SUMMARY
Biotic stress has a major impact on the process of natural selection in plants. As plants have evolved under variable environmental conditions, they have acquired a diverse spectrum of defensive strategies against pathogens and herbivores. Genetic variation in the expression of plant defence offers valuable insights into the evolution of these strategies. The ‘zigzag’ model, which describes an ongoing arms race between inducible plant defences and their suppression by pathogens, is now a commonly accepted model of plant defence evolution. This review explores additional strategies by which plants have evolved to cope with biotic stress under different selective circumstances. Apart from interactions with plant‐beneficial micro‐organisms that can antagonize pathogens directly, plants have the ability to prime their immune system in response to selected environmental signals. This defence priming offers disease protection that is effective against a broad spectrum of virulent pathogens, as long as the augmented defence reaction is expressed before the invading pathogen has the opportunity to suppress host defences. Furthermore, priming has been shown to be a cost‐efficient defence strategy under relatively hostile environmental conditions. Accordingly, it is possible that selected plant varieties have evolved a constitutively primed immune system to adapt to levels of disease pressure. Here, we examine this hypothesis further by evaluating the evidence for natural variation in the responsiveness of basal defence mechanisms, and discuss how this genetic variation can be exploited in breeding programmes to provide sustainable crop protection against pests and diseases.
SCOPE
Until recently, basal resistance against pathogens and herbivorous insects in plants was not well defined and was often referred to as either polygenic or horizontal resistance. Because basal resistance is generally too weak to halt colonization by pathogens and herbivorous insects, it was often regarded as less suitable than race‐specific resistance for exploitation in agriculture. Recent advances in the molecular regulation of plant immunity have provided new insights into the evolution of plant immunity, which suggest that basal resistance and race‐specific resistance are the result of an ongoing arms race between pathogens and host plants. This so‐called ‘zigzag’ model defines basal resistance as the amount of residual resistance that is activated by virulent pathogens on susceptible host plants after active defence suppression by disease‐promoting pathogen effectors (Jones and Dangl, 2006). In addition, it has been known for several years that basal resistance in plants can be boosted by selected environmental signals. Many of these induced resistance phenomena are based on priming of the basal resistance response, causing a faster and stronger activation of these defences on subsequent pathogen or insect attack (Conrath et al., 2006). This review investigates how the priming of basal resistance fits in the current evolutionary model of plant immunity, and explores the hypothesis that natural selection in some plant communities could lead to ‘constitutive priming’ of basal defence mechanisms. Subsequently, we discuss how traits conferring constitutively primed basal resistance can be exploited in agriculture to obtain durable, broad‐spectrum crop protection.
INDUCED PLANT DEFENCE
In their struggle for survival, plants rely on defensive mechanisms to resist exploitation by hostile microbes and insects. In addition to pre‐existing defence barriers, such as thorns, trichomes, cuticles and cell walls, plants possess an inducible immune system that controls the activation of defence mechanisms after recognition of an attacker. This induced defence consists of multiple layers that become activated at successive stages of colonization by the plant attacker (Ton et al., 2009). To establish a successful parasitic interaction, pathogens and herbivores need to penetrate the host tissue. Plant viruses mostly depend on vectors to penetrate plant tissues, but many fungi, oomycetes and aphids can penetrate cell walls directly, whereas pathogenic bacteria often depend on natural openings, such as stomata or wound sites. At this stage of infection, a rapid closure of the stomata can form a first pre‐invasive defence barrier (Melotto et al., 2008). After successful entry of the plant tissue, plant attackers often face an early‐acting, post‐invasive defence barrier that is marked by localized defence responses, such as the accumulation of reactive oxygen species, defence gene induction and deposition of callose‐rich papillae (Eulgem et al., 1999; Flors et al., 2005; Torres et al., 2006). On further colonization, plants undergo a large‐scale transcriptional reprogramming that coincides with the biosynthesis of salicylic acid (SA), jasmonic acid (JA) and complementary long‐distance signals, which regulate a broad spectrum of local and systemic defence mechanisms (Heil and Ton, 2008).
Plants respond to microbes through the recognition of conserved microbial features, such as flagellin, chitin, glycoproteins or lipopolysaccharides, which are referred to as ‘pathogen‐associated molecular patterns’ (PAMPs, synonymously called MAMPs for ‘microbe‐associated molecular patterns’). Defence responses to herbivores are triggered by herbivore‐associated molecular patterns (HAMPs; Mithofer and Boland, 2008). Besides PAMPs and HAMPs, there are endogenous plant elicitors that are released on tissue damage. These damaged‐associated molecular patterns (DAMPs) mediate defence responses to both herbivores and pathogens (Heil, 2009). Although the perception behind HAMPs remains unknown (Mithofer and Boland, 2008), PAMPS and DAMPS are thought to be recognized by plasma membrane‐localized pattern‐recognition receptors (PRRs; Gomez‐Gomez and Boller, 2000; Huffaker et al., 2006; Miya et al., 2007; Scheer and Ryan, 2002). The immune response triggered by these defence elicitors is commonly referred to as PAMP‐triggered immunity (PTI), although the term ‘pattern‐triggered immunity’ would be more appropriate as it more collectively reflects responses to PAMPs, MAMPs, DAMPs and HAMPs. PTI is a nonspecific resistance that is sufficient to fend off the majority of potentially hostile microbes (Boller and He, 2009; Lipka et al., 2005). Virulent pathogens, however, are capable of suppressing PTI through the use of pathogen effectors (Fu et al., 2007; He et al., 2006; Zhang et al., 2007). This effector‐triggered susceptibility (ETS) reduces the plant's immune response to basal levels of resistance, which are insufficient to provide effective protection against disease.
To counteract ETS, selected plant varieties have evolved resistance (R) proteins, which can detect pathogen effectors directly, or can guard the targets of pathogen effectors, thereby indirectly recognizing the activity of effectors (McDowell and Woffenden, 2003). The activation of R proteins often gives rise to a hypersensitive response (HR) that can block virulent pathogens at relatively early stages of infection. This so‐called effector‐triggered immunity (ETI) is extremely effective against biotrophic pathogens, and has therefore been studied extensively over recent decades (Lukasik and Takken, 2009). However, a major limitation of ETI is that it only protects against specific races of biotrophic pathogens, whereas it can be ineffective or even disease promoting in response to necrotrophic pathogens (Kliebenstein and Rowe, 2008). Moreover, avirulent biotrophs are under constant selective pressure to break ETI, which limits the durability of this defence strategy. Pathogens can break ETI by evolving alternative effectors that suppress ETI, or that are no longer recognized by R proteins (Abramovitch et al., 2006; Cui et al., 2009; Fu et al., 2007; Houterman et al., 2009). Consequently, ETI is reverted to basal resistance, imposing further selection pressure on the host plant to evolve improved R proteins that are capable of recognizing the newly evolved effectors. The resulting arms race between plants and their (a)virulent pathogens manifests as an ongoing oscillation in the effectiveness of plant defence, and is referred to as the zigzag model (Jones and Dangl, 2006).
PRIMING: AN ALTERNATIVE DEFENCE STRATEGY TO COPE WITH BIOTIC STRESS
Although ETI can be extremely effective against biotrophic pathogens, plants can also counteract pathogens through a sensitization of their basal immune system. This priming of defence causes a faster and stronger induction of defensive mechanisms on subsequent attack (Conrath et al., 2006; Frost et al., 2008). There are a variety of environmental signals that can trigger defence priming, many of which indicate upcoming stress. Well‐known examples are localized pathogen attack, which gives rise to systemic acquired resistance (SAR; Jung et al., 2009; Kohler et al., 2002), or herbivory‐induced volatiles, which can prime JA‐dependent wound responses (Engelberth et al., 2004; Frost et al., 2008; Ton et al., 2007). On the other hand, plant‐beneficial micro‐organisms, such as nonpathogenic rhizobacteria or mycorrhizal fungi, can also trigger priming (Van Wees et al., 2008). Furthermore, many biologically induced priming phenomena can be mimicked through the application of plant‐derived or xenobiotic compounds, such as SA (Mur et al., 1996), methyl jasmonate (MeJA; Kauss et al., 1994), vitamin B (Ahn et al., 2007), cytokinins (Dervinis et al., 2010) or β‐aminobutyric acid (BABA; Jakab et al., 2001).
Similar to PTI, priming of defence is effective against a broad spectrum of plant attackers, suggesting that primed resistance is at least partially based on an augmented expression of PTI mechanisms. However, both pathogen‐induced SAR and rhizobacteria‐mediated induced systemic resistance (ISR) have been shown to reduce lesion formation by avirulent pathogens (Hoffland et al., 1996; Ross, 1961), suggesting that priming can also boost ETI mechanisms. As basal resistance has been defined as the sum of resistance by PTI and ETI, minus the susceptibility by ETS (Jones and Dangl, 2006), priming of defence can best be defined as an augmented capacity to express basal resistance mechanisms (Fig. 1). If the augmented basal resistance response precedes the delivery of pathogen effectors, priming can provide full immunity against otherwise virulent pathogens (Fig. 1A). Indeed, this has been found for some forms of defence priming (Conrath et al., 2006; Zimmerli et al., 2000). In most cases, however, primed defence expression slows down the colonization by virulent pathogens to a larger extent than does basal resistance (Conrath et al., 2006).
Figure 1.
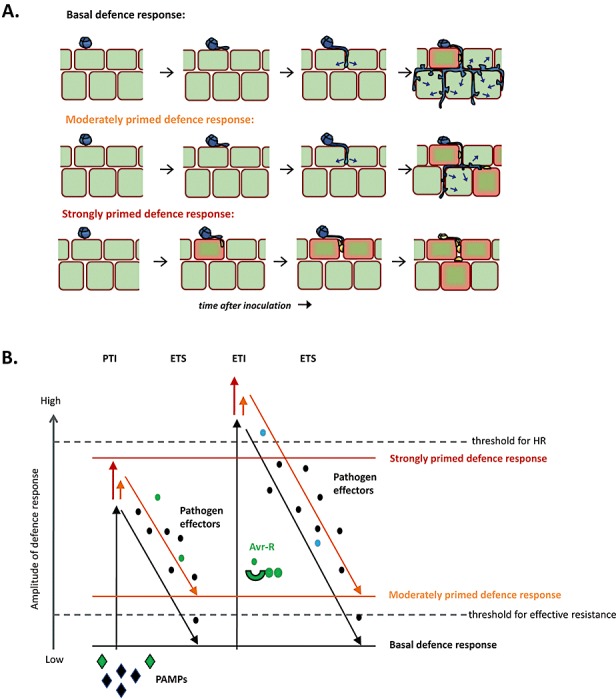
Priming of basal resistance provides protection against virulent pathogens. (A) Basal resistance against virulent pathogens results from a residual level of host defence after defence suppression by disease‐promoting pathogen effectors (blue arrows). Priming of basal resistance leads to a faster and stronger induction of basal defence mechanisms, providing enhanced resistance against the invading pathogen. In most cases, priming of basal resistance cannot prevent the delivery of pathogen effectors entirely, and thereby only slows down the introgression of the pathogen (‘moderately primed defence response’). However, if the primed defence response precedes the delivery of pathogen effectors, this defence strategy can prevent pathogen infection and provide full protection against otherwise virulent pathogens (‘strongly primed defence response’). Red plant cells indicate the expression of basal defence mechanisms. (B) The ‘zigzag’ model describes basal resistance as the sum of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) and weak effector‐triggered immunity (ETI) minus effector‐triggered susceptibility (ETS) (Jones and Dangl, 2006). Apart from newly evolved R proteins that recognize effectors or their activities, ETS can be counteracted by the priming of defence, causing faster and stronger induction of basal defence mechanisms after pathogen attack. A moderately primed defence response merely augments the PTI/ETI response, but would still allow ETS to take place (shown in orange), whereas a strongly primed defence reaction can prevent ETS entirely (shown in red).
Most priming‐inducing stimuli can trigger defence mechanisms directly if applied in higher doses. For instance, relatively high soil drench concentrations of BABA trigger PR‐1 gene induction directly in Arabidopsis, whereas lower concentrations of BABA merely prime the induction of PR‐1 (van Hulten et al., 2006). Furthermore, transient induction of direct defence can give rise to a longer lasting priming of defence (Bruce et al., 2007; Heil and Ton, 2008). Hence, many induced resistance phenomena are based on a combination of direct defence and priming, and their relative contribution depends on the dose of the resistance‐inducing stimulus and the time point after induction.
COSTS AND BENEFITS OF PRIMING
The full development of an inducible defence response requires energy and therefore involves costs on growth and reproduction. Apart from allocation costs, costs can also arise from the toxicity of the defence to the plant's own metabolism, or when the defence response affects the plant's interaction with beneficial organisms (Heil, 2002; Heil and Baldwin, 2002). It is commonly accepted that plants only express inducible defences if the benefits (i.e. protection against the attackers) outweigh the associated costs (Heil, 2002; Walters and Boyle, 2005).
Previously, we have conducted a laboratory study to compare the costs and benefits of defence priming versus direct induction of defence in Arabidopsis (van Hulten et al., 2006). By using low doses of BABA to induce priming and high doses of either BABA or the functional SA analogue benzothiadiazole (BTH) to induce defence expression directly, we found that priming is associated with relatively minor costs on plant growth and seed set. Moreover, the protective benefits of priming outweighed its costs under conditions of high disease pressure. It was thus concluded that priming is a cost‐efficient defence strategy in disease‐imposing environments. Interestingly, the outcome of this laboratory study was recently tested under agronomic field conditions by Walters et al. (2009), who subjected saccharin‐primed barley to varying degrees of disease by the hemibiotrophic fungus Rhynchosporium secalis and monitored fitness levels by plant growth and grain yield. As predicted, primed plants displayed significantly higher fitness than unprimed plants, thereby extending our laboratory demonstration that priming is a beneficial defence strategy in hostile environments.
PLANT DEFENCE STRATEGIES AND THEIR ADAPTIVE VALUES IN HOSTILE ENVIRONMENTS
Many naturally occurring plant species can be subdivided into genetically distinct geographical varieties. Although these ecotypes are sufficiently similar to be considered as one species, they differ genetically in some traits as a result of variant selection pressures from their environments of origin. In this context, we evaluated possible adaptive values of various defensive strategies in comparison with priming of defence under different environmental conditions (Fig. 2).
Figure 2.
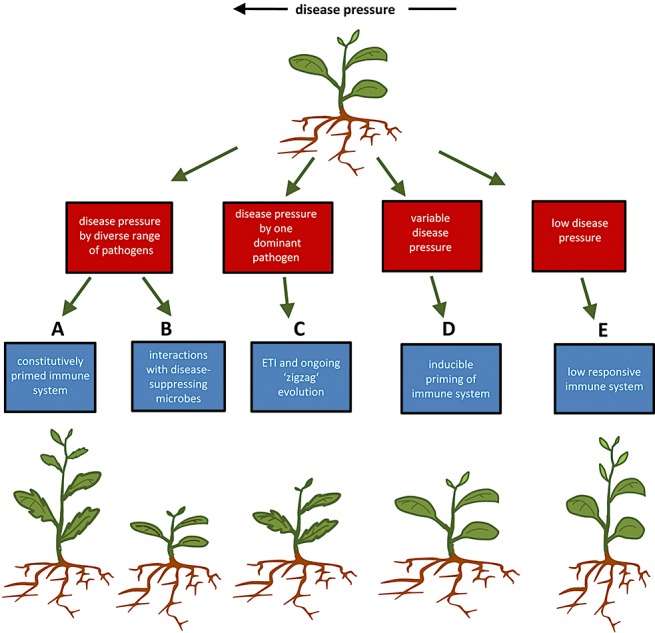
Model of plant defence strategies and their adaptive values under different biotic stress conditions. Plants in environments with relatively high disease pressure from a wide array of different attackers benefit from constitutive priming of basal resistance mechanisms, which provide broad‐spectrum protection against pests and diseases (strategy A). However, this defence strategy may affect the plant's ability to associate with plant‐beneficial microbes, such as mycorrhizae or nitrogen‐fixing bacteria. In this situation, plants would benefit more from an increased ability to attract and associate with disease‐suppressing plant‐beneficial microbes (strategy B). Plants in environments with a constant pressure from one dominant biotrophic pathogen benefit from effector‐triggered immunity (ETI; strategy C). ETI can be broken and give rise to an ongoing ‘zigzag’ evolution, as described by Jones and Dangl (2006). Inducible defence priming on perception of stress‐indicating signals provides a cost‐efficient adaptation to environments with variable degrees of disease pressure (strategy D). Because priming of defence and induction of defence are both associated with costs on plant growth and reproduction, a relatively unresponsive immune system would be beneficial in environments with relatively low disease pressure (strategy E).
The concept that priming of defence provides benefits in hostile environments suggests that plants in these environments are under pressure to evolve a constitutively enhanced responsiveness of basal defence mechanisms. As priming protects against a wide variety of diseases and pests (Conrath et al., 2006), this selection pressure would be most pronounced under pressure by a wide range of different pathogens and herbivores (Fig. 2; strategy A). There are, however, alternative defence strategies that could provide similar or even greater benefits, depending on the nature of the environment. For instance, PAMPs from plant‐beneficial microbes have been demonstrated to trigger plant defence reactions (van Loon et al., 2008), suggesting that plants with primed defence responsiveness to PAMPs may risk compromising their interaction with plant‐beneficial micro‐organisms. Indeed, various studies have reported negative impacts of SA‐dependent resistance on rhizobial and mycorrhizal symbioses with legumes (Faessel et al., 2009; Jin et al., 2009; Stacey et al., 2006). Hence, there could be a counteracting selection against constitutive priming to maintain associations with mycorrhiza or nitrogen‐fixing bacteria. Therefore, an increased ability to attract and interact with micro‐organisms that are capable of suppressing pathogens directly through nutrient competition or antibiosis (Handelsman and Stabb, 1996; Weller et al., 2002) could be an alternative defence strategy in hostile environments (Fig. 2; strategy B). In support of this, Rudrappa et al. (2008) demonstrated that Arabidopsis can attract disease‐suppressing rhizobacteria through the exudation of l‐malic acid, which is further boosted by above‐ground infection by Pseudomonas syringae pv. tomato. Secondly, priming rarely provides complete protection against one pathogen or pathogen race, whereas ETI typically does. Hence, ETI would be more efficient in environments with disease pressure from one predominant pathogen species (Fig. 2; strategy C). Thirdly, although priming is less costly than direct induction of defence, it is still associated with minor costs under conditions of low disease pressure (van Hulten et al., 2006). Consequently, plants exposed to variable levels of disease pressure would benefit from an inducible priming response (Fig. 2; strategy D). As variable degrees of disease pressure are the reality in many natural plant environments, priming mainly manifests as an inducible resistance response. Finally, the selection for certain defensive strategies is probably also influenced by the plant's abiotic environment. For instance, the plant hormone abscisic acid (ABA) not only controls tolerance to abiotic stress, but also plays an important regulatory role in the fine tuning of resistance against diseases and pests (Ton et al., 2009). Consequently, a change in defence responsiveness to biotic stress that is based on a modified ABA response will also have consequences for plant fitness under abiotic stress conditions.
Most plants are capable of expressing combinations of different defence strategies, and the importance of each of these strategies depends on the environment. For instance, many plant‐beneficial micro‐organisms have the ability to protect plants through a combination of direct disease suppression and induction of defence priming in the host plant (ISR; Van Wees et al., 2008). Colonization by these microbes causes a constitutive level of systemic priming that is phenotypically similar to genetically acquired priming, thus combining the advantages of two defence strategies: direct disease suppression by plant‐beneficial microbes and constitutive defence priming. Furthermore, the expression of one defence strategy can give rise to the induction of another. For example, localized expression of PTI results in the development of SAR (Mishina and Zeier, 2007), which is largely based on the priming of defence (Jung et al., 2009; Kohler et al., 2002).
NATURAL VARIATION IN BASAL DEFENCE RESPONSIVENESS
If constant pressure from a wide range of attackers would select for constitutively primed basal resistance (Fig. 2; strategy A), it can be expected that naturally occurring plant species display genetic variation in the responsiveness of basal resistance mechanisms. The vast majority of studies on natural variation in plant defence have focused on ETI (Holub, 2007; de Meaux and Mitchell‐Olds, 2003; Van Poecke et al., 2007), which is likely to be the result of the robustness and reproducibility of the ETI phenotype. There are also numerous studies on the natural variation in basal resistance against pathogens and herbivores, many of which are based on the genetic model plant species Arabidopsis (Koornneef et al., 2004). However, relatively few of these have linked this variation to actual resistance mechanisms. The natural variation in the basal resistance of Arabidopsis to insects often originates from differences in pre‐existing pools of glucosinolates (Kliebenstein et al., 2001; Koornneef et al., 2004). Glucosinolates enable the rapid production of biocidal isothiocyanates after herbivore attack and could therefore be viewed as a constitutively primed defence mechanism. The natural variation in the basal resistance of Arabidopsis against pathogens, on the other hand, seems to stem from more diverse mechanisms rather than from glucosinolates. For instance, Denby et al. (2004) reported that the natural variation in basal resistance against the necrotroph Botrytis cinerea correlates with the responsiveness of pathogen‐ and acifluorfen‐induced camalexin, an indole‐derived phytoalexin. Further genetic dissection of this basal resistance in a mapping population of recombinant inbred lines (RILs) revealed multiple small‐to‐medium‐effect quantitative trait loci (QTLs), but it remained unclear as to what extent these loci influence the responsiveness of camalexin induction itself. Similarly, Llorente et al. (2005) used a RIL population to dissect the natural variation in basal resistance against the necrotrophic fungus Plectosphaerella cucumerina, which identified three different QTLs. The most influential QTL was caused by a polymorphism in the ERECTA gene, a leucine‐rich region (LRR) receptor‐like kinase protein that influences the responsiveness of pathogen‐induced callose deposition. Natural variation in basal resistance against P. syringae pathogens has been reported by different groups (Kover and Schaal, 2002; Kover et al., 2005; Perchepied et al., 2006; Ton et al., 1999). Genetic dissection of this variation between accessions Bayreuth and Shahdara revealed two major QTLs, one of which was found to regulate the responsiveness of SA‐inducible defence genes (Perchepied et al., 2006). The studies by Llorente et al. (2005) and Perchepied et al. (2006) illustrate that one gene can have a major contribution to the natural variation in the responsiveness of basal resistance mechanisms.
The development of DNA arrays has made it possible to measure natural variation in the abundance of large numbers of gene transcripts: ‘expression level polymorphism’ (ELP) analysis. If applied to a genetically characterized mapping population, ELP analysis can link natural variation in transcriptome responses to regulatory loci, called expression (e)QTLs. A first genomic comparison of the transcriptional response to exogenously applied SA between seven Arabidopsis accessions revealed that, on average, 2234 genes were differentially expressed in pair‐wise comparisons (Kliebenstein et al., 2006). This variation correlated positively with genomic sequence diversity, suggesting that single nucleotide polymorphisms have relatively little influence on the genetic variation in SA‐induced gene expression. Further in‐depth analysis of these data identified accession Mt‐0 as hyperresponsive and accession Cvi‐0 as hyporesponsive to SA (van Leeuwen et al., 2007). Although these studies made an important step towards a better understanding of the evolution of basal resistance, it remains unknown which genomic regions are responsible for this variation, and how far this variation actually impacts basal pathogen resistance. Further ELP analysis of well‐characterized mapping populations would be necessary to identify the regulatory genes responsible for both natural variation in gene network responses and basal defence responsiveness.
For basal defence responses involving secondary defence metabolites, considerable debate has surrounded the evolutionary mechanisms involved in the creation of the necessary pathways (Firn, 2010). It is emerging that, for many associated biosynthetic pathways, operons or clusters of coregulated genes can be responsible for the sequential production of the necessary enzymes (Field and Osbourn, 2008). These pathways typically result in the production of penultimate nonbiocidal products, e.g. a glucosinolate (see above) or the glucoside of a benzoxazinoid, which provide a primed capacity to produce the biocide on subsequent pathogen or herbivore attack (Field and Osbourn, 2008; Frey et al., 2009). Interestingly, these secondary defence metabolites also play a role in defence against fungal pathogens (Bednarek and Osbourn, 2009) and have been found to regulate PAMP‐induced depositions of callose‐rich papillae in Arabidopsis (Clay et al., 2009). Clusters of genes in the biosynthesis of secondary defence metabolites can comprise paralogous genes from gene duplication, but also nonhomologous genes that are organized functionally with concomitant clustering and coregulation. Quite how such a pathway evolves before giving rise to an adaptive advantage is not fully understood (Firn, 2010), but it seems obvious that the final biochemical conversion from a nonbiocidal storage product to a biocidal defence product is critical in the regulation of these chemical defences. It remains a future challenge to determine how far genetic variation in the timing, activity and localization of hydrolytic enzymes mediating these conversions contributes to differences in basal resistance against diseases and pests. If so, it will be equally interesting to investigate whether these enzymes are targeted by pathogen and/or insect effectors.
NATURAL SELECTION FOR CONSTITUTIVELY PRIMED BASAL RESISTANCE?
Although the above examples justify the general conclusion that natural variation in responsiveness of basal resistance mechanisms is prevalent, this does not necessarily prove that hostile environments select for constitutively primed immune systems. In order to demonstrate that constitutive defence priming has evolved from inducible priming under constant levels of disease pressure, more evidence is required from both the molecular level and the plant community level (Shindo et al., 2007). For instance, patterns of single nucleotide polymorphisms in alleles contributing to natural variation in basal resistance could provide indications of past selective pressures. If the degree of nucleotide diversity deviates from the estimated diversity under neutral selection, this could be interpreted as evidence for environmental selection pressures. Typically, reduced levels of nucleotide polymorphisms indicate selective sweeps, during which newly evolved gene variants outcompete others (Nielsen, 2005). On the other hand, enhanced levels of nucleotide polymorphisms suggest balancing selection, which maintains ancient genetic variation (Mitchell‐Olds and Schmitt, 2006). Although these methods provide useful indications for past selective forces on genes, fitness assays under different disease pressures would still be necessary to establish what gene variants provide which adaptive phenotypes. As outlined by Holub (2007), a major challenge for the future is to apply currently available genetic resources for Arabidopsis (i.e. fully genotyped recombinant mapping populations or association mapping populations) to field experimentation. For instance, Arabidopsis mapping populations in which defence traits segregate could be grown under different field conditions with varying degrees of disease pressure by one or multiple pathogens and/or herbivores. Subsequent fitness evaluation may reveal defence‐regulating QTLs that provide selective benefits under specified environmental conditions. With more and more Arabidopsis accessions being genome‐sequenced, another promising approach arises from genome‐wide association mapping approaches, which are based on associations between phenotypes and DNA sequence variants within individuals or isogenic populations (Atwell et al., 2010; Nordborg and Weigel, 2008). In particular, if defence phenotypes can be related to ecological stress parameters from the accessions' geographical origins, this technique has the potential to assign measurable ecological significance to defence regulatory alleles.
EXPLOITATION OF NATURAL VARIATION IN PLANT DEFENCE
How can natural variation be exploited to improve crop protection against diseases and pests? Although Arabidopsis cannot be considered to be a crop, the availability of genetically characterized mapping populations and genome‐sequenced accessions provides easy access to the genetic basis of the defence strategies evolved within this species. A potential disadvantage of this approach is that certain defence traits of Arabidopsis are specific for Brassicaceae. For instance, glucosinolates play an important role in the defence of Arabidopsis against unadapted insects and pathogens (Arany et al., 2008; Bednarek et al., 2009; Tierens et al., 2001), and alleles contributing to the genetic variation in the biosynthesis of these secondary metabolites are of little relevance for non‐brassicaceous crops, such as cereals. Moreover, Arabidopsis is a pioneering plant species that was originally selected by plant geneticists for its short generation time, which, by itself, can be regarded as an adaptive strategy to cope with environmental stress, but is not necessarily a desirable trait in crops.
As an alternative strategy, genetic variation in defence traits amongst ancestral plants of crops can be explored as a means to introduce new combinations of alleles into modern crop plants. An extensively researched example comes from the development of breeding programmes that aim to introgress traits from ancestral wheat varieties into modern elite varieties, where limited genetic diversity in defence traits is exhibited as a result of repeated selection for high yield (Skovmand et al., 2001; Trethowan and Mujeeb‐Kazi, 2008). Unfortunately, many wild wheat ancestors and landraces are at risk of disappearing as a result of displacement from their natural habitats by agronomically superior cultivars and overgrazing by livestock. To combat this loss of valuable diversity, extensive collections of rare species are now being assembled for exploitation in wheat breeding programmes (Skovmand et al., 2002). These programmes have enabled a number of race‐specific R genes to be introduced into hexaploid wheat species. However, as discussed above, R genes do not always provide durable disease protection, as the resulting ETI can be broken by pathogen evolution (Jones and Dangl, 2006). A notable exception has been the identification of the WKS1 gene, which confers partial and temperature‐dependent resistance in mature wheat against multiple races of the stripe rust, Puccinia striiformis (Fu et al., 2009). The WKS1 gene is absent in commercial wheat varieties and was introgressed from the ancestral wheat accession Triticum turgidum L. ssp. Dicoccoides (Uauy et al., 2005). Interestingly, WKS1‐dependent resistance manifests as a rapid formation of autofluorescent cells around the sides of P. striiformis infection (Fu et al., 2009), indicating that WKS1 provides primed responsiveness to pathogen attack. The gene encodes a protein kinase with a putative START domain. This class of protein has been reported to play a role in lipid binding and sensing (Alpy and Tomasetto, 2005), suggesting that WKS1 is involved in the transduction of pathogen‐ or plant‐derived lipid signals. Interestingly, lipid signals have also been implicated as critical signals in SAR‐related defence priming. Although the exact signalling function of the WKS1 protein remains to be investigated, the study by Fu et al. (2009) clearly illustrates how genetic variation in basal resistance within an ancestral plant species can be exploited to provide durable disease protection in a commercially important crop.
Apart from breeding strategies to introgress basal resistance genes from ancestral crop species, biotechnological strategies can also be considered. As was recently outlined by Gust et al. (2010), a promising strategy to improve crop resistance would be to transfer PRRs from naturally occurring plant species into crops. This transgenic approach would boost PTI responsiveness, provided that the heterologously expressed PRRs connect onto the appropriate defence signalling pathways. Analogous to the priming of basal resistance, a primed PTI response would provide broad‐spectrum disease resistance if the augmented defence response precedes the opportunity to express ETS by the invading pathogen (Fig. 1A).
CONCLUSIONS AND FUTURE PROSPECTS
Plants have employed diverse evolutionary strategies to cope with biotic stress. Apart from the well‐characterized ‘zigzag’ evolution of R protein‐mediated ETI, there is evidence for alternative defence strategies that can be equally or more effective, depending on the environmental stress conditions. In addition to interactions with disease‐suppressing micro‐organisms, plants can also obtain broad‐spectrum resistance through priming of basal resistance mechanisms. Understanding the mechanisms behind these naturally evolved defence strategies will be a prerequisite for their exploitation in integrated approaches to pest and disease management.
ACKNOWLEDGEMENTS
We thank Uwe Conrath for helpful suggestions. This review was developed from a presentation given at the British Society for Plant Pathology's 2009 Presidential Meeting. Research activities by Jurriaan Ton are supported by a Biotechnology and Biological Sciences Research Council (BBSRC) Institute Career Path Fellowship (no. BB/E023959/1).
REFERENCES
- Abramovitch, R.B. , Janjusevic, R. , Stebbins, C.E. and Martin, G.B. (2006) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. USA, 103, 2851–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, I.P. , Kim, S. , Lee, Y.H. and Suh, S.C. (2007) Vitamin B1‐induced priming is dependent on hydrogen peroxide and the NPR1 gene in Arabidopsis. Plant Physiol. 143, 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpy, F. and Tomasetto, C. (2005) Give lipids a START: the StAR‐related lipid transfer (START) domain in mammals. J. Cell Sci. 118, 2791–2801. [DOI] [PubMed] [Google Scholar]
- Arany, A. , De Jong, T. , Kim, H. , Van Dam, N. , Choi, Y. , Verpoorte, R. and Van Der Meijden, M. (2008) Glucosinolates and other metabolites in the leaves of Arabidopsis thaliana from natural populations and their effects on a generalist and a specialist herbivore. Chemoecology, 18, 65–71. [Google Scholar]
- Atwell, S. , Huang, Y.S. , Vilhjalmsson, B.J. , Willems, G. , Horton, M. , Li, Y. , Meng, D. , Platt, A. , Aaron, M. , Tarone, A.M. , Hu, T.T. , Jiang, R. , Muliyati, N.W. , Zhang, X. , Amer, M.A. , Baxter, I. , Brachi, B. , Chory, J. , Dean, C. , Debieu, M. , De Meaux, J. , Ecker, J.R. , Faure, N. , Kniskern, J.M. , Jones, J.D.G. , Michael, T. , Nemri, A. , Roux, F. , Salt, D.E. , Tang, C. , Todesco, M. , Traw, M.B. , Weigel, D. , Marjoram, P. , Borevitz, J.O. , Bergelson, J. and Nordborg, M. (2010) Genome‐wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature, 465, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek, P. and Osbourn, A. (2009) Plant–microbe interactions: chemical diversity in plant defense. Science, 324, 746–748. [DOI] [PubMed] [Google Scholar]
- Bednarek, P. , Pislewska‐Bednarek, M. , Svatos, A. , Schneider, B. , Doubsky, J. , Mansurova, M. , Humphry, M. , Consonni, C. , Panstruga, R. , Sanchez‐Vallet, A. , Molina, A. and Schulze‐Lefert, P. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad‐spectrum antifungal defense. Science, 323, 101–106. [DOI] [PubMed] [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, T.J.A. , Matthes, M.C. , Napier, J.A. and Pickett, J.A. (2007) Stressful ‘memories’ of plants: evidence and possible mechanisms. Plant Sci. 173, 603–608. [Google Scholar]
- Clay, N.K. , Adio, A.M. , Denoux, C. , Jander, G. and Ausubel, F.M. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science, 323, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath, U. , Beckers, G.J. , Flors, V. , Garcia‐Agustin, P. , Jakab, G. , Mauch, F. , Newman, M.A. , Pieterse, C.M.J. , Poinssot, B. , Pozo, M.J. , Pugin, A. , Schaffrath, U. , Ton, J. , Wendehenne, D. , Zimmerli, L. and Mauch‐Mani, B. (2006) Priming: getting ready for battle. Mol. Plant–Microbe Interact. 19, 1062–1071. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Xiang, T. and Zhou, J.M. (2009) Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol. 11, 1453–1461. [DOI] [PubMed] [Google Scholar]
- Denby, K.J. , Kumar, P. and Kliebenstein, D.J. (2004) Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana . Plant J. 38, 473–486. [DOI] [PubMed] [Google Scholar]
- Dervinis, C. , Frost, C. , Lawrence, S. , Novak, N. and Davis, J. (2010) Cytokinin primes plant responses to wounding and reduces insect performance. J. Plant Growth Regul. in press. Available at 10.1007/s00344-009-9135-2. [DOI] [Google Scholar]
- Engelberth, J. , Alborn, H.T. , Schmelz, E.A. and Tumlinson, J.H. (2004) Airborne signals prime plants against insect herbivore attack. Proc. Natl. Acad. Sci. USA, 101, 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Schmelzer, E. , Hahlbrock, K. and Somssich, I.E. (1999) Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faessel, L. , Nassr, N. , Lebeau, T. and Walter, B. (2009) Chemically‐induced resistance on soybean inhibits nodulation and mycorrhization. Plant Soil, 329, 1–2, 259–268. [Google Scholar]
- Field, B. and Osbourn, A.E. (2008) Metabolic diversification—independent assembly of operon‐like gene clusters in different plants. Science, 320, 543–547. [DOI] [PubMed] [Google Scholar]
- Firn, R. (2010) Nature's Chemicals: The Natural Products That Shaped Our World. Oxford: Oxford University Press. [Google Scholar]
- Flors, V. , Ton, J. , Jakab, G. and Mauch‐Mani, B. (2005) Abscisic acid and callose: team players in defence against pathogens? J. Phytopathol. 153, 377–383. [Google Scholar]
- Frey, M. , Schullehner, K. , Dick, R. , Fiesselmann, A. and Gierl, A. (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry, 70, 1645–1651. [DOI] [PubMed] [Google Scholar]
- Frost, C.J. , Mescher, M.C. , Carlson, J.E. and De Moraes, C.M. (2008) Plant defense priming against herbivores: getting ready for a different battle. Plant Physiol. 146, 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, D. , Uauy, C. , Distelfeld, A. , Blechl, A. , Epstein, L. , Chen, X. , Sela, H. , Fahima, T. and Dubcovsky, J. (2009) A kinase‐START gene confers temperature‐dependent resistance to wheat stripe rust. Science, 323, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Z.Q. , Guo, M. , Jeong, B.R. , Tian, F. , Elthon, T.E. , Cerny, R.L. , Staiger, D. and Alfano, J.R. (2007) A type III effector ADP‐ribosylates RNA‐binding proteins and quells plant immunity. Nature, 447, 284–288. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1012. [DOI] [PubMed] [Google Scholar]
- Gust, A.A. , Brunner, F. and Nurnberger, T. (2010) Biotechnological concepts for improving plant innate immunity. Curr. Opin. Biotechnol. 21, 204–210. [DOI] [PubMed] [Google Scholar]
- Handelsman, J. and Stabb, E.V. (1996) Biocontrol of soilborne plant pathogens. Plant Cell, 8, 1855–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P. , Shan, L. , Lin, N.‐C. , Martin, G.B. , Kemmerling, B. , Nürnberger, T. and Sheen, J. (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell, 125, 563–575. [DOI] [PubMed] [Google Scholar]
- Heil, M. (2002) Ecological costs of induced resistance. Curr. Opin. Plant Biol. 5, 345–350. [DOI] [PubMed] [Google Scholar]
- Heil, M. (2009) Damaged‐self recognition in plant herbivore defence. Trends Plant Sci. 14, 356–363. [DOI] [PubMed] [Google Scholar]
- Heil, M. and Baldwin, I.T. (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–67. [DOI] [PubMed] [Google Scholar]
- Heil, M. and Ton, J. (2008) Long‐distance signalling in plant defence. Trends Plant Sci. 13, 264–272. [DOI] [PubMed] [Google Scholar]
- Hoffland, E. , Hakulinen, J. and Van Pelt, J.A. (1996) Comparison of systemic resistance induced by avirulent and nonpathogenic Pseudomonas species. Phytopathology, 86, 757–762. [Google Scholar]
- Holub, E.B. (2007) Natural variation in innate immunity of a pioneer species. Curr. Opin. Plant Biol. 10, 415–424. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Ma, L. , Van Ooijen, G. , De Vroomen, M.J. , Cornelissen, B.J. , Takken, F.L. and Rep, M. (2009) The effector protein Avr2 of the xylem‐colonizing fungus Fusarium oxysporum activates the tomato resistance protein I‐2 intracellularly. Plant J. 58, 970–978. [DOI] [PubMed] [Google Scholar]
- Huffaker, A. , Pearce, G. and Ryan, C.A. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA, 103, 10 098–10 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hulten, M. , Pelser, M. , Van Loon, L.C. , Pieterse, C.M. and Ton, J. (2006) Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA, 103, 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab, G. , Cottier, V. , Toquin, V. , Rigoli, G. , Zimmerli, L. , Métraux, J.‐P. and Mauch‐Mani, B. (2001) β‐Aminobutyric acid‐induced resistance in plants. Eur. J. Plant Pathol. 107, 29–37. [Google Scholar]
- Jin, L. , Wang, S. , Wang, X. and Shen, Y. (2009) Seed size influences arbuscular mycorrhizal symbiosis across leguminous host‐plant species at the seedling stage. Symbiosis, 49, 111–116. [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jung, H.W. , Tschaplinski, T.J. , Wang, L. , Glazebrook, J. and Greenberg, J. (2009) Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Kauss, H. , Jeblick, W. , Ziegler, J. and Krabler, W. (1994) Pretreatment of parsley (Petroselinum crispum L.) suspension‐cultures with methyl jasmonate enhances elicitation of activated oxygen species. Plant Physiol. 105, 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J. and Rowe, H.C. (2008) Ecological costs of biotrophic versus necrotrophic pathogen resistance, the hypersensitive response and signal transduction. Plant Sci. 174, 551–556. [Google Scholar]
- Kliebenstein, D.J. , Kroymann, J. , Brown, P. , Figuth, A. , Pedersen, D. , Gershenzon, J. and Mitchell‐Olds, T. (2001) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J. , West, M.A. , Van Leeuwen, H. , Kim, K. , Doerge, R.W. , Michelmore, R.W. and St. Clair, D.A. (2006) Genomic survey of gene expression diversity in Arabidopsis thaliana . Genetics, 172, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, A. , Schwindling, S. and Conrath, U. (2002) Benzothiadiazole‐induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 128, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M. , Alonso‐Blanco, C. and Vreugdenhil, D. (2004) Naturally occurring genetic variation in Arabidopsis thaliana . Annu. Rev. Plant Biol. 55, 141–172. [DOI] [PubMed] [Google Scholar]
- Kover, P.X. and Schaal, B.A. (2002) Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc. Natl. Acad. Sci. USA, 99, 11 270–11 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover, P.X. , Wolf, J.B. , Kunkel, B.N. and Cheverud, J.M. (2005) Genetic architecture of Arabidopsis thaliana response to infection by Pseudomonas syringae . Heredity, 94, 507–517. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen, H. , Kliebenstein, D.J. , West, M.A. , Kim, K. , Van Poecke, R. , Katagiri, F. , Michelmore, R.A. , Doerge, R.W. and St. Clair, D.A. (2007) Natural variation among Arabidopsis thaliana accessions for transcriptome response to exogenous salicylic acid. Plant Cell, 19, 2099–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V. , Dittgen, J. , Bednarek, P. , Bhat, R. , Wiermer, M. , Stein, M. , Landtag, J. , Brandt, W. , Rosahl, S. , Scheel, D. , Llorente, F. , Molina, A. , Parker, J. , Somerville, S. and Schulze‐Lefert, P. (2005) Pre‐ and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science, 310, 1180–1183. [DOI] [PubMed] [Google Scholar]
- Llorente, F. , Alonso‐Blanco, C. , Sanchez‐Rodriguez, C. , Jorda, L. and Molina, A. (2005) ERECTA receptor‐like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina . Plant J. 43, 165–180. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. , Bakker, P.A. , Van Der Heijdt, W.H. , Wendehenne, D. and Pugin, A. (2008) Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol. Plant Microbe Interact. 21, 1609–1621. [DOI] [PubMed] [Google Scholar]
- Lukasik, E. and Takken, F.L. (2009) STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12, 427–436. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M. and Woffenden, B.J. (2003) Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol. 21, 178–183. [DOI] [PubMed] [Google Scholar]
- De Meaux, J. and Mitchell‐Olds, T. (2003) Evolution of plant resistance at the molecular level: ecological context of species interactions. Heredity, 91, 345–352. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. and He, S.Y. (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46, 101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Mitchell‐Olds, T. and Schmitt, J. (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature, 441, 947–952. [DOI] [PubMed] [Google Scholar]
- Mithofer, A. and Boland, W. (2008) Recognition of herbivory‐associated molecular patterns. Plant Physiol. 146, 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. , Kawakami, N. , Kaku, H. and Shibuya, N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA, 104, 19 613–19 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur, L.A.J. , Naylor, G. , Warner, S.A.J. , Sugars, J.M. , White, R.F. and Draper, J. (1996) Salicylic acid potentiates defence gene expression in tissue exhibiting acquired resistance to pathogen attack. Plant J. 9, 559–571. [Google Scholar]
- Nielsen, R. (2005) Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218. [DOI] [PubMed] [Google Scholar]
- Nordborg, M. and Weigel, D. (2008) Next‐generation genetics in plants. Nature, 456, 720–723. [DOI] [PubMed] [Google Scholar]
- Perchepied, L. , Kroj, T. , Tronchet, M. , Loudet, O. and Roby, D. (2006) Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana . PLoS ONE, 1, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A.F. (1961) Systemic acquired resistance induced by localized virus infections in plants. Virology, 14, 340–358. [DOI] [PubMed] [Google Scholar]
- Rudrappa, T. , Czymmek, K.J. , Pare, P.W. and Bais, H.P. (2008) Root‐secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, J.M. and Ryan, C.A., Jr (2002) The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA, 99, 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, C. , Bernasconi, G. and Hardtke, C.S. (2007) Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann. Bot. 99, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovmand, B. , Reynolds, M.P. and DeLacy, I.H. (2001) Mining wheat germplasm collections for yield enhancing traits. Euphytica, 119, 25–32. [Google Scholar]
- Skovmand, B.S. , Rajaram, S.J.M. , Ribaut, J.M. and Hede, A.R. (2002) Wheat genetic resources In: Bread Wheat: improvement and production, FAO Plant Production and Protection Series, Rome (Curtis B.C., Rajaram S. and Gomez Macpherson H., eds), 30, 567. ISBN: 9251048096. [Google Scholar]
- Stacey, G. , McAlvin, C.B. , Kim, S.Y. , Olivares, J. and Soto, M.J. (2006) Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula . Plant Physiol. 141, 1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierens, K.F.M. , Thomma, B.P.H.J. , Brouwer, M. , Schmidt, J. , Kistner, K. , Porzel, A. , Mauch‐Mani, B. , Bruno, P.A. , Cammue, P.A. and Broekaert, W.F. (2001) Study of the role of antimicrobial glucosinolate‐derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 125, 1688–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J. , Pieterse, C.M.J. and Van Loon, L.C. (1999) Identification of a locus in Arabidopsis controlling both the expression of rhizobacteria‐mediated induced systemic resistance (ISR) and basal resistance against Pseudomonas syringae pv. tomato . Mol. Plant–Microbe Interact. 12, 911–918. [DOI] [PubMed] [Google Scholar]
- Ton, J. , D'Alessandro, M. , Jourdie, V. , Jakab, G. , Karlen, D. , Held, M. , Mauch‐Mani, B. and Turlings, T.C. (2007) Priming by air‐borne signals boosts direct and indirect resistance in maize. Plant J. 49, 16–26. [DOI] [PubMed] [Google Scholar]
- Ton, J. , Flors, V. and Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Jones, J.D. and Dangl, J.L. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethowan, R.M. and Mujeeb‐Kazi, A. (2008) Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci. 48, 1255–1265. [Google Scholar]
- Uauy, C. , Brevis, J.C. , Chen, X. , Khan, I. , Jackson, L. , Chicaiza, O. , Distelfeld, A. , Fahima, T. and Dubcovsky, J. (2005) High‐temperature adult‐plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content locus Gpc‐B1. Theor. Appl. Genet. 112, 97–105. [DOI] [PubMed] [Google Scholar]
- Van Poecke, R.M. , Sato, M. , Lenarz‐Wyatt, L. , Weisberg, S. and Katagiri, F. (2007) Natural variation in RPS2‐mediated resistance among Arabidopsis accessions: correlation between gene expression profiles and phenotypic responses. Plant Cell, 19, 4046–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees, S.C. , Van der Ent, S. and Pieterse, C.M. (2008) Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. [DOI] [PubMed] [Google Scholar]
- Walters, D.R. and Boyle, C. (2005) Induced resistance and allocation costs: what is the impact of pathogen challenge? Physiol. Mol. Plant Pathol. 66, 40. [Google Scholar]
- Walters, D.R. , Paterson, L. , Walsh, D.J. and Havis, N.D. (2009) Priming for plant defense in barley provides benefits only under high disease pressure. Physiol. Mol. Plant Pathol. 73, 95–100. [Google Scholar]
- Weller, D.M. , Raaijmakers, J.M. , McSpadden Gardener, B.B. and Thomashow, L.S. (2002) Microbial populations responsible for specific soil suppressiveness to pathogens. Annu. Rev. Phytopathol. 40, 309–348. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. , Long, C. , Lan, L. , Chai, J. , She Chen, S. , Tang, X. and Zhou, J.M. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zimmerli, L. , Jakab, G. , Métraux, J.‐P. and Mauch‐Mani, B. (2000) Potentiation of pathogen‐specific defense mechanisms in Arabidopsis by β‐aminobutyric acid. Proc. Natl. Acad. Sci. USA, 97, 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]


