SUMMARY
Maize (Zea mays L.) is a major crop susceptible to Aspergillus flavus infection and subsequent contamination with aflatoxins, the potent carcinogenic secondary metabolites of the fungus. Protein profiles of maize genotypes resistant and susceptible to A. flavus infection and/or aflatoxin contamination have been compared, and several resistance‐associated proteins have been found, including a pathogenesis‐related protein 10 (PR10). In this study, RNA interference (RNAi) gene silencing technology was employed to further investigate the importance of PR10. An RNAi gene silencing vector was constructed and introduced into immature Hi II maize embryos through both bombardment and Agrobacterium infection procedures. PR10 expression was reduced by 65% to more than 99% in transgenic callus lines from bombardment. The RNAi‐silenced callus lines also showed increased sensitivity to heat stress treatment. A similar reduction in PR10 transcript levels was observed in seedling leaf and root tissues developed from transgenic kernels. When inoculated with A. flavus, RNAi‐silenced mature kernels produced from Agrobacterium‐mediated transformation showed a significant increase in fungal colonization and aflatoxin production in 10 and six, respectively, of 11 RNAi lines compared with the non‐silenced control. Further proteomic analysis of RNAi‐silenced kernels revealed a significant reduction in PR10 production in eight of 11 RNAi lines that showed positive for transformation. A significant negative correlation between PR10 expression at either transcript or protein level and kernel aflatoxin production was observed. The results indicate a major role for PR10 expression in maize aflatoxin resistance.
INTRODUCTION
The infection of maize (Zea mays L.) by Aspergillus flavus and the subsequent accumulation of the toxic and highly carcinogenic secondary metabolites, aflatoxins (Squire, 1981), are serious agricultural problems, especially under drought conditions (Diener et al., 1987; Payne, 1998). Aflatoxin contamination significantly reduces the value of grain and poses health hazards to humans (Hsieh, 1989; Payne, 1998) and domestic animals (Nichols, 1983). In the past two decades, maize genotypes resistant to aflatoxin production have been identified through field screening (Campbell and White, 1995; Widstrom et al., 1987). However, the lack of identifiable molecular markers in these resistant lines has hindered the incorporation of resistance into lines with good agronomic characteristics.
Several studies have demonstrated a role for kernel proteins in aflatoxin resistance (Brown et al., 1995; Chen et al., 1998; Guo et al., 1997; Huang et al., 1997b), especially the role of constitutively expressed proteins as a first layer of defence (Chen et al., 2001). A proteomics approach has been employed to identify constitutively expressed resistance‐associated proteins from maize endosperm and embryo tissues (2002, 2007). Over a dozen protein spots, either unique or significantly up‐regulated in resistant lines, have been identified through these comparisons. These proteins can be grouped into three categories based on peptide sequence homology: storage, stress‐responsive, or antifungal proteins (2002, 2007).
One of the identified antifungal proteins is a pathogenesis‐related protein 10 (PR10) (Chen et al., 2006). PR10 proteins are a family of intracellular, acidic proteins with a molecular mass of 16–19 kDa; they lack a signal peptide and are resistant to proteases (van Loon et al., 1994; Warner et al., 1992). The PR10 gene has been identified in a number of other plant species, including asparagus (Warner et al., 1992), lily (Huang et al., 1997a) and rice (Midoh and Iwata, 1996). Some members of this family also share sequence similarities to major food allergens of celery (Breiteneder et al., 1993) and apple (Hoffmann‐Sommergruber et al., 1997), and to a ginseng ribonuclease (Moiseyev et al., 1997). The biological function(s) of this class of proteins remains unclear, although some plant PR10 proteins possess ribonuclease and/or antimicrobial activities (Bantignies et al., 2000; Chen et al., 2006; Liu et al., 2006; Park et al., 2004).
Maize PR10 gene expression is induced during kernel development in an aflatoxin‐resistant line, GT‐MAS : gk, but not in susceptible Mo17, in response to fungal inoculation (Chen et al., 2006). Other evidence linking PR10 to host resistance has also been reported. A barley PR10 gene was specifically induced in resistant cultivars on infection by Rhynchosporium secalis, but not in near‐isogenic susceptible plants (Steiner‐Lange et al., 2003). In cowpea, a PR10 homologue was specifically up‐regulated in resistant epidermal cells inoculated with the rust fungus Uromyces vignae Barclay (Mould et al., 2003). A PR10 transcript was also induced in rice during infection by Magnaporthe grisea (McGee et al., 2001). Other studies also suggest a role for PR10 in stress tolerance: overexpression of a pea PR10 (ABR17) in Arabidopsis enhanced its response to salt stress (Krishnaswamy et al., 2008; Srivastava et al., 2006). In addition, PR10 also interacts with phytohormones, such as cytokinins and brassinosteroids (Mogensen et al., 2002), and elevated levels of cytokinins and increased lateral branching and early flowering were observed in ABR17‐transgenic A. thaliana (Srivastava et al., 2007), suggesting a possible role in signal transduction and cell development.
To further define the particular role of the PR10 protein, to provide further evidence for a correlation of PR10 with resistance and to establish the potential use of PR10 for marker‐assisted breeding or for the enhancement of aflatoxin resistance through genetic engineering, an RNA interference (RNAi) approach was employed in this study to knock down PR10 expression in maize. Interference of double‐stranded RNA (dsRNA) with the expression of specific genes has been widely described (Fire et al., 1998; Gura, 2000). This post‐transcriptional gene silencing is a sequence‐specific RNA degradation process triggered by a dsRNA, providing a powerful tool to study functions of unknown genes in many organisms (Wesley et al., 2001).
The objectives of this study were to silence the endogenous expression of PR10 in maize using RNAi through genetic engineering, and to examine the changes in A. flavus colonization and aflatoxin production of the resulting PR10‐silenced transgenic kernels. An RNAi vector capable of producing a dsRNA was constructed through Gateway homologous recombination (Hartley et al., 2000), and was introduced into immature maize embryos through both bombardment and Agrobacterium infection procedures. The transgenic callus lines from bombardment showed a 65%–99% reduction in PR10 expression. RNAi‐silenced transgenic kernels, in which fungal colonization and aflatoxin accumulation were significantly increased, also showed a significant reduction in PR10 protein level, suggesting a direct role for PR10 in maize aflatoxin resistance.
RESULTS
Construction of a PR10 RNAi vector
The RNAi vector construction scheme is outlined in Fig. 1. The correct assembly of pBS‐PR10‐RNAi, containing the inverted repeat of the PR10 5′ arm and PR10 3′ arm separated by the PR10 intron, was first verified through restriction enzyme digestion. The clones demonstrating correct restriction patterns were confirmed through DNA sequencing. The DNA region containing the PR10‐RNAi cassette was subcloned into pTF102 and verified through digestion and DNA sequencing. This construct is capable of producing a 371‐bp PR10 dsRNA transcript with an 81‐bp single‐strand loop in the middle, once the transcript is processed in the host plant.
Figure 1.
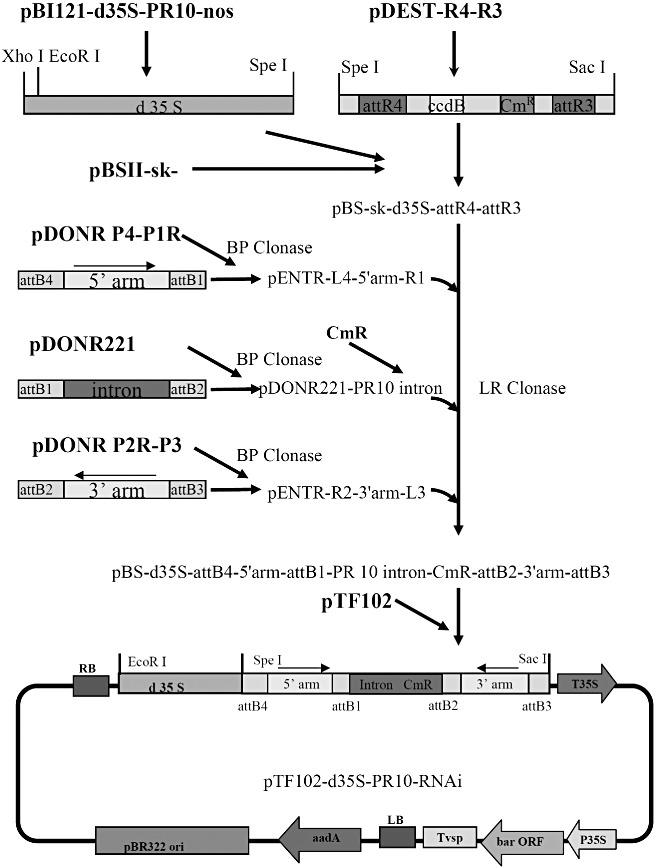
Construction scheme of pathogenesis‐related protein 10 (PR10) gene silencing vector. The double 35S promoter and the DNA region containing the attR4‐attR3 cassette were amplified by polymerase chain reaction (PCR) separately from their corresponding vectors, and cloned into the corresponding restriction sites in pBluesript II SK‐ vector. The DNA regions corresponding to the PR10 5′arm, intron and 3′ arm were amplified by PCR with primers containing unique homologous recombination sites, cloned into their corresponding entry vectors. A chloramphenicol resistance gene (CmR) selection marker was then inserted into the middle of the PR10 intron before the LR clonase reaction to assemble the RNAi cassette into the pBluescript vector to produce the pBS‐d35S‐attB4‐5′arm‐attB1‐PR10 intron‐CmR‐attB2‐3′arm‐attB3 vector (named pBS‐PR10‐RNAi). The RNAi cassette was then cloned into the pTF102 vector through ligation to produce the final pTF102‐PR10‐RNAi vector.
Efficiency of gene silencing in callus lines selected from bombardment
Fifteen callus lines, each representing an independent transformation event, were produced from transformation through particle bombardment and tested for the efficiency of PR10 gene silencing. Eleven of the 15 callus lines were confirmed to be positive for the transgene through polymerase chain reaction (PCR) (Fig. 2A). The remaining four lines (1, 3, 5 and 15) were negative for the transgene. Real‐time PCR analysis revealed that PR10 transcript levels varied among the positively transformed callus lines, but were all significantly lower than the level in either the non‐transformed control Neg 43 (data not shown) or the callus line 5, which was negative for PR10. When the level was normalized to that of callus line 5, the reduction in PR10 expression ranged from 65% (line 13) to 99% (line 10) (Fig. 2B).
Figure 2.
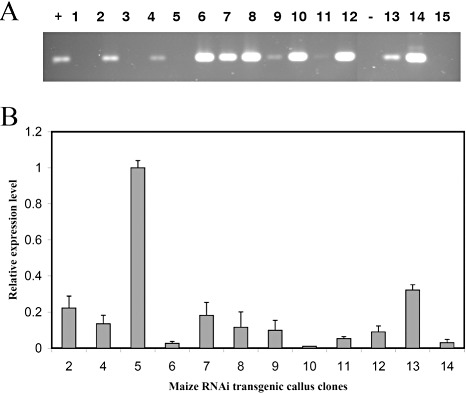
Analysis of transgenic maize callus clones from particle bombardment. (A) Polymerase chain reaction (PCR) verifying the transformation of pathogenesis‐related protein 10 (PR10) RNAi vector in callus clones. The numbers refer to individual callus clones. +, positive control using PR10 RNAi plasmid DNA; −, negative control using genomic DNA isolated from non‐transformed callus clone Neg 43. (B) Real‐time PCR determining the level of PR10 expression in callus clones. The level is relative to that of the control clone 5, which is negative for transformation and had the same level of expression as in Neg 43. The bars with the same letters are not significantly different in their expression levels at P= 0.05.
Responses of transgenic callus lines to heat stress
The effect of reduced PR10 expression in transgenic callus lines (4, 6, and 8) on heat stress tolerance was examined. These callus lines grew more slowly than their corresponding non‐treated callus tissues. The weight changes for the heat‐stressed callus lines 4, 6 and 8 after 2 weeks were 26.9%, 21.8% and 33.7%, respectively, compared with their corresponding controls, which were 53.4%, 59.2% and 57.7%, respectively (Table 1). However, heat stress did not cause significant growth reduction in the non‐transformed control callus line (Neg 43) (Table 1).
Table 1.
Effect of heat stress treatment on the growth of pathogenesis‐related protein 10 (PR10) RNAi transgenic callus lines.
| Callus line | Weight change (%) (±standard deviation)* | |
|---|---|---|
| Control (28 °C) | Heat stressed (37 °C) | |
| Neg 43 | 50.3 ± 6.5 | 51.4 ± 4.0 |
| 4 | 53.4 ± 9.1 | 26.9 ± 6.3† |
| 6 | 59.2 ± 7.6 | 21.8 ± 6.8† |
| 8 | 57.7 ± 6.8 | 33.7 ± 3.7† |
The percentage weight change was the difference between the final callus weight 2 weeks after treatment and the initial callus weight divided by the initial callus weight.
Indicates significant reduction in growth compared with the same callus line kept at 28 °C.
Production of transgenic kernels through Agrobacterium infection
Forty‐five transgenic lines representing 20 independent transformation events (one to three plants/event) were regenerated and pollinated with B73 to produce the mature kernels. The number of mature kernels produced per ear varied from as many as 201 to as few as one. The average weight of mature transgenic kernels also differed significantly from one ear to another, ranging from 0.305 g to 0.022 g, with a mean value of 0.175 g. All of these kernels germinated and developed into normal seedlings after planting despite variations in kernel weight and size.
Evaluation of transgenic kernels for transformation and PR10 expression
Twelve transgenic lines varying in kernel weight and size were investigated further, together with two Hi II controls (HT40917‐1, HT40917‐3) (Table 2). Genomic DNA and total RNA were isolated from the young leaf and root tissues of germinated seedlings to confirm the transformation and to determine the level of PR10 expression. All transformants except A44S1‐15‐3 were confirmed to be positive for the presence of the transgene using PCR (Table 3). PR10 expression in root tissue was significantly higher than that in leaf tissue in both controls and transgenic lines, ranging from 16‐fold to over 1000‐fold. The PR10 transcript level varied greatly among the transgenic lines. In leaf tissue, the PR10 transcript level was significantly reduced in the following RNAi transgenic lines, A44S1‐3‐1 (by 81.3%), A44S1‐3‐4 (by 88.1%), A44S1‐5‐2 (by 64.6%), A44S1‐5‐3 (by 61.8%) and A44S1‐35‐4 (by 86.2%), compared with that in controls (HT40917‐1 and HT40917‐3) (Table 3). In root tissue, the following RNAi transgenic lines showed a significant reduction in PR10 expression compared with the controls: A44S1‐1‐1 (by 87.4%), A44S1‐3‐4 (by 85.5%), A44S1‐5‐3 (by 64.4%), A44S1‐35‐3 (by 95.8%), A44S1‐35‐4 (by 58.7%) and A44S1‐42‐1 (by 80.7%). The degree of reduction in PR10 expression in leaf and root tissue was not always the same within each line. Of the 12 lines examined, only three lines (A44S1‐3‐4, A44S1‐5‐3 and A44S1‐35‐4) showed a significant reduction in PR10 expression in both leaf and root tissues. Line A44S1‐15‐3 (negative for the transgene) showed a level of PR10 expression in leaf tissue similar to that in controls, whereas, in root tissue, the level was higher than that in controls. In addition, PR10 expression in A44S1‐1‐4, A44S1‐6‐1 and A44S1‐36‐1 was similar to that in controls in both tissues (Table 3).
Table 2.
Summary of number of kernels produced per ear and mean dry kernel weight at harvest of 12 putative PR10 RNAi transgenic maize lines.
| Line name | Number of kernels per ear | Kernel weight (g)* |
|---|---|---|
| A44S1‐1‐1 | 80 | 0.155 cd |
| A44S1‐1‐4 | 132 | 0.155 cd |
| A44S1‐3‐1 | 149 | 0.133 d |
| A44S1‐3‐4 | 156 | 0.172 c |
| A44S1‐5‐2 | 215 | 0.139 d |
| A44S1‐5‐3 | 201 | 0.139 d |
| A44S1‐6‐1 | 88 | 0.167 c |
| A44S1‐15‐3 | 73 | 0.239 a |
| A44S1‐35‐3 | 102 | 0.141 d |
| A44S1‐35‐4 | 104 | 0.150 cd |
| A44S1‐36‐1 | 127 | 0.194 b |
| A44S1‐42‐1 | 70 | 0.222 a |
| HT40917‐1† | 174 | 0.159 cd |
| HT40917‐3† | 95 | 0.168 c |
Average kernel weight was based on the measurement of 30 kernels, except for those lines with less than 30 kernels. Means within the column followed by the same letter were not significantly different based on Waller–Dunncan K‐ratio t‐test (at P= 0.05).
HT40917‐1 and HT40917‐3 are non‐transgenic Hi II plants pollinated with B73. They were used as negative controls.
Table 3.
Evaluation of 12 putative transgenic maize lines for transformation and PR10 expression at transcript and protein levels.
| Line name | Transformation* | PR10 expression in leaf†,‡ | PR10 expression in root†,‡ | PR10 level in mature kernels§ | ||
|---|---|---|---|---|---|---|
| ΔCt | Changes (%) | ΔCt | Changes (%) | |||
| A44S1‐1‐1 | Y | 19.250 cd | −47.8 | 15.905 b | −87.4 | −11.2% |
| A44S1‐1‐4 | Y | 17.480 e | 78.2 | 12.752 efg | 11.9 | −30.3% |
| A44S1‐3‐1 | Y | 20.735 ab | −81.3 | 13.705 de | −42.2 | −78.8%¶ |
| A44S1‐3‐4 | Y | 21.380 a | −88.1 | 15.697 b | −85.5 | −65.4%¶ |
| A44S1‐5‐2 | Y | 19.813 bc | −64.6 | 13.475 def | −32.1 | −85.1%¶ |
| A44S1‐5‐3 | Y | 19.703 bc | −61.8 | 14.407 cd | −64.4 | −61.3%¶ |
| A44S1‐6‐1 | Y | 18.320 de | −0.5 | 13.585 def | −37.2 | −39.2% |
| A44S1‐15‐3 | N | 17.053 e | 139.5 | 11.185 h | 231.7 | −6.5% |
| A44S1‐35‐3 | Y | 19.038 cd | −39.5 | 17.480 a | −95.8 | −75.7%¶ |
| A44S1‐35‐4 | Y | 21.173 a | −86.2 | 14.190 cd | −58.7 | −63.2%¶ |
| A44S1‐36‐1 | Y | 19.228 cd | −49.1 | 13.670 de | −40.8 | −64.5%¶ |
| A44S1‐42‐1 | Y | 19.255 cd | −47.9 | 15.287 bc | −80.7 | −84.2%¶ |
| HT40917‐1 | N | 18.425 de | −7.4 | 12.912 efg | 0.0 | 0.0 |
| HT40917‐3 | N | 18.210 de | 7.4 | 12.922 efg | −0.5 | ND |
Transformation was verified through polymerase chain reaction (PCR) with PR10if (GTTCAACTTCACCTCAG G) and PR10ir (AAGCTGAACGGCATGACT) primers corresponding to the chloramphenical resistance gene (CmR) region spanned by the intron using the genomic DNA isolated from seedling leaves of the corresponding transgenic lines. Kernels from non‐transgenic Hi II plants were used as a negative control (HT40917‐1). ‘Y’ and ‘N’ indicate positive and negative for transgene, respectively.
The PR10 transcript level was determined using real‐time PCR with 18S as an internal normalizer. The percentage change in target gene is calculated as the difference in relative expression between RNAi‐silenced transgenic and control, expressed as a percentage of the control. The relative expression level is calculated on the basis of the threshold cycle (Ct) values of the target gene and the 18S normalizer as follows: relative expression level = power (2, ΔCt), where ΔCt=Ct (18S rRNA)−Ct (target). The data presented here are means combined from two experiments.
Means within the column followed by the same letter were not significantly different based on Waller–Dunncan K‐ratio t‐test (at P= 0.05).
PR10 protein reduction was determined using Progenesis gel analysis software based on four gels (two repeated experiments with two replicated gels each).
Indicates the reduction was significant compared with the control (HT40917‐1) based on least‐significant difference (P= 0.05). ND, not determined.
Changes in PR10 protein production in transgenic kernels
The level of PR10 protein production in mature F1 kernels of control HT40917‐1 and 12 transgenic lines (including one that was negative for transgene) was examined using two‐dimensional gel electrophoresis. A significant reduction (≥60%) in the putative PR10 protein level was observed in eight RNAi transgenic lines, with an average reduction of 72.1% (Table 3). The remaining three RNAi transgenic lines showed a reduction in PR10 production in the range 11.2%–39.2% of the control. The line (A44S1‐15‐3) (negative for the PR10 gene) showed an average of a 6.5% reduction in PR10 level (Fig. 3). In order to confirm that the putative protein spot was the PR10 protein, this putative PR10 protein spot (spot ‘d’ in Fig. 3) was recovered from preparative two‐dimensional gels of A44S1‐1‐1 and A44S1‐6‐1 lines, together with several other protein spots (‘a’, ‘b’, ‘c’, ‘e’ and ‘f’ in Fig. 3) close by, which also showed changes in protein levels in some of the RNAi lines. The peptide sequences and sequence homology analysis confirmed that spot ‘d’ was PR10, as was part of spot ‘e’.
Figure 3.
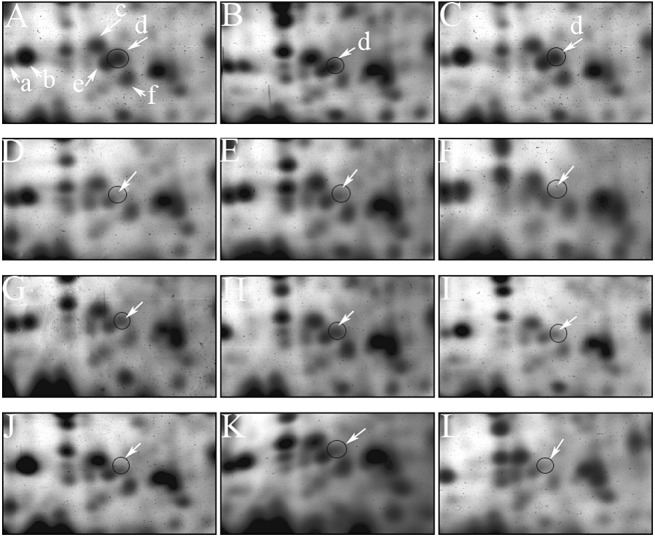
Protoemic confirmation of pathogenesis‐related protein 10 (PR10) levels in RNAi‐silenced transgenic maize kernels compared with non‐silenced control (HT40917‐1) kernels. Sections of two‐dimensional gels showing the level of PR10 protein (spot ‘d’, indicated with a circle) in the non‐silenced control line HT40917‐1 (A) compared with other transgenic lines (B, A44S1‐15‐3; C, A44S1‐1‐1; D, A44S1‐3‐1; E, A44S1‐3‐4; F, A44S1‐5‐2; G, A44S1‐5‐3; H, A44S1‐6‐1; I, A44S1‐35‐3; J, A44S1‐35‐4; K, A44S1‐36‐1; L, A44S1‐42‐1). Other proteins showing changes in their levels are labelled with letters (a, starch synthase; b, ADP‐glucoase pyrophosphorylase small subunit; c, a 40S ribosomal protein and a nucleic acid‐binding protein; e, PR10 and glyoxalase; f, unknown).
Changes in fungal growth and aflatoxin production in RNAi transgenic kernels
The kernel screening assay was performed on the above 12 transgenic lines, together with the control HT40917‐1. A44S1‐35‐4 and A44S1‐42‐1 showed the highest level of fungal colonization at the end of the 7‐day incubation, followed by A44S1‐35‐3 and the susceptible control P3142 used in the kernel screening assay (Fig. 4), whereas A44S1‐15‐3, A44S1‐3‐1, the control HT40917‐1 and the resistant control MI82 showed very low levels of fungal colonization (Fig. 4). The visual ratings of fungal colonization of these lines are summarized in Table 4. Aflatoxin analysis of infected kernels found six lines (A44S1‐3‐1, A44S1‐5‐3, A44S1‐35‐3, A44S1‐35‐4, A44S1‐36‐1 and A44S1‐42‐1) that supported significantly higher levels of aflatoxin production than either the non‐transformed HT40917‐1 control or line A44S‐15‐3, which was confirmed to be negative for the transgene (Table 4). All the other five transgenic lines supported levels of aflatoxin production similar to that of the Hi II control (HT40917‐1) (Table 4).
Figure 4.

Variation in Aspergillus flavus colonization of RNAi‐silenced transgenic maize kernels compared with non‐silenced Hi II × B73 (HT40917‐1) control kernels. Maize genotypes MI82 and Pioneer 3142 (P3142) are controls resistant and susceptible to A. flavus infection and aflatoxin production, respectively.
Table 4.
Fungal colonization and aflatoxin production in RNAi‐silenced transgenic kernels after inoculation with Aspergillus flavus.
| Line name | Colonization rating*,† | Aflatoxin (ppb)*,‡ |
|---|---|---|
| A44S1‐1‐1 | 2.5 de | 658 de |
| A44S1‐1‐4 | 2.8 cd | 102 e |
| A44S1‐3‐1 | 2.1 ef | 987 cd |
| A44S1‐3‐4 | 2.5 de | 484 de |
| A44S1‐5‐2 | 2.4 def | 490 de |
| A44S1‐5‐3 | 1.9 fg | 2399 c |
| A44S1‐6‐1 | 2.8 cd | 365 de |
| A44S1‐15‐3§ | 2.3 def | 6 e |
| A44S1‐35‐3 | 3.2 bc | 3440 bc |
| A44S1‐35‐4 | 3.9 a | 11120 a |
| A44S1‐36‐1 | 3.0 bcd | 7880 ab |
| A44S1‐42‐1 | 3.7 a | 7600 ab |
| HT40917‐1§ | 1.6 g | 94.0 e |
| P3142§ | 3.2 bc | 4425 bc |
| MI82§ | 1.4 g | 889 d |
Means within the column followed by the same letter were not significantly different based on Waller–Dunncan K‐ratio t‐test (at P= 0.05).
Fungal colonization was visually rated 7 days after inoculation and incubation at kernel screening assay conditions as follows: 0, mycelium visible on kernel surface, but no sporulation; 1, 1%–20% of the kernel surface covered by conidiophores bearing conidia; 2, 21%–40%; 3, 41%–60%; 4, 61%–80%; 5, 81%–100%. The means of ratings for individual kernels from two repeated experiments are presented here.
Aflatoxin data were log‐transformed to equalize variations before statistical analysis using SAS.
A44S1‐15‐3 was negative for the PR10 RNAi construct. HT40917‐1 and HT40917‐3 were non‐transgenic Hi II plants pollinated with B73 as negative controls. P3145 and MI82 were aflatoxin‐susceptible and aflatoxin‐resistant control lines used in the kernel screening assay.
Correlation between PR10 expression and aflatoxin production or fungal colonization
When comparing the change in PR10 at both the transcript and protein levels with the changes in fungal colonization or aflatoxin production, significant correlations were found between log‐transformed toxin production and PR10 expression at either transcript (correlation coefficient r, −0.793 to −0.798) or protein (r=−0.714) level, and between square root‐transformed toxin and changes in PR10 protein in kernels (Table 5). These data therefore clearly demonstrate a connection between PR10 expression and maize aflatoxin resistance.
Table 5.
Linear regression analysis between pathogenesis‐related protein 10 (PR10) expression at transcript and protein level vs. aflatoxin production and fungal colonization in transgenic maize kernels.
| Variable 1 | Variable 2 | Correlation coefficient (r 2) | R value |
|---|---|---|---|
| Percentage change in transcript in leaf | Toxin | 0.1582 | −0.398 |
| Sqrt toxin | 0.2776 | −0.527 | |
| Log toxin | 0.6286 | −0.793* | |
| Colonization | 0.0232 | −0.152 | |
| Percentage change in transcript in root | Toxin | 0.0985 | −0.314 |
| Sqrt toxin | 0.2170 | −0.466 | |
| Log toxin | 0.6368 | −0.798* | |
| Colonization | 0.0809 | −0.284 | |
| Percentage change in protein in kernel | Toxin | 0.2037 | −0.541 |
| Sqrt toxin | 0.3299 | −0.574* | |
| Log toxin | 0.5106 | −0.714* | |
| Colonization | 0.1674 | −0.409 |
Correlation is statistically significant. The significance of the correlation coefficient r is determined on the basis of the 95% critical values (CV =±0.555) with a sample size of 13 and d.f. = 11 (n− 2).
DISCUSSION
Resistance to A. flavus infection and aflatoxin production in maize is a multigene‐controlled trait (Paul et al., 2003). A previous study has indicated that constitutively expressed kernel proteins play at least as important a role as inducible proteins in contributing to maize kernel aflatoxin resistance (Chen et al., 2001). Further studies have revealed that the expression of certain storage, stress‐related and antifungal proteins is associated with aflatoxin resistance (2002, 2007). A recent study of one such antifungal protein, PR10, indicated its potentially important role in resistance, thereby rendering PR10 a candidate for further investigation (Chen et al., 2006).
In this study, both particle bombardment and Agrobacterium‐mediated transformation methods were used to introduce the PR10 RNAi vector into immature maize embryos. The former was used to provide a rapid assessment of the efficacy of the RNAi vector in gene silencing. The latter, which results in transgenic materials with fewer copies of foreign genes and is easier to regenerate, was chosen for the generation of transgenic kernels to determine the effect of reduced PR10 protein production on maize resistance to A. flavus infection and aflatoxin contamination. The ideal material for verifying the importance of PR10 in host resistance through RNAi gene silencing would be a known aflatoxin‐resistant maize inbred line. However, a recent evaluation of the responses of two aflatoxin‐resistant maize lines Mp420 (Scott and Zummo, 1988) and GT‐MAS : gk (Widstrom et al., 1987) to tissue culture by Frame et al. (2006) found that only type I callus was produced under various cultural conditions with or without Agrobacterium tumefaciens infection. Therefore, the Hi II line, which produces type II callus and has been successfully transformed and regenerated in earlier studies (Fromm et al., 1990; Gordon‐Kamm et al., 1990), was selected in this study as the starting material for transformation. As a result of the heterogeneous nature of the Hi II genetic background (Armstrong et al., 1991), callus clones or kernels from at least 12 independent transformation events were evaluated for phenotypical and gene expression changes to take into account the potential variations in genetic background.
The callus lines from particle bombardment showed a 65%–99% reduction (with an average of over 90%) of PR10 expression, confirming that the PR10 RNAi vector was working as designed. When the callus lines with reduced PR10 expression were subjected to heat stress treatment, their growth also showed a significant reduction relative to the non‐silenced control, consistent with a putative role of PR10 in stress tolerance (Krishnaswamy et al., 2008; Srivastava et al., 2006).
Mature transgenic kernels from 12 independent transformation events were first evaluated for the presence of the transgene, and only A44S1‐15‐3 was found to be negative. PR10 expression was always significantly higher in root tissue than in leaf tissue. Variation in the level of PR10 expression among the transgenic lines was also clearly visible, with A44S1‐35‐4 and A44S1‐3‐4 showing the lowest level of PR10 expression in leaf tissue (87% reduction) and A44S1‐35‐3 in root tissue (95% reduction). It is not clear why the expression of PR10 in the root tissue of A44S1‐15‐3 was higher than that of the control lines.
Eight of the 12 lines showed a significant reduction in PR10 protein levels, with A44S1‐5‐2 and A44S1‐42‐1 showing the largest reduction (85%), as determined through proteomics, and the identity of the PR10 protein was confirmed through peptide sequencing. In addition to the consistent reduction of PR10 production in RNAi‐silenced kernels, the levels of several other protein spots near the putative PR10 also varied among the transgenic lines based on the two‐dimensional protein profiles (Fig. 4). These proteins were sequenced and one (spot ‘e’) contained two proteins based on peptide sequences: a glyoxalase family protein and PR10. This could be a result of the close proximity of this spot to PR10 (spot ‘d’), which made it difficult to resolve completely, even with large‐format, two‐dimensional gels. This may also explain why the volume of spot ‘e’ was reduced in several of the transgenic lines. The identities of the other sequenced spots did not appear to be related to PR10, and their protein level changes did not correlate with PR10, indicating that the variations might be a result of genetic background differences. A recent study by Colditz et al. (2007) reported an antagonistic induction of other PR proteins when a PR10‐like protein was silenced in Medicago truncatula. However, in this study, no consistent induction of other proteins was observed during the proteome comparisons of the transgenic lines. In our study, kernels were not infected before proteomic analysis, in contrast with the study by Colditz et al. (2007).
When RNAi‐silenced transgenic mature maize kernels were inoculated with A. flavus, significant increases in fungal colonization and aflatoxin production were observed compared with controls. In general, the degree of fungal colonization agrees well with the aflatoxin levels observed inside inoculated kernels (correlation coefficient r, 0.695). However, several lines with high colonization ratings (A44S1‐6‐1, A44S1‐1‐4) contained low levels of aflatoxin, whereas A44S1‐36‐1 (with a colonization rating similar to A44S1‐6‐1) contained a significantly higher level of aflatoxin in the inoculated kernels. This is not unusual, as maize lines with heavy fungal colonization have been shown to support little or no aflatoxin production or vice versa (Brown et al., 2001). Of the eight lines with a significant reduction in PR10, seven showed significant increases in fungal colonization. Six also showed a significant increase in aflatoxin production. Correlation analysis was consistent with a connection between PR10 level and maize aflatoxin resistance. The observed high variation in aflatoxin production among the RNAi transgenic lines was possibly a result of the variation in PR10 protein levels and its antifungal properties, as indicated by the high correlation coefficient (r=−0.798) between PR10 level and maize aflatoxin. However, although maize aflatoxin resistance is often antifungal in nature, other factors, such as the physical barrier limiting fungal colonization or the host proteins inhibitory to fungal toxin biosynthetic pathway genes or enzymes, must also be considered (1995, 2001; Chen et al., 2004; Gembeh et al., 2001).
In summary, the present study constructed a PR10 RNAi gene silencing vector and demonstrated the efficacy of the vector in silencing PR10 expression in both callus tissues and transgenic kernels. Transgenic callus clones with reduced PR10 expression exhibited increased susceptibility to heat stress. Transgenic kernels with significant increases in aflatoxin production showed significant reductions in PR10 levels. These data indicate an important role for PR10 expression in maize aflatoxin resistance.
EXPERIMENTAL PROCEDURES
Construction of RNAi gene silencing vector for PR10 gene
The double 35S promoter was amplified from a pBI121‐d35S‐PR10‐nos vector (Chen et al., 2006) using PCR with primers d35Sf‐Xho (CCCTCGAGgAatTCGGTATCG) and d35Sr‐SpeI (CCCGCaCtaGtAATTGTAAATAG). The nucleotide changes (lower case) were used to incorporate an EcoRI site after a XhoI site at the 5′ end and to change an NcoI site into an SpeI site at the 3′ end of the original construct. This PCR product was then digested with XhoI and SpeI and gel purified before being ligated into XhoI and SpeI double‐digested pBluescript SK‐ to generate the pBS‐d35S vector. The DNA fragment containing attR4‐ccdB‐CmR‐attR3 was amplified from the pDEST™ R4‐R3 vector of a MultiSite Gateway Vector Construction Kit (Invitrogen, Carlsbad, CA, USA) with primers (R4R3f‐SpeI, AACtagTATGACCATGATTACGCCAAG; R4R3r‐SacI, AACGAgctCCAGTGAATTATCA) to incorporate SpeI at the 5′ end and SacI at the 3′ end. The amplified PCR product was digested with SpeI and SacI before ligating into the corresponding sites of the pBS‐d35S vector to generate pBS‐d35S‐attR4‐attR3 (Fig. 1).
The 5′ and 3′ arms of the PR10 gene were also amplified using PCR with homologous recombination sites (italic) attached to the end of the gene‐specific primers. The region selected for the 5′ and 3′ arms was conserved between the two PR10 genes identified so far from maize (Xie et al., 2009). Briefly, the 5′ arm was amplified with attB4‐PRf (GGGGACAACTTTGTATAGAAAAGTTGAGTCATGCCGTTCAGCTT) and attB1‐PRr (GGGGACTGCTTTTTTGTACAAACTGTAATCTCCGTACACAATACCC) with the PR10 cDNA clone as a template, and the 3′ arm was amplified with attB2‐PRf (GGGGACAGCTTTCTTGTACAAAGTGGAATCTCCGTACACAATACCC) and attB3‐PRr (GGGGACAACTTTGTATAATAAAGTTGAGTCATGCCGTTCAGCTT) in a similar manner. The 5′ and 3′ arms were ligated into pDONR P4‐P1R and pDONR P2R‐P3 (Invitrogen), respectively, through BP clonase reactions, according to the manufacturer's instruction. The resulting vectors were named pENTR‐L4‐5′arm‐R1 and pENTR‐R2‐3′arm‐L3, respectively.
An intron used to stabilize the inverted repeat sequences of the 5′ and 3′ arms was amplified from the PR10 genomic DNA clone (Chen et al., 2006) with the primers attB1‐PRf (GGGGACAAGTTTGTACAAAAAAGCAGGCTGTTCAACTTCACCTCAGG) and attB2‐PRr (GGGGACCACTTTGTACAAGAAAGCTGGGTAAGCTGAACGGCATGACT). This PCR product, after being confirmed through DNA sequencing, was cloned into the pDONR221 vector using the BP clonase reaction, according to the manufacturer's instruction. The resulting vector was named pDONR221‐PR 10 intron, containing SphI and ClaI restriction sites in the middle of the intron. A chloramphenical resistance gene (CmR) was then amplified from the pDEST‐R4‐R3 vector using primers PRi‐Sphf (CCGcATgCTCTAGATTACG) and PRi‐Clar (AGTatcGATCCGTCGAGATTT), and was ligated into the SphI and ClaI‐digested pDONR221‐PR 10 intron vector to generate the pDONR221‐PR 10‐intron‐CmR vector, after digesting the PCR product with the corresponding enzymes. The insertion of the antibiotic resistance marker CmR was used for easy selection of downstream target clones, as well as for increased stability of the inverted repeat of the 5′ and 3′ arms.
The MultiSite Gateway LR recombination reaction was performed with the four vectors pBS‐d35S‐attR4‐attR3, pENTR‐L4‐5′arm‐R1, pDONR221‐PR 10‐intron‐CmR and pENTR‐R2‐3′arm‐L3, according to the manufacturer's instructions, except that the reaction mixture was transformed into TOP10 Escherichia coli cells and selected on LB plates containing 100 µg/mL ampicillin and 30 µg/mL chloramphenicol. The resulting vector pBS‐PR10‐RNAi (pBS‐d35S‐attB4‐5′arm‐attB1‐PR 10 intron‐CmR‐attB2‐3′arm‐attB3) was then verified through restriction digestion and sequencing before digesting the vector with EcoRI and SacI to remove the d35S‐attB4‐5′arm‐attB1‐PR 10 intron‐CmR‐attB2‐3′arm‐attB3 cassette, which was then ligated into the corresponding sites of pTF102 (Frame et al., 2002), to generate the final RNAi vector pTF102‐PR10‐RNAi for use in maize transformation.
Maize transformation
Immature zygotic embryos of the maize Hi II hybrid, with the ability to produce close to 100% type II callus (A188 X B73 origin) (Armstrong et al., 1991), were used in this study. The embryos were transformed with the pTF102‐PR10‐RNAi construct through both particle bombardment and Agrobacterium‐mediated methods at the Iowa State University Plant Transformation Facility (Ames, IA, USA). For particle bombardment, plasmid pTF102‐PR10 RNAi DNA was coated onto 0.6‐µm gold particles (Bio‐Rad, Hercules, CA, USA) following the protocol outlined by Frame et al. (2000). F2 immature zygotic embryos (1.5–2.0 mm) of the maize Hi II hybrid genotype were aseptically dissected from glasshouse‐grown ears harvested 10–13 days post‐pollination, and were placed embryo axis side down (about 45 embryos per plate) onto N6‐based medium (Chu et al., 1975), with modifications (Frame et al., 2000), for 3 days in the dark at 28 °C to induce callus initiation before bombardment. Selection of bialaphos‐resistant putative callus events from bombarded immature embryos was conducted according to Frame et al. (2000) before analysis for the level of target gene expression in independent transformation events was carried out. For Agrobacterium‐mediated transformation, the above plasmid DNA was introduced into Agrobacterium strain EHA101. Immature embryo infection, co‐cultivation and resting, and selection and regeneration of transgenic callus events on bialaphos‐containing medium, were performed according to Frame et al. (2002).
Confirmation of transformation
Genomic DNA was isolated from callus clones recovered from bombardment or young leaves developed from transgenic kernels regenerated after Agrobacterium transformation, according to the instructions from the Redextract‐N‐Amp Plant PCR kit (Sigma, St. Louis, MO, USA). Genomic DNA extracted from the nontransformed callus clone Neg 43 was used as a negative control. The isolated genomic DNA was then used as a template to confirm the transformation through PCR with PR10if (GTTCAACTTCACCTCAGG) and PR10ir (AAGCTGAACGGCATGACT) primers corresponding to the CmR region spanned by the intron.
Determination of PR10 expression in callus, root and leaf tissues using real‐time PCR
Total RNA was isolated from Neg 43, callus clones recovered after bombardment, or young leaves and roots developed from transgenic kernels regenerated after Agrobacterium transformation, according to the instructions from the RNeasy Plant mini kit (Qiagen, Valencia, CA, USA). The optional DNase treatment was included to remove trace DNA contamination prior to reverse transcription with random primers using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster, CA, USA). The resulting cDNAs were then used in real‐time PCR with PR10‐Frt2 (TCGATCACTGAAGTAGTAATGGCC) and PR10‐Rrt2 (GAGCCACCGGCGACG) primers and a TaqMan probe (FAM‐CGCCAACAGCTGGACCCTCG‐TAMRA) to quantify PR10 transcript levels in the control and transformed calli, roots, or leaves. The expression of 18S RNA was used as an internal control to normalize the level of target gene expression. The percentage change in target gene was calculated as the difference in relative expression between the RNAi‐silenced transgenic and the control, expressed as a percentage of the control. The relative expression level was calculated on the basis of the threshold cycle (Ct) values of the target gene and 18S normalizer as follows: relative expression level = power (2, ΔCt), where ΔCt=Ct (18S rRNA)−Ct (target). This experiment was conducted twice, each time with three replicates. The data presented here are the means combined from the two repeats.
Evaluation of transgenic callus tissues for changes in heat stress tolerance
To test whether PR10 expression is involved in the enhancement of plant stress tolerance, three transgenic callus lines (4, 6 and 8) with significant reduction in PR10 expression and the non‐silenced Neg 43 control were examined for changes in their stress response. These callus lines were transferred onto fresh N6 medium without bialaphos and subjected to heat stress treatment at 37 °C for 72 h before moving back to the normal 28 °C culture conditions. The percentage of weight change after 2 weeks compared with the initial weight was used to determine the effect of heat stress treatment on callus growth. The data presented here were the average from two repeated studies.
Evaluation of transgenic maize kernels for changes in resistance to A. flavus colonization and aflatoxin production
For each transgenic ear, the weight was determined for all kernels (if the total number of kernels was less than 30) or 30 randomly selected kernels. The mean kernel weight between each transgenic plant was separated using least‐significant differences (P= 0.05). The kernel screening assay (Brown et al., 1995) was used to evaluate the changes in kernel resistance of 12 transgenic lines (including A44S1‐15‐3, which was confirmed to be negative for the target gene) to A. flavus infection and aflatoxin production compared with the non‐silenced control line (HT40917‐1). Mature kernels from control and RNAi‐transformed plants were inoculated with A. flavus conidia and incubated according to Brown et al. (1995). At the end of incubation, fungal colonization on the surface of inoculated kernels, an index of fungal growth, was rated according to a 0–5 scale, with a value of zero for kernels having visible mycelium on the kernel surface, but no sporulation, and a value of five for kernels having 81–100% of their kernel surface covered by conidiophores bearing conidia (Guo et al., 1995). After visual rating of fungal colonization, the kernels were oven dried at 55 °C for 3 days and used to determine aflatoxin levels according to Brown et al. (1995). Aflatoxin data were log‐transformed to equalize variations before statistical analysis using SAS software (SAS Institute Inc., Cary, NC, USA) to determine whether there was a significant difference in aflatoxin production between the RNAi‐silenced transgenic and control kernels. This experiment was conducted twice, each time with seven replicates (three kernels per replicate). The data presented here are the means combined from the two experiments.
PR10 production in transgenic maize kernels
To determine whether the changes in kernel aflatoxin resistance were associated with a reduction in PR10 protein levels in the mature kernels, the above 12 lines, together with the HT40917‐1 control, were further analysed through proteomics. The endosperm tissue, in which the PR10 protein was first identified (Chen et al., 2006), was used for protein extraction according to Chen et al. (2007). Protein extraction was conducted twice, and each extract was subjected to two‐dimensional gel electrophoresis in triplicate, and stained with silver (Chen et al., 2007). Gel analysis was performed using Progenesis SameSpots (Nonlinear Dynamic, Durham, NC, USA) to determine the changes in PR10 protein production. The putative PR10 protein spot and other spots that showed a significant reduction in protein levels were then recovered from Coomassie‐stained gels and sequenced using electrospray ionization‐tandem mass spectrometry to determine their identities.
ACKNOWLEDGEMENTS
We thank Drs Deepak Bhatnagar, Jeffery W. Cary, Marc Cohn and Ahmad M. Fakhoury for critical review of the manuscript, and Wanida Seehachai, Nicole Hazard and Dr Yurong Xie for technical assistance. The authors also wish to thank Drs Kan Wang and Bronwyn R. Frame (Center for Plant Transformation, Plant Science Institute, Iowa State University, Ames, IA, USA) for providing the pTF102 vector, for their help in the transformation of the RNAi vector and for their input in discussions. This study was supported by USDA Cooperative Agreement 58‐6435‐6‐055, the USDA‐ARS Aflatoxin Elimination Workshop and USDA National Research Initiative Competitive Grant 2002‐35201‐12541. Published with the approval of the Director of the Louisiana Agricultural Experiment Station as manuscript number 07‐38‐0441.
REFERENCES
- Armstrong, C.L. , Green, C.E. and Phillips, R.L. (1991) Development and availability of germplasm with high Type II culture formation response. Maize Genet. Coop. News Lett. 65, 92–93. [Google Scholar]
- Bantignies, B. , Seguin, J. , Muzac, I. , Dedaldechamp, F. , Gulick, P. and Ibrahim, R. (2000) Direct evidence for ribonucleolytic activity of a PR‐10‐like protein from white lupin roots. Plant Mol. Biol. 42, 871–881. [DOI] [PubMed] [Google Scholar]
- Breiteneder, H. , Ferreira, F. , Hoffmann‐Sommergrube, K. , Ebner, C. , Breitenbach, M. , Rumpold, H. , Kraft, D. and Scheiner, O. (1993) Four recombinant isoforms of Cora a 1, the major allergen of hazel pollen, show different IgE‐binding properties. Eur. J. Biochem. 212, 355–362. [DOI] [PubMed] [Google Scholar]
- Brown, R.L. , Cleveland, T.E. , Payne, G.A. , Woloshuk, C.P. , Campbell, K.W. and White, D.G. (1995) Determination of resistance to aflatoxin production in maize kernels and detection of fungal colonization using an Aspergillus flavus transformant expressing Escherichia coli β‐glucuronidase. Phytopathology, 85, 983–989. [Google Scholar]
- Brown, R.L. , Chen, Z.‐Y. , Menkir, A. , Cleveland, T.E. , Cardwell, K. , Kling, J. and White, D.G. (2001) Resistance to aflatoxin accumulation in kernels of maize inbreds selected for ear rot resistance in West and Central Africa. J. Food Prot. 64, 396–400. [DOI] [PubMed] [Google Scholar]
- Campbell, K.W. and White, D.G. (1995) Evaluation of corn genotypes for resistance to Aspergillus ear rot, kernel infection, and aflatoxin production. Plant Dis. 79, 1039–1045. [Google Scholar]
- Chen, Z.‐Y. , Brown, R.L. , Lax, A.R. , Guo, B.Z. , Cleveland, T.E. and Russin, J.S. (1998) Resistance to Aspergillus flavus in corn kernels is associated with a 14 kDa protein. Phytopathology, 88, 276–281. [DOI] [PubMed] [Google Scholar]
- Chen, Z.‐Y. , Brown, R.L. , Cleveland, T.E. , Damann, K.E. and Russin, J.S. (2001) Comparison of constitutive and inducible maize kernel proteins of genotypes resistant or susceptible to aflatoxin production. J. Food Prot. 64, 1785–1792. [DOI] [PubMed] [Google Scholar]
- Chen, Z.‐Y. , Brown, R.L. , Damann, K.E. and Cleveland, T.E. (2002) Identification of unique or elevated levels of kernel proteins in aflatoxin‐resistant maize genotypes through proteome analysis. Phytopathology, 92, 1084–1094. [DOI] [PubMed] [Google Scholar]
- Chen, Z.‐Y. , Brown, R.L. , Damann, K.E. and Cleveland, T.E. (2004) Identification of a maize kernel stress‐related protein and its effect on aflatoxin accumulation. Phytopathology, 94, 938–945. [DOI] [PubMed] [Google Scholar]
- Chen, Z.‐Y. , Brown, R.L. , Rajasekaran, K. , Damann, K.E. and Cleveland, T.E. (2006) Evidence for involvement of a pathogenesis‐related protein in maize resistance to Aspergillus flavus infection/aflatoxin production. Phytopathology, 96, 87–95. [DOI] [PubMed] [Google Scholar]
- Chen, Z.‐Y. , Brown, R.L. , Damann, K.E. and Cleveland, T.E. (2007) Identification of maize kernel endosperm proteins associated with resistance to aflatoxin contamination by Aspergillus flavus . Phytopathology, 97, 1094–1103. [DOI] [PubMed] [Google Scholar]
- Chu, C.C. , Wang, C.C. , Sun, C.S. , Hsu, C. , Yin, K.C. , Chu, C.Y. and Bi, F.Y. (1975) Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen source. Sci. Sin. 18, 659–668. [Google Scholar]
- Colditz, F. , Niehaus, K. and Krajinski, F. (2007) Silencing of PR‐10‐like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches . Planta, 226, 57–71. [DOI] [PubMed] [Google Scholar]
- Diener, U.L. , Cole, R.J. , Sanders, T.H. , Payne, G.A. , Lee, L.S. and Klich, M.A. (1987) Epidemiology of aflatoxin formation by Aspergillus flavus . Annu. Rev. Phytopathol. 25, 249–270. [Google Scholar]
- Fire, A. , Xu, S. , Montgomery, M.K. , Kostas, S.A. , Driver, S.E. and Mello, C.C. (1998) Potent and specific genetic interference by double stranded RNA in Caenorhabditis elegans . Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Frame, B.R. , Zhang, H. , Cocciolone, S.M. , Sidorenko, L.V. , Dietrich, C.R. , Pegg, S.E. , Zhen, S. , Schnable, P.S. and Wang, K. (2000) Production of transgenic maize from bombarded type II callus: effect of gold particle size and callus morphology on transformation efficiency. In Vitro Cell. Dev. Biol. Plant, 36, 21–29. [Google Scholar]
- Frame, B.R. , Shou, H. , Chikwamba, R.K. , Zhang, Z. , Xiang, C. , Fonger, T.M. , Pegg, S.E. , Li, B. , Nettleton, D.S. , Pei, D. and Wang, K. (2002) Agrobacterium tumefaciens‐mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame, B.R. , McMurray, J.M. , Fonger, T.M. , Main, M.L. , Taylor, K.W. , Torney, F.J. , Paz, M.M. and Wang, K. (2006) Improved Agrobacterium‐mediated transformation of three maize inbred lines using MS salts. Plant Cell Rep. 25, 1024–1034. [DOI] [PubMed] [Google Scholar]
- Fromm, M.E. , Morrish, F. , Armstrong, C. , Williams, R. , Thomas, J. and Klein, T.M. (1990) Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Bio/Technology, 8, 833–839. [DOI] [PubMed] [Google Scholar]
- Gembeh, S.V. , Brown, R.L. , Grimm, C. and Cleveland, T.E. (2001) Identification of chemical components of corn kernel pericarp wax associated with resistance to Aspergillus flavus infection and aflatoxin production. J. Agric. Food Chem. 49, 4635–4641. [DOI] [PubMed] [Google Scholar]
- Gordon‐Kamm, W.J. , Spencer, T.M. , Mangano, M.L. , Adams, T.R. , Daines, R.J. , Start, W.G. , O'Brien, J.V. , Chambers, S.A. , Adams, W.R. , Willetts, N.G. , Rice, T.B. , Mackey, C.J. , Krueger, R.W. , Kausch, A.P. and Lemaux, P.G. (1990) Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell, 2, 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B.Z. , Russin, J.S. , Brown, R.L. , Cleveland, T.E. and Widstrom, N.W. (1995) Resistance to aflatoxin contamination in corn as influenced by relative humidity and kernel germination. J. Food Prot. 59, 267–281. [DOI] [PubMed] [Google Scholar]
- Guo, B.Z. , Chen, Z.‐Y. , Brown, R.L. , Lax, A.R. , Cleveland, T.E. , Russin, J.S. , Mehta, A.D. , Selitrennikoff, C.P. and Widstrom, N.W. (1997) Germination induces accumulation of specific proteins and antifungal activities in corn kernels. Phytopathology, 87, 1174–1178. [DOI] [PubMed] [Google Scholar]
- Gura, T. (2000) A silence that speaks volumes. Nature, 404, 804–808. [DOI] [PubMed] [Google Scholar]
- Hartley, J.L. , Temple, G.F. and Brasch, M.A. (2000) DNA cloning using in vitro site‐specific recombination. Genome Res. 10, 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann‐Sommergruber, K. , Vanek‐Krebitz, M. , Radauer, C. , Wen, J. , Ferreira, F. , Scheiner, O. and Breiteneder, H. (1997) Genomic characterization of members of the Bet v 1 family: genes coding for allergens and pathogenesis‐related proteins share intron positions. Gene, 197, 91–100. [DOI] [PubMed] [Google Scholar]
- Hsieh, D.P.H. (1989) Potential human health hazards of mycotoxins In: Mycotoxins and Phycotoxins (Natori S., Hashimoto K. and Ueno Y., eds), pp. 69–80. Amsterdam: Elsevier. [Google Scholar]
- Huang, J.C. , Chang, F.C. and Wang, C.S. (1997a) Characterization of a lily tapetal transcript that shares sequence similarity with a class of intracellular pathogenesis‐related (IPR) proteins. Plant Mol. Biol. 34, 681–686. [DOI] [PubMed] [Google Scholar]
- Huang, Z. , White, D.G. and Payne, G.A. (1997b) Corn seed proteins inhibitory to Aspergillus flavus and aflatoxin biosynthesis. Phytopathology, 87, 622–627. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy, S.S. , Srivastava, S. , Mohammadi, M. , Rahman, M.H. , Deyholos, M.K. and Kav, N.N.V. (2008) Transcriptional profiling of pea ABR17 mediated changes in gene expression in Arabidopsis thaliana . BMC Plant Biol. 8, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Huang, B. , Lin, J. , Fei, J. , Chen, Z. , Pang, Y. , Sun, X. and Tang, K. (2006) A novel pathogenesis‐related protein (SsPR10) from Solanum surattense with ribonucleolytic and antimicrobial activity is stress‐ and pathogen‐inducible. J. Plant Physiol. 163, 546–556. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. , Pierpoint, W.S. , Boller, T. and Conejero, V. (1994) Recommendations for naming plant pathogenesis‐related proteins. Plant Mol. Biol. Rep. 12, 245–264. [Google Scholar]
- McGee, J.D. , Hamer, J.E. and Hodges, T.K. (2001) Characterization of a PR‐10 pathogenesis‐related gene family induced in rice during infection with Magnaporthe grisea . Mol. Plant–Microbe Interact. 14, 877–886. [DOI] [PubMed] [Google Scholar]
- Midoh, H. and Iwata, M. (1996) Cloning and characterization of a probenazole‐inducible gene for an intracellular pathogenesis‐related protein in rice. Plant Cell Physiol. 37, 9–18. [DOI] [PubMed] [Google Scholar]
- Mogensen, J.E. , Wimmer, R. , Larsen, J.N. and Spangfort, M.D. (2002) The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 277, 23 684–23 692. [DOI] [PubMed] [Google Scholar]
- Moiseyev, G.P. , Fedoreyeva, L.I. , Zhuravlev, Y.N. , Yasnetskaya, E. , Jekel, P.A. and Beintema, J.J. (1997) Primary structures of two ribonucleases from ginseng calluses: new members of the PR‐10 family of intracellular pathogenesis‐related plant proteins. FEBS Lett. 407, 207–210. [DOI] [PubMed] [Google Scholar]
- Mould, M.J. , Xu, T. , Barbara, M. , Iscove, N.N. and Heath, M.C. (2003) cDNAs generated from individual epidermal cells reveal that differential gene expression predicting subsequent resistance or susceptibility to rust fungal infection occurs prior to the fungus entering the cell lumen. Mol. Plant–Microbe Interact. 16, 835–845. [DOI] [PubMed] [Google Scholar]
- Nichols, T.E. (1983) Economic impact of aflatoxin in corn. South. Coop. Ser. Bull. 279, 67–71. [Google Scholar]
- Park, C.J. , Kim, K.J. , Shin, R. , Park, J.M. , Shin, Y.C. and Paek, K.H. (2004) Pathogenesis‐related protein 10 from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 37, 186–198. [DOI] [PubMed] [Google Scholar]
- Paul, C. , Naidoo, G. , Forbes, A. , Mikkilineni, V. , White, D. and Rocheford, T. (2003) Quantitative trait loci for low aflatoxin production in two related maize populations. Theor. Appl. Genet. 107, 263–270. [DOI] [PubMed] [Google Scholar]
- Payne, G.A. (1998) Process of contamination by aflatoxin‐producing fungi and their impact on crops In: Mycotoxins in Agriculture and Food Safety (Sinha K.K. and Bhatnagar D., eds), pp. 279–306. New York: Marcel Dekker. [Google Scholar]
- Scott, G.E. and Zummo, N. (1988) Sources of resistance in maize to kernel infection by Aspergillus flavus in the field. Crop Sci. 28, 504–507. [Google Scholar]
- Squire, R.A. (1981) Ranking animal carcinogens, a proposed regulatory approach. Science, 214, 877–880. [DOI] [PubMed] [Google Scholar]
- Srivastava, S. , Rahman, M.H. , Shah, S. and Kav, N.N.V. (2006) Constitutive expression of the pea ABA‐responsive 17 (ABR17) cDNA confers multiple stress tolerance in Arabidopsis thaliana . Plant Biotechnol. J. 4, 529–549. [DOI] [PubMed] [Google Scholar]
- Srivastava, S. , Emery, R.J.N. , Rahman, M.H. and Kav, N.N.V. (2007) A crucial role for cytokinins in pea ABR17‐mediated enhanced germination and early seedling growth of Arabidopsis thaliana under saline and low temperature stresses. J. Plant Growth Regul. 26, 26–37. [Google Scholar]
- Steiner‐Lange, S. , Fischer, A. , Boettcher, A. , Rouhara, I. , Liedgens, H. , Schmelzer, E. and Knogge, W. (2003) Differential defense reactions in leaf tissues of barley in response to infection by Rhynchosporium secalis and to treatment with a fungal avirulence gene product. Mol. Plant–Microbe Interact. 16, 893–902. [DOI] [PubMed] [Google Scholar]
- Warner, S.A.J. , Scott, R. and Draper, J. (1992) Characterization of a wound‐induced transcript from the monocot asparagus that shares similarity with a class of intracellular pathogenesis‐related (PR) proteins. Plant Mol. Biol. 19, 555–561. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V. , Helliwell, C.A. , Smith, N.A. , Wang, M.B. , Rouse, D.T. , Liu, Q. , Gooding, P.S. , Singh, S.P. , Abbott, D. , Stoutjesdijk, P.A. , Robinson, S.P. , Gleave, A.P. , Green, A.G. and Waterhouse, P.M. (2001) Construct design for efficient, effective and high‐throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Widstrom, N.W. , McMillian, W.W. and Wilson, D.M. (1987) Segregation for resistance to aflatoxin contamination among seeds on an ear of hybrid maize. Crop Sci. 27, 961–963. [Google Scholar]
- Xie, Y.‐R. , Chen, Z.‐Y. , Brown, R.L. and Bhatnagar, D. (2009) Expression and functional characterization of two pathogenesis‐related protein 10 genes from Zea mays . J. Plant Physiol. (in press). [DOI] [PubMed]


