission of plant viruses by insect vectors. III. The insect's saliva. Proc. R. Soc. Lond., B, 127, 526–543. [Google Scholar]
- Storey, H.H. and McClean, A.P.D. (1930) The transmission of streak disease between maize, sugarcane and wild grasses. Ann. Appl. Biol. 17, 691–719. [Google Scholar]
- Vanderschuren, H. , Stupak, M. , Futterer, J. , Gruissem, W. and Zhang, P. (2007) Engineering resistance to geminiviruses—review and perspectives. Plant Biotechnol. J. 5, 207–220. [DOI] [PubMed] [Google Scholar]
- Varsani, A. , Oluwafemi, S. , Windram, O.P. , Shepherd, D.N. , Monjane, A.L. , Owor, B.E. , Rybicki, E.P. , Lefeuvre, P. and Martin, D.P. (2008b) Panicum streak virus diversity is similar to that observed for Maize streak virus. Arch. Virol. 153, 601–604. [DOI] [PubMed] [Google Scholar]
- Varsani, A. , Shepherd, D.N. , Monjane, A.L. , Owor, B.E. , Erdmann, J.B. , Rybicki, E.P. , Peterschmitt, M. , Briddon, R.W. , Markham, P.G. , Oluwafemi, S. , Windram, O.P. , Lefeuvre, P. , Lett, J.‐M. and Martin, D.P. (2008a) Recombination, decreased host specificity and increased mobility may have driven the emergence of Maize streak virus as an agricultural pathogen. J. Gen. Virol. 89, 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Walt, E. , Martin, D.P. , Varsani, A. , Polston, J.E. and Rybicki, E.P. (2008a) Experimental observations of rapid Maize streak virus evolution reveal a strand‐specific nucleotide substitution bias. Virol. J. 5, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Walt, E. , Palmer, K. , Martin, D.P. and Rybicki, E.P. (2008b) Viable chimaeric viruses confirm the biological importance of sequence specific maize streak virus movement protein and coat protein interactions. Virol. J. 5, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Walt, E. , Rybicki, E.P. , Varsani, A. , Polston, J.E. , Billharz, R. , Donaldson, L. , Monjane, A.L. and Martin, D.P. (2009) Rapid host adaptation by extensive recombination. J. Gen. Virol. 90, 734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambugu, F. and Wafula, J. (2000) Advances in maize streak virus disease research in Eastern and Southern Africa In: Workshop Report, 15–17 September, 1999, KARI and ISAAA Africa Center, ISAAA Brief No 16. Ithaca, NY: International Service for the Acquisition of Agri‐Biotech Applications. [Google Scholar]
- Welz, H.G. , Schechert, A. , Pernet, A. , Pixley, K.V. and Geiger, H.H. (1998) A gene for resistance to the maize streak virus in the African CIMMYT maize inbred line CML 202. Mol. Breed. 4, 147–154. [Google Scholar]
- Willment, J.A. , Martin, D.P. , Palmer, K.E. , Schnippenkoetter, W.H. , Shepherd, D.N. and Rybicki, E.P. (2007) Identification of long intergenic region sequences involved in Maize streak virus replication. J. Gen. Virol. 88, 1831–1841. [DOI] [PubMed] [Google Scholar]
- Willment, J.A. , Martin, D.P. and Rybicki, E.P. (2001) Analysis of the diversity of African streak mastreviruses using PCR‐generated RFLPs and partial sequence data. J. Virol. Methods, 93, 75–87. [DOI] [PubMed] [Google Scholar]
- Willment, J.A. , Martin, D.P. , Van der Walt, E. and Rybicki, E.P. (2002) Biological and genomic sequence characterization of Maize streak virus isolates from wheat. Phytopathology, 92, 81–86. [DOI] [PubMed] [Google Scholar]


