SUMMARY
Loss‐of‐function alleles of plant‐specific MLO (Mildew Resistance Locus O) genes confer broad‐spectrum powdery mildew resistance in monocot (barley) and dicot (Arabidopsis thaliana, tomato) plants. Recessively inherited powdery mildew resistance in pea (Pisum sativum) er1 plants is, in many aspects, reminiscent of mlo‐conditioned powdery mildew immunity, yet the underlying gene has remained elusive to date. We used a polymerase chain reaction (PCR)‐based approach to amplify a candidate MLO cDNA from wild‐type (Er1) pea. Sequence analysis of the PsMLO1 candidate gene in two natural er1 accessions from Asia and two er1‐containing pea cultivars with a New World origin revealed, in each case, detrimental nucleotide polymorphisms in PsMLO1, suggesting that PsMLO1 is Er1. We corroborated this hypothesis by restoration of susceptibility on transient expression of PsMLO1 in the leaves of two resistant er1 accessions. Orthologous legume MLO genes from Medicago truncatula and Lotus japonicus likewise complemented the er1 phenotype. All tested er1 genotypes showed unaltered colonization with the arbuscular mycorrhizal fungus, Glomus intraradices, and with nitrogen‐fixing rhizobial bacteria. Our data demonstrate that PsMLO1 is Er1 and that the loss of PsMLO1 function conditions durable broad‐spectrum powdery mildew resistance in pea.
INTRODUCTION
Fungi are responsible for many of the world's most devastating plant diseases and are a major threat for agriculture with regard to both product yield and quality. Members of the order Erysiphales, phylum Ascomycota, comprise the causal agents of the widespread powdery mildew disease of higher plant species. The disease is particularly prevalent in temperate and humid climates, where it frequently causes significant yield losses and quality reductions in agricultural settings, including greenhouse and field farming. This applies to key cereals (e.g. barley and wheat), horticultural crops (e.g. grapevine, pea and tomato) and economically important ornamentals (e.g. roses). Limited access to natural resources of resistance to powdery mildews, rapid changes in virulence in the pathogen populations and the time‐consuming introgression of suitable resistance genes into elite varieties account for the widespread use of fungicides to control the disease. The challenge is compounded by increasing problems caused by the evolution and spread of fungicide resistance, which is especially dramatic amongst the most economically important powdery mildews (Wyand and Brown, 2005).
In the monocot barley (Hordeum vulgare) and the dicots Arabidopsis thaliana and tomato (Solanum lycopersicum), loss‐of‐function mutations in MLO (Mildew Resistance Locus O) genes confer highly effective broad‐spectrum powdery mildew resistance (Bai et al., 2008; Büschges et al., 1997; Consonni et al., 2006). Resistance conditioned by mlo alleles acts early and typically terminates fungal pathogenesis before invasion of the first host cell (Jørgensen, 1992; Lyngkjaer et al., 2000). This type of immunity was originally discovered in barley following the mutagenesis of wild‐type germplasm, as well as via a natural source of resistance in Ethiopian landraces (Jørgensen, 1992; Piffanelli et al., 2004). In barley, mlo resistance is of great agronomic importance given that approximately one‐half of the current European acreage occupied by spring varieties is of the mlo genotype and thus highly resistant against Blumeria graminis f.sp. hordei, the causal agent of barley powdery mildew disease (Lyngkjaer et al., 2000). In tomato, powdery mildew resistance conferred by ol‐2 is also a result of a loss of MLO function, conditioned by a natural polymorphism resulting in a small deletion within the MLO coding region (Bai et al., 2008).
MLO genes code for a plant‐specific type of integral membrane protein with as yet unknown biochemical function(s) (Devoto et al., 1999; Panstruga, 2005b). The genes occur in small‐ to medium‐sized families with approximately 10–20 members per plant species (Devoto et al., 2003; Feechan et al., 2008; Konishi et al., 2010; Liu and Zhu, 2008); however, only members of a specific phylogenetic clade play a role in powdery mildew susceptibility/resistance (Bai et al., 2008). These members are characterized by the presence of clade‐specific peptide motifs in cytoplasmic regions of the heptahelical MLO protein, of which the D/E‐F‐S/T‐F tetrapeptide at the distal end of the C‐terminal cytoplasmic tail is probably the most diagnostic (Feechan et al., 2008; Panstruga, 2005a). In Arabidopsis and barley, these MLO family members are part of larger sets of coexpressed genes that define a conserved functional module in antifungal immunity (Humphry et al., 2010). It has been hypothesized previously that MLO proteins may serve as targets for defence suppression or as modulators of plant defence (Panstruga and Schulze‐Lefert, 2003). Although the exact mechanism of mlo‐conditioned resistance remains obscure, it seems to rely largely on the plant's basal defence machinery (Collins et al., 2003; 2006, 2010; Humphry et al., 2006).
In pea (Pisum sativum), two recessively inherited genes (er1 and er2) represent the major natural sources of resistance against the respective powdery mildew pathogen, Erysiphe pisi (Heringa et al., 1969; Kumar and Singh, 1981; Vaid and Tyagi, 1997). Both er1 and er2 originate from wild accessions (Harland, 1948; Heringa et al., 1969), have been introgressed into elite pea varieties and, in particular, er1 has been found to provide durable broad‐spectrum protection from powdery mildew disease (Cousin, 1997; Tiwari et al., 1997). The analysis of a range of pea accessions revealed the presence of er1 in multiple lines of different geographical origin (Fondevilla et al., 2006; Tiwari et al., 1997), and it has also been identified recently in chemically induced pea mutants (Pereira and Leitao, 2010; Pereira et al., 2010). The er1 gene has been mapped to pea linkage group 6 (Dirlewanger et al., 1994; Janila and Sharma, 2004; Timmerman et al., 1994; Tiwari et al., 1998; Tonguc and Weeden, 2010), whereas er2 maps to linkage group 3 (Katoch et al., 2010). At the cellular level, resistance conferred by er1 is characterized by a failure of successful invasion of epidermal pea cells, whereas er2 resistance acts post‐penetration and is associated with the execution of host cell death (Fondevilla et al., 2006). Taken together, the features of er1 resistance (monogenic trait, recessive inheritance, broad‐spectrum resistance, durability under agricultural conditions and pre‐invasive termination of fungal pathogenesis) are reminiscent of mlo‐conditioned resistance in barley, Arabidopsis and tomato (Bai et al., 2008; Büschges et al., 1997; Consonni et al., 2006). We thus previously hypothesized that er1 resistance could be caused by loss of MLO gene function (Bai et al., 2008).
In this study, we used a polymerase chain reaction (PCR)‐based approach to obtain a MLO candidate gene for pea Er1 plants. Sequence analysis revealed deleterious nucleotide polymorphisms in PsMLO1 in four pea accessions harbouring er1 resistance. Complementation analysis based on transient gene expression in detached pea leaves of the er1 genotype corroborated that PsMLO1 is Er1. The availability of mlo mutants in a legume provided the exciting possibility to test whether bacterial (rhizobial) and fungal (arbuscular mycorrhizal, AM) symbionts, which, like powdery mildews, have an intracellular lifestyle, also require MLO for invasion of plant cells.
RESULTS
Reverse transcription‐polymerase chain reaction (RT‐PCR)‐based amplification of PsMLO1 cDNA
We have previously cloned the tomato MLO gene, SlMLO1, based on the presence of candidate expressed sequence tag (EST) sequences in public databases (Bai et al., 2008). In the case of pea, however, blast searches did not reveal any EST sequence with significant homology to any MLO gene. We thus opted for a PCR‐based approach by taking advantage of known sequence information from other plant species. First, we performed an alignment of diverse dicotyledonous MLO coding sequences employing genes with a suspected orthologous relationship to Arabidopsis AtMLO2 (Fig. S1, see Supporting Information). Aligned genes were derived from A. thaliana (AtMLO2), Solanum lycopersicum (tomato; SlMLO1), Capsicum annuum (pepper; CaMLO1), Lotus japonicus (LjMLO1) and Medicago truncatula (MtMLO1). On the basis of this multiple sequence alignment, we selected conserved regions to deduce oligonucleotide sequences for PCR‐based amplification of the respective part of the orthologous pea MLO cDNA. In total, we designed three oligonucleotides (PsMLO1–PsMLO3; Table S1, see Supporting Information), whose sequences at the nonconserved positions were adjusted according to the legume MtMLO1 sequence as, of the species under consideration, M. truncatula is that which is most closely related to P. sativum (Young et al., 2003).
We performed RT‐PCR with two distinct primer combinations (PsMLO1 + PsMLO2, PsMLO1 + PsMLO3) using pea RNA derived from the susceptible wild‐type cultivar JI 502 as a template. In each case, we obtained a single distinct product of the expected size range (data not shown). blastx analysis of the sequenced PCR products against the National Center for Biotechnology Information nonredundant (NCBInr) database revealed that the respective cDNA fragments encode part of a MLO‐like protein. We designed new pairs of internal oligonucleotides and performed 5′ and 3′ rapid amplification of cDNA ends (RACE) to obtain full‐length sequence information of this cDNA. The in silico‐assembled overlapping nucleotide sequences from 5′ and 3′ RACE reactions yielded a cDNA that encodes a predicted protein (designated PsMLO1) with high sequence relatedness to CaMLO1, LjMLO1, AtMLO2 and SlMLO1 (Fig. 1). To determine whether PsMLO1 represents a genuine orthologue of these proteins, we performed phylogenetic analysis. The resulting dendrogram indicates that PsMLO1 resides in the same phylogenetic clade as AtMLO2, HvMLO and SlMLO1 (Fig. 2), each of which has been shown previously to be required for powdery mildew susceptibility (Bai et al., 2008; Büschges et al., 1997; Consonni et al., 2006). Taken together, we identified a cDNA that represented the full‐length transcript of an Er1 candidate gene.
Figure 1.

Multiple amino acid sequence alignment of MLO (Mildew Resistance Locus O) proteins. Amino acid sequences of Arabidopsis (AtMLO2), Capsicum annuum (CaMLO1), Lotus japonicus (LjMLO1), tomato (SlMLO1) and pea (PsMLO1) were aligned with clustalw using the default parameters. The positions of the seven transmembrane domains (TM1–TM7), as inferred from the experimentally determined topology of barley MLO (Devoto et al., 1999), are indicated by the bars above and below the sequence alignment. The C‐terminal D/E‐F‐S/T‐F tetrapeptide, which is diagnostic for the phylogenetic clade harbouring proteins with a function in powdery mildew interactions (Panstruga, 2005a), is boxed.
Figure 2.
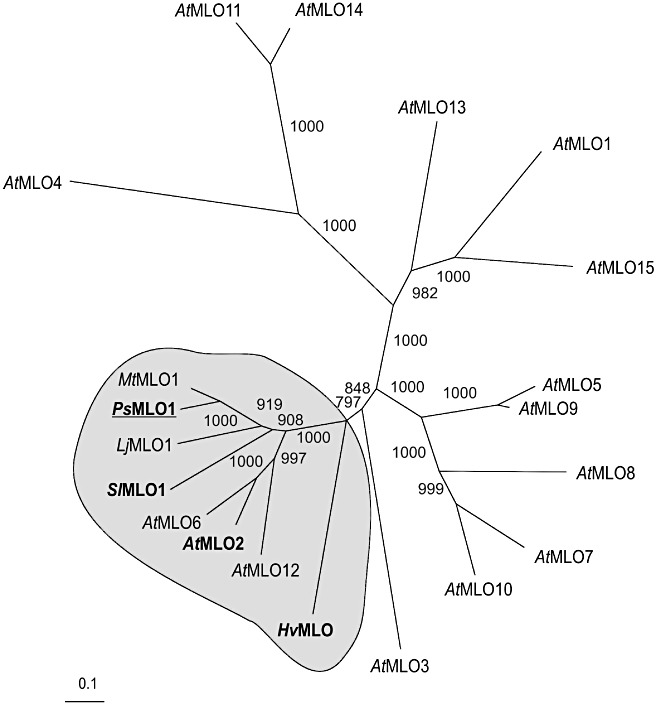
Phylogenetic relationship of selected MLO (Mildew Resistance Locus O) proteins. Phylogenetic (neighbour‐joining) tree of Arabidopsis (AtMLO1–AtMLO15), barley (HvMLO), tomato (SlMLO1), Medicago truncatula (MtMLO), Lotus japonicus (LjMLO1) and pea (PsMLO1) MLO proteins. The clade harbouring presumptive (co‐)orthologues is highlighted in grey. Proteins known to play an important role in conferring powdery mildew susceptibility are highlighted in bold; the pea MLO protein is underlined. The numbers at the edges designate the bootstrap support based on 1000 replicates. The scale bar indicates the number of amino acid exchanges per site.
PsMLO1 is defective in four independent er1 lines
We focused our subsequent molecular analysis on two pea accessions and two pea cultivars originating from various regions of the world that have been reported previously to harbour er1 resistance: JI 210, JI 1559, JI 1951 and JI 2302 (Tiwari et al., 1997) (Table 1). First, we macroscopically and microscopically analysed powdery mildew infection phenotypes of these lines and compared them with the susceptible (Er1 genotype) cultivar JI 502 using our domestic E. pisi isolate. Consistent with a previous report (Fondevilla et al., 2006), we found near‐complete pre‐invasive immunity in these lines (Fig. 3). Next, we determined the nucleotide sequence of PsMLO1 cDNAs of the four resistant lines by direct sequencing of full‐length RT‐PCR products. This revealed single‐nucleotide deletions resulting in frame shifts in lines JI 210 and JI 1951, a nucleotide substitution resulting in a premature stop codon in JI 1559, as well as multiple overlapping sequence traces in JI 2302. The latter often results from a defect in intron splicing, leading to the co‐presence of multiple distinct transcript variants. We thus cloned and sequenced individual PsMLO1 cDNAs derived by RT‐PCR from RNA of line JI 2302. On the basis of nine clones, this analysis revealed three distinct types of mis‐spliced cDNAs, two of which are characterized by the occurrence of ectopic sequences in the cDNA that are not present in the genomic wild‐type sequence (Fig. 4A). We assumed that these extra sequences stem from the insertion of an unrelated DNA fragment into the PsMLO1 genomic sequence. Consistent with this hypothesis, conventional and long‐range PCR amplification of the genomic region under investigation failed only in accession JI 2302, possibly owing to the large size of the inserted DNA fragment (Fig. 4B,C). blastn analysis against the NCBInr database revealed that the part of the insertion that is present in aberrant cDNAs of line JI 2302 is nearly identical (∼93% identity) to a sequence that occurs five times in a pea genomic BAC clone (GenBank accession number CU655882). In addition, an ∼170‐bp stretch of this sequence is highly similar (∼86% identity) to part of the long terminal repeats (LTRs) of the pea Ogre retrotransposon (GenBank accession number AY299398). The Ogre retroelement is an exceptionally large (22 kb) and transcriptionally active Ty/gypsy‐like LTR‐type retrotransposon that makes up 5% of the pea genome (Neumann et al., 2003). We conclude that PsMLO1 function is disrupted through insertional mutation in accession JI 2302, possibly by integration of a (retro‐)transposon that is similar to the Ogre element. Taken together, the identification of four independent mutational events in powdery mildew‐resistant er1 lines of distinct geographical origin suggests that PsMLO1 is identical to Er1 (Table 1).
Table 1.
Pea lines used in this study.
| JI accession number* | Alternative name | Country of origin | Genotype | Mutational event at PsMLO1 † | er1 allele designation | Reference |
|---|---|---|---|---|---|---|
| JI 502 | cv. Rondo | The Netherlands | Er1 Er2 | None | n.a. | Heringa et al. (1969) |
| JI 1559 | Mexique 4 | Mexico | er1 Er2 | C680G (Ser → stop) | er1‐1 | Tiwari et al. (1997) |
| JI 2302 | Stratagem | USA | er1 Er2 | Insertion of unknown size and identity | er1‐2 | Tiwari et al. (1997) |
| JI 210 | Unnamed landrace | India | er1 Er2 | ΔG862 (frame shift) | er1‐3 | Tiwari et al. (1997) |
| JI 1951 | Unnamed landrace | China | er1 Er2 | ΔA91 (frame shift) | er1‐4 | Tiwari et al. (1997) |
| JI 2480 | SVP951, CGN3027 | Peru | Er1 er2 | None | n.a. | Tiwari et al. (1997) |
Accession code of John Innes Pisum collection (http://data.jic.bbsrc.ac.uk/cgi‐bin/germplasm/pisum/).
According to the PsMLO1 coding sequence.
n.a., not applicable.
Figure 3.
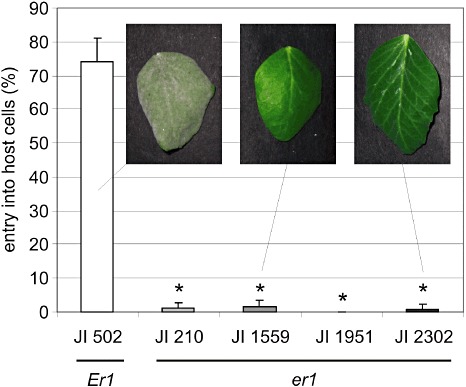
Pre‐invasive resistance terminates powdery mildew pathogenesis in er1 pea accessions. Leaves of accessions JI 502 (wild‐type), JI 210, JI 1559, JI 1951 and JI 2302 (er1 genotypes; Table 1) were inoculated with Erysiphe pisi conidia and harvested at 48 h post‐inoculation. Powdery mildew penetration success was quantified as the average of at least 100 interaction sites per leaf on four distinct individuals per line. Error bars indicate standard deviation. Inset photographs depict the macroscopic infection phenotypes of selected genotypes at 7 days post‐inoculation. Asterisks indicate a statistically significant difference from JI 502 (P < 0.000001; two‐tailed Student's t‐test).
Figure 4.
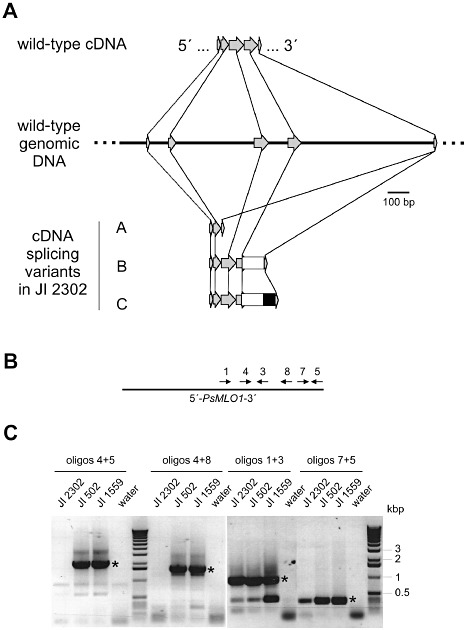
Evidence for the presence of a large DNA insertion in PsMLO1 in line JI 2302. (A) Aberrant transcripts and deduced organization of the genomic PsMLO1 locus in pea line JI 2302. The scheme depicts the organization of part of the wild‐type genomic PsMLO1 locus and its derived cDNA, as well as aberrant splice variants found in line JI 2302. Light grey arrows indicate exons; black lines indicate introns. The white and black boxes symbolize ectopic DNA sequences, most probably derived from the insertional event in PsMLO1 in line JI 2302. (B, C) Lack of polymerase chain reaction (PCR)‐based amplification of particular PsMLO1 fragments in line JI 2302. (B) Scheme depicting the relative position and orientation of oligonucleotides 1, 3, 4, 5, 7 and 8 with respect to the PsMLO1 genomic sequence (full line). (C) Agarose electropherograms of PCR products obtained with genomic DNA of lines JI 2302, JI 502 and JI1559 (templates, plus water control) and various oligonucleotide combinations. Please note the presence of JI 2302‐derived PCR products using oligonucleotide combinations 1 + 3 and 7 + 5, but the absence of amplicons using oligonucleotide combinations 4 + 5 and 4 + 8. Asterisks in the water control lane mark the expected product size for the respective primer combination.
Complementation of er1 resistance by transient gene expression
To further support our assumption that PsMLO1 is Er1, we aimed to complement the defect in er1 plants by transient expression of PsMLO1 in er1 genotypes. We opted for an established assay based on particle bombardment‐mediated transformation of single leaf epidermal cells to test the functionality of PsMLO1 (Panstruga, 2004; Shirasu et al., 1999). We generated a construct that drives the expression of epitope‐tagged PsMLO1 under control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter. This construct, together with a plasmid harbouring the β‐glucuronidase (GUS) reporter gene, was ballistically transformed into leaf epidermal cells of er1 powdery mildew‐resistant accessions JI 1559 and JI 2302. Transformation of the GUS reporter gene alone did not allow fungal entry into epidermal cells in the resistant accessions (Fig. 5 and Table S2, see Supporting Information). In contrast, transient expression of PsMLO1 conferred substantial levels (58%–80%) of host cell entry in these lines, indicating that each of these genes is capable of complementing the er1 resistance phenotype (Fig. 5 and Table S2). Successful host cell penetration mediated by the expression of PsMLO1 was restricted to transformed (GUS‐stained) cells, whereas nontransformed cells retained resistance against the fungal pathogen (Fig. 5C). Transient expression of epitope‐tagged versions of the phylogenetically closely related legume MLO genes LjMLO1 and MtMLO1 likewise complemented the er1 geneotype, although with somewhat lower efficiencies (LjMLO1, 28%–54%; MtMLO1, 52%–69%; Table S2). Collectively, these findings further corroborate the claim that PsMLO1 is Er1.
Figure 5.
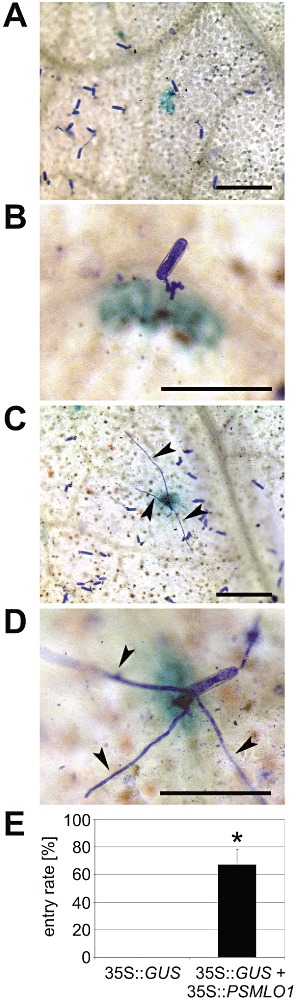
Single‐cell complementation of the er1 phenotype by transient expression of PsMLO1. Pea leaves (line JI 2302; er1 genotype) were bombarded with either 35S::GUS alone or co‐bombarded with 35S::GUS plus 35S::PsMLO1. Leaves were inoculated with Erysiphe pisi conidiospores at 4 h after bombardment and stained for β‐glucuronidase (GUS) activity at 48 h post‐inoculation. (A–D) Micrographs of failed (A, B) and successful (C, D) penetration attempts of E. pisi on GUS‐stained epidermal cells. Successful host cell entry and haustorium formation are indicated by the growth of secondary hyphae (arrowheads). Please note that successful invasion in (C) is restricted to the GUS‐stained cell, whereas the surrounding cells retain resistance. Size bars are 200 µm in (A, C) and 100 µm in (B, D). (E) Quantitative assessment of E. pisi entry into GUS‐stained cells. Results represent the mean ± standard deviation from three independent experiments and are based on the data shown in Table S2. The asterisk indicates a statistically significant difference from 35S::GUS (P < 0.001; two‐tailed Student's t‐test).
Mutations in PsMLO1 do not affect the invasion of plant cells by arbuscular mycorrhiza or nitrogen‐fixing rhizobial bacteria
On the basis of the previously determined resistance spectrum of barley and Arabidopsis mlo mutants against various phytopathogens, mlo resistance seems to be rather specific to powdery mildew fungi (Consonni et al., 2006; Jarosch et al., 1999; Jørgensen, 1977). However, despite the fact that a common mechanistic basis for biotrophic parasitism and endosymbiosis has been discussed (Parniske 2000), the effect of mutations in Mlo genes on mutualistically beneficial plant–microbe interactions, which involve an intracellular lifestyle, has been less thoroughly examined (Ruiz‐Lozano et al., 1999). As the legume species pea is able to engage in both rhizobial (bacterial) and mycorrhizal (fungal) symbiosis, it represents a well‐suited model to study the effect of mlo mutations on the invasion of cells in symbiosis. In legume root nodules, rhizobial bacteria are hosted intracellularly in host‐derived membrane compartments, whereas AM fungi form so‐called intracellular arbuscules that are surrounded by a host membrane. We infected roots of pea lines JI 210, JI 502, JI 1559, JI 1951 and JI 2302 with Rhizobium leguminosarum bv. viciae pMW1071(nodX), which harbours an extrachromosomal copy of the nodX gene that broadens the host range of this symbiotic bacterium. In roots of all tested pea lines, nodules were formed and, except for JI 210, the nodule number was 10 or more per root system (Fig. 6A). For each genotype, several nodules were well developed and had a red colour (owing to leghaemoglobin accumulation), indicating that they were functional. Sectioning also showed that infected cells of nodules were, like wild‐type nodular cells, fully filled with rhizobial bacteria. A reduced number of nodules in the root system of line JI 210 was not seen on inoculation with a different Rhizobium strain (R. leguminosarum bv. viciae CIAM 1026) (Safronova and Novikova, 1996), suggesting that the inability to colonize JI 210 is not caused by the mutation in PsMLO1 but, rather, an incompatibility with R. leguminosarum bv. viciae pMW1071(nodX). Similar to the interaction with the bacterial symbionts, quantitative analysis of the interaction with Glomus intraradices, a fungus that establishes symbiotic relationships with a broad range of plant hosts, revealed essentially unaltered colonization and formation of intracellular arbuscules in all four tested er1 genotypes (Fig. 6B).
Figure 6.
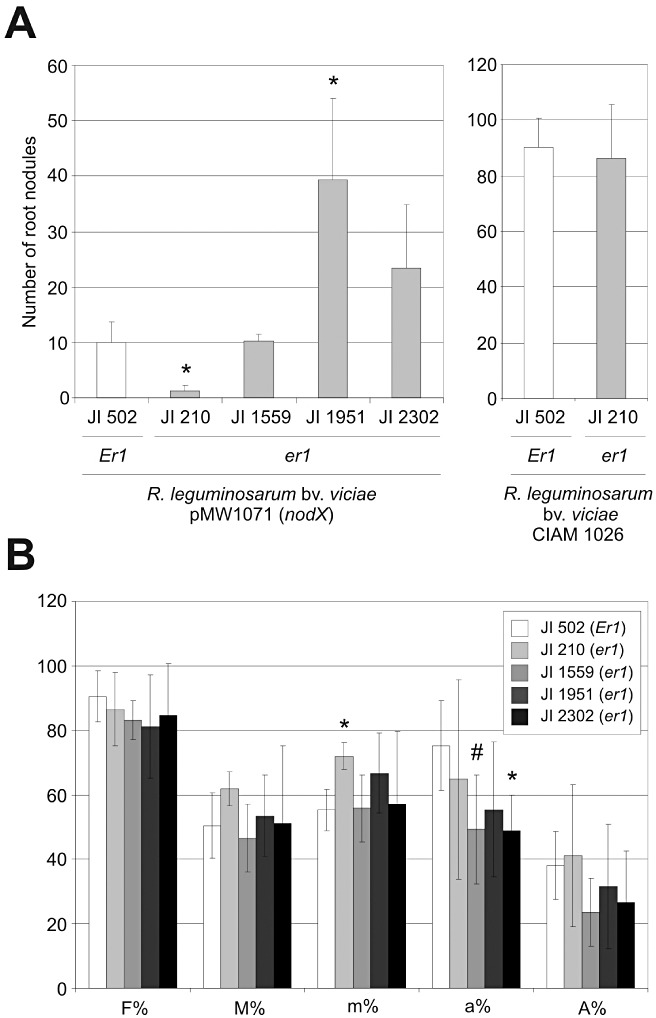
Pea er1 genotypes exhibit unaltered symbiotic interactions. Pea lines JI 502 (wild‐type; Er1 genotype), JI 210, JI 1559, JI 1951 and JI 2302 (er1 genotypes; Table 1) were analysed for root nodule formation (A) and mycorrhization by Glomus intraradices (B). (A) Roots were infected with either Rhizobium leguminosarum bv. viciae pMW1071 (nodX) (left panel) or Rhizobium leguminosarum bv. viciae CIAM 1026 (right panel). Data represent the average ± standard deviation from four to seven plants per genotype. Asterisks indicate a statistically significant difference from JI 502 (P < 0.01; two‐tailed Student's t‐test). (B) Data represent the average ± standard deviation from three to six plants per genotype with approximately 75 cm of root system per plant analysed. F%, frequency of mycorrhiza in the root system; M%, intensity of mycorrhizal colonization in the root system; m%, intensity of the mycorrhizal colonization in the root fragments; a%, arbuscule abundance in mycorrhizal parts of the root fragments; A%, arbuscule abundance in the root system (according to Trouvelot et al. (1986)). Asterisks and the hash sign indicate a statistically significant difference from JI 502 (P < 0.01 and P < 0.05, respectively; two‐tailed Student's t‐test).
DISCUSSION
In this study, we have shown that resistance in pea lines JI 210, JI 1559, JI 1951 and JI 2302 is caused by loss of function in PsMLO1. This claim is supported by detrimental lesions (JI 210, JI 1559 and JI 1951) and a large insertion (JI 2302) in the PsMLO1 gene of the accessions, as well as by complementation of the resistance phenotype (i.e. restoration of susceptibility) by ectopic MLO gene expression. As lines JI 210, JI 1559, JI 1951 and JI 2302 harbour the er1 resistance gene, we propose that PsMLO1 is Er1. We did not find any sequence polymorphism in the PsMLO1 cDNA of line JI 2480, which reportedly harbours the er2 resistance gene. Features of powdery mildew resistance in line JI 2480 differ at the cellular level from resistance in lines JI 210, JI 1559, JI 1951 and JI 2302. For example, in contrast with these lines, powdery mildew resistance in JI 2480 acts at the post‐invasive level, is associated with host cell death and is temperature sensitive (Fondevilla et al., 2006). We thus conclude that resistance in line JI 2480 is unlikely to be mediated by a loss of PsMLO1 function, but is probably a result of the malfunction of a distinct gene. This conclusion is consistent with previous linkage analysis, which genetically separated er1‐ from er2‐conditioned resistance (Heringa et al., 1969; Tiwari et al., 1997). Similarly, a recently discovered gene (Er3) in Pisum fulvum, a wild relative of P. sativum, confers dominantly inherited post‐penetration resistance to E. pisi (Fondevilla et al., 2007), maps elsewhere in the genome (Fondevilla et al., 2008) and therefore is unlikely to relate to PsMLO1.
We were unable to analyse in detail the molecular nature of the insertion in line JI 2302. The size of the insertion exceeds the capacity of conventional and long‐range PCR amplification, suggesting that it is at least several kilobases in size. In addition, blast searches suggest a sequence relationship of part of the inserted DNA to the LTRs of the Ogre retrotransposon. It is therefore likely that the large insertion in pea line JI 2302 represents a (retro‐)transposon, which are known to range in size from less than 100 bp (in the case of miniature transposable elements, MITEs) to 25 kb (in the case of some exceptional LTR‐type retrotransposons (Wicker et al., 2007). Despite the seemingly considerable size of the insertion, the cellular splicing machinery is capable of removing most of it during transcript processing, as evidenced by the presence of rather small ectopic DNA sequences in part of the aberrant PsMLO1 cDNAs derived from line JI 2302 (Fig. 4). This polymorphism, as well as the other mutational events at PsMLO1 in er1 mutant plants revealed in this study, promise to be useful in marker‐assisted selection of powdery mildew‐resistant seedlings in future pea breeding programmes.
Alleles of er1 derive from pea accessions/cultivars that originate from geographically diverse habitats (USA, Mexico, China and India; Table 1). The lesions in PsMLO1 of lines JI 210, JI 1559, JI 1951 and JI 2302 are each of natural origin and thus probably caused by spontaneous mutation events. Powdery mildew resistance conferred by natural mlo alleles has also been found in the case of barley (Piffanelli et al., 2004) and tomato (Bai et al., 2008). However, in these instances, the polymorphisms leading to resistance were either the presence of a complex repeat array perturbing Mlo transcript accumulation or a 19‐bp deletion in the cDNA leading to premature termination of translation, respectively (Bai et al., 2008; Piffanelli et al., 2004). In this study, we found single‐base‐pair deletions resulting in frame shifts in two cases, a nucleotide substitution resulting in a premature stop codon, and a large DNA insertion disrupting the PsMLO1 gene. It is striking that, compared with the collection of induced barley mlo mutants, where the majority of alleles are characterized by single amino acid substitutions (Büschges et al., 1997; Piffanelli et al., 2002; Reinstädler et al., 2010), all natural mlo mutants identified to date are caused by more dramatic alterations of the coding sequence. This could indicate that a complete absence of the MLO protein provides an evolutionary advantage compared with the presence of a nonfunctional MLO protein variant. At present, it is unclear whether the occurrence of natural mlo alleles is linked to geographical regions in which the powdery mildew pathogen currently resides or was a recent prevalent parasite. Accessions of further plant species, such as M. truncatula (barrel medic) and Beta vulgaris (beet), exhibit powdery mildew resistance at the penetration level and thus might represent further candidates for natural mlo mutants (Fernández‐Aparicio et al., 2009; Prats et al., 2007).
Resistance conferred by er1 alleles has been reported to be highly effective and durable, but occasionally incomplete under field conditions (Fondevilla et al., 2006). This is surprising given the fact that mlo resistance in barley null alleles is known to be complete and PsMLO1 lesions in pea lines JI 210, JI 1559, JI 1951 and JI 2302 are predicted to result in null mutations (Table 1). In A. thaliana, lesions in AtMLO2, one of three closely related MLO genes, are insufficient to confer full powdery mildew resistance and only a respective triple mutant mediates complete immunity (Consonni et al., 2006). Thus, in pea, other MLO paralogues may also contribute to powdery mildew susceptibility. However, in our experience, the above‐mentioned pea lines showed rather complete resistance in mature leaves (Fig. 3). Likewise, in controlled inoculation experiments employing detached leaves and a range of E. pisi isolates, the above‐mentioned pea mutants consistently showed strong resistance (Tiwari et al., 1997). This suggests that the reported incompleteness of powdery mildew resistance under field conditions might be a result of extraordinary biotic or abiotic stress conditions, rather than the principal genetic constitution of the mutant lines. An example of the latter is the temporary partial breakdown of barley mlo resistance as a consequence of the sudden relief of drought stress (Baker et al., 1998).
In barley and Arabidopsis, in addition to powdery mildew resistance, mutations in MLO result in a developmental abnormality that is manifested as early leaf senescence (2006, 2010; Piffanelli et al., 2002). We did not observe this undesired side‐effect in the tested er1 pea lines under our growth conditions, and also found no respective report in the literature. This is reminiscent of the situation in tomato, where powdery mildew resistance conditioned by a loss of SlMLO1 function was also not associated with any obvious detrimental phenotype (Bai et al., 2008). There are several possible explanations for this seeming discrepancy. The expression of early leaf senescence might be modulated by as yet unidentified growth parameters (e.g. soil composition, light quality, water quality, use of fertilizers, etc.). Alternatively, the extent of this undesired phenomenon may vary between species and could be below the limit of visual detection in pea and tomato. Finally, the occurrence of early leaf senescence in mlo mutant plants could be a species‐specific phenomenon. The last two scenarios would explain why natural mlo alleles appear to be more common in pea than in barley or Arabidopsis. It thus still remains to be investigated whether—and, if so, to what extent and in which conditions—er1 plants suffer from any pleiotropic effects.
We observed cell‐autonomous complementation of two er1 genotypes (JI 1559 and JI 2302) by transient expression of PsMLO1 as well as MLO genes from the closely related legume species L. japonicus and M. truncatula (LjMLO1 and MtMLO1; Fig. 5 and Table S2). We noted a gene‐specific gradation of complementation efficiency in both tested pea lines, ranging from PsMLO1 (58%–80% entry rate), via MtMLO (46%–69% entry rate) to LjMLO (28%–54% entry rate). These incremental differences in complementation efficiency correlate with the phylogenetic relationship of the three MLO genes (Fig. 2) and the respective legume species (Young et al., 2003). Previously, we have reported similarly staggered complementation rates of the barley mlo mutant by transient expression of orthologous MLO genes from the monocots barley, wheat and rice (Elliott et al., 2002). Together, these data suggest that, with respect to powdery mildew susceptibility, orthologues from closely related taxa can substitute for MLO function in a given plant species. However, complementation efficiency seems to decrease with phylogenetic distance. A lack of full complementation by transiently expressed PsMLO1 (compare the ∼80% entry rate of E. pisi in JI 502 with ∼60% in PsMLO1‐transformed cells; Table S2) could be a result of the presence of the C‐terminal haemagglutinin (HA) tag encoded by the constructs used in the assay, which might interfere with PsMLO function. However, in the case of barley MLO, C‐terminal tagging with green fluorescent protein (GFP) does not interfere with MLO functionality in transient gene expression studies (Shirasu et al., 1999).
Powdery mildew‐resistant barley and Arabidopsis mlo mutants have been analysed previously for their interaction with a range of other phytopathogens. This revealed largely unaltered infection phenotypes for barley mlo mutants in response to the causal agents of stripe rust (Puccinia striiformis), leaf rust (Puccinia hordei), stem rust (Puccinia graminis f.sp. tritici), scald (Rhynchosporium secalis) and the take‐all fungus (Gaeumannomyces graminis) (Jørgensen, 1977). Likewise, Arabidopsis mlo2 mlo6 mlo12 triple mutants exhibited unaltered susceptibility to the bacterial pathogen Pseudomonas syringae (causing the bacterial speck disease) and the oomycete Hyaloperonospora arabidopsidis (formerly known as Hyaloperonospora parasitica; the causal agent of downy mildew disease) (Consonni et al., 2006). However, barley mlo mutants were found to exhibit increased susceptibility to the rice blast pathogen Magnaporthe oryzae (formerly designated Magnaporthe grisea) (Jarosch et al., 1999). In contrast with pathogenic interactions, (endo‐)symbiotic interactions of mlo mutants have been less thoroughly studied. This is possibly a result of the fact that barley is not a well‐developed model system to study symbiosis, and Arabidopsis completely lacks rhizobial or mycorrhizal symbiosis. A preliminary study in barley employing a single mlo allele revealed a possible inhibitory effect of the mlo mutation on the colonization intensity (development within the root cortex) and arbuscule formation of the AM fungus Glomus mossae (Ruiz‐Lozano et al., 1999). The identification of a collection of pea mlo mutant alleles now allows us to study the establishment of intracellular symbiotic infection structures in the context of a plant species that is able to engage in both common types of endosymbiotic interaction, namely rhizobial and AM symbiosis. In contrast with the preliminary data from barley (Ruiz‐Lozano et al., 1999), we observed unaltered numbers of arbuscules in all tested er1 genotypes (Fig. 6B). Similarly, all er1 genotypes showed the formation of wild‐type‐like infected root nodules (Fig. 6). The variation in the number of root nodules probably can be attributed to the diverse genetic backgrounds of the tested er1 plants. Together, these data strongly suggest that PsMLO1 function is dispensable for the establishment of an intracellular lifestyle of the microbe in both mycorrhizal and rhizobial symbiosis.
EXPERIMENTAL PROCEDURES
Plant growth
The seeds of the pea lines used in this study were obtained from the John Innes Pisum collection (http://www.jic.ac.uk/germplas/pisum/). Pea plants for powdery mildew infection experiments and molecular analyses were grown in a controlled environment at 22 °C, 70% relative humidity and a 12‐h light (80–90 µmol/m2/s2)/dark cycle. The MPIZ isolate of E. pisi was propagated on an anonymous susceptible pea variety.
RNA extraction, RT‐PCR and RACE
RNA from pea leaves was extracted using the Qiagen RNAeasy Mini Kit (Qiagen, Hilden, Germany). First‐strand cDNAs were synthesized by oligo‐dT priming with SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The oligonucleotides used in this study are listed in Table S1. The 5′ and 3′ RACE reactions were performed with the FirstChoice RLM‐RACE Kit (Ambion, Austin, TX, USA) following the kit protocol (60 °C annealing temperature, 35 cycles). RACE fragments were cloned via TOPO® cloning (Invitrogen) into vectors pCR4 (Invitrogen; 5′ RACE) and pCR2.1 (Invitrogen; 3′ RACE), respectively. Full‐length PsMLO1 cDNA was amplified (55 °C annealing temperature, 40 cycles) using the oligonucleotides PsMLO‐GWF and PsMLO‐GWR (which were designed on the basis of the sequence information obtained from the RACE products) and cloned by Gateway® BP reaction into the Gateway® entry vector pDONR201 (Invitrogen). The PsMLO1 coding region of the various pea accessions was amplified by RT‐PCR (55 °C annealing temperature, 35 cycles) using oligonucleotides PsMLO1‐GWF and PsMLO5 (Table S1). PCR products were subjected to direct DNA sequencing, except for line JI 2302, for which amplicons were cloned into vector pCRII‐Blunt Topo (Invitrogen) to enable DNA sequencing of individual clones. Genomic fragments of PsMLO1 (Fig. 4C) were amplified by PCR (55 °C annealing temperature, 35 cycles) using the indicated oligonucleotide combinations.
Phylogenetic analysis
For the phylogenetic analysis of MLO proteins, the phylip 3.63 software package was used (http://evolution.gs.washington.edu/phylip.html; Felsenstein, 1989). MLO protein sequences were aligned by clustalw and the Protdist and Neighbor programs were employed to establish the neighbour‐joining phylogenetic tree. To calculate the bootstrap support, the Seqboot, Protdist, Neighbor and Consense algorithms were applied sequentially, using 1000 replicates each. GenBank accession numbers of AtMLO1–AtMLO15 can be found in Devoto et al. (2003). The GenBank accession numbers of the MLO genes encoding the respective proteins are as follows: HvMLO (Z83834), LjMLO1 (AY967410), MtMLO1 (HQ446457), PsMLO1 (FJ463618) and SlMLO1 (AY967408).
Transient gene expression
The procedure for transient gene expression was largely adopted from related experiments in barley (Elliott et al., 2002). Briefly, detached pea leaves of 3–4‐week‐old plants were placed on agar (Merck KGaA, Darmstadt, Germany), supplemented with 85 µm benzimidazole to delay leaf senescence, in Petri dishes. MLO genes (PsMLO1, LjMLO1, MtMLO1) lacking a stop codon were cloned into the pDONR201 entry vector by Gateway® BP reactions and subsequently shuttled by Gateway® LR reaction into the plant expression vector pAM‐PAT‐GWY‐3xHA, a Gateway®‐compatible derivative of the binary vector pPAM (GenBank accession number AY027531). This vector enables CaMV 35S promoter‐driven expression of C‐terminally tagged (3 × HA tag) proteins. Plasmids harbouring reporter (GUS) and test (MLO) genes were coated on gold particles (1 µm) by calcium precipitation. Gene transfer was performed using a helium‐driven PDS1000 gene gun system equipped with a Hepta adapter (Bio‐Rad Laboratories, Hercules, CA, USA). Bombarded leaves were inoculated with a high density of E. pisi (isolate MPIZ) conidiospores using a settling tower 4 h post‐bombardment, and staining for GUS activity was performed at 48 h post‐inoculation. Specimens were cleared in ethanol and epiphytic powdery mildew infection structures were stained with Evan's Blue (Merck KGaA). Fungal entry rates were assessed by light microscopy on the basis of the presence of a haustorium and/or secondary hyphae. We noted that transient gene expression analysis in pea was less efficient than in barley owing to the smaller size of epidermal leaf cells and the lower inoculation density that we were able to achieve with E. pisi. In combination, these conditions led to a reduced number of transformed (GUS‐stained) cells that were attacked by powdery mildew sporelings, thereby reducing the number of quantifiable cells.
Analysis of symbiotic interactions
For the analysis of rhizobial symbiosis, pea seedlings were germinated on agar plates, transferred after 1 week to pots with vermiculite [grown at 18 °C in a 16‐h light (130–160 µmol/m2/s2)/8‐h dark regime] and, 1 week later, inoculated with either R. leguminosarum bv. viciae pMW1071(nodX) or R. leguminosarum bv. viciae CIAM 1026 (Safronova and Novikova, 1996). The former, nodX‐containing, rhizobium was used as some pea varieties have a Sym2 allele that requires the nodX modification of the nod factor (Kozik et al., 1995). Plants were harvested at 3 weeks after inoculation and analysed by counting the number of root nodules per plant. For the analysis of AM symbiosis, pea seeds were initially incubated on agar plates for 2 days at 4 °C in the dark and germinated during 5 days at 18 °C in a 16‐h light (130–160 µmol/m2/s2)/8‐h dark regime. Mycorrhizal colonization of pea plants was analysed in a sand/hydrobeads culture [1 g of Ca3(PO4)2 per 1 kg of sand/hydrobeads mix (1:1, v/v)], supplemented with Hoagland solution (once per week) lacking phosphorus. Pea seedlings were transferred to pots with mycorrhizae, and the mycorrhization test was conducted at 21 °C in a 16‐h light (130–160 µmol/m2/s2)/8‐h dark regime. Mycorrhiza (Glomus intraradices BEG144) stock was propagated on chive (Allium schoenoprasum) and mixed with sand prior to transfer of the pea seedlings. Roots of pea plants were harvested at 3 weeks after inoculation, the latest time point that allows the detection of arbuscules on the roots before major developmental transitions (flowering) in some of the er1 lines. Root samples were cleared in 2% (w/v) KOH and stained with trypan blue (0.05%, w/v) to detect intraradical mycelium; 1‐cm fragments (∼50) of randomly selected roots were mounted on glass slides and assessed by light microscopy. The levels of mycorrhizal colonization and arbuscule abundance were scored according to Trouvelot et al. (1986) using the computer program ‘Mycocalc’ (http://www2.dijon.inra.fr/mychintec/Mycocalc‐prg/download.html).
GenBank accession numbers
The sequences of PsMLO1 and MtMLO1 were deposited under GenBank accession numbers FJ463618 and HQ446457, respectively.
Supporting information
Fig. S1 Sequence alignment of MLO coding sequences. Nucleotide sequences of LjMLO1 (GenBank accession number AY967410), SlMLO1 (AY967408), CaMLO1 (AY934528) and AtMLO2 (AF369563) were aligned by CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Conserved nucleotide stretches used for the design of oligonucleotide primers PsMLO1 (forward primer), PsMLO2 and PsMLO3 (reverse primers) are highlighted in light blue; the actual oligonucleotide sequences (highlighted in yellow) are shown below the respective regions. In the case of nucleotide positions that were divergent between the four MLO sequences, the oligonucleotide sequences were adjusted to the LjMLO sequence.
Table S1 Oligonucleotides used in this study.
Table S2 Single‐cell complementation of the er1 phenotype by transient expression of MLO genes.
Supporting info item
Supporting info item
Supporting info item
ACKNOWLEDGEMENTS
We wish to thank Mike Ambrose (John Innes Centre, Norwich, UK) for making available pea germplasm from the John Innes Pisum collection. This work was supported by funds from the Max‐Planck Society. MH was partially supported by a postdoctoral fellowship from the Alexander‐von‐Humboldt Foundation.
REFERENCES
- Bai, Y.L. , Pavan, S. , Zheng, Z. , Zappel, N.F. , Reinstädler, A. , Lotti, C. , De Giovanni, C. , Ricciardi, L. , Lindhout, P. , Visser, R. , Theres, K. and Panstruga, R. (2008) Naturally occurring broad‐spectrum powdery mildew resistance in a central American tomato accession is caused by loss of Mlo function. Mol. Plant–Microbe Interact. 21, 30–39. [DOI] [PubMed] [Google Scholar]
- Baker, S.J. , Newton, A.C. , Crabb, D. , Guy, D.C. , Jefferies, R.A. , Mackerron, D.K.L. , Thomas, W.T.B. and Gurr, S.J. (1998) Temporary partial breakdown of mlo‐resistance in spring barley by sudden relief of soil water‐stress under field conditions: the effects of genetic background and mlo allele. Plant Pathol. 47, 401–410. [Google Scholar]
- Büschges, R. , Hollricher, K. , Panstruga, R. , Simons, G. , Wolter, M. , Frijters, A. , van Daelen, R. , van der Lee, T. , Diergaarde, P. , Groenendijk, J. , Töpsch, S. , Vos, P. , Salamini, F. and Schulze‐Lefert, P. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell, 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Collins, N.C. , Thordal‐Christensen, H. , Lipka, V. , Bau, S. , Kombrink, E. , Qiu, J.L. , Hückelhoven, R. , Stein, M. , Freialdenhoven, A. , Somerville, S.C. and Schulze‐Lefert, P. (2003) SNARE‐protein‐mediated disease resistance at the plant cell wall. Nature, 425, 973–977. [DOI] [PubMed] [Google Scholar]
- Consonni, C. , Humphry, M.E. , Hartmann, H.A. , Livaja, M. , Durner, J. , Westphal, L. , Vogel, J. , Lipka, V. , Kemmerling, B. , Schulze‐Lefert, P. , Somerville, S.C. and Panstruga, R. (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. [DOI] [PubMed] [Google Scholar]
- Consonni, C. , Bednarek, P. , Humphry, M. , Francocci, F. , Ferrari, S. , Harzen, A. , van Themaat, E.V.L. and Panstruga, R. (2010) Tryptophan‐derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 152, 1544–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin, R. (1997) Peas (Pisum sativum L.). Field Crops Res. 53, 111–130. [Google Scholar]
- Devoto, A. , Piffanelli, P. , Nilsson, I. , Wallin, E. , Panstruga, R. , von Heijne, G. and Schulze‐Lefert, P. (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274, 34 993–35 004. [DOI] [PubMed] [Google Scholar]
- Devoto, A. , Hartmann, H.A. , Piffanelli, P. , Elliott, C. , Simmons, C. , Taramino, G. , Goh, C.S. , Cohen, F.E. , Emerson, B.C. , Schulze‐Lefert, P. and Panstruga, R. (2003) Molecular phylogeny and evolution of the plant‐specific seven‐transmembrane MLO family. J. Mol. Evol. 56, 77–88. [DOI] [PubMed] [Google Scholar]
- Dirlewanger, E. , Isaac, P.G. , Ranade, S. , Belajouza, M. , Cousin, R. and de Vienne, D. (1994) Restriction fragment length polymorphism analysis of loci associated with disease resistance genes and developmental traits in Pisum sativum L. Theor. Appl. Genet. 88, 17–27. [DOI] [PubMed] [Google Scholar]
- Elliott, C. , Zhou, F.S. , Spielmeyer, W. , Panstruga, R. and Schulze‐Lefert, P. (2002) Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol. Plant–Microbe Interact. 15, 1069–1077. [DOI] [PubMed] [Google Scholar]
- Feechan, A. , Jermakow, A.M. , Torregrosa, L. , Panstruga, R. and Dry, I.B. (2008) Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct. Plant Biol. 35, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics, 5, 164–166. [Google Scholar]
- Fernández‐Aparicio, M. , Prats, E. , Emeran, A.A. and Rubiales, D. (2009) Characterization of resistance mechanisms to powdery mildew (Erysiphe betae) in beet (Beta vulgaris). Phytopathology, 99, 385–389. [DOI] [PubMed] [Google Scholar]
- Fondevilla, S. , Carver, T.L.W. , Moreno, M.T. and Rubiales, D. (2006) Macroscopic and histological characterisation of genes er1 and er2 for powdery mildew resistance in pea. Eur. J. Plant Pathol. 115, 309–321. [Google Scholar]
- Fondevilla, S. , Torres, A.M. , Moreno, M.T. and Rubiales, D. (2007) Identification of a new gene for resistance to powdery mildew in Pisum fulvum, a wild relative of pea. Breed. Sci. 57, 181–184. [Google Scholar]
- Fondevilla, S. , Rubiales, D. , Moreno, M.T. and Torres, A.M. (2008) Identification and validation of RAPD and SCAR markers linked to the gene Er3 conferring resistance to Erysiphe pisi DC in pea. Mol. Breed. 22, 193–200. [Google Scholar]
- Harland, S.C. (1948) Inheritance of immunity to mildew in Peruvian forms of Pisum sativum . Heredity, 2, 263–269. [DOI] [PubMed] [Google Scholar]
- Heringa, R.J. , Vannorel, A. and Tazelaar, M.F. (1969) Resistance to powdery mildew (Erysiphe polygoni D.C.) in peas (Pisum sativum L.). Euphytica, 18, 163–169. [Google Scholar]
- Humphry, M. , Consonni, C. and Panstruga, R. (2006) mlo‐based powdery mildew immunity: silver bullet or simply non‐host resistance? Mol. Plant Pathol. 7, 605–610. [DOI] [PubMed] [Google Scholar]
- Humphry, M. , Bednarek, P. , Kemmerling, B. , Koh, S. , Stein, M. , Gobel, U. , Stüber, K. , Pislewska‐Bednarek, M. , Loraine, A. , Schulze‐Lefert, P. , Somerville, S. and Panstruga, R. (2010) A regulon conserved in monocot and dicot plants defines a functional module in antifungal plant immunity. Proc. Natl. Acad. Sci. USA, 107, 21 896–21 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janila, P. and Sharma, B. (2004) RAPD and SCAR markers for powdery mildew resistance gene er in pea. Plant Breed. 123, 271–274. [Google Scholar]
- Jarosch, B. , Kogel, K.H. and Schaffrath, U. (1999) The ambivalence of the barley Mlo locus: mutations conferring resistance against powdery mildew (Blumeria graminis f. sp. hordei) enhance susceptibility to the rice blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 12, 508–514. [Google Scholar]
- Jørgensen, J.H. (1977) Spectrum of resistance conferred by ML‐O powdery mildew resistance genes in barley. Euphytica, 26, 55–62. [Google Scholar]
- Jørgensen, J.H. (1992) Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica, 63, 141–152. [Google Scholar]
- Katoch, V. , Sharma, S. , Pathania, S. , Banayal, D.K. , Sharma, S.K. and Rathour, R. (2010) Molecular mapping of pea powdery mildew resistance gene er2 to pea linkage group III. Mol. Breed. 25, 229–237. [Google Scholar]
- Konishi, S. , Sasakuma, T. and Sasanuma, T. (2010) Identification of novel Mlo family members in wheat and their genetic characterization. Genes Genet. Syst. 85, 167–175. [DOI] [PubMed] [Google Scholar]
- Kozik, A. , Heidstra, R. , Horvath, B. , Kulikova, O. , Tikhonovich, I. , Ellis, T.H.N. , Vankammen, A. , Lie, T.A. and Bisseling, T. (1995) Pea lines carrying sym1 or sym2 can be nodulated by Rhizobium strains containing nodx—sym1 and sym2 are allelic. Plant Sci. 108, 41–49. [Google Scholar]
- Kumar, H. and Singh, R.B. (1981) Genetic analysis of adult plant resistance to powdery mildew in pea (Pisum sativum L.). Euphytica, 30, 147–151. [Google Scholar]
- Liu, Q. and Zhu, H. (2008) Molecular evolution of the MLO gene family in Oryza sativa and their functional divergence. Gene, 409, 1–10. [DOI] [PubMed] [Google Scholar]
- Lyngkjaer, M.F. , Newton, A.C. , Atzema, J.L. and Baker, S.J. (2000) The barley mlo‐gene: an important powdery mildew resistance source. Agronomie, 20, 745–756. [Google Scholar]
- Neumann, P. , Pozarkova, D. and Macas, J. (2003) Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol. Biol. 53, 399–410. [DOI] [PubMed] [Google Scholar]
- Panstruga, R. (2004) A golden shot: how ballistic single cell transformation boosts the molecular analysis of cereal–mildew interactions. Mol. Plant Pathol. 5, 141–148. [DOI] [PubMed] [Google Scholar]
- Panstruga, R. (2005a) Discovery of novel conserved peptide domains by ortholog comparison within plant multi‐protein families. Plant Mol. Biol. 59, 485–500. [DOI] [PubMed] [Google Scholar]
- Panstruga, R. (2005b) Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 33, 389–392. [DOI] [PubMed] [Google Scholar]
- Panstruga, R. and Schulze‐Lefert, P. (2003) Corruption of host seven‐transmembrane proteins by pathogenic microbes: a common theme in animals and plants? Microbes Infect. 5, 429–437. [DOI] [PubMed] [Google Scholar]
- Parniske, M. (2000) Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 3, 320–328. [DOI] [PubMed] [Google Scholar]
- Pereira, G. and Leitao, J. (2010) Two powdery mildew resistance mutations induced by ENU in Pisum sativum L. affect the locus er1 . Euphytica, 171, 345–354. [Google Scholar]
- Pereira, G. , Marques, C. , Ribeiro, R. , Formiga, S. , Damaso, M. , Sousa, M.T. , Farinho, M. and Leitao, J.M. (2010) Identification of DNA markers linked to an induced mutated gene conferring resistance to powdery mildew in pea (Pisum sativum L.). Euphytica, 171, 327–335. [Google Scholar]
- Piffanelli, P. , Zhou, F.S. , Casais, C. , Orme, J. , Jarosch, B. , Schaffrath, U. , Collins, N.C. , Panstruga, R. and Schulze‐Lefert, P. (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli, P. , Ramsay, L. , Waugh, R. , Benabdelmouna, A. , D'Hont, A. , Hollricher, K. , Jorgensen, J.H. , Schulze‐Lefert, P. and Panstruga, R. (2004) A barley cultivation‐associated polymorphism conveys resistance to powdery mildew. Nature, 430, 887–891. [DOI] [PubMed] [Google Scholar]
- Prats, E. , Llamas, M.J. and Rubiales, D. (2007) Characterization of resistance mechanisms to Erysiphe pisi in Medicago truncatula . Phytopathology, 97, 1049–1053. [DOI] [PubMed] [Google Scholar]
- Reinstädler, A. , Müller, J. , Czembor, J.H. , Piffanelli, P. and Panstruga, R. (2010) Novel induced mlo mutant alleles in combination with site‐directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant Biol. 10, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Lozano, J.M. , Gianinazzi, S. and Gianinazzi‐Pearson, V. (1999) Genes involved in resistance to powdery mildew in barley differentially modulate root colonization by the mycorrhizal fungus Glomus mosseae . Mycorrhiza, 9, 237–240. [Google Scholar]
- Safronova, V.I. and Novikova, N.I. (1996) Comparison of two methods for root nodule bacteria preservation: lyophilization and liquid nitrogen freezing. J. Microbiol. Methods, 24, 231–237. [Google Scholar]
- Shirasu, K. , Nielsen, K. , Piffanelli, P. , Oliver, R. and Schulze‐Lefert, P. (1999) Cell‐autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 17, 293–299. [Google Scholar]
- Timmerman, G.M. , Frew, T.J. , Weeden, N.F. , Miller, A.L. and Goulden, D.S. (1994) Linkage analysis of er‐1, a recessive Pisum sativum gene for resistance to powdery mildew fungus (Erysiphe pisi D.C.). Theor. Appl Genet. 88, 1050–1055. [DOI] [PubMed] [Google Scholar]
- Tiwari, K.R. , Penner, G.A. and Warkentin, T.D. (1997) Inheritance of powdery mildew resistance in pea. Can. J. Plant Sci. 77, 307–310. [Google Scholar]
- Tiwari, K.R. , Penner, G.A. and Warkentin, T.D. (1998) Identification of coupling and repulsion phase RAPD markers for powdery mildew resistance gene er‐1 in pea. Genome, 41, 440–444. [Google Scholar]
- Tonguc, M. and Weeden, N.F. (2010) Identification and mapping of molecular markers linked to er1 gene in pea. Plant Mol. Biol. Biotechnol. 1, 1–5. [Google Scholar]
- Trouvelot, A. , Kough, J. and Gianinazzi‐Pearson, V. (1986) Mesure des Taux de Mycorhization VA d'un Systeme Radiculaire. Recherche de Methode d'estimation Ayant une Signification Fonctionnelle, pp. 217–221. Paris: Institut National de la Recherche Agronomique Press. [Google Scholar]
- Vaid, A. and Tyagi, P.D. (1997) Genetics of powdery mildew resistance in pea. Euphytica, 96, 203–206. [Google Scholar]
- Wicker, T. , Sabot, F. , Hua‐Van, A. , Bennetzen, J.L. , Capy, P. , Chalhoub, B. , Flavell, A. , Leroy, P. , Morgante, M. , Panaud, O. , Paux, E. , SanMiguel, P. and Schulman, A.H. (2007) A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982. [DOI] [PubMed] [Google Scholar]
- Wyand, R.A. and Brown, J.K.M. (2005) Sequence variation in the CYP51 gene of Blumeria graminis associated with resistance to sterol demethylase inhibiting fungicides. Fung. Genet. Biol. 42, 726–735. [DOI] [PubMed] [Google Scholar]
- Young, N.D. , Mudge, J. and Ellis, T.H.N. (2003) Legume genomes: more than peas in a pod. Curr. Opin. Plant Biol. 6, 199–204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Sequence alignment of MLO coding sequences. Nucleotide sequences of LjMLO1 (GenBank accession number AY967410), SlMLO1 (AY967408), CaMLO1 (AY934528) and AtMLO2 (AF369563) were aligned by CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Conserved nucleotide stretches used for the design of oligonucleotide primers PsMLO1 (forward primer), PsMLO2 and PsMLO3 (reverse primers) are highlighted in light blue; the actual oligonucleotide sequences (highlighted in yellow) are shown below the respective regions. In the case of nucleotide positions that were divergent between the four MLO sequences, the oligonucleotide sequences were adjusted to the LjMLO sequence.
Table S1 Oligonucleotides used in this study.
Table S2 Single‐cell complementation of the er1 phenotype by transient expression of MLO genes.
Supporting info item
Supporting info item
Supporting info item


