SUMMARY
Previous evidence has indicated that the P25 protein encoded by Potato virus X (PVX) inhibits either the assembly or function of the effector complexes in the RNA silencing‐based antiviral defence system (Bayne et al., Cell‐to‐cell movement of Potato Potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482). This finding prompted us to investigate the possibility that P25 targets the Argonaute (AGO) effector nuclease of RNA silencing. Co‐immunoprecipitation and Western blot analysis indicated that there is a strong interaction between P25 and AGO1 of Arabidopsis when these proteins are transiently co‐expressed in Nicotiana benthamiana. P25 also interacts with AGO1, AGO2, AGO3 and AGO4, but not with AGO5 and AGO9. As an effective suppressor, the amount of AGO1 accumulated in the presence of P25 was dramatically lower than that infiltrated with HcPro, but was restored when treated with a proteasome inhibitor MG132. These findings are consistent with the idea that RNA silencing is an antiviral defence mechanism and that the counter‐defence role of P25 is through the degradation of AGO proteins via the proteasome pathway. Further support for this idea is provided by the observation that plants treated with MG132 are less susceptible to PVX and its relative Bamboo mosaic virus.
INTRODUCTION
RNA silencing in eukaryotes serves a dual role as a regulator of gene expression and as a defence against viruses and transposable elements (Baulcombe, 2004; Brodersen and Voinnet, 2006; Voinnet, 2005). Viruses encode suppressor proteins to counteract this silencing‐based defence system at various levels (Voinnet, 2005; Voinnet et al., 1999). Thus, the suppressors P19 of Tomato bushy stunt virus (Vargason et al., 2003), P21 of Beet yellow virus (Ye and Patel, 2005) and P69 of Turnip yellow mosaic virus (Chen et al., 2004) bind double‐stranded (ds)RNAs that are intermediates in RNA silencing, whereas other suppressors disrupt or decrease the activity of core proteins in the silencing pathway. The suppressor P38 of Turnip crinkle virus targets an enzyme in RNA silencing (Dicer‐like 4: DCL4) (Deleris et al., 2006) that processes long dsRNA into short interfering (si)RNA. These siRNAs are normally the specificity determinant of the Argonaute (AGO) nuclease in silencing. There are also suppressors, including the 2b of Cucumber mosaic virus (CMV) (Zhang et al., 2006) and the P0 of Polerovirus, that target AGO directly (Baumberger et al., 2007). P0 is an F‐box protein component of E3 ubiquitin ligases (Pazhouhandeh et al., 2006) but, unlike most proteins in its class, it mediates the degradation of its target through a mechanism that is independent of the proteasome (Baumberger et al., 2007).
The silencing suppressor P25 of Potato virus X (PVX) (Voinnet et al., 2000) is a multifunctional protein (Bayne et al., 2005; Morozov and Solovyev, 2003) with nucleotide binding and RNA helicase activity (Kalinina et al., 2002; Morozov and Solovyev, 2003; Morozov et al., 1999) and the ability to induce plasmodesmatal gating (Angell et al., 1996; Howard et al., 2004). Its role as a viral movement protein is thought to be a function of the gating and suppressor functions: gating opens the channels for viral movement (Angell et al., 1996) and the suppressor function blocks an antiviral signal that normally interferes with the virus as it moves between cells (Bayne et al., 2005).
To explain the finding that the P25 suppressor does not prevent primary siRNA production, it has been proposed to act in RNA silencing pathways at the level of the AGO effector nuclease (Bayne et al., 2005). To test this hypothesis, we investigated the effect of P25 on AGO1 in Nicotiana benthamiana. We show here that P25 and AGO proteins interact, and that the level of AGO1 decreases in the presence of P25 through a process that can be blocked by the proteasome inhibitor MG132. On the basis of these data, we propose that P25 interacts with AGO1 and targets its degradation through the proteasome pathway. This process would suppress the antiviral defence mechanism based on RNA silencing and would thereby release a constraint on viral RNA accumulation.
RESULTS AND DISCUSSION
The expression level of AGO1 is reduced in the presence of P25
To investigate viral silencing suppressors, we used a transient assay system in N. benthamiana in which transgenes were introduced by the infiltration of Agrobacterium tumefaciens. These strains carry transgene DNA on plasmids that are transferred into the plant nuclear DNA and expressed in planta. The suppressors tested were P25, P25‐CT7 (P25 with C‐terminal T7 tag) (Bayne et al., 2005) and HcPro derived from Tobacco etch virus (Anandalakshmi et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998). We confirmed that each suppressed the silencing of a green fluorescent protein (GFP) transgene in the presence of a GFP inverted repeat RNA, as reported previously (Bayne et al., 2005; Dunoyer et al., 2004) (Fig. S1, see Supporting Information).
To test the hypothesis that P25 affects AGO proteins, we co‐infiltrated N. benthamiana leaves with Agrobacterium strains to express Flag‐tagged AGO1 (Baumberger and Baulcombe, 2005) together with the viral suppressors. A Flag‐tagged heterologous protein (Rx) and untagged β‐glucuronidase (GUS) were used as control proteins without any involvement in RNA silencing. Total proteins were harvested 3 days after Agrobacterium infiltration, and the epitope‐tagged proteins were immunoprecipitated with agarose beads containing anti‐Flag or anti‐T7 antibody. Proteins were then detected by Western blot analysis using either anti‐Flag or anti‐T7 antibody (Fig. 1A).
Figure 1.
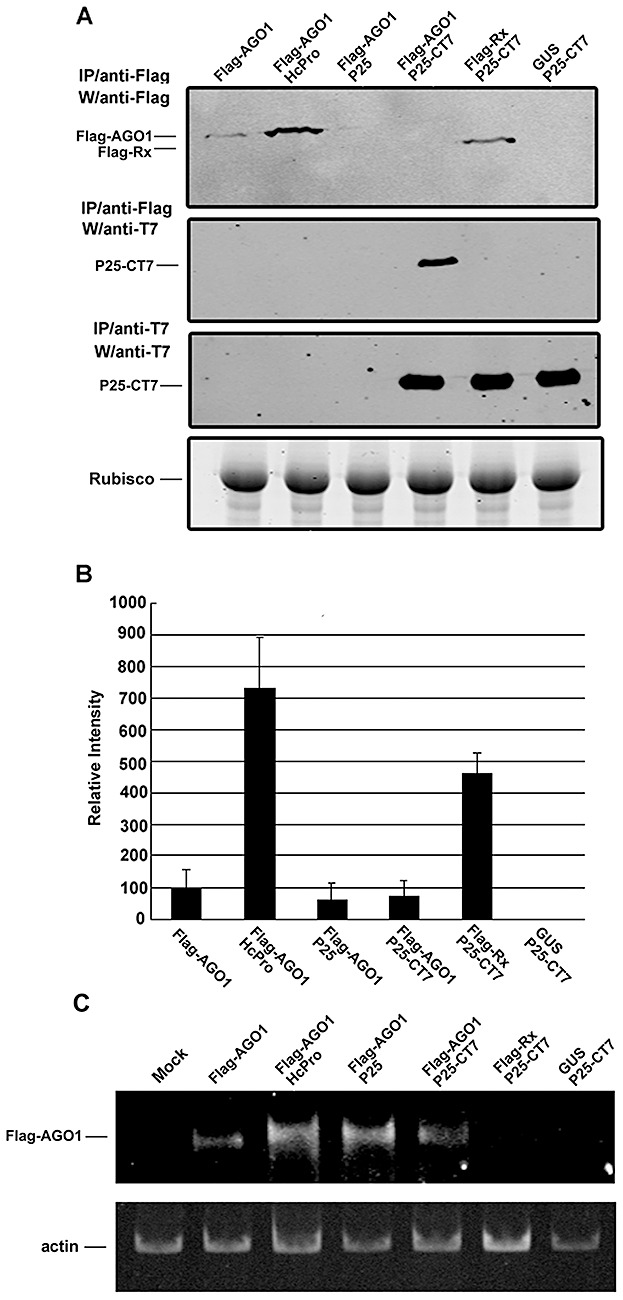
Interaction between P25 and Argonaute1 (AGO1) protein resulting in lower levels of AGO1 accumulation. (A) Immunoprecipitation of transiently expressed Flag‐tagged or T7‐tagged proteins from the total protein extracts of Agrobacterium‐infiltrated Nicotiana benthamiana leaves. Construct 35S:FLAG‐AGO1 (AGO1) was infiltrated alone or with 35S:HcPro (HcPro), 35S:P25 (P25) or 35S:P25‐T7 (P25‐CT7), indicated above each lane. Two controls 35S:Flag‐Rx (Rx) and 35S:GUS (GUS) were co‐infiltrated with 35S:P25‐T7 (P25‐CT7). Total proteins were extracted 3 days post‐infiltration. The Flag‐ or T7‐tagged proteins were immunoprecipitated (IP) with affinity gels or agarose beads containing anti‐Flag or anti‐T7 antibody, respectively. Western blot analysis (W) was performed using the antibody indicated to the left of each panel. Ribulose‐1,5‐bisphosphate carboxylase/oxygenase (Rubisco) protein stained with Coomassie blue served as a loading control. (B) The average of the accumulation levels of AGO1 from five independent experiments together with the standard error. (C) Flag‐AGO1 RNA accumulation in N. benthamiana leaves infiltrated with various constructs indicated at the top of each lane and inspected by reverse transcription‐polymerase chain reaction (RT‐PCR). The expression of the actin gene was used as a control.
The expression levels of AGO1 in N. benthamiana plants were seven‐fold higher when co‐expressed with suppressor HcPro (Fig. 1) than when expressed alone. This increase is probably a result of HcPro having suppressed the self‐silencing of the Flag‐AGO1 transgene in the transient assay and the silencing of this transgene by the endogenous miRNA168 (Vaucheret et al., 2004, 2006). However, the levels of AGO1, when co‐expressed with silencing suppressor P25 or P25‐CT7, were lower than when expressed alone (Fig. 1A,B), although both proteins were functional as suppressors of silencing (Fig. S1). The low level of expression of Flag‐AGO1 was unrelated to the presence of the epitope tag, because Flag‐Rx was expressed at a high level in the presence of P25‐CT7 (Fig. 1A,B), and maintained at the same levels in the presence or absence of P25 or P25‐CT7 (Fig. S2, see Supporting Information).
These data on the accumulation of AGO1 protein are consistent with the possibility that P25 might suppress silencing through an effect on the stability of AGO1. This interpretation was further reinforced by RNA analysis, which showed that the expression of Flag‐AGO1 mRNA, assessed by reverse transcription‐polymerase chain reaction (RT‐PCR), was similar in the presence of HcPro, P25 or P25‐CT7 (Fig. 1C), and higher than in the absence of these suppressors. Thus, in the absence of silencing suppressors, the transiently expressed AGO1 feeds back to suppress its own mRNA through RNA silencing. However, in the presence of the suppressors of silencing, the feedback loop is broken and there is overaccumulation of AGO1 mRNA.
P25 interacts selectively with AGO proteins
To determine whether P25 can interact with AGO1, as does the 2b suppressor of CMV (Zhang et al., 2006), we assayed for the co‐immunoprecipitation of Flag‐AGO1 and P25‐CT7. The Flag and T7 antibodies were highly specific because there was no evidence for co‐immunoprecipitation of FLAG‐ or CT7‐tagged proteins when P25‐CT7 was co‐expressed with Flag‐Rx or GUS (Fig. 1A). An interaction of P25‐CT7 and Flag‐AGO1 could be detected in the anti‐Flag immunoprecipitated extract from plants co‐expressing P25‐CT7 and Flag‐AGO1 (Fig. 1A, second panel from the top), but not in the reciprocal reaction with anti‐T7 (data not shown). To explain this discrepancy, we propose that all or most of Flag‐AGO1 was bound to P25‐CT7, but only a small proportion of P25‐CT7 was bound to Flag‐AGO1. In this scenario, the discrepancy would result because FLAG‐AGO1 immunoprecipitation would enrich for the P25‐CT7 signal more effectively than the reciprocal reaction would enrich for FLAG‐AGO1. Alternatively, AGO1 could form a complex with other proteins, including P25; therefore, the AGO1 complex could immunoprecipitate P25 by an indirect interaction; P25 might fail to immunoprecipitate AGO1 because it might not be interacting directly.
The AGO gene family in Arabidopsis has 10 members. Co‐expression of P25‐CT7 or HcPro with other Flag‐tagged AGOs revealed that P25 can interact with AGO2, AGO3 and AGO4 in addition to AGO1, but not with AGO5 or AGO9 (Fig. 2B). These results suggest that AGO5 and AGO9 might form different complexes with distinct partners not including the P25 interactors.
Figure 2.
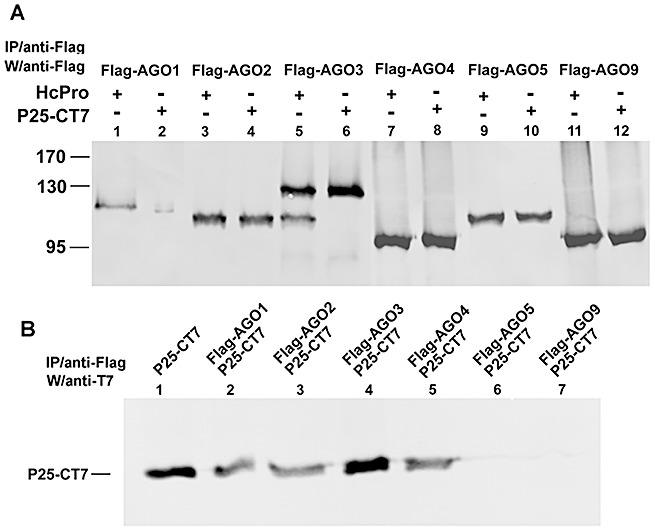
P25 interacts with Argonaute1 (AGO1), AGO2, AGO3 and AGO4, but not with AGO5 and AGO9. Immunoprecipitation of transiently expressed Flag‐tagged proteins from the total protein extracts of Agrobacterium‐infiltrated Nicotiana benthamiana leaves. (A) Constructs of 35S:FLAG‐AGOs (AGO1 in lanes 1 and 2; AGO2 in lanes 3 and 4; AGO3 in lanes 5 and 6; AGO4 in lanes 7 and 8; AGO5 in lanes 9 and 10; AGO9 in lanes 11 and 12) were co‐infiltrated with 35S:HcPro (HcPro) or 35S:P25‐T7 (P25‐CT7), indicated above each lane. Total proteins were extracted 72 h post‐infiltration. (B) The Flag‐tagged proteins were immunoprecipitated (IP) with the anti‐Flag antibody, except lane 1, and the samples were probed with anti‐T7 antibody. Lane 1 is the plant extract of transiently expressed P25‐CT7 loaded as a positive control.
However, of the P25‐interacting isoforms, only AGO1 was less abundant when co‐infiltrated with P25 (Fig. 2A). This result suggests that P25 is unlike the 2b protein (Zhang et al., 2006) that causes suppression of silencing as a consequence of binding to AGO1. With P25, binding to AGO is not sufficient for the suppression of silencing, and there must be a second step in which P25 targets AGO protein degradation.
Interestingly, in the controls for these experiments, a short AGO3 fragment was produced in the presence of HcPro (Fig. 2A, lane 5). As this shorter fragment can be detected with antibody against Flag at the N‐terminus of Flag‐AGO3, a cleavage site for HcPro (a potyvirus‐encoded protease) could be located towards the C‐terminus of AGO3. Such a site AKVGG905–909 in AGO3 is very similar to that (YKVGG) of Tobacco vein mottling virus (Domier et al., 1986). The alanine–tyrosine difference would affect the protease targeting mechanism (Carrington and Herndon, 1992) and the partial cleavage of AGO3 by HcPro (Fig. 2A).
Proteasome inhibitor MG132 prevents destabilization of AGO1 in the presence of P25
The P0 Polerovirus suppressor of silencing binds to AGO1 and destabilizes it in a proteasome‐independent manner (Bortolamiol et al., 2007). To determine whether P25 acts in the same way as P0, we tested the effect of the proteasome‐specific inhibitor MG132 (Hershko and Ciechanover, 1992) on P25‐CT7‐mediated destabilization of AGO1. MG132 (100 µm) was infiltrated into N. benthamiana leaves 2 h before co‐infiltration with Agrobacterium strains to express P25 and AGO1, and the total proteins were extracted after 24 h. Figure 3A shows that the expression level of P25‐CT7 and the interaction of P25‐CT7 with AGO1 were unaffected by MG132 (lanes 5 and 6). However, MG132 enhanced the accumulation of AGO1 by about 10‐fold in the presence of P25 or P25‐CT7 (Fig. 3A, lanes 5–8 and Fig. 3B), but had no effect in the presence of HcPro or when AGO1 was expressed alone (Fig. 3A, lanes 1–4 and Fig. 3B). These results suggest that the low levels of AGO1 in the presence of P25 may be the result of an interaction between these proteins that leads to AGO1 degradation through a proteasome‐dependent pathway. However, the amount of P25 co‐precipitated remains the same with or without MG132. It is possible that P25 interacts at the region near the Flag tag of AGO1 that can be co‐precipitated even after degradation.
Figure 3.
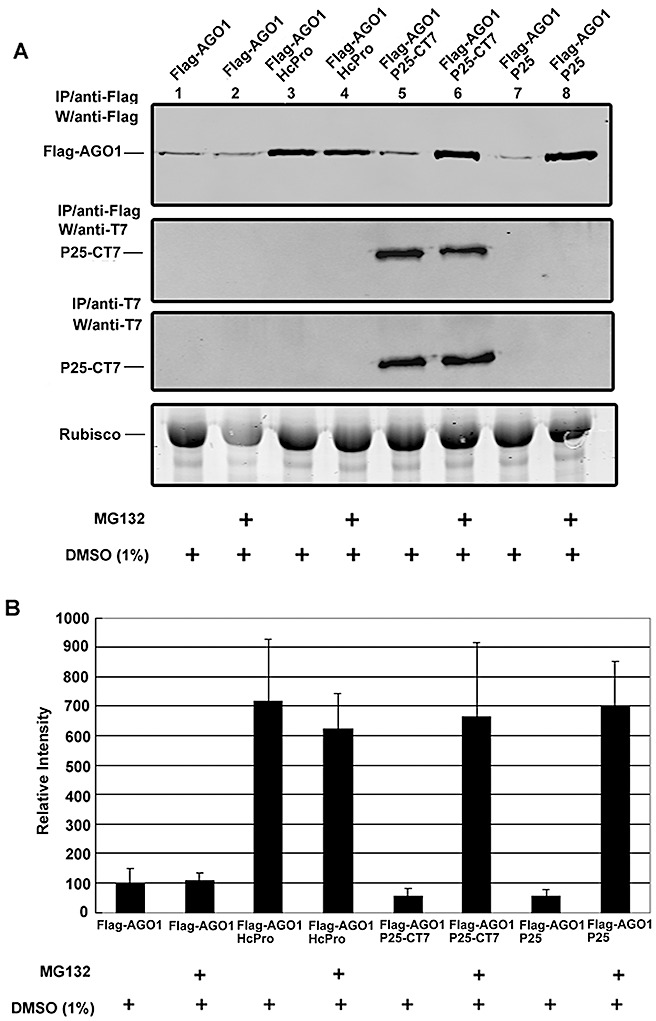
Proteasome inhibitor MG132 blocks the P25‐mediated destabilization of AGO1 in Nicotiana benthamiana. (A) 35S:FLAG‐AGO1 (AGO1) co‐expressed with 35S:HcPro (HcPro), 35S:P25‐T7 (P25‐CT7) or35S:P25 (P25) in N. benthamiana leaves, and then treated with 100 µm of MG132 or 1% dimethylsulphoxide (DMSO) as indicated at the bottom of each lane. The levels of FLAG‐AGO1 protein were analysed 24 h later by Western blotting. The Flag‐ or T7‐tagged proteins were immunoprecipitated (IP) with affinity gels or agarose beads containing anti‐Flag or anti‐T7 antibody, respectively. Western blot analysis (W) was performed using the antibody indicated to the left of each panel. Ribulose‐1,5‐bisphosphate carboxylase/oxygenase (Rubisco) protein stained with Coomassie blue served as a loading control. (B) The average of the accumulation levels of AGO1 from three independent experiments together with the standard error.
Proteasome degradation is normally dependent on the ubiquitination of the targeted protein. However, we failed to detect the ubiquitination of AGO1 when co‐expressed with P25 or P25‐CT7, and it could be that an efficient ubiquitination–deubiquitination cycle occurs so that only a small proportion of the AGO1 protein is ubiquitinated at any one time. Alternatively, it could be that AGO1 is only monoubiquitinated, so that the number of ubiquitin residues on AGO1 is below the limit of detection. A further possibility is that the P25‐mediated degradation is through a ubiquitin‐independent proteasome pathway, as with the activation of NF‐κB (Rape and Jentsch, 2002) and the degradation of ornithine decarboxylase (Zhang and Coffino, 2004).
Proteasome inhibitor MG132 interferes with virus accumulation in plants
We predicted that, if the P25‐mediated destabilization of AGO1 is required for the suppression of an antiviral RNA silencing pathway, its suppression would enhance resistance to PVX and related viruses, but not to unrelated viruses with silencing suppressors acting in a different manner. To test this hypothesis, we applied MG132 30 min before virus infection to prevent P25‐mediated degradation of AGO1 in infected N. benthamiana. The accumulation of viral coat protein was then analysed on Western blots at 12, 24 and 36 h post‐inoculation. The results in Fig. 4 show that the plants treated with MG132 exhibited increased resistance to potexviruses [Bamboo mosaic virus (BaMV) and PVX], but remained fully susceptible to CMV, as predicted. Presumably, the interaction of the silencing suppressor 2b of CMV with AGO1 (Zhang et al., 2006) interferes with silencing through a mechanism that is proteasome‐independent.
Figure 4.
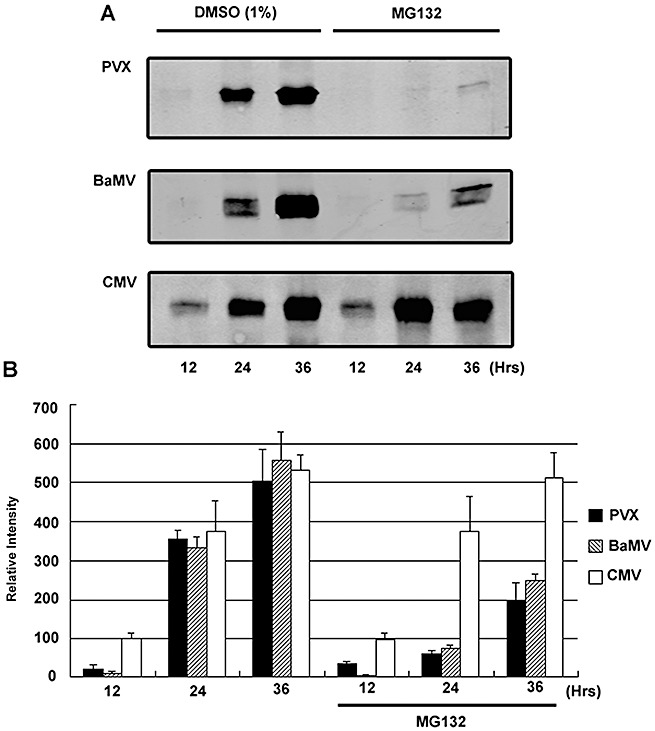
Potexvirus accumulation is reduced by proteasome inhibitor MG132. (A) Western blot analysis of the coat protein accumulation from Nicotiana benthamiana leaves inoculated with Potato virus X (PVX), Bamboo mosaic virus (BaMV) and Cucumber mosaic virus (CMV). Proteasome inhibitor MG132 was infiltrated into the leaves immediately after virus inoculation. Total proteins were harvested after 12, 24 and 36 h, indicated at the bottom of each lane. The coat protein accumulations of the viruses were detected with antiserum against PVX, BaMV and CMV. (B) The average of the coat protein accumulation at 12, 24 and 36 h, with or without proteasome inhibitor MG132, from three independent experiments is indicated, together with the standard error.
To further investigate whether the presence of P25 produced by viral infection can reduce the accumulation of AGO1, we transfected PVX RNA onto N. benthamiana leaves which were transiently expressing FLAG‐AGO1. Western blotting revealed that there was no significant difference in AGO1 accumulation on inoculated and noninoculated leaves (data not shown). However, when protoplasts were derived from N. benthamiana leaves expressing FLAG‐AGO1 and transfected with PVX RNA, there was a reduction in AGO1 after 48 hpi (Fig. 5).
Figure 5.
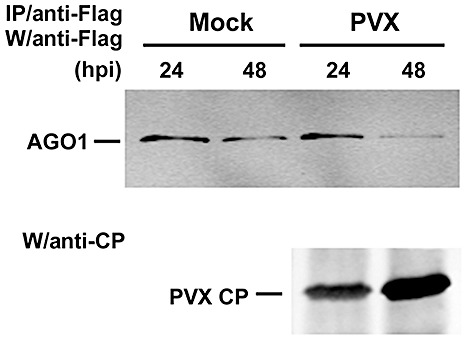
Argonaute1 (AGO1) is destabilized during Potato virus X (PVX) inoculation in Nicotiana benthamiana protoplasts. Immunoprecipitation of transiently expressed Flag‐tagged AGO1 from the total protein extracts of PVX‐inoculated protoplasts derived from Agrobacterium‐infiltrated N. benthamiana leaves. The levels of FLAG‐AGO1 (AGO1) protein were analysed 24 and 48 h post‐inoculation (hpi) by Western blotting. The Flag‐tagged proteins were immunoprecipitated (IP) with affinity gels containing anti‐Flag antibody. Western blot analysis (W) was performed using the antibody indicated. CP, coat protein.
To explain the contrasting results with protoplasts and leaves (4, 5), we propose that AGO1 is destabilized in infected cells at the specific stage of the infection cycle at which P25 accumulates abundantly. The transfected protoplasts would be infected in a synchronized manner, so that all cells would exhibit the decrease in AGO1 at the same time. In contrast, in the infected leaves, the cells would be at different stages of the infection cycle, and the decrease in AGO1 in some cells would be masked by the high levels in other cells at a different stage of the infection cycle.
At present, we do not know how the P25 interaction leads to the destabilization of AGO1 or, indeed, whether the interaction with AGO1 is direct or indirect. At present, we favour a direct interaction because AGO1 forms an active effector of silencing without being complexed to other proteins (Baumberger and Baulcombe, 2005). P25 could be a part of a ubiquitin ligase complex and, through the interaction with AGO1, leads to degradation in the ubiquitin proteasome pathway. However, we cannot formally rule out an indirect interaction.
EXPERIMENTAL PROCEDURES
Plants and constructs
Wild‐type N. benthamiana and the GFP transgenic lines 16c (Brigneti et al., 1998) were grown in a growth chamber at 28 °C under a 16‐h light/8‐h dark cycle. Constructs of pBIN61‐P25, ‐HcPro, ‐FlagRx, ‐GUS, ‐P25‐CT7, ‐FlagAGO1, ‐GFIR, ‐FlagAGO4, ‐FlagAGO4, pN‐FlagAGO2, ‐FlagAGO3 and ‐FlagAGO9 have been described previously (Baumberger et al., 2007).
GFP imaging
Hand‐held, high‐intensity UV lamps (B‐100SP UVP) and a Pentex N73 digital camera (Pentax, Golden, CO, USA) with HAKUBA YG filter (HAKUBA, Tokyo, JAPAN) were used for 16c plants infiltrated by Agrobacterium carrying a GF‐IR construct (Baumberger et al., 2007; Ruiz et al., 1998; Schwach et al., 2005).
Agrobacterium‐mediated transient expression (agroinfiltration)
Constructs in the pBIN61 binary vector (Bendahmane et al., 2000) were transformed into A. tumefaciens strain C58C1 carrying the virulence helper plasmid pCH32 (Bendahmane, 1999). Agrobacterium cells were inoculated into 1 mL of 2YT medium supplemented with 50 µg/mL kanamycin and 5 µg/mL tetracycline and grown at 28 °C overnight. They were then subcultured to 10 mL of 2YT medium containing 50 µg/mL kanamycin, 5 µg/mL tetracycline, 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES) buffer and 450 µm acetosyringone overnight. Cells were precipitated and resuspended to a final optical density at 600 nm (OD600) of 0.5 in a solution containing 10 mm MgCl2, 10 mm MES, pH 5.6, and 450 µm acetosyringone. Cultures were incubated at room temperature for 3 h before agroinfiltration. For the co‐infiltration of Agrobacterium cultures carrying other constructs, equal volumes of both cultures were mixed before agroinfiltration.
Co‐immunoprecipitation experiments and Western blot analyses
Flag‐AGO1 and P25‐CT7 immunoprecipitation was performed with proteins from transiently expressed leaves of N. benthamiana using anti‐Flag M2 affinity gel (Sigma, St. Louis, MO, USA) and T7 tag antibody agarose beads (Novagen, Madison, WI, USA), respectively. Purified proteins derived from affinity gel or agarose beads were boiled for 5 min in Laemmli sample buffer and resolved in an 8% or 12% sodium dodecylsulphate‐polyacrylamide gel. Proteins were then electroblotted onto a nitrocellulose membrane (Protran; Schleicher & Schuell, Dassel, Germany), and incubated with a primary rabbit polyclonal antiserum against PVX, CMV and BaMV coat protein, rabbit anti‐Flag polyclonal antibody (Sigma), and rabbit anti‐T7 polyclonal antibody (ICL, Newberg, OR, USA). The final detection was performed using secondary fluorescent anti‐rabbit IgG (Rockland, Gilbertsville, PA, USA). Data analysis was carried out with LI‐COR (Omaha, NE, USA) Odyssey.
RT‐PCR
Total RNAs were extracted from N. benthamiana leaves with hot phenol and precipitated with LiCl; then mRNAs were purified by oligo(dT) coupled to paramagnetic beads (Dynal A.S., Oslo, Norway). Purified mRNAs were subjected to reverse transcription with reverse primer AGO‐RT (5′‐CTTTCTCGTCTTTGACCTCCA‐3′) and following the recommendations of the manufacturer. PCR was performed with 5 pmol each of forward (5′‐GTCATCGTCATCCTTGTAGTCCAT‐3′) and reverse (5′‐GAAGGAGGTGAAGGCTCTGG‐3′) primers. The PCR product of actin mRNA was used as a normalization control as described previously (Lin et al., 2007).
Protoplast isolation and viral RNA inoculation
The preparation of N. benthamiana protoplasts and viral RNA inoculation have been described previously (Tsai et al., 1999). In brief, protoplasts were prepared from the leaves of 6‐week‐old N. benthamiana plants that had been agroinfiltrated with the constructs to transiently express HcPro and Flag‐AGO1 for 1 day. About 5 × 105 cells were inoculated with 1 µg of PVX virion RNA or water (control) by the polyethylene glycol method (Tsai et al., 1999) and incubated in culture medium (1 µm CuSO4, 10 mm MgSO4, 1 µm KI, 0.2 mm KPO4, 10 mm KNO3, pH 6.5, 10 mm CaCl2, 0.03% cephoridins, 0.001% gentamycin, in 0.55 m mannitol–MES solution) at 25 °C under constant light. Cells were harvested at 24 h or 48 h post‐inoculation using liquid nitrogen to freeze–thaw twice after removing the medium. The cells were then vortexed for 30 s with 500 µL of cold buffer (20 mm Tris/HCl, pH 7.5, 300 mm NaCl, 2 mm MgCl2) supplemented with 5 mm dithiothreitol (DTT) and protease inhibitor. The debris was removed by centrifugation at 16 000 g for 10 min twice, and the supernatant was subsequently incubated with anti‐Flag agarose beads at 4 °C for 4 h. The agarose beads were then washed with 500 µL cold buffer with 0.5% NP‐40 once, and then with 500 µL wash buffer [40 mm N‐2‐hydroxyethylpiperazine‐N′‐2‐ethanesulphonic acid (Hepes)‐KOH, pH 7.5, 100 mm KOAc and 5 mm Mg(OAc)2] once. Finally, the precipitated proteins were run on an 8% sodium dodecylsulphate‐polyacrylamide gel and analysed by Western blotting using rabbit anti‐Flag antibody as the primary antibody.
Supporting information
Fig. S1 The silencing suppressor activity assay in green fluorescent protein (GFP) transgenic Nicotiana benthamiana line 16c. Normal healthy and GFP transgenic 16c N. benthamiana leaves under UV illumination are shown as reference. Control leaf is indicated as GFIR and GFIR+GUS for parts infiltrated with Agrobacterium strains carrying pBin61‐GFIR and pBin61GFIR plus pBin61‐GUS, respectively. The three leaves in the right panel were treated in the same manner as the control leaf but co‐infiltrated with different silencing suppressors, indicated as P25‐CT7 (pBin61‐P25CT7; PVX P25 with T7 tag at C‐terminus), P25 (pBin61‐P25) and HcPro (pBin61‐HcPro; derived from Potyvirus). The suppressor activity of P25 and HcPro was observed under UV light at 5 days post‐infiltration.
Fig. S2 The expression level of Rx protein is not affected by P25 or P25‐CT7. Immunoprecipitation of transiently expressed Flag‐tagged Rx protein from the total protein extracts of Agrobacterium‐infiltrated Nicotiana benthamiana leaves. Construct 35S:Flag‐Rx (Rx) was infiltrated alone or co‐infiltrated with 35S:P25 (P25) or 35S:P25‐T7 (P25‐CT7) indicated above each lane. Total proteins were extracted 3 days post‐infiltration. The Flag‐tagged proteins were immunoprecipitated (IP) with agarose beads containing anti‐Flag antibody. Western blot analysis (W) was performed using the anti‐Flag antibody indicated.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
This work was supported by grants from the National Science Council Project NSC 95‐2752‐B‐005‐012‐PAE and NSC 96‐2752‐B‐005‐012‐PAE, the Gatsby Charitable Foundation and by a Royal Society Research Professorship to DCB.
REFERENCES
- Anandalakshmi, R. , Pruss, G.J. , Ge, X. , Marathe, R. , Smith, T.H. and Vance, V.B. (1998) A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA, 95, 13 079–13 084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angell, S.M. , Davies, C. and Baulcombe, D.C. (1996) Cell‐to‐cell movement of potato virus X is associated with a change in the size exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii . Virology, 215, 197–201. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Baumberger, N. and Baulcombe, D.C. (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits micro RNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA, 102, 11 928–11 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N. , Tsai, C.H. , Lie, M. , Havecker, E. and Baulcombe, D.C. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614. [DOI] [PubMed] [Google Scholar]
- Bayne, E.H. , Rakitina, D.V. , Morozov, S.Y. and Baulcombe, D.C. (2005) Cell‐to‐cell movement of Potato Potexvirus X is dependent on suppression of RNA silencing. Plant J. 44, 471–482. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. (1999) Zero‐background plasmid vector for BAC library construction. BioTechniques, 26, 228–232. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Querci, M. , Kanyuka, K. and Baulcombe, D.C. (2000) Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Bortolamiol, D. , Pazhouhandeh, M. , Marrocco, K. , Genschik, P. and Ziegler‐Graff, V. (2007) The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615–1621. [DOI] [PubMed] [Google Scholar]
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ji, L.H. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brodersen, P. and Voinnet, O. (2006) The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. and Herndon, K.L. (1992) Characterization of the potyviral HC‐pro autoproteolytic cleavage site. Virology, 187, 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Xiang, W.L. , Xie, D. , Peng, J.R. and Ding, S.W. (2004) Viral virulence protein suppresses RNA silencing‐mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell, 16, 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris, A. , Gallego‐Bartolome, J. , Bao, J. , Kasschau, K.D. , Carrington, J.C. and Voinnet, O. (2006) Hierarchical action and inhibition of plant Dicer‐like proteins in antiviral defense. Science, 313, 68–71. [DOI] [PubMed] [Google Scholar]
- Domier, L.L. , Franklin, K.M. , Shahabuddin, M. , Hellmann, G.M. , Overmeyer, J.H. , Hiremath, S.T. , Siaw, M.F. , Lomonossoff, G.P. , Shaw, J.G. and Rhoads, R.E. (1986) The nucleotide sequence of tobacco vein mottling virus RNA. Nucleic Acids Res. 14, 5417–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer, P. , Lecellier, C.H. , Parizotto, E.A. , Himber, C. and Voinnet, O. (2004) Probing the microRNA and small interfering RNA pathways with virus‐encoded suppressors of RNA silencing. Plant Cell, 16, 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hershko, A. and Ciechanover, A. (1992) The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807. [DOI] [PubMed] [Google Scholar]
- Howard, A.R. , Heppler, M.L. , Ju, H.J. , Krishnamurthy, K. , Payton, M.E. and Verchot‐Lubicz, J. (2004) Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology, 328, 185–197. [DOI] [PubMed] [Google Scholar]
- Kalinina, N.O. , Rakitina, D.V. , Solovyev, A.G. , Schiemann, J. and Morozov, S.Y. (2002) RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology, 296, 321–329. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (1998) A counterdefensive strategy of plant viruses: suppression of post‐transcriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Lin, J.W. , Ding, M.P. , Hsu, Y.H. and Tsai, C.H. (2007) Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Res. 35, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov, S.Y. and Solovyev, A.G. (2003) Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84, 1351–1366. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. , Solovyev, A.G. , Kalinina, N.O. , Fedorkin, O.N. , Samuilova, O.V. , Schiemann, J. and Atabekov, J.G. (1999) Evidence for two nonoverlapping functional domains in the potato virus X 25K movement protein. Virology, 260, 55–63. [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh, M. , Dieterle, M. , Lechner, E. , Berry, B. , Brault, V. , Hemmer, O. , Kretsch, T. , Richards, K.E. , Genschik, P. and Ziegler‐Graff, V. (2006) F‐box‐like domain in the polerovirus protein PO is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA, 103, 1994–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape, M. and Jentsch, S. (2002) Taking a bite: proteasomal protein processing. Nat. Cell Biol. 4, 113–116. [DOI] [PubMed] [Google Scholar]
- Ruiz, M.T. , Voinnet, O. and Baulcombe, D.C. (1998) Initiation and maintenance of virus‐induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F. , Vaistij, F.E. , Jones, L. and Baulcombe, D.C. (2005) An RNA‐dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.H. , Cheng, C.P. , Peng, C.W. , Lin, B.Y. , Lin, N.S. and Hsu, Y.H. (1999) Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J. Virol. 73, 2703–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J.M. , Szittya, G. , Burgyan, J. and Tanaka Hall, T.M. (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell, 115, 799–811. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. , Vazquez, F. , Crete, P. and Bartel, D.P. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H. , Mallory, A.C. and Bartel, D.P. (2006) AGO1 Homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell, 22, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O. (2005) Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Pinto, Y.M. and Baulcombe, D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc. Natl. Acad. Sci. USA, 96, 14 147–14 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O. , Lederer, C. and Baulcombe, D.C. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana . Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Ye, K. and Patel, D.J. (2005) RNA silencing suppressor p21 of Beet yellows virus forms an RNA binding octameric ring structure. Structure, 13, 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. and Coffino, P. (2004) Repeat sequence of Epstein–Barr virus‐encoded nuclear antigen 1 protein interrupts proteasome substrate processing. J. Biol. Chem. 279, 8635–8641. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.R. , Pei, Y. , Lin, S.S. , Tuschl, T. , Patel, D.J. and Chua, N.H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The silencing suppressor activity assay in green fluorescent protein (GFP) transgenic Nicotiana benthamiana line 16c. Normal healthy and GFP transgenic 16c N. benthamiana leaves under UV illumination are shown as reference. Control leaf is indicated as GFIR and GFIR+GUS for parts infiltrated with Agrobacterium strains carrying pBin61‐GFIR and pBin61GFIR plus pBin61‐GUS, respectively. The three leaves in the right panel were treated in the same manner as the control leaf but co‐infiltrated with different silencing suppressors, indicated as P25‐CT7 (pBin61‐P25CT7; PVX P25 with T7 tag at C‐terminus), P25 (pBin61‐P25) and HcPro (pBin61‐HcPro; derived from Potyvirus). The suppressor activity of P25 and HcPro was observed under UV light at 5 days post‐infiltration.
Fig. S2 The expression level of Rx protein is not affected by P25 or P25‐CT7. Immunoprecipitation of transiently expressed Flag‐tagged Rx protein from the total protein extracts of Agrobacterium‐infiltrated Nicotiana benthamiana leaves. Construct 35S:Flag‐Rx (Rx) was infiltrated alone or co‐infiltrated with 35S:P25 (P25) or 35S:P25‐T7 (P25‐CT7) indicated above each lane. Total proteins were extracted 3 days post‐infiltration. The Flag‐tagged proteins were immunoprecipitated (IP) with agarose beads containing anti‐Flag antibody. Western blot analysis (W) was performed using the anti‐Flag antibody indicated.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


