SUMMARY
Previously, we have shown that encapsidated Potato virus X (PVX) RNA was non‐translatable in vitro, but could be converted into a translatable form by binding of the PVX movement protein TGBp1 to one end of the virion or by coat protein (CP) phosphorylation. Here, a mutagenic analysis of PVX CP and TGBp1 was used to identify the regions involved in TGBp1–CP binding and translational activation of PVX RNA by TGBp1. It was found that the C‐terminal (C‐ter) 10/18 amino acids region was not essential for virus‐like particle (VP) assembly from CP and RNA. However, the VPs assembled from the CP lacking C‐ter 10/18 amino acids were incapable of TGBp1 binding and being translationally activated. It was suggested that the 10‐amino‐acid C‐ter regions of protein subunits located at one end of a polar helical PVX particle contain a domain accessible to TGBp1 binding and PVX remodelling. The non‐translatable particles assembled from the C‐ter mutant CP could be converted into a translatable form by CP phosphorylation. The TGBp1–CP binding activity was preserved unless a conservative motif IV was removed from TGBp1. By contrast, TGBp1‐dependent activation of PVX RNA translation was abolished by deletions of various NTPase/helicase conservative motifs and their combinations. The motif IV might be essential for TGBp1–CP binding, but insufficient for PVX RNA translation activation. The evidence to discriminate between these two events, i.e. TGBp1 binding to the CP‐helix and TGBp1‐dependent RNA translation activation, is discussed.
INTRODUCTION
Potato Virus X (PVX) is the type member of the genus Potexvirus, family Flexiviridae. About 1300 identical protein subunits in a filamentous PVX virion form a helical array with the positive RNA packed between the turns of the helix (Tollin and Wilson, 1988). The PVX coat protein (CP) consists of 237 amino acids and is glycosylated (Baratova et al., 2004; Tozzini et al., 1994). The PVX RNA contains five genes (Huisman et al., 1988; Skryabin et al., 1988). The 5′‐proximal gene codes for the 165‐kDa replicase, and the 3′‐proximal CP gene is preceded by three partially overlapping genes termed the ‘triple gene block’ (TGB) coding for three movement proteins (MPs) referred to as TGBp1, TGBp2 and TGBp3, respectively. The TGB‐coded MPs together with CP are involved in PVX cell‐to‐cell movement (Chapman et al., 1992; Verchot et al., 1998; for reviews, see Batten et al., 2003; Morozov and Solovyev, 2003; Verchot‐Lubicz, 2005).
Two different models have been proposed to explain the nature of the infectious potexvirus transport that moves from cell to cell over the infected plant: (1) it has been suggested (Allison and Shalla, 1974; Oparka et al., 1996; Santa Cruz et al., 1998) that filamentous virions are involved in cell‐to‐cell movement of PVX; (2) by contrast, it has been reported that in vitro assembled non‐virion RNP (CP‐RNA‐TGBp1) complexes moved from cell to cell in microinjection experiments, in vivo (Lough et al., 1998), and that RNA encapsidation was not needed for potexvirus spread (Lough et al., 2000). Recently, we examined the structure of complexes assembled in vitro from PVX RNA, TGBp1 and CP. The single‐tailed particles (STPs) with the 5′‐terminal region of PVX RNA encapsidated in a helical head‐like structure and TGBp1 bound to the end of the head were revealed (Karpova et al., 2006a). Strong evidence was provided on structural similarity between the native virions and STP heads. Apparently, the STPs represent incompletely assembled PVX virions. We suggested that translatable complexes of TGBp1 with the extremity of the STP and/or of PVX virions may represent the transport form of PVX infection (Karpova et al., 2006a).
Previously we have shown that encapsidated PVX RNA was completely non‐translatable in vitro, but that helical particles could be ‘remodelled’, i.e. converted to a translatable form by two ways. First, this was by a selective binding of the TGBp1 to one end of a polar helical PVX virion (Atabekov et al., 2000, 2001). Similar to native PVX, the STPs were non‐translatable, but could be translationally activated by binding of TGBp1 to the end of the helical STP head containing the 5′‐end of the RNA (Karpova et al., 2006a). We have recently shown that binding of TGBp1 to CP subunits located at one extremity of the polar helical particles induced their remodelling, comprising a linear destabilization of the CP helix, which was transmitted along the whole particle (Rodionova et al., 2003). Second, translation of encapsidated PVX RNA could be triggered by in situ phosphorylation of the PVX CP by Ser/Thr‐specific protein kinases (Atabekov et al., 2001). The N‐terminal (N‐ter) CP peptide located at the surface of the viral particle contains the major phosphorylation site and, therefore, plays a key role in translational activation of the PVX RNA by CP phosphorylation. Significantly, the N‐ter 19‐amino‐acid CP region is dispensable for TGBp1–CP interaction, PVX virion remodelling and activation of RNA translation (Atabekov et al., 2001). On the other hand, deletion of the C‐terminal deleted (C‐terdel) 18‐amino‐acid region from PVX CP led to the inhibition of the viral cell‐to‐cell movement (Fedorkin et al., 2001). A similar result was observed when the white clover mosaic potexvirus (WClMV) with 31‐amino‐acid‐deleted CP was examined (Forster et al., 1992). Significantly, the C‐terminal deleted CP (C‐terdelCP) of WClMV could be assembled, producing full‐length virions in the protoplasts inoculated with the movement‐deficient mutant virus (Forster et al., 1992).
The present paper describes a mutagenic analysis of PVX CP and TGBp1, and associated assays to identify the determinants of the physical TGBp1–CP interactions that lead to PVX‐RNA encapsidation and RNA translational activation. The CP C‐terminal 10‐amino‐acid region and the TGBp1 NTPase/helicase motif IV were identified as being necessary for CP–TGBp1 interaction. Evidence is also presented to show that separate TGBp1 domains specify CP interaction and RNA translational activation.
RESULTS
Assembly of C‐terminal‐deleted PVX CP mutants with viral RNA and suppression of RNA translation
Formation of complexes and reassembly of virus‐like particles from PVX CP and viral RNA could be readily detected by different methods, including electron microscopy (EM), atomic force microscopy (AFM) and a sensitive indirect test based on the ability of PVX CP to suppress viral RNA translation in vitro (Karpova et al., 2006a,b).
The following CP forms were obtained and used for assembly and translation activation experiments: (1) native PVX CP (nCP); (2) full‐length recombinant PVX CP (rCP); recombinant C‐terminal‐deleted PVX CPs with 10, 18, 19, and 20 amino acids deleted: (3) C‐terdelrCPΔ10 or rCPΔ10; (4) C‐terdelrCPΔ18 or rCPΔ18; (5) C‐terdelrCPD19 or rCPD19; and (6) C‐terdelrCPΔ20 or rCPΔ20.
It should be noted that 6xHis‐tagged preparations of the PVX rCP were not functional in reassembly with viral RNA (data to be given in a forthcoming paper). Therefore, the removal of 6xHis‐tag from the rCP was performed with the help of a specific site for cleavage of fusion proteins with factor Xa protease (Jenny et al., 2003).
EM analyses showed that nCP isolated from the virions (Fig. 1a) as well as the full‐length rCP (Fig. 1b), the rCPΔ10 mutant (Fig. 1c) and the rCPΔ18 mutant (not shown in Fig. 1) could be assembled with PVX RNA. It should be emphasized that the CP mutants with a deletion longer than 18 amino acids (rCPΔ19 and rCPΔ20) could not produce virus‐like particles upon incubation with viral RNA (data not shown). Similar results were obtained when AFM was used to test the virus‐like particle assembly in analogous experiments (data not shown).
Figure 1.
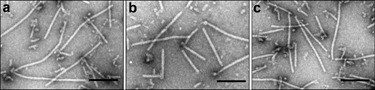
Electron micrographs of virus‐like particles assembled in vitro from PVX RNA and: native PVX CP (a); bacterially expressed 6xHis lacking full‐length rCP (b); C‐terdel mutant protein rCPΔ10 (c). The samples were stained with 2% uranyl acetate. Scale bar, 100 nm.
Next, the translation‐inhibiting activities of the native PVX nCP, bacterially expressed full‐length rCP and C‐terdelrCP mutants were compared in a cell‐free translation system. In accordance with our data mentioned above, the control experiments (Fig. 2, lanes 2–4) showed that non‐translatable products assembled from the native PVX nCP (lane 2) could be converted into a translatable form by binding of TGBp1 (lane 3) or by CP phosphorylation (lane 4). The low translational level observed in lane 2 (as well as lanes 6, 9 and 10) can be attributed to the fact that a minor portion of PVX RNA molecules remained free of CP and, therefore, were translatable (Karpova et al., 2006a). Similarly to native PVX, the particles assembled from PVX RNA and bacterially expressed full‐length rCP were non‐translatable (lane 6), but could be converted into a translatable form by TGBp1 binding (lane 7) or by CP phosphorylation (lane 8). Therefore, translation‐regulating features of the native CP isolated from PVX (Fig. 2, lanes 2–4) and those of the bacterially expressed rCP (Fig. 2, lanes 6–8) were functionally similar.
Figure 2.

Translational features of viral RNA encapsidated in virus‐like particles assembled from PVX RNA and different forms of C‐terminal deletion rCP mutants. Translation in WGE of: PVX RNA (lane 1); particles assembled from PVX RNA and native CP (lane 2); TGBp1 added to particles assembled as in ‘2’ (lane 3); particles assembled as in ‘2’ were phosphorylated by PKC (lane 4); particles assembled from PVX RNA and 6xHis‐taqgged CP (lane 5); particles assembled from PVX RNA and full‐length rCP (lane 6); TGBp1 added to particles assembled as in ‘6’ (lane 7); particles assembled as in ‘6’ were phosphorylated by PKC (lane 8); particles assembled from PVX RNA and C‐terdelrCPΔ10 (or rCPΔ18) (lane 9); TGBp1 was added to particles assembled as in ‘9’ (lane 10); particles assembled as in ‘9’ were phosphorylated by PKC (lane 11); particles assembled from PVX RNA and C‐terdelrCPΔ20 (lane 12). The arrow indicates the largest (165‐kDa) protein coded by the 5′‐proximal replicase gene of PVX.
Analogously to nCP and rCP, the C‐terdelrCPΔ10 mutant suppressed the PVX RNA translation (Fig. 2, lane 9) and the C‐terdelrCPΔ18 mutant inhibited translation as well (data not shown). By contrast, Fig. 2 (lane 12) illustrates the inability of the PVX rCPΔ20 mutant to suppress viral RNA translation. This was in agreement with our EM and AFM observations related to the inability of the C‐terdelrCP mutants with deletion longer than 18 amino acids to assemble virus‐like particles.
Furthermore, a control sample was presented (Fig. 2, lane 5) to confirm that translation‐inhibiting ability of the full‐length rCP was abolished by the presence of the N‐terminal 6xHis‐tag.
A key role of the PVX CP C‐terminal region in TGBp1‐dependent activation of viral RNA translation
Contrary to nCP and full‐length rCP, the PVX RNA encapsidated in the rCPΔ10 mutant (Fig. 2, lane 9) or rCPΔ18 (not shown) could not be translationally activated by incubation with TGBp1 (Fig. 2, lane 10), whereas, both of them could be converted into a translatable form by phosphorylation (e.g. see Fig. 2, lane 11). The inability of TGBp1 to activate translation of the virus‐like particles assembled from rCPΔ10 and rCPΔ18 could be due to the inability of TGBp1 binding to these mutant CPs. Alternatively, TGBp1 could bind, but was unable to ‘remodel’, i.e. to induce a linear destabilization of the PVX CP helix. The far‐Western blot analyses indicated that both native PVX nCP and bacterially expressed full‐length rCP (Fig. 3, lanes 1 and 2, respectively) interacted with the TGBp1 efficiently, whereas no TGBp1–rCPΔ10 complexes could be revealed (Fig. 3, lane 3). Thus, deletion of 10/18 C‐terminal amino acids of PVX CP abolished the ability of TGBp1 to bind to the virus‐like particles. It is reasonable to suggest that the PVX CP C‐terminal region contains the element(s) for TGBp1 molecules binding to one extremity of the polar PVX CP helix.
Figure 3.
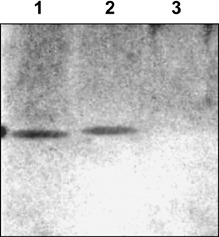
Far‐Western blot analysis of TGBp1–CP complex formation. 1, native PVX CP, 1 µg (control); 2, bacterially expressed full‐length rCP, 1 µg; 3, bacterially expressed C‐terdel mutant rCPΔ10, 1 µg. Separated in 8–20% SDS‐PAGE, CPs were transferred to nitrocellulose membrane and incubated with TGBp1 (5 µg/mL), rabbit anti‐TGBp1 antibodies and anti‐rabbit antibodies conjugated to horseradish peroxidase (Sigma); the reaction was visualized by the ECL system (Amersham Biosciences).
Examination of the functional activity of TGBp1 mutants
The 25‐kDa PVX TGBp1 is a multifunctional protein (for a review, see Verchot‐Lubicz, 2005). It has been found that TGBp1 contains NTPase/helicase domain and is related to helicases of superfamily I (SF‐I) (for a review, see Morozov and Solovyov, 2003). The PVX TGBp1 was shown to exhibit ATP/GTPase and RNA helicase activities in vitro (Kalinina et al., 1996, 2002). As already noted, TGBp1 can bind to one end and destabilize the PVX helical particles, converting them into a translatable form (Atabekov et al., 2000; , Karpova et al., 2006a). All these activities were examined in experiments with bacterially expressed 6xHis‐tagged TGBp1.
To understand further the mechanism of TGBp1–CP binding, a series of TGBp1 mutants were constructed. The arrangement of NTPase/helicase conservative motifs in the full‐length TGBp1 and the TGBp1 deletion mutants used in this work is schematically represented in Fig. 4a. Native PVX was incubated with the TGBp1 mutants in order to determine their ability: (1) to activate in vitro translation of encapsidated PVX RNA; (2) to produce the TGBp1–PVX complexes, which were revealed by immunoelectron microscopy (IEM) and far‐Western blot overlay analyses. The mutants presented in Fig. 4 could be subdivided into three types. (1) Only the full‐length TGBp1 (control, no. 19) and the mutant no.23 (S to N substitution in the GKS site of the NTPase domain) retained both activities: their capability for PVX binding and RNA translational activation. (2) Mutant nos. 2, 6, 15, 32 and 45 retained the PVX‐binding ability, but lost the ability to activate translation of encapsidated PVX RNA. Surprisingly, the ability of TGBp1 binding to PVX was not markedly affected by the size and location of a deletion (4, 5). IEM analyses (Fig. 5) indicated that TGBp1 molecules were located at the extremity of PVX virions. In a control mixture lacking TGBp1, the labelling with secondary gold‐conjugated antibodies was entirely absent (data not shown). The same was the case in a control mixture lacking antibodies to TGBp1. (3) Unlike all other mutants, the TGBp1 no. 11 lost both activities (4, 5). This result may be explained by assuming that the inability of mutant no. 11 to form complexes with PVX was due to the absence of motif IV from this protein. Remarkably, all other TGBp1 mutants containing this motif (including the smallest mutant no. 2) retained the ability to form the complexes with PVX CP (4, 5). It could be presumed that motif IV is essential for TGBp1–PVX binding. These data supported the hypothesis that binding of TGBp1 to the end of the virion did not depend markedly on the conformation of the TGBp1 mutant, whereas the ability of TGBp1 to destabilize the virion and to trigger PVX RNA translation was very sensitive and, therefore, could be abolished by any one of the deletions used (Fig. 4b). These data allowed us to hypothesize that the TGBp1 domains that specify CP interaction and translation activation are separate.
Figure 4.

Schematic representation of NTPase/helicase conservative motifs in TGBp1 deletion mutants (a) and comparative properties of TGBp1 mutants (b).
Figure 5.

Visualization by IEM of TGBp1 mutants binding to the end of PVX particles: (a) no. 19 (wt); (b) no. 2; (c) no. 23; (d) no. 45; (e) no. 11. PVX‐bound TGBp1 molecules indicated by arrows were revealed by gold‐labelled antiglobulin AbSEC. The size of gold particles was 12 nm. Scale bars represent 100 nm.
Presumably, a specific conformation of the full‐length TGBp1 was essential for PVX CP‐helix remodelling and activation of RNA translation. If this were the case, different factors capable of changing the TGBp1 conformation should abolish such activity. For instance, it could be presumed that phosphorylation of TGBp1 might serve as a factor capable of changing its conformation and activity. In order to test this proposal we phosphorylated full‐length TGBp1 (no. 19 in Fig. 4) using protein kinase C (PKC) or a mixture of casein kinases (CKI + CKII). It was found that phosphorylation of TGBp1 by PKC resulted in dramatic changes of antigenic specificity (Fig. 6a) and loss of its translation‐activating ability (cf. Fig. 6b, lanes 2 and 3); generally similar results were obtained with CKI + CKII (data not presented).
Figure 6.

Effect of phosphorylation on antigenic properties of TGBp1 (a) and its ability to activate the PVX RNA translation (b). (a) Immunoprecipitation with polyclonal TGBp1‐specific antibodies. The native TGBp1 or TGBp1 in situ phosphorylated by PKC (TGBp1‐P) was incubated with polyclonal antibodies to TGBp1 (IgG‐TGBp1); the precipitate was subjected to SDS‐PAGE. Purified TGBp1 taken as a control (lane 1); the TGBp1 precipitated with IgG‐TGBp1 (lane 2); the TGBp1‐P incubated with IgG‐TGBp1 (lane 3); control‐TGBp1 incubated with heterospecific antibodies (polyclonal antibodies to Tobacco Mosaic Virus) (lane 4). (b) Translation in WGE: native PVX (lane 1); PVX incubated with TGBp1 (lane 2) and PVX incubated with TGBp1‐P (lane 3).
In a series of experiments, possible in vitro complementation of TGBp1 translation activation ability was examined via co‐incubation in wheat germ extract (WGE) of PVX with TGBp1 mutants arranged in pairs (e.g. no. 11 + no. 15; no. 11 + no. 32; no. 15 + no. 45). No positive result was obtained in these experiments.
DISCUSSION
Previously we have shown that encapsidated PVX RNA could be converted into a translatable form in two ways: (1) by binding of the PVX‐coded TGBp1 to one end of the PVX virion (Atabekov et al., 2000) or to the end of non‐translatable STPs (Karpova et al., 2006a) containing the 5′‐end of the RNA and (2) by in situ phosphorylation of the PVX CP (Atabekov et al., 2001; Karpova et al., 2006a,b). Here we showed (Fig. 2) that, as with native PVX, the particles assembled from PVX RNA and nCP or bacterially expressed full‐length rCP were non‐translatable (lane 6) and could be converted into a translatable form by TGBp1 binding (lane 7) or via CP phosphorylation (lane 8). Taken together, these results and the EM observations indicated that functional characteristics of the full‐length rCP and of the nCP isolated from PVX were similar.
Apparently, removal of 18 C‐terminal amino acids from PVX CP molecules was critical for the CP–RNA interaction; no virus‐like particles could be assembled using C‐terminal deletion mutants rCPΔ19 and rCPΔ20.
Most significantly, translation activation by TGBp1 of the virus‐like particles assembled from C‐terdel mutants rCPΔ10 and rCPΔ18 was abolished. On the other hand, these particles were readily converted into a fully translatable form by in situ CP phosphorylation.
The precise mechanism of PVX CP–TGBp1 interaction is not known. It has been reported that the N‐terminus of PVX the CP is exposed on the virion surface (Baratova et al., 1992; Koenig and Torrance, 1986; Sober et al., 1988). Previously, we showed that the N‐terminal region of CP does not interact with TGBp1 and is not needed for PVX RNA translational activation (Atabekov et al., 2001).
Conversely, evidence is provided here that the C‐proximal N‐Ala‐Glu‐Ala‐Val‐Val‐Thr‐Leu‐Pro‐Pro‐Pro‐C region of PVX CP is essential for the TGBp1‐triggered activation of PVX translation (Fig. 2, lanes 9 and 10). It is reasonable to suggest that the 10‐amino‐acid C‐ter region may represent the site specifically recognized by TGBp1 molecules in terminal CP subunits of the virion for binding and PVX remodelling. The model proposed by Baratova et al. (1992) indicates that the C‐terminal domain of PVX CP is not accessible from the outer surface of the virus particle to the ‘hot’ tritium atoms. However, it should be borne in mind that the TGBp1 molecules bind only to one end of the PVX particle that contains the 5′‐end of the RNA (Karpova et al., 2006b) and there are 8.9 CP subunits per turn of the primary PVX CP helix (Parker et al., 2002). Apparently, fewer than ten of about 1300 CP subunits may be accessible for the TGBp1 molecules, which bind only to a certain domain of several CP subunits exposed at one end of the CP helix, being buried along the length of the virus particle.
In the next series of experiments, native PVX was incubated with TGBp1 mutants in order to determine their ability to produce the TGBp1–PVX complexes and to activate in vitro translation of PVX RNA. Surprisingly, the ability of TGBp1 binding to PVX was not markedly affected by the size and location of a deletion. The majority of the TGBp1 deletion mutants retained the PVX‐binding ability, whereas all of them lost the ability to activate translation of PVX RNA. However, the TGBp1–CP binding activity was preserved unless a conservative motif IV N‐Phe‐Tyr‐Leu‐Glu‐Thr‐Ser‐Phe‐Arg‐Val‐Pro‐C of TGBp1 (Morozov et al., 1999) was removed. Collectively, these results indicate that: (1) two events should be distinguished in the course of TGBp1‐mediated activation of PVX RNA translation. The first event (TGBp1–PVX binding) apparently does not depend critically on the TGBp1 conformation, but requires motif IV of the TGBp1 molecule, which could serve as a possible site involved in CP recognition upon TGBp1 binding with the CP helix. By contrast, a unique conformation of the full‐length TGBp1 seems to be essential for PVX CP helix remodelling and activation of RNA translation. (ii) The NTPase activity of TGBp1 is not needed for PVX remodelling. This hypothesis is in line with the PVX‐binding ability of mutant no. 23 (Fig. 4), which contained an S to N substitution in the GKS site of NTPase. Our previous data also indicated that the TGBp1–PVX complex is ATP‐independent (Atabekov et al., 2000). Therefore, the ATPase and helicase activities of TGBp1 are not required for the TGBp1 binding to the end of the PVX helix.
It was mentioned above that deletion of the C‐ter 18‐amino‐acid region from PVX CP led to the inhibition of the viral cell‐to‐cell movement (Fedorkin et al., 2001). This effect could be due to the inability of the TGBp1 movement protein to bind in vivo to the helically arranged transport form of virus infection (presumably, the virions and/or the STPs) comprising rCPΔ18. Evidence was provided to suggest that phosphorylation of the parental PVX particles by cytoplasmic PKs in primary inoculated cells renders PVX RNA translatable in vivo, whereas translational activation of progeny virions destined for plasmodesmata trafficking is triggered by TGBp1 (Atabekov et al., 2001). It is possible that as CP phosphorylation is the only means of translational activation, given that TGBp1 lost the ability to bind to the CP helix, it will be not favourable to PVX movement.
EXPERIMENTAL PROCEDURES
PVX preparations
PVX (Russian strain) was isolated from systemically infected Datura stramonium L. plants by differential centrifugation.
RNA and CP of PVX preparations
To obtain the PVX CP preparations, the method of salt deproteinization (Goodman, 1975) was used. RNA was isolated with phenol and analysed by 1% agarose gel electrophoresis.
Expression and purification of recombinant proteins
All PVX TGBp1 and PVX CP variants were RT‐PCR‐amplified using PVX Russian strain genomic RNA as a template, and PCR products were cloned as BamHI–XbaI fragments at the corresponding sites into pQE30 expression vector (Qiagen Hilden, Germany). The constructs were transformed into Escherichia coli M15[pREP4] cells (Qiagen) following standard protocols. Resulting cultures were grown at 37 °C with vigorous shaking in the presence of 100 µg/mL ampicillin and 25 µg/mL kanamycin. At an OD600 of 0.6, recombinant protein expression was induced by adding isopropyl‐β‐d‐thiogalactopyranoside (IPTG) to a final concentration of 1 mm. The cultures were grown further for 4 h at 37 °C, and cells were harvested by centrifugation. Bacterially expressed proteins were isolated by affine chromatography on Ni‐NTA agarose as described previously (Karpova et al., 1997).
Assembly of viral RNA with native and bacterially expressed CP
The incubations was performed in 10–20 µL of 10 mm Tris/HCl, pH 7.6, at 20 °C as described (Karpova et al., 2006a,b) at the molar ratio RNA/CP of 1 : 700. Under these conditions a minor portion of PVX RNA molecules remain free of CP and, therefore, are translatable (Karpova et al., 2006a). The TGBp1 was added at the molar ratio PVX RNA/TGBp1 of 1 : 100.
In vitro translation
Cell‐free translation in WGE was carried out in the presence of [35S]methionine for 60 min at 25 °C according to the manufacturer's protocol (Promega). The concentration of RNA was 40 µg/mL. Radioactive translation products were analysed by SDS‐PAGE and localized by autoradiography (Rodionova et al., 2003).
Electron microscopy
Specimens were prepared and examined as reported previously (Karpova et al., 2006a).
Immunoelectron microscopy
The purified virion preparations were adsorbed to Formvar film attached to 200‐mesh nickel EM grids. Specimens were blocked in 1% bovine serum albumin in PBS for 20 min and floated on 0.1 µg/µL TGBp1 for 20 min. Grids were washed with PBS, incubated with polyclonal antibodies to TGBp1 (AbTGBp1) and then with secondary gold‐conjugated antibodies (AbSEC). The size of gold particles was 12 nm. After immunolabelling, adsorbed material was visualized by staining with 2% aqueous uranyl acetate solution, the grids were washed with distilled water and air‐dried. In control (lacking TGBp1) mixtures, labelling of complexes with AbSEC was entirely absent.
Phosphorylation of TGBp1
Phosphorylation of TGBp1 was performed by PKC (Promega) as described previously by Karpova et al. (1999) with some modifications. The reaction mixture (10 µL) contained TGBp1 (1 µg/µL); 5 mL of 5× activation buffer (1.6 mg/mL phosphatidylserine, 0.16 mg/µL diacylglycerol, 100 mm Tris/HCl, pH 7.5, 50 mm MgCl2); 5 µL of 5× coactivation buffer (1.25 mm EGTA, 2 mm CaCl2, 0.5 mg/mL BSA); 0.5 µL of [γ‐32P]ATP (5000 Ci/mmol, 400 MBq/mL); and 0.5 µL of PKC (Promega). The reaction was incubated at room temperature for 15 min. Phosphorylation of TGBp1 by casein kinases I and II (Promega) was done in strict compliance with the Promega protocol.
Immunoprecipitation of TGBp1 and phosphorylated TGBp1 with polyclonal TGBp1‐specific antibodies
Immunoprecipitation was carried out as described by Atabekov et al. (2001). The control or phosphorylated TGBp1 preparations were incubated with polyclonal antibodies to TGBp1 (IgG‐TGBp1) or with heterospecific antibodies (polyclonal antibodies to Tobacco Mosaic Virus) in PBS for 2 h at 20 °C; then Protein A Sepharose (1–2 mg/mL) was added and incubation was continued overnight. The precipitates obtained by centrifugation at 12 000 g for 10 min were subjected to SDS‐PAGE.
Far Western blots
Far Western blotting was performed as reported by Hall (2004) (details at http://www.utoronto.ca/krause/Farwestern.html). Following separation in 8–20% SDS‐PAGE, CPs were transferred to nitrocellulose membrane and incubated with TGBp1 (5 µg/mL), rabbit anti‐TGBp1 antibodies and anti‐rabbit antibodies conjugated to horseradish peroxidase (Sigma); the reaction was visualized via the ECL system (Amersham Biosciences).
ACKNOWLEDGEMENTS
We are grateful to Professor I. V. Yaminsky for AFM analyses of the PVX assembly. This work was funded in part by the Federal Agency of Science and Innovation (FASI) (contract no. 02.512.11.2141 and no. 02.512.11.2103), and the Russian Foundation for Basic Research (Grant 06‐04‐48‐176).
REFERENCES
- Allison, A.V. and Shalla, T.A. (1974) The ultrastructure of local lesions induced by potato virus X. A sequence of cytological events in the course of infection. Phytopathology, 64, 784–793. [Google Scholar]
- Atabekov, J.G. , Rodionova, N.P. , Karpova, O.V. , Kozlovsky, S.V. and Poljakov, V.Y. (2000) The movement protein‐triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology, 271, 259–263. [DOI] [PubMed] [Google Scholar]
- Atabekov, J.G. , Rodionova, N.P. , Karpova, O.V. , Kozlovsky, S.V. , Novikov, V.K. and Arkhipenko, M.V. (2001) Translational activation of encapsidated potato virus X RNA by coat protein phosphorylation. Virology, 286, 466–474. [DOI] [PubMed] [Google Scholar]
- Baratova, L.A. , Fedorova, N.V. , Dobrov, E.N. , Lukashina, E.V. , Kharlanov, A.N. , Nasonov, V.V. , Serebryakova, M.V. , Kozlovsky, S.V. , Zayakina, O.V. and Rodionova, N.P. (2004) N‐Terminal segment of potato virus X coat protein subunits is glycosylated and mediates formation of a bound water shell on the virion surface. Eur. J. Biochem. 271, 3136–3145. [DOI] [PubMed] [Google Scholar]
- Baratova, L.A. , Grebenshchikov, N.I. , Dobrov, E.N. , Gedrovich, A.V. , Kashirin, I.A. , Shishkov, A.V. , Efimov, A.V. , Jarvekulg, L. , Radavsky, Y.L. and Saarma M. (1992) The organization of potato virus X coat proteins in virus particles studied by tritium planigraphy and model building. Virology, 188, 175–180. [DOI] [PubMed] [Google Scholar]
- Batten, J.S. , Yoshinary, S. and Hemenway, C. (2003) Potato virus X: a model system for virus replication, movement and gene expression. Mol. Plant Pathol. 4, 125–131. [DOI] [PubMed] [Google Scholar]
- Chapman, S.N. , Hills, G. , Watts, J. and Baulcombe, D.C. (1992) Mutational analysis of the coat protein gene of potato virus X. Effect of virion morphology and viral pathogenicity. Virology, 191, 223–230. [DOI] [PubMed] [Google Scholar]
- Fedorkin, O. , Solovyev, A. , Yelina, N. , Zamyatnin, A. Jr , Zinovkin, R. and Makinen, K. , Schiemann, J. and Yu Morozov, S. (2001) Cell‐to‐cell movement of potato virus X involves distinct functions of the coat protein. J. Gen. Virol. 82, 449–458. [DOI] [PubMed] [Google Scholar]
- Forster, R.L. , Beck, D.L. , Guilford, P.J. , Voot, D.M. , Van Dolleweerd, C.J. and Andersen, M.T. (1992) The coat protein of white clover mosaic potexvirus has a role in facilitating cell‐to‐cell transport in plants. Virology, 191, 480–484. [DOI] [PubMed] [Google Scholar]
- Goodman, R.M. (1975) Reconstitution of potato virus X in vitro. I. Properties of the dissociated protein structural subunits. Virology, 68, 287–298. [DOI] [PubMed] [Google Scholar]
- Hall, R.A. (2004) Studying protein–protein interactions via blot overlay or Far Western blot. Methods Mol. Biol. 261, 167–174. [DOI] [PubMed] [Google Scholar]
- Huisman, M.J. , Linthorst, H.J. , Ju, H.‐J. , Bol, J.F. and Cornelissen, J.C. (1988) The complete nucleotide sequence of Potato virus X and its homologies at the amino acid level with various plus‐stranded RNA viruses. J. Gen. Virol. 69, 1789–1798. [DOI] [PubMed] [Google Scholar]
- Jenny, R.J. , Mann, K.G. and Lundblad, R.L. (2003) A critical review of the methods for cleavage of fusion proteins with thrombin and factor Xa. Protein Expr. Purif. 31, 1–11. [DOI] [PubMed] [Google Scholar]
- Kalinina, N.O. , Fedorkin, O.V. , Samuilova, O.V. , Maiss, E. , Korpela, T. , Morozov, S.Y. and Atabekov, J.G. (1996) Expression and biochemical analyses of the recombinant potato virus X 25K movement protein. FEBS Lett. 397, 75–78. [DOI] [PubMed] [Google Scholar]
- Kalinina, N.O. , Rakitina, D.A. , Solovyev, A.G. , Schiemann, J. and Morozov, S.Y. (2002) RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology, 296, 321–329. [DOI] [PubMed] [Google Scholar]
- Karpova, O.V , Rodionova, N.P , Ivanov, K.I , Kozlovsky, S.V , Dorokhov, Y.L and Atabekov, J.G. (1999) Phosphorylation of tobacco mosaic virus movement protein abolishes its translation repressing ability. Virology, 261, 20–24. [DOI] [PubMed] [Google Scholar]
- Karpova, O.V. , Arkhipenko, M.V. , Zayakina, O.V. , Nikitin, N.A. , Kiselyova, O.I. , Kozlovsky, S.V. , Rodionova, N.P. and Atabekov, J.G. (2006b) Regulation of RNA translation in Potato virus X RNA‐coat protein complexes: the key role of the N‐terminal segment of the protein. Mol. Biol. (Russia), 4, 628–634. [Google Scholar]
- Karpova, O.V. , Ivanov, K.I. , Rodionova, N.P. , Dorokhov, Y.L. and Atabekov, J.G. (1997) Nontranslatability and dissimilar behavior in plants and protoplasts of viral RNA and movement protein complexes formed in vitro. Virology, 230, 11–21. [DOI] [PubMed] [Google Scholar]
- Karpova, O.V. , Zayakina, O.V. , Arkhipenko, M.V. , Sheval, E.V. , Kiselyova, O.I. , Poljakov, V.Y. , Yaminsky, I.V. , Rodionova, N.P. and Atabekov, J.G. (2006a) Potato virus X mediated assembly of single‐tailed ternary complex ‘coat protein‐RNA – movement protein’. J. Gen. Virol. 87, 2731–2740. [DOI] [PubMed] [Google Scholar]
- Koenig, R. and Torrance, L. (1986) Antigenic analysis of potato virus X by means of monoclonal antibodies. J. Gen. Virol. 67, 2145–2151. [DOI] [PubMed] [Google Scholar]
- Lough, T.G. , Netzler, N.E. , Emerson, S.J. , Sutherland, P. , Carr, F. , Beck, D.L. , Lucas, W.J. and Forster, R.L. (2000) Cell‐to‐cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant–Microbe Interact. 13, 962–974. [DOI] [PubMed] [Google Scholar]
- Lough, T.G. , Shash, K. , Xoconostle‐Cazares, B. , Hofstra, K.R. , Beck, D.L. , Balmori, E. , Forster, R.L. and Lucas, W.J. (1998) Molecular dissection of the mechanism by which potexvirus triple gene block proteins mediate cell‐to‐cell transport of infectious RNA. Mol. Plant–Microbe Interact. 11, 801–814. [Google Scholar]
- Morozov, S.Y. , Solovyev, A.G. , Kalinina, N.O. , Fedorkin, O.N. , Samuilova, O.V. , Schiemann, J. and Atabekov, J.G. (1999) Evidence for two nonoverlapping functional domains in the potato virus X 25K movement protein. Virology, 260, 55–63. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. and Solovyev, A.G. (2003) Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84, 1351–1366. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J. , Roberts, A.J. , Roberts, I.M. , Prior, D.A.M. and Santa Cruz, S. (1996) Viral coat protein is targeted but does not gate, plasmodesmata during cell‐to‐cell movement of potato virus X. Plant J. 10, 805–813. [Google Scholar]
- Parker, L. , Kendall, A. and Stubbs, G. (2002) Surface features of potato virus X from fiber diffraction. Virology, 300, 291–295. [DOI] [PubMed] [Google Scholar]
- Rodionova, N.P. , Karpova, O.V. , Kozlovsky, S.V. , Zayakina, O.V. , Arkhipenko, M.V. and Atabekov, J.G. (2003) Linear remodeling of helical virus by movement protein binding. J. Mol. Biol. 333, 565–572. [DOI] [PubMed] [Google Scholar]
- Santa Cruz, S. , Roberts, A.G. , Prior, D.A. , Chapman, S. and Oparka K.J. (1998) Cell‐to‐cell and phloem‐mediated transport of potato virus X: the role of virions. Plant Cell, 10, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin, K.G. , Morozov, S.Y. , Kraev, A.S. , Rozanov, M.M. , Chernov, B.K. and Atabekov, J.G. (1988) Conserved and variable elements in RNA genomes of potexviruses. FEBS Lett. 240, 33–40. [DOI] [PubMed] [Google Scholar]
- Sober, J. , Jarvekulg, L. , Toots, I. , Rodavsky, J. , Villems, R. , and Saarma, M. (1988) Antigenic characterization of potato virus X with monoclonal antibodies. J. Gen. Virol. 69, 1799–1807. [Google Scholar]
- Tollin, P and Wilson, H.R. (1988) Particle structure In: The Plant Viruses, Vol. 4. The filamentous Plant Viruses (Milne R.C., ed.), pp. 51–83. New York: Plenum Press. [Google Scholar]
- Tozzini, A.C. , Ek, B. , Palva, E.T. and Hopp, H.E. (1994) Potato virus X coat protein: a glycoprotein. Virology, 202, 651–658. [DOI] [PubMed] [Google Scholar]
- Verchot, J. , Angell, S. and Baulcombe, G.C. (1998) In vitro translation of the triple gene block of Potato virus X requires two subgenomic mRNAs. J. Virol. 72, 8316–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. (2005) A new cell‐to‐cell transport model for potexviruses. Mol. Plant–Microbe Interact. 18, 283–290. [DOI] [PubMed] [Google Scholar]


