SUMMARY
On infection by pathogens, plants initiate defence responses that are able to curtail infection locally. These responses are mediated either by receptor‐like proteins that recognize pathogen‐associated molecular patterns or by the protein products of disease resistance (R) genes. At the same time, primary defence responses often result in the generation of signals that induce what is known as systemic acquired resistance (SAR), such that defence responses are enhanced on secondary pathogen challenge in distal tissues. R protein‐mediated SAR induction is normally accompanied by a type of programmed cell death known as the hypersensitive response (HR) and, in some instances, cell death alone has been implicated in the induction of SAR. This has raised the question of whether R protein‐mediated signalling per se induces SAR or whether SAR is an indirect result of the induction of HR. Using the Rx gene of potato, which confers resistance to Potato Virus X in the absence of cell death, we have shown that the HR is dispensable for R protein‐mediated induction of SAR and that Rx‐induced SAR is mediated by the same salicylate‐dependent pathway induced by other R proteins.
INTRODUCTION
Plants have evolved multiple mechanisms to recognize pathogens and to induce both local and systemic responses. One of the first lines of defence is mediated by the recognition of highly conserved pathogen‐associated molecular patterns (PAMPs) associated with large groups of pathogens, such as lipopolysaccharides (LPSs) and highly conserved bacterial proteins, e.g. flagellin and Ef‐Tu. For the most part, this PAMP‐triggered immunity (PTI) appears to be mediated by transmembrane receptor‐like proteins, and is presumed to be sufficient to protect plants against most bacterial, fungal and oomycete pathogens (Chisholm et al., 2006). However, the best‐characterized basal defence against viral pathogens is RNA silencing, which targets viruses based on the physical characteristics of their RNAs (Ding and Voinnet, 2007). However, host‐adapted pathogens are able to interfere with and overcome PTI and/or RNA silencing (Chisholm et al., 2006; Diaz‐Pendon and Ding, 2008), necessitating a second line of defence based on much more specific recognition of pathogens or pathogen isolates. This recognition is mediated by the products of plant disease resistance (R) genes and results in a much more dramatic defence response, often culminating in a type of programmed cell death known as the hypersensitive response (HR) (Mur et al., 2008). Many R proteins recognize so‐called effector proteins (pathogen proteins secreted into the host cell) and, as such, R gene‐mediated responses are often referred to as effector‐mediated immunity (ETI) (Chisholm et al., 2006). When a given effector protein is recognized by an R protein, it is referred to as an avirulence (Avr) protein, as it renders the pathogen non‐virulent. However, most Avr proteins are thought to play a role in virulence on plants that do not possess a matching R gene, in part by suppressing PTI. R proteins also recognize Avr proteins from viruses. Like effector proteins, in the absence of a cognate R gene, viral Avr proteins also play roles in virulence by interfering with host processes and/or by playing structural or enzymatic roles (Diaz‐Pendon and Ding, 2008; Kang et al., 2005; Schoelz, 2006). The most common type of R gene encodes nucleotide‐binding and leucine‐rich repeat (NB‐LRR) proteins. There are two major classes of NB‐LRR proteins that are distinguished by the domains present at their N‐termini: those that possess a TIR (Toll, and interleukin‐1 receptor homology) domain and those that do not (Meyers et al., 1999). Many non‐TIR‐NB‐LRR proteins possess an N‐terminus that is predicted to be α‐helix rich and often, but not always, encodes a predicted coiled‐coil (CC) motif. As such, non‐TIR‐NB‐LRR proteins are often referred to as CC‐NB‐LRRs. Plant NB‐LRR proteins have been shown to confer resistance to viruses, bacteria, oomycetes, fungi and arthropods (Martin et al., 2003). Given that R proteins are able to counter several different types of pathogen, they probably activate multiple signalling pathways. These induced responses stop pathogen infection locally, but also induce signals that enhance defence responses to pathogens in distal systemic tissues, a phenomenon known as systemic acquired resistance (SAR) (Vlot et al., 2008). SAR responses are associated with the transcriptional up‐regulation of a number of genes, including those encoding pathogenesis‐related (PR) proteins in both local and systemic tissues (Durrant and Dong, 2004).
Although classical SAR experimental systems are associated with cell death induced either as a symptom of infection or by the activation of an R protein (Durrant and Dong, 2004), it has recently been reported that PTI responses can also induce SAR in Arabidopsis in the absence of cell death (Mishina and Zeier, 2007). As such, it is unclear whether SAR induced by R protein activation is a direct consequence of R protein signalling or whether it is an indirect consequence induced by dying cells which might, in turn, release signals that activate PTI‐like mechanisms in neighbouring cells.
The N gene of Nicotiana glutinosa, encoding a TIR‐NB‐LRR protein, confers resistance to tobacco mosaic virus (TMV) and has been introgressed into commercial tobacco (N. tabacum) (Whitham et al., 1994). Infection of N genotype tobacco leads to a typical HR‐type resistance response which, in turn, induces SAR, such that secondary TMV infections will induce HR lesions significantly reduced in size, consistent with a faster resistance response. This well‐established model system is highly robust and has been used to demonstrate the requirement for salicylate (SA) in the induction of SAR, and the involvement of methyl salicylate (MeSA) as a mobile signal in SAR in tobacco (Delaney et al., 1994; Gaffney et al., 1993; Park et al., 2007; Vernooij et al., 1994).
The potato Rx gene encodes a typical CC‐NB‐LRR protein and confers resistance to potato virus X (PVX) (Bendahmane et al., 1999). Unlike the N gene, however, Rx confers what is known as extreme resistance (ER) in potato and, as a transgene, in tobacco (Bendahmane et al., 1999). Extreme resistance is seen with a number of R genes conferring resistance to viruses; it is manifested as a complete lack of both macroscopic and microscopic HR lesions and is accompanied by the absence of detectable virus accumulation (Kang et al., 2005). However, many R genes that induce ER, including Rx, also induce HRs depending on the strain of virus, host genetic background and environmental conditions (Baures et al., 2008; Bendahmane et al., 1999; Kang et al., 2005). Thus, it is likely that there is no qualitative difference in the responses induced by Rx, but rather that the response is very rapid and efficient, such that virus accumulation is inhibited before the onset of secondary responses, such as HR.
We found that Rx‐mediated defence responses leading to ER ware able to induce SAR against subsequent TMV infections, and that this response showed the hallmarks of classic SAR responses. We conclude that the activation of Rx leads to the direct induction of SAR‐inducing signals and that R protein‐mediated SAR is not a by‐product of HR.
RESULTS AND DISCUSSION
To determine whether HRs are required to induce SAR, we made use of tobacco plants expressing the Rx gene from its own promoter in the cultivar Samsun (NN) (Bendahmane et al., 1999). The lower leaves of plants were subjected to either a mock treatment or infected with TMV or PVX. As expected, TMV induced HR lesions on inoculated leaves, whereas PVX did not (Fig. 1A). All plants were subsequently infected with TMV on the upper leaves 5 days later. As expected, plants previously infected with TMV showed a reduction in HR lesion size compared with mock‐inoculated plants, indicating the induction of SAR (Fig. 1B,C). Likewise, plants that had been infected previously with PVX showed similarly reduced lesion sizes (Fig. 1B,C), indicating that the Rx‐mediated resistance response is able to induce SAR to a degree similar to that induced by N.
Figure 1.
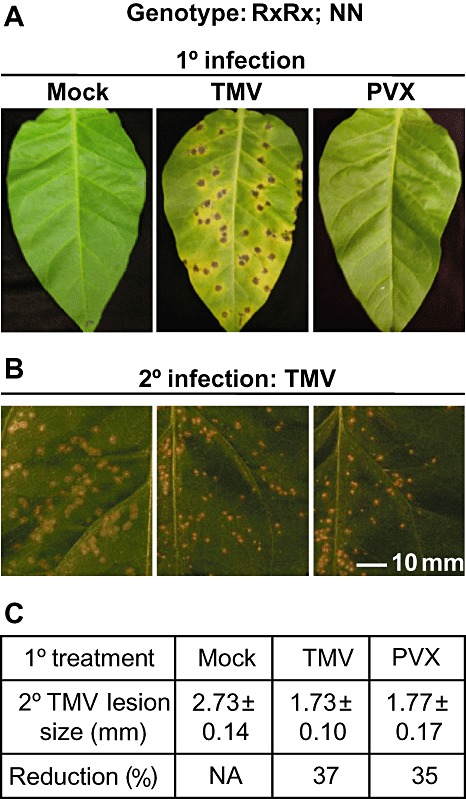
Extreme resistance induces systemic acquired resistance (SAR) signals. (A) Tobacco plants expressing both the N and Rx genes were rub‐inoculated on three lower leaves with buffer (Mock), tobacco mosaic virus (TMV) or potato virus X (PVX). Representative leaves from this primary (1°) infection are shown 5 days post‐infection (dpi). (B) Three systemic leaves immediately above the 1° infections were challenged with TMV (2° infection) 5 days after the 1° infection. Five days after the 2° infection, leaves were photographed and lesion sizes were determined. (C) Lesions sizes in millimetres (±standard deviation) from the experiment shown in (B). Reduction (%) is the percentage reduction in the size of 2° lesions on plants that received a 1° infection of TMV or PVX vs. mock 1° infected plants. N/A, not applicable. Experiments were performed at least four times with similar results.
Previous studies have shown important roles for SA and MeSA in the induction and signalling of SAR in the tobacco–TMV pathosystem (Vlot et al., 2008). To determine whether the Rx‐mediated induction of SAR is similar in mechanism to that induced by N, we investigated the requirement for these molecules in Rx‐induced SAR. To this end, Rx transgenic tobacco was crossed to transgenic tobacco (cultivar Xanthi nc [NN]) expressing either the bacterial nahG gene (NahG) or an RNA hairpin targeting the tobacco SA‐binding protein 2 (SABP2) transcript for degradation by RNAi (RNAi::SABP2), or to wild‐type plants. NahG encodes a salicylate hydroxylase that metabolizes SA, whereas SABP2 is an esterase required for the conversion of the SAR signal MeSA to SA in healthy systemic tissue (Forouhar et al., 2005; Kumar and Klessig, 2003; Park et al., 2007). The RNAi::SABP2 transgene reduces the SABP2 transcript by greater than 75% (Kumar and Klessig, 2003). As all three transgenes are dominant, the resulting F1 progeny were used for SAR analysis. Plants were subjected to a primary infection (mock, TMV or PVX) followed by a secondary infection with TMV as described above. Tobacco plants expressing nahG can generate the SAR‐inducing signal in primary infected tissue, but are unable to perceive or process the signal in systemic tissue (Delaney et al., 1994; Gaffney et al., 1993; Vernooij et al., 1994). As expected, Rx/NahG/NN genotype plants failed to develop SAR after a primary infection with TMV, as the size of the lesions formed after secondary infection with TMV were as large as those formed in plants that had received a mock primary infection (Fig. 2A). Likewise, plants subjected to primary infection with PVX showed no reduction in HR lesion size on secondary infection with TMV compared with those receiving mock primary infection (Fig. 2A). Infection of F1 progeny of crosses between Rx and RNAi::SABP2 plants with TMV following a primary infection with either TMV, PVX or mock treatment showed that the Rx‐induced SAR is similarly compromised in the absence of SABP2 (Fig. 2B). The lack of SAR development in the F1 progeny described above was not caused by differences in genetic background or gene dosage, as the progeny of Rx transgenic plants crossed with wild‐type Xanthi nc (NN) plants showed a similar degree of SAR‐induced HR lesion size reduction as the Rx transgenic parent (Fig. 2C).
Figure 2.
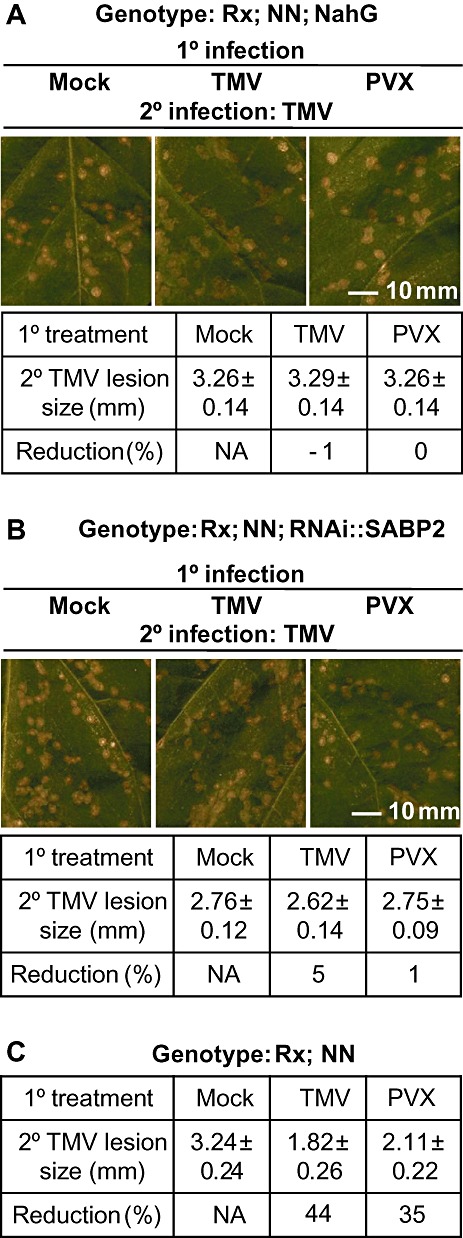
The systemic acquired resistance (SAR) signal induced by extreme resistance is dependent on salicylate (SA)‐mediated signalling. F1 progeny from the crossing of Rx transgenic tobacco with NahG (A) or RNAi::SABP2 (B) transgenic tobacco or wild‐type tobacco (C) were assessed for their ability to develop SAR. Plants were rub‐inoculated on three lower leaves with buffer (Mock), tobacco mosaic virus (TMV) or potato virus X (PVX). Three systemic leaves immediately above the 1° infections were challenged with TMV (2° infection) 5 days after the 1° infection. Five days after the 2° infection, leaves were photographed (A and B) and lesion sizes were determined. Lesions sizes in millimetres (±standard deviation) are shown in table form (A–C). Reduction (%) is the percentage reduction in the size of 2° lesions on plants that received a 1° infection of TMV or PVX vs. mock 1° infected plants. N/A, not applicable. Experiments were performed at least three times with similar results.
One of the hallmarks of N‐induced SAR is the rapid induction of SA‐responsive PR genes in both inoculated and healthy systemic leaves (Durrant and Dong, 2004). Cell death is not required for the local induction of PR genes in another plant–virus interaction. Plants resistant to cauliflower mosaic virus, which respond to infection with chlorotic lesions, have been shown to induce PR proteins in inoculated leaves, although SAR has not been tested in this system (Cole et al., 2001). As an additional demonstration that Rx‐ and N‐mediated responses are similar in mechanism, we assessed the expression of the tobacco acidic PR‐1 gene in Rx transgenic plants inoculated with PVX. Although there was little PR‐1 expression in uninfected (day 0) or mock‐infected plants, expression was significantly induced in inoculated leaves 1 day after PVX infection and in systemic leaves 5 days after PVX infection (Fig. 3). Thus, Rx‐mediated induction of SAR appears to utilize the same SA signals and induces the same type of defence‐related genes as in N‐induced SAR, suggesting a similar mode of action.
Figure 3.
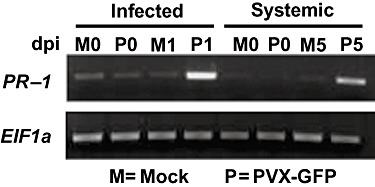
Extreme resistance induces expression of the PATHOGENESIS‐RELATED‐1 (PR‐1) gene. Leaves of Rx transgenic tobacco were either mock‐inoculated (M) or infected with potato virus X (PVX) (P). RNA was extracted from 1° inoculated leaves at 0 (M0 and P0) and 1 (M1 and P1) day post‐infection (dpi), and from systemic leaves at 0 (M0 and P0) and 5 (M5 and P5) dpi. RNA was subsequently used for reverse transcription followed by polymerase chain reaction (PCR) using primers targeting the tobacco acidic PR‐1 gene. The constitutively expressed translation elongation factor 1α gene (EF1α) was used as a control. Two independent experiments yielded similar results.
As with other types of resistance responses, SAR is likely to be inducible by multiple stimuli and signalling pathways. Infection with Pseudomonas syringae can induce SAR by inducing an HR which is mediated by the products of R genes. At the same time, Pseudomonas can also induce SAR by inducing necrosis during the normal course of infection (Durrant and Dong, 2004). The latter is presumably initiated not by R proteins, but by other, as yet undefined, types of signalling pathway. In addition, SAR is induced in plants exposed to non‐host bacteria as well as the purified PAMP LPS and the flagellin‐derived peptide flg22 (Mishina and Zeier, 2007). Thus, both ETI‐ and PTI‐mediated signalling are capable of inducing SAR, although the latter may often be suppressed by pathogens that employ effector proteins to disrupt PTI.
ETI and PTI are mediated by different types of receptor‐like protein, and the physiological responses they induce differ greatly in magnitude. Both mechanisms appear to utilize at least some common components, such as SA signalling, and both induce PR gene expression (Mishina and Zeier, 2007). However, the fact that cell death is not required for PTI‐mediated SAR induction does not rule out a role for HR in SAR induction by ETI‐mediated responses. Whether ETI‐induced SAR is a direct consequence of R protein signalling or an indirect consequence of HR remains an open question. Indeed, it is possible that ETI responses may induce SAR indirectly in a manner analogous to a paradigm known in animal immunity as the ‘danger model’, whereby PAMP receptors induce inflammatory responses in response to self‐molecules released by dying cells (Matzinger, 2002). As such, R protein signalling could activate SAR indirectly by inducing an HR which, in turn, activates PTI‐like mechanisms that respond to molecular patterns released by dying cells. Indeed, plants with lesion mimic phenotypes resulting from mutation or transgene expression often possess SAR‐like enhanced disease resistance, suggesting that certain types of cell death alone are sufficient to induce non‐cell autonomous defences (Lorrain et al., 2003; Mittler and Rizhsky, 2000). As Rx induces SAR but not HR, this indicates, for the first time, that cell death is not intrinsically necessary for SAR induced by NB‐LRR proteins. Thus, although we do not rule out the possible existence of SAR‐inducing danger signals generated by dying cells, it would appear that NB‐LRR protein signalling leads directly to the generation of SAR signals.
The induction of systemic signals by Rx must also be very strong. In the case of the N–TMV interaction, a relatively large number of cells are eventually infected within individual infection foci. This would allow for a relatively large base of production either in cells undergoing a resistance response and/or in cells adjacent to dying cells. However, PVX infection in Rx plants results in the infection of only isolated individual cells, such that only several hundred cells would be infected on a given inoculated leaf (see Experimental procedures). Nonetheless, Rx induced the same level of systemic resistance as N, as measured by the reduction in lesion size after secondary infection by TMV.
Although we have not addressed the exact nature of the SAR‐inducing signals generated by the Rx response, it would appear that they are similar to other R protein‐mediated responses, in that SA and MeSA are both required in healthy systemic tissue (Fig. 2). The primary resistance response to PVX mediated by Rx, however, is not affected by NahG and thus appears to be independent of SA (Bendahmane et al., 2000) (data not shown). In contrast, NahG affects the primary defence responses mediated by the tobacco N and tomato Tm2 2 genes, as TMV inoculation results in large, spreading necrotic lesions in nahG transgenic plants (Brading et al., 2000; Delaney et al., 1994). In virus–R gene interactions, distinct HR lesions result because the initially infected individual cells are not able to contain the virus, which subsequently spreads to neighbouring cells. However, cells at the outer edge of the infection front undergo a stronger defence response, which enables containment of the virus (Wright et al., 2000). Thus, rather than having an intrinsically antiviral activity, SA serves to potentiate defence responses both systemically and locally (Cordelier et al., 2003; Ryals et al., 1996). This suggests that R proteins probably initiate bifurcating signalling pathways that lead to cell‐autonomous antiviral resistance on the one hand and the production of SA on the other, with the latter leading to the potentiation of the former in adjacent cells and systemic tissues.
Our data are in agreement with previous studies indicating that ER induced by Rx is not qualitatively different from HR‐type antiviral responses (Baures et al., 2008; Bendahmane et al., 1999), and provide an explanation for a lack of necessity for SA for Rx‐mediated resistance. That is, SA is not required because the rapid response induced by Rx does not need to be potentiated in adjacent cells. Like other R proteins, however, Rx initiates the same SA‐mediated signalling pathway(s) leading to the production of SAR signal(s). However, our results definitively rule out a requirement for cell death in SAR induction induced by NB‐LRR proteins, and show that R protein signalling alone is sufficient for SAR induction. The question of whether NB‐LRR proteins induce SAR by the same mechanism as PAMP receptors, however, requires further investigation.
EXPERIMENTAL PROCEDURES
Plant materials
Nicotiana tabacum (tobacco) expressing the Rx gene from its genomic promoter in the cultivar Samsun (NN), as well as the SABP2‐silenced line 1‐2 (RNAi::SABP2) and NahG‐10 (NahG) transgenic lines in the tobacco cultivar Xanthi nc (NN), have been described previously (Bendahmane et al., 1999; Gaffney et al., 1993; Kumar and Klessig, 2003). The experiments in 1, 3 were performed using Rx transgenic plants. Pollen from SABP2‐silenced line 1‐2, NahG‐10 and non‐transgenic Xanthi nc (NN) was used to pollinate Rx‐transgenic flowers and the resulting F1 hybrid seeds were used for the experiments shown in Fig. 2.
Pathogen preparation and experiments
PVX inoculum was prepared from N. benthamiana plants previously agro‐infected with PVX‐green fluorescent protein (GFP) (Peart et al., 2002). Infected systemic leaves were collected and macerated in 0.1 m sodium phosphate buffer, pH 5.2, approximately 1 week post‐infection. The suspension was centrifuged at 13 000 g for 15 min at 4 °C. The supernatant was collected and frozen at −80 °C. The concentration of PVX was determined by checking the GFP fluorescent foci under UV illumination on infection of non‐transgenic tobacco, and found to result in approximately 150 PVX infections per 25 cm2. Uninfected N. benthamiana plants were similarly processed for mock infections.
To induce SAR, primary TMV, PVX or mock inoculations were carried out on approximately 6‐week‐old tobacco plants, as described previously (Guo et al., 2000). Five days after primary infection, three systemic tissues of all plants were challenged with TMV infection. The secondary lesion sizes were measured 5 days after primary infection, at which point the establishment of SAR was determined by measuring and comparing the lesion sizes of TMV‐infected leaves with a vernier caliper. In each experiment, 50 lesions were measured on each of three leaves of a plant and the standard deviation was calculated. The percentage reduction in lesion size was calculated as the difference in diameter between the secondary lesions on plants that received a primary infection of TMV or PVX vs. mock primary infected plants, divided by the diameter of the secondary lesions on plants that received a mock primary infection. Each experiment was performed on at least three separate occasions, with both control (mock) and experiment (TMV and PVX) performed at the same time on any given genotype.
Reverse transcriptase‐polymerase chain reaction (RT‐PCR)
Total RNA was extracted from leaves using Trizol reagent (Invitrogen; Carlsbad, California) according to the manufacturer's instructions. DNase‐treated total RNA (2 µg) was used for reverse transcription with a SuperscriptII kit (Invitrogen). The RT product was subjected to PCR using primers (5′‐TAGTCATGGGATTTGTTCTC‐3′ and 5′‐TCAGATCATACATCAAGCTG ‐3′) designed to amplify the tobacco acidic PR‐1 gene (accession number X06361) using the following conditions: one cycle at 94 °C (4 min), one cycle at 80 °C (2 min), touchdown cycles (94 °C for 15 s, 62 °C → 56 °C for 15 s and 72 °C for 30 s) (one cycle for each temperature) and 25 cycles at 94 °C (15 s), 55 °C (15 s) and 72 °C (30 s), followed by extension at 72 °C (7 min). The same samples were used in PCR using primers (5′‐AAGTATGCCTGGGTGCTTG ‐3′ and 5′‐AGGGACAGTACCAATTCCACC‐3′) designed to amplify the elongation factor 1α gene (EF1α) employing essentially the same conditions as above, except for the temperatures for the touchdown cycles (67 °C → 61 °C for 15 s) and the 20‐cycle amplification (94 °C for 15 s, 60 °C for 15 s and 72 °C for 30 s).
REFERENCES
- Baures, I. , Candresse, T. , Leveau, A. , Bendahmane, A. and Sturbois, B. (2008) The rx gene confers resistance to a range of potexviruses in transgenic Nicotiana plants. Mol. Plant–Microbe Interact. 21, 1154–1164. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane, A. , Querci, M. , Kanyuka, K. and Baulcombe, D.C. (2000) Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21, 73–81. [DOI] [PubMed] [Google Scholar]
- Brading, P.A. , Hammond‐Kosack, K.E. , Parr, A. and Jones, J.D. (2000) Salicylic acid is not required for Cf‐2‐ and Cf‐9‐dependent resistance of tomato to Cladosporium fulvum . Plant J. 23, 305–318. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cole, A.B. , Kiraly, L. , Ross, K. and Schoelz, J.E. (2001) Uncoupling resistance from cell death in the hypersensitive response of Nicotiana species to cauliflower mosaic virus infection. Mol. Plant–Microbe Interact. 14, 31–41. [DOI] [PubMed] [Google Scholar]
- Cordelier, S. , De Ruffray, P. , Fritig, B. and Kauffmann, S. (2003) Biological and molecular comparison between localized and systemic acquired resistance induced in tobacco by a Phytophthora megasperma glycoprotein elicitin. Plant Mol. Biol. 51, 109–118. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , Gaffney, T. , Gut‐Rella, M. , Kessmann, H. , Ward, E. , and Ryals, J. (1994) A central role of salicylic acid in plant disease resistance. Science, 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Diaz‐Pendon, J.A. and Ding, S.‐W. (2008) Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E. and Dong, X. (2004) Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Forouhar, F. , Yang, Y. , Kumar, D. , Chen, Y. , Fridman, E. , Park, S.W. , Chiang, Y. , Acton, T.B. , Montelione, G.T. , Pichersky, E. , Klessig, D.F. , and Tong, L. (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA, 102, 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T. , Friedrich, L. , Vernooij, B. , Negrotto, D. , Nye, G. , Uknes, S. , Ward, E. , Kessmann, H. , and Ryals, J. (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science, 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Guo, A. , Salih, G. and Klessig, D.F. (2000) Activation of a diverse set of genes during the tobacco resistance response to TMV is independent of salicylic acid; induction of a subset is also ethylene independent. Plant J. 21, 409–418. [DOI] [PubMed] [Google Scholar]
- Kang, B.C. , Yeam, I. and Jahn, M.M. (2005) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Kumar, D. and Klessig, D.F. (2003) High‐affinity salicylic acid‐binding protein 2 is required for plant innate immunity and has salicylic acid‐stimulated lipase activity. Proc. Natl. Acad. Sci. USA, 100, 16 101–16 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S. , Vailleau, F. , Balague, C. and Roby, D. (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. [DOI] [PubMed] [Google Scholar]
- Martin, G.B. , Bogdanove, A.J. and Sessa, G. (2003) Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- Matzinger, P. (2002) The danger model: a renewed sense of self. Science, 296, 301–305. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C. , Dickerman, A.W. , Michelmore, R.W. , Sivaramakrishnan, S. , Sobral, B.W. and Young, N.D. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide‐binding superfamily. Plant J. 20, 317–332. [DOI] [PubMed] [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Mittler, R. and Rizhsky, L. (2000) Transgene‐induced lesion mimic. Plant Mol. Biol. 44, 335–344. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Lloyd, A.J. , Ougham, H. and Prats, E. (2008) The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. [DOI] [PubMed] [Google Scholar]
- Park, S.W. , Kaimoyo, E. , Kumar, D. , Mosher, S. and Klessig, D.F. (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science, 318, 113–116. [DOI] [PubMed] [Google Scholar]
- Peart, J.R. , Lu, R. , Sadanandom, A. , Malcuit, I. , Moffett, P. , Brice, D.C. , Schauser, L. , Jaggard, D.A. , Xiao, S. , Coleman, M.J. , Dow, M. , Jones, J.D. , Shirasu, K. , and Baulcombe, D.C. (2002) Ubiquitin ligase‐associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA, 99, 10 865–10 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A. , Neuenschwander, U.H. , Willits, M.G. , Molina, A. , Steiner, H.Y. and Hunt, M.D. (1996) Systemic acquired resistance. Plant Cell, 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoelz, J.E. (2006) Viral determinants of resistance and susceptibility In: Natural Resistance Mechanisms of Plant to Viruses (Loebenstein G. and Carr J. eds), pp. 13–43. Dordrecht: Springer. [Google Scholar]
- Vernooij, B. , Friedrich, L. , Morse, A. , Reist, R. , Kolditz‐Jawhar, R. , Ward, E. , Uknes, S. , Kessmann, H. , and Ryals, J. (1994) Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell, 6, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C. , Klessig, D.F. and Park, S.W. (2008) Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11, 436–442. [DOI] [PubMed] [Google Scholar]
- Whitham, S. , Dinesh‐Kumar, S.P. , Choi, D. , Hehl, R. , Corr, C. and Baker, B. (1994) The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin‐1 receptor. Cell, 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wright, K.M. , Duncan, G.H. , Pradel, K.S. , Carr, F. , Wood, S. , Oparka, K.J. , and Cruz, S.S. (2000) Analysis of the N gene hypersensitive response induced by a fluorescently tagged tobacco mosaic virus. Plant Physiol. 123, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]


