SUMMARY
Genes at the M locus in flax (Linum usitatissimum) that confer resistance to flax rust (Melampsora lini) occur in complex haplotypes containing up to 15 related genes or gene fragments. We have cloned two additional functional resistance genes at this locus, M1 and M3, by transposon tagging and candidate gene approaches, and investigated the genetic relationships between four genes (M, M1, M3 and M4) by recombination analysis. M1 and M3, like M, are members of the nucleotide binding site, leucine‐rich repeat (NBS‐LRR) family. Comparisons of the predicted M1 and M3 amino acid sequences with M and L6 reveal that: (i) M1 contains four additional LRRs, probably as a result of an unequal crossover event between duplicated regions; (ii) M1 shares large segments of exact identity with M and M3, indicative of intragenic recombination events; and (iii) a large number of amino acid differences are scattered throughout the M, M1 and M3 proteins. Recombination analysis (here and in previous studies) has revealed that M readily recombines with M1, M3 and M4, whereas these three genes fail to recombine despite large family sizes (>5800) in two test‐cross families, suggesting that they may occupy allelic positions in the gene cluster. Several restriction fragment length polymorphism markers within or near the M locus were mapped with respect to seven crossover events between M and M1. The results of this and previous studies provide evidence of structural differences between: (i) homoeologous loci in the different genomes of flax; (ii) different haplotypes at the M locus; (iii) different resistance genes in the M group; and (iv) the flanking regions downstream of M locus resistance genes.
INTRODUCTION
In plants, the major class of disease resistance genes are those that encode nucleotide binding site–leucine‐rich repeat (NBS‐LRR) proteins. These genes are often located in clusters: for example, about two‐thirds of the approximately 150 NBS‐LRR genes in Arabidopsis thaliana and a high proportion of the approximately 500 NBS‐LRR genes in rice occur in clusters (Monosi et al., 2004; Meyers et al., 2003; Richly et al., 2002). Within a species, clusters or haplotypes at a particular locus frequently show extensive structural variation, as revealed (when whole genome sequencing is not available) by sequencing BAC clones that cover the locus or by DNA gel blot analyses combined with sequencing of the different members in the haplotype. Examples include the RPP5/RPP4 locus in A. thaliana (van der Biezen et al., 2002; Noel et al., 1999), the rp1 locus in maize (Collins et al., 1999; Ramakrishna et al., 2002; Smith and Hulbert, 2005; Smith et al., 2004; Sun et al., 2001) and the RGC2 locus in lettuce (Kuang et al., 2004; Meyers et al., 1998). The haplotype variation includes differences in the size and number of NBS‐LRR genes (which may not all be complete or functional genes), as well as differences in indels, tandem repeats, the location of unique sequence DNA within the clusters and the presence/absence of transposons. The mechanisms proposed to account for the structure and structural differences of these complex haplotypes include unequal crossing over, a duplication method (unknown) not involving crossing over, intrastrand recombination between different NBS‐LRR genes, replication slippage, illegitimate recombination, the use of filler sequences to repair double‐strand breaks and transposon insertion/excision.
The genes in flax and linseed (Linum usitatissimum) that confer resistance to flax rust (Melampsora lini) map to five loci: K, L, M, N and P (Flor, 1956; Flor and Comstock, 1972; Hoes and Kenaschuk, 1986; Islam and Mayo, 1990; Wicks and Hammond, 1978; Zimmer and Comstock, 1973). Genes at the L, M, N and P loci all encode resistance proteins of the Toll interleukin 1 receptor–nucleotide binding site–leucine‐rich repeat (TIR‐NBS‐LRR) class, although the P locus encoded proteins have an additional C‐terminal domain of 150 amino acids downstream of the LRR region (Anderson et al., 1997; 2001a, 2001b; Ellis et al., 1999; Lawrence et al., 1995). The M, N and P loci each contain a cluster of TIR‐NBS‐LRR genes, whereas only a single gene has ever been identified at the L locus. Thus, the 13 resistance genes at the L locus are alternative forms of the same gene (alleles), whereas different resistance genes at the M, N and P loci could each be separate genes in the cluster, or alternative forms of one particular gene in the cluster, or some could be separate genes and some alternative forms of the same gene. Of the seven M group resistance genes, only one, the M gene, has been cloned (Anderson et al., 1997). The cloning of the M gene has not facilitated the cloning of the other six resistance genes in the M group, because each gene occurs in a different cluster of up to 15 related genes, pseudogenes or gene fragments. The M gene has 86% nucleotide identity to the L6 coding and intron sequences, and L6 fragments cross‐hybridize to up to 15 fragments from the M locus (Anderson et al., 1997; Ellis et al., 1995; Lawrence et al., 1995; this paper). Flax is an ancient tetraploid, and so L and M may be homoeologous loci in the different genomes.
Before the gene cloning era, relationships between resistance genes at a locus were investigated by genetic studies, which sought to characterize the phenotypic outcome(s) of recombination events within the locus. Test‐crosses involving 27 pairwise combinations of L locus genes yielded either no recombinants at all or only double‐susceptible recombinants (Flor, 1965; Islam and Shepherd, 1991; Islam et al., 1989; Shepherd and Mayo, 1972), an observation consistent with the different genes at this locus being alternative forms of the same gene. Family sizes in these studies were between 1500 and 5000 in all but a few cases. In contrast, of five pairwise combinations of genes at the M locus investigated in test‐crosses, three combinations, all involving the M gene (MM1, MM3 and MM4), yielded both double‐resistant and double‐susceptible recombinants, whereas two combinations not involving the M gene (M1M4 and M3M4) yielded no recombinants (Flor, 1965; Shepherd and Mayo, 1972). Family sizes in the last two test‐crosses were comparatively small (776 and 1471, respectively). Thus, M specificity can coexist on the same strand as M1, M3 and M4, suggesting that the gene determining M specificity may be a different gene in the M locus cluster from those determining M1, M3 and M4. However, these test‐cross studies provide little evidence regarding the relationships between M1, M3 and M4 and no evidence concerning the remaining genes M2, M5 and M6.
In this article, we report further investigations into the relationships between genes in the M group using both approaches that have been employed previously. In the first approach, two additional resistance genes in the M group, M1 and M3, plus an additional gene in the M1 haplotype, were cloned and sequenced. In the second approach, recombination analysis, progeny from three test‐crosses involving parent lines heterozygous for MM1, M1M3 and M3M4 were screened for double‐resistant and double‐susceptible recombinants. In addition, four restriction fragment length polymorphism (RFLP) markers at or near the M locus were mapped with respect to M and M1.
RESULTS
Cloning of the M3 gene
The M3 gene for rust resistance in flax was cloned after it had been tagged with the maize transposable element Activator (Ac). To maximize the chances of tagging M3, a flax line was developed (see Experimental procedures) that contained many Ac elements, one of which was closely linked (1 map unit) to M3, because many (preferably 10 or more) Ac elements are necessary for Ac activity in flax (Ellis et al., 1992) and because Ac preferentially transposes to linked locations (Dooner and Belachew, 1989; Jones et al., 1990). This line, which also contained the L6, N and P2 resistance genes, was crossed as female to Hoshangabad (no known resistance genes), and the resulting progeny were inoculated as seedlings with rust strain Sp‐y (virulent to L6, N and P2, but avirulent to M3) to identify rust‐susceptible individuals which lacked a functional M3 gene. Of the 13 400 progeny screened, 19 rust‐susceptible mutant plants were identified.
A comparison of the DNA gel blot patterns of the susceptible mutants with that of their parent lines, using a probe from the 3′ end of Ac (3′Ac probe), revealed that 14 of the mutants possessed one or more Ac–flax DNA junction fragments not present in their parent lines, indicating that they contained newly transposed Ac elements. Eleven of these mutants were judged likely to contain a deletion, because a pollen squash test (see Experimental procedures) indicated that 50% or more of their pollen grains had not developed fully and, in some cases, because the plants failed to set seed. The remaining three mutants (X168, X173 and X175) with new Ac junction fragments were crossed to the flax line Cass (homozygous for M3), and progeny possessing the new Ac junction fragment(s) were crossed to Hoshangabad (no resistance genes) to produce test‐cross families, which were scored for M3 (by inoculating with rust strain Sp‐y) and for the new junction fragments (by gel blot analyses with the 3′Ac probe). In the X173 test‐cross, in which two new Ac junction fragments were segregating, all 19 individuals expressing M3 resistance lacked the smaller of the two new junction fragments, whereas all 29 susceptible individuals possessed this fragment, consistent with this Ac element being allelic to the M3 gene and potentially tagging the gene.
To determine whether an Ac element had indeed tagged the M3 gene in mutant X173, fragments flanking the critical Ac element were cloned from EcoRI‐digested DNA from a progeny plant of X173 homozygous for this Ac element and lacking any other Ac element. The 4.5‐kb Ac element has a single EcoRI restriction site 2.5 kb from the 5′ end. A 10‐kb fragment recovered using the 5′Ac probe contained 2.5 kb of the 5′ end of Ac and 7.5 kb of flax DNA that contained a complete TIR‐NBS‐LRR gene and 3 kb downstream of this gene. The Ac element had inserted 40 nucleotides upstream of the start of the TIR‐NBS‐LRR coding region. The 5.5‐kb fragment recovered using the 3′Ac probe contained 2 kb of the 3′ end of Ac and 3.5 kb of flax DNA upstream of the TIR‐NBS‐LRR gene. The 3‐kb region downstream of the putative M3 gene contained an 850‐bp segment with very high homology to the Mxb‐1 probe derived from the downstream region of the M gene, which is M locus specific (Table 1; Anderson et al., 1997). This probe hybridizes to two fragments (one 11 kb and the other 8 kb) in EcoRI‐digested Cass (M3) DNA. The larger of these fragments, which corresponds to the predicted size of the uninterrupted putative M3 fragment (7.5 plus 3.5 kb), was cloned. The sequence of this fragment is identical to that of the two Ac‐associated flax DNA fragments in X173. This 11‐kb fragment contains 3.0 kb of DNA upstream and 3.5 kb downstream of the putative M3 gene. The 3.0‐kb upstream region shows high sequence identity to the upstream region of M, except for the distal 1.6‐kb region which contains two open reading frames (ORFs) encoding a tetratricopeptide repeat domain.
Table 1.
Restriction fragment length polymorphism (RFLP) markers at or near the M locus in flax.
| Probe | Description of RFLP(s) and mapping information | |
|---|---|---|
| Name | Description | |
| X22‐B (= X22A‐1) | A HindIII/XhoI fragment from plasmid pUC X22‐B Bcl which contains DNA adjacent to a T‐DNA insert in a transgenic line of Forge. The T‐DNA contains an Activator (Ac) transposable element plus a neomycin phosphotransferase (NPT‐II) gene (Ellis et al., 1995) | Probe hybridizes to a single fragment. In a test‐cross family of 52 individuals, no recombinants were observed between M and RFLP marker (T‐DNA not present). In test‐crosses segregating for NPT‐II in the M‐linked T‐DNA and either the M, M1 or M3 resistance genes, the number of recombinants/family size was: |
| M—NPT‐II 6/339 | ||
| M1—NPT‐II 1/320 | ||
| M3—NPT‐II 2/316 | ||
| Mxb‐1 | An Xba1 fragment of ∼700 bp derived from a region ∼1 kb downstream of the M gene (Anderson et al., 1997) | Probe hybridizes (EcoRI digest) to single fragment in Dakota (M), Williston Brown (M1), Victory A (M4) and Hoshangabad and to two fragments in Ward (M2), Cass (M3) and the M6 line. Apparently M locus‐specific marker |
| M3.2/5 | A 791‐bp polymerase chain reaction (PCR)‐amplified fragment derived from a gene ∼2 kb upstream of M3. | Probe hybridizes to two fragments: one is located near the M locus (no recombinants with M in 52 test‐cross progeny), the other is located near the L locus (two recombinants with L6 in 52 test‐cross progeny) |
| Amplified using primers M3.2 (5′‐GAAAGAAGCAATAGATGAACTG‐3′) and M3.5 (5′‐GATTCGGAAAGGGAGACCG‐3′) | ||
| Lu‐2 | A HindIII/BglII fragment of ∼1.6 kb located ∼2 kb upstream of the L6 gene (Ellis et al., 1995). HindIII end of fragment has homology to glycogenin glucosyltransferase gene in rice | Probe hybridizes to two fragments, one of which is located at the L locus and the other at the M locus (Ellis et al., 1995) |
| Lu‐3 | An EcoRI/BglII fragment of ∼1.1 kb from the leucine‐rich repeat (LRR) region of the L6 gene (Ellis et al., 1995; Lawrence et al., 1995) | Probe hybridizes to up to 15 restriction fragments. Fragments originate from only two locations, the L and M loci (Anderson et al., 1997). Single polymorphic fragment maps to L locus, multiple polymorphic fragments map to M locus (Ellis et al., 1995; this paper) |
The 11‐kb fragment containing the putative M3 gene was transferred to the flax line Ward by Agrobacterium‐mediated transformation. Seven of 11 transgenic lines (as determined by gel blot analysis) were resistant to rust strain CH5 (virulent on Ward, but avirulent to M3), indicating that the TIR‐NBS‐LRR transgene can confer resistance. The specificity of the resistance conferred by the transgene was tested by inoculating transgenic Ward plants with eight rust strains selected from a family of rust strains obtained by selfing strain CH5, which segregate for avirulence/virulence on Cass (M3) (Lawrence et al., 1981). The transgenic plants were resistant to four strains avirulent to M3 and susceptible to four strains virulent to M3. Thus, the resistance conferred by the cloned gene displays the same specificity as M3, thereby confirming that it is the M3 gene. The nucleotide sequence of the M3 gene predicts a protein product of 1293 amino acids (Fig. 1) and contains three predicted introns in the same locations as those in the M gene (Anderson et al., 1997).
Figure 1.
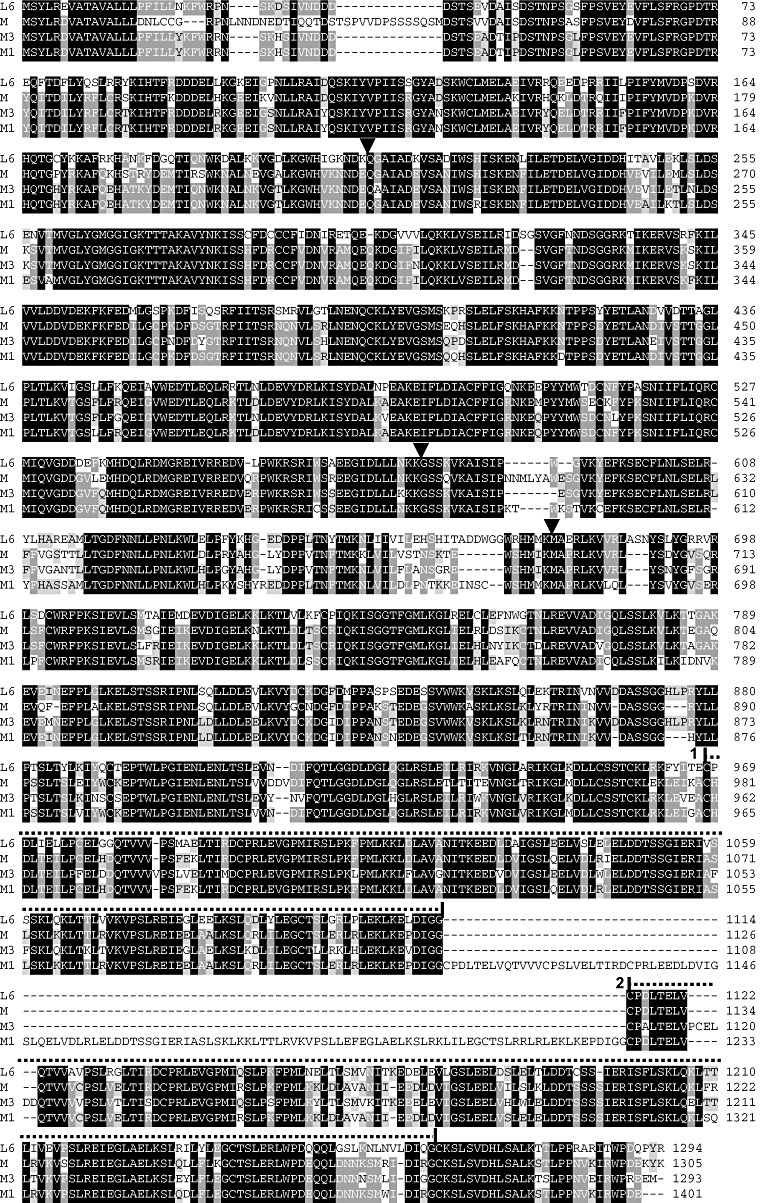
Comparison of the predicted amino acid sequences of L6, M, M3 and M1. Black, medium and light shading indicates four, three and two identical amino acids, respectively. The three black arrow heads indicate intron locations in the encoding genes. Exons 1 and 2 encode the Toll interleukin 1 receptor (TIR) and nucleotide binding site (NBS) domains, respectively. The two large repeats in the LRR region are indicated by dotted lines labelled 1 and 2 above the relevant sequence.
Cloning of the M1 gene
An attempt was made to clone the M1 gene by Ac tagging. As for M3, a line homozygous for M1 and a closely linked Ac element, which also contained multiple other Ac elements, was developed. This line was extensively crossed as female to Hoshangabad, and the resulting progeny were inoculated with rust strain CH5‐132 (avirulent to M1) to identify M1 mutants. Amongst 23 100 progeny screened, 45 M1 mutants were identified. One of the mutants was exceedingly weak and died as a seedling. Of the remaining 44 mutants, 33 contained one or more newly transposed Ac elements, as indicated by the presence of Ac–flax DNA junction fragments in gel blots not present in their parent plants. Based on the pollen squash test, 32 were judged likely to be deletion mutants because 50% or more of their pollen grains did not develop fully. These mutants all lacked an M1‐associated restriction fragment (EcoRI digest) identified using the Mxb‐1 probe (data not shown), thereby confirming their deletion status. The remaining mutant (X221) possessed a single newly transposed Ac element that did not map to the M1 locus. Interestingly, one of the parent plants used in this study yielded an extremely high mutation frequency, with 18 mutants identified from 1900 progeny (9.4 per 1000), compared with 27 mutants from 21 200 progeny (1.27 per 1000) from the other 17 parent lines. The mutants derived from this line also showed a larger number of newly transposed Ac elements (3.3 per mutant) than those derived from the other lines (1.2 per mutant), suggesting a very high level of Ac activity. Such high transposition levels have not been observed in previous Ac‐tagging strategies in flax (Anderson et al., 1997; Dodds et al., 2001b; Lawrence et al., 1995).
As the Ac tagging strategy was unsuccessful for M1, we used unique sequences flanking the cloned M3 gene (probes M3.2/5 and Mxb‐1; see Table 1) to recover related fragments from the M1 cluster. The Mxb‐1 probe hybridizes to a single 9.5‐kb fragment in EcoRI‐digested DNA from the flax line Williston Brown, which possesses the M1 gene. This fragment was cloned and sequenced and found to contain an apparently full‐length TIR‐NBS‐LRR gene, and included 1 kb upstream and 3.8 kb downstream of the gene (which contained the Mxb‐1 region). However, this gene could not be M1 because this fragment was not present in the four double‐resistant (M+M1) recombinant individuals recovered from an MM1 test‐cross, but was present in the three double‐susceptible recombinants that lacked both M and M1(see below). This gene was designated M1‐comp1 (M1‐companion gene 1). The nucleotide sequence of the M1‐comp1 gene predicts a protein product of 1554 amino acids (not shown).
Probe M3.2/5, derived from the region upstream of M3 encoding the tetratricopeptide repeat domain, hybridizes to two fragments in gel blot analysis, one at the L locus and one at the M locus (Table 1). Using this probe, a 9.5‐kb fragment from Williston Brown DNA (EcoRI digest) that originated from the M1 locus was cloned and sequenced. The fragment, which was present in the four M+M1 recombinants and absent from the three recombinants that lacked both of these genes, contained an apparently functional TIR‐NBS‐LRR gene and included approximately 3 kb upstream of the gene and approximately 2 kb downstream of the gene. Therefore, this fragment was introduced into the flax line Ward. Nineteen of 32 transgenic plants (as determined by gel blot analysis) were resistant to rust strain CH5‐132 (virulent to Ward, but avirulent to M1), indicating that the TIR‐NBS‐LRR transgene can confer rust resistance. Transgenic plants were also resistant to four strains from the CH5 family avirulent to M1, but were susceptible to four strains virulent to M1, thereby confirming that the cloned gene expresses M1 specificity. The nucleotide sequence of the M1 gene predicts a protein product of 1401 amino acids (Fig. 1) with three predicted introns in the same locations as those in the M gene (Anderson et al., 1997).
Recombination analyses
We used recombination analysis to determine the relative positions of the M, M1, M3 and M4 resistance genes in their respective haplotypes. In the first experiment, two F1 plants, obtained by crossing Dakota (M) to Williston Brown (M1), were crossed as female to Hoshangabad (no known resistance genes) to produce test‐cross progeny; these were inoculated with rust strain CH5‐87 (avirulent to M, virulent to M1) followed, 6 days later, by a second inoculation with strain CH5‐132 (virulent to M, avirulent to M1). Amongst 2495 progeny, 2488 showed parental phenotypes segregating 1193 (resistant to CH5‐87, susceptible to CH5‐132) to 1295 (resistant to CH5‐132, susceptible to CH5‐87), a result that just differs from the expected 1 : 1 ratio (χ 2 1= 4.18, P < 0.05). Of the remaining seven putative recombinants, four were resistant to both tester strains and three were susceptible to both strains (Table 2). All seven possessed restriction fragments (probes Lu‐2 and X22‐B; Table 1) unique to the male parent Hoshangabad (data not shown), confirming that the four double‐resistant individuals were recombinants and not heterozygotes arising from accidental selfing of the M/M1 female parent. Lines homozygous for each recombination event were selected from selfed progeny of the test‐cross recombinants by identifying individuals that lacked the Hoshangabad M locus‐specific markers in gel blot analyses (X22‐B/DraI or Lu‐2/XbaI). Having individuals homozygous for the recombinant strand greatly facilitated the mapping of restriction fragment length polymorphism (RFLP) markers with respect to the M and M1 genes (see below). This analysis also confirmed that the three double‐susceptibles were derived from crossover events rather than the mutation of one of the parent genes, as each possessed fragments specific to the Dakota (M) parent, as well as fragments specific to the Williston Brown (M1) parent at or near the M locus. Thus, with the Mxb‐1 probe, all three possessed a Williston Brown (M1)‐specific EcoRI fragment, whereas, with probe X22‐B, all three possessed a Dakota (M)‐specific DraI fragment (data not shown). Twenty selfed progeny from each of four putative MM1/MM1 homozygotes (each derived from a separate double‐resistant recombinant) were resistant to both rust strains CH5‐87 (avirulent to M, virulent to M1) and CH5‐132 (virulent to M, avirulent to M1), thereby confirming that the parent lines were homozygous for M and M1 on the same strand.
Table 2.
Recombinant test‐cross progeny derived from parent lines heterozygous for rust resistance genes at the M locus crossed to lines with no known M locus resistance genes.
| Genotype of heterozygous parent | Number of test‐cross progeny | Recombinants* | Reference | |
|---|---|---|---|---|
| MxMy | – | |||
| MM1 | 1 274 | 1† | 4 | Flor (1965) |
| 2 495 | 4 | 3 | This paper | |
| MM3 | 16 188 | 16 | 11 | Flor (1965) |
| 2 300 | 2 | — | Shepherd and Mayo (1972) | |
| MM4 | 1 504 | 1 | 3 | Flor (1965) |
| M1M3 | 5 838 | — | — | This paper |
| M1M4 | 776 | — | — | Flor (1965) |
| M3M4 | 1 471 | — | — | Flor (1965) |
| 6 306 | — | 1‡ | This paper | |
MxMy, recombinant expressing both parental resistance specificities; –, recombinant expressing neither parental specificity.
Plant died—not ascertained whether it was a recombinant or a heterozygous self.
Examination of linked markers suggests that this individual arose not from a crossover recombination event, but from either a mutation of the M4 gene or from a conversion recombination event that altered M4 without an associated crossover (see text).
To examine the recombination relationship of the M1 and M3 resistance genes, five F1 plants obtained by crossing Cass (M3) to Williston Brown (M1) were crossed as female to Hoshangabad. The resulting test‐cross progeny were inoculated with rust strain CH5‐87 (avirulent to M3, virulent to M1) followed, 6 days later, with strain CH5‐132 (avirulent to M1, virulent to M3). No recombinants of either kind (double‐resistant or double‐susceptible) were recovered amongst 5838 progeny screened (Table 2). The parental‐type progeny segregated 2916 (resistant to CH5‐87, susceptible to CH5‐132) to 2922 (resistant to CH5‐132, susceptible to CH5‐87), a close fit to the expected 1 : 1 ratio.
In a third test‐cross, the recombination relationship of the M3 and M4 genes was examined. Six F1 plants obtained by crossing Cass (M3) to Victory A (M4) were crossed as female to Hoshangabad. The resulting test‐cross progeny were inoculated with rust strain CH5‐96 (avirulent to M3, virulent to M4) followed, 6 days later, with rust strain CH5‐104 (avirulent to M4, virulent to M3). Amongst the 6306 progeny tested, one non‐parental type was identified: this plant was susceptible to both tester rust strains (Table 2). The parental‐type progeny segregated 3037 (resistant to CH5‐96, susceptible to CH5‐104) to 3268 (resistant to CH5‐104, susceptible to CH5‐96), which differs significantly from the expected 1 : 1 ratio (χ 2 1= 8.46, P < 0.01). To examine the event (crossover, conversion or mutation) that gave rise to the single double‐susceptible individual, a progeny individual homozygous for the recombinant or mutant strand (designated R/M‐A) was investigated by gel blot analysis using the following M locus probe/restriction enzyme combinations, X22‐B/AccI, M3.2/5/EcoRI, Mxb‐1/EcoRI and Lu‐3, in combination with AccI, SacI and XbaI (Table 1). R/M‐A possessed all 11 of the Victory A (M4)‐specific fragments identified in these gel blots and lacked all 10 of the Cass (M3)‐specific fragments (data not shown). As these markers most probably flank the M4 gene (see below), the double‐susceptible individual most probably arose from either a mutation in the M4 gene or a conversion event that altered M4 without an associated crossover.
Mapping of M locus resistance genes and RFLP markers
Figure 2 shows a map (not to scale) of the relative positions of four M locus resistance genes and several RFLP markers at or near the M locus which were mapped with respect to seven crossover events between M and M1; it is consistent with previous observations (Ellis et al., 1995). Because M recombines with M1, M3 and M4 (Table 2), it has been placed at a different site on the map to M1, M3 and M4. The lack of recombination between M1, M3 and M4, despite relatively large family sizes in two cases (Table 2), suggests that these genes may be alternative forms of the same gene within the array. For this reason, these genes have been placed at the same site in the map.
Figure 2.
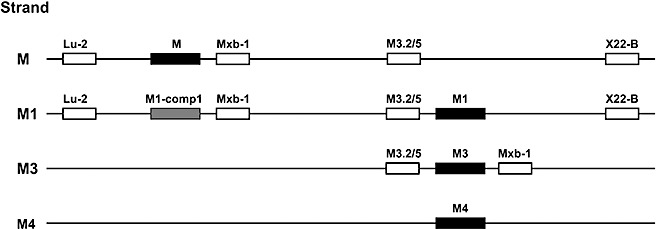
Map of resistance genes and closely associated restriction fragment length polymorphism (RFLP) markers at the M locus in flax on chromosomes from the M, M1, M3 and M4 genomes. The markers Lu‐2, Mxb‐1, M3.2/5 and X22‐B are described in Table 2. M1‐comp1 is a Toll interleukin 1 receptor–nucleotide binding site–leucine‐rich repeat (TIR‐NBS‐LRR) gene cloned from the M1 genome. The placement of the Mxb‐1 marker just downstream of the M, M1‐comp1 and M3 genes and the M3.2/5 marker just upstream of M1 and M3 comes from sequencing cloned fragments. The Lu‐2 and X22‐B markers on the M and M1 strands, the M3.2/5 marker on the M strand and the M1‐comp1/Mxb‐1 pair on the M1 strand are positioned relative to seven crossover events between M and M1: potential inaccuracies exist in the placement of these markers relative to others on the same side of the crossover events (see Results).
The Lu‐2 and X22‐B markers have been placed on the map flanking M and M1 in the order Lu‐2 . . . M . . . M1 . . . X22‐B, because the four MM1 recombinant strands each contained the Lu‐2 fragment (XbaI digest) associated with the M strand and the X22‐B fragment (SacI digest) associated with the M1 strand, whereas the three double‐susceptible recombinant strands each contained the Lu‐2 fragment associated with the M1 strand and the X22‐B fragment associated with the M strand (Fig. 3a,b). These are the observations expected if the marker order is Lu‐2 . . . M . . . M1 . . . X22‐B and each of the seven recombinants from the MM1 test‐cross is derived from a single crossover event between M and M1. This marker order is consistent with that of Ellis et al. (1995), who proposed that Lu‐2 and X22‐B flank M based on an analysis of a deletion mutant that had lost M specificity together with several M locus LRR fragments and the X22‐B fragment, but had retained the Lu‐2 fragment.
Figure 3.
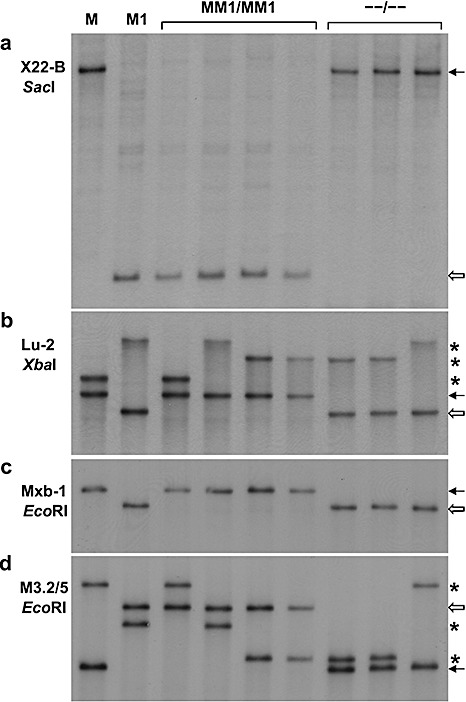
Southern blots of test‐cross F1 parents Dakota and Williston Brown, with the M and M1 genes, respectively, and four lines homozygous for recombinant strands possessing both M and M1 (MM1/MM1) and three lines homozygous for recombinant strands lacking both M and M1 (–/–). (a–d) Different blots with the probe and restriction enzyme combination indicated. Black arrows indicate fragments in recombinants derived from the M parent. White arrows indicate fragments derived from the M1 parent. Asterisks indicate fragments presumed to be derived from the L locus from either of the F1 parents M and M1 or the other test‐cross parent Hoshangabad. In (a) and (d), the four MM1 recombinants all possess a fragment in common with M1 and the three –/– recombinants a fragment in common with M. In (b) and (c), the four MM1 recombinants all possess a fragment in common with M and the three –/– recombinants a fragment in common with M1.
The Mxb‐1 fragment is shown on the map closely associated with M, as it is located approximately 1 kb downstream of the M gene (Anderson et al., 1997), but the orientation of M with respect to M1, M3 and M4 is not known. As expected, the MM1 recombinant strands all possess the single Mxb‐1 fragment on the M strand (Fig. 3c). As these four MM1 recombinant strands all lacked the single Mxb‐1 fragment associated with the M1 strand, whereas the three double‐susceptible recombinants all possessed this fragment (Fig. 3c), the Mxb‐1 fragment on the M1 strand has been placed at the same location on the M1 strand as it is on the M strand (Fig. 2). However, a location of the Mxb‐1 fragment on the M1 strand nearer to or even beyond the Lu‐2 marker on the M1 strand would also be consistent with the observed results. The M1‐comp1 gene has been placed on the map of the M1 strand closely associated with the Mxb‐1 fragment, as it occurs just upstream of this sequence. This places it at a similar site on the M1 strand as M is on the M strand. As M and M1‐comp1 each have the unique Mxb‐1 fragment located just downstream of them, they may be alternative forms of the same gene in the different haplotypes.
The M3.2/5 fragment, which is apparently part of a gene encoding a tetratricopeptide repeat domain, is shown on the map (Fig. 2) closely associated with M1, as it is located just upstream of M1. The M3.2/5 probe hybridizes to two fragments, only one of which is located at the M1 locus. As expected, the four MM1 recombinants all possessed the M3.2/5 fragment associated with the M1 strand and lacked the fragment associated with the M strand, whereas the three double‐susceptible recombinant strands showed a reciprocal arrangement (Fig. 3d). Therefore, the M3.2/5 fragment has been placed at the same location on the M strand as it is on the M1 strand in Fig. 2. However, a location nearer to the X22‐B marker would also be consistent with the observed results. No recombinants involving the M3 and M4 genes were available for DNA marker analysis; however, the M3.2/5 and Mxb‐1 markers are shown flanking the M3 gene (Fig. 2) because these markers occurred upstream and downstream, respectively, of M3 in the cloned sequence (see above). As the orientation of the resistance genes is unknown, the position of close flanking markers, such as M3.2/5 and Mxb‐1, could be reversed. The map also assumes that all resistance genes have the same orientation.
To investigate the position of the M and M1 genes in the cluster of up to 15 TIR‐NBS‐LRR genes at the M locus, gel blot analyses of individuals homozygous for each of the seven MM1 recombinant strands were carried out with probe Lu‐3 (Table 1) in combination with six restriction enzymes (BglII, AccI, XbaI, DraI, SacI and BamHI). The AccI blot was particularly informative as the two parent lines had only four fragments in common, with the M parent Dakota having 10 unique fragments and the M1 parent Williston Brown having nine unique fragments (Fig. 4). An examination of the gel blots of the recombinant lines revealed two features of interest. First, the three double‐susceptible recombinants all had identical patterns, as did the four double‐resistant recombinants, except, in all seven cases, for a single polymorphic fragment (marked by * in Fig. 4) presumed to originate from the L locus, a conclusion supported by the observation that the polymorphic fragment in four of the seven recombinants was not present in either of the M or M1 parent lines, but was present in the Hoshangabad line used in the test‐cross (data not shown). Second, both recombinant types possessed a mixture of the polymorphic fragments present in the M and M1 parent lines, with the double‐resistant lines possessing four and two and the double‐susceptible lines possessing five and six of the M and M1 parent fragments, respectively. These observations suggest that recombination events between M and M1 occur in a highly restricted region and are consistent with M and M1 being located at adjacent positions within the cluster with all of the polymorphic fragments located on either side of these genes (it should be noted here that no information is available with regard to the location of the four non‐polymorphic fragments). An alternative possibility is that sequence and structural divergence between the haplotypes may restrict recombination to a small region within the M and M1 clusters; in such a case, uniformity of pattern within each of the recombinant types would be expected, even if all of the polymorphic fragments were located between M and M1.
Figure 4.
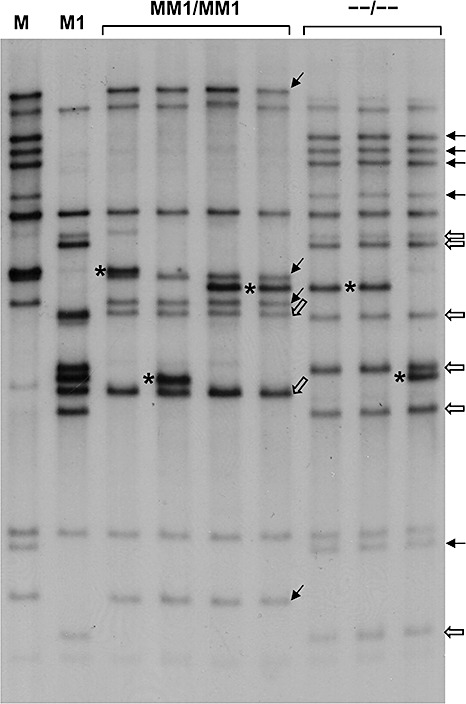
Southern blots of test‐cross F1 parents Dakota and Williston Brown, with the M and M1 genes, respectively, and four lines homozygous for recombinant strands possessing both M and M1 (MM1/MM1) and three lines homozygous for recombinant strands lacking both M and M1 (–/–). Probe Lu‐3 [from the leucine‐rich repeat (LRR) region of the L6 gene] with Acc1 digestion. Black arrows indicate fragments in recombinants derived from the M parent. White arrows indicate fragments derived from the M1 parent. Asterisks indicate fragments presumed to be derived from the L locus from either of the F1 parents M and M1 or the other test‐cross parent Hoshangabad.
DISCUSSION
Sequence comparisons of M, M1 and M3
We used transposon tagging and homology‐based cloning to identify the M1 and M3 rust resistance genes of the M locus in flax. These genes encode TIR‐NBS‐LRR proteins and are members of the same gene family as the previously identified M gene and the related L locus resistance genes. A comparison of the predicted amino acid composition of the M, M1 and M3 protein products, together with that of the L6 protein product (Fig. 1), reveals several features of interest. First, as noted previously, (Anderson et al., 1997; Ellis et al., 1999), most of the L group proteins and the M protein contain two large repeats, each of approximately 150 amino acids, at the C‐terminal end of the LRR region, and unequal crossing over between the repeat encoding regions in the gene has apparently been the cause of some structural variants observed in this group of proteins. Thus, the L1 protein contains only one repeat, whereas the L2 protein contains four. Furthermore, three mutant alleles of M recovered during the Ac tagging of M were found to contain only one repeat (Anderson et al., 1997; Ellis et al., 1999). Here, we see further evidence of unequal exchange in this region leading to sequence diversity. Thus, although the M3 protein contains two repeats, M1 contains a novel arrangement with three copies of this repeat, although the additional repeat has been modified by the loss of the second LRR unit. The additional repeat is likely to have arisen by an unequal crossover event, as one part is identical and the other part is closely related to parts of the two (non‐exact) repeats that flank it. In detail, the first 29 amino acids of the addition are identical in sequence to the first 29 amino acids of the second repeat. The next 26 amino acids of repeat 2 (or 27 amino acids of repeat 1), which represent one LRR, are absent from the addition. The sequence of the remaining 86 amino acids of the addition matches that of the last 86 amino acids of the first repeat, except for seven amino acid differences, but nevertheless includes all the differences that distinguish repeat 1 from repeat 2. This arrangement may have resulted from either a recent unequal exchange with another member of the M locus gene family, or a more ancient unequal exchange within the ancestral gene itself, followed by subsequent sequence divergence. The (presumptive) deletion event that resulted in the loss of the second LRR unit in the addition occurred between a GAGG motif in the second LRR unit and an identical motif in the third LRR unit. Therefore, an illegitimate recombination event between these motifs is a probable cause of the deletion. Such events appear to have played a major role in generating diversity in the LRR regions of plant resistance genes (Wicker et al., 2007). Overall, the protein products of the three M locus homologues are more similar to each other (82%–87% amino acid identity—excluding the four additional LRRs in M1 from the comparison) than to L6 (75%–79% identity).
Another important process in resistance loci evolution is sequence exchange between either allelic genes, such as those at the L locus in flax (Ellis et al., 1997), or between allelic and/or paralogous genes in the cluster, such as the N and P loci in flax (2001a, 2001b) and the Cf‐4/Cf‐9 locus in tomato (Parniske et al., 1997). This characteristic is also observed amongst the M locus genes. For instance, the M and M1 protein sequences are identical over a 216‐amino‐acid region (which, in M1, flanks the addition), but differ substantially throughout the rest of their sequences (Fig. 1). Similarly, the M1 and M3 proteins are identical for the first 210 amino acids, but then diverge extensively, with 161 amino acid differences (excluding the large duplication in M1) scattered throughout the remainder of their protein products (Fig. 1). Thus, M1 contains a segment of 210 amino acids identical to M3 and a segment (split by a duplication) of 216 amino acids identical to M. These observations suggest that these genes consist of a mosaic of fragments shared with other genes as a result of intragenic recombination events. The sequence divergence in the 5′ and 3′ flanking regions of M, M1 and M3, as well as their different locations in the cluster as suggested by recombination analysis (Fig. 2), indicate that sequence exchanges are not limited to strictly allelic genes within the cluster, as observed for the N and P loci in flax (2001a, 2001b).
Previously, it has been noted (Anderson et al., 1997) that substantial differences occur between L6 and M in the region between the conserved hydrophobic N‐terminal sequence of 15 amino acids and the TIR domain, with the 37 amino acids in the M protein showing very little sequence similarity to the 22 amino acids in L6. As shown in Fig. 1, M1 and M3 closely resemble L6 in this region. Overall, the sequences of M, M1 and M3 provide no information with regard to which amino acid differences in their products are responsible for their specificity differences, because of the very large number of amino acid differences scattered throughout their products (Fig. 1).
Structural differences at rust resistance loci
Flax (2n= 30) is an ancient tetraploid, and L and M may be homoeologous loci in the different genomes given their close sequence relationship and the presence of related flanking sequences associated with each of these loci, including a DNA segment with homology to a glycogen glucosyltransferase gene in rice (the Lu‐2 probe) and a DNA segment encoding a tetratricopeptide repeat domain (the M3.2/5 probe). Assuming that the regions are homoeologous, a clear structural difference that has arisen between the genomes is that the L locus contains a single TIR‐NBS‐LRR gene, whereas the M locus contains up to 15 copies of the TIR‐NBS‐LRR gene, although some of these may not be complete copies but gene fragments, as one gene fragment of 720 bp occurs downstream of M3 (see below). A possible mechanism with regard to how multiple copies at the M locus arose (discussed by Ellis et al., 1995) is that an initial duplication occurred as a result of unequal crossing over between repeated DNA segments (possibly transposable elements) that flanked the original ancestral gene, with subsequent amplification occurring as a result of further unequal crossing over between TIR‐NBS‐LRR genes. This mechanism would also amplify sequences between the genes; therefore, the detection of single copy sequences (as determined by gel blot analyses) in the haplotype clusters, such as the Mxb‐1 probe region (located just downstream of the M, M3 and M1‐comp1 genes) and the M3.2/5 probe region (located just upstream of M1 and M3), suggests that such intragenic regions may be either lost over time, perhaps by intrastrand homologous recombination between linked repeated sequences or by illegitimate recombination events (Devos et al., 2002) or, if deletions do not occur, accumulate so many changes that a probe derived from one no longer hybridizes to the other region(s). Alternatively, unique sequences could be introduced if used as filler DNA in double‐strand break repair (Kirik et al., 2000).
Evidence of structural differences between different haplotypes at the M locus comes from the following findings: (i) the M, M1 and M4 haplotypes possess only a single copy of the M3.2/5 fragment, whereas the M2, M3 and M6 haplotypes possess two copies; (ii) the M3 gene possesses a M3.2/5 fragment just upstream of it and a Mxb‐1 fragment just downstream of it, whereas, in the M1 haplotype, these fragments are upstream and downstream, respectively, of two different genes (M1 and M1‐comp1); and (iii) of the nine polymorphic fragments detected on the M strand by probe Lu‐3 (Fig. 4), four are located on the M side of the seven crossover events and five on the M1 side, whereas, of the eight polymorphic fragments in the M1 haplotype, six are located on the M side of the crossovers and two on the M1 side. Structural changes are also evident in the 2‐kb regions downstream of M and M3, which both contain the 850‐bp Mxb‐1 segment. These regions are quite different, except for 150 bp immediately downstream of the genes and the presence of the Mxb‐1 segment. However, the Mxb‐1 segment begins 150 bp downstream of M3, but 550 bp downstream of M. In M3, the Mxb‐1 segment is followed by a 720‐bp segment encoding a series of LRRs, apparently a fragment of a TIR‐NBS‐LRR gene. Overall, the structural differences described above between genomes, haplotypes, genes and downstream gene regions suggest that the L and M loci have a very ancient origin.
Recombination of M, M1, M3 and M4 genes
The test‐cross analyses (summarized in Table 2) reveal that M readily recombines with M1, M3 and M4, whereas these three genes fail to recombine, despite large family sizes (>5800) for two of the three pair combinations tested (M1M3 and M3M4). If the M1, M3 and M4 haplotypes each have the same number and arrangement of TIR‐NBS‐LRR genes, the lack of recombination between M1, M3 and M4 might be accounted for by proposing that these genes are alternative forms of the same gene within the different haplotypes (alleles). Some support for this comes from the finding that M1 and M3 each have a region encoding a tetratricopeptide repeat domain (M3.2/5 probe) just upstream of them. Against this, however, is the finding that, although the M1 and M4 haplotypes each have a single copy of the Mxb‐1 fragment, the M3 haplotype has two copies; one of the two copies in the M3 haplotype occurs just downstream of M3, but the single copy in the M1 haplotype is not located downstream of M1, but downstream of another gene in the cluster, M1‐comp1. These observations suggest that the M1 and M3 haplotypes show structural differences, thereby making it difficult to identify genes in the different haplotypes as being allelic. It is possible that structural differences between these haplotypes restrict recombination between these genes, but this still leaves the question as to why these genes failed to give rise to double‐susceptible recombinants as a result of intragenic recombination, as observed in 17 of the 27 pairwise combinations of L group genes examined in test‐crosses (for a review, see Islam and Shepherd, 1991), where the recombinant gene failed to express either of the parental specificities. Subsequent analysis of some of these recombinants revealed that many resulted from a crossover within the NBS domain, which led to the proposal that the splitting up of co‐evolved TIR and NBS‐LRR partners in the parental alleles resulted in loss of function (Luck et al., 2000). However, in the case of M1 and M3, a crossover in the NBS region may not result in a loss of function as these genes have identical TIR regions. Intragenic crossovers would also not result, or be unlikely to result, in a loss of specificity if the specificity differences between alleles are determined by single nucleotide differences, or by several nucleotide differences localized to a small region of the gene. Another possibility is that no intragenic recombination occurred because the genes are highly divergent and have no significant regions of exact homology which promote recombination—this could apply to M1 and M3, which are highly divergent except for a region of high homology at their 5′ ends (Fig. 1).
In addition to the test‐cross recombination studies listed in Table 2, an extensive recombination analysis of M locus genes was undertaken by Mayo and Shepherd (1980) using a special F2 analysis. F2 progeny from F1 plants of genotype MM3/M1 or MM3/M4 were successively screened with two rust strains to identify individuals recombinant for M and M3 (both tester strains were virulent to M1 and M4). Any recombinants recovered were then tested with a third rust strain to determine whether they did, or did not, possess the M1 or M4 gene. With the MM3/M1 parent, the results are consistent with M1 being located at the same site as M3 (in agreement with our test‐cross studies) or distal to M3. With the MM3/M4 parent, one putative recombinant strand of 23 tested, which possessed M but not M3 or M4, could have arisen from a crossover between M4 and M3, assuming a gene order M‐M4‐M3 with M4 very close to M3. This result potentially separates M3 and M4, whereas, in the test‐cross study, M3 and M4 were not separated. However, the exceptional strand could also have arisen from a mutation event (of the M3 gene) or an intragenic recombination event (between M3 and M4), as we observed one such event amongst our M3/M4 test‐cross progeny (Table 2). Thus, it remains open as to whether or not M3 and M4 can be separated (very rarely) by recombination. Nevertheless, the cloning of the M1 and M3 resistance genes described here now allows the use of these genes in combination in transgenic plants despite the lack of natural recombination.
EXPERIMENTAL PROCEDURES
Plant and fungal materials
The flax lines Dakota (M), Williston Brown (M1), Cass (M3), Victory A (M4) and Ward are all members of Flor's original differential series (Flor, 1956) and have been described by Islam and Mayo (1990). The line Forge is homozygous for four rust resistance genes L6, M, N and P2 (Lawrence et al., 1993). The line Hoshangabad contains no known rust resistance genes (Mayo and Shepherd, 1980).
Rust strain Sp‐y is virulent to the four resistance genes in Forge (L6, M, N and P2), but avirulent to M3. The four strains CH5‐87, CH5‐132, CH5‐96 and CH5‐104 are selfed progeny of strain CH5 (Lawrence et al., 1981). The reactions of lines with resistance genes M, M1, M3 and M4 to these strains that are relevant to this study are as follows (R, resistant; S, susceptible).
| CH5‐87 | CH5‐132 | CH5‐96 | CH5‐104 | |
|---|---|---|---|---|
| M | R | S | ||
| M1 | S | R | ||
| M3 | R | S | R | S |
| M4 | S | R |
Rust inoculation of test‐cross plants
As the reaction of each test‐cross plant to two different rust strains had to be recorded, it was necessary to identify each plant individually. This was achieved by laying a piece of metal mesh containing 12 × 9 = 108 squares (each square 2.5 cm × 2.5 cm) on top of the soil in a plastic crate 34 cm × 26 cm × 12 cm deep. One seed was planted inside each square of the mesh, except for the four corner squares. Plants were inoculated with the first rust strain when approximately 8 cm tall, with inoculation of the second rust strain carried out 6 days later. To prevent plants from lodging between the first and second inoculations, dowelling pieces (18 cm × 1 cm diameter), each with a brass screw projecting at right angles 1 cm from one end, were inserted to the full depth of the soil in the four corner squares of the mesh. The mesh was then raised and supported by the projecting screws. Using the raised metal mesh to keep plants upright ensured that the new‐growth tip leaves formed after the first inoculation were fully exposed to the spores of the second inoculation. Plants were scored (resistant or susceptible) approximately 12 days after each inoculation using score sheets drawn up to contain a grid of 12 × 9 squares to match the ‘gridded’ plants in the crates. Putative recombinant plants were transplanted to large pots. Those susceptible to both strains had their infected leaves removed and their stems painted with fungicide to eliminate the rust.
For inoculation, a plastic cover with two sleeves in one side, supported by a metal frame, was placed over 12 crates in a large metal tray on a bench. Inoculation was achieved by mixing 100–120 mg of urediniospores with 4.3 g of talc and dusting this spore–talc mixture over the plants inside the cover, as described previously (Lawrence, 1988). After inoculation, the plants and the inside of the cover were misted with water and left overnight at a temperature below 24 °C.
Tagging of M3 and M1 with Ac: stock development
A line possessing an Ac element closely linked to M3 was produced by crossing Cass (M3) to Solo M‐T1, which possesses a T‐DNA containing an Ac element and a neomycin phosphotransferase gene (NPT‐II) closely linked (1–2 map units) to M. An F1 plant was crossed to Hoshangabad and the resulting test‐cross progeny were scored for M3 by inoculation with rust strain CH5‐96 and for NPT‐II in the cotyledons using a dot blot assay based on that of McDonnell et al. (1987). One recombinant possessing M3 and NPT‐II was identified amongst 316 progeny. The M3 gene and its closely linked Ac element in this recombinant individual were then backcrossed to a transgenic line of Forge, D97, (Lawrence et al., 1993) which contains approximately 15 copies of Ac. A line homozygous for M3 and its closely linked Ac, and containing multiple other Ac elements, was selected amongst the selfed progeny of the final backcross line (the L6, N and P2 genes are also present in this line). This line was crossed as female to Hoshangabad (no known resistance genes) to produce the progeny that were screened with rust strain Sp‐y (virulent to L6, N and P2, but avirulent to M3) to identify rust‐susceptible M3 mutant individuals that may contain an Ac‐tagged M3 gene.
For M1, one recombinant individual possessing M1 and a closely linked Ac element was identified amongst 319 (Williston Brown × Solo M‐T1) × Hoshangabad test‐cross progeny. The M1 gene and its closely linked Ac element were then backcrossed into line X157‐3G, another high‐Ac copy number line (∼15 copies). X157‐3G was derived by three generations of selfing from a line that contained an Ac‐tagged P2 resistance gene (mutant X157—see Dodds et al., 2001b), with selection in each selfing generation for high‐Ac copy number individuals which also had one or more newly transposed Ac elements. A line homozygous for M1 and its closely linked Ac element was selected amongst the selfed progeny of the second backcross line (M1 was the only resistance gene in this line). Rust strain CH5‐132, avirulent to M1, but virulent to M, was used to identify the presence of M1 throughout.
DNA extraction and Southern blot analysis
Flax DNA was extracted essentially as described by Taylor and Powell (1982). In brief, 4.5 g of leaf and stem tissue, ground in liquid nitrogen, was incubated in 5.5 mL of a 2% (w/v) cetyltrimethylammonium bromide (CTAB)‐based extraction buffer at 65 °C for 15 min, treated twice with chloroform–isoamyl alcohol (24 : 1), followed by the addition of a 1% (w/v) CTAB‐based DNA precipitation solution. The precipitate was resuspended in 2 mL of 1 m CsCl for 24 h, with 30 µL of 10 mg/mL ethidium bromide added, before layering onto a cushion of 2 mL of 5.7 m CsCl in a Beckman polyallomer 13 mm × 51 mm tube and purifying in a step gradient by centrifugation at 35 000 rpm for 16 h. The ethidium bromide‐bound DNA band was removed with a syringe and cleaned of ethidium bromide by three successive isopropanol (CsCl‐saturated) treatments. Then, 1 mL of H2O was added and the DNA was precipitated with 3 vol of 70% (v/v) ethanol, spooled onto a glass capillary tube and redissolved in 500 µL 10 mM Tris, 1 mM Ethylenediaminetetra‐acetic acid pH 8.0 (TE) and 50 µL of 3 m NaOAc. Subsequently, the DNA was re‐precipitated with ethanol, the pellet was washed with 70% (v/v) ethanol and redissolved in 300 µL of TE.
For Southern blot analysis, 5 µg of flax genomic DNA was digested with the appropriate restriction enzyme for 7 h in a reaction volume of 50 µL. DNA fragments were separated by 1% (w/v) agarose gel electrophoresis for 16 h and transferred to nylon membranes (Pall) in 400 mm NaOH for 24 h. After a brief wash in 2 × standard saline citrate (SSC) (1 × SSC: 150 mm NaCl, 150 mm sodium citrate), the membrane was rotated in 40 mL of prehybridization mix [50% (v/v) formamide, 10% (w/v) dextran sulphate, 4 × SSC, 0.125% (w/v) SDS, 50 mm phosphate buffer pH 6.5, 5 × Denhart's reagent, 0.0525% (w/v) sheared salmon sperm DNA) for 24 h at 42 °C. The prehybridization mix was then removed and replaced with 40 mL of hybridization mix (same as prehybridization mix except for 5 × SSC, 15 mm phosphate buffer pH 6.5 and 0.75 × Denhart's reagent) containing the radioactive probe. The filter was rotated in the hybridization mix for 24 h at 42 °C, followed by four 15‐min washes at room temperature [two in 2 × SSC and 0.1% (w/v) SDS and two in 0.1 × SSC and 0.1% (w/v) SDS]. The dried membrane was then wrapped in polyethylene film and exposed to radiographic film. The probes used are described in Table 1, except for the 5′ Ac probe, which is a 405‐bp BamHI/NruI fragment that commences 182 bp from the 5′ end of Ac, and the 3′ Ac probe [polymerase chain reaction (PCR) generated], which contains 1 kb of the 3′ terminal end Ac.
Cloning of flax genomic fragments and transformation of flax
Flax genomic DNA digested with EcoRI was cloned into the Lambda DASH II/EcoRI vector according to the manufacturer's instructions Stratagene (La Jolla, California, USA). The resulting lambda libraries were screened with 32P‐labelled DNA probes and hybridizing plaques were purified.
The EcoRI fragments were subcloned from λ phage DNA into the plasmid vector pBluescript and sequenced. The M3 and M1 genes were subcloned into the binary Agrobacterium transformation vector pTNotTreg containing a plant‐selectable marker gene providing resistance to spectinomycin sulphate (Dodds et al., 2001b). These binary vectors were transferred from Escherichia coli to the disarmed Agrobacterium tumefaciens strain GV3101Pmp90 (Koncz and Schell, 1986) by triparental mating (Ditta et al., 1980). The flax EcoRI fragments in the binary vectors were checked by DNA sequencing. Transformation of the flax line Ward was carried out as described by Anderson et al. (1997).
Pollen squash test to identify deletion mutants
The anthers in a flower, from which the petals had been removed, were dabbed into a small drop of orcein stain (1% orcein in 45% acetic acid) on a glass microscope slide to release pollen grains. The droplet was spread with a needle and any large anther pieces were removed. A cover slip was added, the slide was enclosed between two folded 90‐mm filter papers and the pollen grains were squashed by pressing down (fairly firmly) on the cover slip with the thumb two or three times, whilst ensuring that no sideways slippage of the cover slip occurred. Under a microscope, the number of normal pollen grains (large and accompanied by abundant exuded cytoplasm) vs. the number of aberrant pollen grains (small with no or only a trace amount of exuded cytoplasm) was scored. With ‘wild‐type’ plants, 95%–100% of the pollen grains are normal. When a mutant plant possessed approximately 50% (or more) aberrant pollen grains, this was taken as an indication that the mutation was caused by a deletion and that the aberrant pollen grains were those that received the chromosome with the deletion.
ACKNOWLEDGEMENTS
Excellent technical assistance was provided by Patricia Moore, Kim Newell and Valerie Ryle.
GenBank accession numbers M1 GQ141888 M3 GQ141889 M1‐comp1 GQ141890
REFERENCES
- Anderson, P.A. , Lawrence, G.J. , Morrish, B.C. , Ayliffe, M.A. , Finnegan, E.J. and Ellis, J.G. (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine‐rich repeat coding region. Plant Cell, 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Biezen, E.A. , Freddie, C.T. , Kahn, K. , Parker, J.E. and Jones, J.D.G. (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR‐NB‐LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Collins, N. , Drake, J. , Ayliffe, M. , Sun, Q. , Ellis, J. , Hulbert, S. and Pryor, T. (1999) Molecular characterization of the maize Rp1‐D rust resistance haplotype and its mutants. Plant Cell, 11, 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M. , Brown, J.K.M. and Bennetzen, J.L. (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis . Genome Res. 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta, G. , Stanfield, S. , Corbin, D. and Helinski, D.R. (1980) Broad host range DNA cloning system for Gram‐positive bacteria: construction of a gene bank of Rhizobium meliloti . Proc. Natl. Acad. Sci. USA, 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. and Ellis, J.G. (2001a) Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27, 439–453. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Pryor, T. and Ellis, J.G. (2001b) Six amino acid changes confined to the leucine‐rich repeat β‐strand/β‐turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell, 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K. and Belachew, A. (1989) Transposition pattern of the maize element Ac from the bz‐m2 (Ac) allele. Genetics, 122, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G. , Finnegan, E.J. and Lawrence, G.J. (1992) Developing a transposon tagging system to isolate rust‐resistance genes from flax. Theor. Appl. Genet. 85, 46–54. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. , Finnegan, E.J. and Anderson, P.A. (1995) Contrasting complexity of two rust resistance loci in flax. Proc. Natl. Acad. Sci. USA, 92, 4185–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. , Ayliffe, M.A. , Anderson, P.A. , Collins, N. , Finnegan, E.J. , Frost, D. , Luck, J. and Pryor, A.J. (1997) Advances in the molecular genetic analysis of the flax‐flax rust interaction. Annu. Rev. Phytopathology, 35, 271–291. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G. , Lawrence, G.J. , Luck, J.E. and Dodds, P.N. (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene‐for‐gene specificity. Plant Cell, 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1956) The complementary genic systems in flax and flax rust. Adv. Genet. 8, 29–54. [Google Scholar]
- Flor, H.H. (1965) Tests for allelism of rust‐resistance genes in flax. Crop Sci. 5, 415–418. [Google Scholar]
- Flor, H.H. and Comstock, V.E. (1972) Identification of rust‐conditioning genes in flax cultivars. Crop Sci. 12, 800–804. [Google Scholar]
- Hoes, J.A. and Kenaschuk, E.O. (1986) Gene K1 of Raja flax: a new factor for resistance to rust. Phytopathology, 76, 1043–1045. [Google Scholar]
- Islam, M.R. and Mayo, G.M.E. (1990) A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104, 89–100. [Google Scholar]
- Islam, M.R. and Shepherd, K.W. (1991) Present status of genetics of rust resistance in flax. Euphytica, 55, 255–267. [Google Scholar]
- Islam, M.R. , Shepherd, K.W. and Mayo, G.M.E. (1989) Recombination among genes at the L group in flax conferring resistance to rust. Theor. Appl. Genet. 77, 540–546. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. , Carland, F.M. , Lim, E. , Ralston, E.J. and Dooner, H.K. (1990) Preferential transposition of the maize element Activator to linked locations in tobacco. Plant Cell, 2, 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik, A. , Salomon, S. and Puchta, H. (2000) Species‐specific double‐strand break repair and genome evolution in plants. EMBO J. 19, 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C. and Schell, J. (1986) The promoter of TL‐DNA gene 5 controls the tissue‐specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Kuang, H. , Woo, S‐S. , Meyers, B.C. , Nevo, E. and Michelmore, R.W. (2004) Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell, 16, 2870–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, G.J. , Mayo, G.M.E. and Shepherd, K.W. (1981) Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology, 71, 12–19. [Google Scholar]
- Lawrence, G.J. (1988) Melampsora lini, rust of flax and linseed. Adv. in Plant Pathology, 6, 313–331. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J. , Finnegan, E.J. and Ellis, J.G. (1993) Instability of the L6 gene for rust resistance in flax is correlated with the presence of a linked Ac element. Plant J. 4, 659–669. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J. , Finnegan, E.J. , Ayliffe, M.A. and Ellis, J.G. (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N . Plant Cell, 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, J.E. , Lawrence, G.J. , Dodds, P.N. , Shepherd, K.W. and Ellis, J.G. (2000) Regions outside of the leucine‐rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell, 12, 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo, G.M.E. and Shepherd, K.W. (1980) Studies of genes controlling specific host–parasite interactions in flax and its rust. I. Fine structure analysis of the M group in the host. Heredity, 44, 211–227. [Google Scholar]
- McDonnell, R.E. , Clark, R.D. , Smith, W.A. and Hinchee, M.A. (1987) A simplified method for the detection of neomycin phosphotransferase II activity in transformed plant tissues. Plant Mol. Biol. Rep. 5, 380–386. [Google Scholar]
- Meyers, B.C. , Chin, D.B. , Shen, K.A. , Sivaramakrishnan, S. , Lavelle, D.O. , Zhang, Z. and Michelmore, R.W. (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell, 10, 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C. , Kozik, A. , Griego, A. , Kuang, H. and Michelmore, R.W. (2003) Genome‐wide analysis of NBS‐LRR‐encoding genes in Arabidopsis. Plant Cell, 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosi, B. , Wisser, R.J. , Pennill, L. and Hubert, S.H. (2004) Full genome analysis of resistance gene homologues in rice. Theor. Appl. Genet. 109, 1434–1447. [DOI] [PubMed] [Google Scholar]
- Noel, L. , Moores, T.L. , Van Der Biezen, E.A. , Parniske, M. , Daniels, M.J. , Parker, J.E. and Jones, J.D.G. (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis . Plant Cell, 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Parniske, M. , Hammond‐Kosack, K.E. , Goldstein, C. , Thomas, C.W. , Jones, D.A. , Harrison, K. , Wulff, B.B.H. and Jones, J.D.G. (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf‐4/9 locus of tomato. Cell, 91, 821–832. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W. , Emberton, J. , Ogden, M. , SanMiguel, P. and Bennetzen, J.L. (2002) Structural analysis of the maize Rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell, 14, 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E. , Kurth, J. and Leister, D. (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol. Biol. Evol. 19, 76–84. [DOI] [PubMed] [Google Scholar]
- Shepherd, K.W. and Mayo, G.M.E. (1972) Structure of genes in plants conferring resistance to obligate parasites. Science, 175, 375–379. [DOI] [PubMed] [Google Scholar]
- Smith, S.M. and Hulbert, S.H. (2005) Recombination events generating a novel Rp1 race specificity. Mol. Plant–Microbe Interact. 18, 220–228. [DOI] [PubMed] [Google Scholar]
- Smith, S.M. , Pryor, A.J. and Hulbert, S.H. (2004) Allelic and haplotype diversity at the Rp1 rust resistance locus of maize. Genetics, 167, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q. , Collins, N.C. , Ayliffe, A. , Smith, S.M. , Drake, J. , Pryor, T. and Hulbert, S.H. (2001) Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics, 158, 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B. and Powell, A. (1982) Isolation of plant DNA and RNA. Bethesda Research Laboratories Focus publication 4, 4–6. [Google Scholar]
- Wicker, T. , Yahiaoui, N. and Keller, B. (2007) Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J. 51, 631–641. [DOI] [PubMed] [Google Scholar]
- Wicks, Z.W. and Hammond, J.J. (1978) Screening of flax species for new sources of genes resistant to Melampsora lini (Ehrenb.) Lev. Crop Sci. 18, 7–10. [Google Scholar]
- Zimmer, D.E. and Comstock, V.E. (1973) New genes for rust resistance in flax. Phytopathology, 63, 777–780. [Google Scholar]


