SUMMARY
SGT1 (suppressor of G2 allele of Skp1), an interactor of SCF (Skp1‐Cullin‐F‐box) ubiquitin ligase complexes that mediate protein degradation, plays an important role at both G1–S and G2–M cell cycle transitions in yeast, and is highly conserved throughout eukaryotes. Plant SGT1 is required for both resistance (R) gene‐mediated disease resistance and nonhost resistance to certain pathogens. Using virus‐induced gene silencing (VIGS) in Nicotiana benthamiana, we demonstrate that SGT1 positively regulates the process of cell death during both host and nonhost interactions with various pathovars of Pseudomonas syringae. Silencing of NbSGT1 in N. benthamiana plants delays the induction of hypersensitive response (HR)‐mediated cell death against nonhost pathogens and the development of disease‐associated cell death caused by the host pathogen P. syringae pv. tabaci. Our results further demonstrate that NbSGT1 is required for Erwinia carotovora‐ and Sclerotinia sclerotiorum‐induced disease‐associated cell death. Overexpression of NbSGT1 in N. benthamiana accelerates the development of HR during R gene‐mediated disease resistance and nonhost resistance. Our data also indicate that SGT1 is required for pathogen‐induced cell death, but is not always necessary for the restriction of bacterial multiplication in planta. Therefore, we conclude that SGT1 is an essential component affecting the process of cell death during both compatible and incompatible plant–pathogen interactions.
INTRODUCTION
Plants have evolved intricate defence mechanisms against a variety of environmental stresses, including attacks from a vast number of potential pathogens. There are at least two classes of plant innate immune responses (Chisholm et al., 2006; Jones and Dangl, 2006). The primary immune response of plants to pathogen perception is the recognition of pathogen molecules, called pathogen‐associated molecular patterns (PAMPs) or microbe‐associated molecular patterns (MAMPs), such as flagellin (Gomez‐Gomez and Boller, 2002) and elongation factor Tu (Kunze et al., 2004), by plant pattern recognition receptors (PRRs), resulting in PAMP‐triggered immunity (PTI). This class of immune response is called basal resistance. The second class of plant immunity involves direct or indirect recognition of the specific effector(s) secreted from pathogens by the host surveillance system, resulting in so‐called ‘gene‐for‐gene resistance’ or effector‐triggered immunity (ETI), which occurs, for example, during AvrPto–Pto and AvrRpt2–RIN4 interactions (Mudgett, 2005; Pedley and Martin, 2003). Plant resistance and pathogen virulence have co‐evolved as a ‘zigzag’ system (Chisholm et al., 2006; Jones and Dangl, 2006).
Generally, plant immunity triggered by ETI involves rapid plant cell death, known as the hypersensitive response (HR), resulting in an unsuccessful infection of the host plant (Greenberg, 1997; Greenberg and Yao, 2004). As the process of HR‐mediated cell death involves signal transduction pathways similar to those of programmed cell death (PCD), it is generally considered as a form of PCD (Lam et al., 2001). Significant progress has been made in our understanding of the signal transduction pathways leading to HR. Many resistance (R) genes in plants and corresponding avirulence (Avr) genes or specific effectors in pathogens have been identified, and their direct or indirect interactions often lead to HR, which limits the spread of the pathogen from the initial infection site (Martin et al., 2003).
Some effectors contributing to pathogen virulence serve as suppressors of PTI or ETI, so that the pathogens can successfully colonize the host plants and cause disease (Bent and Mackey, 2007). For example, in the absence of resistance protein Pto, the avirulence effector avrPtoB functions to promote bacterial virulence and cause plant disease by the inhibition of PCD through host E3 ubiquitin ligase activity (Abramovitch et al., 2006; Jamir et al., 2004; Rosebrock et al., 2007). Pseudomonas syringae virulence effector HopM1 suppresses host immunity by destroying an immunity‐associated protein, AtMIN7, in Arabidopsis thaliana via the host proteasome to cause infection in plants (Nomura et al., 2006). The disease symptoms developed during compatible interactions of various pathovars of P. syringae with their respective hosts are often associated with cell death. The molecular basis of cell death derived from compatible host–pathogen interactions is still poorly understood and has been suggested to be genetically controlled (Greenberg and Yao, 2004). A MAPKKKα has been shown to function as a positive regulator of bacterial speck disease‐associated cell death in tomato (del Pozo et al., 2004).
SGT1 (suppressor of G2 allele of Skp1) physically associates with SCF (Skp1‐Cullin‐F‐box) ubiquitin ligase complex and plays an essential role in yeast kinetochore function (Kitagawa et al., 1999). SGT1 is a critical signalling component required for R gene‐mediated resistance in several plant species against various plant pathogens, including fungi, bacteria and viruses (Austin et al., 2002; Azevedo et al., 2002; Liu et al., 2004; Tör et al., 2002). In Nicotiana benthamiana, it has been shown that SGT1 is required for nonhost resistance against various bacterial pathogens (Peart et al., 2002). Taken together, these studies indicate that SGT1 plays an important role in both PTI‐ and ETI‐mediated PCD during the plant resistance response. However, it is also important to note that SGT1 has also been shown to be involved in cell death that promotes the pathogenesis of the necrotrophic fungal pathogen, Botrytis cinerea(EI Oirdi and Bouarab, 2007), and the hemibiotrophic fungal pathogen, Fusarium culmorum (Cuzick et al., 2009).
In this study, we used virus‐induced gene silencing (VIGS) in N. benthamiana to investigate the involvement of SGT1 in the process of cell death during compatible interaction, and further characterized the involvement of SGT1a and SGT1b in cell death in A. thaliana using nonhost pathogens. We demonstrated that SGT1‐mediated cell death during plant–bacterial interactions, at least in N. benthamiana and A. thaliana, did not always correlate with the bacterial population in planta, and therefore a delay in SGT1‐mediated cell death did not always result in increased accumulation of all nonhost bacterial pathogens. Furthermore, we showed that overexpression of SGT1 expedited cell death in N. benthamiana during ETI and PTI as well as compatible and incompatible (nonhost) plant–bacterial interactions. Therefore, we conclude that SGT1 is a critical component that positively regulates the process of PCD during both compatible and incompatible plant–pathogen interactions.
RESULTS
SGT1 is required for plant cell death during incompatible interaction, but does not always limit in planta bacterial growth
Nicotiana benthamiana is an ideal plant species for VIGS to assess the functions of candidate genes in development and environmental stress (Lu et al., 2003b). To examine the role of SGT1 in pathogen‐induced PCD, we silenced NbSGT1 in N. benthamiana by VIGS using the Tobacco rattle virus (TRV) (Liu et al., 2002). NbSGT1 gene silencing efficiency was examined by real‐time quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) 2 weeks post‐inoculation with Agrobacterium containing TRV::NbSGT1. The transcript level of NbSGT1 in the gene‐silenced plants was at least 10‐fold lower than that in nonsilenced control plants (Fig. 1A), indicating that NbSGT1 was efficiently silenced in N. benthamiana by VIGS.
Figure 1.
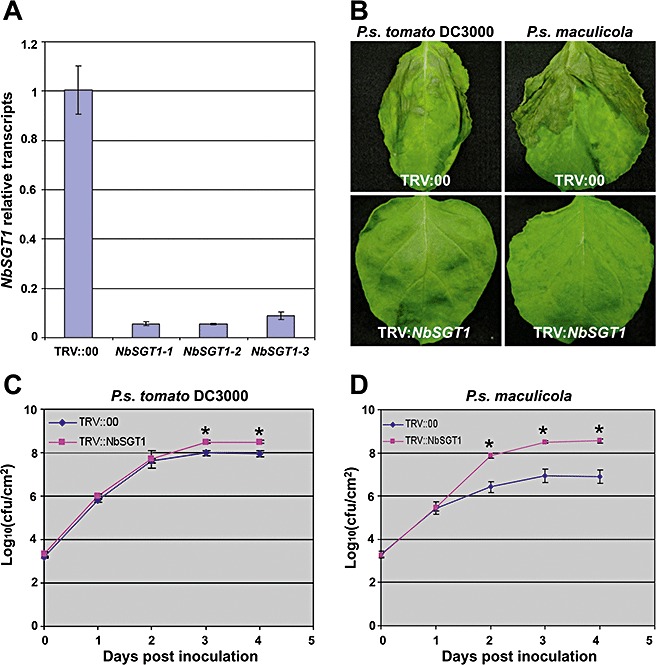
Silencing of NbSGT1 in Nicotiana benthamiana results in delayed cell death induced by nonhost bacterial pathogens. Three‐week‐old N. benthamiana plants were inoculated with disarmed Agrobacterium containing TRV::NbSGT1 or TRV::00 (vector control). (A) Relative expression of NbSGT1 determined by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) in N. benthamiana plants 2 weeks after Agrobacterium inoculation. Three plants (NbSGT1‐1, NbSGT1‐2, NbSGT1‐3) silenced with TRV::NbSGT1 were used to examine the silencing effect. The plants silenced by TRV:00 were used as controls. The error bars were derived from three technical replicates. (B) Cell death phenotypes in NbSGT1‐silenced and control N. benthamiana plants induced by nonhost pathogenic bacteria. The NbSGT1‐silenced and control plants were vacuum infiltrated with the nonhost pathogens Pseudomonas syringae pv. tomato DC3000 and P. syringae pv. maculicola at a concentration of 105 colony‐forming units/mL. Photographs were taken at 3 days post‐inoculation (dpi). (C, D) Growth of bacteria in NbSGT‐silenced and control N. benthamiana plants. Bacterial numbers were examined by plating serial dilutions of leaf extracts every 24 h after inoculation. Error bars represent standard deviations of four replicates. Asterisk indicates significant difference at α= 0.01 by Student's t‐test.
Previous studies have shown that nonhost resistance‐associated HR develops in N. benthamiana 1–2 days after inoculation with nonhost pathogens, such as P. syringae pv. maculicola, at a concentration of 108 colony‐forming units (cfu)/mL (Kang et al., 2004; Peart et al., 2002). To investigate the general role of SGT1 in PCD (HR or disease‐associated cell death) and in planta bacterial growth, we first examined the effects of SGT1 silencing on the development of PCD during nonhost resistance against pathovars of P. syringae that do not cause disease on N. benthamiana. As compatible pathogens also cause rapid cell death in N. benthamiana when inoculated at high levels of inoculum, low levels of inoculum are preferred for the differentiation of compatible and incompatible interactions (Wei et al., 2007). We used low concentrations of two different incompatible pathogens, including P. syringae pv. tomato DC3000 and P. syringae pv. maculicola, to challenge NbSGT1‐silenced N. benthamiana plants. Two to three weeks after Agrobacterium inoculation, the plants were vacuum infiltrated with the nonhost pathogens at a concentration of 1 × 105 cfu/mL. HR was observed in the leaves of non‐silenced control plants, but not in NbSGT1‐silenced plants, at 3 days post‐inoculation (dpi) with P. syringae pv. tomato DC3000 and P. syringae pv. maculicola (Fig. 1B). The bacterial population in planta was determined by plating serial dilutions of plant leaf extracts every 24 h after inoculation. During the first 24 h after infiltration, there was no significant difference in the number of nonhost bacteria tested in both SGT1‐silenced and control plants (Fig. 1C, D). However, by 3 dpi, there was a significant reduction in the bacterial populations of P. syringae pv. maculicola in control plants, but a rapid increase in NbSGT1‐silenced plants (Fig. 1D). This result demonstrates that NbSGT1 is required for cell death, which, in turn, may limit the growth of P. syringae pv. maculicola in N. benthamiana.
Interestingly, P. syringae pv. tomato DC3000 multiplied rapidly (close to 108 cfu/cm2 by 3 and 4 dpi) in control N. benthamiana plants (Fig. 1C). This result was consistent with those of a previous study (Wei et al., 2007). As this pathogen did not cause disease on N. benthamiana when inoculated at 104 cfu/cm2, whereas the host pathogen P. syringae pv. tabaci did cause disease under the same conditions, P. syringae pv. tomato DC3000 was considered to be an incompatible pathogen (Wei et al., 2007). However, the population of P. syringae pv. tomato DC3000 in NbSGT1‐silenced plants was only about three‐fold greater than that in control plants at 3 dpi and later (Fig. 1C), although the development of cell death in NbSGT1‐silenced plants was significantly slower than that in nonsilenced control plants inoculated with P. syringae pv. tomato DC3000 (Fig. 1B). These data demonstrate that NbSGT1 is a critical component for cell death induction during incompatible interaction with P. syringae pv. tomato DC3000. However, HR associated with incompatible interaction with P. syringae pv. tomato DC3000 was not effective in controlling in planta bacterial growth (Fig. 1B). Furthermore, silencing of SGT1 did not result in enhanced bacterial growth (Fig. 1C), suggesting that HR and bacterial growth can be delineated during this specific incompatible interaction. However, the population of nonhost pathogen P. syringae pv. maculicola in SGT1‐silenced plants increased to 100‐fold greater than that in the vector control (Fig. 1D), which was consistent with a previous study (Peart et al., 2002). Taken together, these results indicate that NbSGT1 is required for nonhost pathogen‐associated HR/PCD, but may not function as a major factor limiting the growth of all nonhost pathogens.
To further test whether nonhost bacterial‐induced cell death and in planta bacterial multiplication were correlated, we used Arabidopsis sgt1 mutants. Arabidopsis has two SGT1 paralogues, AtSGT1a and AtSGT1b, which share 87% similarity at the amino acid level. However, previous studies have shown that only AtSGT1b is required for certain R gene‐mediated resistance responses because of greater expression of AtSGT1b, although AtSGT1a and AtSGT1b are functionally redundant during embryo development (Austin et al., 2002; Azevedo et al., 2006; Holt et al., 2005; Tör et al., 2002). We examined the requirement of these two SGT1 paralogues in PCD on pathogen infection. We first examined the effect of AtSGT1a and AtSGT1b on plant cell death during the development of HR induced by nonhost pathogens. Arabidopsis sgt1a and sgt1b mutants, as well as wild‐type plants, were inoculated with the nonhost pathogen P. syringae pv. tabaci by leaf infiltration at a concentration of 2.0 × 107 cfu/mL. As the mutants sgt1a and sgt1b have a different genetic background, A. thaliana ecotype Col‐0 was used as control for the sgt1a mutant and Col‐5 for the sgt1b mutant. As shown in Fig. 2A, the leaves of wild‐type A. thaliana Col‐5 inoculated with P. syringae pv. tabaci showed cell death (HR) by 1 dpi, whereas the Arabidopsis sgt1b mutant did not show any HR by 1 dpi. However, the Arabidopsis sgt1a mutant showed a similar degree of plant cell death (HR) as its wild‐type counterpart by 1 dpi. These data indicate an important role for AtSGT1b in the process of HR (PCD) during nonhost plant–bacterial interactions.
Figure 2.
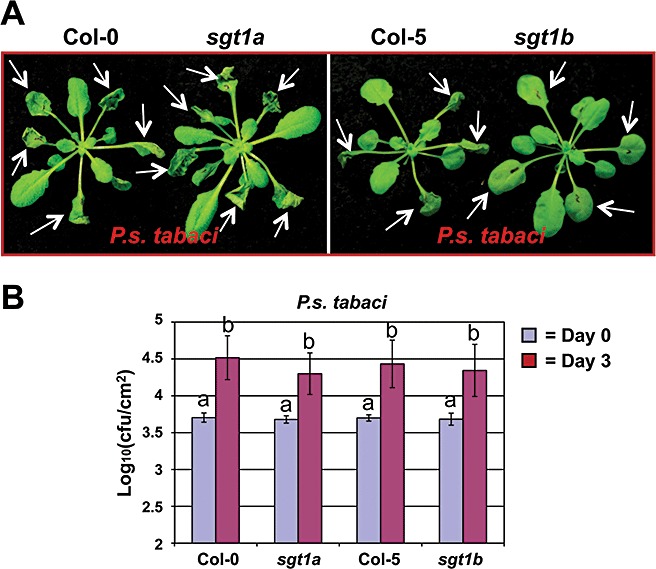
Cell death induced by the nonhost pathogen Pseudomonas syringae pv. tabaci in sgt1 mutants and wild‐type Arabidopsis. (A) Leaves of 4‐week‐old plants were infiltrated with the nonhost pathogen P. syringae pv. tabaci at a concentration of 2 × 107 colony‐forming units/mL. White arrows indicate infiltrated leaves. Photographs were taken at 2 days post‐inoculation (dpi). (B) Growth of bacteria in sgt1 mutants and corresponding wild‐type Arabidopsis plants. Leaves of 4‐week‐old plants were infiltrated with the nonhost pathogen P. syringae pv. tabaci at a concentration of 1 × 106 cfu/mL. Bacterial cell numbers were examined by plating serial dilutions of leaf extracts at 3 dpi. Error bars represent the standard deviations of four replicates. Letters indicate a significant difference at α= 0.01 using Student's t‐test.
We then measured bacterial growth to determine whether or not there was a correlation between cell death and bacterial growth in sgt1 mutants. Interestingly, the bacterial number of nonhost pathogen P. syringae pv. tabaci did not show a significant difference in mutants and their wild‐types, although the development of cell death in the sgt1b mutant was delayed as indicated above (Fig. 2B). It has been reported that Arabidopsis sgt1a and sgt1b mutants do not enhance the growth of the host bacterial pathogen P. syringae pv tomato DC3000 (Holt et al., 2005). These data further confirm our conclusion that AtSGT1b‐mediated cell death induced by nonhost pathogens is independent of in planta bacterial populations.
SGT1 is required for cell death in N. benthamiana during compatible interaction with bacterial pathogens
Plant cell death during a compatible host–pathogen interaction is considered to be a genetically programmed control process (Greenberg and Yao, 2004; del Pozo et al., 2004). Although our results and previous studies have shown that SGT1 is required for HR development during nonhost resistance and R gene‐mediated resistance (Peart et al., 2002), the function of SGT1 in cell death during compatible plant–bacterial pathogen interactions has not been studied. It is believed that both resistant and susceptible plant responses share signalling components that lead to PCD (Greenberg and Yao, 2004; Lincoln et al., 2002; del Pozo and Lam, 2003; del Pozo et al., 2004). To determine whether SGT1 is required for PCD during compatible plant–pathogen interactions, the host pathogen P. syringae pv. tabaci was used to inoculate NbSGT1‐silenced N. benthamiana plants and control plants by vacuum infiltration. As it has been demonstrated that P. syringae pv. syringae B728a is highly virulent on N. benthamiana (Vinatzer et al., 2006), we also included this strain to investigate compatible plant–pathogen interactions. At 4 dpi, disease‐associated cell death developed in the leaves of non‐silenced control plants, but no cell death was observed in NbSGT1‐silenced plants inoculated with P. syringae pv. tabaci (Fig. 3A). The occurrence of disease‐associated cell death in NbSGT1‐silenced N. benthamiana inoculated with P. syringae pv. tabaci was delayed for up to 2 weeks when compared with control plants (Fig. 3B). To further demonstrate the involvement of SGT1 in the compatible interaction, we also checked the effect of NbSGT1 silencing on cell death caused by two other compatible bacterial pathogens, P. syringae pv. syringae B728a and a broad host range pathogen Erwinia carotovora. Cell death was observed in the leaves of the nonsilenced control plants, but not in NbSGT1‐silenced plants, at 2 and 5 dpi with P. syringae pv. syringae B728a and E. carotovora, respectively (3, 4). These data indicate that SGT1 also plays a critical role in the development of cell death during more than one compatible plant–bacterial interaction.
Figure 3.
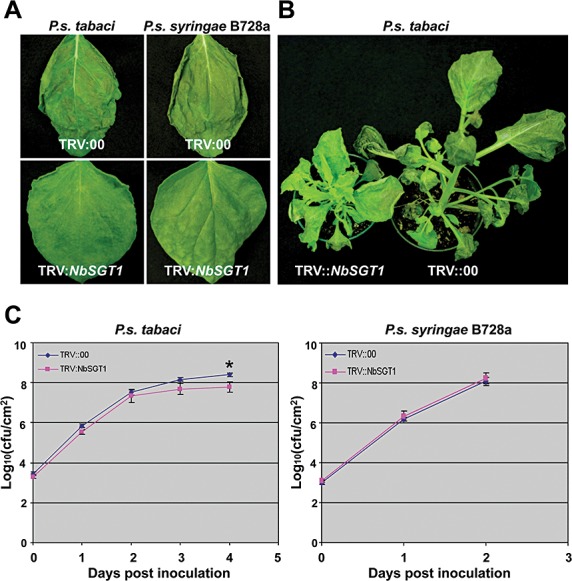
Silencing of NbSGT1 in Nicotiana benthamiana results in delayed disease‐associated cell death induced by Pseudomonas syringae pv. tabaci and P. syringae pv. syringae B728a. (A) Cell death phenotypes in NbSGT1‐silenced and control N. benthamiana plants induced by the virulent pathogens P. syringae pv. tabaci and P. syringae pv. syringae B728a. Two weeks after inoculation with Agrobacterium containing TRV::NbSGT1 or empty vector, the plants were vacuum infiltrated with the virulent pathogens P. syringae pv. tabaci and P. syringae pv. syringae B728a at a concentration of 105 colony‐forming units (cfu)/mL. Photographs were taken at 4 days post‐inoculation (dpi). (B) Disease symptoms (cell death) of NbSGT1‐silenced and control N. benthamiana plants. Two weeks after Tobacco rattle virus (TRV) inoculation, plants were vacuum infiltrated with P. syringae pv. tabaci at a concentration of 105 cfu/mL. Photographs were taken at 14 dpi. (C) Growth of bacteria in NbSGT1‐silenced and control N. benthamiana plants. Bacterial numbers were examined by plating serial dilutions of leaf extracts every 24 h after inoculation. The bacterial population in plant leaves inoculated with P. syringae pv. syringae B728a from 3 dpi was not examined as all inoculated wild‐type plant leaves were completely dead. Error bars represent the standard deviations of four replicates. Asterisk indicates a significant difference at α= 0.01 by Student's t‐test.
Figure 4.
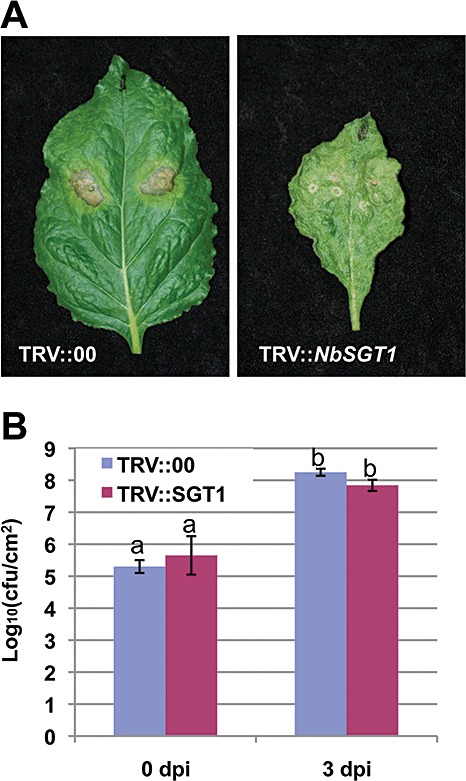
Erwinia carotovora disease symptoms (cell death) and bacterial growth in NbSGT1‐silenced (TRV::NbSGT1) and control (TRV::00) Nicotiana benthamiana plants. (A) Disease (cell death) phenotypes were monitored following infiltration with E. carotovora at a concentration of 1 × 105 colony‐forming units/mL. The inoculated plants were incubated in a glasshouse at 26 °C and 16 h daylight. Photographs were taken at 5 days post‐inoculation (dpi). (B) Bacterial populations in NbSGT1‐silenced and control plants. Error bars represent the standard deviations of three replicates. Letters indicate a significant difference at α= 0.01 using Student's t‐test.
To examine the relationship between cell death and bacterial population during compatible interactions, we monitored bacterial growth after vacuum inoculation. The populations of P. syringae pv. tabaci in NbSGT1‐silenced plants were similar at 2 dpi and only slightly (two‐ to five‐fold) less than that in control plants at 3 and 4 dpi (Fig. 3C). Pseudomonas syringae pv. syringae B728a grew rapidly in both NbSGT1‐silenced and control plants (Fig. 3C), in which the population of P. syringae pv. syringae B728a reached 108 cfu/cm2 by 2 dpi after vacuum inoculation (Fig. 3C), although no cell death was observed in NbSGT1‐silenced plants at that time (Fig. 3A). As wild‐type plant leaves inoculated with P. syringae pv. syringae B728a were completely dead, bacterial numbers in planta from 3 dpi were not examined. The multiplication of E. carotovora was also not severely compromised in NbSGT1‐silenced plants when compared with control plants (Fig. 4B). These data confirm that NbSGT1 is required for cell death during N. benthamiana responses to all tested host and nonhost bacterial pathogens, but not necessarily required to restrict bacterial growth.
To confirm the above results from vacuum/syringe inoculations and to mimic the natural infection and disease development (cell death), NbSGT1‐silenced and control N. benthamiana plants were spray inoculated with P. syringae pv. tabaci at a concentration of 2 × 108 cfu/mL. The control plants developed typical wild fire disease symptoms at 9 dpi, whereas, at the same time point, NbSGT1‐silenced plants did not show any symptoms (Fig. 5A). The small visible spots of cell death were not observed in NbSGT1‐silenced plants until 15 dpi, during which time the control plants showed severe cell death (Fig. 5B). At 32 dpi, the control N. benthamiana plants were completely dead, whereas SGT1‐silenced plants were still alive (Fig. 5D). Bacterial growth in planta was also monitored by bacterial plating assay. The bacterial cell numbers of P. syringae pv. tabaci in NbSGT1‐silenced plants (1.60 × 108 cfu/cm2) were not significantly different from those of control plants (1.63 × 108 cfu/cm2) at 9 dpi. Interestingly, at 15 dpi, the bacterial populations in NbSGT1‐silenced plants were increased two‐fold to 3.27 × 108 cfu/cm2, whereas the bacterial numbers in control plants were decreased significantly by nearly four‐fold to 4.28 × 107 cfu/cm2. Bacterial populations in plant leaves at 24 and 32 dpi were not examined for comparison as the inoculated control N. benthamiana leaves were completely dead. The decrease in bacterial cell number in control plants could be caused by cell death. These results further demonstrate that NbSGT1 is required for cell death during compatible plant–bacterial interactions, but is not involved in resistance (bacterial multiplication) to the host bacterial pathogen.
Figure 5.
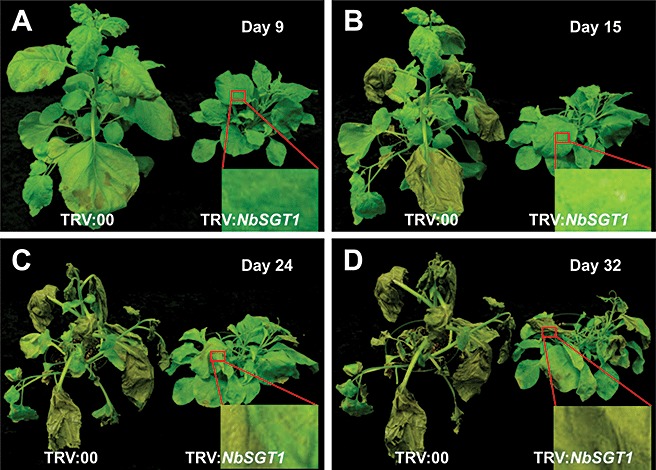
Cell death‐associated disease symptoms developed in NbSGT1‐silenced and control Nicotiana benthamiana plants. The plants were spray inoculated with Pseudomonas syringae pv. tabaci at a concentration of 2 × 108 colony‐forming units/mL (Silwet, 0.02%). The inoculated plants were incubated at room temperature. Photographs were taken 9 (A), 15 (B), 24 (C) and 32 days post‐inoculation (dpi) (D).
SGT1 is required for cell death during the development of disease symptoms in N. benthamiana during compatible interactions with fungal pathogens
To test whether NbSGT1 plays a general role in disease‐associated cell death, we monitored disease development in control and NbSGT1‐silenced plants on inoculation with Sclerotinia sclerotiorum (Fig. 6) and Phytophthora infestans (Fig. S2, see Supporting Information). Silencing of NbSGT1 significantly reduced cell death and disease symptom development by the necrotrophic fungal pathogen S. sclerotiorum (Fig. 6). To further understand whether NbSGT1 is required for cell death associated with hemibiotrophic fungal pathogens, we used an isolate of Ph. infestans that has been reported previously to be pathogenic on N. benthamiana (Becktell et al., 2006). When the sporangial suspensions were inoculated on N. benthamiana leaves, disease‐associated cell death was observed on control but not on NbSGT1‐silenced plants (Fig. S2). However, it is important to note that, in some of the experiments, Ph. infestans sporangial suspensions induced HR‐like cell death on control plants which was not associated with in planta mycelial growth (Fig. S2). Nicotiana benthamiana is generally considered to be a nonhost to Ph. infestans (Kamoun et al., 1998). In our hands, the results were variable and both compatible and incompatible interactions of Ph. infestans on N. benthamiana were observed (Fig. S2). It is possible that the sporangial concentration or plant age might alter the outcome of the interaction. In either case, NbSGT1‐silenced plants showed no cell death (Fig. S2). These data, taken together with the results from S. sclerotiorum (Fig. 6), suggest that NbSGT1 may play a general role in disease‐associated cell death during plant–fungal interactions.
Figure 6.
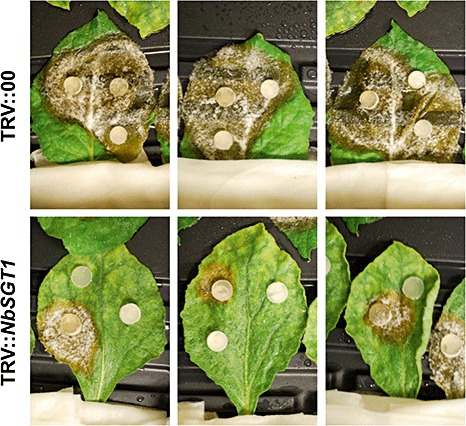
Sclerotinia sclerotiorum disease symptoms on NbSGT1‐silenced (TRV::NbSGT1) and control (TRV::00) Nicotiana benthamiana plants. Control and NbSGT1‐silenced plants were inoculated with potato dextrose agar (PDA) plugs with actively growing S. sclerotiorum cultures. The inoculated plants were incubated at room temperature. Photographs were taken 4 days post‐inoculation.
Overexpression of NbSGT1 in N. benthamiana expedites PCD induced by compatible and incompatible pathovars of P. syringae pathogens
We have shown that SGT1 is required for PCD during compatible and incompatible plant–bacterial interactions. To explore further the function of SGT1 in PCD, we examined the effects of SGT1 overexpression on plant cell death during plant responses to bacterial infection. The full‐length NbSGT1 gene was cloned under the control of the double 35S promoter in a binary vector. Agrobacterium tumefaciens strain GV2260 carrying the construct 35S:NbSGT1 was used to transform N. benthamiana. We selected three homozygous transgenic lines, SGT6521, SGT633 and SGT662, for further characterization. The phenotype of these transgenic lines was similar to that of wild‐type N. benthamiana in terms of development (data not shown). The transcription levels of NbSGT1 were determined by real‐time RT‐PCR. As shown in Fig. 7A, NbSGT1 transcripts in the transgenic lines SGT6521 and SGT662 were at least three‐fold higher than that in wild‐type plants. However, the transgenic line SGT633 did not show a significant increase in NbSGT1 transcript level compared with wild‐type plants. Therefore, the transgenic lines SGT6521 and SGT662 were selected for further analyses.
Figure 7.
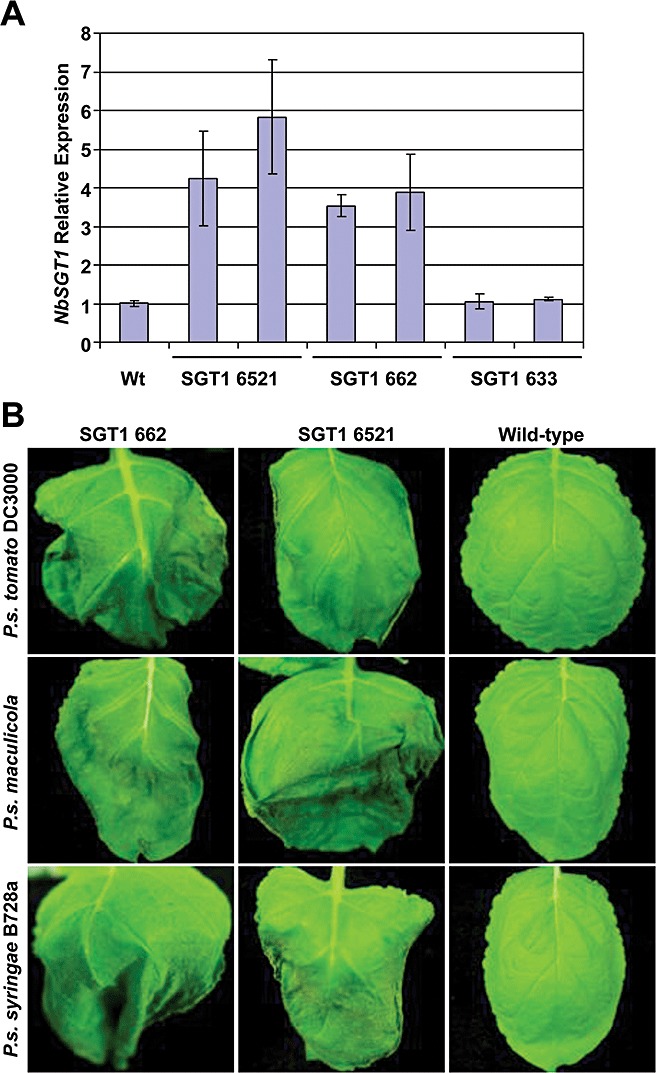
Overexpression of NbSGT1 in Nicotiana benthamiana expedites the cell death induced by various pathogens. (A) Relative expression of NbSGT1 examined by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) in transgenic NbSGT1‐overexpressing lines of N. benthamiana. Error bars were derived from three technical replicates. (B) Early cell death response of N. benthamiana NbSGT1 overexpressors on challenge with various pathogens. Plants were vacuum infiltrated with Pseudomonas syringae pv. tomato DC3000, P. syringae pv. maculicola and P. syringae pv. syringae B728a at a concentration of 105 colony‐forming units/mL. Photographs were taken 30 h post‐inoculation.
We challenged the transgenic lines overexpressing NbSGT1 with the host pathogen P. syringae pv. tabaci, virulent pathogen P. syringae pv. syringae and nonhost pathogens P. syringae pv. tomato DC3000 and P. syringae pv. maculicola by vacuum infiltration at low concentration (1 × 105 cfu/mL). Notably, HR/cell death on the leaves of transgenic plants was observed 30 h after inoculation with nonhost and host pathogens, whereas wild‐type N. benthamiana did not show cell death until 48 h after inoculation with P. syringae pv. syringae, and at 3 days following infiltration with P. syringae pv. tomato DC3000 and P. syringae pv. maculicola (Fig. 7B). The cell death‐associated disease symptoms on the transgenic lines caused by the host pathogen P. syringae pv. tabaci were observed about 12 h earlier than those on wild‐type plants (data not shown). However, the populations of host and nonhost bacterial pathogens tested in this study did not show significant differences between the NbSGT1‐overexpressing lines and the wild‐type control (Fig. S1, see Supporting Information). These data indicate that increased expression of NbSGT1 in N. benthamiana accelerates cell death on infection with both host and nonhost bacterial pathogens when compared with wild‐type plants, but does not affect the growth of host and nonhost pathogens.
It has been shown that NbSGT1 is required for HR induced by multiple gene‐for‐gene interactions (Peart et al., 2002). Therefore, we hypothesized that NbSGT1 overexpression might expedite the development of HR associated with ETI and PTI. The R gene Cf‐9 and its cognate avirulence gene Avr9 were transiently co‐expressed, using Agrobacterium, in fully expanded young leaves of NbSGT1 overexpressors and control plants. As hypothesized, HR due to ETI was observed 5 days after Agrobacterium inoculation in NbSGT1‐overexpressing lines, whereas cell death was not observed in control plant leaves until 7 dpi (Fig. 8A). Similarly, when the Ph. infestans gene encoding elicitor protein INF1 (PAMP) was transiently expressed, HR developed at least 1 day earlier in NbSGT1‐overexpressing lines (Fig. 8B). Overall, these data suggest that NbSGT1 positively regulates the process of cell death during both ETI and PTI.
Figure 8.
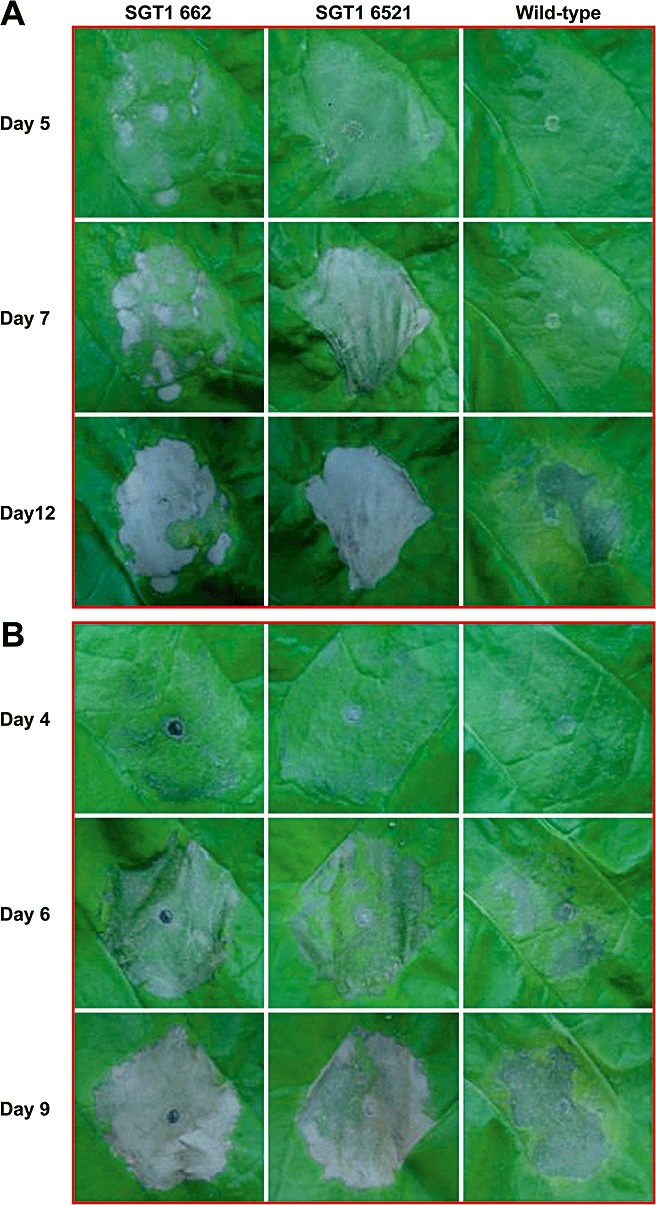
ETI and PTI‐mediated cell death in NbSGT1‐overexpressing lines and wild‐type Nicotiana benthamiana plants. Cell death was elicited by Agrobacterium‐mediated transient co‐expression of a resistance protein Cf9 and corresponding avirulence protein Avr9 (A) or expression of elicitor Inf1 (B). Seven‐week‐old N. benthamiana plant leaves were infiltrated with virulence‐induced Agrobacterium carrying constructs 35S : Inf1 or the mixed bacterial suspension of Agrobacterium with 35S : Cf9 and 35S : Avr9. Photographs were taken on different days as indicated after Agrobacterium infiltration.
DISCUSSION
Plant cell death can occur in both resistant and susceptible plant–pathogen interactions. HR‐associated cell death, a form of PCD, results from plant immunity reactions triggered by host receptor‐mediated perception of pathogen nonrace‐specific elicitors or by the recognition of race‐specific elicitors (Jones and Dangl, 2006; Zipfel and Felix, 2005). It has also been reported that SGT1 is required for cell death mediated by several pairs of Avr–R interactions and other effector‐induced cell death (1, 4) (Peart et al., 2002). However, we have just begun to understand the molecular events involved in plant cell death associated with diseases caused by hemibiotrophs. In this study, we have clearly shown that SGT1 is involved in the process of PCD for both compatible and incompatible plant–pathogen interactions (1, 2, 3, 4, 5, 6). This strongly suggests that cell death during plant responses to resistant and susceptible interactions shares a common step mediated by SGT1. The existence of common steps during HR and susceptible host resistance is also supported by previous studies. del Pozo et al. (2004) identified a MAPKKKα that positively regulates cell death associated with both gene‐for‐gene‐mediated resistance and disease. The expression of the anticaspase baculovirus protein p35 also attenuates cell death during both susceptible and resistance responses to a number of pathogens (Lincoln et al., 2002; del Pozo and Lam, 2003).
Wang et al. (2008) have reported that the overexpression of OsSGT1 in rice increases the basal resistance to Xanthomonas oryzae pv. oryzae PXO99, but not to X. oryzae pv. oryzae strain DY89031. Furthermore, in rice, the overexpression of SGT1 does not result in the acceleration of disease‐associated cell death (Wang et al., 2008). In our study, overexpression of NbSGT1 accelerated cell death induced by nonhost pathogens (Fig. 7B), host pathogens, during ETI and PTI (Fig. 8). However, overexpression of NbSGT1 did not result in cell death in the absence of the pathogen (Fig. 7B). These results suggest that NbSGT1 is a component of the signalling cascade that mechanistically controls cell death associated with compatible and incompatible interactions. It is also important to note that SGT1‐mediated pathways may also vary in different plants and in response to a specific pathogen. It is not clear how increased levels of SGT1 expedite cell death and whether SGT1 is part of the previously reported MAPKKKα‐mediated cell death pathway (del Pozo et al., 2004).
Recently, it has been shown that SGT1b is required for fungal disease development in Arabidopsis and N. benthamiana (Cuzick et al., 2009; EI Oirdi and Bouarab, 2007). Botrytis cinerea has been shown to exploit the SGT1‐mediated HR cell death pathway to initiate its necrotrophic life style (EI Oirdi and Bouarab, 2007). Furthermore, Fusarium culmorum has been shown to require SGT1b to cause full disease symptoms associated with cell death and tissue dehydration (Cuzick et al., 2009). These results suggest that cell death associated with either necrotrophic or hemibiotrophic fungal pathogens may require SGT1. Our results showing the requirement of SGT1 for cell death during susceptible interactions in N. benthamiana to several bacterial and fungal pathogens indicate that cell death during disease development is, at least partially, genetically programmed (Greenberg, 1997). Interestingly, very little or no difference in the virulent bacterial population (P. syringae pv. tabaci, P. syringae pv. syringae B728a and E. carotovora) between NbSGT1‐silenced and nonsilenced control N. benthamiana plants was observed, in contrast with a dramatic difference in the development of cell death‐associated disease symptoms in NbSGT1‐silenced plants (3, 4, 5 and Fig. S1). It is unlikely that the slightly reduced bacterial population in NbSGT1‐silenced plants contributed to a significant delay (1–2 weeks) in the development of disease‐associated cell death. Therefore, our data support the earlier suggestion (Greenberg and Yao, 2004) that pathogen‐induced cell death during disease development is a result of several events, and not solely dependent on the pathogenic bacterial population. Our data also support a number of findings which indicate that apoptotic‐like cell death does not always correlate with pathogen growth (Bent et al., 1992; Liang et al., 2003; Pilloff et al., 2002; Yao et al., 2002). Similarly, HR is considered to be a secondary feature of Rx‐mediated viral resistance, and cell death has been shown to strictly correlate with viral growth (Bendahmane et al., 1999). However, when the viral coat protein was overexpressed in planta, Rx was shown to mediate SGT1‐dependent HR cell death (Bendahmane et al., 1999; Peart et al., 2002). These results suggest that resistance‐associated cell death may or may not correlate with the pathogen load. As HR occurs during incompatible plant–pathogen interactions, it has long been speculated that cell death is directly responsible for restricting pathogen growth and development. Our study shows that the attenuation of pathogen‐induced cell death (HR) by the silencing of SGT1 in N. benthamiana only slightly increases the growth of P. syringae pv. tomato DC3000 (Fig. 1). This is in contrast with the dramatic growth of the nonhost pathogen P. syringae pv. maculicola (Fig. 1B) and P. syringae pv. tomato strain T1 (Peart et al., 2002; Wang et al., 2007), which increased almost 100‐fold in SGT1‐silenced N. benthamiana when compared with nonsilenced control plants. In addition, the acceleration of cell death by the overexpression of SGT1 in N. benthamiana does not confer resistance by reducing the bacterial growth of the nonhost pathogen (Fig. S1). Our results support the earlier observation that cell death is not an obligatory step in achieving the resistance observed in plant–fungus interactions, in which the arrest of fungal development may occur before cell death in heterozygous plants with one copy of the Mlg resistance gene (Görg et al., 1993). In addition, Arabidopsis dnd1 (defence, no death) exhibits strong gene‐for‐gene resistance to avirulent P. syringae without the HR phenotype, and shows a certain degree of constitutive resistance to some virulent bacterial pathogens (2000, 1998). The majority (90%) of VIGS‐identified genes involved in HR associated with ArvPto‐Pto resistance against P. syringae strains failed to suppress bacterial growth in N. benthamiana plants (Lu et al., 2003a). Therefore, it is possible that cell death is a secondary feature of disease resistance and not always necessary for the suppression of pathogen growth. In some plant–pathogen interactions, PCD even promotes pathogen growth, so that pathogen‐secreted toxins, such as AAL toxin secreted from Alternaria alternata f.sp. lycopersici, can kill host cells rapidly (Akamatsu et al., 1997; Greenberg and Yao, 2004). This is also suggested indirectly by the fact that the population of the host pathogen P. syringae pv. tabaci was reduced slightly in NbSGT1‐silenced N. benthamiana plants in which pathogen‐induced cell death was delayed significantly when compared with control plants (Fig. 2).
As SGT1 is required for much R gene‐mediated disease resistance and/or HR (Austin et al., 2002; Azevedo et al., 2002; Peart et al., 2002), it has been proposed that SGT1 might be required for defence signalling (Muskett and Parker, 2003; Tör et al., 2002). However, a recent study has indicated that AtSGT1b is not required for disease resistance signalling per se, as RPS5‐, RPP4‐, RPP8‐ and RPP31‐mediated resistance can be restored in the Arabidopsis rar1 sgt1b double mutant (Holt et al., 2005). It was speculated that SGT1b is required for efficient HR development to limit pathogen spread. Our data indicate that SGT1 is essentially required for PCD during both compatible and incompatible plant–pathogen interactions for all the tested pathogens in this study. However, the requirement of SGT1 for resistance is specific to a particular plant–pathogen interaction. Therefore, our findings support the previous proposal that the function of SGT1 in plant resistance is particularly relevant in cases in which HR plays a key role in limiting pathogen spread (Holt et al., 2005). For resistance in which cell death does not correlate with pathogen growth, SGT1 may have little effect on resistance. This model can explain why SGT1 mutation or silencing results in an alteration in cell death, but little change in pathogen growth or resistance to some pathogens.
On the basis of our experimental results and previous findings, SGT1 appears to positively regulate the process of cell death during nonhost, ETI‐ and PTI‐mediated resistance. In addition, our results have identified a new role for SGT1 in the process of cell death associated with disease development of compatible bacterial and fungal pathogens in N. benthamiana. Therefore, we propose a common role for SGT1 as a shared signalling component of cell death during both immunity (gene‐for‐gene and nonhost) and susceptibility. Furthermore, SGT1, especially SGT1b, might associate with shared or distinct complexes that trigger the cell death pathway leading to HR, necrosis or trailing necrosis associated with a broad range of plant–pathogen interactions. Our identification of a role for SGT1 in disease‐associated cell death will facilitate future studies aimed at the identification of shared and unique molecular events during cell death associated with host and nonhost resistance and disease. The identification of corresponding nonhost pathogen elicitors or host effectors that trigger SGT1‐mediated cell death in response to pathogen attack may provide new insights into the pathways that control cell death during these processes.
EXPERIMENTAL PROCEDURES
Bacterial strains and plant materials
Agrobacterium tumefaciens GV2260 was routinely grown in Luria–Bertani (LB) medium at 28 °C. Pseudomonas syringae pv. tomato strain DC3000, P. syringae pv. maculicola, P. syringae pv. syringae B728a and P. syringae pv. tabaci were grown in King's B (KB) medium at 28 °C. The following concentrations of antibiotics (Sigma, St. Louis, MO, USA) were used individually or combined for selection when necessary, unless otherwise indicated: rifampicin (100 µg/mL), kanamycin (50 µg/mL). Cultures of E. carotovora ssp. carotovora were maintained on KB medium at 28 °C without any supplementary antibiotics.
Bacterial growth was monitored by measuring the optical density at 600 nm (OD600) for the preparation of the inoculum, and the number of viable cells was determined by counting the colonies grown on appropriate plates supplemented with the appropriate antibiotics.
Nicotiana benthamiana seeds were germinated in Metro Mix 350 (SUNGRO Horticulture Distribution Inc., Bellevue, WA, USA) in a growth chamber at 26 °C and 16 h daylight. Fertilizer (20–12–20), together with soluble trace element mix (The Scotts Co., Marysville, OH, USA), was applied to 2‐week‐old seedlings with water in the tray. Three‐week‐old seedlings were transplanted to 4‐in pots containing Professional Blend soil (SUNGRO Horticulture Distribution Inc.) and grown in a glasshouse under the following conditions: 24 ± 2 °C, 70% humidity and 16 h daylight supplemented with 50–100 µE/s/m2 light intensity. The VIGS experiment was conducted 2 days after transplanting.
The T‐DNA knockout lines sgt1a and sgt1b were kindly provided by B.F. Holt (Holt et al., 2005). The seeds of A. thaliana were cold treated for 3–4 days at 4 °C and germinated in Professional Blend soil in a growth chamber at 20–22 °C under short‐day conditions (8 h light). Two‐week‐old seedlings were transplanted into 4‐in pots containing Professional Blend soil and grown under the same conditions as seed germination. Four‐week‐old plants were used for inoculation in all experiments.
TRV‐based VIGS in N. benthamiana
The TRV vector system was used for gene silencing in N. benthamiana as described previously (Liu et al., 2002). Briefly, A. tumefaciens strain GV2260 containing construct TRV2::SGT1 or TRV::00 was mixed with A. tumefaciens strain GV2260 containing TRV1 at a 1:1 ratio and infiltrated into cotyledons and the first two leaves of 3‐week‐old N. benthamiana plants. The transcript levels were examined by real‐time RT‐PCR and the plants were used for bacterial challenge 2–3 weeks after TRV infection.
Agrobacterium‐mediated transient expression in N. benthamiana
The expression of Cf‐9, Avr9 and Inf1 was driven by the cauliflower mosaic virus (CaMV) 35S promoter in the pBtex vector. Transient expression in N. benthamiana mediated by Agrobacterium was performed as described previously (Peart et al., 2002). Briefly, bacterial cells of A. tumefaciens GV2260 containing constructs were harvested by centrifugation, washed twice and resuspended in an induction medium containing acetosyringone (200 µm) at OD600= 1. The virulence genes of A. tumefaciens were induced for at least 4 h at room temperature by shaking at 100 r.p.m. The bacterial cells were collected by centrifugation and resuspended in 10 mm 2‐(N‐morpholino)ethanesulphonic acid (MES) with acetosyringone (200 µm). The concentrations of A. tumefaciens containing Cf‐9 and avr9 were adjusted to OD600= 0.4, respectively, and mixed at a 1:1 ratio, whereas the bacterial suspension of Agrobacterium containing Inf1 was adjusted to a final concentration of OD600= 0.2 for the infiltration of leaves of N. benthamiana.
NbSGT1 overexpression in N. benthamiana
Full‐length NbSGT1 cDNA was amplified by RT‐PCR from N. benthamiana and cloned into the donor vector pDONR207. NbSGT1 in pDONR207 was subcloned into the plant expression vector pYL436 (Rubio et al., 2005) using the GATEWAY cloning system (Invitrogen, Carlsbad, CA, USA), resulting in the plasmid pYL436‐NbSGT1. Transgenic N. benthamiana plants expressing NbSGT1 were created by introducing pYL436‐NbSGT1 into N. benthamiana plants using standard Agrobacterium‐mediated leaf disc transformation (Horsch et al., 1985).
RNA isolation and real‐time RT‐PCR
Total RNA was extracted from the leaves of N. benthamiana using the RNAeasy Kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. The RNA concentration was measured using a NanoDrop® ND‐1000 UV–visible spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA) and adjusted to 1 µg/µL. Reverse transcription was carried out by using 2 µg of total RNA and 0.5 µg of poly(dT)16 primer in a final volume of 20 µL for 1 h at 37 °C employing an Omniscript RT Kit (Qiagen) and RNAase inhibitor (Promega, Madison, WI, USA). The following gene‐specific primers for PCR were designed using PrimerExpress software (Applied Biosystems, ABI, Foster City, CA, USA) and synthesized by Integrated DNA Technologies (Coralville, IA, USA): for SGT1 silencing: NbSGT1F, 5′‐TTTGCCAAGGGAATACCAGCCAA‐3′; NbSGT1R, 5′‐TTCCCATTTCTTCAGCTCCATGC‐3′; for SGT1 overexpression: LeSGT1F, 5′‐CACATCCTGCATCTGAGTTACC‐3′; LeSGT1R, 5′‐CTCTGAATTACAACAGGTTCCGTT‐3′. Primer concentrations were optimized on an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems) and specific amplification was confirmed by melting curve assay and agarose gel electrophoresis. Real‐time quantitative PCR was performed with the ABI 7900HT Fast Real‐Time PCR System (Applied Biosystems) using Power SYBR Green PCR Master Mix (ABI) under the following conditions: 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The quality of amplification was determined using dissociation curve analysis of the 7900HT Real‐Time PCR system. Three biological replicates and three technical replicates were used for each line or treatment. Threshold cycle (C T) values reported by the 7900HT Real‐Time PCR system were used for data analysis. The relative quantification of PCR products was calculated by the comparative C T method (ΔΔC T) using N. benthamiana Actin gene as endogenous control.
Plant inoculation and bacterial population measurement
Pseudomonas syringae pathovars were grown in KB broth with the appropriate antibiotics. Bacterial cells were collected by centrifugation of an overnight culture at 3000 g for 10 min, washed twice, resuspended in 10 mm MgCl2 and diluted to the desired concentration in 10 mm MgCl2 with 0.01% (v/v) Silwet L‐77 (Osi Specialties, Friendship, WV, USA) to facilitate infiltration. The bacterial suspension was used to inoculate gene‐silenced N. benthamiana plants 2 weeks after TRV infection by vacuum infiltration, or to inoculate 4‐week‐old Arabidopsis plants by leaf infiltration with needle‐less syringes. The fully expanded leaves were used for disease assays. The inoculated plants were kept in a growth chamber at 20–22 °C for disease development. Two leaf discs (0.5 cm2 each) of each plant were sampled, homogenized and serially diluted in 10 mm MgCl2 for the measurement of bacterial growth in planta. The bacterial population was calculated as cfu/cm2. Experiments were performed with at least four replicates.
Erwinia carotovora was grown on KB medium, suspended in distilled water and diluted to the desired concentration. The inoculum was infiltrated using a needle‐less syringe into fully expanded leaves of control (TRV::00) and gene‐silenced N. benthamiana plants 2 weeks after TRV infection. The bacterial populations were quantified as described above for P. syringae pathovars.
Fungal inoculum preparation and plant inoculation
Agar plugs (diameter, 5 mm) from growing regions of Sclerotinia sclerotiarum cultures on potato dextrose agar (PDA; Difco, Sparks, MD, USA) medium were used as inoculum. Control and NbSGT1‐silenced N. benthamiana leaves were inoculated with two to three agar plugs per leaf, and fungal inoculated leaves were placed on moist filter paper, sealed and incubated at 22 °C/19 °C for a 16‐h photoperiod at a photon flux density of 150‐200 µmol/m2/s. The disease symptoms were recorded at 5 dpi.
Phytopthora infestans (Becktell et al., 2006) was maintained on Rye A agar (Caten and Jinks, 1968), and then transferred onto Rye B agar supplemented with β‐sitosterol (Sigma Aldrich Inc.) to induce sporulation for 14 days in a dark chamber maintained at 15 °C. Sporangia were collected in distilled water, counted using a haematocytometer and adjusted to 2 × 104 sporangia/mL. An aliquot of 25 µL of the sporangial suspension was spot inoculated onto control and NbSGT1‐silenced N. benthamiana leaves. The leaves were incubated at 15 °C (16‐h light period) and maintained at 100% humidity using moist filter paper. Disease symptoms were monitored periodically until 10 dpi.
Supporting information
Fig. S1 Growth of bacteria in Nicotiana benthamiana leaves.
Fig. S2 Phytopthora infestans‐induced cell death in control (TRV::00) and SGT1‐silenced (TRV::NbSGT1) Nicotiana benthamiana leaves.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
ACKNOWLEDGEMENTS
We thank Drs Ben Holt and Jeff Dangl (University of North Carolina) for the seeds of Arabidopsis sgt1a and sgt1b mutants, Dr Marty Dickman (Texas A & M University) for S. sclerotiarum, Dr William Fry (Cornell University) for Ph. infestans, and Drs Elison Blancaflor and Phil Harries for critical reading of the manuscript. This work was supported by the Samuel Roberts Noble Foundation.
REFERENCES
- Abramovitch, R.B. , Janjusevic, R. , Stebbins, C.E. and Martin, G.B. (2006) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. USA, 103, 2851–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu, H. , Itoh, Y. , Kodama, M. , Otani, H. and Kohmoto, K. (1997) AAL‐toxin‐deficient mutants of Alternaria alternata tomato pathotype by restriction enzyme‐mediated integration. Phytopathology, 87, 967–972. [DOI] [PubMed] [Google Scholar]
- Austin, M.J. , Muskett, P. , Kahn, K. , Feys, B.J. , Jones, J.D.G. and Parker, J.E. (2002) Regulatory role of SGT1 in early R gene‐mediated plant defenses. Science, 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C. , Sadanandom, A. , Kitagawa, K. , Freialdenhoven, A. , Shirasu, K. and Schulze‐Lefert, P. (2002) The RAR1 interactor SGT1, an essential component of R gene‐triggered disease resistance. Science, 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Azevedo, C. , Betsuyaku, S. , Peart, J. , Takahashi, A. , Noël, L. , Sadanandom, A. , Casais, C. , Parker, J. and Shirasu, K. (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 25, 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becktell, M.C. , Smart, C.D. , Haney, C.H. and Fry, W.E. (2006) Host–pathogen interactions between Phytophthora infestans and the solanaceous hosts Calibrachoa ×hybridus, Petunia ×hybrida, and Nicotiana benthamiana . Plant Dis. 90, 24–32. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. and Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F. and Mackey, D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. , Innes, R.W. , Ecker, J.R. and Staskawicz, B.J. (1992) Disease development in ethylene‐insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant–Microbe Interact. 5, 372–378. [DOI] [PubMed] [Google Scholar]
- Caten, C.E. and Jinks, J.L. (1968) Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Can. J. Bot. 46, 329–348. [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Cuzick, A. , Maguire, K. and Hammond‐Kosack, K.E. (2009) Lack of the plant signalling component SGT1b enhances disease resistance to Fusarium culmorum in Arabidopsis buds and flowers. New Phytol. 181, 901–912. [DOI] [PubMed] [Google Scholar]
- EI Oirdi, M. and Bouarab, K. (2007) Plant signalling components EDS1 and SGT1 enhance disease caused by the necrotrophic pathogen Botrytis cinerea . New Phytol. 175, 131–139. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256. [DOI] [PubMed] [Google Scholar]
- Görg, R. , Hollricher, K. and Schulze‐Lefert, P. (1993) Functional analysis and RFLP‐mediated mapping of the Mlg resistance locus in barley. Plant J. 3, 857–866. [Google Scholar]
- Greenberg, J.T. (1997) Programmed cell death in plant–pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. and Yao, N. (2004) The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 6, 201–211. [DOI] [PubMed] [Google Scholar]
- Holt, B.F. , Belkhadir, Y. and Dangl, J.L. (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science, 309, 929–932. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B. , Rogers, S.G. and Fraley, R.T. (1985) Transgenic plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. [DOI] [PubMed] [Google Scholar]
- Jamir, Y. , Guo, M. , Oh, H.S. , Petnicki‐Ocwieja, T. , Chen, S. , Tang, X. , Dickman, M.B. , Collmer, A. and Alfano, J.R. (2004) Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Van West, P. , Vleeshouwers, V.G. , De Groot, K.E. and Govers, F. (1998) Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell, 10, 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, L. , Tang, X.Y. and Mysore, K.S. (2004) Pseudomonas type III effector AvrPto suppresses the programmed cell death induced by two nonhost pathogens in Nicotiana benthamiana and tomato. Mol. Plant–Microbe Interact. 17, 1328–1336. [DOI] [PubMed] [Google Scholar]
- Kitagawa, K. , Skowyra, D. , Elledge, S.J. , Harper, J.W. and Hieter, P. (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell, 4, 21–33. [DOI] [PubMed] [Google Scholar]
- Kunze, G. , Zipfel, C. , Robatzek, S. , Niehaus, K. , Boller, T. and Felix, G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell, 16, 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E. , Kato, N. and Lawton, M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature, 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Liang, H. , Yao, N. , Song, L.T. , Luo, S. , Lu, H. and Greenberg, L.T. (2003) Ceramides modulate programmed cell death in plants. Genes Dev. 17, 2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, J.E. , Richael, C. , Overduin, B. , Smith, K. , Bostock, R. and Gilchrist, D.G. (2002) Expression of the antiapoptotic baculovirus p35 gene in tomato blocks programmed cell death and provides broad‐spectrum resistance to disease. Proc. Natl. Acad. Sci. USA, 99, 15 217–15 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Marathe, R. and Dinesh‐Kumar, S.P. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N‐mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Liu, Y.L. , Burch‐Smith, T. , Schiff, M. , Feng, S.H. and Dinesh‐Kumar, S.P. (2004) Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108. [DOI] [PubMed] [Google Scholar]
- Lu, R. , Malcuit, I. , Moffett, P. , Ruiz, M.T. , Peart, J. , Wu, A.J. , Rathjen, J.P. , Bendahmane, A. , Day, L. and Baulcombe, D.C. (2003a) High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Martin‐Hernandez, A.M. , Peart, J.R. , Malcuit, I. and Baulcombe, D. (2003b) Virus‐induced gene silencing in plants. Methods, 30, 296–303. [DOI] [PubMed] [Google Scholar]
- Martin, G.B. , Bogdanove, A.J. and Sessa, G. (2003) Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- Mudgett, M.B. (2005) New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu. Rev. Plant Biol. 56, 509–531. [DOI] [PubMed] [Google Scholar]
- Muskett, P. and Parker, J. (2003) Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect. 5, 969–976. [DOI] [PubMed] [Google Scholar]
- Nomura, K. , DebRoy, S. , Lee, Y.H. , Pumplin, N. , Jones, J. and He, S.Y. (2006) A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science, 313, 220–223. [DOI] [PubMed] [Google Scholar]
- Peart, J.R. , Lu, R. , Sadanandom, A. , Malcuit, I. , Moffett, P. , Brice, D.C. , Schauser, L. , Jaggard, D.A. , Xiao, S. , Coleman, M.J. , Dow, M. , Jones, J.D. , Shirasu, K. and Baulcombe, D.C. (2002) Ubiquitin ligase‐associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA, 99, 10 865–10 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley, K.F. and Martin, G.B. (2003) Molecular basis of Pto‐mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243. [DOI] [PubMed] [Google Scholar]
- Pilloff, R.K. , Devadas, S.K. , Enyedi, A. and Raina, R. (2002) The Arabidopsis gain‐of‐function mutant dll1 spontaneously develops lesions mimicking cell death associated with disease. Plant J. 30, 61–70. [DOI] [PubMed] [Google Scholar]
- Del Pozo, O. and Lam, E. (2003) Expression of the baculovirus p35 protein in tobacco affects cell death progression and compromises N gene‐mediated disease resistance response to tobacco mosaic virus. Mol. Plant–Microbe Interact. 16, 485–494. [DOI] [PubMed] [Google Scholar]
- Del Pozo, O. , Pedley, K.F. and Martin, G.B. (2004) MAPKKK alpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23, 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock, T.R. , Zeng, L. , Brady, J.J. , Abramovitch, R.B. , Xiao, F. and Martin, G.B. (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature, 448, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, V. , Shen, Y. , Saijo, Y. , Liu, Y. , Gusmaroli, G. , Dinesh‐Kumar, S.P. and Deng, X.W. (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778. [DOI] [PubMed] [Google Scholar]
- Tör, M. , Gordon, P. , Cuzick, A. , Eulgem, T. , Sinapidou, E. , Mert‐Turk, F. , Can, C. , Dangl, J.L. and Holub, E.B. (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell, 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer, B.A. , Teitzel, G.M. , Lee, M.W. , Jelenska, J. , Hotton, S. , Fairfax, K. , Jenrette, J. and Greenberg, J.T. (2006) The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non‐host plants. Mol. Microbiol. 62, 26–44. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Kang, L. , Anand, A. , Lazarovits, G. and Mysore, K.S. (2007) Monitoring in planta bacterial infection at both cellular and whole‐plant levels using the green fluorescent protein variant GFPuv. New Phytol. 174, 212–223. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Gao, M. , Li, Q. , Wang, L. , Wang, J. , Jeon, J.S. , Qu, N. , Zhang, Y. and He, Z. (2008) OsRAR1 and OsSGT1 physically interact and function in rice basal disease resistance. Mol Plant–Microbe Interact. 21, 294–303. [DOI] [PubMed] [Google Scholar]
- Wei, C.F. , Kvitko, B.H. , Shimizu, R. , Crabill, E. , Alfano, J.R. , Lin, N.C. , Martin, G.B. , Huang, H.C. and Collmer, A. (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1‐1 is able to cause disease in the model plant Nicotiana benthamiana . Plant J. 51, 32–46. [DOI] [PubMed] [Google Scholar]
- Yao, N. , Imai, S. , Tada, Y. , Nakayashiki, H. , Tosa, Y. , Park, P. and Mayama, S. (2002) Apoptotic cell death is a common response to pathogen attack in oats. Mol. Plant–Microbe Interact. 15, 1000–1007. [DOI] [PubMed] [Google Scholar]
- Yu, I. , Fengler, K.A. , Clough, S.J. and Bent, A.F. (2000) Identification of Arabidopsis mutants exhibiting an altered hypersensitive response in gene‐for‐gene disease resistance. Mol. Plant–Microbe Interact. 13, 277–286. [DOI] [PubMed] [Google Scholar]
- Yu, I.C. , Parker, J. and Bent, A.F. (1998) Gene‐for‐gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA, 95, 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. and Felix, G. (2005) Plants and animals: a different taste for microbes? Curr. Opin. Plant Biol. 8, 353–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Growth of bacteria in Nicotiana benthamiana leaves.
Fig. S2 Phytopthora infestans‐induced cell death in control (TRV::00) and SGT1‐silenced (TRV::NbSGT1) Nicotiana benthamiana leaves.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


