SUMMARY
Small RNA (sRNA)‐guided processes, referred to as RNA silencing, regulate endogenous and exogenous gene expression. In plants and some animals, these processes are noncell autonomous and can operate beyond the site of initiation. Viroids, the smallest self‐replicating plant pathogens known, are inducers, targets and evaders of this regulatory mechanism and, consequently, the presence of viroid‐derived sRNAs (vd‐sRNAs) is usually associated with viroid infection. However, the pathways involved in the biogenesis of vd‐sRNAs are largely unknown. Here, we analyse, by high‐throughput pyrosequencing, the profiling of the Hop stunt viroid (HSVd) vd‐sRNAs recovered from the leaves and phloem of infected cucumber (Cucumis sativus) plants. HSVd vd‐sRNAs are mostly 21 and 22 nucleotides in length and derived equally from plus and minus HSVd RNA strands. The widespread distribution of vd‐sRNAs across the genome reveals that the totality of the HSVd RNA genome contributes to the formation of vd‐sRNAs. Our sequence data suggest that viroid‐derived double‐stranded RNA functions as one of the main precursors of vd‐sRNAs. Remarkably, phloem vd‐sRNAs accumulated preferentially as 22‐nucleotide species with a consensus sequence over‐represented. This bias in size and sequence in the HSVd vd‐sRNA population recovered from phloem exudate suggests the existence of a selective trafficking of vd‐sRNAs to the phloem tissue of infected cucumber plants.
INTRODUCTION
RNA silencing is a pan‐eukaryotic sequence‐specific mechanism that plays a relevant role in the control of genome stability, development and response to biotic and abiotic stress (Baulcombe, 2004; Vaucheret, 2006). In plants, these RNA‐dependent regulatory phenomena share four basic steps: (i) double‐stranded RNA (dsRNA) generation; (ii) cleavage of dsRNAs into 21–24‐nucleotide small RNA (sRNA) duplexes; (iii) stabilization of sRNAs by 2′‐O‐methylation of the 3′‐overhang ends; and (iv) loading of sRNAs into RNA‐induced silencing complexes (RISCs) that mediate the association with partially or fully complementary RNA or DNA targets (Ruiz‐Ferrer and Voinnet, 2009). The dsRNA can originate by any of several mechanisms, including intramolecular base pairing of self‐complementary regions from endogenous and/or exogenous RNAs, and transcription of transposons and (sense or inverted repeat) transgenes. In plants, dsRNA is also synthesized by RNA‐dependent RNA polymerases (RDRs) using single‐stranded RNAs (ssRNAs) with aberrant molecular features as templates (Ruiz‐Ferrer and Voinnet, 2009; Voinnet, 2008). Arabidopsis encodes six RDRs (RDR1–6), although dsRNA generation activity has been described or suggested only for RDR1, RDR2 and RDR6 (Qi et al., 2009; Schiebel et al., 1998; Wassenegger and Krczal, 2006). dsRNA is processed into sRNAs by RNase III‐type enzymes, called Dicer‐like proteins (DCLs). In Arabidopsis, four DCLs (DCL1–4) act in a hierarchical manner to generate 21–24‐nucleotide sRNAs, the key guide of the regulatory processes mediated by RNA silencing (Ramachandran and Chen, 2008).
In plants, the effects of RNA silencing extend beyond its sites of induction by cell‐to‐cell movement (local spreading) and also over longer distances to apical tissues (systemic spreading) as a result of the movement of signalling molecules (Brosnan et al., 2007; Dunoyer and Voinnet, 2008). Although it has been established that systemic movement shows a source‐to‐sink pattern through the phloem (Palauqui et al., 1997), the mechanism underlying the long‐distance transmission of RNA silencing and the precise identity of the transported signal have remained elusive. The identification of sRNAs in the phloem of cucurbit (Yoo et al., 2004) and brassica (Buhtz et al., 2008) species argues for their long‐distance mobility, probably mediated by specific RNA‐binding proteins (Yoo et al., 2004). Although the accumulation of different sRNAs in vascular tissue leads to the suggestion that they could be mobile silencing signals (Aung et al., 2006; Bari et al., 2006; Buhtz et al., 2008; Válóczi et al., 2006), direct evidence of their involvement in long‐distance RNA silencing spread is still lacking.
Viroids are the smallest (246–401 nucleotides) self‐replicating plant pathogens known to date. Their genome consists of single‐stranded, covalently closed, circular and highly structured RNA (Daròs et al., 2006; Ding, 2009; Flores et al., 2005; Tsagris et al., 2008). These plant‐pathogenic RNAs are unable to code for proteins, and therefore their life cycle is strictly dependent on host factors. Viroids are classified into two families: the Pospiviroidae, whose replication takes place in the nucleus, and the Avsunviroidae, which replicates (and accumulates) in the chloroplast (Daròs et al., 2006; Ding, 2009; Flores et al., 2005; Tsagris et al., 2008). The detection of viroid‐derived sRNAs (vd‐sRNAs) in diverse hosts indicates that viroids are potential activators of RNA silencing in infected plants (Carbonell et al., 2008; Gómez and Pallás 2007; Itaya et al., 2001; Markarian et al., 2004; Martinez de Alba et al., 2002; Papaefthimiou et al., 2001; St‐Pierre et al., 2009; Vogt et al., 2004; Wang et al., 2004). Moreover, the demonstration that mature forms of Potato spindle tuber viroid (PSTVd) and Hop stunt viroid (HSVd) can simultaneously elicit and resist this plant defence mechanism in tomato (Itaya et al., 2007) and Nicotiana benthamiana (Gómez and Pallás 2007) plants has provided a unified picture in which these noncoding RNAs are inductors, potential targets and evaders of RNA silencing at the same time. In addition, the possibility that viroid‐induced RNA silencing could be associated with symptom expression in infected plants is currently accepted as a plausible hypothesis to explain the pathogenesis process induced by nuclear viroids (2008, 2009; Markarian et al., 2004; Matoušek et al., 2007; Papaefthimiou et al., 2001; Wang et al., 2004). Although it is highly probable that viroid‐induced RNA silencing could be translocated systemically through the plant, experimental approaches addressing this question are currently nonexistent. Likewise, although the association between diseases induced by nuclear viroids and the accumulation of specific sRNAs has been established for several host–viroid interactions, little is known about how the RNA silencing machinery targets the viroidal genome to produce vd‐sRNAs (Ding, 2009).
Here, we report the first high‐resolution map of sRNAs derived from HSVd from leaf and phloem tissue of infected cucumber plants to understand the biogenesis pathways of vd‐sRNAs and their potential as signalling molecules of systemic RNA silencing. Deep sequencing analysis of viroid‐specific, 21–24‐nucleotide sRNA species shows the following: (i) vd‐sRNAs are predominantly 21 nucleotides in length; (ii) sense and antisense vd‐sRNAs accumulate at similar levels in infected plants; and (iii) vd‐sRNA reads spread throughout the entire plus and minus HSVd RNA. Remarkably, vd‐sRNAs of 22 nucleotides were shown to accumulate differentially in the phloem sap of infected cucumber plants, suggesting the existence of a selective trafficking of sRNAs from nonvascular cells to phloem.
RESULTS
Characterization of vd‐sRNAs
The profiling of vd‐sRNAs can help to decipher the mechanisms and components involved in their biogenesis. Consequently, we used an sRNA high‐throughput sequencing approach to analyse and characterize the viroid‐specific sRNA population in HSVd‐infected cucumber (Cucumis sativus) plants, including leaves and phloem. The HSVd–cucumber interaction was selected because HSVd, previously reported as an RNA silencing inducer in N. benthamiana plants (Gómez et al., 2008; Gómez and Pallás, 2007), infects systemically cucumber plants. In addition, cucumber constitutes a well‐known biological model from which analytical quantities of phloem sap can be collected easily (Gómez et al., 2005; Yoo et al., 2004).
Systemically infected (5–6 weeks post‐inoculation) and healthy cucumber plants were used as the source material for RNA purification and sRNA library construction. Viroid mature forms were readily detected by Northern blot assays in cucumber leaves and phloem exudates using an HSVd‐specific probe (Fig. 1A), suggestive of a widespread distribution of HSVd in infected cucumber plants. RNA hybridization assays also revealed a significant accumulation of HSVd‐specific sRNAs in leaves and phloem exudate of infected cucumber plants (Fig. 1B). These results provide evidence that, similar to that previously reported in N. benthamiana (Gómez and Pallás, 2007), HSVd induces an RNA silencing response in infected cucumber plants. The clear detection of vd‐sRNAs in phloem exudate indicates that these RNAs accumulate in this plant tissue at sufficient levels to be characterized easily (Fig. 1B). We observed that both viroid RNA and ribulose bisphosphate carboxylase/oxygenase (rubisco)‐specific mRNA accumulated to high levels in leaves (Fig. 1C). In contrast, viroid RNA, but not rubisco mRNA, accumulated in phloem exudates (Fig. 1C). These controls discarded the possibility that our phloem samples were contaminated with the content of photosynthetic cells unrelated to mature transporting sieve elements.
Figure 1.
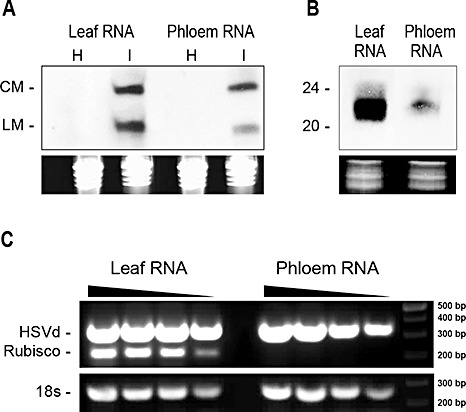
Analysis of cucumber RNAs. (A) Northern blot assays of total RNA extracted from leaves and phloem exudates of infected plants grown at 30 °C. The circular (CM) and linear (LM) monomeric forms of Hop stunt viroid (HSVd) were detected in both tissues. (B) Small‐RNA enriched RNAs extracted from leaves and phloem were analysed by Northern blot assays to detect HSVd‐specific small interfering RNAs (siRNAs). RNAs were quantified by spectrometry and their concentration equalled. Ethidium bromide‐stained gels are shown as RNA load controls (in A and B). (C) Total RNAs extracted from cucumber leaves and phloem were analysed by reverse transcriptase‐polymerase chain reaction (RT‐PCR) for the presence of the photosynthetic related mRNA ribulose bisphosphate carboxylase/oxygenase (rubisco) and electrophoresed in 1% agarose gel. Fragments of ∼300 bp of HSVd and 18s RNA (∼200 bp) were amplified by RT‐PCR in both tissues, whereas rubisco mRNA (fragment of ∼220 bp amplified) was not amplified in the phloem sample.
A pool of sRNAs enriched from total RNA preparations was extracted from the leaves and phloem sap of infected and healthy cucumber plants, ligated to 5′‐ and 3′‐adapters, amplified by reverse transcriptase‐polymerase chain reaction (RT‐PCR) and subjected to multiplexed pyrosequencing. Adapters were designed to specifically ligate sRNAs containing 5′‐monophosphate and 3′‐hydroxyl ends, consistent with DCL‐catalysed cleavage products (Kasschau et al., 2007). A total of 136 592 and 78 374 raw sequences were obtained from infected and healthy cucumbers, respectively. The removal of sequence tags with incomplete matching to the 5′‐ or 3′‐adapter, or resulting from adapter self‐ligation, yielded a set of 127 347 (93.23%) and 72 932 (93.05%) high‐quality sequence tags, each containing an sRNA insert with perfectly matched adapters in the expected configuration (Fig. 2A). The retrieval of a small proportion of ribosomal RNA (rRNA)‐derived noncoding RNAs has been suggested recently as a good indicator of RNA sample quality in conventional sequencing of Peach latent mosaic viroid (PLMVd)‐derived siRNAs (St‐Pierre et al., 2009). In consequence, plant endogenous sRNAs recovered from infected and healthy cucumber were analysed by pairwise alignment using the available database of rRNA sequences of members of the Cucumis genus. The small proportion of unique sRNAs fully homologous to rRNA recovered from infected (11.80%) and healthy (7.95%) cucumber plants (Fig. 2B) was assumed to be an indirect indicator of RNA sample integrity.
Figure 2.
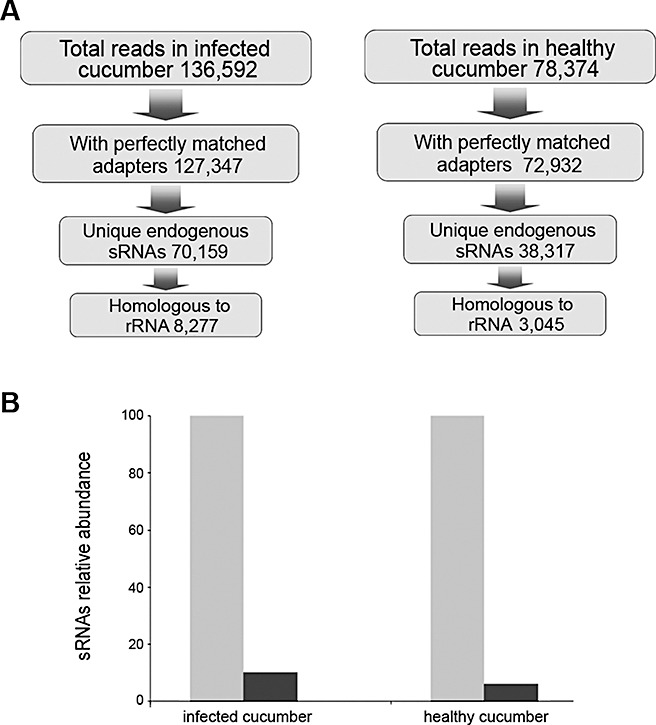
Analysis of small RNAs retrieved in RNA samples extracted from cucumber leaves and phloem. (A) A flowchart showing the stepwise computational extraction of unique endogenous small RNAs recovered from leaves and phloem of infected and healthy plants. (B) Graphic representation of the relative accumulation of the sequenced small RNAs fully homologous to ribosomal RNA.
A total of 4343 (4.91%) sequences recovered from the dataset of infected cucumber leaves (88 321 sequences) was perfectly complementary to HSVd and considered as vd‐sRNAs (Fig. 3A and Table S1, see Supporting Information). Importantly, no HSVd fully homologous sequences were recovered from healthy cucumber leaves, confirming the integrity of the RNA samples. vd‐sRNAs of between 17 and 26 nucleotides were recovered from our leaf library, although reads in the range of 20–24 nucleotides constituted 95.6% (4154) of the total (Fig. 3A). These vd‐sRNAs were mainly of 21 (53.1%) and 22 (21.6%) nucleotides in size (Fig. 3B), whereas vd‐sRNAs of 24 nucleotides corresponded to 15.1% of the total population. vd‐sRNA species of 20, 23 and 25 nucleotides represented 6.85% of the reads. The residual fraction, including vd‐sRNAs of <20 or >25 nucleotides in length (∼3%), was discarded for further analysis. The sequenced pool of 20–25‐nucleotide vd‐sRNAs encompassed 90 166 nucleotides, which represented almost 300 more nucleotides than the total genomic length (297 nucleotides) of HSVd used in this study. This observation suggests that our sequence set accurately reflects the entire vd‐sRNA population in the infected tissue and provides a comprehensive scenario to study vd‐sRNA biogenesis. In addition, the strong bias in size distribution was consistent with the concept that viroid RNA might be targeted by distinct DCLs in a hierarchical fashion to produce different vd‐sRNA sizes, as described for plant‐ and virus‐derived sRNAs (Ramachandran and Chen, 2008; Ruiz‐Ferrer and Voinnet, 2009).
Figure 3.
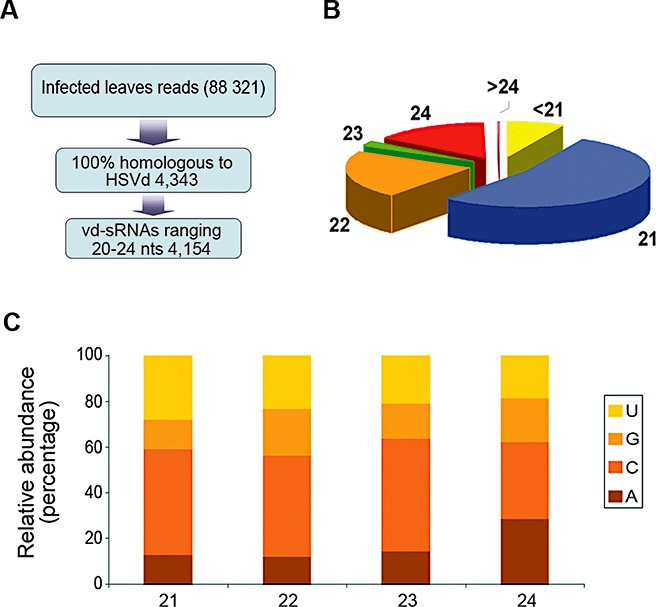
Characterization of viroid‐derived small RNAs (vd‐sRNAs) recovered from Hop stunt viroid (HSVd)‐infected cucumber leaves by deep sequencing. (A) A flowchart showing the stepwise computational extraction of HSVd‐derived sRNA reads from our sRNA library of infected cucumber leaves. (B) Representation of the size distribution of the sequenced vd‐sRNAs. (C) Histogram illustrating the relative abundance of the four different 5′‐terminal nucleotides in the most representative species of vd‐sRNAs.
RNA silencing‐associated sRNAs interact with ARGONAUTE (AGO)‐containing RISCs to guide them to their target molecules. In these complexes, sRNAs are responsible for target specificity, whereas AGOs determine the RNA silencing effects (Kim, 2008; Ruiz‐Ferrer and Voinnet, 2009). It has been reported recently that, in Arabidopsis, the selective loading of sRNAs into specific AGOs is influenced by their 5′‐terminal nucleotide (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008). Assuming that, in cucumber plants, the corresponding orthologue to the protein family AGO exists, we analysed the relative abundance of sequenced vd‐sRNAs according to their 5′‐terminal nucleotide (Fig. 3C) in order to infer potential interactions with distinct AGO complexes. In contrast with the observations for diverse plant virus‐specific sRNAs, which display a clear tendency to begin with U or A (Donaire et al., 2009; Qi et al., 2009), vd‐sRNAs with a C at their 5′‐end were the most abundant (∼45%), those with U or A at their 5′‐end were similarly recovered (∼22% and 20%, respectively) and those with G at their 5′‐end were under‐represented (∼11%) in the sequenced pool. These results suggest that vd‐sRNAs may be potentially loaded into diverse AGO‐containing silencing complexes in cucumber plants.
dsRNA plays a relevant role as one of the main precursors of vd‐sRNAs
To examine the genomic distribution of the vd‐sRNA set, all sequences recovered from cucumber leaves were mapped along the HSVd genome. The 5′‐ends of the vd‐sRNAs (21–24 nucleotides) were plotted against the HSVd sequence according to their polarity and abundance (Fig. 4A and Fig. S1, see Supporting Information). Several features were revealed from this analysis. First, the polarity distribution of the vd‐sRNAs indicated that sense (44.7%) and antisense (55.2%) species were similarly represented in the sequenced pool, providing evidence that the vd‐sRNAs originated from both plus and minus HSVd RNA strands to a similar extent. Our results were in good agreement with the fact that sense and antisense polarities were also equally represented in a population of PSTVd‐derived sRNAs (Machida et al., 2007). Interestingly, in a recent study addressed to characterize sRNAs from grape, Carra et al. (2009) found that the sense and antisense polarities of the siRNAs recovered from the Grapevine yellow speckle viroid were equally represented. In contrast, previous findings obtained by low‐scale sequencing of Citrus exocortis viroid (CEVd)‐ and PSTVd‐derived sRNAs showed that the amount of sense vd‐sRNAs (∼80%) was significantly higher than that of antisense vd‐sRNAs (∼20%) (Markarian et al., 2004; Martín et al., 2007). Second, sense and antisense vd‐sRNAs spread along the entire HSVd genome in an overlapping configuration, with virtually all nucleotide positions in the HSVd RNA being covered by vd‐sRNAs in both polarities (Fig. 4A). Third, vd‐sRNAs from both polarities displayed a heterogeneous distribution pattern along the HSVd sequence, as observed for most plant viruses (Donaire et al., 2009; Qi et al., 2009).
Figure 4.
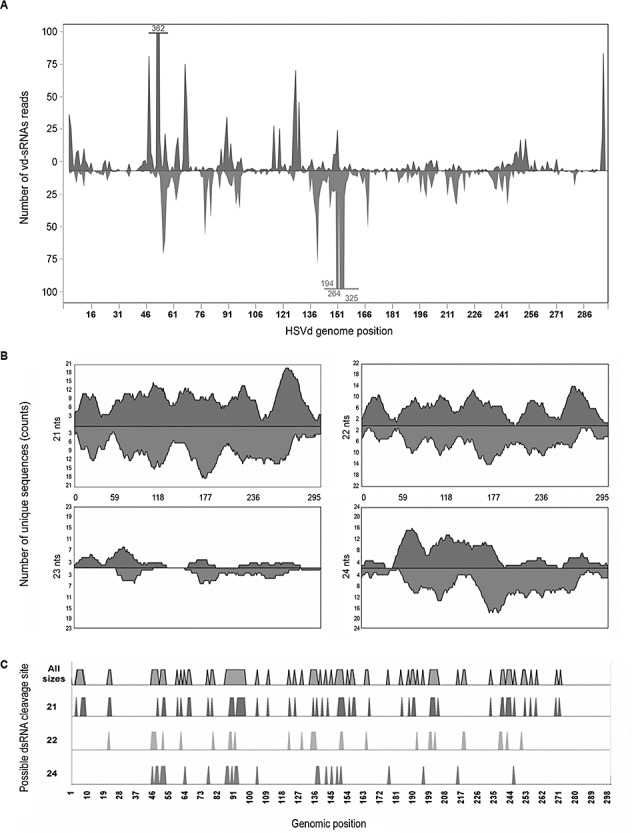
A genome view of Hop stunt viroid (HSVd)‐derived small RNAs (sRNAs) recovered from infected cucumber plants. (A) The viroid‐derived sRNAs (vd‐sRNAs) were plotted according to the position of their 5′‐extremity onto the sequence of HSVd RNA used to infect cucumber plants (Y09352), in either sense (above the x‐axis) or antisense (below the x‐axis) configuration. The values on the y‐axis represent the abundance of vd‐sRNAs in the library. The nucleotide positions 1–297 of HSVd RNA are represented on the x‐axis. (B) A high‐resolution view of the HSVd genome showing the number of plotted unique vd‐sRNAs, classified according to size class (21, 22, 23 and 24 nucleotides). On the y‐axis, the times that a single HSVd nucleotide was present in a sense (above the x‐axis) or antisense (below the x‐axis) unique vd‐sRNA is represented. (C) Sense and antisense pairs of vd‐sRNAs with perfect complementary strands, forming putative Dicer‐like protein (DCL) products generated from dsRNA substrates, were mapped onto the HSVd genome according to the size of the species.
To gain further insight into vd‐sRNA biogenesis, the sequence complexity of HSVd‐derived sRNAs was analysed for different‐sized species. Unique vd‐sRNAs from each of the most representative classes (21–24 nucleotides) were individually plotted onto the HSVd genome, and their sequence complexity was estimated as the number of unique vd‐sRNA sequences that hit each of the single nucleotide positions along the HSVd genome. Using a sliding window of 21, the maximum value of the sequence complexity was 21, which corresponded to the hypothetical formation of vd‐sRNAs starting at any of the 21 nucleotide positions within the window. The highest sequence complexity was observed for the 21‐nucleotide‐sized species, followed by the 22‐ and 24‐nucleotide size classed, whereas the 23‐nucleotide size presented the lowest sequence complexity (Fig. 4B). Interestingly, values close to the highest sequence complexity were found for both sense and antisense 21‐nucleotide vd‐sRNAs in at least one genomic region. Our data indicate that the viroid genome is not uniformly targeted by the different components of the RNA silencing machinery involved in vd‐sRNA biogenesis. The production of 21‐nucleotide vd‐sRNA species is, in general, more efficient at any position of the viroid genome compared with that of the 22‐ and 24‐nucleotide species. Furthermore, regions with higher sequence diversity for a given size class can be found throughout the HSVd genome.
Finally, we tested whether our dataset contained pair(s) of perfectly complementary vd‐sRNAs with two‐nucleotide overhangs at their 3′‐ends. Sense and antisense vd‐sRNAs forming a duplex might be diagnostic of a DCL‐mediated cleavage event on dsRNA substrates. We observed that a high frequency of sense 21‐ (54.7%), 22‐ (47.0%) and 24‐nucleotide (50.3%) vd‐sRNAs showed perfect complementarity with their antisense counterparts (Fig. 4C), suggesting that dsRNAs might be one of the main precursors for these highly represented vd‐sRNAs.
Phloem‐derived vd‐sRNA accumulates preferentially as 22‐nucleotide species
In order to obtain a comprehensive picture of the vd‐sRNA species present in the phloem tissue, we analysed independently the 39 026 sequences of sRNAs recovered from the phloem sap of infected cucumber plants. A total of 1278 (3.27%) vd‐sRNAs perfectly complementary to HSVd were recovered from phloem exudates (herein referred to as Pvd‐sRNAs) (Fig. 5A). Our data indicate that Pvd‐sRNAs represented in the sequenced pool are of both sense (54.2%) and antisense (45.8%) orientation.
Figure 5.
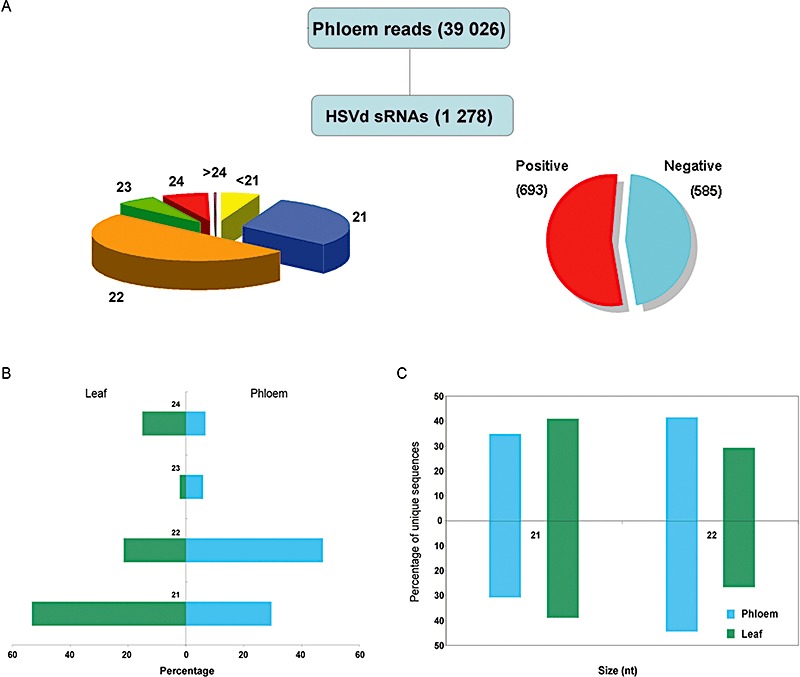
Analysis of viroid‐derived small RNAs (vd‐sRNAs) recovered from the phloem sap of infected cucumber plants. (A) Size distribution of the vd‐sRNA population detected in the phloem of infected cucumber plants. The proportional accumulation of sense and antisense vd‐sRNAs is also illustrated. (B) Relative abundance of the most representative size classes of sense and antisense vd‐sRNAs recovered from leaves (left) and phloem (right). (C) Accumulation values in leaves and phloem of unique sense and antisense 21‐ and 22‐nucleotide vd‐sRNAs.
A striking feature of vd‐sRNAs derived from phloem exudates concerns their differential size distribution. Although vd‐sRNA from leaf tissue was predominantly 21 nucleotides long, Pvd‐sRNA of 22 nucleotides was the most representative sized species (47%) compared with the 21‐nucleotide size class (21.7%) (Fig. 5B). A similar bias in size distribution was observed when unique 21‐ and 22‐nucleotide vd‐sRNA species were analysed (Fig. 5C). Although the 23‐ and 24‐nucleotide size species were poorly represented in both phloem and leaf libraries, a substantial increase in 23‐nucleotide vd‐sRNAs and a decrease in 24‐nucleotide species were observed in phloem exudates when compared with vd‐sRNAs from cucumber leaves (Fig. 5B). In conclusion, we observed a clear bias in the composition of vd‐sRNAs from phloem with respect to that observed in infected cucumber leaves. Interestingly, an increase in the relative accumulation of the 22‐nucleotide class in phloem (17.62%) compared with leaves (13.24%) was observed for endogenous sRNAs in infected plants (Fig. 7A, right panel), whereas this ratio was inverted in healthy plants, for which a decrease of approximately 50% was observed in the relative accumulation of 22‐nucleotide sRNAs in the phloem (15.07%) relative to leaves (32.72%) (Fig. 7A, left panel).
Figure 7.
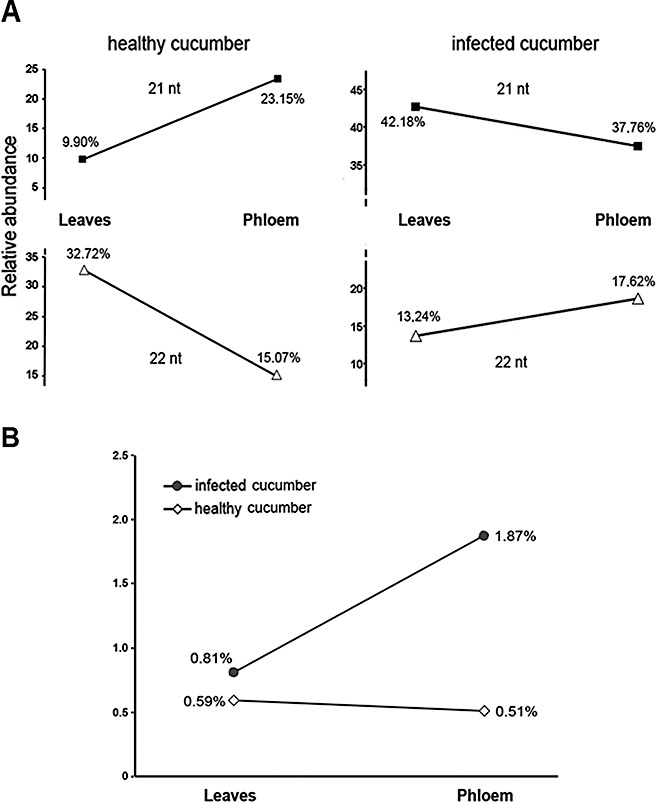
Comparative analysis of the cucumber endogenous small RNAs (sRNAs) accumulated in the leaves and phloem of healthy and infected plants. A pool of sRNAs enriched from total RNA preparations was extracted from the phloem of healthy cucumber plants, processed and subjected to multiplexed pyrosequencing, as explained in Experimental procedures. A total of 1783 unique high‐quality sequences, each containing endogenous sRNAs, was obtained. (A) Graph shows a comparative analysis of the relative abundance of the two most representative size classes (21 and 22 nucleotides) of endogenous sRNAs recovered from leaves and phloem in healthy (left panel) and infected (right panel) cucumber plants. (B) Graph shows a comparative analysis of the relative abundance of the endogenous sRNAs carrying the sequence signature AGAnGCG recovered from the leaves and phloem of healthy and infected plants.
A consensus sequence is over‐represented in phloem vd‐sRNAs
Finally, we investigated the genomic distribution and origin of Pvd‐sRNAs obtained from phloem exudates. The 5′‐end of each Pvd‐sRNA (21–24 nucleotides) was plotted against the HSVd sequence according to their polarity and abundance (Fig. 6A and Fig. S2, see Supporting Information). Although Pvd‐sRNAs were derived from all regions along the HSVd genome in both orientations, a high proportion of Pvd‐sRNAs (34.1%) was restricted around two well‐differentiated hot‐spots, designated as phloem hot‐spots 1 (phs‐1) and 2 (phs‐2) (Fig. 6A and Fig. S2, see Supporting Information). The 5′‐termini of the 297 (23.3% of phloem library) Pvd‐sRNA reads clustering in phs‐1 localized between positions 197 and 207 of HSVd RNA. phs‐2, which encompassed 138 Pvd‐sRNA reads (10.8% of library), was restricted to positions 251–255. Interestingly, as illustrated in Fig. 7B, high‐resolution mapping revealed that phs‐1 and phs‐2 did not correlate with any of the remaining vd‐sRNA hot‐spots observed in the dataset obtained from leaves. Furthermore, phs‐1 and phs‐2 colocalized in the HSVd genome with regions of high sequence complexity estimated from unique Pvd‐sRNAs (Fig. S3, see Supporting Information), which argued against the possibility that the observed differences could be a result of over‐representation of specific Pvd‐sRNA sequences owing to a bias in the sequencing procedure. Together, our data analysis, including size and genome distribution, suggests that the population of vd‐sRNAs recovered from the phloem is substantially different to that observed in infected cucumber leaves.
Figure 6.
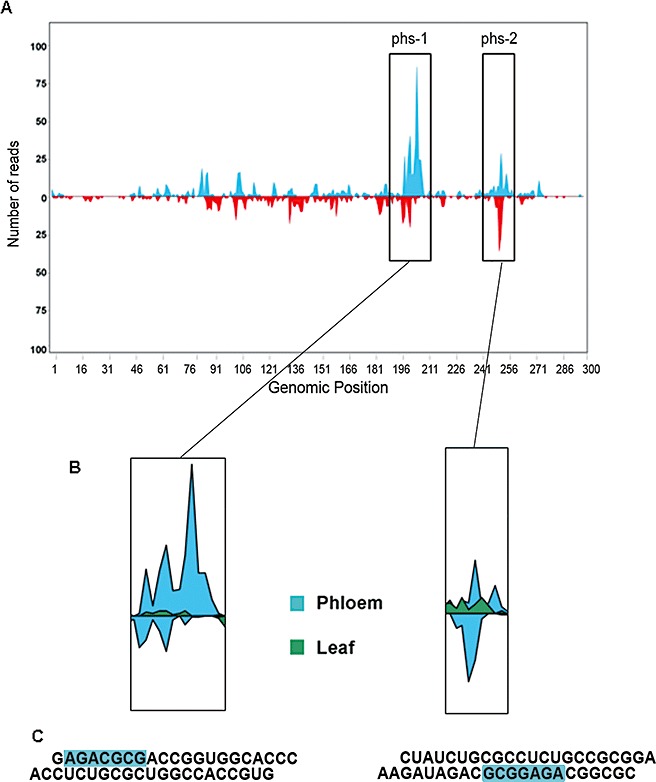
Sequence analysis of the Hop stunt viroid (HSVd)‐derived small RNAs (sRNAs) highly accumulated in phloem tissue. (A) The viroid‐derived sRNAs (vd‐sRNAs) recovered from the phloem of infected cucumber plants were plotted according to the position of their 5′‐extremity onto HSVd RNA, in either the sense (above the x‐axis) or antisense (below the x‐axis) configuration. (B) High‐resolution genome view of the vd‐sRNA‐accumulating hot‐spots observed in the phloem sRNAs. The maps of the vd‐sRNAs recovered from leaves and phloem were overlapped to analyse the relative abundances of each sRNA in the different tissues. (C) Two representative phloem vd‐sRNAs clustering in hot‐spots 1 and 2, respectively, were identified and aligned according to their sequence. The common sequence signature AGAnGCG conserved in the highly accumulated phloem vd‐sRNAs is boxed. The phloem vd‐sRNAs are illustrated according to the sequence of the plus strand HSVd RNA.
A detailed analysis of the sequence composition within the subset of Pvd‐sRNAs from phs‐1 and phs‐2 revealed a common AGAnGCG sequence signature that was not observed for Pvd‐sRNAs derived from other regions of the HSVd genome (Fig. 6C and Fig. S4, see Supporting Information). This sequence was not exclusive for 22‐nucleotide species as the two highest 21‐nucleotide vd‐sRNAs represented (∼50%) in the phloem dataset contained a AGAnGCG signature (Fig. S3, see Supporting Information). This common sequence signature was also over‐represented in the nonviroid‐related sRNAs recovered from infected phloem (1.87%) relative to the endogenous sRNAs recovered from leaves (0.81%) (Fig. 7B). These findings suggest the existence, in infected cucumber plants, of a selective predisposition to accumulate in phloem sRNAs, preferentially 22 nucelotides long, carrying this consensus sequence.
DISCUSSION
Infection by members of the Pospiviroidae family is associated with the presence of vd‐sRNAs (e.g. Itaya et al., 2001; Papaefthimiou et al., 2001). However, the mechanisms potentially involved in the biogenesis of vd‐sRNAs are still a matter of speculation. Previously, it was proposed that dsRNA viroid replication intermediates (Denti et al., 2004; Wang et al., 2004) and/or highly structured regions present in the viroid mature forms (Itaya et al., 2007; Markarian et al., 2004; Martín et al., 2007; St‐Pierre et al., 2009) could serve as potential DCL substrates for the biosynthesis of vd‐sRNAs. Nevertheless, whether the biogenesis of vd‐sRNAs is a DCL‐dependent process remains to be demonstrated experimentally. Taking into account these previous data, alternative mechanisms of sRNA biosynthesis, such as unprimed RNA synthesis directed by RDR activity, as described in Caenorhabditis elegans (Pak and Fire, 2007; Sijen et al., 2007), cannot be excluded.
In this paper, we used deep sequencing to obtain an exhaustive picture of the diversity and abundance of viroid‐specific sRNAs in infected tissue. Our sequence analysis represented a high‐resolution map for vd‐sRNAs derived from a nuclear viroid (HSVd), providing an essential tool to understand the biogenesis pathways of RNA silencing involved in their generation.
The profiling of the sequenced vd‐sRNAs from infected cucumber plants indicated that the majority of vd‐sRNAs could be classified as 21‐, 22‐ and 24‐nucleotide size classes, which coincided with the three major classes predicted by the coordinated hierarchical action of DCL4, DCL2 and DCL3 (Bouche et al., 2006; Deleris et al., 2006). The presence of 5′‐monophosphate ends in vd‐sRNAs, inferred from the initial ligation with T4 RNA ligase, suggests that the majority of the sRNAs reads in our sequenced pool are true DCL products. In addition, the ubiquitous distribution of sense and antisense vd‐sRNA species and their similar ratio observed in our libraries can be easily envisioned as a consequence of DCL (or orthologous proteins present in cucumber plants) processing of perfectly complementary viroid‐derived dsRNAs. Multiple sense/antisense pairs with 3′‐overhangs were identified among the sequences in our HSVd dataset, suggesting that they corresponded to the complementary strands originating from single DCL‐mediated processing events.
Our results on the polarity distribution are in apparent contradiction with those of previous studies, reporting a predominant accumulation of sense vd‐sRNAs in tomato plants infected with members of the Pospiviroidae family (Itaya et al., 2007; Martín et al., 2007). We believe that this difference is probably a result of the relatively small number of vd‐sRNA sequences analysed in those studies. However, the possibility that differences in polarity distribution could be a consequence of functional differences between the uncharacterized tomato and cucumber RNA silencing mechanisms cannot be excluded.
These results together indicate that the entire viroid genome is targeted by the RNA silencing machinery to produce vd‐sRNAs, and suggest a model whereby vd‐sRNAs of both polarities may be mostly produced through DCL‐mediated processing of long, viroid‐derived dsRNA. Therefore, this argues against, but does not exclude, a model in which local secondary structures within the positive‐strand HSVd RNA are the principal precursors of vd‐sRNA biosynthesis (Itaya et al., 2007; Martín et al., 2007). The wide spectrum of different vd‐sRNAs suggests that the process by which these sRNAs are generated is more complex than previously thought, probably involving different biogenesis pathways and subcellular compartments.
The viroid‐derived sRNAs recovered in our libraries showed a strong bias for sequences beginning with a 5′‐cytosine, indicative of sRNAs with high binding affinity for AGO5 in Arabidopsis (Mi et al., 2008; Takeda et al., 2008). The role of AGO5 in RNA silencing remains unclear and its activity in plant defence has never been established (Brodersen and Voinnet, 2006; Chapman and Carrington, 2007; Ruiz‐Ferrer and Voinnet, 2009). The potential preference of vd‐sRNAs for an AGO without a demonstrated antiviral role is consistent with the recently suggested model by which the loading of vd‐sRNAs to AGOs with little or no intrinsic activity in the defence mechanism could contribute to prevent the RISC‐mediated degradation of viroid mature forms (Ruiz‐Ferrer and Voinnet, 2009). This mechanism, in conjunction with the apparent resistance of structured viroid genomes to RISC activity (Gómez and Pallás, 2007; Itaya et al., 2007), could explain how viroid mature forms can overcome RNA silencing‐mediated degradation in infected plants.
In plants, RNA silencing spreads systemically to distant tissues, travelling through sieve elements following a source to sink pattern (Palauqui et al., 1997; Voinnet et al., 1998). Although it is commonly accepted that it should be an RNA molecule (Jorgensen, 2002), the identity of the phloem‐transported signal and the mechanism whereby the signal is translocated are currently unknown (Dunoyer and Voinnet, 2008; Kalantidis et al., 2008). To provide a more comprehensive picture of the sRNAs that occur in the plant tissue responsible for long‐distance communication, we performed a detailed analysis of the vd‐sRNAs recovered from the phloem of infected cucumber plants. Unlike that observed in leaves, the phloem sap of infected cucumber appears to contain a preponderant accumulation of 22‐nucleotide vd‐sRNAs. This divergence in the general profile of ‘phloem’ Pvd‐sRNAs is relevant, considering that the 21‐nucleotide size class is the highest represented species in infected leaves. As a result, the ratio of the 21‐ to 22‐nucleotide classes within the pool of vd‐sRNAs was inverted from 1.8 : 1 in leaves to 1 : 2.3 in phloem. To date, DCL activity has not been described within the phloem. If this activity does not exist in that tissue, it would be reasonable to assume that Pvd‐sRNAs could be generated in nonvascular tissues and subsequently transported to sieve elements (Yoo et al., 2004). In consequence, the bias in the relative accumulation of the 22‐nucleotide class in the phloem of infected plants could be considered as an indirect indicator of selective trafficking of 22‐nucleotide vd‐sRNAs from their sites of biogenesis to the phloem. Interestingly, it has been reported recently that 22‐nucleotide RNAs are able to direct the systemic silencing of a green fluorescent protein (GFP) construct from transgenic stocks to wild‐type scions in Arabidopsis plants (Brosnan et al., 2007). However, we cannot exclude the possibility that this bias in the accumulation of the 22‐nucleotide class in the phloem could be a result of the differential processing of the 22‐nucleotide sRNAs in the companion cells of infected cucumber plants. Further studies, providing a detailed analysis of the relative accumulation of sRNAs in phloem and nonphloem tissues of infected and healthy cucumber plants, are required to elucidate this question.
High‐resolution mapping of phloem‐derived vd‐sRNAs revealed a genome distribution that differed from the genome‐wide spreading of vd‐sRNA recovered from infected leaves. vd‐sRNAs from phloem exudates were mainly clustered into two well‐defined regions that did not correlate with any of the conspicuous hot‐spots of vd‐sRNA accumulation identified in the leaf dataset. Interestingly, the most highly represented Pvd‐sRNAs possessed a common AGAnGCG sequence signature in the vd‐sRNAs. It is therefore tempting to speculate that this sequence signature may favour the translocation of vd‐sRNAs into the phloem, possibly by acting as a more efficient target for specific sRNA‐binding proteins involved in sRNAs trafficking from companion cell to sieve element (Yoo et al., 2004).
In general, our results permit us to envision a scenario in which the accumulation of vd‐sRNAs in the phloem could be regulated at two different, but complementary, levels, size and sequence, with vd‐sRNAs of 22 nucleotides that contain the signature sequence being preferentially uploaded into vascular tissue. Additional studies are necessary to establish the importance and exact physiological role of this selective trafficking to the phloem, and to elucidate whether it can also be extended to plant endogenous and/or virus‐derived small RNAs.
EXPERIMENTAL PROCEDURES
Plant material
Forty cucumber (Cucumis sativus cv. Suyo) plants were agroinoculated with Agrobacterium tumefaciens strain C58C1 transformed with a binary pMOG800 vector carrying a head‐to‐tail infectious dimeric HSVd cDNA (Y09352) (Gómez and Pallás, 2006), as described previously (Gómez et al., 2008). Ten noninfected cucumber plants were used as negative controls. The plants were maintained in environmentally controlled growth chambers at 30 °C for 16 h with fluorescent light and at 25 °C for 8 h in darkness, and analysed at 35 days post‐inoculation (dpi).
RNA isolation
Pools of leaves (∼50 g) obtained from different infected (40) and healthy (10) cucumber plants were used. Total RNA was extracted from this pool of infected and healthy cucumber leaves (∼10 g) using TRI reagent (Sigma, St. Louis, MO, USA) according to the manufacturer's instructions. Phloem RNA sampling from infected and healthy cucumber plants was performed as described previously (Gómez and Pallás, 2004; Gómez et al., 2005). Phloem sap (∼100 µL) was collected from groups of two (healthy) or four (infected) plants by multiple cuts in petioles or shoot apices directly onto 1 mL of TRI reagent (Sigma) and processed according to the manufacturer's instructions. Pools of approximately 1 mL of phloem sap obtained from healthy and infected cucumber plants were used in this assay. The total RNA preparations were quantified by spectrometry and their concentration was equalled.
Northern blot assays
Total RNA was electrophoresed under denaturing conditions in 5% polyacrylamide gels with 0.25 × TBE (Tris 89 mM, Boric acid 89mM, EDTA 2mM) and 8 m urea (Pallás et al., 1987). RNA was blotted onto nylon membranes (Roche Diagnostics GmbH, Mannheim, Germany) and hybridized as described previously (Gómez and Pallás, 2001). The low‐molecular‐weight RNA (<200 nucleotide) fraction was enriched using total RNA and MIRACLE (miRNA Isolation Kit, Stratagene, La Jolla, CA, USA) according to the manufacturer's instructions. Approximately 25 µg of low‐molecular‐weight RNA was loaded onto 20% polyacrylamide gels with 0.25 × TBE and 8 m urea. RNA was transferred to a nylon membrane (Roche Diagnostics GmbH). Hybridization was performed at 32.5 °C for 14–16 h, using a digoxigenin‐labelled, negative‐strand HSVd RNA as a probe. The detection was performed as described previously (Gómez and Pallás, 2007).
vd‐RNA amplification and sequencing
Total RNAs (∼300 µg) extracted from leaves and phloem exudates of HSVd‐infected cucumber plants were used for the construction of sRNA libraries, as described previously (Kasschau et al., 2007), with the following minor modifications. The 3′‐adapter was replaced by a pre‐activated 5′‐adenylated oligo (5′‐rAppCTGTAGGCACCATCAAT3ddC‐3′) (Integrated DNA Technologies, Coralville, IO, USA) to avoid the circularization of sRNAs. The chimeric RNA/DNA oligonucleotide 5′‐adapters have been described previously (Kasschau et al., 2007). A new adapter variant was generated by modification of the four‐nucleotide identifier (3‐1, ATCGTAGACGCCUGAUA). After each ligation step, sRNA was purified using 17% denaturing polyacrylamide gel electrophoresis (PAGE). The purified ligated sRNA was reverse transcribed and the cDNA was amplified using Taq DNA polymerase (Perkin‐Elmer, Applied Biosystems Division, Norwalk, CT, USA) and 3′‐PCR FusionB and 5′‐PCR FusionA primers (Kasschau et al., 2007). PCR primers contained the ‘A’ and ‘B’ tag sequences used by 454 Life Science during sequencing. DNA amplicons were gel purified using 12% native polyacrylamide and eluted in 0.3 m NaCl, as described previously (Donaire et al., 2008). The quantity and quality of DNA amplicons were measured using an ND‐1000 spectrophotometer (Nanodrop, Thermo Scientific Inc., Wilmington, DE, USA) and Experion Automated Electrophoresis System (Bio‐Rad, Hercules, CA, USA), respectively. The same quantity of DNA amplicon from each library was pooled and sequenced by 454 Life Science Technology (Lifesequencing, Branfor, CT, USA; http://www.lifesequencing.com). The sRNA sequences discussed in this article are archived at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under Accession code SRP001408.
Data mining of the sRNA pool and sequence analysis
Library identification, adapter trimming and cleaning of the reads, as well as library comparison, were performed by Perl scripts locally developed by the Bioinformatics Service at the Instituto de Biología Molecular y Celular de Plantas (IBMCP), Valencia, Spain (http://www.ibmcp.upv.es). Eventual identification of HSVd‐specific sRNAs, as well as their location in the viroid genome, was performed using a local installation of the blast program. Data analysis was performed using STatGRAPHICS plus 5.1 software (http://www.statgraphics.com).
Supporting information
Fig. S1 The most represented viroid‐derived small RNA (vd‐sRNA) species (21–24 nucleotides) recovered from infected cucumber plants were plotted individually according to the position of their 5′‐extremity onto Hop stunt viroid (HSVd) RNA in either sense (above the x‐axis) or antisense (below the x‐axis) configuration. The values on the y‐axis represent the abundance of vd‐sRNAs in the library. The nucleotide positions 1–297 of HSVd are represented on the x‐axis.
Fig. S2 The sense and antisense viroid‐derived small RNAs (vd‐sRNAs) specifically recovered from leaves (left panel) and phloem exudate (right panel) of infected cucumber plants were classified according to their size class and plotted separately onto the Hop stunt viroid (HSVd) genome. Graphic contents are as described in Fig. 3.
Fig. S3 Analysis of the most represented phloem 22‐nucleotide viroid‐derived small RNAs (vd‐sRNAs). (A) High‐resolution view of the vd‐sRNAs clustering in the sRNA‐accumulating hot‐spots 1 and 2 (shown above). (B) Sense and antisense unique 22‐nucleotide vd‐sRNAs were plotted onto the Hop stunt viroid (HSVd) genome. (C) Sequence complexity of the 22‐nucleotide vd‐sRNAs in the surrounding regions of the hot‐spots. The values of sequence complexity were obtained as explained in the text.
Fig. S4 Identification of the specific signature sequence in the phloem viroid‐derived small RNAs (Pvd‐sRNAs). The vd‐sRNAs clustering in the sRNA‐accumulating hot‐spots 1 and 2 were identified and aligned according to their sequence. The common sequence signature AGAnGCG conserved in the highly accumulated Pvd‐sRNAs is boxed. The Pvd‐sRNAs are illustrated according to the sequence of the plus strand Hop stunt viroid (HSVd) RNA. Pvd‐sRNAs in black correspond to the sequenced strand; vd‐sRNAs in grey represent their predicted complementary strand.
Table S1 Sequences of the 5621 Hop stunt viroid (HSVd)‐derived small RNAs recovered from leaves and phloem sap of infected cucumber plants. The sense and antisense viroid‐derived small RNAs (vd‐sRNAs) are marked with ‘+’ and ‘−’, respectively.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We thank Dr J. Forment (Bioinformatics Service of IBMCP) for his valuable contribution to the sequence analysis. This work was supported by grants BIO2008‐03528 and BIO2006‐13107 to VP and CL, respectively, from the Spanish granting agency Dirección General de Investigación Científica y Técnica, and by grant GV05‐238 from the Generalitat Valenciana to GG. GM and LD are the recipients of a fellowship from the Ministry of Science and Innovation. GG is the recipient of a contract from Consejo Superior de Investigaciones Científicas.
REFERENCES
- Aung, K. , Lin, S.I. , Wu, C.C. , Huang, Y.T. , Su, C.I. and Chiou, T.J. (2006) pho2, a phosphate over accumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari, R. , Pant, B.D. , Stitt, M. and Scheible, W.R. (2006) PHO2, microRNA399, and PHR1 define a phosphate‐signaling pathway in plants. Plant Physiol. 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Bouche, N. , Lauressergues, D. , Gasciolli, V. and Vaucheret, H. (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25, 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P. and Voinnet, O. (2006) The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280. [DOI] [PubMed] [Google Scholar]
- Brosnan, C.A. , Mitter, N. , Christie, M. , Smith, N.A. , Waterhouse, P.M. and Carrol, B.J. (2007) Nuclear gene silencing directs reception of long‐distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA, 104, 14741–14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz, A. , Springer, F. , Chappell, L. , Baulcombe, D. and Kehr, J. (2008) Identification and characterization of small RNAs from the phloem of Brassica napus . Plant J. 53, 739–749. [DOI] [PubMed] [Google Scholar]
- Carbonell, A. , Martínez de Alba, A.E. , Flores, R. and Gago, S. (2008) Double‐stranded RNA interferes in a sequence‐specific manner with the infection of representative members of the two viroid families. Virology, 371, 44–53. [DOI] [PubMed] [Google Scholar]
- Carra, A. , Mica, E. , Gambino, G. , Pindo, M. , Moser, C. , Pè, M. and Schubert, A. (2009) Cloning and characterization of small non‐coding RNAs from grape. Plant J. 59, 750–753. [DOI] [PubMed] [Google Scholar]
- Chapman, E.J. and Carrington, J.C. (2007) Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8, 884–896. [DOI] [PubMed] [Google Scholar]
- Daròs, J.A. , Elena, S.F. and Flores, R. (2006) Viroids: an Ariadne's thread into the RNA labyrinth. EMBO Rep. 7, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris, A. , Gallego‐Bartolome, J. , Bao, J. , Kasschau, K.D. , Carrington, J.C. and Voinnet, O. (2006) Hierarchical action and inhibition of plant Dicer‐like proteins in antiviral defense. Science, 313, 68–71. [DOI] [PubMed] [Google Scholar]
- Denti, M.A. , Boutla, A. , Tsagris, M. and Tabler, M. (2004) Short interfering RNAs specific for Potato spindle tuber viroid are found in the cytoplasm but not in the nucleus. Plant J. 37, 762–769. [DOI] [PubMed] [Google Scholar]
- Ding, B. (2009) Biology of viroid–host interactions. Annu. Rev. Phytopathol. 47, 105–131. [DOI] [PubMed] [Google Scholar]
- Donaire, L. , Barajas, D. , Martinez‐Garcia, B. , Martinez‐Priego, L. , Pagan, I. and Llave, C. (2008) Structural and genetic requirements for the biogenesis of Tobacco rattle virus‐derived small interfering RNAs. J. Virol. 82, 5167–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaire, L. , Wang, Y. , Gonzalez‐Ibeas, D. , Mayer, K. , Aranda, M. and Llave, C. (2009) Deep‐sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology, 30, 203–214. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P. and Voinnet, O. (2008) Mixing and matching: the essence of plant systemic silencing? Trends Genet. 24, 151–154. [DOI] [PubMed] [Google Scholar]
- Flores, R. , Hernández, C. , Martínez de Alba, A.E. , Daròs, J.A. and Di Serio, F. (2005) Viroids and viroid–host interactions. Ann. Rev. Phytopathol. 43, 117–139. [DOI] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2001) Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant–Microbe Interact. 14, 910–913. [DOI] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2004) A long‐distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with Hop stunt viroid RNA. J. Virol. 78, 10104–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2006) Hop stunt viroid is processed and translocated in transgenic N. benthamiana plants. Mol. Plant Pathol. 7, 511–517. [DOI] [PubMed] [Google Scholar]
- Gómez, G. and Pallás, V. (2007) Mature monomeric forms of Hop stunt viroid resist RNA silencing in transgenic plants. Plant J. 51, 1041–1049. [DOI] [PubMed] [Google Scholar]
- Gómez, G. , Torres, H. and Pallás, V. (2005) Identification of translocatable RNA‐binding phloem proteins from melon, potential components of the long‐distance RNA transport system. Plant J. 41, 107–116. [DOI] [PubMed] [Google Scholar]
- Gómez, G. , Martinez, G. and Pallas, V. (2008) Viroid‐induced symptoms in Nicotiana benthamiana plants are dependent on RDR6 activity. Plant Physiol. 148, 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, G. , Martinez, G. and Pallás, V. (2009) Interplay between viroid‐induced pathogenesis and RNA silencing pathways. Trends Plant Sci. 14, 264–269. [DOI] [PubMed] [Google Scholar]
- Itaya, A. , Folimonov, A. , Matsuda, Y. , Nelson, R.S. and Ding, B. (2001) Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant–Microbe Interact. 14, 1332–1334. [DOI] [PubMed] [Google Scholar]
- Itaya, A. , Zhong, X. , Bundschuh, R. , Qi, Y. , Wang, Y. , Takeda, R. , Harris, A.R. , Molina, C. , Nelson, R.S. and Ding, B. (2007) A structured viroid RNA is substrate for Dicer‐like cleavage to produce biologically active small RNAs but is resistant to RISC‐mediated degradation. J. Virol. 81, 2980–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A. (2002) RNA traffics information systemically in plants. Proc. Natl. Acad. Sci. USA, 99, 11561–11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantidis, K. , Schumacher, H.T. , Alexiadis, T. and Helm, J.M. (2008) RNA silencing movement in plants. Biol. Cell, 100, 13–26. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. , Fahlgren, N. , Chapman, E.J. , Sullivan, C.M. , Cumbie, J.S. , Givan, S.A. and Carrington, J.C. (2007) Genome‐wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, V.N. (2008) Sorting out small RNAs. Cell, 133, 25–26. [DOI] [PubMed] [Google Scholar]
- Machida, S. , Yanahata, N. , Watanuki, H. , Owens, R.A. and Sano, T. (2007) Successive accumulation of two size classes of viroid‐specific small RNA in Potato spindle tuber viroid‐infected tomato plants. J. Gen. Virol. 88, 3452–3457. [DOI] [PubMed] [Google Scholar]
- Markarian, N. , Li, H.W. , Ding, S.W. and Semancik, J.S. (2004) RNA silencing as related to viroid induced symptom expression. Arch. Virol. 149, 397–406. [DOI] [PubMed] [Google Scholar]
- Martín, R. , Arenas, C. , Daròs, J.A. , Covarrubias, A. , Reyes, J.L. and Chua, N. (2007) Characterization of small RNAs derived from Citrus exocortis viroid in infected tomato plants. Virology, 367, 135–146. [DOI] [PubMed] [Google Scholar]
- Martinez de Alba, A.E. , Flores, R. and Hernandez, C. (2002) Two chloroplastic viroids induce the accumulation of the small RNAs associated with post‐transcriptional gene silencing. J. Virol. 76, 13094–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoušek, J. , Kozlová, P. , Orctová, L. , Schmitz, A. , Pesina, K. , Bannach, O. , Diermann, N. , Steger, G. and Riesner, D. (2007) Accumulation of viroid‐specific small RNAs and increase in nucleolytic activities linked to viroid‐caused pathogenesis. Biol. Chem. 388, 1–13. [DOI] [PubMed] [Google Scholar]
- Mi, S. , Cai, T. , Hu, Y. , Chen, Y. , Hodges, E. , Ni, F. , Wu, L. , Li, S. , Zhou, H. , Long, C. , Chen, S. , Hannon, G.J. and Qi, Y. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell, 133, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, T.A. , Howell, M.D. , Cuperus, J.T. , Li, D. , Hansen, J.E. , Alexander, A.L. , Chapman, E.J. , Fahlgren, N. , Allen, E. and Carrington, J.C. (2008) Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans‐acting siRNA formation. Cell, 133, 128–141. [DOI] [PubMed] [Google Scholar]
- Pak, J. and Fire, A. (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science, 315, 241–244. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C. , Elmayan, T. , Pollien, J.M. and Vaucheret, H. (1997) Systemic acquired silencing: transgene‐specific post‐transcriptional silencing is transmitted by grafting from silenced stocks to non‐silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallás, V. , Navarro, A. and Flores, R. (1987) Isolation of a viroid‐like RNA from Hop different to Hop stunt viroid . J. Gen. Virol. 68, 3201–3205. [Google Scholar]
- Papaefthimiou, I. , Hamilton, A.J. , Denti, M.A. , Baulcombe, D.C. , Tsagris, M. and Tabler, M. (2001) Replicating Potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post‐transcriptional gene silencing. Nucleic Acids Res. 29, 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X. , Bao, F.S. and Xie, Z. (2009) Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA‐dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE, 4, e4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, V. and Chen, X. (2008) Small RNA metabolism in Arabidopsis. Trends Plant Sci. 13, 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. and Voinnet, O. (2009) Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. [DOI] [PubMed] [Google Scholar]
- Schiebel, W. , Pélissier, T. , Riedel, L. , Thalmeir, S. , Schiebel, R. , Kempe, D. , Lottspeich, F. , Sänger, H.L. and Wassenegger, M. (1998) Isolation of an RNA‐directed RNA polymerase‐specific cDNA clone from tomato. Plant Cell, 10, 2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T. , Steiner, F.A. , Thijssen, K.L. and Plasterk, R.H. (2007) Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science, 315, 244–247. [DOI] [PubMed] [Google Scholar]
- St‐Pierre, P. , Hassen, F.I. , Thompson, D. and Perreault, J.P. (2009) Characterization of the siRNAs associated with peach latent mosaic viroid infection. Virology, 383, 178–182. [DOI] [PubMed] [Google Scholar]
- Takeda, A. , Iwasaki, S. , Watanabe, T. , Utsumi, M. and Watanabe, Y. (2008) The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49, 493–500. [DOI] [PubMed] [Google Scholar]
- Tsagris, M. , Martinez de Alba, A.E. , Gozmanova, M. and Kalantidis, K. (2008) Viroids. Cell Microbiol. 10, 2168–2179. [DOI] [PubMed] [Google Scholar]
- Válóczi, A. , Várallyay, E. , Kauppinen, S. , Burgyán, J. and Havelda, Z. (2006) Spatio‐temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 47, 140–151. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. (2006) Post‐transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771. [DOI] [PubMed] [Google Scholar]
- Vogt, U. , Pelissier, T. , Putz, A. , Razvi, F. , Fischer, R. and Wassenneger, M. (2004) Viroid‐induced RNA silencing of GFP–viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 38, 107–118. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2008) Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13, 317–328. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Vain, P. , Angel, S. and Baulcombe, D. (1998) Systemic spread of sequence specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promotorless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Wang, M.B. , Bian, X.Y. , Wu, L.M. , Liu, L.X. , Smith, N.A. , Isenegger, D. , Wu, R.M. , Masuta, C. , Vance, V.B. , Watson, J.M. , Rezaian, A. , Dennis, E.S. and Waterhouse, P.M. (2004) On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA, 101, 3275–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M. and Krczal, G. (2006) Nomenclature and functions of RNA‐directed RNA polymerases. Trends Plant Sci. 11, 142–151. [DOI] [PubMed] [Google Scholar]
- Yoo, B.C. , Kragler, F. , Varkonyi‐Gasic, E. , Haywood, V. , Archer‐Evans, S. , Lee, Y.M. , Lough, T.J. and Lucas, W.J. (2004) A systemic small RNA signaling system in plants. Plant Cell, 16, 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The most represented viroid‐derived small RNA (vd‐sRNA) species (21–24 nucleotides) recovered from infected cucumber plants were plotted individually according to the position of their 5′‐extremity onto Hop stunt viroid (HSVd) RNA in either sense (above the x‐axis) or antisense (below the x‐axis) configuration. The values on the y‐axis represent the abundance of vd‐sRNAs in the library. The nucleotide positions 1–297 of HSVd are represented on the x‐axis.
Fig. S2 The sense and antisense viroid‐derived small RNAs (vd‐sRNAs) specifically recovered from leaves (left panel) and phloem exudate (right panel) of infected cucumber plants were classified according to their size class and plotted separately onto the Hop stunt viroid (HSVd) genome. Graphic contents are as described in Fig. 3.
Fig. S3 Analysis of the most represented phloem 22‐nucleotide viroid‐derived small RNAs (vd‐sRNAs). (A) High‐resolution view of the vd‐sRNAs clustering in the sRNA‐accumulating hot‐spots 1 and 2 (shown above). (B) Sense and antisense unique 22‐nucleotide vd‐sRNAs were plotted onto the Hop stunt viroid (HSVd) genome. (C) Sequence complexity of the 22‐nucleotide vd‐sRNAs in the surrounding regions of the hot‐spots. The values of sequence complexity were obtained as explained in the text.
Fig. S4 Identification of the specific signature sequence in the phloem viroid‐derived small RNAs (Pvd‐sRNAs). The vd‐sRNAs clustering in the sRNA‐accumulating hot‐spots 1 and 2 were identified and aligned according to their sequence. The common sequence signature AGAnGCG conserved in the highly accumulated Pvd‐sRNAs is boxed. The Pvd‐sRNAs are illustrated according to the sequence of the plus strand Hop stunt viroid (HSVd) RNA. Pvd‐sRNAs in black correspond to the sequenced strand; vd‐sRNAs in grey represent their predicted complementary strand.
Table S1 Sequences of the 5621 Hop stunt viroid (HSVd)‐derived small RNAs recovered from leaves and phloem sap of infected cucumber plants. The sense and antisense viroid‐derived small RNAs (vd‐sRNAs) are marked with ‘+’ and ‘−’, respectively.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Supporting info item
Supporting info item


