SUMMARY
The cereal ear blight fungal pathogen Fusarium culmorum can infect Arabidopsis floral tissue, causing disease symptoms and mycotoxin production. Here we assessed the effect of seven mutants and one transgenic overexpression line, residing in either the salicylic acid (SA), jasmonic acid (JA) or ethylene (ET) defence signalling pathways, on the outcome of the Fusarium–Arabidopsis floral interaction. The bacterial susceptiblity mutant eds11 was also assessed. Flowering plants were spray inoculated with F. culmorum conidia to determine the host responses to initial infection and subsequent colonization. Enhanced susceptibility and higher concentrations of deoxynivalenol mycotoxin were observed in buds and flowers of the npr1 and eds11 mutants than in the wild‐type Col‐0 plants. An effect of the other two defence signalling pathways on disease was either absent (ET/JA combined), absent/minimal (ET) or inconclusive (JA). Overall, this study highlights a role for NPR1 and EDS11 in basal defence against F. culmorum in some floral organs. This is the first time that any of these well‐characterized defence signalling mutations have been evaluated for a role in floral defence in any plant species.
Ear blight disease of cereal crops, also known as head scab disease, is caused by several Fusarium species, including F. culmorum and F. graminearum, and results in considerable losses to yield, grain quality and safety (Goswami and Kistler, 2004; Parry et al., 1995). Developing grains become contaminated with various mycotoxins, for example deoxynivalenol (DON) (Hohn et al., 1998). In wheat the molecular basis of resistance is poorly understood, but is quantitative trait loci (QTL) based and Fusarium species non‐specific. Arabidopsis has been extensively used for molecular genetic investigations of numerous pathosystems. We have previously demonstrated that both F. culmorum and F. graminearum can infect Arabidopsis floral tissue, causing disease symptoms and mycotoxin production (Cuzick et al., 2008; Urban et al., 2002).
In Arabidopsis many functional plant resistance (R) genes have been identified and cloned as well as components of the downstream defence signalling network of both R‐gene and basal resistance, and those of importance to the induction of systemic responses. Salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) have been identified as key plant defence signalling molecules (reviewed in Glazebrook, 2005; Hammond‐Kosack and Parker, 2003). Genetic dissection of these signalling pathways by the research community has generated a collection of Arabidopsis mutants which exhibit either enhanced susceptibility or enhanced resistance to one or more pathogenic species. Here, we assess the levels of disease caused by F. culmorum in floral tissues of Arabidopsis ecotype Col‐0, in single gene mutants compromised in the SA, JA or ET pathway, and a transgenic line overexpressing the ERF1 protein known to integrate signals from the combined JA and ET signalling pathways.
Arabidopsis defence gene activation and defence signalling has extensively been studied in root or leaf tissue, as part of the local or systemic response to pathogen attack. To determine whether the known defence signalling genes were also expressed in floral tissues the Gene Atlas tool from GENEVESTIGATOR (http://www.genevestigator.ethz.ch) (Zimmermann et al., 2004) was queried for tissue‐specific expression of the SA pathway genes NPR1 and SID2, the JA pathway genes COI1 and JAR1, and the ET pathway genes EIN2, ERF1, ETO1 and ETR1 (Fig. 1; see Supplementary Experimental Procedures for further details). The signal intensity data recovered were the average of different microarray experiments using a variety of treatments and genotypes (including mutants) on the same standardized Affymetrix GeneChips. Figure 1A encompasses the ‘overall’ plant tissues whilst Fig. 1B focuses on the breakdown of the individual ‘floral’ subcomponents.
Figure 1.
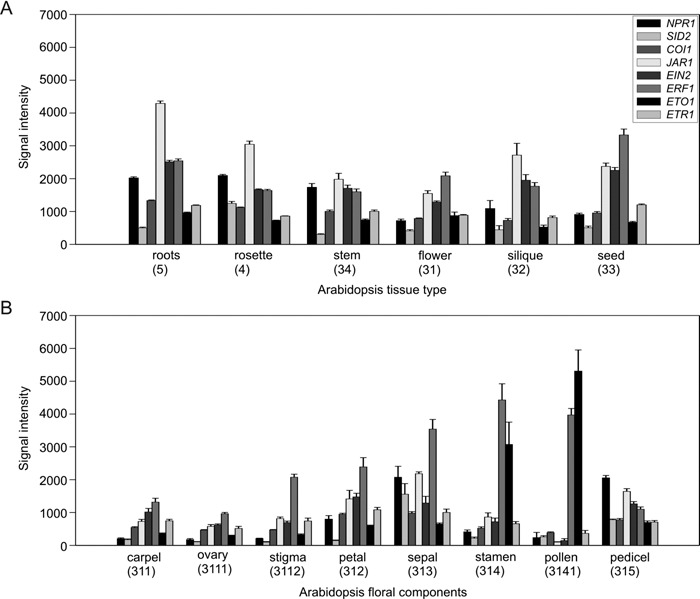
GENEVESTIGATOR analysis of the tissue‐specific expression profiles of the eight selected defence signalling genes. (A) Expression levels for individual genes for the six indicated plant tissues. (B) Expression levels of the same genes for eight specific flower tissue components. These data were extracted from the Gene Atlas Tool of the GENEVESTIGATOR microarray database (Zimmermann et al., 2004). Bars represent the mean signal intensities and error bars represent the standard error. The numbers in brackets below each tissue name refer to the GENEVESTIGATOR Arabidopsis anatomy nomenclature.
The defence signalling genes of interest were all found to be expressed in the overall ‘flower’ at levels higher than the default threshold signal intensity. However, the gene expression was found to differ greatly between the ‘floral’ subcomponents with SID2 transcript abundance being below this threshold for carpel, ovary, stigma and petal, NPR1 for ovary, and JAR1 and EIN2 for pollen. By contrast, NPR1 expression was highest in pedicel, expression of ETO1 was highest in pollen, and ERF1 was highest in stamen, sepal, petal, stigma, ovary and carpel. Therefore, all the defence signalling genes investigated within this study were expressed in Arabidopsis floral tissues.
The selected Arabidopsis defence signalling mutants and the single transgenic ERF1‐overexpressing line seed stocks were obtained from the following sources: coi1‐16 from John Turner (University of East Anglia, UK); sid2‐2/eds16‐1 from Fred Ausubel (Harvard University, Cambridge, MA); npr1‐1 from Xinnian Dong (Duke University, Durham, NC); and wild‐type Col‐0, ein2‐1 (N3071), 35S:ERF1 (N6142), etr1‐1 (N237), eto1‐1 (N3072) and jar1‐1(N8072) from the Nottingham Arabidopsis Stock Centre, UK. Plants were grown as described previously (Cuzick et al., 2008). For plant inoculations the F. culmorum strain 98/11 was propagated on synthetic nutrient‐poor agar (SNA) plates, and then transferred from confluent SNA plates to potato dextrose agar plates for 4 days to enable bulk conidia production (Urban et al., 2002). Floral spray inoculation of plants was done as described previously (Cuzick et al., 2008). Each flowering plant was sprayed with approximately 500 000 conidia (0.5 mL of 1 × 106 conidia mL−1). The numerical Fusarium–Arabidopsis disease individual floral component (FAD‐I value) scoring system was used to assess disease at 8 days post‐inoculation (dpi) on two floral subcomponents: (1) unopened and opened flowers and (2) new siliques that were fully open flowers at inoculation (Cuzick et al., 2008). Data were statistically analysed using mixed models (see Supplementary Experimental Procedures for further details). Representative floral images of inoculated plants were generated using a Leica MZFLIII dissecting microscope using light or UV light with a violet filter (excitation at 425/40 nm and emission at 475 nm) to visualize the differential plant cell responses (Fig. 2 and Supporting Information Fig. S2).
Figure 2.

The npr1 and eds11 buds and flowers are more susceptible to disease caused by Fusarium culmorum than wild‐type Col‐0. Arabidopsis floral tissues were spray inoculated with conidia and representative plants were photographed at day 11 in the light at either low (A, D and G) or high magnification (C, F and I) or at low magnification under UV light (B, E and H). Under UV light the chlorophyll present in the healthy green tissues autofluoresced red, whereas green autofluorescence was visible from the colonized tissues (white arrows). Scale bars = 0.5 mm. (A–C) Wild‐type Col‐0 had visible aerial mycelium (m) on the floral tissues and some buds and flowers were dried (d). The apical stem region remained green (C). (D–F) The npr1 mutant exhibited more disease than the wild‐type with extensive drying of buds and flowers. Occasionally plants exhibited constriction (c) of the apical stem tissue (F). (G–I) The eds11 mutant also had more disease than the wild‐type, and exhibited constriction of the apical stem tissue (I).
Although the entire plant received inoculum, disease development occurred only in the floral tissue of all genotypes tested. This floral‐specific infection had previously been observed (Urban et al., 2002). Interestingly, the floral spray inoculation data revealed that the new siliques of all the mutants tested in this study showed wild‐type levels of disease (Table 1). By contrast, in the flowers, the disease levels of the mutants in the SA defence signalling pathway differed from each other. The sid2 mutant, which has previously been reported to have reduced levels of SA (Wildermuth et al., 2001), exhibited wild‐type levels of disease in the flowers. By contrast, mutation of the NPR1 gene, genetically located downstream of SA production, resulted in enhanced susceptibility. In the wild‐type Col‐0 infections, aerial mycelium was visible on the buds and flowers with occasional drying of buds and flowers (Fig. 2A–C). In contrast, the npr1 mutant exhibited extensive drying of buds and flowers with aerial mycelium and occasional stem constriction within the apical stem region connected to the buds and flowers (Fig. 2D–F). In numerous pathosystems the npr1 mutation has been shown to cause enhanced disease susceptibility to many pathogen types, including oomycetes, fungi and bacteria (reviewed in Glazebrook, 2005; Hammond‐Kosack and Parker, 2003). The role of the NPR1 protein in pathogen defence has primarily been explored in leaf and root pathosystems. GENEVESTIGATOR analysis revealed that NPR1 expression was high in the pedicels and sepals, but far lower in the siliques. These differences in gene expression might be functionally relevant to floral defence because both the pedicels and the sepals were more susceptible to F. culmorum in the npr1 mutant, whereas the siliques remained as per the wild‐type. In Arabidopsis leaves and wheat ears, overexpression of the wild‐type Arabidopsis NPR1 gene led to increased resistance to F. graminearum (Makandar et al., 2006). However, in both studies NPR1 overexpression did not prevent initial F. graminearum infection, just the subsequent internal hyphal spread. In addition, Arabidopsis plants engineered to overexpress constitutively a specific transcription factor which led to the activation of several genes associated with systemic acquired resistance, including PR‐1, were found to be highly resistant to F. graminearum, when conidia were infiltrated into the leaves (Savitch et al., 2007). In this later study the transgenic Arabidopsis line NahG, which has highly reduced SA levels, was also reported to be more susceptible. This again implicates the involvement of the SA defence pathway in leaf tissue against F. graminearum. In contrast, assessment of the sid2 mutant in our study revealed wild‐type levels of disease. The sid2 mutation is in the isochorismate synthase 1 (ICS1) gene and results in reduced levels of SA (Wildermuth et al., 2001). In the sid2 mutant, SA biosynthesis may still occur via the phenylalanine ammonium lyase pathway. The NahG line expressing the bacterial salicylate hydroxylase gene was excluded from this study because of known pleiotrophic effects (Glazebrook et al., 2003; Heck et al., 2003). There is also the formal possibility that the NPR1 protein in floral tissue may be operating independently of SA to confer basal defence against F. culmorum.
Table 1.
Fusarium culmorum disease formation on the floral tissue of various defence signalling mutants and the corresponding wild‐type ecotype Col‐0.
| Arabidopsis genotype | Tissue type | |||||||
|---|---|---|---|---|---|---|---|---|
| Flowers | New siliques | Replication | ||||||
| Mean* | SEM | P‐value† | Mean | SEM | P‐value | N‡ | M§ | |
| Col‐0 (wt)¶ | 1.85 | 0.27 | n.a.** | 1.90 | 0.24 | n.a. | 244 | 19 |
| npr1 | 3.21 | 0.42 | 0.003 | 1.90 | 0.36 | n.s.†† | 103 | 6 |
| sid2/eds16 | 0.98 | 0.45 | n.s. | 1.90 | 0.38 | n.s. | 112 | 5 |
| coi1 | 0.15 | 0.57 | 0.006 | 0.62 | 0.50 | n.s. | 43 | 4 |
| jar1 | 0.83 | 0.46 | 0.039 | 1.53 | 0.40 | n.s. | 77 | 5 |
| ein2 | 2.77 | 0.43 | 0.045 | 2.21 | 0.38 | n.s. | 89 | 6 |
| 35S:ERF1 | 2.15 | 0.51 | n.s. | 1.56 | 0.45 | n.s. | 52 | 5 |
| eto1 | 0.74 | 0.64 | n.s. | 1.02 | 0.57 | n.s. | 28 | 3 |
| etr1 | 2.22 | 0.55 | n.s. | 2.37 | 0.48 | n.s. | 44 | 4 |
| eds11 | 3.62 | 0.48 | 0.001 | 2.74 | 0.41 | n.s. | 92 | 5 |
Estimated mean disease values determined using mixed model analysis for flowers and new siliques.
Represents the disease comparison between the Arabidopsis mutant genotype and the wild‐type.
A P‐value < 0.05 is considered to be statistically significant. ‡Total number of plants inoculated per Arabidopsis genotype.
Number of experiments in which the Arabidopsis mutant genotype was tested against the wild‐type.
Degrees of freedom for the flowers = 40 and for the new siliques = 34.
Not applicable.
††Not significant.
In Arabidopsis the JA and ET defence signalling pathways have been implicated in resistance to necrotrophic fungal pathogens such as Botrytis cinerea and Alternaria brassicicola (reviewed in Glazebrook, 2005). In the F. culmorum–Arabidopsis floral pathosystem the two JA‐insensitive mutants, coi1 and jar1, both exhibited reduced disease levels in the flowers in comparison with the wild‐type (Table 1). However, further observations revealed that the apparent increased resistance in the flowers seen in both JA mutants appeared to be primarily due to disease escape. The apical inflorescences of these two mutants were longer than in the wild‐type plants (see Supporting Information Fig. S1). A less acute form of this escape phenotype had previously been observed in wild‐type Col‐0 (Urban et al., 2002). In Arabidopsis JA is required for male fertility (Park et al., 2002). This reduced pollen fertility also makes the interpretation of the results difficult because F. culmorum requires the presence of pollen for successful infection (Strange et al., 1974; Strange and Smith, 1971; Urban et al., 2002). The coi1‐16 mutant used in this study is fertile when grown at 16 °C (Ellis and Turner, 2002). However, in the present study, under the conditions used for F. culmorum inoculation, numerous small siliques were observed, indicating reduced fertility (Fig. S1). Therefore, reduced fertility of coi1‐16 is the most likely explanation for the very low levels of disease in the flowers. The new siliques in comparison exhibited some disease, although there was a trend towards less disease than in the wild‐type. Although the jar1 mutant is male fertile, its inflorescences are less compact than in the wild‐type. Therefore, assessment of these two mutants proved to be inconclusive in defining a role for JA in the F. culmorum–Arabidopsis floral pathosystem.
Overall there were no differences in disease levels with the two ET mutants eto1 and etr1 in comparison with the wild‐type. The disease levels on the ein2 mutant exhibited a trend towards more disease than on the wild‐type Col‐0 plants. However, this was only weakly significant in six independent experiments. The ERF1 overexpression line did not influence F. culmorum floral disease levels in comparison with the wild‐type and had a wild‐type floral morphology. The ERF1 protein functions at the convergent point of the ET and JA signalling pathways, downstream of the biosynthesis of both molecules (Lorenzo et al., 2003). Therefore, the combined ET and JA defence pathways are not likely to be involved in basal resistance to F. culmorum in Arabidopsis floral tissue. Previously, the Arabidopsis 35S:ERF1 overexpression line was reported to display enhanced resistance to the necrotrophic root and vascular invading pathogen Fusarium oxysporum (Berrocal‐Lobo and Molina, 2004). By contrast, an Arabidopsis mutant which exhibits reduced resistance to F. oxysporum, called rfo1, has also been described (Diener and Ausubel, 2005). Preliminary F. culmorum floral spray data of the rfo1 mutant revealed wild‐type disease levels (data not shown). F. culmorum is capable of infecting cereal roots and stem bases (Parry et al., 1995). Collectively these observations suggest that different basal defence requirements either exist within specific tissue types, or alternatively operate against distinct but taxonomically related species.
To explore in greater detail the differential response of the bud/flower and silique tissue to Fusarium infection observed in some of the mutants spray inoculated with F. culmorum conidia, a minimal subset of mutants which represent the three main defence pathways, i.e. npr1, jar1 and ein2, were selected for point inoculation of apically wounded green siliques when ~14 mm in length. The apically wounded silique inoculations were done as described previously (Cuzick et al., 2008). A 1‐µL droplet of inoculum consisting of ~2000 conidia for strain 98/11 was placed on the wound. Typically 20 siliques (from six plants) were assessed per genotype. Data were statistically analysed using mixed models (see Supplementary Experimental Procedures for further details). There were no significant differences identified in disease progression between the wild‐type and npr1, jar1 and ein2 mutant plants (Fig. S2, P = 0.370, d.f. = 19). The average overall mean distance of disease progression at 8 dpi was 20.57 mm (SD = 0.44). These data correlate with the spray inoculation data for the new siliques, suggesting that F. culmorum defence in buds and flowers is more sensitive to alterations in the selected defence signalling pathways than new or older green siliques.
Following a preliminary floral spray screen with F. culmorum of other defence signalling mutants, the eds11 mutation was assessed in detail for disease levels. The eds11 mutation was originally identified when screening for mutants exhibiting enhanced bacterial disease susceptibility to the virulent bacterial pathogen Pseudomonas syringae pv. maculicola (Psm) ES4326 (Volko et al., 1998). The eds11‐1 seed was obtained from F. Ausubel (Harvard University). In the F. culmorum–Arabidopsis floral pathosystem the eds11 mutant exhibited significantly enhanced susceptibility in the flowers (Table 1). Again, disease levels in the new siliques were as per the wild‐type. The F. culmorum‐infected eds11 flowers were engulfed in aerial mycelium, dried out and occasional stem constriction within the main apical flower stem was seen (Fig. 2G–I). Therefore, the eds11 mutant exhibited comparable levels of disease with the npr1 mutant. The identification of enhanced susceptibility of the eds11 mutation to floral infection by F. culmorum was particularly intriguing. This mutant had previously been shown to exhibit enhanced leaf basal disease susceptibility to the virulent bacterial pathogen Psm ES4326 and was subsequently found to retain the ability to mount a hypersensitive response and to restrict the growth of the avirulent strain Psm ES4326/avrRpt2. This mutant was also more susceptible to the virulent P. syringae pv. tomato DC3000, and the multi‐host bacterial pathogen P. aeruginosa UCBPP‐PA14. However, no phenotypic differences were seen with either the leaf spotting bacterial pathogen Xanthomonas camprestris pv. raphani 1946, which is a necrotroph, or the obligate biotrophic fungal powdery mildew pathogen Erysiphe orontii isolate MGH (Volko et al., 1998). The eds11 mutant is a recessive mutation in a nuclear gene. However, this gene has not been isolated. The eds11 mutation complemented nine other previously characterised eds mutations as well as the npr1 mutation, indicating that EDS11 is not an allele of any of these genes (Volko et al., 1998). This new finding highlights that EDS11 has a role in both bacterial and fungal basal resistance, and has identified that EDS11 has a functional role in the defence of floral tissue.
A feature of F. culmorum and F. graminearum infection on wheat and Arabidopsis is the production of mycotoxins, such as the B‐type trichothecene DON (Cuzick et al., 2008; Hohn et al., 1998; Urban et al., 2002). We previously reported that DON is not required for F. graminearum floral infection of the Arabidopsis ecotype Ler‐0 (Cuzick et al., 2008). Currently it is not known whether F. culmorum requires DON for pathogenesis on Arabidopsis floral tissue. DON levels in enhanced F. culmorum floral susceptiblilty mutants npr1 and eds11 as well as wild‐type Col‐0 were quantified by using competitive ELISA as described previously (Cuzick et al., 2008). In each experiment, the floral tissue from six plants was pooled at least 8 dpi. Each experiment was done at least twice. Infected wild‐type Col‐0 flowers had a DON mean value of 0.9 p.p.m. (SD = 0.40), whereas the npr1 mutant had 4.4 p.p.m. (SD = 2.73) and the eds11 mutant had 3.2 p.p.m. (SD = 2.97). The water‐only sprayed plants exhibited low background DON levels (<0.2–0.3 p.p.m.) possibly due to non‐specific binding of plant components in the competitive ELISA reaction. These results suggest that increased DON levels may reflect the higher fungal biomass or alternatively that these two host mutations directly or indirectly led to greater DON accumulation. In the future, the use of Fusarium reporter strains containing a constitutive promoter or the TRI5 promotor fused to a suitable reporter protein to monitor the onset of mycotoxin production (Jansen et al., 2005) may help to explore further whether the DON levels accumulating are proportional to the fungal biomass present in each host genotype.
In the present study, the three classic Arabidopsis defence signalling pathways were quantitatively assessed for their contribution to basal defence in floral tissue to F. culmorum. This is the first time that the defensive role of these important gene mutations has been explored in floral tissue of any species. As a consequence, the results can be related only to susceptibility and resistance data generated in other Arabidopsis tissues. Altered disease levels were only observed within the buds and flower tissues of some mutants, whereas the silique tissues always exhibited wild‐type disease. We report that the Arabidopsis defence signalling mutations npr1 and eds11 independently increased susceptibility to F. culmorum in the flowers and both resulted in higher accumulation of DON mycotoxin than in wild‐type plants. By contrast, the interaction outcome was not altered in the 35S:ERF1 transgenic line, whereas the effects of the three ET defence signalling mutants were absent or minimal. The role of JA alone in this pathosystem remains inconclusive because the reduced fertility and/or the abnormal floral development of these mutants complicated the interpretation of the disease results obtained. Thus far, only one other mutation has been shown to enhance the susceptibility of Arabidopsis floral tissue to F. culmorum infection. This was esa1, originally identified with enhanced susceptibility to Alternaria brassicicola (Van Hemelrijck et al., 2006). However, this gene has still to be isolated.
The use of the Fusarium–Arabidopsis floral pathosystem to study defence signalling pathways in Arabidopsis has proven to be considerably more efficient than testing the equivalent eight mutations and one transgenic line in wheat through stable transgenesis. The results both correlate with and extend previously published data (Makandar et al., 2006). Genes conferring broad‐spectrum basal resistance, such as NPR1 and possibly EDS11, are ideal candidates for tissue‐specific targeted over‐ and/or inducible expression. This approach may lead to durable resistance against economically important pathogens of crop plants.
Supporting information
Fig. S1 Comparison of the floral morphology of the Arabidopsis ecotype Col‐0 (left) and the corresponding coi1‐16 jasmonic acid mutant (right) which is altered in fertility. The coi1‐16 mutant exhibits a longer inflorescence and predominantly stunted siliques with no or reduced quantities of seed in comparison with the wild‐type Col‐0 when grown at 22 °C. Similarly, the jar1‐1 mutant exhibited longer inflorescences than the wild‐type (data not shown). Scale bar = 1 cm.
Fig. S2 The wounded siliques of npr1, jar1 and ein2 mutants plants are as susceptible to Fusarium culmorum infection as wild‐type Col‐0. (A) A representative Col‐0 silique 8 dpi at the tip (white arrow). The fungus has colonized the entire silique (si) and part of the pedicel (p) tissue. Colonized tissues were bleached and white surface mycelium was visible. The green healthy stem (s) has not been colonized at this time‐point. Scale bar = 2 mm. (B) Close‐up images of (A) taken with a dissecting microscope under visble light (upper panel) and UV light (lower panel). The fully colonized silique and partially colonized pedicel tissue autofluoresce green under UV light, whereas the non‐colonized healthy pedicel and stem tissues autofluoresce red, indicative of healthy chlorophyll. Scale bar = 2 mm. (C) Quantification of disease progression from the apically wounded silique tips, through the silique and into the pedicel tissue. Bars represent the mean distance of disease progression and error bars the upper 95% confidence limit. No significant differences in disease progression were observed (P = 0.370, d.f. = 19).
Supplementary Experimental Procedures Data mining from the GENEVESTIGATOR microarray database
The Gene Atlas program of the microarray database GENEVESTIGATOR (Zimmermann et al., 2004) (http://www.genevestigator.ethz.ch) was used to search the expression levels of the defence signalling genes NON‐EXPRESSER OF PR GENES 1 (NPR1, At1g64280), SALICYLIC ACID INDUCTION DEFICIENT 2 (SID2, At1g74710), CORONATINE INSENSITIVE 1 (COI1, At2g39940), JASMONATE RESISTANT 1 (JAR1, At2g46370), ETHYLENE INSENSITIVE 2 (EIN2, At5g03280), ETHYLENE RESPONSE FACTOR 1 (ERF1, At5g47880), ETHYLENE OVERPRODUCER 1, (ETO1, At3g51770) and ETHYLENE RESPONSE 1 (ETR1, At1g66340) in different plant tissues. For chip type ‘ATH1:22k array’ was selected. The database searches were done on 19th November 2007. The signal intensity data available from GENEVESTIGATOR was an average of different microarray experiments using a variety of treatments and genotypes (including mutants) on the same standardised Affymetrix GeneChips. Significant gene expression was determined with average signal intensities of 200 and above. Only a subset of the specific tissues annotated by GENEVESTIGATOR using the Gene Ontology Consortium (http://www.geneontology.org/) were investigated.
Statistical analysis of treated plants
For the floral spray experiments a total of 19 independent experiments, consisting of randomised block designs, were done to compare fusarium disease severities between each Arabidopsis mutant and the wild‐type Col‐0 genotype. Each genotype was tested in at least three independent experiments. For the statistical analysis plot means were used (some transformations were evaluated but these did not improve the residuals) to fit the following linear mixed model:
yijk = µ + Expt i + Tray (Expt)ij + Genotype k + (Expt x Genotype)ik + eijk
where Expt is the experiment, Tray(Expt) is the block (or tray) within each experiment, Genotype identifies the mutant and wild‐type lines, Expt x Genotype is the interaction experiment x genotype. All components were assumed random (with the exception of Genotype effect), in order to consider inference to a much larger sample of experiments, and because not all of the mutant genotypes were present in each experiment. This model was fitted using restricted maximum likelihood (REML) with the inclusion of additional weights that corresponded to the number of plants per plot. Mutant plant disease scores were compared against Col‐0 using an asymptotic Wald test. All parameter estimates and tests were obtained using GenStat 8.0 (Payne et al., 2005) considering a significance level of 5%.
For the apically wounded silique experiments data was collected over two experiments from the Arabidopsis wild‐type Col‐0 and the npr1, jar1 and ein2 mutants. Wild‐type Col‐0 plants were present in both experiments to enable a comparison with the mutants. The response variables evaluated were DIS (the distance of disease), and P_Dis which corresponds to the affected proportion of the total silique and pedicel length. A logit transformation of the latter variable was evaluated but no improvement was noted in the analysis. The analysis of variance was obtained from fitting the following linear mixed model:
yijkl = µ + Expt i + Fusarium j + Indiv k + e ijkl
where, y is the score measurement for an individual plant, Expt is the fixed effect of experiment, Fusarium is the fixed effect of fusarium strain and Indiv is the random effect of an individual plant. The interaction experiment x fusarium was not incorporated because only one fusarium strain was present in both experiments. The random effect Indiv was included in order to consider the presence of several measurements on the same individual plant. Mean Fusarium disease progression differences were evaluated between the Arabidopsis genotypes. Each response variable was evaluated and tested for the significance of the factor Fusarium using a significance level of 5%.
References
Payne, R. W., Harding, S. A., Murray, D. A., Soutar, D. M., Baird, D. B., Welham, S. J., Kane, A. F., Gilmour, A. R., Thompson, R., Webster, R. and Tunnicliffe Wilson, G. (2005) The Guide to GenStat Release 8, Part 2: Statistics. Oxford: VSN International.
Zimmermann, P., Hirsch‐Hoffmann, M., Hennig, L. and Gruissem, W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We would like to thank Ian Pearman and Julian Franklin for maintenance of controlled environment facilities. Microscopy was done in the Bioimaging facility at Rothamsted Research. Rothamsted Research receives grant‐aided support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK. This study was additionally supported by a BBSRC responsive mode grant (BBS/B/12261).
REFERENCES
- Berrocal‐Lobo, M. and Molina, A. (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum . Mol. Plant–Microbe Interact. 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Cuzick, A. , Urban, M. and Hammond‐Kosack, K. (2008) Fusarium graminearum gene deletion mutants map1 and tri5 reveal similarities and differences in the pathogenicity requirements to cause disease on Arabidopsis and wheat floral tissue. New Phytol. 177, 990–1000. [DOI] [PubMed] [Google Scholar]
- Diener, A.C. and Ausubel, F.M. (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease‐resistance gene, is not race specific. Genetics, 171, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C. and Turner, J.G. (2002) A conditionally fertile coi1 allele indicates cross‐talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta, 215, 549–556. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. , Chen, W.J. , Estes, B. , Chang, H.S. , Nawrath, C. , Metraux, J.P. , Zhu, T. and Katagiri, F. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. [DOI] [PubMed] [Google Scholar]
- Goswami, R.S. and Kistler, H.C. (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. [DOI] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Parker, J.E. (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- Heck, S. , Grau, T. , Buchala, A. , Metraux, J.P. and Nawrath, C. (2003) Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis‐Pseudomonas syringae pv. tomato interaction. Plant J. 36, 342–352. [DOI] [PubMed] [Google Scholar]
- Hohn, T.M. , McCormick, S.P. , Alexander, N.J. , Desjardins, A.E. and Proctor, R.H. (1998) Function and biosynthesis of trichothecenes produced by Fusarium species In: Molecular Genetics of Host‐Specific Toxins in Plant Disease (Kohmoto K. and Yoder O.C., eds), pp. 17–24. Dordrecht: Kluwer Academic. [Google Scholar]
- Jansen, C. , Von Wettstein, D. , Schafer, W. , Kogel, K.H. , Felk, A. and Maier, F.J. (2005) Infection patterns in barley and wheat spikes inoculated with wild‐type and trichodiene synthase gene disrupted Fusarium graminearum . Proc. Natl Acad. Sci. USA, 102, 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O. , Piqueras, R. , Sanchez‐Serrano, J.J. and Solano, R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell, 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makandar, R. , Essig, J.S. , Schapaugh, M.A. , Trick, H.N. and Shah, J. (2006) Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1 . Mol. Plant–Microbe Interact. 19, 123–129. [DOI] [PubMed] [Google Scholar]
- Park, J.H. , Halitschke, R. , Kim, H.B. , Baldwin, I.T. , Feldmann, K.A. and Feyereisen, R. (2002) A knock‐out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. [DOI] [PubMed] [Google Scholar]
- Parry, D.W. , Jenkinson, P. and McLeod, L. (1995) Fusarium ear blight (scab) in small grain cereals—a review. Plant Pathol. 44, 207–238. [Google Scholar]
- Savitch, L.V. , Rajagopal, S. , Allard, G.C. and Jas, S. (2007) The GLK1 ‘regulon’ encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis . Biochem. Biophys. Res. Commun. 359, 234–238. [DOI] [PubMed] [Google Scholar]
- Strange, R.N. and Smith, H. (1971) A fungal growth stimulant in anthers which predisposes wheat to attack by Fusarium graminearum . Physiol. Plant Pathol. 1, 141–150. [Google Scholar]
- Strange, R.N. , Majer, J.R. and Smith, H. (1974) The isolation and identification of choline and betaine as the two major components in anthers and wheat germ that simulate Fusarium graminearum in vitro . Physiol. Plant Pathol. 4, 277–290. [Google Scholar]
- Urban, M. , Daniels, S. , Mott, E. and Hammond‐Kosack, K.E. (2002) Arabidopsis is susceptible to the cereal ear blight fungal pathogens Fusarium graminearum and Fusarium culmorum . Plant J. 32, 961–973. [DOI] [PubMed] [Google Scholar]
- Van Hemelrijck, W. , Wouters, P.F.W. , Brouwer, M. , Windelinckx, A. , Goderis, I.J.W.M. , De Bolle, M.F.C. , Thomma, B.P.H.J. , Cammue, B.P.A. and Delaure, S.L. (2006) The Arabidopsis defense response mutant esa1 as a model to discover novel resistance traits against Fusarium diseases. Plant Sci. 171, 585–595. [Google Scholar]
- Volko, S.M. , Boller, T. and Ausubel, F.M. (1998) Isolation of new Arabidopsis mutants with enhanced disease susceptibility to Pseudomonas syringae by direct screening. Genetics, 149, 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P. , Hirsch‐Hoffmann, M. , Hennig, L. and Gruissem, W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Comparison of the floral morphology of the Arabidopsis ecotype Col‐0 (left) and the corresponding coi1‐16 jasmonic acid mutant (right) which is altered in fertility. The coi1‐16 mutant exhibits a longer inflorescence and predominantly stunted siliques with no or reduced quantities of seed in comparison with the wild‐type Col‐0 when grown at 22 °C. Similarly, the jar1‐1 mutant exhibited longer inflorescences than the wild‐type (data not shown). Scale bar = 1 cm.
Fig. S2 The wounded siliques of npr1, jar1 and ein2 mutants plants are as susceptible to Fusarium culmorum infection as wild‐type Col‐0. (A) A representative Col‐0 silique 8 dpi at the tip (white arrow). The fungus has colonized the entire silique (si) and part of the pedicel (p) tissue. Colonized tissues were bleached and white surface mycelium was visible. The green healthy stem (s) has not been colonized at this time‐point. Scale bar = 2 mm. (B) Close‐up images of (A) taken with a dissecting microscope under visble light (upper panel) and UV light (lower panel). The fully colonized silique and partially colonized pedicel tissue autofluoresce green under UV light, whereas the non‐colonized healthy pedicel and stem tissues autofluoresce red, indicative of healthy chlorophyll. Scale bar = 2 mm. (C) Quantification of disease progression from the apically wounded silique tips, through the silique and into the pedicel tissue. Bars represent the mean distance of disease progression and error bars the upper 95% confidence limit. No significant differences in disease progression were observed (P = 0.370, d.f. = 19).
Supplementary Experimental Procedures Data mining from the GENEVESTIGATOR microarray database
The Gene Atlas program of the microarray database GENEVESTIGATOR (Zimmermann et al., 2004) (http://www.genevestigator.ethz.ch) was used to search the expression levels of the defence signalling genes NON‐EXPRESSER OF PR GENES 1 (NPR1, At1g64280), SALICYLIC ACID INDUCTION DEFICIENT 2 (SID2, At1g74710), CORONATINE INSENSITIVE 1 (COI1, At2g39940), JASMONATE RESISTANT 1 (JAR1, At2g46370), ETHYLENE INSENSITIVE 2 (EIN2, At5g03280), ETHYLENE RESPONSE FACTOR 1 (ERF1, At5g47880), ETHYLENE OVERPRODUCER 1, (ETO1, At3g51770) and ETHYLENE RESPONSE 1 (ETR1, At1g66340) in different plant tissues. For chip type ‘ATH1:22k array’ was selected. The database searches were done on 19th November 2007. The signal intensity data available from GENEVESTIGATOR was an average of different microarray experiments using a variety of treatments and genotypes (including mutants) on the same standardised Affymetrix GeneChips. Significant gene expression was determined with average signal intensities of 200 and above. Only a subset of the specific tissues annotated by GENEVESTIGATOR using the Gene Ontology Consortium (http://www.geneontology.org/) were investigated.
Statistical analysis of treated plants
For the floral spray experiments a total of 19 independent experiments, consisting of randomised block designs, were done to compare fusarium disease severities between each Arabidopsis mutant and the wild‐type Col‐0 genotype. Each genotype was tested in at least three independent experiments. For the statistical analysis plot means were used (some transformations were evaluated but these did not improve the residuals) to fit the following linear mixed model:
yijk = µ + Expt i + Tray (Expt)ij + Genotype k + (Expt x Genotype)ik + eijk
where Expt is the experiment, Tray(Expt) is the block (or tray) within each experiment, Genotype identifies the mutant and wild‐type lines, Expt x Genotype is the interaction experiment x genotype. All components were assumed random (with the exception of Genotype effect), in order to consider inference to a much larger sample of experiments, and because not all of the mutant genotypes were present in each experiment. This model was fitted using restricted maximum likelihood (REML) with the inclusion of additional weights that corresponded to the number of plants per plot. Mutant plant disease scores were compared against Col‐0 using an asymptotic Wald test. All parameter estimates and tests were obtained using GenStat 8.0 (Payne et al., 2005) considering a significance level of 5%.
For the apically wounded silique experiments data was collected over two experiments from the Arabidopsis wild‐type Col‐0 and the npr1, jar1 and ein2 mutants. Wild‐type Col‐0 plants were present in both experiments to enable a comparison with the mutants. The response variables evaluated were DIS (the distance of disease), and P_Dis which corresponds to the affected proportion of the total silique and pedicel length. A logit transformation of the latter variable was evaluated but no improvement was noted in the analysis. The analysis of variance was obtained from fitting the following linear mixed model:
yijkl = µ + Expt i + Fusarium j + Indiv k + e ijkl
where, y is the score measurement for an individual plant, Expt is the fixed effect of experiment, Fusarium is the fixed effect of fusarium strain and Indiv is the random effect of an individual plant. The interaction experiment x fusarium was not incorporated because only one fusarium strain was present in both experiments. The random effect Indiv was included in order to consider the presence of several measurements on the same individual plant. Mean Fusarium disease progression differences were evaluated between the Arabidopsis genotypes. Each response variable was evaluated and tested for the significance of the factor Fusarium using a significance level of 5%.
References
Payne, R. W., Harding, S. A., Murray, D. A., Soutar, D. M., Baird, D. B., Welham, S. J., Kane, A. F., Gilmour, A. R., Thompson, R., Webster, R. and Tunnicliffe Wilson, G. (2005) The Guide to GenStat Release 8, Part 2: Statistics. Oxford: VSN International.
Zimmermann, P., Hirsch‐Hoffmann, M., Hennig, L. and Gruissem, W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632.
Please note: Blackwell Publishing are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


