SUMMARY
Bacterial lipopolysaccharides (LPS) are triggers of defence responses in plants, and induce local as well as systemic acquired resistance. Arabidopsis thaliana plants pretreated with LPS show an increased resistance to the virulent bacterial plant pathogen Pseudomonas syringae pv. tomato DC3000. To investigate the mobilization and transport of LPS in Arabidopsis leaves, fluorescently labelled LPS (Alexa Fluor® 488 conjugate) from Salmonella minnesota was used. Leaves were pressure infiltrated with fluorescein‐labelled LPS and fluorescence microscopy was used to follow the movement and localization of LPS as a function of time. The observation of leaves 1 h after supplementation with fluorescein‐labelled LPS revealed a fluorescent signal in the intercellular space. Capillary zone electrophoresis was used for the detection and analysis of the labelled LPS in directly treated leaves and systemic leaves. In addition, gel electrophoresis was used to confirm LPS mobilization. The results indicated that LPS mobilization/translocation occurs through the xylem from local, treated leaves to systemic, untreated leaves. Consequently, care should be taken when ascribing the observed biochemical responses and induced resistance from LPS perception as being uniquely local or systemic, as these responses might overlap because of the mobility of LPS in the plant vascular system.
INTRODUCTION
Plants constantly monitor for pathogen challenge as part of their innate immunity (Nurnberger et al., 2004; Sanabria et al., 2008). To this end, plants have evolved receptors capable of recognizing conserved components of invading microbial pathogens, called pathogen‐associated molecular patterns (PAMPs). PAMPs, such as bacterial flagellin or lipopolysaccharides (LPS), are invariant epitopes within molecules that are fundamental to the pathogen's fitness, widely distributed among different microbes, absent from the host and recognized by a wide array of potential hosts (Schwessinger and Zipfel, 2008). They are able to trigger innate immune responses, and many of these molecules have been shown to act as general elicitors of defence responses in various plant species (Boller and Felix, 2009; Zipfel, 2008). Complex and largely unresolved perception systems for these elicitors exist on the plant cell surface. These lead to immediate primary defence responses and subsequent cellular system responses (Gerber et al., 2008).
Graham and coworkers first identified LPS as the cell wall component of Pseudomonas solanacearum, which is responsible for induced disease resistance in tobacco, and investigated the interaction of LPS with mesophyll cell walls (Graham et al., 1977). LPS was observed as a laminated micellar aggregate that was tightly associated with the cell wall, and induced ultrastructural changes in the host cell similar to those associated with disease resistance induced by whole heat‐killed cells (Graham et al., 1977). Later, the induction of plant defence was confirmed by several groups working on different plant models (Mishina and Zeier, 2007; Newman et al., 2002; Zeidler et al., 2004). To date, no LPS receptor has been identified, and little is known about the fate of LPS molecules after the perception by the plant. Only Gross et al. (2005) have monitored the fate of fluorescein isothiocyanate (FITC)‐labelled LPS from Xanthomonas campestris following its interaction with the plant cell surface. Their data suggest an energy‐dependent internalization, possibly involving two different receptors. However, currently, there is no further evidence supporting the existence of different LPS receptors, especially not of the low‐affinity type. Interestingly, there are indications that the structure of LPS and associated lipid A can influence recognition in plants. The acylation and phosphorylation pattern of lipid A seems to influence strongly its ability to trigger the innate immune response in Arabidopsis thaliana (Silipo et al., 2008). Furthermore, the LPS of Sinorhizobium meliloti suppressed defence‐associated gene expression in cell cultures of the host plant Medicago truncatula, pointing to a differential recognition of LPS from diverse sources (Tellstrom et al., 2007).
In a previous study, LPS from an endophytic strain of Burkholderia cepacia (LPSB.cep.) was used to investigate the biochemical processes activated in tobacco cells in response to LPS perception and resulting signal transduction (Gerber and Dubery, 2004). LPS was found to trigger a rapid influx of Ca2+ into the cytoplasm of tobacco cells, the production of reactive oxygen and nitrogen species (ROS and NO) during an oxidative burst reaction, as well as K+/H+ exchange and alkalinization of the extracellular culture medium (Gerber et al., 2004). These responses are typically associated with the initiation of hypersensitive response (HR)‐related cell death. However, LPS, as an elicitor of PAMP‐triggered immunity (Jones and Dangl, 2006), does not trigger a cell death programme in tobacco. Previously, it has been demonstrated that LPS has specific effects on the reversible protein phosphorylation events underlying the perception systems involved in the interaction of plant cells with LPS (Gerber and Dubery, 2004; Gerber et al., 2006, 2008). For instance, an extracellular signal‐regulated kinase (ERK)‐like mitogen‐activated protein kinase (MAPK) was phosphorylated in response to LPS treatment. Evidence was also provided for the phosphorylation of an LPS‐responsive ERK‐like MAPK in tobacco (Piater et al., 2004). Together, these results indicate that the perception and signal transduction responses during LPS elicitation of tobacco cells require an intricate balance between the actions of certain protein kinases and protein phosphatases.
Moreover, gene expression studies in LPS‐treated A. thaliana plants revealed the induction of an array of defence‐ or stress‐associated genes (Zeidler et al., 2004). Defence gene expression was almost completely eliminated when Atnoa1 (NO‐associated protein 1) mutant plants were treated with LPS, suggesting a functional link between LPS perception, NO production and gene expression (Zeidler et al., 2004). Collectively, the results indicate that LPS induces specific alterations in plant defence responses, and suggest the existence of an important signalling and response system in plant–pathogen interactions that could be part of a broad‐spectrum defence mechanism against pathogens.
In contrast with the downstream events following LPS perception, little is known about LPS mobilization and transport in plants. LPS is a major constituent of the Gram‐negative outer membrane, estimated at 105 molecules/µm2 (Dow et al., 2000). LPS might be released from the bacterial cell wall into the apoplast by living cells or by dying or dead cells after disintegration of the wall. In addition, LPS is an important component of vesicles, released from the outer membrane of many Gram‐negative bacteria, including the plant pathogen X. campestris. These outer membrane vesicles are capable of transporting compounds involved in cell–cell signalling (Sidhu et al., 2008).
LPS from Gram‐negative bacteria has been used as a model PAMP to investigate the induction of an extensive array of plant innate immune responses (Newman et al., 2007). LPS can activate plant signalling pathways, act as an elicitor to induce and potentiate basal defence responses, suppress HR in dicot plants and restrict pathogen growth in treated plants (Dow et al., 2000; Newman et al., 2007). In addition, LPS from endophytic or pathogenic bacteria, as well as rhizobacteria, has been described as an inducer of systemic acquired resistance (SAR) and induced systemic resistance (ISR), respectively (Bakker et al., 2007; Coventry and Dubery, 2001; Mishina and Zeier, 2007). In SAR and ISR, plant defences are preconditioned by previous infection or exposure to microbe‐derived molecules, which results in resistance against subsequent challenges. Previously, we have found that LPS induces a number of pathogenesis‐related (PR) proteins in local and systemic leaves (Zeidler et al., 2004). PR proteins are regarded as important markers for SAR (Hunt et al., 1996; Ward et al., 1991). However, it is still unclear whether systemic PR gene expression is induced by long‐distance translocation of plant resistance signals or by LPS itself. In this context, it is of great interest that the systemic movement of the Xanthomonas effector molecule cyclic β‐glucan through the plant has been observed (Rigano et al., 2007).
In this study, we address the question of to what extent LPS is mobile in planta. We investigate the uptake of fluorescently labelled LPS in A. thaliana leaf tissue in order to obtain more knowledge about the translocation of LPS released into the plant apoplast.
RESULTS AND DISCUSSION
Induction of SAR and PR genes by LPS
Systemic leaves of LPS‐pretreated A. thaliana plants were challenged with either virulent Pseudomonas syringae pv. tomato DC3000 (Pst) or avirulent Pst AvrRpt2. The Pseudomonas effector AvrRpt2 is recognized by the A. thaliana disease‐resistance protein RPS2 (Ausubel et al., 1995). LPSB.cep.‐pretreated A. thaliana plants showed enhanced resistance against Pst in relation to control plants, 2 and 5 days after infection (Fig. 1a). The growth of Pst AvrRpt2 was not significantly attenuated on pretreatment with LPSB.cep. relative to the control treatment (data not shown). A previous report has demonstrated that pretreatment of pepper leaves with LPS from X. campestris pv. campestris (Xcc) has a similar limiting effect on the growth of X. axonopodis pv. vesicatoria (Xav) (Newman et al., 2002). However, in our case, the effects of LPS treatment on the growth of bacteria in compatible plant–bacterial interactions were less pronounced; however, the fact that, in the Newman study, local leaves were infected, whereas, in our study, systemic leaves were used, makes a direct comparison difficult. Enhanced systemic resistance against phytopathogens was also obtained by treatment of A. thaliana with the chemical elicitor 1,2‐benzisothiazol‐3(2H)‐one‐1,1‐dioxide (BIT), 4 days prior to inoculation of Pst DC3000 or with the bacterial elicitors flagellin and harpin (Dong et al., 1999; Mishina and Zeier, 2007; Zipfel et al., 2004). LPS‐induced suppression of bacterial growth in A. thaliana leaves was not as pronounced as after harpin or flagellin treatment. This effect could be an indirect consequence of the absence of HR in LPS‐primed plants (Newman et al., 2002, 2007). HR is a form of programmed cell death (PCD) that occurs at the site of pathogen entry and around the infection site. Its classification is based mainly on morphological criteria of the resultant cell death lesions, as well as the functional suppression of pathogen growth (Beers and McDowell, 2001). It is accompanied by the induction of plant defence responses that serve to confine the pathogen and protect the plant (Lam et al., 2001). Harpin and flagellin induce HR in various plant species, but only if applied at high concentrations (Che et al., 2000; Krause and Durner, 2004). LPS did not induce, and could even prevent HR, resulting in weaker plant protection against subsequent infection with phytopathogens compared with harpin or flagellin (Newman et al., 2007).
Figure 1.
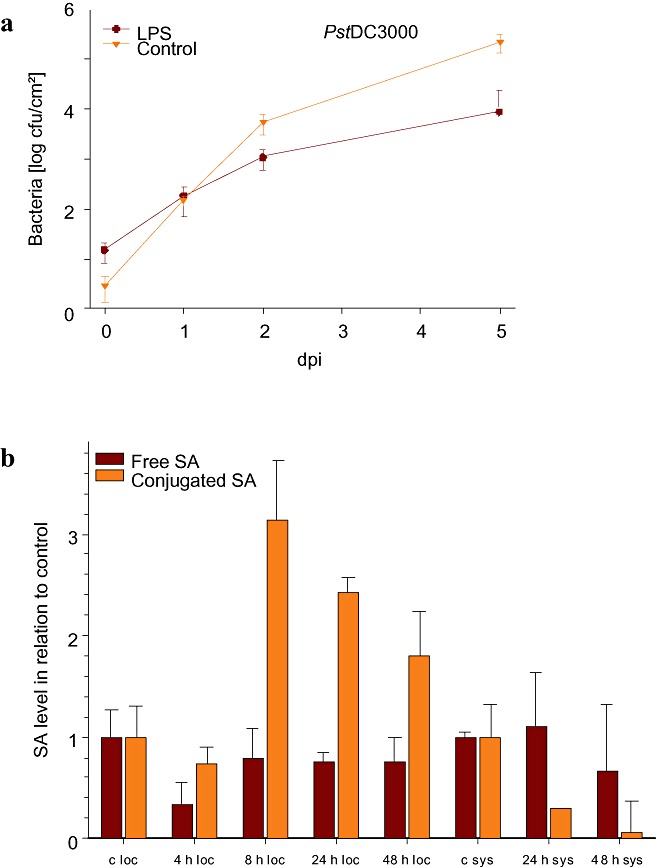
Lipopolysaccharides (LPS) induce systemic acquired resistance against Pseudomonas syringae DC3000 (Pst) and the accumulation of salicylic acid (SA). (a) Arabidopsis thaliana Col‐0 plants were pretreated with LPS from an endophytic strain of Burkholderia cepacia (LPSB.cep.) for 2 days, and systemic leaves were then inoculated with Pst. The diagrams indicate the number of colony‐forming Pst bacteria extracted from systemic leaves 0, 1, 2 and 5 days after infection (dpi). (b) SA accumulation in local and systemic leaves of LPSB.cep.; treated and untreated control plants were analysed at the indicated time points. Values (nmol/g fresh weight) are displayed in relation to control leaves and represent a mean of three biological replicates. c, control; loc, treated leaves; sys, systemic leaves.
For the activation of PR gene expression and the development of SAR, elevated levels of salicylic acid (SA) are necessary (Ryals et al., 1994, 1996). Recently, LPS‐containing cell walls, the pyoverdine siderophores and the flagella of Pseudomonas putida WCS358, P. fluorescens WCS374 and P. fluorescens WCS417—all known to induce ISR—were tested for their effects on tobacco suspension cells. In this study, LPS was able to induce ROS, alkalinization of the extracellular culture medium, elevation of cytoplasmic Ca2+ and defence‐related gene expression in tobacco suspension cells (van Loon et al., 2008). These results, together with LPS‐elicited SAR induction, prompted us to analyse the SA levels in LPS‐treated plants. Although treatment of A. thaliana leaves with LPSB.cep. did not result in significant changes in the content of free SA, the accumulation of conjugated SA was increased in treated tissue after 8, 24 and 48 h (see Fig. 1b). Interestingly, although Mishina and Zeier (2007) reported a 1.5‐fold increase in salicylic acid/3‐glucoside (SAG) in systemic leaves of A. thaliana after treatment with LPS from P. aeruginosa, we found that treatment with LPSB.cep. caused a reduction in conjugated SA in systemic leaves. A possible explanation for this striking difference may be the different source and quality of LPS preparations, which, in some cases, contain up to 3% bacterial protein and RNA. Our preparation contained no detectable impurities (Fig. 5), and did not induce local cell death (HR).
Figure 5.
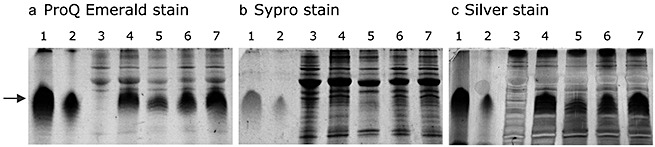
Visualization of lipopolysaccharide (LPS) in midribs of Arabidopsis thaliana leaves with sodium dodecylsulphate‐polyacrylamide gel electrophoresis. Gels were stained with an LPS‐specific stain (a), a protein‐specific stain (b) or a nonspecific stain (c). Lanes: 1, LPS standard (100 µg/mL); 2, LPS standard (10 µg/mL); 3, untreated control; 4, whole treated leaf directly after LPS inoculation; 5, midrib after 1 h; 6, midrib after 6 h; 7, midrib after 24 h.
LPS mobility in leaves
LPSB.cep. induces an NO burst and upregulates an array of defence genes in A. thaliana plants and cells (Zeidler et al., 2004). The fate of LPS in plant tissues is unclear. Previously, FITC has been used to monitor the fate of these signal molecules in intact tobacco cells. In that study, LPS bound rapidly to the cell wall and was then internalized into the cells in a temperature‐ and energy‐dependent manner (Gross et al., 2005). These observations suggest the specific endocytosis of LPS into tobacco cells. The possibility for a receptor‐mediated endocytosis, comparable with that in the mammalian system, has been discussed, but is still unproven (Gross et al., 2005).
In order to monitor the localization of LPS in A. thaliana plants, fluorescently labelled LPS molecules from Salmonella minnesota (LPSS.min.) were used. It should be noted that LPSB.cep. is a potent inducer of systemic PR gene expression and SAR, and other LPS preparations, including that from S. minnesota, might be weaker with regard to the induction of defence reactions (Zeidler et al., 2004). For the localization of LPS by fluorescence microscopy, A. thaliana leaves were pressure infiltrated with 100 µg/mL fluorescein‐labelled LPSS.min.. The observation of leaves 1 h after supplementation with fluorescein‐labelled LPSS.min. revealed a fluorescent signal in the intercellular space of the infiltrated leaf area (Fig. 2a–c). After 4 h (Fig. 2d–f), the LPS fluorescence was visible in the midrib of the leaves. After 6 h, this fluorescence had spread to smaller leaf veins near the midrib (Fig. 2g–i) and was finally detectable in lateral veins after 24 h (Fig. 2j–l). For a more detailed analysis of LPSS.min. fluorescence in the vascular bundle, cross‐sections of the midribs were made; 3 h after supplementation with fluorescein‐labelled LPS, a green fluorescent signal was observed in the xylem (Fig. 3). This is interesting as the transport of defence signals is usually associated with the phloem (Gomez and Stuefer, 2006; Robert and Friml, 2009). However, the transport of signal peptides through the xylem has been suggested previously, and signal peptide activity has been detected after the treatment of cell cultures with purified xylem sap (Neumann, 2007). The possibility for the uptake and diffusion of external molecules into leaves have been shown previously using two fluorescent dyes as model xenobiotics in broad bean plants (Liu and Gaskin, 2004). Other studies have demonstrated the internalization of fluorescently labelled LPS in tobacco cells (Gross et al., 2005). Interestingly, we could not observe the intracellular accumulation of fluorescent LPS, as described for X. campestris pv. campestris LPS, in nonhost plant cells of Nicotiana tabacum (Gross et al., 2005).
Figure 2.
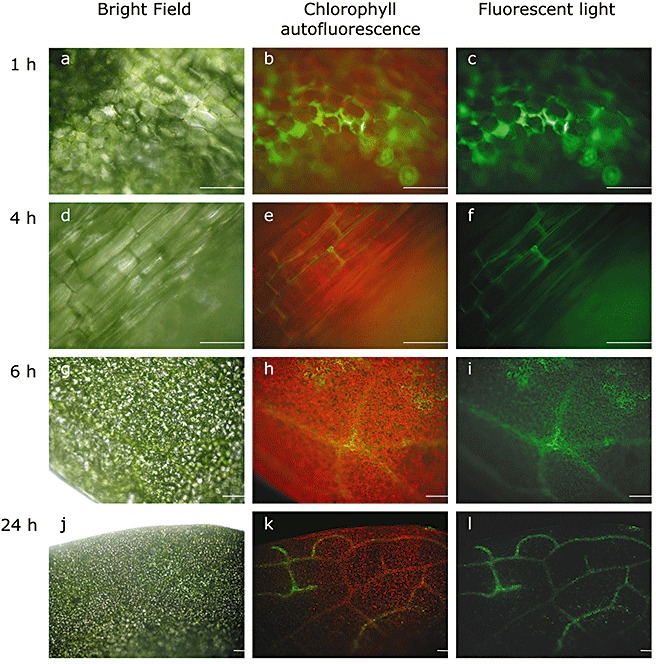
Investigation of lipopolysaccharide (LPS) mobilization in Arabidopsis thaliana leaves using fluorescein‐labelled LPS from Salmonella minnesota (LPSS.min.). After pressure infiltration of 100 µg/mL fluorescein‐labelled LPSS.min., images were obtained from the abaxial leaf side at the indicated time points under bright field (a, d, g, j) and fluorescent (green light filter, 505–530 nm; c, f, i, l) light. Chlorophyll autofluorescence was captured with a long‐pass filter (585 nm; b, e, h, k). Scale bar, 10 µm.
Figure 3.
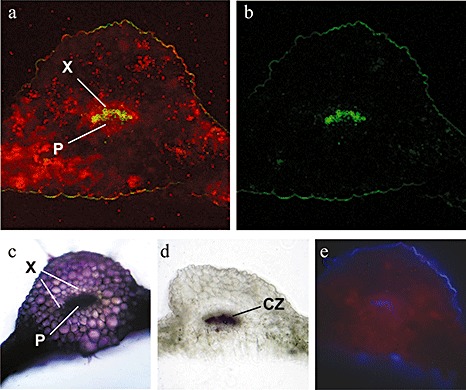
Investigation of lipopolysaccharide (LPS) mobilization in cross‐sections of Arabidopsis thaliana leaves using fluorescently labelled LPS from Salmonella minnesota (LPSS.min.). After pressure infiltration of 100 µg/mL fluorescein‐labelled LPSS.min., images of cross‐sections were obtained after 3 h under fluorescent light (green light filter, 435–485 nm; red light filter, 653–695 nm). (a) LPS fluorescence (green) and autofluorescence of chloroplasts (red); (b) green fluorescence of injected LPS; (c) toludine blue staining shows pectin and pectic substances (pink to purple); (d) phloroglucinol test shows lignin (red–violet); (e) autofluorescence of ferulic acid bound to lignin‐ or cutin‐containing cells (blue). Staining was performed to characterize tissue‐specific properties. CZ, cambial zone; P, phloem; X, xylem.
For confirmation that the detected fluorescence originated from fluorescein‐labelled LPS rather than nonbound fluorescein or other nonspecific fluorescing compounds, an independent approach was used (Fig. 4). Capillary zone electrophoresis is the most efficient separation technique available for the analysis of both large and small molecules (Xu, 1996). We used capillary zone electrophoresis for the detection and analysis of fluorescently labelled LPSS.min. in local and systemic A. thaliana leaves. Defined amounts of sample were introduced by controlling either the injection voltage or injection pressure, resulting in a narrow sample zone, which is surrounded by separation buffer. As an electric field is applied, each component in the sample zone migrates according to its (own) apparent mobility. The distribution of fluorescently labelled LPS from S. minnesota was observed in treated and systemic leaves. For the investigation of local leaves, midribs were excised and extracts were separated by capillary zone electrophoresis. To obtain standard separation data, the first run was performed with LPSS.min. stock solution (Fig. 4a). For the detection of LPSS.min., samples of untreated leaf veins (Fig. 4c) were compared with treated veins, 1 h (Fig. 4e) and 6 h (Fig. 4g) after injection. The LPS peak (dark red arrow) was visible 1 h and increased 6 h after treatment. These results are in accordance with the microscopic analyses, in which fluorescent LPS entered the vasculature at 4 h after treatment.
Figure 4.
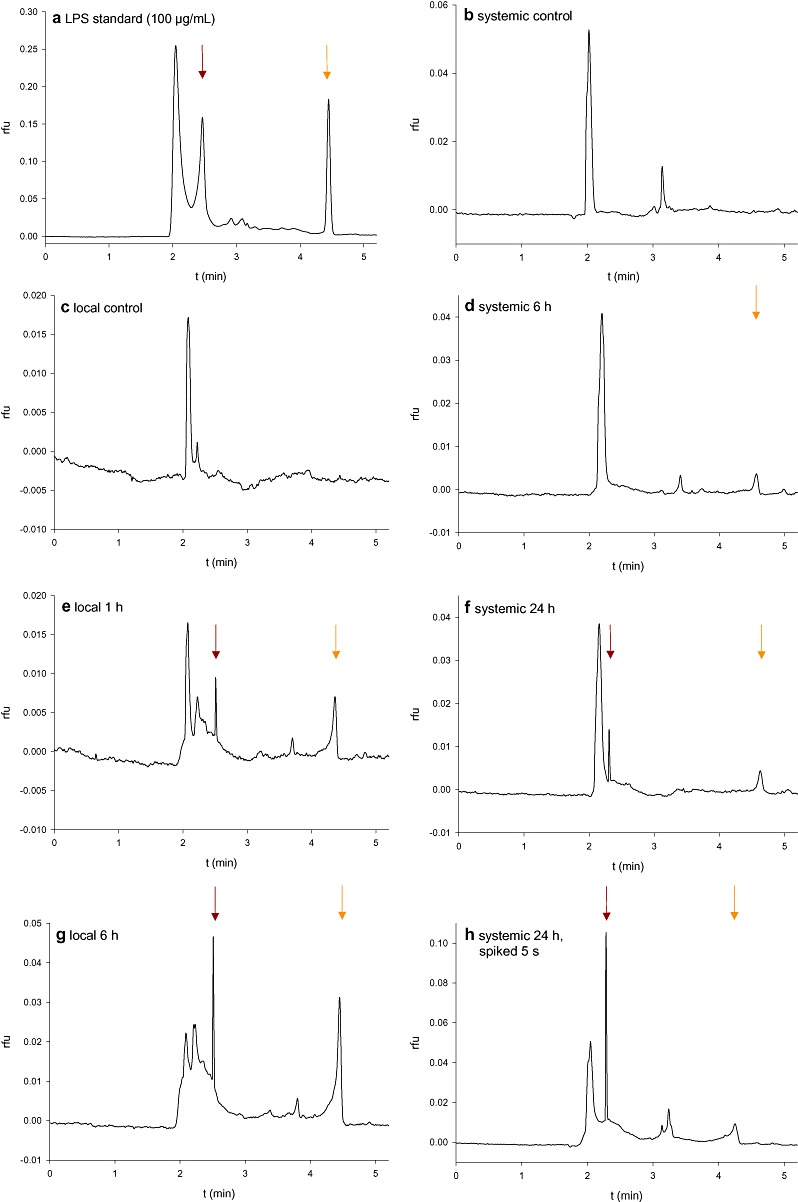
Investigation of lipopolysaccharide (LPS) mobilization using capillary zone electrophoresis. After treatment of Arabidopsis leaves with 100 µg/mL fluorescently labelled LPS from Salmonella minnesota (LPSS.min.), leaves were harvested at the indicated time points and veins (local leaves) or whole leaves (systemic leaves) were investigated. The LPS signal (dark red arrows) became visible after 1 and 6 h in local midribs and after 24 h in systemic leaves. An additional peak indicative of nonbound fluorescein was also observed (orange arrow). rfu, relative fluorescence units.
Systemic leaves were tested in the same way. No LPS was detected in systemic control leaves (Fig. 4b) or in systemic leaves 6 h (Fig. 4d) after application. However, after 24 h (Fig. 4f), an LPS signal (dark red arrow) appeared, which could be intensified by spiking for 5 s with LPS stock solution (5 µg/mL; Fig. 4h). In addition to capillary zone electrophoresis, sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) was used for the detection of LPS mobilization in A. thaliana leaves. This is the most suitable separation technique for LPS, during which the heterogeneous mixture of polymers separates into a characteristic ladder or banding pattern (Palva and Makela, 1980). Only the midribs of local leaves of LPSS.min.‐treated Arabidopsis were analysed as the method was not sufficiently sensitive for the detection of LPS in systemic leaves. After gel electrophoresis, LPS and proteins were stained with different techniques. The ProQ Emerald stain is specific for LPS and visualized the characteristic LPS pattern (dark red arrow) in midrib extracts of LPS‐treated leaves at time points of 1, 6 and 24 h (Fig. 5a, lanes 5–7). Equal loading was checked by staining the protein content of the samples with Sypro Ruby (Fig. 5b). Finally, silver stain was used for the detection of LPS and proteins together (Fig. 5c). Overall, using three different techniques, our results were strongly suggestive that LPS molecules are transported throughout the plant from inoculated leaves into systemic leaves.
The intercellular signal transduction mechanisms leading to the development of SAR are not well understood. Several molecules are currently being discussed as possible systemic SAR signals (Vlot et al., 2008). The original hypothesis, that SA could be the systemic signal for SAR, has been disputed (Vernooij et al., 1994; Vlot et al., 2009). Other molecules currently under discussion include methyl salicylate, azelaic acid, jasmonic acid derivatives and other lipid‐derived signals (e.g. Shah, 2009). It has been difficult to prove that any one signal is responsible for the systemic induction of SAR, and there are indications that multiple signals could be involved (Vlot et al., 2008). Flg22, the active epitope of flagellin, has been shown to elicit SAR in A. thaliana (Mishina and Zeier, 2007), but the mobility of bacterial peptides inside the plant has not been examined to date. It seems possible that peptides or LPS originating from pathogens or symbionts may be transported into the systemic leaves via the vascular system. In any case, the results of our study suggest that great care should be applied when analysing the data of experiments in which LPS is used for the induction of SAR and ISR. It is difficult to ascribe the observed biochemical responses to LPS as uniquely local or systemic, as these responses may overlap because of the mobility of LPS in the plant vascular system.
EXPERIMENTAL PROCEDURES
Plants
Arabidopsis thaliana (Col) was grown as described previously (Gerber et al., 2004). The lower leaves of A. thaliana were pressure inoculated with LPS or buffer A (2.5 mm MgCl2 and 1 mm CaCl2) using a 1‐mL needleless syringe. Inoculated leaves were labelled and harvested after 4, 8, 24 and 48 h, and used for ‘local’ analysis. Upper, noninoculated leaves were harvested at 24 and 48 h, and used for ‘systemic’ analysis. Plant material was stored at −80 °C until RNA preparation.
LPS preparation
LPS (1 mg/mL) were dissolved in water containing 2.5 mm MgCl2 and 1 mm CaCl2, shaken for 3 h on a mixer (Thermomixer Comfort, Eppendorf, Hamburg, Germany) at 1400 r.p.m., and stored at 4 °C until further use. If there is no other description, experiments were performed with LPS from an endophytic strain of B. cepacia (ASP B 2D), purified using the phenol–water method as described previously (Coventry and Dubery, 2001), or, for control, with buffer A containing 0.25 mm CaCl2 and 0.1 mm MgCl2.
Determination of SA
Free and conjugated SA were determined with some modifications following a standard protocol, using a high‐performance liquid chromatography (HPLC) system equipped with an autosampler, an RP‐18 Nucleosil‐Column and a fluorescence detector (excitation, 305 nm; emission, 407 nm) (Huang et al., 2004).
Quantification of bacterial growth
Two days before infection with P. syringae pv. tomato DC3000, plants were pretreated with either LPS to induce a possible resistance, or with buffer A (control). Three systemic leaves per plant were pressure infiltrated from the abaxial side with bacterial suspension using a 1‐mL needleless syringe. The concentration of the bacterial inoculum was equivalent to an optical density at 600 nm (OD600) of 0.0002, which correlates with 105 colony‐forming units/mL. Bacterial virulence was measured in an assay for bacterial multiplication within the host tissue as described previously (Zeidler et al., 2004).
Treatment of leaf tissue with fluorescent LPS
LPSS.min. (Alexa Fluor® 488 conjugate; Invitrogen, Darmstadt, Germany) were used to investigate LPS mobilization in Arabidopsis leaves. For this purpose, 100 µg of lyophilized LPS were dissolved in 1 mL double‐distilled H2O and incubated for 10 min at 37 °C and 1400 r.p.m. Leaves were then pressure inoculated with a needleless syringe from the abaxial side and analysed at the indicated time points using a fluorescence microscope (Axioskop, Zeiss, Jena, Germany) equipped with a digital camera (G2 powershot, Canon, Krefeld, Germany). Cross‐sections of LPS‐treated leaves were cut with a razor blade. The filter settings for fluorescein‐labelled LPS were as follows: green light filter, 505–530 nm. Chlorophyll autofluorescence was captured with a long‐pass filter (585 nm; Fig. 2b,e,h,k).
Capillary zone electrophoresis
Leaves were pressure infiltrated with fluorescently labelled LPSS.min. as described above, and incubated in the dark. After 1, 6 and 24 h, five to six leaves per time point were sampled; the veins of treated leaves were cut out, whereas systemic leaves were used as whole leaves. Veins or whole leaves were ground in liquid nitrogen using a mortar and pestle. Fine powder was dissolved in a concentration of 2 mg fresh weight/µL double‐distilled H2O. The mixture was incubated for 10 min at 37 °C and 1400 r.p.m., followed by centrifugation at 13 200 r.p.m. for 5 min. The supernatant was centrifuged again to remove all solid particles, and stored at −80 °C in the dark until use.
Capillary zone electrophoresis measurements were performed with a Beckman Coulter (Krefeld, Germany) P/ACE 5510 CE system, equipped with a fluorescence detector (excitation, 488 nm; emission, 520 nm), an autosampler and a power supply. Data acquisition was obtained by a computer with corresponding software (Gold Software Version 8.10). An uncoated fused silica capillary (inside diameter, 75 µm; outside diameter, 375 µm; length to detector, 50 cm; total length, 57 cm; Polymicro Technologies, Phoenix, AZ, USA), liquid cooled and filled with adequate buffer, was used for separation. The capillary was washed before and between each run, first with 0.1 m NaOH for 5 min and then with double‐distilled H2O for 2 min. Finally, the capillary was filled with separation buffer (20 mm carbonate buffer, 20 mm SDS), which was changed after every run. Samples were automatically applied by hydrodynamic injection for 2–5 s. Sample separation was performed at 32 °C and 25 kV for 6–7 min.
Analysis of LPS in SDS‐PAGE gels
For the separation of LPS, an SDS‐PAGE gel, consisting of a 12.5% separating and 4% stacking gel, was used, as described previously (Palva and Makela, 1980). LPS was stained with the ProQ Emerald 300 dye (Molecular Probes, Invitrogen, Darmstadt, Germany), which reacts with periodate‐oxidized carbohydrate groups, creating a bright green fluorescent signal. After LPS had been separated by standard SDS‐PAGE, the gels were immersed in 100 mL fixing solution for 45 min according to the manufacturer's instructions. This step was repeated once, and the gels were washed twice with 100 mL washing solution for 10 min. The carbohydrates were then oxidized with 25 mL oxidizing solution for 30 min. Gels were stained in freshly prepared ProQ Emerald 300 staining solution for 2 h. ProQ Emerald stain was visualized using a 300‐nm UV transilluminator.
Staining with Sypro® Ruby protein gel stain
The Sypro Ruby protein gel stain is an ultrasensitive, luminescent dye for the detection of proteins separated by PAGE (Bio‐Rad Laboratories, Munich, Germany). This dye could be used for the detection of proteins after staining the LPS gel with ProQ Emerald. Therefore, the gels were incubated in 50 mL Sypro Ruby protein gel stain overnight with gentle agitation on an orbital shaker. Proteins were visualized using a 300‐nm UV transilluminator.
Silver staining
After staining the LPS gel with ProQ Emerald and Sypro Ruby, the gels were additionally stained with silver nitrate to detect LPS and protein together. LPS and proteins in the gels were oxidized twice in 100 mL oxidizing solution for 30 min. The gels were then washed three times with double‐distilled H2O and stained as described previously (Tsai and Frasch, 1982).
REFERENCES
- Ausubel, F. , Katagiri, F. , Mindrinos, M. and Glazebrook, J. (1995) Use of Arabidopsis thaliana defense‐related mutants to dissect the plant response to pathogens. Proc. Natl. Acad. Sci. USA, 92, 4189–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, P.A. , Pieterse, C.M. and Van Loon, L.C. (2007) Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology, 97, 239–243. [DOI] [PubMed] [Google Scholar]
- Beers, E.P. and McDowell, J.M. (2001) Regulation and execution of programmed cell death in response to pathogens, stress and developmental cues. Curr. Opin. Plant Biol. 4, 561–567. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Che, F.S. , Nakajima, Y. , Tanaka, N. , Iwano, M. , Yoshida, T. , Takayama, S. , Kadota, I. and Isogai, A. (2000) Flagellin from an incompatible strain of Pseudomonas avenae induces a resistance response in cultured rice cells. J. Biol. Chem. 275, 32 347–32 356. [DOI] [PubMed] [Google Scholar]
- Coventry, H.S. and Dubery, I.A. (2001) Lipopolysaccharides from Burkholderia cepacia contribute to an enhanced defense capacity and the induction of pathogenesis‐related proteins in Nicotiana tabacum . Physiol. Mol. Plant Pathol. 58, 149–158. [Google Scholar]
- Dong, H. , Delaney, T.P. , Bauer, D.W. and Beer, S.V. (1999) Harpin induces disease resistance in Arabidopsis through the systemic acquired resistance pathway mediated by salicylic acid and the NIM1 gene. Plant J. 20, 207–215. [DOI] [PubMed] [Google Scholar]
- Dow, M. , Newman, M.A. and Von Roepenack, E. (2000) The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. and Dubery, I.A. (2004) Protein phosphorylation in Nicotiana tabacum cells in response to perception of lipopolysaccharides from Burkholderia cepacia . Phytochemistry, 65, 2957–2966. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. , Zeidler, D. , Durner, J. and Dubery, I.A. (2004) Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia . Planta, 218, 647–657. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. , Laukens, K. , Witters, E. and Dubery, I.A. (2006) Lipopolysaccharide‐responsive phosphoproteins in Nicotiana tabacum cells. Plant Physiol. Biochem. 44, 369–379. [DOI] [PubMed] [Google Scholar]
- Gerber, I.B. , Laukens, K. , De Vijlder, T. , Witters, E. and Dubery, I.A. (2008) Proteomic profiling of cellular targets of lipopolysaccharide‐induced signalling in Nicotiana tabacum BY‐2 cells. Biochim. Biophys. Acta, 1784, 1750–1762. [DOI] [PubMed] [Google Scholar]
- Gomez, S. and Stuefer, J.F. (2006) Members only: induced systemic resistance to herbivory in a clonal plant network. Oecologia, 147, 461–468. [DOI] [PubMed] [Google Scholar]
- Graham, T.L. , Sequeira, L. and Huang, T.S. (1977) Bacterial lipopolysaccharides as inducers of disease resistance in tobacco. Appl. Environ. Microbiol. 34, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, A. , Kapp, D. , Nielsen, T. and Niehaus, K. (2005) Endocytosis of Xanthomonas campestris pathovar campestris lipopolysaccharides in non‐host plant cells of Nicotiana tabacum . New Phytol. 165, 215–226. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Stettmaier, K. , Michel, C. , Hutzler, P. , Mueller, M.J. and Durner, J. (2004) Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana . Planta, 218, 938–946. [DOI] [PubMed] [Google Scholar]
- Hunt, M.D. , Neuenschwander, U.H. , Delaney, T.P. , Weymann, K.B. , Friedrich, L.B. , Lawton, K.A. , Steiner, H.‐Y. and Ryals, J.A. (1996) Recent advances in systemic acquired resistance research – a review. Gene, 179, 89–95. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Krause, M. and Durner, J. (2004) Harpin inactivates mitochondria in Arabidopsis suspension cells. Mol. Plant–Microbe Interact. 17, 131–139. [DOI] [PubMed] [Google Scholar]
- Lam, E. , Kato, N. and Lawton, M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature, 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Liu, Z. and Gaskin, R.E. (2004) Visualisation of the uptake of two model xenobiotics into bean leaves by confocal laser scanning microscopy: diffusion pathways and implication in phloem translocation. Pest Manag. Sci. 60, 434–439. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. , Bakker, P.A. , Van Der Heijdt, W.H. , Wendehenne, D. and Pugin, A. (2008) Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol. Plant–Microbe Interact. 21, 1609–1621. [DOI] [PubMed] [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Neumann, P.M. (2007) Evidence for long‐distance xylem transport of signal peptide activity from tomato roots. J. Exp. Bot. 58, 2217–2223. [DOI] [PubMed] [Google Scholar]
- Newman, M.A. , Von Roepenack‐Lahaye, E. , Parr, A. , Daniels, M.J. and Dow, J.M. (2002) Prior exposure to lipopolysaccharide potentiates expression of plant defenses in response to bacteria. Plant J. 29, 487–495. [DOI] [PubMed] [Google Scholar]
- Newman, M.A. , Dow, J.M. , Molinaro, A. and Parrilli, M. (2007) Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J. Endotoxin Res. 13, 69–84. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Palva, E.T. and Makela, P.H. (1980) Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur. J. Biochem. 107, 137–143. [DOI] [PubMed] [Google Scholar]
- Piater, L.A. , Nürnberger, T. and Dubery, I.A. (2004) Identification of a lipopolysaccharide responsive erk‐like MAPkinase in tobacco leaf tissue. Mol. Plant Pathol. 5, 331–341. [DOI] [PubMed] [Google Scholar]
- Rigano, L.A. , Payette, C. , Brouillard, G. , Marano, M.R. , Abramowicz, L. , Torres, P.S. , Yun, M. , Castagnaro, A.P. , Oirdi, M.E. , Dufour, V. , Malamud, F. , Dow, J.M. , Bouarab, K. and Vojnov, A.A. (2007) Bacterial cyclic beta‐(1,2)‐glucan acts in systemic suppression of plant immune responses. Plant Cell, 19, 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, H.S. and Friml, J. (2009) Auxin and other signals on the move in plants. Nat. Chem. Biol. 5, 325–332. [DOI] [PubMed] [Google Scholar]
- Ryals, J. , Uknes, S. and Ward, E. (1994) Systemic acquired resistance. Plant Physiol. 104, 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A. , Neuenschwander, U.H. , Willits, M.G. , Molina, A. , Steiner, H.‐Y. and Hunt, M.D. (1996) Systemic acquired resistance. Plant Cell, 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria, N. , Goring, D. , Nurnberger, T. and Dubery, I. (2008) Self/nonself perception and recognition mechanisms in plants: a comparison of self‐incompatibility and innate immunity. New Phytol. 178, 503–514. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. and Zipfel, C. (2008) News from the frontline: recent insights into PAMP‐triggered immunity in plants. Curr. Opin. Plant Biol. 11, 389–395. [DOI] [PubMed] [Google Scholar]
- Shah, J. (2009) Plants under attack: systemic signals in defence. Curr. Opin. Plant Biol. 12, 459–464. [DOI] [PubMed] [Google Scholar]
- Sidhu, V.K. , Vorholter, F.J. , Niehaus, K. and Watt, S.A. (2008) Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris . BMC Microbiol. 8, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo, A. , Sturiale, L. , Garozzo, D. , Erbs, G. , Jensen, T.T. , Lanzetta, R. , Dow, J.M. , Parrilli, M. , Newman, M.A. and Molinaro, A. (2008) The acylation and phosphorylation pattern of lipid A from Xanthomonas campestris strongly influences its ability to trigger the innate immune response in Arabidopsis. Chembiochem, 9, 896–904. [DOI] [PubMed] [Google Scholar]
- Tellstrom, V. , Usadel, B. , Thimm, O. , Stitt, M. , Kuster, H. and Niehaus, K. (2007) The lipopolysaccharide of Sinorhizobium meliloti suppresses defense‐associated gene expression in cell cultures of the host plant Medicago truncatula . Plant Physiol. 143, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.M. and Frasch, C.E. (1982) A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119, 115–119. [DOI] [PubMed] [Google Scholar]
- Vernooij, B. , Friedrich, L. , Morse, A. , Reist, R. , Kolditz‐Jawhar, R. , Ward, E. , Uknes, S. , Kessmann, H. and Ryals, J. (1994) Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell, 6, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C. , Klessig, D.F. and Park, S.W. (2008) Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11, 436–442. [DOI] [PubMed] [Google Scholar]
- Vlot, A.C. , Dempsey, D.A. and Klessig, D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. [DOI] [PubMed] [Google Scholar]
- Ward, E.R. , Uknes, S.J. , Williams, S.C. , Dincher, S.S. , Wiederhold, D.L. , Aleander, D.C. , Al‐Goy, P. , Métraux, J.P. and Ryals, J.A. (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell, 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. (1996) Tutorial: capillary electrophoresis. Chem. Educ. 1, 1–14. [Google Scholar]
- Zeidler, D. , Zahringer, U. , Gerber, I. , Dubery, I. , Hartung, T. , Bors, W. , Hutzler, P. and Durner, J. (2004) Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA, 101, 15 811–15 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2008) Pattern‐recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]


