SUMMARY
Three different flavonoids—naringenin, quercetin and kaempferol—accumulate in root galls of Arabidopsis thaliana after infection with the obligate biotrophic pathogen Plasmodiophora brassicae. In addition, high‐performance liquid chromatography and thin layer chromatography analysis indicated that these flavonoids and their glycosides were induced in galls rather than in healthy roots. The transcripts of selected genes involved in the biosynthesis of flavonoids were up‐regulated during the time course of the disease. Some, such as chalcone synthase and chalcone isomerase, were up‐regulated at both times investigated in this study, whereas up‐regulation was observed only at later times for others, such as a flavonol synthase‐like gene. Plants with mutations in different flavonoid biosynthesis genes were slightly more tolerant to clubroot at low infection pressure. However, flavonoid treatment of either leaves or roots did not reduce gall development. The possibility that flavonoids might influence auxin levels by regulating auxin transport or auxin degradation in roots was investigated by measuring auxin levels and response in roots of flavonoid‐deficient mutants and the wild‐type after inoculation with P. brassicae, as well as the antioxidative potential of flavonoids in the peroxidase‐catalysed degradation of indole‐3‐acetic acid. In addition, the auxin transport rate from the shoots to the roots was measured in infected wild‐type or flavonoid mutant plants compared with controls. In conclusion, our results indicate a role of flavonoids in the modulation of auxin efflux in root galls.
INTRODUCTION
The infection of cruciferous hosts with the obligate biotroph Plasmodiophora brassicae Wor. leads to cell elongation and cell division in infected roots and hypocotyls, resulting in typical hypertrophied roots (clubroot). The disease is among the most damaging within this plant family. Plasmodiophora brassicae infects a range of economically important crop plants within the Brassicaceae, e.g. Brassica napus, Brassica oleracea L., Brassica pekinensis and Brassica rapa, and also forms galls on Arabidopsis thaliana (Mithen and Magrath, 1992; Siemens et al., 2002). The life cycle of P. brassicae consists of the primary phase, which is restricted to root hairs of the host, and the secondary phase, which occurs in the cortex and stele of roots and hypocotyls, and leads to abnormal development of the roots (Ingram and Tommerup, 1972). During the secondary phase, the infected root acts as a strong metabolic sink, redirecting assimilates and other nutrients to the pathogen (Evans and Scholes, 1995).
Plant growth hormones are involved in symptom development (reviewed in Ludwig‐Müller and Schuller, 2008). Although there is consensus that indole‐3‐acetic acid (IAA) is increased during gall development, the specific times and types of auxin (i.e. free IAA or IAA conjugates) differ from study to study, perhaps because different experimental conditions and host plants were chosen (reviewed in Ludwig‐Müller et al., 2009). Auxin‐responsive promoters or elements, coupled to a suitable reporter gene, can be used to characterize the auxin response within a given tissue (Gee et al., 1991; Hagen et al., 1991). Siemens et al. (2006) used the auxin‐responsive DR5 promoter to show an increased auxin response during the development of clubroot in Arabidopsis. Higher IAA levels might be caused by an increase in biosynthesis via the indole glucosinolate and indole‐3‐acetonitrile (IAN) pathway (reviewed in Ludwig‐Müller et al., 2009). IAN is converted by nitrilase to IAA in clubbed roots (Rausch et al., 1981) and nitrilase mRNA levels increase in Arabidopsis (Grsic‐Rausch et al., 2000; Neuhaus et al., 2000) and in turnip (Brassica rapa) (Ishikawa et al., 2007). Immunolocalization has shown that the nitrilase protein is predominantly induced in cells harbouring sporulating plasmodia of the pathogen (Grsic‐Rausch et al., 2000). Furthermore, mutants in the nit1 gene are more tolerant to clubroot (Grsic‐Rausch et al., 2000), making the nitrilase pathway a good candidate for the generation of IAA in infected roots.
In addition to synthesis, IAA levels are controlled by conjugation, degradation and transport (Ljung et al., 2002). IAA is transported by a polar transport mechanism (Geisler and Murphy, 2006; Muday and DeLong, 2001; Muday and Murphy, 2002). The polarity of auxin transport is controlled by the localization of auxin transport proteins, where efflux and influx carriers show asymmetric distributions (reviewed in Kramer and Bennett, 2006). Mutant analysis has indicated that flavonoids are involved in auxin transport. In the transparent testa 4 (tt4) mutant, which is flavonoid‐deficient, IAA transport rates are increased (Brown et al., 2001; Buer and Muday, 2004) and the localization of IAA efflux carriers is altered (Peer et al., 2004). The correct localization of IAA efflux carriers and, subsequently, the correct IAA distribution can be successfully restored by treatment with exogenous flavonoids (Buer and Muday, 2004; Murphy et al., 2000; Peer et al., 2004). Flavonoids have some structural similarity to the synthetic auxin transport inhibitor N‐(1‐naphthyl)phthalamic acid (NPA), which is believed to bind a regulatory protein of the auxin efflux machinery (Muday and DeLong, 2001). Flavonoid effects on auxin transport were first suggested by Jacobs and Rubery (1988) in experiments in which they displaced NPA specifically from its binding protein using different flavonoids, including quercetin, apigenin and kaempferol. In addition, naringenin was effective as an auxin transport inhibitor as shown in later work (Peer et al., 2004). Evidence is emerging that flavonoids may also be involved in the modulation of auxin efflux during plant–microbe interactions. For example, Schwalm et al. (2003) examined the pattern of auxin and flavonoid distribution in Agrobacterium tumefaciens tumours of Trifolium repens, and found that flavonoid formation is probably the endogenous regulator of the basipetal auxin flux and thus contributes to auxin levels in tumours. In Medicago truncatula, auxin efflux carriers of the PIN family were necessary for root nodule formation (Huo et al., 2006). Flavonoid‐deficient roots were unable to initiate root nodules, even though normal root hair curling occurred after inoculation with rhizobia (Wasson et al., 2006). Nodule formation was restored by the addition of naringenin. The flavonoid‐deficient roots had an increased auxin transport rate (Wasson et al., 2006). The biosynthesis of kaempferol was induced in rhizobia‐inoculated M. truncatula roots, and exogenously applied kaempferol was able to restore nodulation in a flavonoid‐deficient plant (Zhang et al., 2009).
Another possibility could be that flavonoids are regulators of auxin breakdown by peroxidase, as shown in cells undergoing nodule organogenesis in white clover (Mathesius, 2001), because several peroxidases can act as nonspecific IAA oxidases. Inhibition of this activity by flavonoids could result in decreased IAA degradation. Interestingly, IAA oxidation has been reported to result in increased radical production (Kim et al., 2006), which could be detoxified by flavonoids, and IAA is typically increased in root galls (Ludwig‐Müller et al., 2009).
In addition to being modulators of auxin transport or degradation, flavonoids are a group of secondary products with a vast array of biological functions, including stress protection, such as UV radiation (Winkel‐Shirley, 2002) and plant defence (Dixon, 2001; Ryder et al., 1987). Microarray data collected by Siemens et al. (2006) indicated an increase in the transcripts involved in flavonoid biosynthesis in Arabidopsis clubroots. In the present study, we have investigated the contribution of flavonoids to clubroot development by: (i) investigating flavonoid accumulation in root galls of Arabidopsis by a fluorescent staining method, and by high‐performance liquid chromatography (HPLC) and thin layer chromatography (TLC) analysis; (ii) examining the expression pattern of selected transcripts involved in flavonoid biosynthesis during disease development; and (iii) analysing several mutants altered in flavonoid levels for changes in clubroot formation (see Fig. 1). Furthermore, several possible roles of flavonoids during gall formation, such as defence compounds, antioxidants or auxin transport modulators, are addressed. Our results indicate that flavonoids might be involved in auxin accumulation and thus influence the outcome of gall size, which is an important factor for the reproduction of the pathogen.
Figure 1.
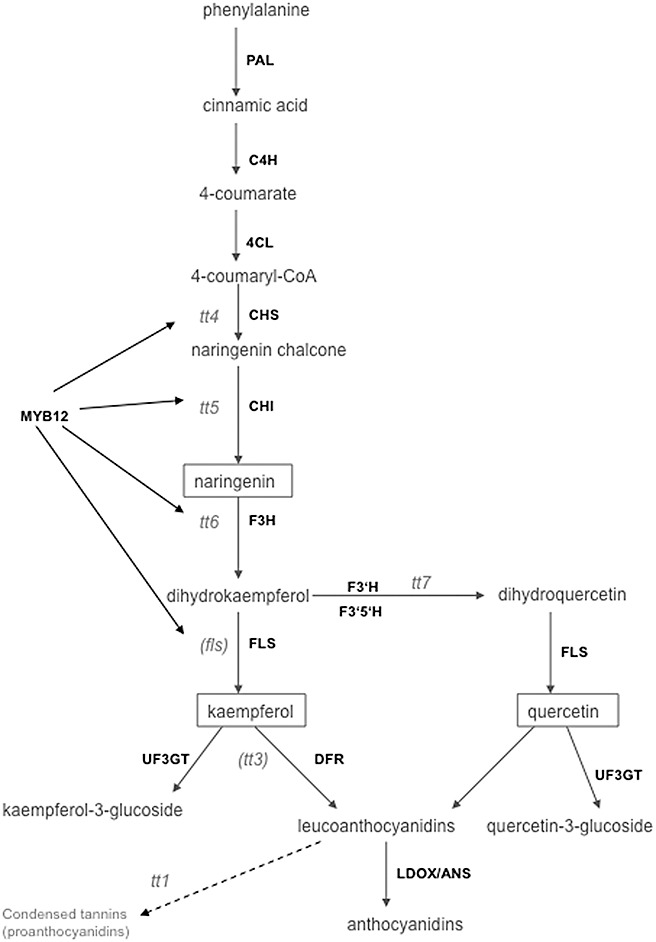
Biosynthesis of flavonoids and anthocyanins in Arabidopsis thaliana. Mutants for the respective genes used in this study (with the exception of fls and tt3) are given. Transgenic plants overexpressing MYB12 (myb12‐OX) and a knockout in the same gene (myb12‐KO) were also studied. C4H, cinnamic acid 4‐hydroxylase; CHI, chalcone isomerase; CHS, chalcone synthase; 4CL, 4‐coumarate‐CoA ligase; DFR, dihydroflavonol reductase; F3H, flavanone‐3‐hydroxylase; F3′H, flavonoid‐3′‐hydroxylase; F3′5′H, flavonoid‐3′,5′‐hydroxylase; FLS, flavonol synthase; LDOX/ANS, anthocyanin synthase; PAL, phenylalanine ammonium lyase; UF3GT, flavonol‐3‐O‐glucosyltransferase.
RESULTS
Flavonoid synthesis genes are differentially regulated during clubroot disease
Transcriptome analysis, which was carried out to check P. brassicae‐infected root samples of Arabidopsis at two time points after inoculation using the Affymetrix ATH1 microarray (Siemens et al., 2006), indicated the differential regulation of a variety of transcripts involved in flavonoid biosynthesis (compiled in Table S1, see Supporting Information). Chalcone‐related genes (for the biosynthesis pathway, see Fig. 1) were up‐regulated at two time points. Most of the dihydroflavonol‐ and anthocyanin‐related genes were also up‐regulated, which corroborates the observation that anthocyanins show higher accumulation in hypocotyls and rosette leaves of infected plants than in controls (Fig. S1, see Supporting Information). Some transcription factors involved in the control of flavonoid biosynthesis—the zinc finger protein TT1, the bHLH family protein TT8 and the MYB family transcription factors MYB12 and MYB75—were also up‐regulated, especially at the later time (Table S1).
Confirmation of the expression patterns for selected genes was performed with semiquantitative reverse transcription‐polymerase chain reaction (RT‐PCR) analysis (Fig. 2). From each gene family, a representative member which showed up‐regulation in the microarray experiment (Table S1) was analysed at two different time points after inoculation [10 and 24 days post‐inoculation (dpi)]; for the UDP‐glucosyltransferase family (AtUF3GT), two genes (‘AtUF3GTa’ and ‘AtUF3GTb’ in Fig. 2) were chosen. Chalcone synthase (AtCHS), chalcone isomerase (AtCHI), dihydroflavonol reductase (AtDFR), anthocyanin synthase (AtANS) and flavonol‐3‐O‐glucosyltransferase AtUF3GTa[for AGI numbers, see Table S4 (Supporting Information)] were all up‐regulated at both times, confirming the microarray results. Flavanone‐3‐hydroxylase (AtF3H), flavonol synthase‐like (AtFLS‐like) and AtUF3GTb were induced at the second, but not at the first, time point investigated after inoculation. Development of the pathogen was demonstrated by the amplification of P. brassicae Actin 2 (Siemens et al., 2009). For the genes AtCHS, AtCHI, AtF3H, AtFLS1, AtDFR and AtANS, a role in flavonoid biosynthesis has been demonstrated previously (Owens et al., 2008; and references cited therein). FLS1 was also up‐regulated at time point 2 (TP2) in the microarray (Table S1), but the general expression level was so low that a different gene (AtFLS‐like, At3g19010) was chosen for RT‐PCR expression analysis. More recently, Preußet al. (2009) observed that FLS3 also showed the respective enzymatic activity. However, FLS3 was actually downregulated at TP2 according to the microarray data (Table S1). Of the two UDP‐glucosyltransferases (Fig. 2), one (AtUT3GTa) was shown to be active in the glycosylation of quercetin (UGT73C6; Lim et al., 2004); the second (AtUT3GTb) was annotated as ‘UDP‐glucose:flavonoid 3‐O‐glucosyltransferase’ (Table S1).
Figure 2.
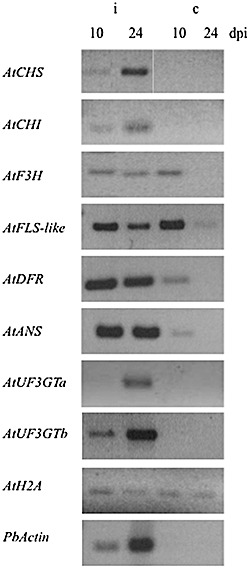
Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the expression of selected flavonoid biosynthesis genes during clubroot infection after different time points (dpi, days post‐inoculation; controls were of the same age). ANS, anthocyanin synthase (At4g22870); CHI, chalcone isomerase (tt5, At3g55120); CHS, chalcone synthase (tt4, At5g13930); DFR, dihydroflavonol reductase; F3H, flavanone‐3‐hydroxylase (tt6, At3g51240); FLS‐like, flavonol synthase‐like (At3g19010); UF3GT, flavonol‐3‐O‐glucosyltransferase (a, At2g36790; b, At5g54060). The different samples were standardized on host AtHistone2A (AtH2A, At1g52740) expression, and infection was shown using the expression of PbActin (AY452179).
Accumulation of flavonoids in Arabidopsis roots infected with P. brassicae
Flavonoids accumulated during gall formation in Arabidopsis roots of ecotype Columbia (Col) (Fig. 3) and Landsberg erecta (Ler) (data not shown). In particular, naringenin and quercetin were present in most root galls, as indicated by the cyan/yellow–orange fluorescence, respectively, after staining with diphenylboric acid 2‐amino‐ethylester (DPBA) (Fig. 3D–H,N), whereas the green fluorescence indicating kaempferol (Fig. 3N) was found only in a few root galls (Fig. 3I). However, the colours cannot be attributed to individual compounds when different flavonoids overlap in their patterns. A cross‐section showed the accumulation specifically in the hypertrophied part and not in the middle of the roots (Fig. 3J), but other uncharacterized fluorescing substances were also present, as indicated by the red colour. Uninfected roots showed only the bright blue fluorescence of sinapate derivatives (Fig. 3B,C). The unstained root samples were characterized by a darker blue colour (Fig. 3A). The CHS‐ and CHI‐deficient mutants tt4 (Fig. 3) and tt5 (data not shown) showed only accumulation of sinapate derivatives, but not flavonoids, in both infected and control roots (Fig. 3K,L). A green colour was observed in galls of the kaempferol‐accumulating mutant tt7 (Peer et al. 2001; for a further description of the flavonoid levels of different mutants, see below) (Fig. 3M).
Figure 3.
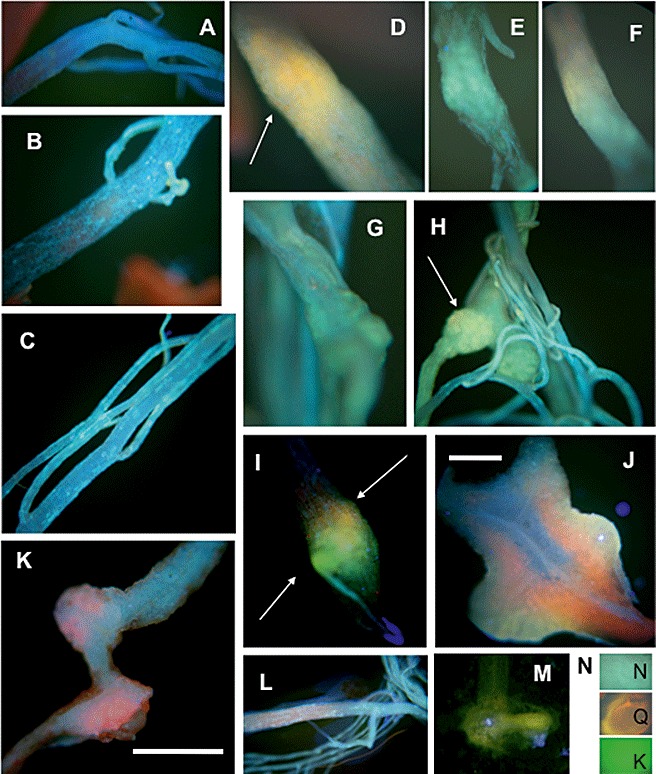
Accumulation of flavonoids in Arabidopsis control roots and root galls after infection with Plasmodiophora brassicae as shown by staining with 0.5% diphenylboric acid 2‐amino‐ethylester (DPBA) for 2 h. Fluorescence was visualized by excitation with a filter (G365/FT395/LP420 nm). The infected samples were taken at 21 days post‐inoculation (dpi) and control samples were taken at the same age. (A) Control roots of the wild‐type stained with H2O only (the photograph looked the same for infected roots). (B, C) Control roots of the wild‐type showing a blue stain caused by sinapate derivatives. (D–I) Infected roots showing the accumulation of kaempferol (green), naringenin (cyan) and quercetin (orange), taking the colours as preliminary indicators of the respective flavonoid; arrows point to samples for each flavonoid. (J) Cross‐sections showing that flavonoid staining occurs throughout the whole gall and not within single cells. (K) Infected root of the tt4 mutant. (L) Control root of the tt4 mutant. (M) Young gall of the tt7 mutant showing the green coloration typical for kaempferol. (N) Samples of flavonoid standards (K, kaempferol; N, naringenin; Q, quercetin) when incubated directly with the staining solution on a microscopic slide for better comparison. Bar, 0.5 cm (except in J, 0.1 cm).
The compounds present in root galls were further analysed for the aglycones and glycosylated flavonoids present in Arabidopsis by HPLC and TLC (Fig. 4; Fig. S4, see Supporting Information). Naringenin was increased by about seven fold in root galls at 23 dpi as shown by HPLC analysis, quercetin approximately two fold and kaempferol did not show any differences (Fig. 4). Although the increase in naringenin did not change over the time of infection, quercetin increased even more in galls at a later infection time (28 dpi, data not shown). Sample chromatograms of the separation of flavanone and flavonol aglycones are shown in Fig. S2 (see Supporting Information) and sample spectra of standards and peaks from chromatograms in Fig. S3 (see Supporting Information). Acid hydrolysis of glycosylated flavonoids increased the levels of naringenin and, in particular, those of quercetin and kaempferol, and this was again more visible in galls, showing that glycosides were also induced in clubroots. This is in accordance with the induction of UGT transcripts in root galls compared with controls (Fig. 2, Table S1). Further analysis of individual compounds will require liquid chromatography‐mass spectrometry (LC‐MS) analysis.
Figure 4.
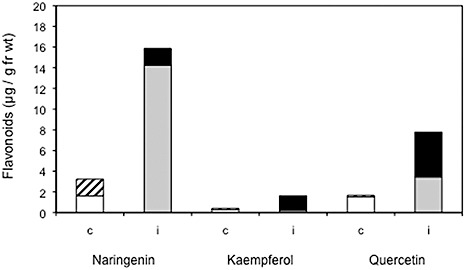
Accumulation of flavonoids in Arabidopsis control roots and root galls after infection with Plasmodiophora brassicae determined by high‐performance liquid chromatography (HPLC) analysis. The control samples were taken at 35 days post‐germination (dpg), and the infected samples at the same age [21 days post‐inoculation (dpi)]. White histogram, aglycones (control); hatched histogram, glycosides (control); grey histogram, aglycones (infected); black histogram, glycosides (infected).
TLC analysis showed that no flavonoids were detected in the control extracts on the basis of their colour and Rf values (Fig. S4). However, all extracts from infected roots contained naringenin, quercetin and kaempferol, although these were only visible after acid hydrolysis, indicating the presence of glycosides. The same extracts were analysed using a different solvent to separate flavonoid glycosides (data not shown). One band, which might correspond to quercetin‐3‐glycoside, was detected only in infected roots. Interestingly, light blue spots, most probably representing sinapate derivatives, decreased in infected roots (Fig. S4), perhaps indicating a redirection of the phenylpropanoid pathway towards flavonoids.
Mutants associated with the flavonoid biosynthetic pathway are slightly altered in their response to clubroot
To elucidate the role of flavonoid accumulation, we investigated a set of flavonoid mutants in flavonoid synthesis (tt4, tt5, tt6, tt7) or in transcription factors (tt1, myb12‐OX, myb12‐KO) for possible alterations in root gall size after infection with P. brassicae. According to the literature, tt4 does not accumulate any flavonoids and tt5 shows accumulation of naringenin chalcone (Peer et al. 2001). In tt6, naringenin would be expected to accumulate, but this was not found (Peer et al. 2001). However, because of the leaky mutation, the end products quercetin and kaempferol were detected. Finally, tt7 accumulated kaempferol in all tissues to a higher extent than in the wild‐type (Peer et al. 2001). The myb12‐KO lines showed root‐specific alterations in flavonoid patterns, e.g. drastically reduced flavonol levels (among them several glycosides) in the root and the hypocotyl–root transition zone (Stracke et al., 2007). On the other hand, quercetin and kaempferol derivatives were increased four‐ and three fold, respectively, in myb12‐OX compared with the wild‐type (Mehrtens et al., 2005).
The mutants tt3, tt4, tt5 and tt7 have been tested previously (Alix et al., 2007; Siemens et al., 2002), but only at high inoculation pressure (106 and 107 spores/mL). If flavonoids play a role in defence, the mutants should be more susceptible. At lower infection pressure, one would be able to identify susceptible lines (Siemens et al., 2002), which should have a higher infection rate and disease index (DI). Therefore, we included three of these lines in this investigation to test at lower infection pressure (105 spores/mL). The flavonoid biosynthesis mutants did not show significant differences in terms of infection rate, but tt4, tt5 and tt6 had a lower DI, especially at low infection pressure, whereas tt7 was similar to the wild‐type (Table 1). This is contrary to the expectation that flavonoids will play a role in defence, because most mutant lines were slightly more tolerant. To evaluate this effect further, the percentage of plants in individual disease classes is shown. Although class ‘1 + 2’ denotes less severe gall development, roots in the ‘3 + 4’ class show severe symptoms. In this presentation, tt4, tt5 and tt6 showed more plants in disease class ‘1 + 2’ than did the wild‐type Ler and tt7, again indicative of tolerant plants. However, it should be noted that plasmodia were found in microscopic sections to a similar extent in tt mutants and the wild‐type (data not shown).
Table 1.
Infection of mutants and transgenic lines with altered flavonoid patterns. Comparison of Arabidopsis Ler with various transparent testa (tt) mutants in the flavonoid biosynthetic pathway, as well as comparison of Arabidopsis Col with two mutants in the Myb transcription factor (MYB12) controlling various steps in flavonoid biosynthesis (MYB12 is root specific). myb12‐KO, knockout mutant; myb12‐OX, overexpressor. The parameters used for phytopathological analysis are the infection rate and disease index (DI). For better comparison, the plants in individual disease classes are also shown (0, no infection; 1 + 2, light infection; 3 + 4, severe infection). All experiments were performed at two different spore concentrations (105 and 107 spores/mL). Different letters indicate statistically significant differences between the mutant lines and the respective wild‐type under one specific condition. Bold indicates differences for one line between two treatments. Italic indicates lines that are considered to be more tolerant according to the DI or disease classes. For the 1‐naphthoxyacetic acid (NOA) experiment, only the high spore concentration was used for inoculation to detect possible effects of tolerance; nd, not determined.
| Line | Infection rate (%) | Disease index | Plants in individual disease classes (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 105 mL−1 | 107 mL−1 | 105 mL−1 | 107 mL−1 | 105 mL−1 | 107 mL−1 | |||||
| 0 | 1 + 2 | 3 + 4 | 0 | 1 + 2 | 3 + 4 | |||||
| Ler | 95a | 99a | 67a | 82a | 5a | 41 a | 54 a | 1a | 22a | 77a |
| tt4 | 91a | 91a | 62a | 58 b | 10b | 44a | 46a | 9b | 52b | 39 b |
| tt5 | 79a | 88a | 48 b | 65ab | 21 c | 56a | 23 b | 12b | 37ab | 51 c |
| tt6 | 80a | 86a | 56ab | 63ab | 20 c | 50a | 30 b | 14b | 43b | 43 bc |
| tt7 | 95a | 91a | 78a | 84a | 5a | 24b | 71c | 9b | 15c | 76a |
| Ler + NOA | nd | 100 | nd | 77a | nd | nd | nd | 1a | 20a | 79a |
| tt4+ NOA | nd | 89 | nd | 53 b | nd | nd | nd | 10b | 62d | 28 d |
| Col | 80a | 94a | 66a | 83a | 20 a | 37a | 44a | 6a | 37a | 57a |
| myb12‐KO | 79a | 100a | 47 b | 88a | 21 a | 57b | 22 b | 4a | 41a | 55a |
| myb12‐OX | 68 b | 90a | 56 ab | 76a | 32 b | 40a | 28 b | 0a | 35a | 65a |
The transcription factor MYB12 controls four different steps in flavonoid biosynthesis (Fig. 1), but it is mostly root‐specific (Stracke et al., 2007), indicating that a shoot‐derived effect can be ruled out. Surprisingly, DI was reduced for both lines, the mutant and the overexpressor, at low infection pressure (Table 1). In addition, when disease classes were compared, the myb12‐KO line had more roots in disease classes with low infection symptoms, especially at low spore number, which is similar to the results obtained with tt5 and tt6. This indicated that the loss of flavonoids caused a slight tolerance, but only at lower infection pressure. In addition, the myb12‐OX line was the only line with a lower infection rate at low infection pressure. Flavonoids might therefore be an additional, although not essential, factor in gall formation. The transcription factor mutant tt1 was not affected (data not shown), which was to be expected because tt1 encodes a seed‐specific transcription factor (Sagasser et al., 2002).
In addition, we noted that tt mutants, as shown for tt4 (Fig. S5, see Supporting Information), when subjected to additional abiotic stress, i.e. high temperature, were more susceptible than the wild‐type to clubroot. The inflorescence of tt4 plants was much smaller relative to the Ler ecotype (Fig. S5), whereas the flavonoid‐deficient root mutant myb12‐KO was not affected, in contrast with ecotype Col. Therefore, shoot flavonoids might be necessary to protect the plants from additional stress factors.
Are flavonoids acting as antioxidants in root galls?
Flavonoids could act as antioxidants in root galls to protect the plant tissue from oxidative stress. We therefore performed a stain to detect H2O2 in root galls of the wild‐type and flavonoid mutants using 3,3′‐diaminobenzidine (DAB). No difference in the accumulation of H2O2 was found in the galls of all lines tested, as indicated by the dark colour development (data not shown).
Flavonoids may also be regulators of auxin degradation by peroxidases/IAA oxidases. Many peroxidase genes were up‐regulated according to the microarray data (Table S2, see Supporting Information). The inhibition of peroxidase activity by flavonoids could result in decreased IAA degradation, and subsequently lead to an increase in IAA in galls. Several experiments were performed to evaluate this hypothesis.
-
1
The antioxidative potentials of the three main Arabidopsis flavonoids were tested using the 1,1′‐diphenyl‐2‐picrilhydrazyl (DPPH) radical (Fig. 5A). Although quercetin and kaempferol indeed showed high antioxidative activity, naringenin, which accumulated in large amounts in root galls, did not act as an antioxidant.
-
2
Peroxidase could be inhibited by high concentrations (100 µm) of the three flavonoids present in Arabidopsis (Fig. 5B).
-
3
IAA was degraded by peroxidase (Fig. 5C).
-
4
Peroxidase, IAA and flavonoids were simultaneously incubated and IAA was measured (Fig. 5D). If flavonoid prevents the formation of H2O2, which occurred during peroxidase‐catalysed IAA oxidation, the IAA levels should be stable and should not decrease in the presence of flavonoids. On the contrary, flavonoids accelerated the rate of IAA oxidation by peroxidase (Fig. 5D).
Figure 5.
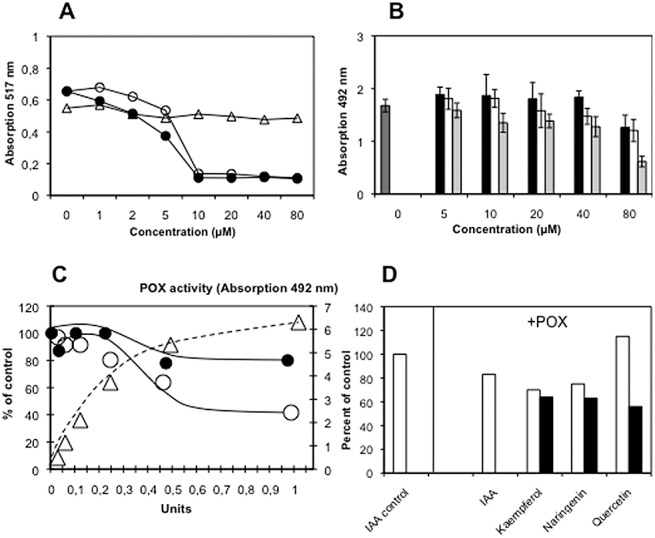
Possible inhibition of peroxidase‐catalysed indole‐3‐acetic acid (IAA) oxidation by flavonoids. (A) Antioxidative activities of quercetin (●), kaempferol (○), and naringenin (▵). (B) Peroxidase (POX) activity under the influence of different flavonoids; dark grey histogram, control; black histogram, quercetin; light grey histogram, kaempferol; white histogram, naringenin. (C) IAA (500 µm) was incubated with different amounts (units) of peroxidase and, after 1 h (●) and 4 h (○), the remaining IAA was determined by high‐performance liquid chromatography (HPLC); POX activity (▵) was determined using o‐phenylenediamine (OPD) as substrate and the absorption at 492 nm is given. (D) Treatment of peroxidase during incubation with IAA with two different concentrations of flavonoids: (□) 100 µm; ( ) 500 µm.
) 500 µm.
Therefore, we conclude that flavonoids do not prevent IAA oxidation, at least not under the in vitro conditions tested here.
Flavonoids might alter auxin distribution in root galls
Monitoring auxin biosynthesis via the nitrilase pathway showed its up‐regulation relatively late during gall development (Fig. 6). The promoter–β‐glucuronidase (GUS) fusion for nitrilase 2 (NIT2::GUS), the isoform mainly up‐regulated in Arabidopsis galls (Grsic‐Rausch et al., 2000), was compared with an auxin‐inducible promoter–GUS fusion (IAA2::GUS) (Marchant et al., 2002) in the development of root galls (Fig. 6). Although IAA2::GUS activity was found early during gall development (Fig. 6A,C,E), NIT2::GUS roots showed only a slight increase in GUS activity at early time points, which strongly increased in older galls (Fig. 6B,D,F).
Figure 6.
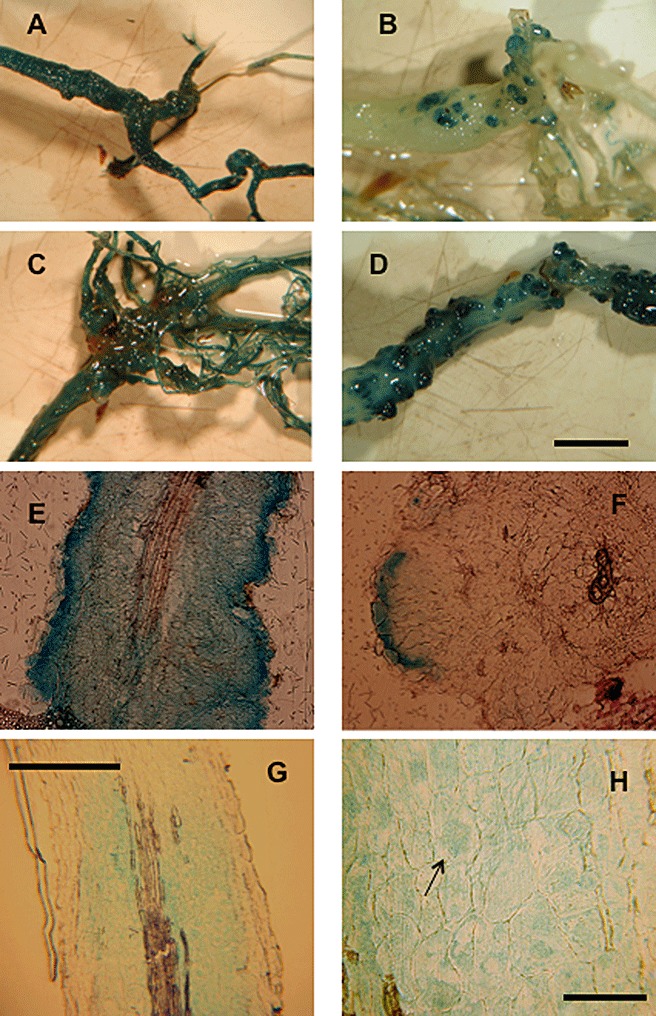
Comparison of IAA2::GUS (A, C, E) and NIT2::GUS (B, D, F) staining during the development of clubroot disease at different times post‐inoculation (days post‐inoculation, dpi). (A, B) Total roots at 23 dpi. (C, D) Total roots at 28 dpi. (E, F) Root sections at 23 dpi. Nitrilase is confined to the areas in which the gall develops and is visible only at later stages of infection, whereas the auxin‐inducible promoter responds very early during infection. (G) Root sections of Arabidopsis auxin‐responsive promoter‐reporter line IAA2::GUS infected with Plasmodiophora brassicae, analysed after 28 dpi and treated with 10 µm kaempferol. (H) Close‐up of (G) with plasmodia containing cells to which β‐glucuronidase (GUS) staining was restricted (one plasmodium is marked by an arrow). Bars: (A–D) 100 µm; (E–G) 50 µm; (H) 10 µm.
Several possibilities exist to explain the increased auxin levels at early time points: (i) hydrolysis from conjugates; (ii) other biosynthetic pathways for IAA synthesis; and (iii) altered transport. Experimental evidence in the literature has indicated correlations between flavonoids and changes in auxin efflux. Several genes coding for proteins that are members of the auxin transport machinery are differentially regulated during clubroot disease (Table S2). A fine tuning of auxin transport might possibly be provided by flavonoids. Therefore, we analysed the effect of flavonoids on the auxin signal in IAA2::GUS (Fig. 6G,H) and DR5::GUS (data not shown) plants. The treatment of roots with flavonoids mainly restricted GUS activity to cells containing plasmodia of the pathogen, as shown in Fig. 6 for kaempferol, and similar results were obtained with naringenin and quercetin and the IAA transport inhibitor NPA (data not shown).
The galls of tt4 and tt5 mutant plants did not show a reduction in free IAA compared with the wild‐type (Fig. 7). Therefore, the additional influence of auxin influx was investigated. As auxin influx is dependent on more than one transport carrier (Vieten et al., 2007), the auxin influx inhibitor 1‐naphthoxyacetic acid (NOA; Parry et al., 2001; Rahman et al., 2002) was used, although at lower concentrations than in the research cited (10 µm vs. 30 µm). When wild‐type (Ler) and tt4 mutant plants were treated after inoculation with NOA, slight but statistically significant differences were observed in the disease development of treated tt4 plants (Table 1). Twice as many plants were in the disease classes 1 and 2 compared with untreated tt4 and Ler under both conditions. However, it should be borne in mind that chemical inhibitors, such as NOA or NPA, alter the development of the whole plant, so that additional effects cannot be ruled out.
Figure 7.
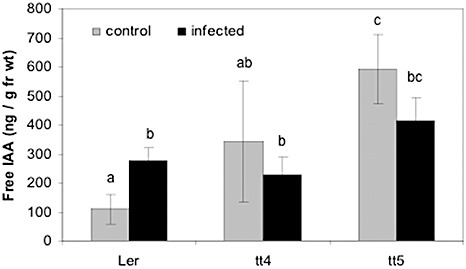
Determination of auxin levels in root galls of Ler, and transparent testa 4 (tt4) and transparent testa 5 (tt5) mutants. Auxin levels were higher in control roots of tt4 and tt5 mutants relative to Ler, but were not elevated in root galls. Under these conditions, no differences in root and shoot indices were found, indicating no tolerance to the clubroot pathogen. Different letters indicate significant differences between samples at the P ≤ 0.05 level. IAA, indole‐3‐acetic acid.
Shoot‐derived auxin is not necessary for gall development
To investigate whether foliar flavonoid treatment would affect gall size, possibly via the inhibition of shoot‐derived auxin transport, we treated control and infected Arabidopsis plants with different flavonoids and the auxin transport inhibitor NPA. Only after NPA application was the leaf rosette retarded in growth, whereas flavonoid‐treated plants showed normal growth (Fig. 8A). Gall size varied from small galls with a large root system attached to large galls with almost no root system, but there were no differences between the treatments (Fig. 8B). In addition, phytopathological analysis did not indicate that the treated plants were more tolerant (Fig. 8C). Plants with an auxin‐inducible promoter (DR5::GUS) were used to monitor auxin responsiveness in the root galls (Fig. 8), but again no differences were visible between dimethylsulphoxide (DMSO)‐treated and flavonoid‐ or NPA‐treated plants. Therefore, the free IAA levels in control and infected roots of flavonoid‐treated plants after foliar application were determined (Fig. 8D). In all samples, the infected roots showed higher IAA levels than the controls, whereas no significant differences were found between the control samples of DMSO‐ and flavonoid‐/NPA‐treated roots. Likewise, no significant differences were seen in the galls of DMSO‐ and flavonoid‐/NPA‐treated plant roots, although there was a trend for NPA‐treated galls to have a slightly lower IAA content.
Figure 8.
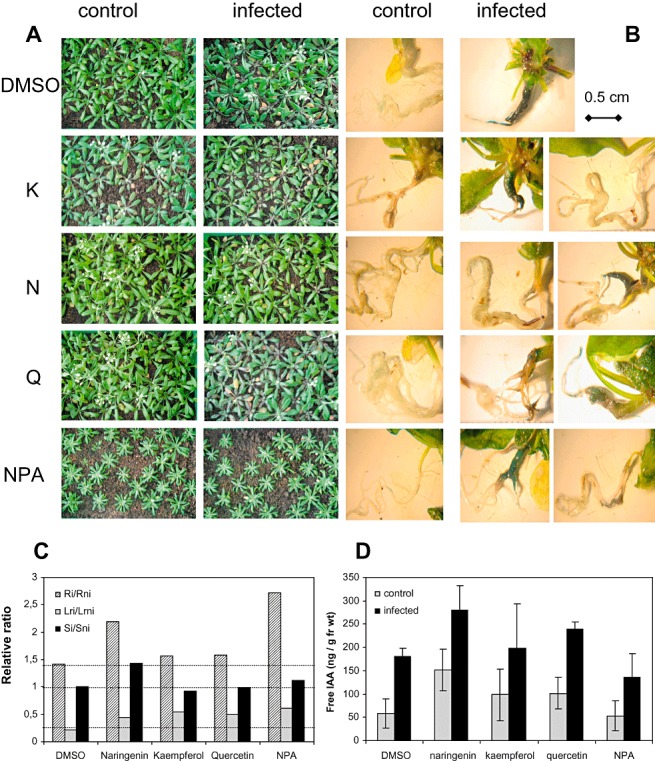
Foliar treatment with flavonoids, and their effects on gall size and indole‐3‐acetic acid (IAA) level determined at 21 days post‐inoculation (dpi). (A) Phenotype of the shoots of Arabidopsis Col (DR5::GUS) plants treated with dimethylsulphoxide (DMSO) as control, the flavonoids kaempferol (K), naringenin (N) and quercetin (Q), and the auxin transport inhibitor N‐(1‐naphthyl)phthalamic acid (NPA). (B) Root phenotype of control and infected plants treated as in (A). Staining for β‐glucuronidase (GUS) activity reveals auxin responsiveness only in the parts of the roots in which gall development occurred. (C) Phytopathological analysis showing that the gall size was not reduced (for a detailed description of the parameters used, see text). (D) Free IAA levels in control and infected roots after treatment.
Furthermore, IAA transport experiments were carried out using tt4 mutant and wild‐type shoot segments from control and infected plants (Fig. 9). Transport rates were determined using d5‐labelled IAA applied via agar blocks to stem segments, and the label that accumulated in receiver agar blocks was measured. No differences between wild‐type Col and the root‐specific mutants (myb12‐KO, myb12‐OX) were found, and the same was true for the IAA transport rate of the tt4 mutant compared with Ler. However, infected stems of tt4 tended to transport less IAA than infected Ler stem segments but, because of the high standard deviation, these differences were not significant. Taken together, these data are not in agreement with a role for shoot‐derived auxin in clubroot development.
Figure 9.
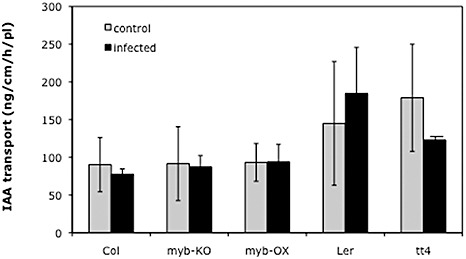
Indole‐3‐acetic acid (IAA) transport in shoots of wild‐type and mutant plants with altered flavonoid contents. IAA transport rates were measured in wild‐type (Ler) and transparent testa 4 (tt4) mutant control and Plasmodiophora brassicae‐infected plants, as well as Col, myb‐KO and myb‐OX lines under the same conditions.
DISCUSSION
Flavonoids accumulate in clubroots
Clubroot disease in Brassicaceae alters many biosynthetic pathways, although their role is not well elucidated (Ludwig‐Müller et al., 2009; Siemens et al., 2006). In the present study, we combined molecular, biochemical, histological and genetic analysis to investigate the role of flavonoids in the development of the disease. First, we showed that many genes in flavonoid/anthocyanin synthesis are differentially expressed during clubroot disease (Table S1, Fig. 2). Second, the major Arabidopsis flavonoids naringenin, kaempferol and quercetin accumulate specifically in the regions in which root galls are formed, as shown by in vivo staining with DPBA (Fig. 3). Further analysis of the flavonoid composition by HPLC and TLC confirmed these observations (Fig. 4, Fig. S4). These findings highlight a possible role of flavonoid accumulation in root galls in the development of disease.
Flavonoids as defence compounds or antioxidants
Flavonoids are known to be stress protectors, signalling molecules for the establishment of symbiosis and defence compounds (Dixon, 2001; Ryder et al., 1987; Wasson et al., 2009). Flavonoid accumulation might also be associated with defence reactions in the case of clubroot development. In this case, mutants and transgenic lines with altered flavonoid content should be more susceptible to P. brassicae infection. Careful monitoring of the disease parameters revealed differences which have not been detected previously (Alix et al., 2007; Siemens et al., 2002). At low infection pressure, several of the mutant lines were slightly more tolerant. This could be interpreted as a role of flavonoids as pathogenicity factors (Table 1). When simultaneous abiotic stress factors were applied, the flavonoid‐deficient mutant tt4 was more affected than Ler in shoot growth (Fig. S5). It should be noted that Buer and Djordjevic (2009) described aberrant phenotypes on various tt mutants when grown under stress conditions. On the other hand, the root‐deficient mutant myb12‐KO was not affected in shoot growth when compared with the wild‐type. Flavonoids might therefore be necessary to protect against simultaneous above‐ground stress factors.
In addition, flavonoids could act as antioxidants that scavenge reactive oxygen species, which could be a result of biotic, but also abiotic, stresses associated with various cellular dysfunctions. Protection against oxidative stress or oxidative burst may be likely, because no indications for a hypersensitive response were found in Arabidopsis galls (e.g. 2002, 2006). Moreover, their role as antioxidants in root galls might be connected with the degradation of IAA. In white clover, flavonoids were postulated to be involved in the regulation of auxin breakdown by peroxidase (Mathesius, 2001). However, the experiments carried out in the present study do not support this hypothesis (Fig. 5). Arguments against flavonoid inhibition of IAA degradation include the following.
-
1
Many flavonoids have antioxidative activity, but naringenin, in particular, which accumulates at high concentrations in root galls, does not show high antioxidative activity with DPPH.
-
2
Horseradish peroxidase catalyses IAA degradation in vitro and horseradish peroxidase can be inhibited by flavonoids in vitro. However, when IAA was simultaneously incubated with flavonoids and peroxidase, IAA was not protected from degradation. On the contrary, flavonoids seem to accelerate the degradation of IAA by horseradish peroxidase in vitro. This is in agreement with observations that formonetin accelerates IAA breakdown in vitro (Mathesius, 2001).
-
3
It has been reported that the degradation of IAA by peroxidase itself creates higher levels of H2O2, which could be detoxified by flavonoids, but we did not detect such an activity with IAA and root extracts containing peroxidase (data not shown).
-
4
No differences in H2O2 after staining with DAB were found in two ecotypes and two transgenic lines with either fewer or more flavonoids in the roots (data not shown).
Flavonoids modulate auxin transport in root galls, but shoot‐derived auxin is not necessary for gall formation
Flavonoids synthesized by plants play a role in signalling between host and microbes. However, they may have a dual function as an auxin transport inhibitor, thus being involved in increasing auxin levels and thereby inducing root nodule formation or growth (Mathesius, 2001). The inhibition of CHS in Medicago truncatula results in reduced flavonoid accumulation and reduced auxin transport which, in turn, reduces nodulation (Wasson et al., 2006). More directly, the reduction of PIN gene expression in M. truncatula also results in reduced nodulation (Huo et al., 2006). Similar hypotheses have been put forward for Agrobacterium tumefaciens infection of plant roots (Schwalm et al., 2003). Auxin induction may also trigger root gall formation caused by root‐knot nematodes in white clover, and has been implicated in the activation of the flavonoid pathway (Hutangura et al., 1999). Several features may be manifested in conjunction with nematode gall formation, nodulation and clubroot formation induced by P. brassicae. Recently, Wasson et al. (2009) have concluded that flavonoids are essential for nodulation, but not root‐knot gall formation, in M. truncatula. Galls on flavonoid‐deficient roots have normal giant cells but are shorter. This is reminiscent of the slightly smaller galls after P. brassicae infection of Arabidopsis flavonoid‐deficient mutants observed in our study. The accumulation of flavonoids in clubroot galls could likewise negatively influence auxin efflux from infected root cells, thus increasing endogenous IAA levels. The treatment of auxin‐responsive promoter–GUS lines with flavonoids confined GUS activity to cells harbouring plasmodia of the pathogen (Fig. 6G,H). Flavonoids interact directly with PGP but probably not PIN proteins (reviewed in Peer and Murphy, 2007). As several auxin efflux carriers of the PIN and PGP families were up‐regulated at the first time point of investigation (Table S2), whereas others were mostly down‐regulated at the second time point (with the exception of one PGP gene), the role of flavonoids might be to direct auxin fluxes within a growing gall. The clubroot phenotype observed would be in accordance with higher auxin efflux in lines tt4, tt5 and tt6, but not tt7, which accumulates kaempferol (Fig. 3M; Peer et al. 2001). Kaempferol might substitute for the other flavonoids not present in this line. However, to draw any further conclusions, more detailed flavonoid analyses are necessary in infected mutant roots.
In addition, the auxin influx facilitators AUX1, LAX1 and LAX3 were up‐regulated at the first time point of infection in the microarray experiment (Table S3, see Supporting Information). At the second time point of infection, all influx carriers were unchanged or down‐regulated. The aux1 mutant did not show any alterations in gall size (Siemens et al., 2002), probably because the two other influx carriers substitute for AUX1. Therefore, it was tested whether the auxin influx inhibitor NOA had an additional effect on gall development. This was the case in tt4 mutant plants, where the addition of NOA reduced disease severity slightly more than in treated wild‐type plants (Table 1). Similar to nodule formation by rhizobia, the formation of nodules on Casuarina glauca by the actinomycete Frankia is dependent on intact auxin transport. The application of NOA disturbed the appropriate development of actinorhizal nodule formation, which correlated with the expression of an AUX1 homologue in nodules of C. glauca (Peret et al., 2007).
In addition, our data do not indicate a role for shoot‐derived auxin in gall formation. First, the flavonoids naringenin, quercetin and kaempferol, which all accumulate in root galls of Arabidopsis (3, 4), did not alter gall size when they were administered to the shoots. Second, the auxin distribution was not altered in the root, as shown by the DR5::GUS expression pattern (Fig. 8). Third, free IAA levels in root galls treated with flavonoids were the same as in nontreated root galls (Fig. 8). Fourth, auxin transport rates were not altered in stem segments from infected plants relative to control stems (Fig. 9). In addition, the tt4 mutants showed the same, or maybe even less, auxin transport than the corresponding wild‐type. This result might be caused by the different plant materials and mutant backgrounds used in different studies (Peer and Murphy, 2007). Interestingly, free IAA levels were higher in tt4 and tt5 roots compared with the wild‐type, but there was no further enhancement of IAA in galls. The mutants might be able to accumulate IAA by different routes, which are sufficient for gall formation. However, Devos et al. (2006) noticed that the alh1 mutant was more tolerant to clubroot. This mutant has a defect in the cross‐talk between ethylene and auxins, probably at the level of auxin transport (Vandenbussche et al., 2003). These workers proposed that this mutant was resistant because host IAA transport was hampered. It was postulated that, in this mutant, IAA cannot reach the site of infection, underlining the need for appropriate IAA transport in order to obtain gall formation. Also, there is evidence that the time of NPA treatment, i.e. early in infection, is important for the reduction of gall growth (Devos and Prinsen, 2006). This implicates a more important role of auxin transport during earlier infection stages in roots but not shoots, whereas the later stages are dominated by biosynthesis to increase auxin levels.
CONCLUSIONS
In conclusion, we have shown that P. brassicae infection leads to an intense accumulation of flavonoids in Arabidopsis root galls. Flavonoids modulate auxin transport through direct and indirect interactions with cellular transport and regulatory mechanisms. Developmental regulation of flavonoid biosynthesis also determines where and when these interactions take place. This makes it difficult to ascertain when flavonoids are functioning primarily as scavengers, defence compounds, regulators of phosphorylation, enzyme inhibitors or modulators of membrane fluidity (Peer and Murphy, 2007). Our results suggest that the flavonoids might be involved in the regulation of auxin efflux from the roots, but we cannot rule out the possibility that they have additional roles as protective substances. Other features, such as sink regulation, protecting agents/antioxidants and regulators of membrane fluidity, might be additionally beneficial for the development of the pathogen. The size of a gall is of importance for the parasite because more volume will provide more space for the development of the resting spores. Therefore, even if flavonoids are not essential factors for clubroot development, they might contribute to gall size in a more subtle way. Future studies should also include an investigation of the auxin transport apparatus in root galls.
EXPERIMENTAL PROCEDURES
Plant material
The ecotypes Col‐0 and Ler of A. thaliana, as well as the tt mutants and myb12 lines used in this study, were originally provided by the Nottingham Arabidopsis Stock Centre (NASC, Nottingham, UK). The IAA2::GUS line was provided by Dr Jennifer Normanly (University of Amherst, MA, USA), the NIT2::GUS line by Dr Bonnie Bartel (Rice University, Houston, TX, USA) and the DR5::GUS line by Dr Tom Guilfoyle (University of Missouri, CO, USA).
Pathogen material
The P. brassicae isolate ‘e3’ used in this study was described by Fähling et al. (2003). Clubroot galls were stored at −20 °C until required. Resting spores were extracted by the homogenization of mature clubroot galls of Chinese cabbage, followed by filtration through gauze (25 µm pore width) and two centrifugation steps (2500 g, 10 min).
Inoculation and cultivation of plants
Fourteen‐day‐old Arabidopsis seedlings that were cultivated in a controlled environment (23 ± 1 °C, 16 h light, 100 µmol photons/s/m2) using a compost–sand (9 : 1, v/v) mixture (pH 5.8) were inoculated by injecting the soil around each plant with 2 mL of a resting spore suspension of the pathogen with the spore concentration indicated in Results. Disease symptoms were assessed at 28 dpi. For quantitative estimation, the plants were cut at the top of the hypocotyl into shoots and roots. The infected roots were measured as galls ignoring occasionally remaining noninfected lateral roots. Root fresh weight was used to calculate the root index Ri/Rni (roots of infected plants/roots of noninfected plants) according to Ludwig‐Müller et al. (1999) and Siemens et al. (2002). Likewise, indices for lateral roots (Li/Lni) and shoots (Si/Sni) were calculated in some experiments. At least 60 Arabidopsis plants were analysed for each line and treatment. The disease was assessed qualitatively on the basis of DI, as described by Siemens et al. (2002). The percentage of plants in different disease classes is also specified (0, no symptoms; 1 + 2, roots with light symptoms; 3 + 4, roots with severe symptoms). The qualitative disease assessment data were analysed first using the Kruskal–Wallis test and subsequently by comparing the mean rank differences as described by Siemens et al. (2002). Harvesting times for other experiments are given in Results. Controls were the same age and were treated with H2O instead of spore suspension.
Chemical treatments
For the treatment of P. brassicae‐infected and control plants with different flavonoids, all flavonoids tested were dissolved in DMSO as 10 mm stock solutions and stored at 4 °C. Stock solutions were diluted in Murashige and Skoog (MS) medium to achieve final concentrations of 10−4 and 10−5 m. Isorhamnetin was purchased from Fluka (Buch, Switzerland) and all other flavonoids from Sigma‐Aldrich (Steinheim, Germany). The plants were also treated with 10−4 m NPA (OlChemIm Ltd., Olomouc, Czech Republic). Disease symptoms were again assessed at 28 dpi or the roots were used for histological analyses (see below). Flavonoids were either sprayed every 4 days onto the shoots of the plants or administered every second day directly to the roots. Treatment with NOA (Sigma‐Aldrich) was performed by spraying plants with a 10−5 m solution three times between the time point of inoculation and harvest (28 dpi).
RT‐PCR analysis
Total RNA was isolated from control and infected roots of Arabidopsis at different times after infection using TRIzol® reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized by RT‐PCR. The primers, annealing temperatures (°C) and cycle number used are listed in Table S4. These sequences are perfect match primers corresponding to a nonhomologous region in other family member genes, either in the coding region or, in the case of CHI, in the 5′‐untranslated region (5′‐UTR) and coding region. To rule out the amplification of genomic DNA, the primers were chosen so that they always spanned an intron in the genomic sequence. Consequently, the resulting PCR amplification product would be larger (data not shown). As control, all cDNA samples were amplified with A. thaliana histone 2A (At1g52740) primers and with P. brassicae actin primers (AY452179) to confirm the presence of the pathogen. PCR was performed according to standard procedures using the following programme for amplification: Initial denaturation at 95 °C for 5 min, followed by the number of cycles given in Table S4 of 95 °C/60 s–x °C/40 s–72 °C/60 s (x stands for the temperature given in Table S4), and final elongation at 72 °C for 10 min.
Determination of GUS activity
The pattern of auxin distribution in IAA‐responsive promoter–GUS lines (IAA2::GUS, DR5::GUS) was determined in roots by histochemical staining with 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X‐Gluc) (Jefferson, 1987). Plants were incubated in 0.1 m NaPO3 buffer, pH 7.4, containing 10 mm Na2EDTA, 0.5 mm K3[Fe(CN)6], 0.5 mm K4[Fe(CN)6], 0.5% (w/v) Triton X‐100 and 50 µm of substrate (X‐Gluc dissolved in DMSO). After 1 h of incubation at 37 °C, the plants were rinsed and placed in 100% acetone for 30 min. After rinsing, the plants were transferred to NaPO3 buffer (pH 7.4) without substrate and kept overnight to block the reaction. Plants were either photographed directly or infected roots were fixed and embedded in Technovit (Heraeus‐Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. From these blocks, 3‐µm root sections were made with a microtome (Minot‐Mikrotom, Leitz, Wetzlar, Germany).
In vivo staining of flavonoids
Flavonoid compound locations were visualized in vivo by the fluorescence of DPBA conjugated to the flavonoid compounds after excitation with blue light using the method described by Buer and Muday (2004). DPBA shows gold fluorescence (emission maximum E max= 543 nm) when bound to quercetin and yellow–green fluorescence (E max= 520 nm) when bound to kaempferol. Plant roots were used directly in the case of controls or thin hand sections were obtained from clubroot tissue. The staining time was 5 min using saturated (0.25%, w/v) DPBA and 0.005% (v/v) Triton X‐100. Seedlings were then washed for 5 min with 100 mm sodium phosphate buffer [plus 0.005% (v/v) Triton X‐100, pH 7.0). Seedlings were mounted on slides in 50% (v/v) glycerol. Fluorescence was visualized by excitation with the following filter (G365/FT395/LP420 nm) on a Zeiss Axioskop 2 (Carl Zeiss, Jena, Germany). Digital images were captured with a camera (Canon, Krefeld, Germany) and exported into Photoshop as TIFF files.
Flavonoid determination
HPLC
Flavonoids were extracted from approximately 1 g fresh weight per sample in liquid nitrogen, and then resuspended in 15 mL of 70% methanol per gram fresh weight, extracted for 2 h at 8 °C, centrifuged for 10 min at 13 000 g, and the supernatant was evaporated to the aqueous phase. The latter was brought to pH 3 with 1 m HCl and then extracted twice with equal volumes of ethyl acetate. The organic fractions were pooled and evaporated to dryness. The residues were taken up in a final volume of 100 µL methanol for HPLC analysis. For the hydrolysis of flavonoid glycosides, two parts of the methanolic extract after centrifugation were mixed with one part of hydrolysis reagent (10 mL H2O, 100 mg butylated hydroxytoluene, 12 mL 2 m HCl) and incubated for 2 h at 85 °C, shaking every 15 min. The extract was then cooled on ice, centrifuged for 10 min at 13 000 g and brought to pH 3 with 1 m NaOH. The extract was then treated further as described for nonglycosylated flavonoids. Rutin hydrolysed completely under these conditions (Ludwig‐Müller et al., 2005). The sample was completely injected onto a high‐performance liquid chromatograph (Jasco, Groβ‐Umstadt, Germany) using a 25 cm × 4 mm RP C18 column (Phenomenex, Aschaffenburg, Germany) for separation. The solvents were 100% acetonitrile (A) and aqueous 2.5% acetic acid (B), and the flow rate was 1 mL/min. The gradient started with 3% A, followed by an increase to 9% A in 5 min, 16% A in 10 min, 50% A in 30 min, staying at 50% A for another 5 min before a washing (100% A) and equilibrating step (starting conditions) followed. Detection was performed with a multiwavelength photodiode array detector (Jasco) at 280 nm (naringenin) and 375 nm (kaempferol and quercetin). Calculations were performed on the basis of a standard curve with authentic standards. The experiment was performed in triplicate and repeated with independently cultivated plant material. Examples of chromatograms and spectra for standards and Arabidopsis extracts are shown in Figs S2 and S3.
TLC
For TLC, the extracts were prepared as described above, including hydrolysis under the same conditions, and analysed on an Alugram R SIL G/UV2 54, 20 cm × 20 cm (Macherey‐Nagel, Düren, Germany) using 80 µL of the extract for the separation of aglycones with toluene–dioxan–ethyl acetate (95 : 25 : 4, v/v/v) as solvent, and 100 µL of the extract for the separation of glycosylated flavonoids with ethyl acetate–formic acid–acetic acid–H2O (100 : 11 : 11 : 27, v/v/v/v) as solvent. TLC plates were sprayed with DPBA reagent (1% DPBA in methanol), followed, after drying, by treatment with 5% polyethyleneglycol for fluorescence enhancement. Flavonoids were visualized under a UV lamp at 366 nm and photographed with a digital camera (Canon). Identification was performed using the colours of individual bands (orange, quercetin derivatives; green, kaempferol derivatives; brownish, naringenin derivatives) and comparison of the Rf values. The standards used were naringenin (N), quercetin (Q), kaempferol (K) for aglycones (Fig. S4), and naringenin‐7‐rhamnoglycoside (naringin), quercetin‐3‐rutinoside (rutin), quercetin‐3‐O‐glucoside (isoquercitrin), quercetin‐3‐d‐galactoside, quercetin‐3‐d‐rhamnoside, kaempferol‐3‐O‐glucoside (astragalin) for glycosides (data not shown).
Auxin determination
For each individual sample, 80–150 mg fresh weight was used and all determinations were performed in triplicate. The samples were homogenized with a mixture of isopropanol and acetic acid (95 : 5, v/v) and 100 ng of 13C6‐IAA was added to each sample. The extracts were then incubated under continuous shaking (500 rpm) for 2 h at 4 °C. The samples were further processed as described by Fitze et al. (2005). Methylation of samples prior to GC‐MS analysis was carried out according to Cohen (1984) with freshly prepared diazomethane. The samples were finally resuspended in 30 µL of ethyl acetate and 1 µL was subjected to further analysis. GC‐MS analysis was carried out on a Varian Saturn 2100 ion‐trap mass spectrometer using electron impact ionization at 70 eV, connected to a Varian CP‐3900 gas chromatograph equipped with a CP‐8400 autosampler (Varian, Walnut Creek, CA, USA). The samples were injected in the splitless mode (splitter opening 1 : 100 after 1 min) onto a Phenomenex ZB‐5 column (30 m × 0.25 mm × 0.25 µm) using He carrier gas at 1 mL/min. The injector temperature was 250 °C and the transfer line temperature was 280 °C. The scan rate was 0.6 s/scan, the multiplier offset voltage was 200 V, the emission current was 30 µA and the trap temperature was 200 °C. For the analysis of IAA, the following programme was used: 60 °C for 1 min, followed by an increase of 25 °C/min to 180 °C, 5 °C/min to 250 °C, 25 °C/min to 280 °C, and then 5 min isothermally at 280 °C. The µSIS mode (Varian Manual) was used to attain higher sensitivity. The calculation of endogenous IAA levels was performed according to the principles of isotope dilution (Cohen et al., 1986) using the quinolinium ions at m/z 130 and 136, representing the endogenous and labelled compound, respectively. The mean values of three independent extractions ± standard error (SE) are given for free IAA. Statistical data analysis was carried out using Student's t‐test and significance at the P≤ 0.05 level is denoted by different letters.
Auxin transport
Arabidopsis plants were grown until stem segments of approximately 8 cm had developed. The plants were cut into 2‐cm segments at the base close to the rosette and the lower parts of the segments were placed between two agar blocks (receiver blocks) in a moist Petri dish. The upper part of the segments was covered with an agar donor block containing 1 mm d5‐IAA. The plates were placed vertically in the dark at 23 °C over the experimental period. The basal receiver blocks were extracted after an incubation time of 3 h using isopropanol–acetic acid (95 : 5, v/v) and quartz sand. To quantify the levels of d5‐IAA, 13C6‐IAA (100 ng) was added to each sample. After extraction for 2 h at 8 °C in the dark, the sample was centrifuged for 15 min at 13 000 g and the supernatant was removed very carefully. If necessary, the remaining extract was centrifuged again as described. The supernatant was then evaporated to dryness under a stream of N2 to the aqueous phase, and then extracted twice at pH 3 with ethyl acetate. The organic fractions were combined, evaporated to dryness under N2 and resuspended in 100 µL of ethyl acetate for methylation (see above). GC‐MS analysis was carried out as described above. The basipetal transport of IAA was calculated as the amount of d5‐IAA found in the receiver block as ng/h/cm. One sample consisted of 10 segments and the analysis was carried out in triplicate. The experiment was repeated with independently cultivated plants.
Peroxidase activity
Horseradish peroxidase (Merck, Darmstadt, Germany) was used to investigate the effect of flavonoids and IAA. The enzymatic activity was measured using o‐phenylenediamine (OPD, Sigma‐Aldrich, Steinheim, Germany) as substrate. The assay was performed with 0.03–0.05 mg protein in 20 mm Tricine buffer, pH 7.0. Prior to use, one tablet of OPD was dissolved in 5 mL of substrate buffer consisting of 20 mm Tricine, pH 7 and 3.6% (v/v) H2O2. Of this mixture, 150 µL were added to each sample in a microtitre plate. The assay was incubated for 15 min at room temperature and stopped with 150 µL of 0.5 m H2SO4. The absorbance was determined at 492 nm in a microtitre plate reader (Magellan Sunrise‐Basic, Tecan, Grödig, Austria) using the software Magellan‐Standard V 5.03. Each experiment was performed three times with independent plant material and each sample was measured in triplicate. Means ± SE are given. IAA and flavonoids were dissolved in methanol, for the diluted assay, so that the methanolic portion never exceeded 1%. IAA was determined by HPLC (see above) using 45% methanol in a 1% aqueous acetic acid solution at a detection wavelength of 280 nm.
In situ H2O2 stain
H2O2 was stained in situ using DAB according to Thordal‐Christensen et al. (1997). Briefly, roots were incubated with a solution of 1.6 mg/mL DAB (Sigma‐Aldrich), pH 3.8, in the dark for 3 h. Destaining was performed by boiling the tissue in 96% ethanol for 10 min. The galls were then washed with water before inspection under a binocular.
Antioxidative activity with the DPPH assay
The scavenging activity of flavonoids on the DPPH radical was measured as the decrease in DPPH absorbance at 517 nm (Blois, 1958), and expressed as a percentage of the absorbance of a control DPPH solution.
Database analysis
The AGI numbers of transcripts for flavonoid biosynthesis genes were located from the TAIR database (AraCyc; http://www.arabidopsis.org/) and transcript levels were compared for the two infection times investigated using the ATH1 array (Affymetrix), as described previously (Siemens et al., 2006, E‐MEXP‐254). Ratios for control to infected roots were calculated for each time point (TP1, TP2) for all values.
Supporting information
Fig. S1 Control and Plasmodiophora brassicae‐infected wild‐type Col plants.
Fig. S2 Sample chromatograms of high‐performance liquid chromatography (HPLC) separations of the standards quercetin, kaempferol and naringenin, as well as selected chromatograms from control and infected roots.
Fig. S3 Samples of flavonoid spectra extracted from high‐performance liquid chromatograms.
Fig. S4 Thin layer chromatography separation of flavonoid aglycones from galls vs. controls at different time points after inoculation.
Fig. S5 Combination of biotic and abiotic stress shows that the transparent testa 4 (tt4) mutant is more susceptible than ecotype Ler to clubroot, as indicated by the smaller inflorescence. For comparison, the root flavonoid‐deficient line myb12‐KO and the respective wild‐type Col are shown.
Table S1 Expression levels of flavonoid‐related genes during clubroot disease according to microarray analysis (ATH1, Affymetrix).
Table S2 Expression levels of peroxidase, catalase and superoxide dismutase genes during clubroot disease according to microarray analysis (ATH1, Affymetrix).
Table S3 Expression levels of auxin transport‐related genes during clubroot disease according to microarray analysis (ATH1, Affymetrix).
Table S4 Primers, annealing temperature and reaction conditions used for the reverse transcription‐polymerase chain reaction (RT‐PCR) experiment for different genes from the flavonoid biosynthetic pathway.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We would like to thank Silvia Heinze for technical assistance and two anonymous reviewers for their valuable comments.
REFERENCES
- Alix, K. , Lariagon, C. , Delourme, R. and Manzanares‐Dauleux, M.J. (2007) Exploiting natural genetic diversity and mutant resources of Arabidopsis thaliana to study the A. thaliana–Plasmodiophora brassicae interaction. Plant Breed. 126, 218–221. [Google Scholar]
- Blois, M.S. (1958) Antioxidant determinations by the use of a stable free radical. Nature, 181, 1199–1200. [Google Scholar]
- Brown, D.E. , Rashotte, A.M. , Murphy, A.S. , Normanly, J. , Tague, B.W. , Peer, W.A. , Taiz, L. and Muday, G.K. (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis export. Plant Physiol. 126, 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C.S. and Djordjevic, M.A. (2009) Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana . J. Exp. Bot. 60, 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C.S. and Muday, G.K. (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell, 16, 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J.D. (1984) Convenient apparatus for the generation of small amounts of diazomethane. J. Chromatogr. 303, 193–196. [Google Scholar]
- Cohen, J.D. , Baldi, B.G. and Slovin, J.P. (1986) 13C6‐[benzene ring]‐indole‐3‐acetic acid. Plant Physiol. 80, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, S. and Prinsen, E. (2006) Plant hormones: a key in clubroot development. Commun. Agric. Appl. Biol. Sci. 71 (3 Pt B), 869–872. [PubMed] [Google Scholar]
- Devos, S. , Laukens, K. , Deckers, P. , Van Der Straeten, D. , Beeckman, T. , Inze, D. , Van Onckelen, H. , Witters, E. and Prinsen, E. (2006) A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis . Mol. Plant–Microbe Interact. 19, 1431–1433. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A. (2001) Natural products and plant disease resistance. Nature, 411, 843–847. [DOI] [PubMed] [Google Scholar]
- Evans, J.L. and Scholes, J.D. (1995) How does clubroot alter the regulation of carbon metabolism in its host? Asp. Appl. Biol. 42, 125–132. [Google Scholar]
- Fähling, M. , Graf, H. and Siemens, J. (2003) Pathotype‐separation of Plasmodiophora brassicae by the host plant. J. Phytopathol. 151, 425–430. [Google Scholar]
- Fitze, D. , Wiepning, A. , Kaldorf, M. and Ludwig‐Müller, J. (2005) Auxins in the development of an arbuscular mycorrhizal symbiosis in maize. J. Plant Physiol. 162, 1210–1219. [DOI] [PubMed] [Google Scholar]
- Gee, M.A. , Hagen, G. and Guilfoyle, T.J. (1991) Tissue‐specific and organ‐specific expression of soybean auxin‐responsive transcripts GH3 and SAURs. Plant Cell, 3, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, M. and Murphy, A.S. (2006) The ABC of auxin transport: the role of p‐glycoproteins in plant development. FEBS Lett. 580, 1094–1102. [DOI] [PubMed] [Google Scholar]
- Grsic‐Rausch, S. , Kobelt, P. , Siemens, J. , Bischoff, M. and Ludwig‐Müller, J. (2000) Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis thaliana . Plant Physiol. 122, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, G. , Martin, G. , Li, Y. and Guilfoyle, T.J. (1991) Auxin‐induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17, 567–579. [DOI] [PubMed] [Google Scholar]
- Huo, X. , Schnabel, E. , Hughes, K. and Frugoli, J. (2006) RNAi phenotypes and the localization of a protein:GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. J. Plant Growth Regul. 25, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutangura, P. , Mathesius, U. , Jones, M.G.K. and Rolfe, B.G. (1999) Auxin induction is a trigger for root gall formation caused by root‐knot nematodes in white clover and is associated with the activation of the flavonoid pathway. Aust. J. Plant Physiol. 26, 221–231. [Google Scholar]
- Ingram, D.S. and Tommerup, I.C. (1972) The life history of Plasmodiophora brassicae Woron . Proc. R. Soc. Lond. B 180, 103–112. [Google Scholar]
- Ishikawa, T. , Okazaki, K. , Kuroda, H. , Itoh, K. , Mitsui, T. and Hori, H. (2007) Molecular cloning of Brassica rapa nitrilases and their expression during clubroot development. Mol. Plant Pathol. 8, 623–637. [DOI] [PubMed] [Google Scholar]
- Jacobs, M. and Rubery, P.H. (1988) Naturally occurring auxin transport regulators. Science, 241, 346–349. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Kim, D.‐S. , Jeon, S.‐E. , Jeong, Y.‐M. , Kim, S.‐Y. , Kwon, S.‐B. and Park, K.‐C. (2006) Hydrogen peroxide is a mediator of indole‐3‐acetic acid/horseradish peroxidase‐induced apoptosis. FEBS Lett. 580, 1439–1446. [DOI] [PubMed] [Google Scholar]
- Kramer, E.M. and Bennett, M.J. (2006) Auxin transport: a field in flux. Trends Plant Sci. 11, 382–386. [DOI] [PubMed] [Google Scholar]
- Lim, E.‐K. , Ashford, D.A. , Hou, B. , Jackson, R.G. and Bowles, D.J. (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 87, 623–631. [DOI] [PubMed] [Google Scholar]
- Ljung, K. , Hull, A.K. , Kowalczyk, M. , Marchant, A. , Celenza, J. , Cohen, J.D. and Sandberg, G. (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole‐3‐acetic acid in Arabidopsis thaliana . Plant Mol. Biol. 49, 249–272. [PubMed] [Google Scholar]
- Ludwig‐Müller, J. and Schuller, A. (2008) What can we learn from clubroots: alterations in host roots and hormone homeostasis caused by Plasmodiophora brassicae . Eur. J. Plant Pathol. 121, 291–302. [Google Scholar]
- Ludwig‐Müller, J. , Pieper, K. , Ruppel, M. , Cohen, J.D. , Epstein, E. , Kiddle, G. and Bennett, R. (1999) Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana L. glucosinolate mutants and the development of the clubroot disease. Planta, 208, 409–419. [DOI] [PubMed] [Google Scholar]
- Ludwig‐Müller, J. , Tokalov, S.V. , Franz, A. and Gutzeit, H.O. (2005) Quercetin metabolism in vital and apoptotic human leukaemia cells. Biol. Chem. 386, 279–283. [DOI] [PubMed] [Google Scholar]
- Ludwig‐Müller, J. , Prinsen, E. , Rolfe, S. and Scholes, J. (2009) Metabolism and plant hormone action during the clubroot disease. J. Plant Growth Regul. 28, 229–244. [Google Scholar]
- Marchant, A. , Bhalerao, R. , Casimiro, I. , Eklöf, J. , Casero, P.J. , Bennett, M. and Sandberg, G. (2002) AUX1 promotes lateral root formation by facilitating indole‐3‐acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell, 14, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius, U. (2001) Flavonoids induced in cells undergoing nodule organogenesis in white clover are regulators of auxin breakdown by peroxidase. J. Exp. Bot. 52, 419–426. [DOI] [PubMed] [Google Scholar]
- Mehrtens, F. , Kranz, H. , Bednarek, P. and Weisshaar, B. (2005) The Arabidopsis transcription factor MYB12 is a flavonol‐specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen, R. and Magrath, R. (1992) A contribution to the life history of Plasmodiophora brassicae: secondary plasmodia development in root galls of Arabidopsis thaliana . Mycol. Res. 96, 877–885. [Google Scholar]
- Muday, G.K. and DeLong, A. (2001) Polar auxin transport: controlling where and how much. Trends Plant Sci. 6, 535–542. [DOI] [PubMed] [Google Scholar]
- Muday, G.K. and Murphy, A.S. (2002) An emerging model of auxin transport regulation. Plant Cell, 14, 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A.S. , Peer, W.A. and Taiz, L. (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta, 211, 315–324. [DOI] [PubMed] [Google Scholar]
- Neuhaus, K. , Grsic‐Rausch, S. , Sauerteig, S. and Ludwig‐Müller, J. (2000) Arabidopsis plants transformed with nitrilase 1 or 2 in antisense direction are delayed in clubroot development. J. Plant Physiol. 156, 756–761. [Google Scholar]
- Owens, D.K. , Alerding, A.B. , Crosby, K.C. , Bandara, A.B. , Westwood, J.H. and Winkel, B.S. (2008) Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 147, 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry, G. , Delbarre, A. , Marchant, A. , Swarup, R. , Napier, R. , Perrot‐Rechenmann, C. and Bennett, M.J. (2001) Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1 . Plant J. 25, 399–406. [DOI] [PubMed] [Google Scholar]
- Peer, W.A. and Murphy, A.S. (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci. 12, 556–563. [DOI] [PubMed] [Google Scholar]
- Peer, W.A. , Brown, D.E. , Tague, B.W. , Muday, G.K. , Taiz, L. and Murphy, A.S. (2001) Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 126, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A. , Bandyopadhyay, A. , Blakeslee, J.J. , Makam, S.N. , Chen, R.J. , Masson, P.H. and Murphy, A.S. (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana . Plant Cell, 16, 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret, B. , Swarup, R. , Jansen, L. , Devos, G. , Auguy, F. , Collin, M. , Santi, C. , Hocher, V. , Franche, C. , Bogusz, D. , Bennett, M. and Laplaze, L. (2007) Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca . Plant Physiol. 144, 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuß, A. , Stracke, R. , Weisshaar, B. , Hillebrecht, A. , Matern, U. and Martens, S. (2009) Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett. 583, 1981–1986. [DOI] [PubMed] [Google Scholar]
- Rahman, A. , Hosokawa, S. , Oono, Y. , Amakawa, T. , Goto, N. and Tsurumi, S. (2002) Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol. 130, 1908–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch, T. , Butcher, D.N. and Hilgenberg, W. (1981) Nitrilase activity in clubroot diseased plants. Physiol. Plant. 52, 467–470. [Google Scholar]
- Ryder, T.B. , Hedrick, S.A. , Bell, J.N. , Liang, X. , Clouse, S.D. and Lamb, C.J. (1987) Organization and differential activation of a gene family encoding the plant defense enzyme chalcone synthase in Phaseolus vulgaris . Mol. Genet. Genomics, 210, 219–233. [DOI] [PubMed] [Google Scholar]
- Sagasser, M. , Lu, G.H. , Hahlbrock, K. and Weisshaar, B. (2002) A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 16, 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalm, K. , Aloni, R. , Langhans, M. , Heller, W. , Stich, S. and Ullrich, C.I. (2003) Flavonoid‐related regulation of auxin accumulation in Agrobacterium tumefaciens‐induced plant tumors. Planta, 218, 163–178. [DOI] [PubMed] [Google Scholar]
- Siemens, J. , Nagel, M. , Ludwig‐Müller, J. and Sacristán, M.D. (2002) The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: parameters for disease quantification and screening of mutant lines. J. Phytopathol. 150, 592–605. [Google Scholar]
- Siemens, J. , Keller, I. , Sarx, J. , Kunz, S. , Schuller, A. , Nagel, W. , Schmülling, T. , Parniske, M. and Ludwig‐Müller, J. (2006) Transcriptome analysis of Arabidopsis clubroots and disease resistance of cytokinin oxidase/dehydrogenase gene overexpressing plants indicate a key role for cytokinin in disease development. Mol. Plant–Microbe Interact. 19, 480–494. [DOI] [PubMed] [Google Scholar]
- Siemens, J. , Graf, H. , Bulman, S. , In, O. and Ludwig‐Müller, J. (2009) Monitoring expression of selected Plasmodiophora brassicae genes during clubroot development in Arabidopsis thaliana . Plant Pathol. 58, 130–136. [Correction added after online publication 17 June 2010: the page extend for reference Siemens et al., 2009 has been corrected] [Google Scholar]
- Stracke, R. , Ishihara, H. , Huep, G. , Barsch, A. , Mehrtens, F. , Niehaus, K. and Weisshaar, B. (2007) Differential regulation of closely related R2R3‐MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Vandenbussche, F. , Smalle, J. , Le, J. , Saibo, N.J.M. , De Paepe, A. , Chaerle, L. , Tietz, O. , Smets, R. , Laarhoven, L.J.J. , Harren, F.J.M. , Van Onckelen, H. , Palme, K. , Verbelen, J.‐P. and Van Der Straeten, D. (2003) The Arabidopsis mutant alh1 illustrates a cross talk between ethylene and auxin. Plant Physiol. 131, 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten, A. , Sauer, M. , Brewer, P.B. and Friml, J. (2007) Molecular and cellular aspects of auxin‐transport‐mediated development. Trends Plant Sci. 12, 160–168. [DOI] [PubMed] [Google Scholar]
- Wasson, A.P. , Pellerone, F.I. and Mathesius, U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell, 18, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson, A.P. , Ramsay, K. , Jones, M.G.K. and Mathesius, U. (2009) Differing requirements for flavonoids during the formation of lateral roots, nodules and root knot nematode galls in Medicago truncatula . New Phytol. 183, 167–179. [DOI] [PubMed] [Google Scholar]
- Winkel‐Shirley, B. (2002) Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Subramanian, S. , Stacey, G. and Yu, O. (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti . Plant J. 57, 171–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Control and Plasmodiophora brassicae‐infected wild‐type Col plants.
Fig. S2 Sample chromatograms of high‐performance liquid chromatography (HPLC) separations of the standards quercetin, kaempferol and naringenin, as well as selected chromatograms from control and infected roots.
Fig. S3 Samples of flavonoid spectra extracted from high‐performance liquid chromatograms.
Fig. S4 Thin layer chromatography separation of flavonoid aglycones from galls vs. controls at different time points after inoculation.
Fig. S5 Combination of biotic and abiotic stress shows that the transparent testa 4 (tt4) mutant is more susceptible than ecotype Ler to clubroot, as indicated by the smaller inflorescence. For comparison, the root flavonoid‐deficient line myb12‐KO and the respective wild‐type Col are shown.
Table S1 Expression levels of flavonoid‐related genes during clubroot disease according to microarray analysis (ATH1, Affymetrix).
Table S2 Expression levels of peroxidase, catalase and superoxide dismutase genes during clubroot disease according to microarray analysis (ATH1, Affymetrix).
Table S3 Expression levels of auxin transport‐related genes during clubroot disease according to microarray analysis (ATH1, Affymetrix).
Table S4 Primers, annealing temperature and reaction conditions used for the reverse transcription‐polymerase chain reaction (RT‐PCR) experiment for different genes from the flavonoid biosynthetic pathway.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


