SUMMARY
The potyviruses Plum pox virus (PPV) and Tobacco vein mottling virus (TVMV) have distinct host ranges and induce different symptoms in their common herbaceous hosts. To test the relevance of the P1 protein in host compatibility and pathogenicity, hybrid viruses were constructed in which the P1 coding sequence of PPV was completely or partially replaced by the corresponding sequences from TVMV. Infections induced by these chimeric viruses revealed that the TVMV P1 and a PPV/TVMV hybrid P1 proteins are functionally equivalent in herbaceous plants to the P1 protein of a PPV isolate adapted to these hosts, in spite of having high sequence divergence. Moreover, the presence of TVMV P1 sequences enhanced the competence of a low‐infectivity PPV‐D‐derived chimera in Nicotiana clevelandii. Conversely, all PPV/TVMV hybrids were unable to infect Prunus persicae, a specific host for PPV, suggesting that TVMV P1 is not functionally competent in this plant. Together, these data highlight the importance of the P1 protein in defining the virus host range.
INTRODUCTION
Plum pox virus (PPV) and Tobacco vein mottling virus (TVMV) belong to the genus Potyvirus in the family Potyviridae. The host ranges of both viruses include several herbaceous species of the genus Nicotiana, including N. clevelandii and N. benthamiana; however, while PPV also infects stone fruit trees of the genus Prunus, TVMV does not (Glasa and Candresse, 2005; Pirone and Shaw, 1988). In addition, while TVMV systemically infects N. tabacum, PPV is only able to establish a local infection in this host (Sáenz et al., 2002). The potyvirus genome consists of a ~10‐kb single‐stranded RNA molecule that is translated into a polyprotein and then proteolitically processed by three self‐encoded proteinases to produce at least ten mature products (Riechmann et al., 1992; Urcuqui‐Inchima et al., 2001). Previous studies suggest that several potyviral proteins play a role in host range definition. For example, transgenic tobacco plants expressing the HCPro protein from a tobacco‐infecting potyvirus, Tobacco etch virus (TEV), were able to overcome the defect in systemic spread of PPV in N. tabacum (Sáenz et al., 2002) and the ability of Turnip mosaic virus (TuMV) to infect Brassica spp. and/or Raphanus sativus is mapped to its P3 protein (Suehiro et al., 2004).
P1 is the most variable potyvirus protein in both sequence and length (Adams et al., 2005), and it is thought that P1 diversification has contributed to the successful adaptation of potyviruses to a wide range of host species (Valli et al., 2007). The C‐terminal region of P1 is moderately conserved and contains a serine protease domain that is responsible for self‐cleavage from the viral polyprotein (Verchot et al., 1991). The potyviral P1 protein has RNA‐binding activity (Brantley and Hunt, 1993; Merits et al., 1998; Soumounou and Laliberté, 1994) and functions in trans as an accessory factor for genome amplification (Verchot and Carrington, 1995). In addition, P1 was shown to enhance the silencing suppression activity of HCPro (Rajamäki et al., 2005; Valli et al., 2006), a process that is thought to contribute to the synergistic interaction between TEV and Potato virus X (Pruss et al., 1997). Direct evidence of a role for the P1 protein in host range definition, however, has not yet been demonstrated.
A successful virus infection cycle requires the highly co‐ordinated activity of different viral proteins and cis‐acting RNA sequences. The complex network of potyviral protein–protein interactions (Choi et al., 2000; Daròs et al., 1999; Guo et al., 2001; Kang et al., 2004; Merits et al., 1999) makes coevolution a possible constraint for the functionality of heterologous potyviral proteins within divergent viral backgrounds. However, interspecies cistron exchanges have been engineered that render fully infectious viruses within the family Potyviridae (Stenger and French, 2004; Tobias et al., 2001; Ullah et al., 2003; Varrelmann et al., 2000).
The aim of the preseny study was to investigate the role of the potyviral P1 protein in host range specificity. To address this, regions of the PPV P1 cistron were replaced with corresponding regions from the TVMV genome, and infectivity of the hybrid viruses was assessed in common hosts of both PPV and TVMV and in PPV exclusive hosts.
RESULTS
PPV/TVMV chimeric viruses that carry a partial or complete TVMV P1 coding sequence are infectious in herbaceous hosts
P1 is the most variable potyviral protein. TVMV P1 has 34 amino acids fewer than PPV P1 and shares 24.1% identity with this protein (Fig. 1a). The difference in length is primarily due to large gaps in the N‐terminus of TVMV P1. The most conserved P1 region among potyviruses is the C‐terminal serine proteinase domain, although conserved motifs are also found in the N‐terminus (FGSFT) and in the middle region (AKAx4VEx1Ix2KRV) of TVMV and PPV P1 proteins (Fig. 1a; Valli et al., 2007). In a series of engineered chimeric clones, the P1 coding sequence of the PPV‐R isolate (pGPPV cDNA clone) was partially (N‐terminus 183 codons, pGPPV‐Tm, RTm) or entirely (pGPPV‐Tc, RTc) replaced by the P1 coding sequence of TVMV (Fig. 1b). The exchanged PPV P1 sequence began at an NcoI site that was engineered around the second AUG of the large viral open reading frame (ORF), the codon used for the initiation of translation during PPV infection (Riechmann et al., 1991). The first AUG of the PPV ORF was also disrupted in the chimeric viruses. In spite of high divergence between the interchanged sequences, in vitro synthesized transcripts of the hybrid clones were infectious in N. clevelandii plants (Fig. 2). N. clevelandii and N. benthamiana had similar symptoms in response to infection with PPV‐R and the chimeras, while symptoms induced by the RTc hybrid were somewhat more severe than PPV‐R and RTm (Fig. 2). ELISA analysis of N. clevelandii and N. benthamiana leaves showed that both hybrid viruses and PPV‐R had accumulated to similar levels (data not shown). In C. foetidum, a local host of PPV that is not susceptible to TVMV, the RTm and RTc hybrids caused necrotic lesions that were indistinguishable from those induced by PPV‐R (Fig. 2). N. tabacum has also been shown to respond differently to PPV and TVMV infection: while TVMV results in systemic infection, PPV is only able to establish a local infection in the tobacco plant (Sáenz et al., 2002). Interestingly, RTm and RTc were unable to induce a systemic infection in tobacco, resembling wild‐type PPV‐R. All three viruses induced similar ring‐shaped symptoms and accumulated to comparable levels in the inoculated leaves of this plant (data not shown). Together, these results show that TVMV P1 sequences do not affect the susceptibility of herbaceous hosts to PPV.
Figure 1.
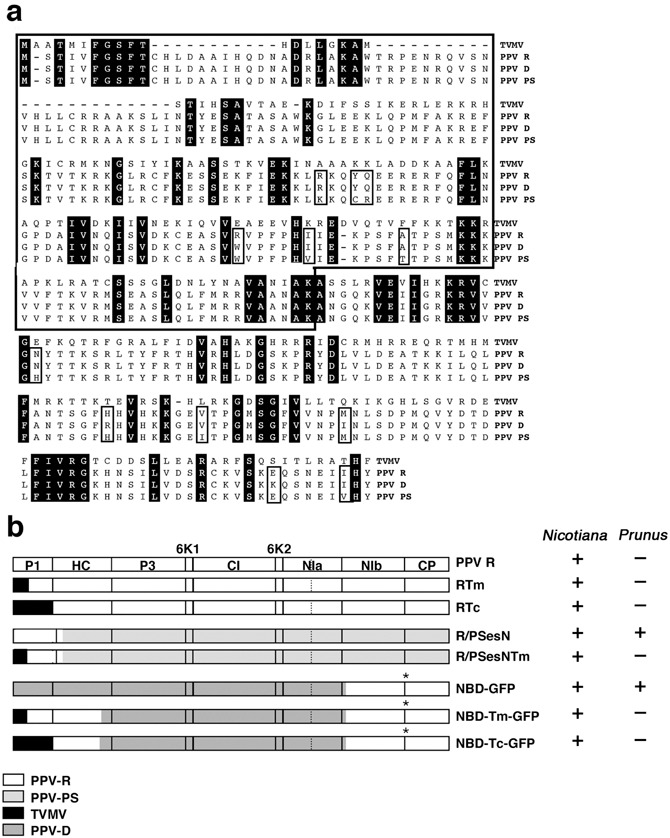
PPV and TVMV P1 protein alignment and PPV/TVMV chimeras. (a) Sequence alignment of PPV and TVMV P1 proteins. Conserved amino acid positions in the sequence alignment are shadowed in black. Open boxes indicate amino acid differences between the PPV isolates. (b) Schematic representation of the different virus constructs used in this work. The pattern assigned to each parental virus is depicted below the constructs. GFP sequences are indicated with an asterisk. The ability of the chimeras to infect Nicotiana species and/or Prunus persicae GF305 is indicated.
Figure 2.

Analysis of the infection of PPV‐R/TVMV chimeras in different herbaceous hosts. Symptoms in the inoculated leaves of N. clevelandii at 15 dpi and C. foetidum at 7 dpi and in the upper non‐inoculated leaves of N. clevelandii and N. benthamiana at 15 dpi.
TVMV P1 sequences impair PPV infectivity in peach trees
As the RTm and RTc chimeras are derived from PPV‐R, a PPV isolate that has lost its ability to infect Prunus species after extensive propagation in herbaceous plants (Dallot et al., 2001), they are not useful for assessing the effect of P1 substitution in PPV natural woody hosts. Thus, we constructed new PPV/TVMV chimeras based on pGPPV‐PS‐E109S232 (PSes), a full‐length cDNA clone derived from the PPV‐PS isolate that is infectious in both herbaceous and woody hosts (Sáenz et al., 2001). In the new chimeras, pGPPV‐R/PS‐1334esN (R/PSesN) and pGPPV‐R/PS‐1334esN‐Tm (R/PSesNTm), the first 1334 nucleotides of PSes were replaced by the corresponding sequences from PPV‐R and RTm, respectively (Fig. 1b). In addition to the P1 coding sequence, this fragment includes the sequence encoding the first 88 amino acids of HCPro that contains four silent nucleotide differences between PPV‐R and PPV PS. In vitro synthesized transcripts from both chimeras were infectious in two common PPV and TVMV hosts, N. clevelandii and N. benthamiana (data not shown). GF305 peach seedlings were manually inoculated with RTm‐, RTc‐, R/PSesNTm‐, R/PSesN‐ or PSes‐infected N. benthamiana plant extracts. Inoculated and upper non‐inoculated leaves were analysed by ELISA for the presence of virus in at least two different experiments (Table 1). Most PSes‐inoculated trees were systemically infected, as shown by faint mottling symptoms and the accumulation of virus in the upper non‐inoculated leaves. As expected, the PPV/TVMV chimeras, RTm and RTc, were unable to infect the trees as PPV‐R is not infectious in this host. Although no symptoms were observed in R/PSesN‐inoculated GF305 seedlings, 40% of the seedlings were infected with this chimera (Table 1). Levels of virus accumulation, however, were considerably lower than in PSes‐infected plants (data not shown). This suggests that the genome fragment of PPV‐R that is present in R/PSesN, with 9‐amino‐acid differences in the P1 sequence, reduces but does not abolish infectivity in peach. When the coding sequence of the TVMV P1 N‐terminus was included in the chimera (R/PSesNTm), however, infectivity was completely abolished. Thus, while the TVMV region is fully functional for infection of herbaceous hosts, it is unable to support PPV infection in a woody host.
Table 1.
Infectivity of PPV/TVMV hybrids in GF305.
| DNA construct* | Inoculated leaves† | Upper non‐inoculated leaves† |
|---|---|---|
| PSes | 45/48 | 47/48 |
| RTm | 0/24 | 0/24 |
| RTc | 0/24 | 0/24 |
| R/PSesN | 9/24 | 11/24 |
| R/PSesNTm | 0/48 | 0/48 |
Inoculated virus.
Number of PPV infected trees as determined by ELISA/total number of inoculated trees.
Manual inoculation of GF305 trees is a poor method for promoting infection. As a result, we used a more efficient inoculation method, biolistic delivery of viral cDNA for transcription in planta, to assess further the effect of P1 exchange on PPV infectivity in Prunus. New PPV/TVMV chimeras were constructed from pICPPVN‐5′BD‐GFP (NBD‐GFP), a derivative of pICPPV‐5′BD‐GFP (BD‐GFP; Salvador et al., 2008) that includes the same NcoI site and AUG mutations that were present in the chimeric viruses described above (Fig. 1b). NBD‐GFP expresses a full‐length hybrid cDNA consisting of PPV‐R and PPV‐D sequences under control of the CaMV 35S promoter (Fig. 1b), and is highly infectious in GF305 peach (Fig. 3). Unlike PPV‐D, NBD‐GFP is also infectious in N. clevelandii although it causes no symptoms in this herbaceous host (Fig. 3a) as shown with BD‐GFP (Salvador et al., 2008). pICPPVN‐5′BD‐Tm‐GFP (NBD‐Tm‐GFP) and pICPPVN‐5′BD‐Tc‐GFP (NBD‐Tc‐GFP) were created by replacing nucleotides 146–2276, the coding sequence of P1 and the first 402 amino acids of HCPro, from NBD‐GFP with the corresponding region from the RTm and RTc chimeras, respectively (Fig. 1b). Micro gold particles coated with DNA from the different chimeras were prepared for biolistic inoculation and three different inoculation experiments were performed in GF305 peach and N. clevelandii. GFP expression was observed in 100% of the inoculated N. clevelandii plants; however, fluorescence intensity was higher in the NBD‐Tm‐GFP and NBD‐Tc‐GFP than the control NBD‐GFP plants, with NBD‐Tc‐GFP showing the highest level of intensity (Fig. 3a). Both PPV/TVMV chimeras showed significantly higher levels of viral accumulation than NBD‐GFP in N. clevelandii‐infected leaves, and NBD‐Tc‐GFP accumulated to higher titres than NBD‐Tm‐GFP (Fig. 3b). Higher GFP expression and virus accumulation correlated with the appearance of extensive mild (NBD‐Tm‐GFP) or strong (NBD‐Tc‐GFP) chlorotic mottling in the upper non‐inoculated leaves of infected plants (Fig. 3). These data indicate that an increase in the extent of the TVMV P1 sequence in the NBD‐GFP‐derived chimeras enhanced their virulence in N. clevelandi.
Figure 3.
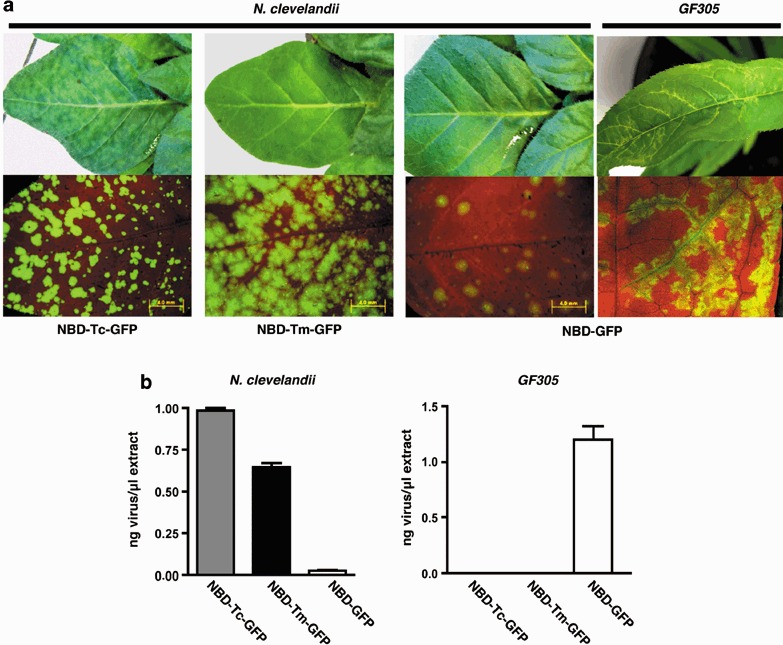
Analysis of NBD‐GFP, NBD‐Tm‐GFP and NBD‐Tc‐GFP infection in N. clevelandii and GF305 peach. (a) Symptoms and GFP expression observed under visible or UV light, respectively, at 21 days post‐inoculation (dpi) for N. clevelandii and at 35 dpi for GF305 peach. (b) Virus accumulation in upper non‐inoculated infected leaves of N. clevelandii (21 dpi) and GF305 peach (35 dpi) plants. Bars represent the average values and standard deviations of four N. clevelandii and eight peach plants.
In contrast, while 100% of the 16 NBD‐GFP‐infected peach GF305 plants had GFP expression and strong vein clearing and distortion (Fig. 3a), none of the 22 peach seedlings infected with the NBD‐Tm‐GFP or NBD‐Tc‐GFP chimeras that contained a portion or the complete TVMV P1 sequence had either symptoms or GFP expression. ELISA analysis confirmed that virus accumulated in the NBD‐GFP‐inoculated peach seedlings and did not accumulate in the NBD‐Tc‐GFP‐ or NBD‐Tm‐GFP‐inoculated plants (Fig. 3b). These results provide further evidence that TVMV P1 is not functional in the PPV‐specific host Prunus persicae.
DISCUSSION
Although abundant information has been gathered about the molecular biology of potyviruses in general and of PPV in particular, there is still a dearth of information on the role of particular mature potyvirus proteins, including P1 and P3 (reviewed by Salvador et al., 2006; Urcuqui‐Inchima et al., 2001). Gene functions appear to be more conserved among plant viruses than gene sequences, as illustrated by the viability of chimeric viruses that are derived from cistron replacements between unrelated viruses even from different families (De Jong and Ahlquist, 1992; Ryabov et al., 1999). These chimeric viruses are used to clarify the roles of particular proteins and the specific domains required for their function. We utilized this approach to explore a potential role for the potyviral P1 protein in host specificity.
It was initially thought that P1 may be a movement protein (Domier et al., 1987). More recently it was shown that P1 functions as an accessory factor for genome amplification and plays little, if any, role in virus movement (Verchot and Carrington, 1995). The importance of P1 in potyvirus infection is highlighted by the finding that chimeric viruses containing foreign sequences between the P1 and HCPro cistrons of PPV (Guo et al., 1998) and TEV (Dolja et al., 1993) undergo deletions that sometimes affected the TEV HCPro N‐terminus but never removed P1 coding sequences. The high divergence of P1 sequences among different potyviruses suggests that the function of P1 may depend on precise interactions with (a) species‐specific host factor(s) (Adams et al., 2005; Valli et al., 2007). Recombination events affecting P1 sequences that may be linked to host adaptation support this possibility (Valli et al., 2007). In addition, a specific point mutation was introduced in P1 during adaptation of a PPV isolate to N. clevelandii (Salvador et al., 2008), and some insertions in the P1 gene from Potato virus A were shown to have host‐specific effects on virus accumulation and symptom severity (Rajamäki et al., 2005).
In this report we show that PPV/TVMV hybrids are efficient at infecting two common herbaceous hosts of PPV and TVMV, N. benthamiana and N. clevelandii. Thus, in spite of high divergence between the PPV and TVMV P1 proteins, structural and/or sequence determinants involved in P1 intramolecular organization and the interaction between P1 and other virus or plant factors are conserved between PPV and TVMV, allowing heterologous contacts to be functional. Interestingly, while substitution of P1 sequences from the N. clevelandii‐adapted PPV‐R isolate with those from the TVMV P1 (chimeric clones RTm and RTc) had little effect on virus virulence in this plant (Fig. 2), replacement with TVMV P1 sequences in a PPV‐D‐derived chimera (NBD‐GFP), which infects herbaceous hosts only poorly, enhanced infection efficiency in N. clevelandii, and complete substitution of the TVMV P1 sequence (NBD‐Tc‐GFP) induced stronger virulence than partial substitution (NBD‐Tm‐GFP) (Fig. 3). Thus, appropriate P1 sequences, such as those from PPV‐R or TVMV, are required for efficient infection of N. clevelandii.
The PPV‐R isolate, which is unable to infect Prunus trees, has 68 nucleotide changes that are translated in 24 amino acid substitutions with respect to the Prunus‐adapted PPV‐D isolate (Salvador et al., 2008). Nine of these nucleotide changes, six of which cause amino acid substitutions, are found in the P1 coding region. Analysis of PPV‐R/D chimeric clones showed that while host range determinants were spread throughout the genome, the 5′ third of the genome was especially relevant for Prunus infectivity (Salvador et al., 2008). However, a chimeric clone with the first 3628 nucleotides from PPV‐R was still slightly infectious in GF305 peach, indicating that both P1 and HCPro from PPV‐R can support Prunus infection. In contrast, both NBD‐Tm‐GFP and NBD‐Tc‐GFP were unable to infect GF305 peach, even using the highly efficient biolistic inoculation method (Fig. 3). This result is consistent with the finding that R/PSesNTm, a PPV‐PS/R chimera with a coding sequence for the TVMV P1 N‐terminus, is unable to infect GF305 peach (Table 1), and both results suggest that TVMV P1 is not functional in Prunus trees, highlighting the relevance of potyviral P1 in host adaptation.
The finding that the RTc and RTm chimeras, which contained either a TVMV or a hybrid TVMV/PPV P1 protein, induced pathology that was more characteristic of PPV than of TVMV, causing necrotic local lesions in C. foetidum and an infection restricted to the inoculated leaves in N. tabacum, suggests that P1 is not the only viral factor involved in host specificity. This finding is consistent with the extensive spread of host adaptation factors of the PPV‐R and D isolates in the PPV genome that was reported by Salvador et al. (2008).
Our results do not define the nature of the host‐specific functions of potyviral P1. P1 proteinase activity catalyses its self‐cleavage at the P1/HCPro junction, and this cleavage rather than the P1 proteolytic activity itself is essential for viral infectivity (Verchot and Carrington, 1995). A plant co‐factor was thought to be required for the proteolytic activity of P1 because in vitro self‐cleavage at the TEV P1 C‐terminus occurred in wheat germ but not reticulocyte lysate (Verchot et al., 1992). PPV and TVMV P1 proteins are also unable to cleave themselves in rabbit reticulocyte lysate, while the PPV/TVMV chimeric P1 from RTm can undergo cleavage (P.S. and J.A.G., unpublished results), suggesting that a mammalian protein, possibly a chaperone, may substitute for the plant co‐factor depending on particular features of the potyviral P1 protein. The possibility exists that specific coevolution of the plant co‐factor and the P1 protein may be involved in the host adaptation of potyviruses. Moreover, some reports suggest that P1 is able to enhance the silencing suppression activity of HCPro (Kasschau and Carrington, 1998; Pruss et al., 1997; Rajamäki et al., 2005; Valli et al., 2006). Compatible interactions between P1 and host factors that ensure correct polyprotein processing and/or facilitate the P1 role in silencing suppression may be essential for overcoming antiviral defence mechanisms and facilitating host susceptibility. Identifying the plant factor(s) that interacts with P1 protein will help to shed light on the molecular basis of the host‐specific functions of this protein during potyviral infection.
EXPERIMENTAL PROCEDURES
Construction of full‐length cDNA clones
The full‐length cDNA clones pGPPV (Riechmann et al., 1990), pICPPV‐5′BD‐GFP (Salvador et al., 2008), pICPPV‐NK‐GFP (Fernández‐Fernández et al., 2001) and pGPPV‐PS‐E109S232 (Sáenz et al., 2000) have been described previously.
As the P1 cistrons of PPV and TVMV lack common restriction enzyme sites, pGPPV‐Tm and pGPPV‐Tc were constructed by the method of gene splicing via overlap extension (Horton et al., 1989) (Fig. 1b). To construct pGPPV‐Tm, TVMV cDNA from pXbS7 (Domier et al., 1989) was used as a template for PCR amplification with primer A, 5′CTAGCCATG G CAACCATTCACTCAG3′, containing an NcoI site (underlined) to facilitate cloning and nucleotides encoding the first amino acids of TVMV P1 (nt 205–226, in bold), and primer B, 5′TTGCCTTGGC TATATTAGCCAC3′, in which the first 10 nucleotides (in italics) and the last 17 nucleotides (in bold) correspond to P1 coding sequences from PPV (nt 699–690) and TVMV (nt 602–586), respectively. A PCR fragment that was partially complementary to the TVMV fragment that was amplified with primers A and B, was amplified using primer B′, 5′CTAATATA GCCAAGGCAAATGG3′, in which the first 13 nucleotides (in bold) and the last 14 nucleotides (in italics) corresponded to the P1 coding sequences from TVMV (nt 590–602) and PPV (nt 690–703), respectively, and primer D, 5′CGCATTAGTTCAC3′ (PPV nucleotides 1378–1390), and pGG5S6N, a partial PPV cDNA clone (PPV nucleotides 1–3628), in which the first AUG of the large ORF was mutated and the surroundings of the second AUG were engineered to display an NcoI restriction site (Simón‐Buela et al., 1997), as a template. The resulting PCR fragments were used as templates for a second round of PCR with primers A and D. The product of this PCR was digested with NcoI and BsaBI and cloned in pGG5S6N, yielding pGG5S6N‐TmP1Δ54. New data available from John Shaw (University of Kentucky, personal communication; Genbank accession number NC_001768.1) suggested that the actual initiation codon of the TVMV polyprotein was in position 154–156. A new TVMV fragment was amplified from pXbS7 by PCR with primers C, 5′CTAGCC ATGGCAGCAACAATGATC3′ (TVMV nucleotides 154–171 in bold), and B. The PCR product was digested with NcoI and MunI and substituted for the corresponding sequence of pGG5S6N‐TmΔ54 to obtain pGG5S6N‐Tm. Finally, pGPPV‐Tm was obtained by introducing the BglI–DraIII fragment of pGG5S6N‐Tm that contained the chimeric P1 coding sequence into pGPPV.
For pGPPV‐Tc cloning, a first round of PCR was conducted to obtain partially overlapping TVMV and PPV fragments on pXbS7 using primers A and E, 5′GCCTGGGTC TGAGAAGTGAGTCG3′ (TVMV nucleotides 978–965, corresponding to the 3′ end of the P1 sequence and the first three nucleotides of HCPro cistron, in bold, and PPV nucleotides 1082–1074, in italics), and on pGG5S6N, using primers E′, 5′CACTTCTCA GACCCAGGCAAAC3′ (TVMV nucleotides 970–978, in bold, and PPV nucleotides 1074–1086, in italics), and D. The PCR products were used for recombinant PCR with primers A and D, and the resulting fragment was cloned in pGG5S6N after NcoI/BsaBI digestion, yielding pGG5S6N‐TcΔ54. In order to obtain a recombinant clone with the correct TVMV initiation codon, pGG5S6N‐TcP1, a MunI–SalI fragment derived from pGG5S6N‐TcΔ54, and an NcoI–MunI fragment of the PCR product that was amplified from pXbS7 using primers C and B (see above) were inserted into NcoI/SalI‐digested pGG5S6N. Finally, pGPPV‐Tc was obtained by a triple ligation of the MunI–SacI and BsaBI–SacI fragments of pGPPV‐Tm and a MunI–BsaBI fragment derived from pGG5S6N‐Tc.
pGPPV‐R/PS‐1334esN and pGPPV‐R/PS‐1334esN‐Tm were obtained by inserting a BglI–NcoI fragment from pGG5S6N and an NcoI–Bsh1361I fragment from pGG5S6N (pGPPV‐R/PS‐1334esN) or pGPPV‐Tm (pGPPV‐R/PS‐1334esN‐Tm) into pGPPV‐PS‐E109S232 that was digested with BglI and Bsh1361I.
pICPPV‐N5′BD‐GFP was constructed by replacing the BglI–CpoI fragment from pICPPV‐5′BD‐GFP with the corresponding fragment from pICPPVN, a derivative of pICPPV (López‐Moya and García, 2000) harbouring the mutations in the first AUG codons of pGG5S6N (J.J. López‐Moya and J.A.G., unpublished results).
pICPPVN‐5′BD‐Tm‐GFP and pICPPVN‐5′BD‐Tc‐GFP were constructed by replacing the NcoI–Bpu1102I fragment of pICPPVN‐5′BD‐GFP (nt 1–2284) with the corresponding fragments from PCR products amplified using 5′AAAATATAAAAACTCAACAC3′ (PPV nt 1–20) and 5′GGAAGCTCAGCATTCCGAG3′ (PPV nt 2284–2266, with a mismatch, in italics, to create a Bpu1102I site, underlined) from pGPPVTmP1 and pGPPVTcP1, respectively.
The accuracy of the constructions was verified by restriction digestion analysis and DNA sequencing of all PCR amplified regions.
Inoculation and protein analysis
The Helios Gene Gun System (Bio‐Rad, Hercules, CA) was used for biolistic inoculation. Microcarrier cartridges were prepared with 1.0‐µm gold particles coated with the different plasmids at a DNA loading ratio of 2 µg/mg of gold and a microcarrier loading quantity of 0.5 mg per shooting, according to the manufacturer's instructions. Helium pressures of 7.5 and 10 bar were used for N. clevelandii and Prunus persicae (peach) cv. GF305, respectively. Each cartridge was shot twice onto two leaves from each plant, and two cartridges were administered for each plant.
For inoculation of viral transcripts, the recombinant plasmids were linearized with PvuII (pGPPV‐R/PS‐1334esN and pGPPV‐R/PS‐1334esN‐Tm) or PstI and PvuII (pGPPV, pGPPV‐Tm and pGPPV‐Tc). The digested DNAs were used as templates for capped transcription with a T7 mMessage mMachine (Ambion) or a CAPScribe (Boehringer) kit according to the manufacturer's instructions. Transcript yield and integrity were analysed using agarose gel electrophoresis. Three primary leaves per plant were dusted with carborundum and mechanically inoculated with 1.5 µL per leaf of the transcription reaction mixture diluted 1:1 with 5 mM sodium phosphate buffer, pH 7.5.
For inoculation with the infected tissue, infected plant leaves were ground with 5 mM sodium phosphate buffer (pH 7.5) with an ice‐cold pestle (1 g, 2 ml). Extracts were centrifuged to eliminate tissue fragments. For each extract, three leaves were dusted with carborundum and inoculated with a total of 15 µL of extract.
Plants were maintained in a greenhouse at 19–23 °C with 16 h of light and supplementary illumination, or in a climate‐controlled chamber at 22 or 16 °C with 14 h of light. Virus infection was monitored by assessing symptoms and visualizing GFP fluorescence using an MZ FLIII (Leica Microsystems) stereomicroscope with excitation and barrier filters of 480/40 and 510 nm, respectively. Images were collected using an OLYMPUS DP 70 digital camera with DP Controller and DP manager software (Olympus Optical Co., Ltd). Virus accumulation was assessed at various days post‐inoculation by double antibody sandwich indirect‐enzyme‐linked immunoadsorbent assay (DASI‐ELISA) using the REALISA kit (Durviz).
ACKNOWLEDGEMENTS
We are grateful to Emilio Rodríguez Cerezo for kindly providing the plasmid pXbS7. We thank Elvira Dominguez for technical assistance. This work was supported by grants BIO2004‐02687 and BIO2007‐67283 from Spanish MEC, CPE03‐022‐C5‐3 from INIA, SAL/0185/2006 from Comunidad de Madrid and QLG2‐CT‐2002‐01673 and SP22‐CT‐2004 from European Union. B.S. was a recipient of an FPI fellowship from MEC.
REFERENCES
- Adams, M.J. , Antoniw, J.F. and Fauquet, C.M. (2005) Molecular criteria for genus and species discrimination within the family Potyviridae . Arch. Virol. 150, 459–479. [DOI] [PubMed] [Google Scholar]
- Brantley, J.D. and Hunt, A.G. (1993) The N‐terminal protein of the polyprotein encoded by the potyvirus tobacco vein mottling virus is an RNA‐binding protein. J. Gen. Virol. 74, 1157–1162. [DOI] [PubMed] [Google Scholar]
- Choi, I.R. , Stenger, D.C. and French, R. (2000) Multiple interactions among proteins encoded by the mite‐transmitted wheat streak mosaic tritimovirus. Virology, 267, 185–198. [DOI] [PubMed] [Google Scholar]
- Dallot, S. , Quiot‐Douine, L. , Sáenz, P. , Cervera, M.T. , García, J.A. and Quiot, J.B. (2001) Identification of Plum pox virus determinants implicated in specific interactions with different Prunus spp. Phytopathology, 91, 159–164. [DOI] [PubMed] [Google Scholar]
- Daròs, J.A. , Schaad, M.C. and Carrington, J.C. (1999) Functional analysis of the interaction between VPg‐proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J. Virol. 73, 8732–8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, W. and Ahlquist, P. (1992) A hybrid RNA virus made by transferring the noncapsid movement protein from a rod‐shaped to an icosahedral virus is competent for systemic infection. Proc. Natl Acad. Sci. USA, 89, 6808–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja, V.V. , Herndon, K.L. , Pirone, T.P. and Carrington, J.C. (1993) Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J. Virol. 67, 5968–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier, L.L. , Franklin, K.M. , Hunt, A.G. , Rhoads, R.E. and Shaw, J.G. (1989) Infectious in vitro transcripts from cloned cDNA of a potyvirus, tobacco vein mottling virus. Proc. Natl Acad. Sci. USA, 86, 3509–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier, L.L. , Shaw, J.G. and Rhoads, R.E. (1987) Potyviral proteins share amino acid sequence homology with picorna‐, como‐, and caulimoviral proteins. Virology, 158, 20–27. [DOI] [PubMed] [Google Scholar]
- Fernández‐Fernández, M.R. , Mouriño, M. , Rivera, J. , Rodríguez, F. , Plana‐Durán, J. and García, J.A. (2001) Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus‐based vector. Virology, 280, 283–291. [DOI] [PubMed] [Google Scholar]
- Glasa, M. and Candresse, T. (2005) Plum pox virus. C.M.I./A.A.B. Descriptions of Plant Viruses No. 410. Commonwealth Mycological Institute and the Association of Applied Biologist. Ferry Lane, Kew, Surrey, England.
- Guo, D.Y. , Rajamaki, M.L. , Saarma, M. and Valkonen, J.P.T. (2001) Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two‐hybrid system. J. Gen. Virol. 82, 935–939. [DOI] [PubMed] [Google Scholar]
- Guo, H.S. , López‐Moya, J.J. and García, J.A. (1998) Susceptibility to recombination rearrangements of a chimeric plum pox potyvirus genome after insertion of a foreign gene. Virus Res. 57, 195–207. [DOI] [PubMed] [Google Scholar]
- Horton, R.M. , Hunt, H.D. , Ho, S.N. , Pullen, J.K. and Pease, L.R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene, 77, 61–68. [DOI] [PubMed] [Google Scholar]
- Kang, S.H. , Lim, W.S. and Kim, K.H. (2004) A protein interaction map of soybean mosaic virus strain G7H based on the yeast two‐hybrid system. Mol. Cells, 18, 122–126. [PubMed] [Google Scholar]
- Kasschau, K.D. and Carrington, J.C. (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell, 95, 461–470. [DOI] [PubMed] [Google Scholar]
- López‐Moya, J.J. and García, J.A. (2000) Construction of a stable and highly infectious intron‐containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 68, 99–107. [DOI] [PubMed] [Google Scholar]
- Merits, A. , Guo, D.Y. and Saarma, M. (1998) VPg, coat protein and five non‐structural proteins of potato A potyvirus bind RNA in a sequence‐unspecific manner. J. Gen. Virol. 79, 3123–3127. [DOI] [PubMed] [Google Scholar]
- Merits, A. , Guo, D.Y. , Jarvekulg, L. and Saarma, M. (1999) Biochemical and genetic evidence for interactions between potato A potyvirus‐encoded proteins P1 and P3 and proteins of the putative replication complex. Virology, 263, 15–22. [DOI] [PubMed] [Google Scholar]
- Pirone, T.P. and Shaw, J.G. (1988) Tobacco vein mottling virus. C.M.I./A.A.B. Descriptions of Plant Viruses No. 325. Commonwealth Mycological Institute and the Association of Applied Biologist. Ferry Lane, Kew, Surrey, England.
- Pruss, G. , Ge, X. , Shi, X.M. , Carrington, J.C. and Vance, V.B. (1997) Plant viral synergism: the potyviral genome encodes a broad‐range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell, 9, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamäki, M.L. , Kelloniemi, J. , Alminaite, A. , Kekarainen, T. , Rabenstein, F. and Valkonen, J.P. (2005) A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology, 342, 88–101. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L. , Laín, S. and García, J.A. (1990) Infectious in vitro transcripts from a plum pox potyvirus cDNA clone. Virology, 177, 710–716. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L. , Laín, S. and García, J.A. (1991) Identification of the initiation codon of plum pox potyvirus genomic RNA. Virology, 185, 544–552. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L. , Laín, S. and García, J.A. (1992) Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73, 1–16. [DOI] [PubMed] [Google Scholar]
- Ryabov, E.V. , Robinson, D.J. and Taliansky, M.E. (1999) A plant virus‐encoded protein facilitates long‐distance movement of heterologous viral RNA. Proc. Natl Acad. Sci. USA, 96, 1212–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz, P. , Cervera, M.T. , Dallot, S. , Quiot, L. , Quiot, J.B. , Riechmann, J.L. and García, J.A. (2000) Identification of a pathogenicity determinant of Plum pox virus in the sequence encoding the C‐terminal region of protein P3+6K1 . J. Gen. Virol. 81, 557–566. [DOI] [PubMed] [Google Scholar]
- Sáenz, P. , Quiot, L. , Quiot, J.‐B. , Candresse, T. and García, J.A. (2001) Pathogenicity determinants in the complex virus population of a Plum pox virus isolate. Mol. Plant–Microbe Interact. 14, 278–287. [DOI] [PubMed] [Google Scholar]
- Sáenz, P. , Salvador, B. , Simón‐Mateo, C. , Kasschau, K.D. , Carrington, J.C. and García, J.A. (2002) Host‐specific involvement of the HC protein in the long‐distance movement of potyviruses. J. Virol. 76, 1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador, B. , Delgadillo, M.O. , Sáenz, P. , García, J.A. and Simón‐Mateo, C. (2008) Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol. Plant‐Microbe Interact. In press. [DOI] [PubMed]
- Salvador, B. , García, J.A. and Simón‐Mateo, C. (2006) Causal agent of sharka disease: Plum pox virus genome and function of gene products. EPPO Bull. 36, 229–238. [Google Scholar]
- Simón‐Buela, L. , Guo, H.S. and García, J.A. (1997) Cap‐independent leaky scanning as the mechanism of translation initiation of a plant viral genomic RNA. J. Gen. Virol. 78, 2691–2699. [DOI] [PubMed] [Google Scholar]
- Soumounou, Y. and Laliberté, J.‐F. (1994) Nucleic acid‐binding properties of the P1 protein of turnip mosaic potyvirus produced in Escherichia coli . J. Gen. Virol. 75, 2567–2573. [DOI] [PubMed] [Google Scholar]
- Stenger, D.C. and French, R. (2004) Functional replacement of Wheat streak mosaic virus HC‐Pro with the corresponding cistron from a diverse array of viruses in the family Potyviridae . Virology, 323, 257–267. [DOI] [PubMed] [Google Scholar]
- Suehiro, N. , Natsuaki, T. , Watanabe, T. and Okuda, S. (2004) An important determinant of the ability of Turnip mosaic virus to infect Brassica spp. and/or Raphanus sativus is in its P3 protein. J. Gen. Virol. 85, 2087–2098. [DOI] [PubMed] [Google Scholar]
- Tobias, I. , Palkovics, L. , Tzekova, L. and Balazs, E. (2001) Replacement of the coat protein gene of plum pox potyvirus with that of zucchini yellow mosaic potyvirus: characterization of the hybrid potyvirus. Virus Res. 76, 9–16. [DOI] [PubMed] [Google Scholar]
- Ullah, Z. , Chai, B.L. , Hammar, S. , Raccah, B. , Gal‐On, A. and Grumet, R. (2003) Effect of substitution of the amino termini of coat proteins of distinct potyvirus species on viral infectivity and host specificity. Physiol. Mol. Plant Pathol. 63, 129–139. [Google Scholar]
- Urcuqui‐Inchima, S. , Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. [DOI] [PubMed] [Google Scholar]
- Valli, A. , López‐Moya, J.J. and García, J.A. (2007) Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae . J. Gen. Virol. 88, 1016–1028. [DOI] [PubMed] [Google Scholar]
- Valli, A. , Martín‐Hernández, A.M. , López‐Moya, J.J. and García, J.A. (2006) RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus (CVYV), a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80, 10055–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrelmann, M. , Palkovics, L. and Maiss, E. (2000) Transgenic or plant expression vector‐mediated recombination of Plum pox virus . J. Virol. 74, 7462–7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. and Carrington, J.C. (1995) Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 69, 3668–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J. , Herndon, K.L. and Carrington, J.C. (1992) Mutational analysis of the tobacco etch potyviral 35‐kDa proteinase: identification of essential residues and requirements for autoproteolysis. Virology, 190, 298–306. [DOI] [PubMed] [Google Scholar]
- Verchot, J. , Koonin, E.V. and Carrington, J.C. (1991) The 35‐kDa protein from the N‐terminus of a potyviral polyprotein functions as a third virus‐encoded proteinase. Virology, 185, 527–535. [DOI] [PubMed] [Google Scholar]


