SUMMARY
A mutant of the root pathogen Fusarium oxysporum f. sp. lycopersici, deficient in class V chitin synthase, has been shown previously to be nonvirulent. In this study, we tested the hypothesis that the cause of its avirulence could be the elicitation of the induced plant defence response, leading to the restriction of fungal infection. Co‐inoculation of tomato plants with the wild‐type strain and the ΔchsV mutant resulted in a significant reduction in symptom development, supporting a protective mechanism exerted by the mutant. The ability of the mutant to penetrate and colonize plant tissues was determined by scanning and transmission electron microscopy, as well as fluorescence microscopy using green fluorescent protein‐ or cherry fluorescent protein‐labelled fungal strains. The extent of wild‐type strain colonization in co‐inoculated plants decreased steadily throughout the infection process, as shown by the quantification of fungal biomass using real‐time polymerase chain reaction. The hypothesis that defence responses are activated by the ΔchsV mutant was confirmed by the analysis of plant pathogenesis‐related genes using real‐time reverse transcriptase‐polymerase chain reaction. Tomato plants inoculated with the ΔchsV mutant showed a three fold increase in endochitinase activity in comparison with wild‐type inoculated plants. Taken together, these results suggest that the perturbation of fungal cell wall biosynthesis results in elicitation of the plant defence response during the infection process.
INTRODUCTION
Fusarium oxysporum is a common soil‐borne fungus that causes vascular wilt disease on a wide range of plants, resulting in severe crop losses throughout the world. According to host specificity, F. oxysporum isolates have been classified in formae speciales (Armstrong and Armstrong, 1981). As an example, the forma specialis lycopersici is pathogenic on tomato plants (Lycopersicon esculentum).
Plant infection by pathogenic fungi involves molecular interactions between plants and microbes. In this scenario, the fungal cell wall plays an important role: on the one hand, it provides the necessary rigidity that enables the fungus to enter the plant cells and, on the other, many elicitors are generated from it, alerting the plants to the presence of a potential pathogen and activating the plant defence response. Indeed, it has been reported that the disruption of genes involved in the synthesis and maintenance of the cell wall results in nonvirulent fungal strains (Martin‐Urdiroz et al., 2008; Soulie et al., 2006; Weber et al., 2006; Werner et al., 2007).
A nonvirulent mutant, ΔchsV, affected in cell wall synthesis, has been isolated previously in F. oxysporum f. sp. lycopersici (Madrid et al., 2003). This nonvirulent strain lacks a functional class V chitin synthase, shows an altered cell wall with balloon‐like structures along the hyphae and has lost the capacity to infect tomato plants. We proposed that, as a consequence of its altered cell wall, the mutant elicits rapid plant defence responses, leading to host restriction of fungal colonization.
To validate this hypothesis, we performed single‐ and co‐inoculation experiments using the wild‐type and/or ΔchsV mutant. The results of these experiments were analysed by three approaches. In the first approach, we evaluated disease symptom development in co‐inoculated plants. A second approach involved time course analysis of plant colonization by means of visualization of F. oxysporum f. sp. lycopersici‐infected root sections by scanning electron microscopy (SEM), transmission electron microscopy (TEM) and fluorescence microscopy, as well as real‐time polymerase chain reaction (PCR) quantification of fungal biomass from each strain within the infected stems. Finally, we analysed whether ΔchsV induces plant defence response genes, which may limit the fungal colonization of host tissues and restrict infection by the wild‐type strain and nonvirulent mutant, similar to the mechanisms of plant disease control carried out by biocontrol agents (Alabouvette et al., 2006; Fravel et al., 2002).
RESULTS
Nonvirulent ΔchsV mutant protects tomato plants against wilt disease caused by the wild‐type strain
To determine whether the nonvirulent mutant ΔchsV could protect tomato plants against wilt disease caused by the F. oxysporum f. sp. lycopersici wild‐type strain, co‐inoculation was carried out by immersing the roots of 2‐week‐old tomato plants in microconidial suspensions of the pathogenic wild‐type strain and the nonvirulent ΔchsV mutant mixed at different ratios. These included the same amount of both strains (5 × 106 wild‐type : 5 × 106ΔchsV) and two fold (2.5 × 106 wild‐type : 5 × 106ΔchsV) or 10‐fold (5 × 105 wild‐type : 5 × 106ΔchsV) more mutant than wild‐type strain. Plants were scored for vascular wilt symptoms at different time intervals after inoculation. The severity of wilt symptoms in plants inoculated with the wild‐type strain alone, at 5 × 106, 2.5 × 106 or 5 × 105 microconidia/mL, increased steadily throughout the experiment, and most plants were dead 25 days after inoculation (data not shown). In contrast, a delay in the development of disease symptoms was observed in plants co‐inoculated with inoculum ratios containing at least double the amount of mutant versus wild‐type strain. The most significant reduction in the disease index was observed at the ratio 0.1 × wild‐type : 1 ×ΔchsV (5 × 105 : 5 × 106 microconidia/mL, respectively) (Fig. 1a). To demonstrate that the reduction in disease progression during co‐infection was not a result of physical competition during attachment to the roots, we performed successive inoculation assays in which tomato roots were first immersed in a microconidial suspension of the wild‐type strain (5 × 105 microconidia/mL for 15 min) and subsequently of the ΔchsV mutant (5 × 106 microconidia/mL for 15 min). In this way, the wild‐type strain was allowed to adhere to the roots without physical interference from the nonvirulent mutant. The results obtained did not differ from those observed during simultaneous co‐inoculation using strains at a ratio of 0.1 × wild‐type : 1 ×ΔchsV for 30 min (5 × 105 : 5 × 106 microconidia/mL, respectively). (Fig. 1a). In addition, no differences in the severity of wilt symptoms were found using wild‐type inoculum at 0.1 × concentration (5 × 105 microconidia/mL) for 15 min or 1 × concentration (5 × 106 microconidia/mL) for 30 min (data not shown).
Figure 1.
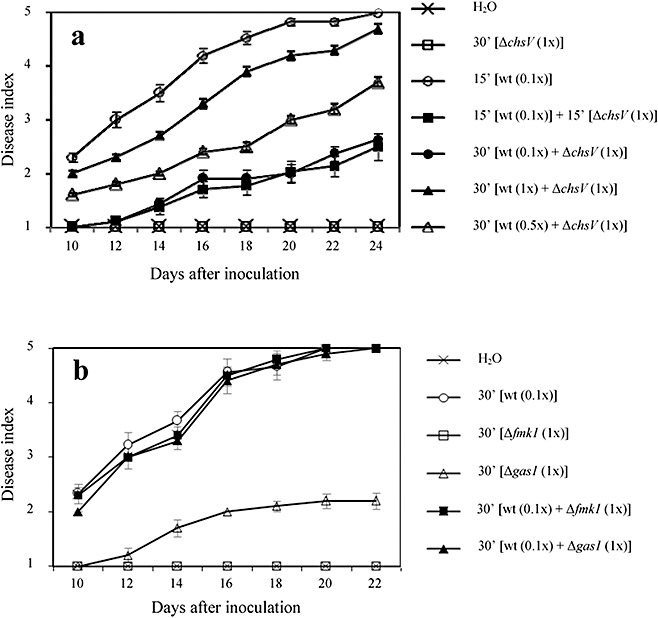
(a) Progression of wilt symptoms in tomato plants inoculated with the wild‐type strain, the ΔchsV mutant or simultaneously with both strains in different conditions. The severity of disease symptoms was recorded at different times after inoculation, using an index ranging from 1 (healthy plant) to 5 (dead plant). (b) Progression of wilt symptoms in tomato plants singly inoculated with the wild‐type strain, the Δgas1 mutant or the Δfmk1 mutant, or co‐inoculated as indicated. Error bars indicate the standard deviations from three independent experiments, each including 15 plants per treatment. See Table 1 for statistical analysis of the data.
To test whether this protective effect was specific for the ΔchsV mutant, tomato plants were co‐inoculated with the wild‐type microconidia and either the nonvirulent Δfmk1 mutant (lacking a functional mitogen‐activated protein kinase) (Di Pietro et al., 2001) or the highly virulence‐reduced Δgas1 mutant (lacking a functional β‐1,3‐glucanosyltransferase) (Caracuel et al., 2005), which is able to adhere to the root surface and to germinate (Fig. S1, see Supporting Information), using the ratio 0.1 × wild‐type : 1 × mutant strain (5 × 105 : 5 × 106 microconidia/mL, respectively). No delay in disease development was observed with any of these mutants (Fig. 1b), confirming the specificity of the protective effect exerted by the ΔchsV mutant. Significant differences among treatments were confirmed by analyses of variance (anovas) of the data (Table 1).
Table 1.
Severity of wilt disease in tomato plants inoculated with different combinations of Fusarium oxysporum f. sp. lycopersici strains.
| Treatment | Days post‐inoculation | |||||||
|---|---|---|---|---|---|---|---|---|
| 10 | 12 | 14 | 16 | 18 | 20 | 22 | 24 | |
| H2O | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A |
| 30′[ΔchsV (1×)]* | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A |
| 15′[wt (0.1×)]† | 2.3 (±0.1) B | 3.0 (±0.1) B | 3.5 (±0.2) B | 4.2 (±0.1) B | 4.5 (±0.1) B | 4.8 (±0.1) B | 4.8 (±0.1) B | 5.0 (±0.0) B |
| 15′[wt (0.1×)]†+ 15′[ΔchsV (1×)]* | 1.0 (±0.0) A | 1.1 (±0.1) A | 1.4 (±0.1) C | 1.7 (±0.2) C | 1.8 (±0.2) C | 2.0 (±0.2) C | 2.1 (±0.2) C | 2.5 (±0.2) C |
| 30′[wt (0.1×)†+ΔchsV (1×)*] | 1.0 (±0.0) A | 1.1 (±0.1) A | 1.4 (±0.1) C | 1.9 (±0.2) C | 1.9 (±0.2) C | 2.0 (±0.2) C | 2.4 (±0.1) C | 2.6 (±0.1) C |
| 30′[wt (1×)†+ΔchsV (1×)*] | 2.0 (±0.1) B | 2.3 (±0.1) C | 2.7 (±0.1) D | 3.3 (±0.1) D | 3.9 (±0.1) D | 4.2 (±0.1) D | 4.3 (±0.1) D | 4.7 (±0.1) B |
| 30′[wt (0.5×)‡+ΔchsV (1×)*] | 1.6 (±0.0) C | 1.8 (±0.0) D | 2.0 (±0.0) C | 2.4 (±0.1) E | 2.5 (±0.1) E | 3.0 (±0.1) E | 3.2 (±0.1) E | 3.7 (±0.1) D |
| 30′[wt (0.1×)]† | 2.3 (±0.2) B | 3.1 (±0.2) B | 3.6 (±0.2) B | 4.5 (±0.2) B | 4.7 (±0.2) B | 5.0 (±0.0) B | 5.0 (±0.0) B | 5.0 (±0.0) B |
| 30′[Δgas1 (1×)]* | 1.0 (±0.0) A | 1.2 (±0.1) A | 1.7 (±0.2) C | 2.0 (±0.0) C | 2.1 (±0.1) C | 2.2 (±0.1) C | 2.2 (±0.1) C | 2.2 (±0.1) C |
| 30′[Δfmk1 (1×)]* | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A | 1.0 (±0.0) A |
| 30′[wt (0.1×)†+Δgas1 (1×)*] | 2.0 (±0.0) B | 3.0 (±0.0) B | 3.3 (±0.2) B | 4.4 (±0.2) B | 4.7 (±0.1) B | 4.9 (±0.1) B | 5.0 (±0.0) B | 5.0 (±0.0) B |
| 30′[wt (0.1×)†+Δfmk1 (1×)*] | 2.3 (±0.2) B | 3.0 (±0.2) B | 3.4 (±0.2) B | 4.5 (±0.2) B | 4.8 (±0.1) B | 5.0 (±0.0) B | 5.0 (±0.0) B | 5.0 (±0.0) B |
Values are means (±standard error) for three independent experiments, each including 15 plants per treatment. Means with the same letter within the same column are not significantly different (Duncan, P≤ 0.05).
Concentration 1× corresponds to 5 × 106 microconidia/mL.
Concentration 0.1× corresponds to 5 × 105 microconidia/mL.
Concentration 0.5× corresponds to 2.5 × 106 microconidia/mL.
To rule out a plant genotype effect, simultaneous co‐inoculations using strains at the ratio of 0.1 × wild‐type : 1 ×ΔchsV (5 × 105 : 5 × 106 microconidia/mL, respectively) for 30 min were performed in three different tomato cultivars, Abramo, Moneymaker and Vemar, all susceptible to F. oxysporum f. sp. lycopersici wild‐type strain 4287 (race 2). In all cases, the results were similar to those observed with cultivar Monika (data not shown).
The ΔchsV mutant is able to penetrate and colonize tomato plant tissues
The ability of the ΔchsV mutant to adhere to the root surface and to penetrate and colonize the root tissues was compared with that of the wild‐type and the nonvirulent mutant ΔchsV using light microscopy, SEM and TEM of root sections from infected plants. Light microscopy revealed that, immediately after inoculation, the conidia of the wild‐type and the nonvirulent mutant strains became attached to the roots (data not shown). Eight hours after inoculation, they had germinated and started to colonize the root surface (Fig. 2a,e) and, 16 h after inoculation (Fig. 2b,f), hyphae were forming a network interwoven with the root hairs. SEM analysis of infected roots 24 h post‐inoculation revealed that, after attachment and germination, both strains formed a dense network randomly distributed around the root surface (Fig. 2c,g). In addition, the nonvirulent ΔchsV strain showed swollen structures in subapical regions (Fig. 2g). Although F. oxysporum does not produce specialized infection structures, we identified short hyphae penetrating the epidermal cells of the roots during the infection process of both the wild‐type and nonvirulent mutant (Fig. 2d,h). TEM analysis showed that, during the first 24 h of infection, hyphae from both the wild‐type and nonvirulent mutant penetrated the epidermis and invaded the cortex and the xylem bundles of tip and middle root (Fig. 3a,d), growing inter‐ or intracellularly (Fig. 3b,e). The invading hyphae appeared to be constricted and often formed a septum at the penetration point (Fig. 3c,f). No structural barriers, such as cell wall appositions, to stop fungal penetration into the root cells were detected during infection with either strain. Likewise, no differences in the accumulation of hydrogen peroxide were observed among plants inoculated with either strain (data not shown).
Figure 2.
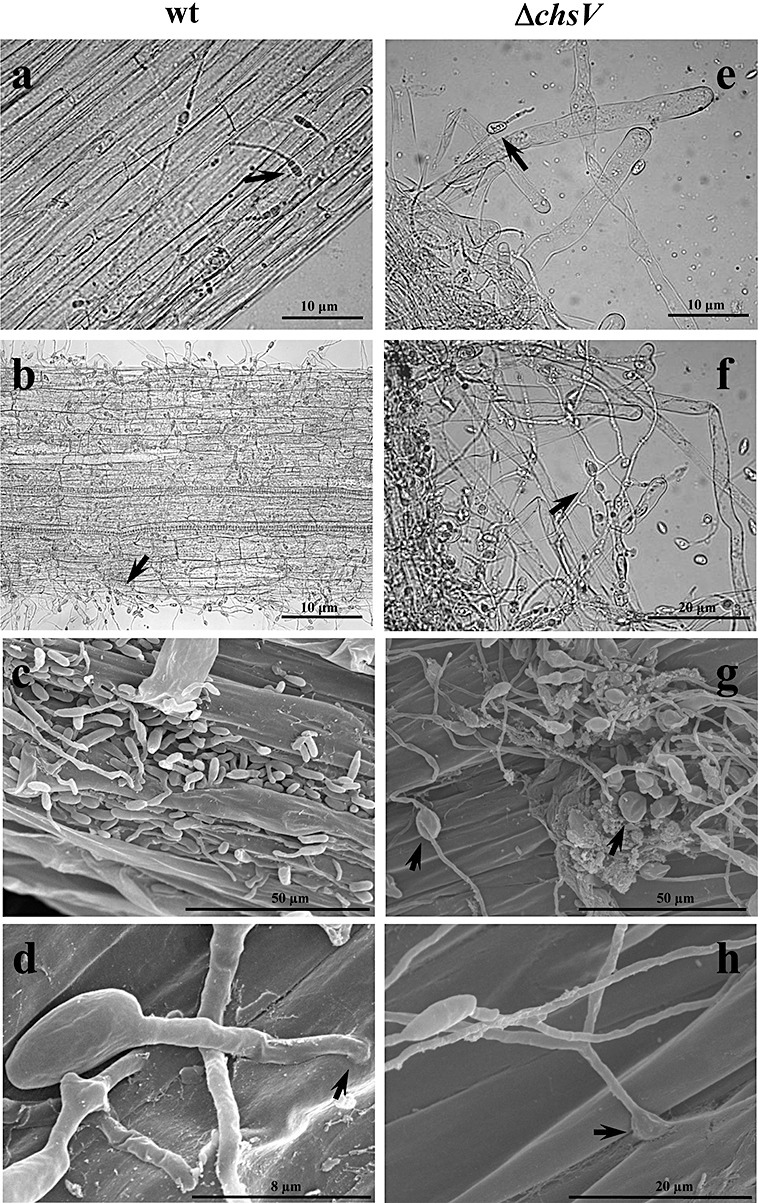
(a, b, e, f) Light microscopy analysis of tomato roots infected with the wild‐type strain (a, b) or the nonvirulent mutant ΔchsV (e, f). Micrographs were taken 8 h (a, e) or 16 h (b, f) after inoculation. Arrows indicate germlings adhering to the root surface. (c, d, g, h) Scanning electron microscopy analysis of tomato roots infected with the wild‐type strain (c, d) or the nonvirulent mutant ΔchsV (g, h). Micrographs were taken 24 h after inoculation. Arrows indicate penetration sites into the root (d, h) and globular structures in the ΔchsV mutant (g).
Figure 3.
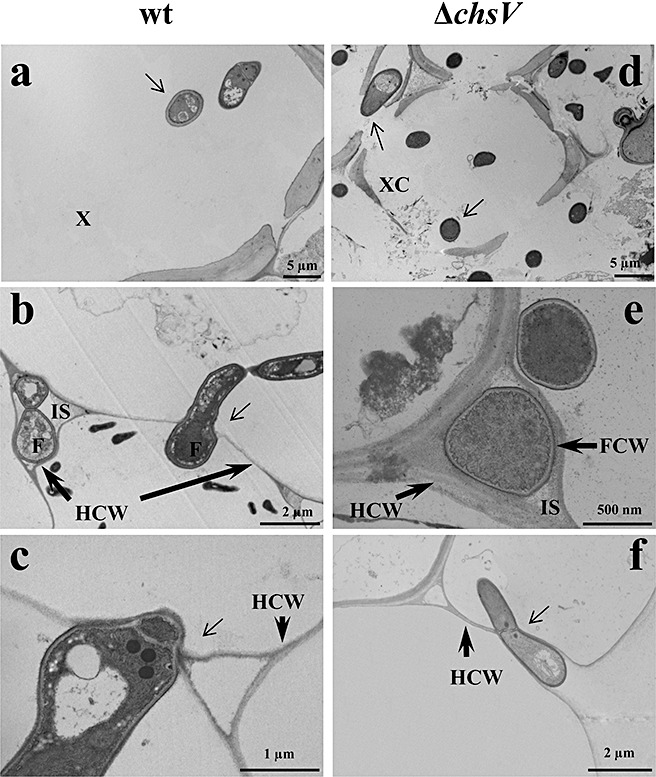
Transmission electron micrographs of tomato roots inoculated with the wild‐type strain (a–c) or the nonvirulent mutant ΔchsV (d–f). Micrographs were taken 24 h after inoculation. F, fungal cell; FCW, fungal cell wall; HCW, host cell wall; IS, intercellular space; XC, xylem cell. Thin arrows indicate fungal cells (a, d) or cell penetration sites (b, c, f).
To follow the colonization process by the wild‐type strain and the ΔchsV mutant, and to detect possible interactions during co‐infection, strains harbouring histone H1 tagged with green fluorescent protein (GFP) or the monomeric cherry red variant (ChFP) of monomeric red fluorescent protein (mRFP), respectively, were used (M. C. Ruiz‐Roldán et al., unpublished). Inoculation of tomato roots separately (concentration, 5 × 106 microconidia/mL) or simultaneously at a ratio of 0.1 × wild‐type : 1 ×ΔchsV (5 × 105 : 5 × 106 microconidia/mL, respectively) was carried out, and sections of infected roots were observed under a microscope using the differential interference contrast (DIC) technique or fluorescence. The presence of green‐ or red‐labelled nuclei in single‐inoculated plants confirmed the colonization of root tissues by both the virulent and nonvirulent strains, respectively (Fig. S2, see Supporting Information). Colonization was massive in infected plants, where labelled nuclei were visualized in all root tissues (Fig. S2c,d). Micrographs obtained using DIC from the infected root sections showed no externally attached hyphae (Fig. S2a,b), leading to the conclusion that the fluorescence‐tagged nuclei observed corresponded to mycelium growing inside the plant tissues.
After co‐inoculation of tomato roots with both strains at a ratio of 0.1 × wild‐type : 1 ×ΔchsV (5 × 105 : 5 × 106 microconidia/mL, respectively), we observed fluorescence‐tagged nuclei from both strains in the infected roots (Fig. 4). Although the inoculum density of the nonvirulent mutant was 10‐fold higher than that of the wild‐type strain, the extent of colonization by the two strains appeared to be similar, at least at the spatial root level analysed in this study. Considering that the observation of fluorescence nuclei is only a qualitative method, it should not be interpreted for the quantification of plant colonization by any of the strains.
Figure 4.
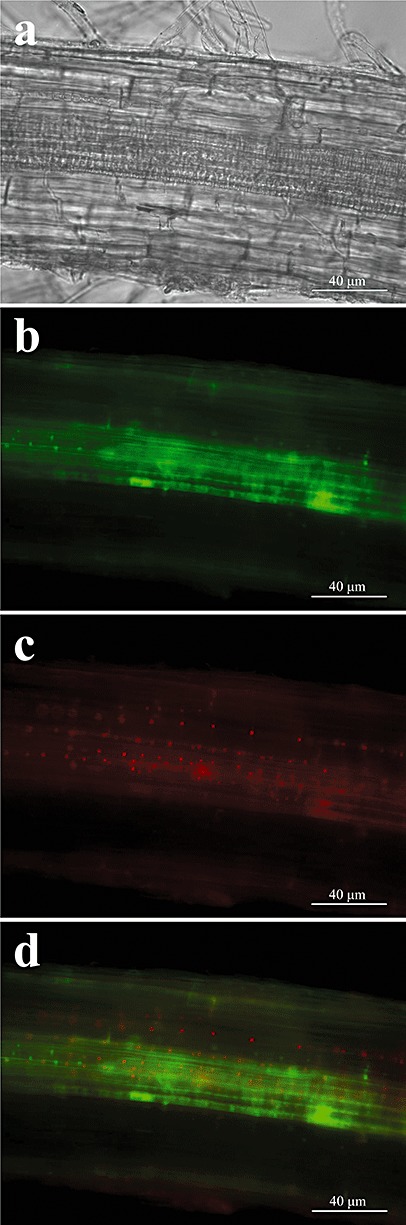
Co‐localization of wild‐type strain [harbouring green fluorescent protein (GFP)‐tagged H1 histone, green] and ΔchsV mutant [harbouring cherry red fluorescent protein (ChFP)‐tagged H1 histone, red] during co‐infection of tomato roots. Differential interference contrast (a) and fluorescence (b, c) micrographs were taken 5 days after co‐inoculation with the wild‐type strain and the ΔchsV mutant in the ratio 5 × 105 : 5 × 106 microconidia/mL, respectively (0.1× : 1×). (d) Images (b) and (c) merged.
ΔchsV mutant restricts colonization of tomato plant tissues by F. oxysporum wild‐type strain
The estimation of the wild‐type or ΔchsV biomass growing inside the infected plants was performed by quantification of specific fungal DNA within stems using real‐time PCR. The amount of fungal biomass in the stems increased during the course of infection in wild‐type inoculated plants, with maximal levels at 5 and 7 days after inoculation (Fig. 5a). By contrast, very low levels of wild‐type strain were detected in co‐inoculated plants (Fig. 5a). However, no significant differences in the amount of DNA amplified from ΔchsV were observed between single and co‐inoculated plants (Fig. 5b) at any time point. The total amount of mutant genomic DNA decreased during the course of infection, with a peak at 3 days after inoculation. Larger amounts of ΔchsV genomic DNA were detected compared with genomic DNA of the wild‐type strain (Fig. 5a,b), probably as a result of the different inoculum concentrations used (5 × 105 microconidia/mL for the wild‐type strain, and 5 × 106 microconidia/mL for the ΔchsV mutant). To avoid differences caused by inoculum concentration and primer efficiencies, quantitative real‐time PCR experiments were performed on plants inoculated with either the wild‐type strain or the ΔchsV mutant at 5 × 106 microconidia/mL, using a primer pair corresponding to the F. oxysporum actin1 gene (Table S2, see Supporting Information). At 3 days after inoculation, the amount of fungal DNA in the stems of plants inoculated with the ΔchsV mutant was significantly higher than that in those inoculated with the same number of spores of the wild‐type strain (Fig. 5c). By contrast, 7 days after inoculation, the wild‐type strain was more abundant than the ΔchsV mutant. The apparent discrepancies observed in the amounts of fungal DNA amplified from plants inoculated with the same strain using different primer pairs are probably a result of the different amplification efficiencies of the three primer sets used (Fig. 5). Differences among treatments within each day were confirmed by anovas of the data (Duncan, P≤ 0.05). These data suggest that, during the first 3 days of infection, the ΔchsV mutant colonizes the stem tissues more efficiently than does the wild‐type strain, but then gradually declines.
Figure 5.
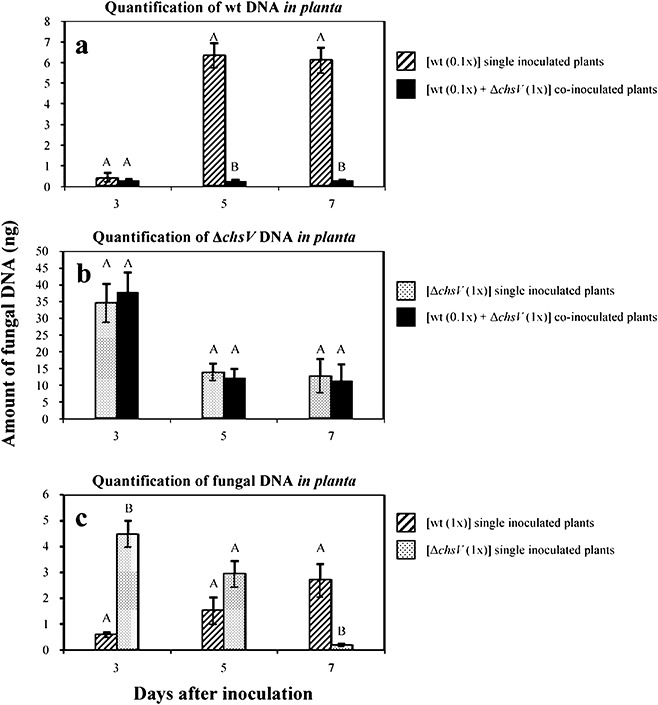
Comparative analysis of fungal biomass during single‐ or co‐inoculation experiments. (a) Quantitative real‐time polymerase chain reaction (PCR), with primer pair CHSV‐3 and CHSV‐26 specific for the wild‐type strain, on total DNA from single‐inoculated (striped bars) (using inoculation density of 5 × 105 microconidia/mL) or co‐inoculated (black bars) (using the ratio 5 × 105 microconidia/mL wild‐type : 5 × 106 microconidia/mL ΔchsV mutant) tomato plants. (b) Quantitative real‐time PCR, with primer pair GPDA‐16 and CHSV‐6 specific for the ΔchsV mutant, on total DNA from single‐inoculated (dotted bars) (using inoculation density of 5 × 106 microconidia/mL) or co‐inoculated (black bars) (using the ratio of 5 × 105 microconidia/mL wild‐type : 5 × 106 microconidia/mL ΔchsV mutant) tomato plants. (c) Quantitative real‐time PCR, with a primer pair specific for the Fusarium oxysporum actin1 gene, on total DNA from plants singly inoculated with the wild‐type strain (striped bars) or the ΔchsV mutant (dotted bars) using an inoculation density of 5 × 106 microconidia/mL in both cases. Data represent nanograms of fungal DNA amplified from 100 ng of DNA extracted from infected stems. Each column represents the mean from three independent inoculation experiments with three replicates each. Standard error bars are indicated. Columns with the same letter within the same day are not significantly different (Duncan, P≤ 0.05).
To test the possibility that the ΔchsV mutant inhibits directly the growth of the wild‐type strain, we performed in vitro fungal inhibition assays by co‐inoculation into either liquid or solid minimal medium (Fig. 6). No reduction in wild‐type growth was observed, in the number of colony‐forming units, distance between colony margins or colony diameter, indicating that the ΔchsV mutant did not produce any fungal growth inhibitor.
Figure 6.
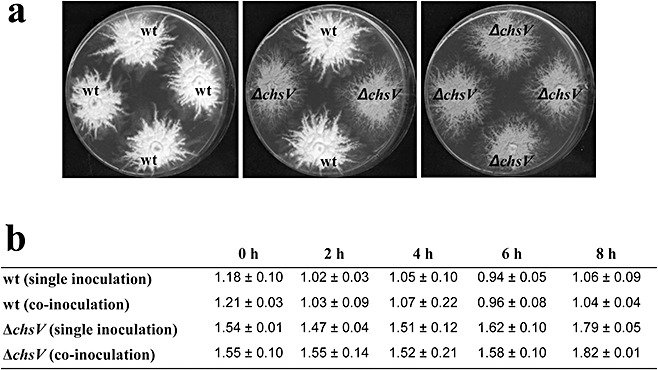
(a) Colony growth of the wild‐type strain and the ΔchsV mutant in solid minimal medium; 103 spores of each strain were inoculated and the plates were incubated at 28 °C for 7 days. (b) Number of colony‐forming units/mL (×105) of the indicated strains after different times of growth on single or co‐inoculated liquid cultures.
Cell wall‐degrading enzyme (CWDE)‐encoding genes are not differentially induced during plant infection
Fungal secreted CWDEs have multiple functions in virulence, including host penetration, nutrient acquisition, pathogen cell wall remodelling and elicitation of the plant defence response (Di Pietro et al., 2009). Fusarium oxysporum produces a remarkable variety of CWDEs belonging to different glycosyl hydrolase (GH) families, such as pectinases, cellulases and xylanases, among others, that are important for degrading plant cell wall components (Di Pietro et al., 2003). To test whether GH genes are differentially expressed in the ΔchsV mutant and the wild‐type strain during the early stages of plant infection, reverse transcriptase (RT)‐PCR was performed using RNA isolated from infected tomato roots 3 days after inoculation. The analysis included 21 genes belonging to different GH families, such as β‐glucuronidase (GH79), β‐1,3‐glucanase (GH64), polygalacturonase (GH28) and chitinase (GH18), among others (gene identification numbers are listed in Table S2). No differences in transcript levels were observed in plants inoculated with either strain. The expression of six representative genes is depicted in Fig. S3 (see Supporting Information).
Plant defence responses are differentially activated by the ΔchsV mutant in single and co‐infected tomato plants
The plant defence reaction in response to co‐infection with the wild‐type and nonvirulent ΔchsV strains was analysed by the quantification of transcript levels of defence‐related genes encoding extracellular invertase (lin6) (Hedley et al., 1994), acidic (gluA) and basic (gluB) glucanases (van Kan et al., 1992), acidic chitinase 3 (chi3), basic chitinase 9 (chi9) (Danhash et al., 1993) and pathogenesis‐related protein 1 (pr‐1) (van Kan et al., 1992) in tomato plant roots at 24 h post‐inoculation, using quantitative real‐time RT‐PCR (Fig. 7a). The expression level of each defence‐related gene was compared among plants infected with either the wild‐type strain (at concentrations of 5 × 105 or 5 × 106 microconidia/mL), nonvirulent ΔchsV mutant (at 5 × 106 microconidia/mL), or co‐inoculated with both strains (5 × 105 microconidia/mL wild‐type : 5 × 106 microconidia/mL ΔchsV), as well as noninoculated control plants, and referred to the relative levels of the constitutive reference gene gapdh encoding the glyceraldehyde‐3‐phosphate‐dehydrogenase (Shih et al., 1991). The expression levels of the six genes detected in plants inoculated with the wild‐type strain did not differ significantly from those observed in the noninoculated controls (Fig. 7a). By contrast, transcript levels of lin6, gluB, chi3, chi9 and pr‐1 were significantly higher in plants inoculated with the ΔchsV mutant and in co‐inoculated plants, compared with plants inoculated with the wild‐type strain or the noninoculated controls. In the case of gluA, transcription levels were significantly reduced in plants inoculated with the ΔchsV mutant, as well as in co‐inoculated plants, in comparison with noninoculated controls or plants inoculated with the wild‐type strain. Differences in transcript levels among treatments were confirmed by anovas of the data (Duncan, P≤ 0.05).
Figure 7.
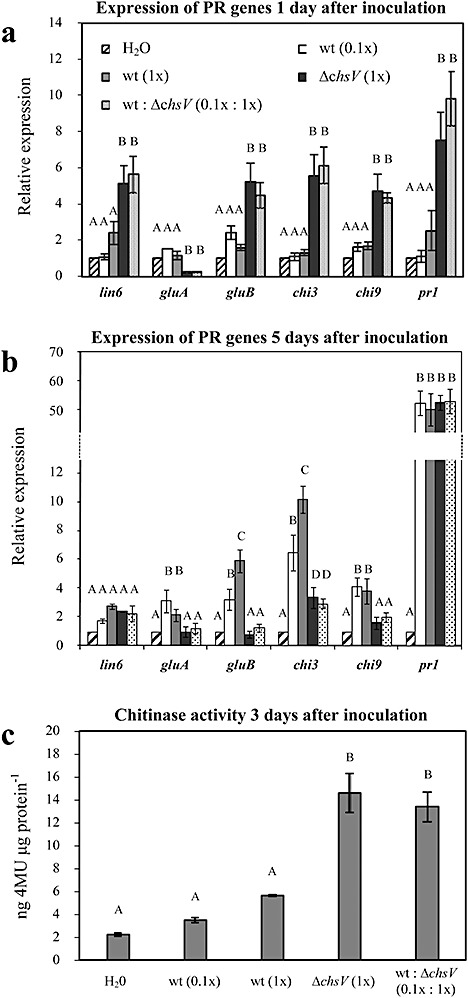
(a, b) Expression of defence‐response genes in tomato plants at 1 (a) or 5 days (b) after inoculation with the wild‐type strain at concentrations of 5 × 105 microconidia/mL (white bars) and 5 × 106 microconidia/mL (grey bars), mutant at 5 × 106 microconidia/mL (black bars) or both strains simultaneously in the ratio of 5 × 105 microconidia/mL wild‐type : 5 × 106 microconidia/mL ΔchsV mutant (dotted bars), and the noninoculated controls (striped bars). Transcript abundance was determined by quantitative reverse transcriptase‐polymerase chain reaction. Expression levels in each sample were normalized to the expression of the tomato gadph gene and were calculated relative to the uninfected control plants by the ΔΔCt method. Error bars indicate the standard error calculated from four independent inoculation experiments with two replicates each. Columns with the same letter within the same gene are not significantly different (Duncan, P≤ 0.05). (c) Endochitinase activity in stems of tomato plants 3 days after inoculation with the indicated strains at the indicated concentrations (0.1×, 5 × 105 microconidia/mL; 1×, 5 × 106 microconidia/mL). Noninoculated plants (H2O) were used as controls. Chitinase activity is expressed as nanograms of 4‐methylumbelliferone (4MU) released from the substrate per microgram of protein. Error bars indicate standard deviations from three independent inoculation experiments. Columns with the same letter are not significantly different (Duncan, P≤ 0.05).
In order to study the persistence of the induced plant defence response, transcript levels of the six defence‐related genes were quantified 3, 5 and 7 days after inoculation. The results obtained 3 days after inoculation were similar to those observed 24 h post‐inoculation (data not shown) but, after 5 days, the expression levels of lin6, gluB, chi3 and chi9 decreased in plants inoculated with the ΔchsV mutant, as well as in co‐inoculated plants, whereas the levels of gluB, chi3 and chi9 increased significantly in plants inoculated with the wild‐type strain (Fig. 7b). The expression of pr‐1 increased through time in all treatments, except for the noninoculated controls. The increase ranged from five fold in plants inoculated with the ΔchsV mutant, or in co‐inoculated plants, to 50‐fold in plants inoculated with the wild‐type strain (Fig. 7b). Differences in transcript levels among treatments and time points were confirmed by anovas of the data (Duncan, P≤ 0.05). The results observed 7 days after inoculation were similar to those obtained 5 days post‐inoculation (data not shown).
Chitinase activity is commonly induced as part of the plant defence response (Dalisay and Kuc, 1995; Heath, 1996). A significant increase in endochitinase activity, with a maximum at 3 days post‐inoculation, was observed in ΔchsV and co‐inoculated plants, in comparison with wild‐type inoculated plants and the noninoculated controls (Fig. 7c). No significant differences were observed in exochitinase activity. Noninoculated controls showed basal levels of chitinase activity that remained constant throughout the experiment (data not shown). Differences in chitinase activity among treatments were confirmed by anovas of the data (Duncan, P≤ 0.05).
DISCUSSION
Colonization of plant tissues by the nonvirulent mutant ΔchsV
In order to clarify the molecular basis for the inability of the ΔchsV mutant to cause wilt disease on tomato plants, we carried out a comparative microscopy analysis of the infection process of the ΔchsV mutant and wild‐type strain. We demonstrated that, after adhesion to the root surface, both strains were able to penetrate the epidermal cells and colonize the root tissue. Interestingly, 3 days after inoculation, the amount of fungal biomass detected in the stems of tomato plants inoculated with the ΔchsV mutant was significantly higher than that in those inoculated with the wild‐type strain, suggesting that the mutant colonizes plant tissues more efficiently than the wild‐type strain during the early stages of infection. However, no differences in the growth rate were found between these strains during in vitro culture. As shown previously, the absence of ChsV does not induce the expression of the genes involved in fungal cell wall synthesis, such as chs1, chs2, chs3, chs7, gas1 or rho1 (Caracuel et al., 2005; Madrid et al., 2003; Martin‐Urdiroz et al. 2008), which might be subjected to possible compensatory mechanisms. Nevertheless, we do not exclude the existence of unidentified additional traits influencing the rapid and abundant colonization of tomato tissues by the ΔchsV mutant.
Our results disagree with previous studies conducted in the ΔchsV mutant, where the growth of this strain was not detected inside inoculated plants (Madrid et al., 2003). The discrepancy between the two studies could be a result of the different accuracy of the methods used.
Protection against tomato wilt disease by the nonvirulent mutant ΔchsV
As a second approach, we explored the hypothesis that the ΔchsV mutant colonizes the plant but, as a consequence of its altered cell wall structure, rapidly elicits an intense and effective plant defence response, leading to a restriction of pathogen colonization. If this assumption is correct, the plant should show reduced disease symptoms on infection with the Fusarium wild‐type strain in the presence of the ΔchsV mutant. We performed co‐inoculation experiments with both strains and observed a significant delay in the appearance of disease symptoms in co‐inoculated plants. This protective effect was only detected with the cell wall‐defective mutant ΔchsV, as other nonvirulent mutants (Δgas1 orΔfmk1) did not protect against infection by the wild‐type strain. It has been shown previously that inoculation with avirulent isolates of F. oxysporum can result in a significant reduction in disease symptoms caused by virulent isolates from the same or a different forma specialis (Huertas‐Gonzalez et al., 1999). However, as most of these strains are soil‐borne nonpathogenic isolates, there is insufficient knowledge about the biochemical and genetic bases that differentiate them from the virulent strains and enable them to protect plants against pathogens. Some authors have proposed the use of nonvirulent mutants as biocontrol agents for fungal pathogenesis. A nonvirulent mutant of the ascomycete Colletotrichum magna (path‐1) was able to colonize cucurbit plants without inducing disease symptoms, and protected plants against C. magna, F. oxysporum (Freeman and Rodriguez, 1993) and C. orbiculare (Redman et al., 1999a). This nonpathogenic mutant is impaired in the production and secretion of pectate lyase and endopolygalacturonase (Wattad et al., 1995), and elicits a more rapid, intense and localized defence response (Redman et al., 1999a). Likewise, the C. magna R1 nonvirulent mutant confers protection to watermelon plants against the wild‐type strain (Redman et al., 1999b). Fusarium oxysporum f. sp. melonis nonvirulent mutant 4/4 significantly reduces the mortality of muskmelon and watermelon plants caused by F. oxysporum f. sp. melonis (race 1,2) and F. oxysporum f. sp. niveum (race 2), respectively (Freeman et al., 2001).
The protective abilities of biocontrol agents generally involve multiple mechanisms expressed successively, simultaneously or synergistically, including microbial antagonism (parasitism, competition for nutrients or for the colonization of plant tissues, and antibiosis) and induced resistance of the plant (reviewed by Alabouvette et al., 2009). On the basis of the results of the in vitro fungal inhibition assays, we exclude the possibility that ΔchsV inhibits the growth of the wild‐type strain. However, the fact that protection was increased with a higher inoculum density of the nonvirulent mutant versus the wild‐type, together with the small amount of wild‐type biomass present in stems of co‐inoculated plants, suggest that both strains are competing for nutrients and/or plant tissues. The direct competition between two strains of F. oxysporum (nonvirulent and virulent) within the host plant has been proposed previously as the cause of the reduced colonization of the carnation Dianthus caryophyllus stem by the pathogenic strain, resulting in a decrease in disease severity (Postma and Luttikholt, 1996).
Activation of the plant defence responses
The induction of tomato defence responses by the ΔchsV mutant could also be one of the reasons for the protective effect exerted by this strain. Indeed, expression analysis of plant defence‐related genes revealed an earlier accumulation of higher levels of transcripts in plants inoculated with the nonvirulent mutant, in comparison with the wild‐type strain and the noninoculated controls, as early as 24 h post‐inoculation. This rapid and intense activation of plant defences on infection by the ΔchsV mutant, together with the higher sensitivity of this mutant to plant defence compounds, would lead to an efficient restriction of fungal growth inside the plant, which was evident 5 days after inoculation with the mutant strain. By contrast, the delayed activation of plant defences in response to wild‐type infection is not sufficient to prevent disease development. In addition, increased endochitinase activity was detected in plants either single or co‐inoculated with the ΔchsV mutant. By contrast, no cell wall appositions or differential accumulation of reactive oxygen species were observed in response to the nonvirulent mutant infection. Alterations in the response to nonvirulent Fusarium isolates in comparison with virulent strains have been reported previously in other systems (van Kan et al., 1992). Plants inoculated with the nonvirulent strain Fo47 showed wall appositions and increased levels of PR‐1 protein, chitinase, β‐1,3‐glucanase and β‐1,4‐glucosidase activities (Benhamou and Garand, 2001; Duijff et al., 1998; Fuchs et al., 1997; Olivain et al., 2003); plants treated with the nonvirulent Fusarium strain CS‐20 showed increased accumulation of phenolic compounds (Panina et al., 2007). Attenuated expression of PR‐5‐ and PR‐7‐encoding genes was detected in F. solani strain Fs‐K‐infected plants (Kavroulakis et al., 2007).
The elicitation of plant defence reactions has also been attributed to the secretion of increased amounts of CWDEs by the pathogen (Di Pietro et al., 2003). However, the ΔchsV mutant did not show the differential expression of several CWDE genes tested during plant infection, indicating that the cell wall rigidity defect has no pleiotropic effect on the expression of CWDEs.
In summary, we have demonstrated that the ΔchsV mutant, lacking a chitin synthase essential for pathogenicity, is able to penetrate tomato roots, grow efficiently inside plant cells and elicit the plant defence response. The activation of plant defences might be caused simply by the increased fungal biomass of the ΔchsV mutant within infected tissues. Presumably, elicitors are readily released from the altered cell wall of the mutant, rapidly alerting the plant to activate its defences, highlighting the importance of the fungal cell wall in the infection process. These elicitors are more abundant at higher inoculum concentrations, and thus the protective effect exerted by the mutant is more effective in these conditions. Furthermore, the rapid response caused by the nonvirulent ΔchsV mutant protects the plant against infection by the pathogenic wild‐type strain. In addition, both strains probably compete for nutrients and/or plant tissues. In this sense, the higher colonization efficiency of the ΔchsV mutant at the beginning of the infection process could be considered as an advantage over the wild‐type strain. Thus, we propose two successive or simultaneous mechanisms to explain the protective effect exerted by the ΔchsV mutant: elicitation of plant defence responses and competition for tissue colonization. Our results highlight the importance of deciphering the molecular dialogue between the plant and pathogen during the early stages of infection in order to understand the bases that govern fungal pathogenicity.
EXPERIMENTAL PROCEDURES
Fungal isolates and culture conditions
Fusarium oxysporum f. sp. lycopersici wild‐type strain 4287 (race 2) was obtained from J. Tello, Universidad de Almería, Spain. The ΔchsV mutant deficient in the class V chitin synthase gene (Madrid et al., 2003), the Δfmk1 mutant deficient in the mitogen‐activated protein kinase 1 (Di Pietro et al., 2001), the Δgas1 mutant deficient in a β‐1,3‐glucanosyltransferase (Caracuel et al., 2005) and the wild‐type harbouring histone H1 tagged with GFP (wild‐type FoH1::GFP) (M. C. Ruiz‐Roldán et al., unpublished) have been described elsewhere. Tagging of histone H1 in the ΔchsV mutant strain was completed by fusion PCR, as described previously (Yang et al., 2004), using a cassette containing a hinge region encoding five Gly‐plus‐Ala repeats in frame at the N‐terminus of ChFP of mRFP (Campbell et al., 2002; Shaner et al., 2004), followed by the phleomycin resistance cassette as a selectable marker (M. C. Ruiz‐Roldán et al., unpublished). The tagging construct was used to transform protoplasts of F. oxysporum f. sp. lycopersici isolate 4287, as reported previously (Di Pietro and Roncero, 1998). Several transformants carrying a copy of ChFP‐tagged hisH1 at the homologous locus (ΔchsV‐FoH1::ChFP) were identified by PCR and southern hybridization. Tomato infection assays were performed using wild‐type FoH1::GFP or ΔchsV FoH1::ChFP in order to confirm their pathotypic behaviour: highly virulent and nonvirulent, respectively (data not shown).
Microconidial suspensions were stored with 30% glycerol at −80 °C. The pathotype of the isolates was confirmed periodically by plant infection assays. For fungal DNA extraction and microconidia production, cultures were grown in potato dextrose broth (PDB) (Difco Laboratories, Detroit, MA, USA) at 28 °C with shaking at 170 rpm for 3 days, as described previously (Di Pietro and Roncero, 1998).
Plant infection
For pathogenicity assays, infection of tomato plants was performed as reported previously (Di Pietro and Roncero, 1998). Briefly, 2‐week‐old tomato seedlings were inoculated with F. oxysporum wild‐type and/or mutant strains by immersing the roots in a suspension of 5 × 106 microconidia/mL (1 × concentration), 2.5 × 106 microconidia/mL (0.5 × concentration) or 5 × 105 microconidia/mL (0.1 × concentration) for 30 min. For co‐inoculation experiments, microconidia of both strains were mixed at the desired concentrations. The inoculum size of each strain during co‐inoculation is indicated as a ratio of wild‐type : mutant strain. As controls, tomato roots were immersed in microconidial suspension from one strain for 15 min, followed by immersion in a suspension of the other strain during an additional 15 min. Infected tomato seedlings were planted in vermiculite and maintained in a growth chamber. Fifteen plants were used for each treatment. The severity of disease symptoms was recorded at different times after inoculation using the following index: 1, no apparent symptoms; 2, beginning of wilt symptoms in leaves; 3, leaves heavily wilted; 4, all leaves completely wilted; 5, dead plant (Huertas‐Gonzalez et al., 1999). Virulence experiments were performed three times. Data were analysed with the software SPSS 15.0 for Windows® (LEAD Technologies, Inc., Charlotte, NC, USA). anova was performed and the Duncan post hoc test was executed to assess the differences among treatments within each day at P≤ 0.05. The arcsine transformation was performed for data normalization. For fluorescence microscopy analysis, tomato roots were immersed in a microconidial suspension at the desired concentration for 30 min, planted in vermiculite and maintained in a growth chamber for 5 days.
For light microscopy, SEM and TEM, tomato roots were immersed in Erlenmeyer flasks containing a suspension of 5 × 106 spores/mL, and maintained in a growth chamber at 80 rpm. At different times after inoculation, 1‐cm tissue sections were excised from three different root zones, the root tip, the middle root (around 3 cm away from the root tip) and the upper root (around 6 cm away from the root tip), and the samples were processed for further analysis.
Inoculations for the analysis of fungal colonization, plant defence‐related gene expression and fungal gene expression were performed by immersing the roots in microconidial suspensions at the desired concentrations, and the flasks were maintained in a growth chamber at 80 rpm for 1, 3, 5 or 7 days. After incubation, roots and stems were collected separately and stored at −80 °C until analysis. Five plants were used per treatment.
Seeds from tomato cv. Abramo, Monika, Moneymaker and Vemar were kindly provided by Syngenta (Almeria, Spain).
Microscopy
Sections of infected roots were wetted with water and placed directly on glass slides to be observed using a Leica DMRB microscope (Leica Microsystems, Wetzlar, Germany) by the DIC technique. Fluorescence was obtained using appropriate filter blocks: GFP for green fluorescence (470/40‐nm excitation, 500‐nm dichromatic mirror, 525/50‐nm barrier filter; Leica) or TX2 for red fluorescence (560/40‐nm excitation, 595‐nm dichromatic mirror, 645/75‐nm barrier filter; Leica). Photographs were recorded with a Leica DC 300F digital camera. Dual colour images were acquired by sequential scanning with settings optimal for GFP, followed by settings optimal for ChFP. The projections of the individual channels were merged in Photoshop 7.0 (Adobe, San Jose, CA, USA) and ImageJ (National Institutes of Health, Bethesda, MA, USA) to facilitate visualization. For each treatment, an average of 10 sections from three different roots was examined. For SEM, 5‐mm sections of infected roots were fixed for 2 h in 2% glutaraldehyde at room temperature and dehydrated through a graded ethanol series (30–100%) followed by 100% acetone for critical point drying. Samples were coated with a thin gold layer and observed with a JEOL 6300 scanning electron microscope. For each treatment, an average of 10 sections from three different roots was examined.
For TEM, 1‐cm sections of infected roots were fixed overnight at 4 °C in a mixture of 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 m sodium cacodylate buffer pH 7.0 (CAB). After fixation, tissues were washed twice for 10 min in CAB and post‐fixed for 45 min in 1% (v/v) osmium tetroxide in CAB. Tissues were then washed again in CAB (twice for 10 min) and dehydrated in a graded ethanol series [30, 50, 70, 80 and 90% (v/v) ethanol; 15 min each change and then three changes of 100% ethanol of 20 min each in duration]. Samples were transferred to two changes of propylene oxide of 20 min each in duration and progressively embedded in Epon 812 (Polaron, Watford, UK). Tissues were then subjected to 12 h in pure resin, followed by a change of fresh resin for 4 h, placed in blocks and polymerized at 65 °C for 48 h. Ultrathin sections (80 nm) were cut with a diamond knife and mounted on copper grids. The samples were stained in 2% aqueous uranyl acetate for 2 min at 37 °C, and then transferred to Reynold's lead citrate for 3 min at room temperature. Micrographs were obtained using a Philips CM 10 electron microscope (Cambridge, UK). For each treatment, an average of five samples from three different roots was investigated. For each sample, 10–15 ultrathin sections were examined.
Cytochemical detection of hydrogen peroxide (H2O2)
The histochemical method used for the localization of H2O2 was based on the generation of cerium perhydroxides described previously (Bestwick et al., 1997). One‐centimetre sections were excised from roots 24 and 48 h post‐inoculation. Noninoculated plants were included as controls. Sections were incubated in freshly prepared 5 mm CeCl3 in 50 mm 3‐(N‐morpholino)propanesulphonic acid (MOPS) at pH 7.2 for 1 h at room temperature. Tissues were then fixed, post‐fixed, dehydrated and embedded as described above for TEM. Unstained 80‐nm sections were examined using a Philips CM 10 electron microscope. For each treatment, an average of five samples from three different roots was investigated. For each sample, 10–15 ultrathin sections were analysed.
Fungal inhibition assay
Inhibition assays in axenic cultures were performed by simultaneous inoculation of 2.5 × 105 spores/mL from both the wild‐type FoH1::GFP strain harbouring the phleomycin resistant cassette and the ΔchsV mutant (resistant to hygromycin) into minimal medium (MM). Cultures were incubated at 28 °C with shaking at 170 rpm, and aliquots were removed 0, 2, 4, 6 or 8 h after inoculation, and plated on MM agar plates containing either 55 µg/mL hygromycin or 5.5 µg/mL phleomycin. Fungal inhibition was estimated by colony counting after 2 days at 28 °C. Control experiments were conducted by the single inoculation of either the wild‐type FoH1::GFP strain or the ΔchsV mutant in MM. In addition, MM agar plates were inoculated with water droplets containing 103 spores from either the wild‐type strain or the ΔchsV mutant. The colony diameter and distance between the colony margins were measured after incubation for 7 days at 28 °C. All experiments were performed three times.
Nucleic acid isolation
Total genomic DNA from mycelia was extracted from each strain and from stems of noninfected and infected plants using the commercially available DNeasy Plant Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Total RNA was isolated from germlings and from roots of noninfected and infected plants using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. The quality of extracted nucleic acids was verified by running aliquots in ethidium bromide‐stained agarose gels [0.7% w/v in Tris‐acetate‐EDTA (TAE) buffer] and further visualized under ultraviolet (UV) light. In addition, they were also quantified spectrophotometrically in a NanoDrop ND‐1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The isolated RNA was treated with deoxyribonuclease I (DNase I, Fermentas, Glen Burnie, MD, USA) and used to synthesize cDNA with the ribonuclease inhibitor RNasin Plus RNase Inhibitor (Promega, Madison, WI, USA) and M‐MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions, using a poly‐dT antisense primer.
In planta quantification of F. oxysporum strains
Plant roots were maintained immersed in microconidial suspensions of the wild‐type strain (5 × 105 microconidia/mL) and the ΔchsV mutant (5 × 106 microconidia/mL), either separately or simultaneously, for 3, 5 or 7 days. To avoid amplification of genomic DNA from external fungal mycelium which had not penetrated the roots, only the stems were collected for DNA analysis. Five plants were used per treatment. Real‐time PCR assays for the quantification of fungal genomic DNA from infected stems were performed using primer pairs CHSV‐3 (5′‐ACAGCTCCAACGAACTCTCTT‐3′) and CHSV‐26 (5′‐GGAGGTACTTGGTCATGTCGT‐3′) for specific detection of the wild‐type strain (Madrid et al., 2003), and GPDA‐16 (5′‐AGGGGCTGTATTAGGTCTC‐3′) and CHSV‐6 (5′‐CCGAGTTTCTGGGTATGACA‐3′) for specific detection of the ΔchsV mutant strain, which define amplicons of 511 and 460 bp, respectively. Primer specificity was corroborated by standard PCR using fungal genomic DNA from both strains as template (data not shown).
Reaction mixtures contained 12.5 µL of iQ SYBR Green Supermix (Bio‐Rad, Hercules, CA, USA), 300 nm of each primer and 100 ng of total DNA extracted from stems in a final volume of 25 µL. Three simultaneous replicated amplifications were carried out for each DNA sample, using 25‐µL aliquots from a 75‐µL mixture. Amplification reactions were performed in 96‐well microtitre plates (Bio‐Rad). PCRs were performed in an iCycler apparatus (Bio‐Rad) using the following cycling protocol: an initial step of denaturation (5 min, 94 °C) followed by 40 cycles of 30 s at 94 °C, 30 s at 62 °C for wild‐type strain detection or 60 °C for ΔchsV strain detection, 45 s at 72 °C and 20 s at 80 °C for measurement of the fluorescence emission. After this, a melting curve programme was run for which measurements were made at 0.5 °C temperature increases every 5 s within a range of 55–95 °C. Finally, PCR products were also visualized under UV light in ethidium bromide‐stained agarose gels (1% in TAE buffer).
The DNA concentration of each sample was extrapolated from standard curves, which were developed by plotting the logarithm of known concentrations (10‐fold dilution series from 10 ng to 1 pg/25 µL reaction) of DNA from F. oxysporum strains against the Ct values. In order to normalize the amplification conditions of the serially diluted DNA samples, 100 ng of DNA from noninoculated plants were added to each sample in the dilution series. The experiment was repeated three times using independent infected tissues. Data were analysed with the software SPSS 15.0 for Windows® (LEAD Technologies, Inc.). anova was performed and the Duncan post hoc test was executed to assess the differences among treatments within each day at P≤ 0.05.
Quantitative PCR of defence‐related genes
Real‐time PCRs were performed in an iCycler apparatus (Bio‐Rad) using iQ SYBR Green Supermix (Bio‐Rad), 2 µL of cDNA template and 300 nm of each gene‐specific primer (Table S1, see Supporting Information) in a final reaction volume of 25 µL. All primer pairs amplified products of 160–200 bp. The following PCR programme was used for all reactions: an initial step of denaturation (5 min, 94 °C), followed by 40 cycles of 30 s at 94 °C, 30 s at 60 °C, 30 s at 72 °C and 20 s at 80 °C for measurement of the fluorescence emission. A melting curve programme was run for which measurements were made at 0.5 °C temperature increments every 5 s within a range of 55–95 °C. Each sample reaction was performed in duplicate for each gene assay. Relative levels of the RT‐PCR products were determined using the ΔΔCt method (Livak and Schmittgen, 2001). Ct values were normalized to the Ct value of the gapdh housekeeping gene. Normalized transcript levels of each gene in infected samples were compared with levels in noninoculated samples. The experiments were repeated four times with independent infected tissues. Data were analysed with the software SPSS 15.0 for Windows® (LEAD Technologies, Inc.). anova was performed and the Duncan post hoc test was executed to assess differences among treatments for each gene at P≤ 0.05.
RT‐PCR analysis of fungal CWDE genes
To determine the expression of Fusarium genes encoding CWDEs, total RNA from roots obtained 1 or 3 days after inoculation, or from germlings grown for 14 h in PDB medium, was used as a template for RT‐PCR. Specific primers were designed for each gene based on the predicted F. oxysporum f. sp. lycopersici protein sequences available at the Broad Institute database (Table S2, see Supporting Information). The F. oxysporum actin 1 gene was used as a control for gene expression, and genomic DNA as control template for size comparison with bands amplified from cDNA of intron‐containing genes. PCR products were electrophoretically separated in 2% agarose gels, stained with ethidium bromide and photographed. Assays were repeated in three independent experiments with similar results.
Detection of chitinase activity
Crude enzyme extracts were obtained from infected plant stems. Four plants per treatment were bulked prior to enzyme extraction. Stems were separated, washed under running water, dried gently and ground with a mortar and pestle under liquid nitrogen. Samples were homogenized in phosphate buffer (0.05 m, pH 6.0) by vortexing, followed by incubation at 4 °C for 2 min. The homogenate was centrifuged twice at 10 000 g and 4 °C, and the supernatant was collected and stored at −20 °C. The protein content was quantified according to the method of Bradford using the Bio‐Rad protein assay (Bio‐Rad, Munchen, Germany), with bovine serum albumin as standard. Endo‐ and exochitinase activities were measured using the Fluorimetric Chitinase Assay Kit (Sigma, Ronkonkoma, NY, USA) following the manufacturer's instructions. Chitinase activity is expressed as nanograms of 4‐methylumbelliferone released from the substrate per microgram of protein. Assays were performed in three independent experiments, each including four plants per treatment. Data were analysed with the software SPSS 15.0 for Windows® (LEAD Technologies, Inc.). anova was performed and the Duncan post hoc test was executed to assess differences among treatments at P≤ 0.05.
Supporting information
Fig. S1 Attachment of Fusarium oxysporum strains to tomato roots.
Fig. S2 Tomato roots inoculated with wild‐type 'harbouring green fluorescent protein (GFP)‐tagged H1 histone, green' or ΔchsV mutant 'harbouring cherry red fluorescent protein (ChFP)‐tagged H1 histone, red'.
Fig. S3 Expression pattern of Fusarium oxysporum cell wall‐degrading enzyme coding genes during the infection of tomato plants with the wild‐type strain, the ΔchsV mutant or with both strains simultaneously.
Table S1 Sequences of plant gene‐specific primers used in quantitative polymerase chain reaction (PCR) expression analysis.
Table S2 Sequences of Fusarium oxysporum gene‐specific primers used in reverse transcriptase‐polymerase chain reaction (RT‐PCR) expression analysis.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
ACKNOWLEDGEMENTS
The authors gratefully acknowledge E. Martínez for technical assistance, Dr A. Muñoz for statistical analyses and Dr A. Di Pietro for critical reading of the manuscript, all from the University of Córdoba, Spain. This research was supported by the Ministerio de Educación y Ciencia of Spain (BIO2004‐0276) and Junta de Andalucía (CVI‐138 and AGR‐209). Y. Pareja‐Jaime was supported by a fellowship from the Ministerio de Educación y Ciencia of Spain.
REFERENCES
- Alabouvette, C. , Olivain, C. and Steinberg, C. (2006) Biological control of plant diseases: the European situation. Eur. J. Plant Pathol. 114, 329–341. [Google Scholar]
- Alabouvette, C. , Olivain, C. , Migheli, Q. and Steinberg, C. (2009) Microbiological control of soil‐borne phytopathogenic fungi with special emphasis on wilt‐inducing Fusarium oxysporum . New Phytol. 184, 529–544. [DOI] [PubMed] [Google Scholar]
- Armstrong, G.M. and Armstrong, J.K. (1981) Formae speciales and races of Fusarium oxysporum causing wilt diseases In: Fusarium: Disease, Biology, and Taxonomy (Nelson P.E., Tousson T.A. and Cook R.J., eds), pp. 391–399. University Park, PA: State University Press. [Google Scholar]
- Benhamou, N. and Garand, C. (2001) Cytological analysis of defense‐related mechanisms induced in pea root tissues in response to colonization by nonpathogenic Fusarium oxysporum Fo47. Phytopathology, 91, 730–740. [DOI] [PubMed] [Google Scholar]
- Bestwick, C.S. , Brown, I.R. , Bennett, M.H.R. and Mansfield, J.W. (1997) Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola . Plant Cell, 9, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E. , Tour, O. , Palmer, A.E. , Steinbach, P.A. , Baird, G.S. , Zacharias, D.A. and Tsien, R.Y. (2002)A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA, 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuel, Z. , Martinez‐Rocha, A.L. , Di Pietro, A. , Madrid, M.P. and Roncero, M.I.G. (2005) Fusarium oxysporum gas1 encodes a putative β‐1,3‐glucanosyltransferase required for virulence on tomato plants. Mol. Plant–Microbe Interact. 18, 1140–1147. [DOI] [PubMed] [Google Scholar]
- Dalisay, R.F. and Kuc, J.A. (1995) Persistence of induced resistance and enhanced peroxidase and chitinase activities in cucumber plants. Physiol. Mol. Plant Pathol. 47, 315–327. [Google Scholar]
- Danhash, N. , Wagemakers, C.A. , Van Kan, J.A. and De Wit, P.J. (1993) Molecular characterization of four chitinase cDNAs obtained from Cladosporium fulvum‐infected tomato. Plant Mol. Biol. 22, 1017–1029. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. and Roncero, M.I.G. (1998) Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum . Mol. Plant–Microbe. Interact. 11, 91–98. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. , Garcia‐Maceira, F.I. , Meglecz, E. and Roncero, M.I.G. (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Di Pietro, A. , Madrid, M.P. , Caracuel, Z. , Delgado‐Jarana, J. and Roncero, M.I.G. (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–326. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. , Roncero, M.I.G. and Ruiz‐Roldan, M.C. (2009) From tools of survival to weapons of destruction: role of cell wall‐degrading enzymes in plant infection In: The Mycota V: Plant Relationships (Deising H., ed.), pp. 181–200. Heidelberg, Berlin: Springer Verlag. [Google Scholar]
- Duijff, B.J. , Pouhair, D. , Olivain, C. , Alabouvette, C. and Lemanceau, P. (1998) Implication of systemic induced resistance in the suppression of fusarium wilt of tomato by Pseudomonas fluorescens WCS417r and by nonpathonic Fusarium oxysporum Fo47. Eur. J. Plant Pathol. 104, 903–910. [Google Scholar]
- Fravel, D. , Olivain, C. and Alabouvette, C. (2002) Fusarium oxysporum and its biocontrol. New Phytol. 157, 493–502. [DOI] [PubMed] [Google Scholar]
- Freeman, S. and Rodriguez, R.J. (1993) Genetic conversion of a fungal plant pathogen to a nonpathogenic, endophytic mutualist. Science, 260, 75–78. [DOI] [PubMed] [Google Scholar]
- Freeman, S. , Zveibil, A. , Vintal, H. and Maymon, M. (2001) Isolation on nonpathogenic mutants of Fusarium oxysporum f. sp. melonis for biological control of Fusarium wilt in cucurbits. Phytopathology, 92, 164–168. [DOI] [PubMed] [Google Scholar]
- Fuchs, J.‐G. , Moënne‐Loccoz, Y. and Défago, G. (1997) Nonpathogenic Fusarium oxysporum strain Fo47 induces resistance to Fusarium wilt in tomato. Plant Dis. 81, 492–496. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (1996) Plant resistance to fungi. Can. J. Plant Pathol. 18, 469–475. [Google Scholar]
- Hedley, P.E. , Machray, G.C. , Davies, H.V. , Burch, L. and Waugh, R. (1994) Potato (Solanum tuberosum) invertase‐encoding cDNAs and their differential expression. Gene, 145, 211–214. [DOI] [PubMed] [Google Scholar]
- Huertas‐Gonzalez, M.D. , Ruiz‐Roldan, M.C. , Di Pietro, A. and Roncero, M.I.G. (1999) Cross protection provides evidence for race‐specific avirulence factors in Fusarium oxysporum . Physiol. Mol. Plant Pathol. 54, 63–72. [Google Scholar]
- Kavroulakis, N. , Ntougias, S. , Zervakis, G.I. , Ehaliotis, C. , Haralampidis, K. and Papadopoulou, K.K. (2007) Role of ethylene in the protection of tomato plants against soil‐borne fungal pathogens conferred by an endophytic Fusarium solani strain. J. Exp. Bot. 58, 3853–3864. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Madrid, M.P. , Di Pietro, A. and Roncero, M.I. (2003) Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47, 257–266. [DOI] [PubMed] [Google Scholar]
- Martin‐Urdiroz, M. , Roncero, M.I. , Gonzalez‐Reyes, J.A. and Ruiz‐Roldan, C. (2008) ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum . Eukaryot. Cell 7, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivain, C. , Trouvelot, S. , Binet, M.N. , Cordier, C. , Pugin, A. and Alabouvette, C. (2003) Colonization of flax roots and early physiological responses of flax cells inoculated with pathogenic and nonpathogenic strains of Fusarium oxysporum . Appl. Environ. Microbiol. 69, 5453–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina, Y. , Fravel, D.R. , Baker, C.J. and Shcherbakova, L.A. (2007) Biocontrol and plant pathogenic Fusarium oxysporum‐induced changes in phenolic compounds in tomato leaves and roots. J. Phytopathol. 155, 475–481. [Google Scholar]
- Postma, J. and Luttikholt, A.J.G. (1996) Colonization of carnation stems by a nonpathogenic isolate of Fusarium oxysporum and its effect on Fusarium oxysporum f. sp. dianthi . Can. J. Bot. 74, 1841–1851. [Google Scholar]
- Redman, R.S. , Freeman, S. , Clifton, D.R. , Morrel, J. , Brown, G. and Rodriguez, R.J. (1999a) Biochemical analysis of plant protection afforded by a nonpathogenic endophytic mutant of Colletotrichum magna . Plant Physiol. 119, 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman, R.S. , Ranson, J.C. and Rodriguez, R.J. (1999b) Conversion of the pathogenic fungus Colletotrichum magna to a nonpathogenic, endophytic mutualist by gene disruption. Mol. Plant–Microbe Interact. 12, 969–975. [Google Scholar]
- Shaner, N.C. , Campbell, R.E. , Steinbach, P.A. , Giepmans, B.N. , Palmer, A.E. and Tsien, R.Y. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shih, M.C. , Heinrich, P. and Goodman, H.M. (1991) Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glyceraldehyde‐3‐phosphate‐dehydrogenase from Arabidopsis thaliana . Gene, 104, 133–138. [DOI] [PubMed] [Google Scholar]
- Soulie, M.C. , Perino, C. , Piffeteau, A. , Choquer, M. , Malfatti, P. , Cimerman, A. , Kunz, C. , Boccara, M. and Vidal‐Cros, A. (2006) Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell. Microbiol. 8, 1310–1321. [DOI] [PubMed] [Google Scholar]
- Van Kan, J.A. , Joosten, M.H. , Wagemakers, C.A. , Van Den Berg‐Velthuis, G.C. and De Wit, P.J. (1992) Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum . Plant Mol. Biol. 20, 513–527. [DOI] [PubMed] [Google Scholar]
- Wattad, C. , Freeman, S. , Dinoor, A. and Prusky, D. (1995) A nonpathogenic mutant of Colletotrichum magna is deficient in extracellular secretion of pectate lyase. Mol. Plant–Microbe Interact. 8, 621–626. [Google Scholar]
- Weber, I. , Assmann, D. , Thines, E. and Steinberg, G. (2006) Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis . Plant Cell, 18, 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, S. , Sugui, J.A. , Steinberg, G. and Deising, H.B. (2007) A chitin synthase with a myosin‐like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola . Mol. Plant–Microbe Interact. 20, 1555–1567. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Ukil, L. , Osmani, A. , Nahm, F. , Davies, J. , De Souza, C.P. , Dou, X. , Perez‐Balaguer, A. and Osmani, S.A. (2004) Rapid production of gene replacement constructs and generation of a green fluorescent protein‐tagged centromeric marker in Aspergillus nidulans . Eukaryot. Cell, 3, 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Attachment of Fusarium oxysporum strains to tomato roots.
Fig. S2 Tomato roots inoculated with wild‐type 'harbouring green fluorescent protein (GFP)‐tagged H1 histone, green' or ΔchsV mutant 'harbouring cherry red fluorescent protein (ChFP)‐tagged H1 histone, red'.
Fig. S3 Expression pattern of Fusarium oxysporum cell wall‐degrading enzyme coding genes during the infection of tomato plants with the wild‐type strain, the ΔchsV mutant or with both strains simultaneously.
Table S1 Sequences of plant gene‐specific primers used in quantitative polymerase chain reaction (PCR) expression analysis.
Table S2 Sequences of Fusarium oxysporum gene‐specific primers used in reverse transcriptase‐polymerase chain reaction (RT‐PCR) expression analysis.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


