SUMMARY
Cutinolytic enzymes are secreted by fungal pathogens attacking the aerial parts of the plant, to facilitate penetration of the outermost cuticular barrier of the host. The role of cutinases in soil‐borne root pathogens has not been studied thus far. Here we report the characterization of the zinc finger transcription factor Ctf1 from the vascular wilt fungus Fusarium oxysporum, a functional orthologue of CTF1α that controls expression of cutinase genes and virulence in the pea stem pathogen Fusarium solani f. sp. pisi. Mutants carrying a Δctf1 loss‐of‐function allele grown on inducing substrates failed to activate extracellular cutinolytic activity and expression of the cut1 and lip1 genes, encoding a putative cutinase and lipase, respectively, whereas strains harbouring a ctf1C allele in which the ctf1 coding region was fused to the strong constitutive Aspergillus nidulans gpdA promoter showed increased induction of cutinase activity and gene expression. These results suggest that F. oxysporum Ctf1 mediates expression of genes involved in fatty acid hydrolysis. However, expression of lip1 during root infection was not dependent on Ctf1, and virulence of the ctf1 mutants on tomato plants and fruits was indistinguishable from that of the wild‐type. Thus, in contrast to the stem pathogen F. solani, Ctf1 is not essential for virulence in the root pathogen F. oxysporum.
INTRODUCTION
The plant cuticle forms a hydrophobic coating that covers nearly all above‐ground parts of terrestrial plants and constitutes the interface between plant and environment. The main structural component of the plant cuticle is cutin, a polyester consisting of hydroxy and epoxy fatty acids of n‐C16 and n‐C18 types (Kolattukudy, 2002; Kolattukudy et al., 1995). Fungal pathogens secrete cutinases and lipases that catalyse the hydrolysis of ester bonds from fatty acid polymers, thus facilitating fungal penetration through the cuticle (Hynes et al., 2006; Kolattukudy, 1985; Voigt et al., 2005). Both cutinases and lipases are esterase‐like enzymes whose active sites share the catalytic triad serine–histidine–aspartic acid. Globally these enzymes are designated carboxylesterases, catalysing the hydrolysis and/or synthesis of long‐chain acylglycerols (ten or more carbon units). The activity of lipases on triglycerides is usually enhanced at the lipid–water interface, an effect known as the interfacial activation phenomenon (Martínez et al., 1992). Cutinases occupy an intermediate position between esterases and lipases, as they are active on soluble and on emulsified triglycerides (Kim et al., 2003; Martínez et al., 1992). Moreover, cutinases hydrolyse model substrates such as p‐nitrophenyl palmitate (pNPP), p‐nitrophenyl butyrate (pNPB) and triglycerides (Martínez et al., 1992).
Regulation of fungal cutinases has been extensively studied in the pea stem pathogen Fusarium solani f. sp. pisi (Nectria haematococca). Three cutinase genes have been reported in this species, cut1, cut2 and cut3, which share a high degree of identity. While cut2 and cut3 are expressed constitutively at basal levels (Li et al., 2002), expression of cut1 is strongly induced by cutin monomers, and this transcriptional activation is mediated by the zinc finger transcription factor CTF1α (Li and Kolattukudy, 1997; Li et al., 2002). In Aspergillus nidulans, two proteins with homology to CTF1α, farA and farB, regulate the expression of genes implicated in metabolism of short‐chain and long‐chain fatty acids (Hynes et al., 2006). FarA and FarB bind in vitro to the same core DNA element CCGAGG that mediates binding of the cutinase transcription factor CTF1α (Li and Kolattukudy, 1997; Li et al., 2002), suggesting that the transcriptional regulation of fatty acid metabolism may be widely conserved in fungi (Hynes et al., 2006).
The role of cutinases in fungal pathogenicity to plants remains controversial. A number of gene knockout studies failed to show an essential role for cutinase in different pathogen species (Stahl and Schäfer, 1992; Sweigard et al., 1992; Yao and Köller, 1995). By contrast, knockout of the cutinase gene cut1 in F. solani f. sp. pisi resulted in decreased virulence on pea (Rogers et al., 1994; Li et al., 2002), and insertion of cut1 into Mycosphaerella, a pathogen that normally requires wounds in the plant surface to cause infection, allowed the transgenic strains to penetrate an intact surface (Dickman et al., 1989). Biochemical studies also support a role for cutinase in virulence. Immunolocalization with antibodies against Cut1 suggested that the cutinase was targeted to the growing tip of germinating spores of F. solani (Podila et al., 1995). Application of cutinase antibodies to germinating spores inhibited infection of pea by the fungus (Maiti and Kolattukudy, 1979). Collectively, these studies support the view that cutinase is involved in virulence of fungal pathogens attacking the aerial parts of the plant.
Interestingly, disruption of the transcriptional regulator Ctf1α eliminated phytopathogenicity of Fusarium solani f. sp. pisi, but supplementation of the mutant with cutinase did not restore pathogenicity, suggesting that Ctf1α participates in the regulation of other genes essential for pathogenicity (Li et al., 2002). The goal of the present work was to determine whether Ctf1 also regulates virulence factors in a pathogen which attacks the host plant through the roots, an organ that lacks a cuticle. To this aim, we have functionally characterized the Fusarium oxysporum transcription factor Ctf1, an orthologue of F. solani Ctf1α and of A. nidulans FarA. F. oxysporum is a soil‐borne pathogen which causes economically important losses on a wide variety of crops and infects its host plants strictly through the roots (Di Pietro et al., 2003). During penetration and root colonization, F. oxysporum secretes a battery of cell‐wall‐degrading enzymes such as polygalacturonases, pectate lyases and xylanases (Di Pietro and Roncero, 1998; García‐Maceira et al., 2001; Gómez‐Gómez et al., 2002; Huertas‐González et al., 1999; Ruiz‐Roldán et al., 1999). F. oxysporum therefore represents an excellent model to study the role of cutinases and lipases in fungal root pathogens. We find that F. oxysporum strains lacking a functional copy of the ctf1 gene are impaired in induction of cutinase activity, as well as in expression of two genes encoding a predicted cutinase and lipase, respectively, during growth in culture. However, in contrast to F. solani, targeted disruption of the ctf1 gene in F. oxysporum did not affect virulence on tomato plants, suggesting that transcriptional activation of cutinolytic genes plays differential roles in soil‐borne and foliar plant pathogens.
RESULTS
Cloning of the ctf1 gene encoding a cutinase transcription factor of F. oxysporum f. sp. lycopersici
The ctf1 gene, encoding a putative cutinase transcription factor of F. oxysporum f. sp. lycopersici, was cloned from a λEMBL3 genomic library and a lambda ZAP cDNA library of isolate 4287, using a probe generated by PCR with primers based on the coding sequence of the ctf1α gene from F. solani f. sp. pisi (Li and Kolattukudy, 1997) (Genbank accession number AAM22470). Sequencing of hybridizing genomic and cDNA clones revealed the presence of an open reading frame (ORF) of 2712 nucleotides, interrupted by five introns of 59, 192, 92, 65 and 56 bp (supplementary Fig. S1). The F. oxysporum ctf1 gene encodes a predicted protein of 904 amino acids with a molecular weight of 100.6 kDa, which shows 88.9 and 57.5% identity, respectively, to the cutinase and fatty acid transcription factors Ctf1α of F. solani (Li et al., 2002) and FarA of A. nidulans (Hynes et al., 2006). The Genbank accession number of ctf1 is EF613328.
The predicted F. oxysporum Ctf1 protein contains a Cys6Zn2 binuclear cluster DNA‐binding motif located close to the N‐terminus (amino acid residues 52–85), a middle homology region (MHR) and three ‘coiled coil’ regions (predicted by the ‘PEPCOIL’ program) (supplementary Fig. S2A), all characteristic of the family of fungal regulatory proteins with Cys6 zinc cluster DNA binding motifs (Schjerling and Holmberg, 1996; Todd and Andrianopoulos, 1997). The algorithm ‘PROSITE’ was used to predict 14 potential consensus phosphorylation sequences for casein kinase II, six potential sites for protein kinase C (PKC), three potential sites for tyrosine kinase, one potential site for cyclic AMP‐dependent protein kinase A (PKA) and two potential sites for mitogen‐activated protein kinase (MAPK) (supplementary Fig. S2B). Three putative nuclear localization signals with the residue patterns KRKK (position 86), KRHR (position 636) and PKRK (position 659) were detected, with a probability of 0.7 of Ctf1 being localized in the nucleus as predicted by the PSORT program (supplementary Fig. S2B). Southern analysis of genomic DNA treated with different restriction enzymes indicated that ctf1 is present as a single copy in the F. oxysporum genome (data not shown). In agreement with this result, a BLAST search of the complete genome database of F. oxysporum (http://www.broad.mit.edu/annotation/genome/fusarium_group.1/MultiHome.html) detected a single gene corresponding to ctf1.
Construction of ctf1 loss‐of‐function and overexpressing alleles
Loss‐of‐function mutants carrying a disrupted copy of the ctf1 gene were generated using one‐step gene replacement. A 2.4‐kb BamHI fragment within the ctf1 coding region was replaced with the hygR cassette (Fig. 1A). This replacement results in disruption of the Cys6Zn2 DNA‐binding motif, which is essential for proper function of CTF1α (Li and Kolattukudy, 1997). The knockout construct was introduced into protoplasts of the wild‐type strain, and 40 hygromycin‐resistant transformants were obtained and analysed by PCR with gene‐specific primers ctf14 and ctf11 (see Table 1). Two transformants produced PCR banding patterns consistent with homologous replacement of the ctf1 gene (data not shown). Southern blot analysis, using as a probe the ctf1 genomic fragment which had been replaced by the hygromycin resistance cassette, confirmed that the hybridizing 8.3‐kb BamHI fragment containing the wild‐type ctf1 allele was absent in transformant Δctf1, indicative of homologous integration‐mediated gene replacement (Fig. 1B).
Figure 1.
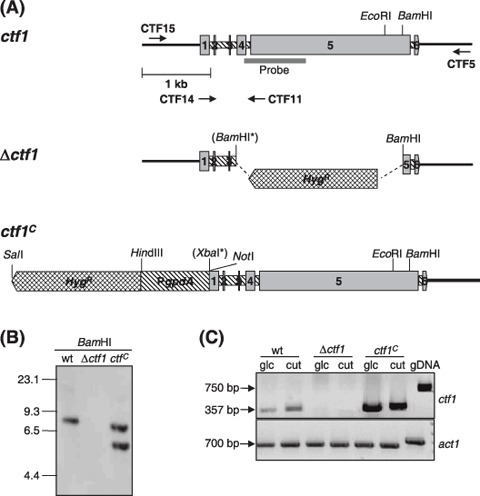
Targeted disruption and overexpression of the F. oxysporum ctf1 gene. (A) Physical maps of the ctf1 locus, the gene replacement vector (Δctf1 allele) and the ctf1C allele (ctf1 coding region fused to PgpdA from A. nidulans). The positions of the ctf1 probe, the HygR resistance cassette and the PgpdA promoter are indicated. The BamHI and XbaI sites introduced by site‐directed mutagenesis are marked with asterisks. (B) Southern hybridization analysis of wild‐type strain 4287 and transformants Δctf1 and ctf1C. Genomic DNA treated with BamHI was hybridized to the labelled ctf1 probe indicated in (A). The Δctf1 mutant shows homologous integration of the vector, whereas the constitutive ctf1C mutant carries an ectopic insertion of the ctf1C allele. Positions of DNA molecular size standards are shown. (C) Total RNAs from wild‐type, Δctf1 and ctf1C strains grown in SM with 1% glucose or 0.2% apple cutin as carbon sources were used for RT‐PCR analysis with ctf1‐specific primers. F. oxysporum genomic DNA was used as a control template. The actin gene (act1) was used as a control for equal amounts of RNA.
Table 1.
Oligonucleotides used in this study.
| Primer | Sequence |
|---|---|
| ctf1 | GGAATGATAATCCGAAGGAGC |
| ctf2 | TGGATCCATGTTCGGCTGCCAC |
| ctf5 | TTGACTGAGGTATCTTGCCTA |
| ctf4 | GCCATCATGTGGTCATCGG |
| ctf11 | GGAATGATAATCCGAAGGAGC |
| ctf14 | GTCGCAGTTGTCGGCACCA |
| ctf15 | TTGTCATTCTCGGCAAAGTATT |
| ctf16 | ATCCGTGGAACAGTCGCAAC |
| ctf17B | TATTCTGCAGGATCCATGCGACGC |
| ctf18B | GCGTCGCATGGATCCTGCAGAATA |
| ctf20X | TCTAGAATGTCGTCAGGCAGCGGCACGC |
| ctf22 | TTCCTCCATCATTGCTGCTTCT |
| cut9d | GT(N)TGGAT(H)CA(R)GG(N)GT(N)GG(N)GG |
| cut10d | T(Y)TG(N)A(R)(R)TT(Y)TT(N)GT(R)TA(N)CC(R)AA |
| cut14 | GACTCCAGCAATCTTCTCCC |
| cut19 | GGCAGCATCACCCGCAACG |
| lip11 | GTTTTACATCCAACACGGCG |
| lip12 | ATCTGGACATAAGAGTTCAGC |
| lip16 | TTGGCGACGATACAGAAGCG |
| lip18 | ACGAGACGACGATTTCCTTTC |
Mutations introduced in the original sequences for creation of BamHI restriction sites (B) are shown in bold. Degenerated primers are designated with a ‘d’, and use the UIPAC code: ‘N’ for A/C/T/G, ‘R’ for A/G; ‘H’ for A/T/C, and ‘Y’ for C/T.
F. oxysporum strains carrying a constitutively overexpressed ctf1C allele were produced by fusing the ctf1 coding and terminator regions to the the A. nidulans gpdA promoter (PgpdA) (Fig. 1A). The entire construct including the hygR cassette was amplified by PCR and introduced into protoplasts of the wild‐type strain. Thirty hygromycin‐resistant transformants were selected, and the presence of the intact PgpdA‐ctf1 allele was confirmed by PCR with primers ctf4 and ctf16 (data not shown). Southern analysis of one of the transformants named ctf1C revealed the presence of two hybridizing bands, one corresponding to the wild‐type ctf1 allele and an additional 6‐kb band, consistent with ectopic insertion of the pPgpdA‐ctf1 construct into the genome (Fig. 1B).
RT‐PCR analysis with gene‐specific primers ctf14 and ctf11 flanking introns 1–4 (see Fig. 1A) confirmed the absence of a ctf1 transcript in the knockout mutant and increased levels of transcript in the ctf1C strain, independently of the carbon source (Fig. 1C). We concluded that the Δctf1 mutant lacks a functional copy of the ctf1 gene, whereas transformant ctf1C constitutively produces high amounts of ctf1 transcript.
Mutations in ctf1 affect fatty acid hydrolysis
Microscopic analysis of the fungal strains grown in liquid media containing different carbon sources, including wheat germ oil, did not reveal any apparent differences in growth and development between the wild‐type, Δctf1 and ctf1C strains (Fig. 2A). By contrast, clear differences between wild‐type and the ctf1 mutants were observed on solid synthetic medium (SM) containing 1% olive oil with or without 0.1% sucrose. Vigorously growing colonies of the wild‐type strain observed under UV light contained many fluorescent droplets surrounding the colony, indicating the presence of degradation products from fatty acid hydrolysis (Fig. 2B). The Δctf1 mutant not only grew poorly, producing much less aerial mycelium than the wild‐type, but also showed a complete absence of fluorescent droplets, indicative of a failure in hydrolysis of fatty acids. Conversely, the transformant ctf1C exhibited significantly increased fatty acid hydrolysis compared with the wild‐type strain. These results suggest that Ctf1 is required for expression of F. oxysporum genes involved in fatty acid hydrolysis.
Figure 2.
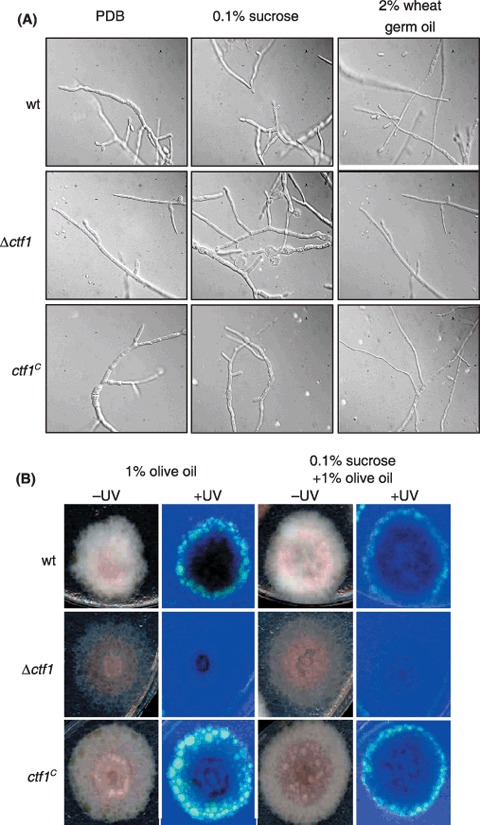
Growth phenotype of ctf1 mutants. (A) Microconidia of the indicated strains were germinated for 12 h in liquid PDB, transferred to SM with the indicated carbon sources, incubated for 24 h at 28 °C and 170 r.p.m. and observed under the microscope. (B) Strains were grown for 5 days on solid SM supplemented with the indicated carbon sources. Products of fatty acid hydrolysis are visible as fluorescence under UV light.
Characterization of F. oxysporum genes involved in fatty acid hydrolysis
To study the role of Ctf1 in regulation of fatty acid hydrolysis of F. oxysporum further, the cut1 and lip1 genes were cloned encoding a putative cutinase and lipase, respectively. cut1 was isolated from a λEMBL3 genomic library of F. oxysporum by screening with a probe obtained by PCR with degenerated primers derived from conserved regions of fungal cutinase genes. The nucleotide sequence revealed an ORF of 690 bp interrupted by one intron of 65 bp (supplementary Fig. S3A). The deduced amino acid sequence of the cut1 gene product had 78.3, 76.5 and 76.1% identity, respectively, to cut1 (P00590), cut2 (AAL18696) and cut3 (AAL18697) of F. solani f. sp. pisi (Li et al., 2002; Soliday et al., 1984) (supplementary Fig. S3B). The Cut1 protein contained the characteristic conserved catalytic triad Ser (120), Asp (175) and His (188) (Egmond and de Vlieg, 2000). A putative signal peptide was detected at the N terminus from amino acid residues 1–16. The mature Cut1 protein has a predicted molecular mass of 21.7 kDa and a pI of 8.1. The GenBank accession number of F. oxysporum cut1 is EF613272.
The lip1 gene encoding a putative extracellular lipase of F. oxysporum was isolated by PCR amplification of F. oxysporum genomic DNA with primers lip16 and lip18, derived from the F. verticilloides genome database (http://www.broad.mit.edu/annotation/genome/fusarium‐group/OrganismList.html#F%20verticillioides). The nucleotide sequence of the lip1 locus revealed an ORF of 1035 bp interrupted by two introns of 52 and 50 bp, respectively (supplementary Fig. S3C). The deduced amino acid sequence was 80% identical to the lipase Fgl1 from F. graminearum (AAQ23181) (Voigt et al., 2005). The predicted Lip1 protein contains the characteristic catalytic triad, comprising a serine, a histidine and an aspartic acid, which is highly conserved in lipases (supplementary Fig. S3D) (Schrag et al., 1991). A putative signal peptide is present at the N terminus, indicating that Lip1 is a secreted lipase. The predicted mature protein has a molecular mass of 34.8 kDa and a pI of 6.6. The Genbank accession number of the lip1 gene is EF613329.
The promoter regions of F. oxysporum cut1 and lip1 each contained one copy of the CCGAGG DNA element (positions –711 and –495, respectively) mediating transcriptional activation by the transcription factors Ctfα and FarA (Hynes et al., 2006).
Ctf1 regulates extracellular esterase and lipase activities and expression of cut1 and lip1 during growth in submerged culture
To test whether Ctf1 regulates extracellular esterase and lipase activities in F. oxysporum, supernatants of the wild‐type strain and the ctf1 mutants grown for various time periods in liquid SM with different carbon sources were analysed using colorimetric assays with pNPB or pNPP, respectively, as the substrate (Fig. 3). Esterase activity remained at basal levels in supernatants of all strains grown in SM without any carbon source (carbon starvation), but increased rapidly and to a similar extent in supernatants of the wild‐type and the ctf1C strain grown on 0.2% apple cutin (Fig. 3A,B). By contrast, no induction of esterase activity on apple cutin was detected in supernatants of the Δctf1 strains (Fig. 3B). Induction of extracellular esterase activity was both delayed and attenuated when glucose was supplemented as a carbon source in addition to apple cutin, probably due to preferential utilization of glucose by the fungus (Fig. 3C). Induction of lipolytic activity on 2% wheat germ oil was slightly increased in the ctf1C strain as compared with the wild‐type, and was not detected in the Δctf1 mutant (Fig. 3D). We conclude that Ctf1 mediates activation of extracellular esterase and lipase activities in F. oxysporum under inducing conditions, but not under carbon starvation or glucose‐repressed conditions.
Figure 3.
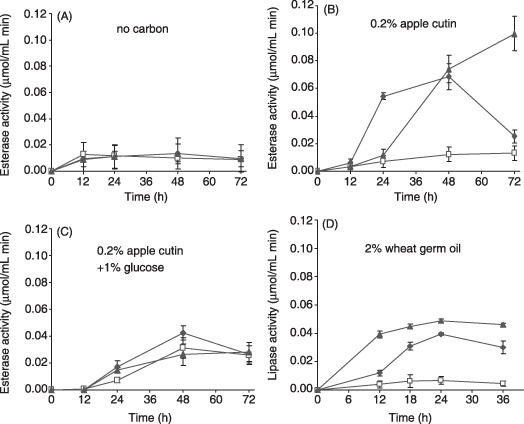
Ctf1 regulates extracellular esterase and lipase activities in liquid cultures. Activities in supernatants were determined by a colorimetric assay using as substrate p‐nitrophenyl butyrate (A–C) or p‐nitrophenyl palmitate (D), respectively. Wild‐type (filled diamonds), Δctf1 (open squares) and ctf1C (filled triangles) strains were grown for 12 h in PDB and transferred for the indicated times to SM without carbon source (A), 0.2% apple cutin (B), 0.2% apple cutin plus 1% sucrose (C) or 2% wheat germ oil (D). Error bars indicate standard deviations from three independent experiments.
To test whether expression of the cut1 gene depends on Ctf1, the level of cut1 transcripts in mycelia grown under inducing conditions was determined by RT‐PCR with gene‐specific primers cut19 and cut14 which flank the intron (see supplementary Fig. S3A). Total RNA was obtained from mycelia of the wild‐type, the Δctf1 and the ctf1Cstrains grown for 20 h in SM supplemented with different carbon sources (1% glucose or 0.2% apple cutin) and subjected to RT‐PCR. Higher levels of RT‐PCR product corresponding to the cut1 transcript were detected in mycelia of the wild‐type grown on apple cutin than in those grown on glucose, suggesting that cut1 transcription is triggered by inducing carbon sources (Fig. 4A). Activation of cut1 by apple cutin was even more pronounced in the ctf1C strain than in the wild‐type. By contrast, only a weak activation of cut1 expression was observed in the Δctf1 mutant. RT‐PCR analysis of mycelia grown for 24 or 48 h on apple cutin confirmed the low level of cut1 induction in the Δctf1 mutant compared with the wild‐type and ctf1C strain (Fig. 4B).
Figure 4.
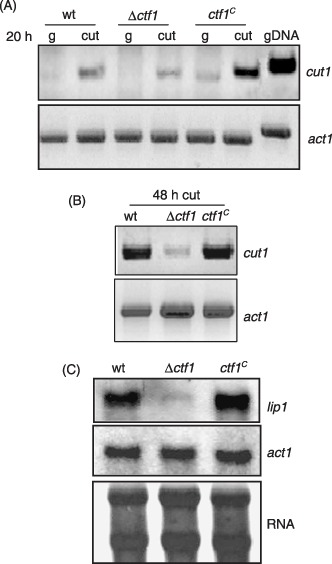
Ctf1 regulates expression of cut1 and lip1 in axenic culture. (A) Transcript levels of the cut1 gene determined by RT‐PCR. The indicated strains were grown for 12 h in PDB and transferred for 20 h to SM with 1% glucose or 0.2% apple cutin as carbon source. (B) Expression of cut1 determined by RT‐PCR in the indicated strains grown for 48 h in SM with 0.2% apple cutin. (C) Northern analysis for lip1 transcript accumulation. Strains were grown for 12 h in PDB and transferred for 24 h to SM with 2% wheat germ oil as carbon source. Upper panel: the filter hybridized to the lip1 probe. Middle panel: the filter hybridized to the act1 probe. Lower panel: total RNA stained on the filter with methylene blue.
Northern hybridization analysis was used to study the effect of mutations in ctf1 on expression of the lip1 gene. Total RNA obtained from mycelia of the strains grown for 24 h on 2% wheat germ oil as the sole carbon source was hybridized to the lip1 probe. Transcript levels of the lip1 gene were slightly higher in the ctf1Cstrain than in the wild‐type, whereas an extremely faint hybridization signal was detected in the Δctf1 mutant (Fig. 4C). We conclude that efficient transcriptional activation of cut1 and lip1 on fatty acid substrates, but not on glucose, is mediated by the Ctf1 transcription factor.
Ctf1 is not a major regulator of lip1 during root infection
Expression of ctf1, cut1 and lip1 during infection of tomato plants by F. oxysporum was determined in total RNA extracted from roots or stems of plants at several time points after inoculation with the different strains, using RT‐PCR with gene‐specific primers (Fig. 5). Both ctf1 and cut1 were expressed at very low levels. A faint signal corresponding to ctf1 was detected in roots inoculated with the ctf1C strain at 7 and 10 days after inoculation. Likewise faint cut1 transcript signals were observed in roots 10 days after inoculation with the three different strains and in stems infected with the wild‐type or the ctf1C strain. By contrast, robust expression of lip1 was detected in plants inoculated with all the strains, particularly in infected roots. No expression signal was detected in non‐infected control roots.
Figure 5.
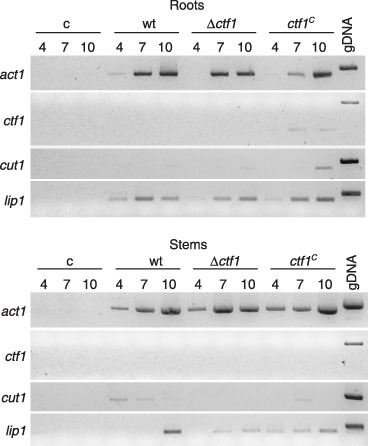
Ctf1 is not required for expression of lip1 during infection of tomato plants. RT‐PCR products showing the expression pattern of ctf1, cut1 and lip1 during infection of tomato plants by the wild‐type strain, the Δctf1 mutant and the ctf1C strain. cDNA generated from total RNA isolated at the indicated time points (days after inoculation) from roots and stems of infected plants were used as templates for PCR with gene‐specific primers (see Experimental procedures). Uninoculated plants were used as control. gDNA refers to PCR with genomic DNA as template.
Ctf1 is not required for virulence of F. oxysporum on tomato
The role of Ctf1 in virulence of F. oxysporum f. sp. lycopersici was determined both by plant and by fruit infection assays. Plant infection assays were performed by immersing the roots of 3‐week‐old tomato plants in microconidial suspensions of the different strains and planting them into minipots with vermiculite. Severity of wilt symptoms in plants inoculated with the wild‐type strain increased steadily throughout the experiment. At the beginning, leaf tips turned yellow and curved, then the stalk weakened and decayed, and most of the plants were dead 20 days after inoculation. Plants inoculated with the Δctf1 and ctf1C mutant strains showed a very similar symptom development as the wild‐type during the entire period of the experiment (Fig. 6). The capacity to grow invasively on tomato fruit tissue was assayed by injecting microconidia of the strains directly into tomato fruits. The wild‐type and the ctf1 mutant strains efficiently colonized and macerated the fruit tissue, producing aerial mycelium on the surface (data not shown). We conclude that Ctf1 is not essential for virulence of F. oxysporum.
Figure 6.
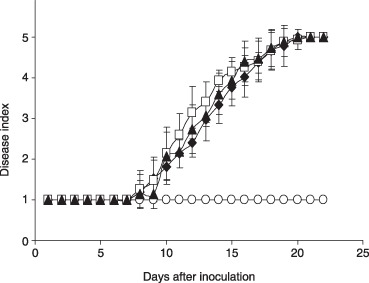
Ctf1 is not required for virulence of F. oxysporum. Incidence of Fusarium wilt caused on tomato plants (cultivar Vemar). Severity of disease symptoms was recorded at different times after inoculation, using an index ranging from 1 (healthy plant) to 5 (dead plant). Symbols refer to plants inoculated with the wild‐type strain 4287 (filled diamonds), loss‐of‐function mutant Δctf1 (open squares), constitutive mutant ctf1C (filled triangles) and the water control (open circles). Error bars represent standard deviations from 20 plants for each treatment. The experiment was repeated twice with similar results.
DISCUSSION
Infection by fungal pathogens causes severe losses in crop plants worldwide. For fungal pathogens attacking the aerial parts of the plant, the first contact between pathogen and host occurs on the plant surface, which is protected by a cuticle layer (Agrios, 1997). By contrast, soil‐borne pathogens enter their host plants through the roots, which lack a cuticle layer. While a role of lipolytic and cutinolytic enzymes during infection of aerial plant pathogens has been established (Li et al., 2002; Rogers et al., 1994; Voigt et al., 2005), the function of these enzymes has not been investigated in soil‐borne pathogens. To test the role of cutin‐degrading enzymes in virulence of the soil‐borne vascular wilt pathogen F. oxysporum, we cloned and mutated the gene encoding the transcription factor Ctf1, the deduced amino acid sequence of which shows a high degree of identity with F. solani f. sp. pisi CTFα, which regulates the inducible cutinase gene cut1 (Li et al., 2002), and with A. nidulans FarA, which controls expression of genes involved in fatty acid metabolism (Hynes et al., 2006). The predicted F. oxysporum Ctf1 protein contains three putative nuclear localization signals, and its amino terminus shows the Zn2(II)Cys6 DNA binding domain, characteristic of regulatory proteins with diverse function. This domain follows the canonical pattern CX2CX6CX8CX2CX6C, with both repeats (CX2CX6C) separated by a variable number of residues (eight in CTFα and CTF1). This group of transcription factors binds to promoter regions at two conserved palindromic triplets GCC(N)xCCG, where N has a variable length (e.g. 11 in Saccharomyces cerevisiae GAL4) (Liang et al., 1996). In F. solani f. sp. pisi, the DNA motifs recognized by CTFα have the opposite orientation and are separated only by two nucleotides (GCCN2GGC; Li et al., 2002). FarA and FarB, two transcription factors regulating fatty acid metabolism in A. nidulans, bind to the conserved motif CCGAGG in the promoter regions of the target genes (Hynes et al., 2006). The promoter of the F. oxysporum ctf1 gene contains a copy of the CCGAGG motif at position –374, suggesting that Ctf1 may regulate its own expression. This hypothesis is supported by the observation that ctf1 transcript levels were higher in mycelia grown on cutin as opposed to those grown on glucose (Fig. 1C).
Mutants of F. oxysporum carrying either a loss‐of‐function or a constitutively expressed allele of ctf1 did not show differences in growth and development in submerged culture even when grown on fatty acid‐containing carbon sources such as olive oil or wheat germ oil. However, clear differences were detected on solid media with olive oil as carbon source. The phenotype of the Δctf1 mutants under these conditions (poor growth, reduced aerial hyphae, absence of UV‐fluorescent droplets) was consistent with the notion that these mutants were impaired in hydrolysis of fatty acids. This suggests that Ctf1 controls expression of the major lipolytic enzymes produced by F. oxysporum under these conditions. As CTFα and FarA regulate genes involved in fatty acid metabolism and therefore affect the production of encoded enzyme activities (Hynes et al., 2006; Li et al., 2002), we tested the role of F. oxysporum Ctf1 in production of extracellular esterase and lipase activities on apple cutin extract and wheat germ oil, respectively. Esterase activity was clearly induced by apple cutin in the wild‐type strain, and this induction was abolished by deletion of ctf1, such that activity remained at basal levels similar to those produced under carbon starvation conditions (Fig. 3). Similar results were obtained for lipase activities. Thus, Ctf1 is required for efficient induction of extracellular lipolytic enzymes in F. oxysporum. By contrast, the differences in enzyme activities between the wild‐type and the constitutive ctf1Cstrain were much smaller, and esterase activity in the ctf1C strain was still subject to induction by fatty acid substrates. These results are consistent with a model in which activation of Ctf1 occurs, at least in part, at the post‐translational level. Similar results have been reported recently for the zinc finger transcription factors XlnR controlling expression of xylanase genes in F. oxysporum (Calero‐Nieto et al., 2007) and Zfr1 regulating fumonisin production in F. verticillioides (Flaherty and Woloshuk, 2004).
Different approaches were used to isolate F. oxysporum cutinase genes, including PCR amplification of genomic DNA or RT‐PCR with mRNA obtained from the wild‐type strain and ctf mutants under different growth conditions, using primers derived from conserved regions of F. solani cutinase genes. All these approaches yielded the same amplified fragment, indicating the existence, in F. oxysporum, of a single gene encoding this structural class of cutinases, as opposed to three (cut1, 2 and 3) in F. solani (Li et al., 2002). The different number of cutinase genes in the two Fusarium species may point to divergent evolutionary mechanisms during adaptation of the infection process in root and aerial plant pathogens. In fungal leaf and stem pathogens, multiple genes encoding cuticle‐degrading enzymes may have evolved from a common ancestor by duplication and may have adapted to the specific requirements encountered during host penetration. The product of the F. oxysporum lip1 gene has 80% identity to F. graminearum Lip1, an extracellular lipase required for virulence on wheat (Voigt et al., 2005). The promoters of both cut1 and lip1 genes contained the element CCGAGG, which mediates binding of the fatty acid hydrolysis‐regulating transcription factor FarA from A. nidulans (Hynes et al., 2006). The key role of Ctf1 in regulation of cut1 and lip1 was substantiated by RT‐PCR and Northern analysis showing that expression of the two genes was nearly abolished in the Δctf1 mutant. Thus, the role of this transcription factor in activation of genes involved in fatty acid hydrolysis appears to be broadly conserved in ascomycetes.
The roles of cutinases and lipases in the infection process of fungal plant pathogens has been highly controversial and may vary substantially between different enzymes and species. Targeted disruption of ctfα in F. solani f. sp. pisi resulted in loss of virulence on pea, but the decreased level of secreted cutinase was not sufficient to fully explain the loss of virulence, as addition of exogenous cutinase did not restore pathogenicity (Li et al., 2002). In the present work we failed to detect significant differences in virulence between wild‐type and Δctf1 mutants, in both tomato plant and fruit infection assays. Thus, Ctf1, a major regulator of cutinase and lipase activity during growth in axenic culture, is not required for pathogenicity of F. oxysporum. This result can be interpreted in two ways. First, neither cutinase nor lipase may act as virulence factors in F. oxysporum. At first, this interpretation appears convincing because F. oxysporum infects roots, an organ which lacks a cuticle. However, the results from RT‐PCR show that the lip1 gene is highly expressed during infection of roots, both in the wild‐type and the Δctf1 strain. Thus, under these conditions lip1 expression appears to be largely independent of the presence of Ctf1. Consequently, Ctf1 appears to regulate lip1 under the in vitro conditions studied, whereas during root infection this function could be fulfilled by another transcriptional regulator. Therefore, a similar role of lip1 in virulence of F. oxysporum as reported for its orthologue fgl1 in F. graminearum (Voigt et al., 2005) cannot yet be ruled out.
EXPERIMENTAL PROCEDURES
Strains and culture conditions
F. oxysporum f. sp. lycopersici 4287 (race 2) was obtained from J. Tello, Universidad de Almería, Spain. The pathotype of the strain was periodically confirmed by plant infection assays. Fungal strains were stored at –80 °C with glycerol as a microconidial suspension (Di Pietro and Roncero, 1998). For extraction of DNA and microconidia production, cultures were grown in potato dextrose broth (PDB, Difco, Detroit, MI) at 28 °C with shaking at 170 r.p.m. as described previously (Di Pietro and Roncero, 1998).
For phenotypic analysis of colony growth, a 5‐µL drop containing 1 × 106 freshly obtained microconidia was transferred on 1.5% (w/v) agar plates of synthetic medium (SM) (Di Pietro and Roncero, 1998), containing 1% (v/v) olive oil or 1% olive oil and 0.1% (w/v) sucrose as carbon source and 0.1% NH4NO3 as the nitrogen source, and incubated at 28 °C for 5 days as described previously (Di Pietro and Roncero, 1998). For microscopic analysis, strains were observed in a Leica DMR microscope using the Nomarsky or phase contrast techniques, or in a Leica MZ FLIII stereomicroscope.
For analysis of gene expression or extracellular cutinase and lipase production, freshly obtained microconidia from wild‐type and mutants strains were inoculated in PDB medium at a final concentration of 5 × 106 mL−1 and germinated for 12 h at 28 °C and 170 r.p.m. Germlings were washed twice in sterile water and transferred to liquid SM (as above without agar) containing either 0.2% (w/v) apple cutin prepared as described (Walton and Kolattukudy, 1972), 1% (w/v) glucose or 2% (v/v) wheat germ oil (Sigma) as the carbon source.
Nucleic acid manipulations and cloning of the ctf1, cut1 and lip1 genes
Total RNA and genomic DNA were extracted following previously reported protocols (Chomczynski and Sacchi, 1987; Raeder and Broda, 1985). Southern and Northern blot analyses and probe labelling were carried out as described (Di Pietro and Roncero, 1998) using the non‐isotopic digoxigenin labelling kit (Roche Diagnostics SL). The expression analysed by RT‐PCR was performed using Superscript II reverse transcriptase (Gibco‐BRL, UK). The primers used to amplify the gene transcripts were ctf11 and ctf14 for ctf1, cut14 and cut19 for cut1, and lip11 and lip18 for lip1 (Table 1). Other routine DNA procedures were performed according to standard protocols (Sambrook and Russell, 2001).
Cloning of the ctf1 gene was achieved by PCR amplification of F. oxysporum 4287 genomic DNA using the primers ctf1 and ctf2 (Table 1) derived from the ctf1α gene of F. solani (Genbank accession number P52958). For isolation of cut1 gene the degenerated primers cut9d and cut10d were used (Table 1), which were derived from conserved sequence domains of fungal cutinases. Cloning of the lip1 gene was done using the primers lip11 and lip12 derived from a putative lipase gene in the F. verticillioides genome sequence database (http://www.broad.mit.edu/annotation/genome/fusarium‐group/OrganismList.html#F%20verticillioides) with homology to the fgl1 gene from F. graminearum (AY292529.1). PCRs were routinely performed with the Long Template PCR system (Roche Diagnostics, SL). The amplified DNA fragments were cloned into the pGEM‐T vector (Promega, Madison, WI) and used to screen a λEMBL3 genomic library of F. oxysporum f. sp. lycopersici isolate 4287. Library screening, subcloning and other routine procedures were performed according to standard protocols (Sambrook and Russell, 2001). The cDNA clone of ctf1 gene was obtained from a lambda ZAP cDNA library of F. oxysporum (4287) (Roldán‐Arjona et al., 1999). Sequencing of both DNA strands of the obtained clones was performed at the Servicio de Secuenciación Automática de DNA, SCAI (University of Córdoba, Spain) using the Dyedeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on an ABI Prism 377 Genetic Analyser apparatus (Applied Biosystems). DNA and protein sequence databases were searched using the BLAST algorithm (Altschul et al., 1990) at the National Center for Biotechnology Information (Bethesda, MD).
Construction of plasmids vectors and fungal transformation
Gene replacement vector pDctf1 was constructed as follows: a BamHI site was introduced at position 515–521 of the ctf1 coding region using the overlapping method (Ho et al., 1989) with the primers ctf17B and ctf18B (Table 1), and the final amplification product was cloned into vector pGEM‐T. This construct was treated with BamHI and the excised internal 2.4‐kb BamHI fragment was replaced with the hygromycin B resistance gene (hph) under the control of a Cochliobolus heterostrophus promoter (Turgeon et al., 1987), resulting in disruption of the ctf1 coding region (supplementary Fig. S1A). The linear fragment containing the interrupted ctf1 allele was PCR‐amplified with primers ctf5 and ctf15, and used to transform protoplasts of F. oxysporum wild‐type strain following the protocol of Di Pietro and Roncero (1998).
The constitutively expressed ctfC allele was obtained by introducing an XbaI site immediately upstream of the ATG start codon of ctf1 using primers ctf20X and ctf22, and cloning the resulting fragment containing the ctf1 coding and 3′ flanking regions into pGEM‐T. The vector pPgpdA‐ctf1 was constructed by excising the ctf1 allele using XbaI and NotI and cloning into pGEM‐T containing the gpdA promoter (Punt et al., 1987). The PgpdA‐ctf1 DNA fragment was excised with HindIII and NotI, and cloned into a vector containing the HygR cassette (Punt and van den Hondel, 1992) (Fig. 1A). The final construct containing the ctf1C allele under the control of the gpdA promoter plus the hyg cassette was amplified using the universal forward and reverse primers. The amplification product was purified and used to transform protoplasts of the 4287 wild‐type strain to hygromycin resistance as described previously (Di Pietro and Roncero, 1998).
Determination of cutinase and lipase activity
For the activity assays, culture supernatants of the wild‐type and mutant strains collected at different times of growth under the conditions described above were used as the enzyme source. Each assay was carried out in triplicate in 96‐well microtitre plates. Esterase assays were performed using as substrate p‐nitrophenyl butyrate (pNPB; Sigma) (Davies et al., 2000; Kolattukudy et al., 1981). A 20‐µL volume of crude supernatant was mixed with 200 µL of reaction buffer containing 50 mM sodium phosphate buffer, pH 7, 0.4% (v/v) Triton X‐100 and 1.76% (v/v) PNB in acetonitrile, and incubated at 37 °C for 30 min.
Lipase activity was determined using as substrate p‐nitrophenyl palmitate (pNPP; Sigma) (Winkler and Stuckman, 1979). A 10‐µL volume of supernatant (concentrated 12.5× with acetone) was added to 240 µL of reaction buffer, containing 2 mM pNPP in isopropanol, 1% gum arabic (Sigma), 0.1% Triton X‐100 (Merck), and 50 mM bis‐Tris‐propane (Sigma), at pH 8 and incubated at 37 °C for 30 min.
Production of p‐nitrophenol in both assays was measured at 405 nm using a multiwell spectrophotometer (SpectraFluor Plus). One unit of enzymatic activity was defined as 1 nmol/min p‐nitrophenol released from the substrate. Calibration curves were made using standard p‐nitrophenol solutions.
Virulence assays
Tomato plant inoculation assays were performed as described previously (Di Pietro and Roncero, 1998). Briefly, 2‐week‐old tomato seedlings (cultivar Vemar) were inoculated with F. oxysporum strains by immersing the roots in a microconidial suspension, planted in vermiculite and maintained in a growth chamber. At different times after inoculation, severity of disease symptoms was recorded using an index ranging from 1 (healthy plant) to 5 (dead plant). Twenty plants were used for each treatment. Assays for invasive growth on tomato fruits (cultivar Daniela) were carried out as described (Di Pietro et al., 2001). Virulence experiments were performed three times with similar results.
Supporting information
Supporting info item
ACKNOWLEDGEMENTS
We are grateful to Dr Keith Johnstone (University of Cambridge) for helpful advice on preparation of cutin from apple peels, and to Isabel Caballero (University of Córdoba) for technical assistance. This research was supported by grants BIO2004‐0276 from the Spanish Ministerio de Educación y Ciencia and CVI‐138 from Junta de Andalucía. A.M.R. received a PhD fellowship from Ministerio de Educación y Ciencia and A.D.P. was a recipient of Ramón y Cajal grant from the Ministerio de Ciencia y Tecnología.
REFERENCES
- Agrios, G.N. (1997) Plant Pathology. San Diego, CA. (EE.UU), Academic Press Inc. [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Calero‐Nieto, F. , Di Pietro, A. , Roncero, M.I.G. and Hera, C. (2007) Role of the transcriptional activator XlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol. Plant–Microbe Interact. 20, 977–985. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. (1987) Single‐step method of RNA isolation by acid guanidinium thiocyanate‐phenol‐chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Davies, K.A. , De Lorono, I. , Foster, S.J. , Li, D. , Johnstone, K. and Ashby, M. (2000) Evidence for a role of cutinase in pathogenicity of Pyrenopeziza brassicae on brassicas. Physiol. Mol. Plant Pathol. 57, 63–75. [Google Scholar]
- Di Pietro, A. and Roncero, M.I.G. (1998) Cloning, expression and role in pathogenicity of pg1, encoding the major endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum f. sp. lycopersici . Mol. Plant–Microbe Interact. 11, 91–98. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A. , García‐Maceira, F. , Meglecz, E. and Roncero, M.I.G. (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Di Pietro, A. , Madrid, M. , Caracuel, Z. , Delgado‐Jarana, J. and Roncero, M.I.G. (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Dickman, M.B. , Podila, G.K. and Kolattukudy, P.E. (1989) Insertion of cutinase gene into a root pathogen enables it to infect intact host. Nature, 342, 446–448. [Google Scholar]
- Egmond, M.R. and De Vlieg, J. (2000) Fusarium solani pisi cutinase. Biochimie, 82, 1015–1021. [DOI] [PubMed] [Google Scholar]
- Flaherty, J.E. and Woloshuk, C.P. (2004) Regulation of fumonisin biosynthesis in Fusarium verticillioides by a zinc binuclear cluster‐type gene, ZFR1. Appl. Environ. Microbiol. 70, 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Maceira, F.I. , Di Pietro, A. , Huertas‐González, M.D. , Ruiz‐Roldán, M.C. and Roncero, M.I.G. (2001) Molecular characterization of an endopolygalacturonase from Fusarium oxysporum expressed during early stages of infection. Appl. Environ. Microbiol. 67, 2191–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gómez, E. , Ruiz‐Roldán, M.C. , Di Pietro, A. , Roncero, M.I.G. and Hera, C. (2002) Role in pathogenesis of two endo‐beta‐1,4‐xylanase genes from the vascular wilt fungus Fusarium oxysporum . Fungal Genet. Biol. 35, 213–222. [DOI] [PubMed] [Google Scholar]
- Ho, S.N. , Hunt, H.D. , Horton, R.M. , Pullen, J.K. and Pease, U. (1989) Site‐directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Huertas‐González, M.D. , Ruíz‐Roldán, M.C. , García‐Maceira, F.I. , Roncero, M.I.G. and Di Pietro, A. (1999) Cloning and characterization of pl1 encoding an in planta‐secreted pectate lyase of Fusarium oxysporum . Curr. Genet. 35, 36–40. [DOI] [PubMed] [Google Scholar]
- Hynes, M.J. , Murray, S.L. , Duncan, A. , Khew, G.S. and Davis, M.A. (2006) Regulatory genes controlling fatty acid catabolism and peroxisomal functions in the filamentous fungus Aspergillus nidulans . Eukaryot. Cell 5, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.H. , Lee, J. and Moon, S.H. (2003) Degradation of an endocrine disrupting chemical, DEHP [di‐(2‐ethylhexyl)‐phthalate], by Fusarium oxysporum f. sp. pisi cutinase. Appl. Microbiol. Biotechnol. 63, 75–80. [DOI] [PubMed] [Google Scholar]
- Kolattukudy, P.E. (1985) Enzymatic penetration of the plant cuticle by fungal pathogens. Annu. Rev. Phytopathol. 23, 223–250. [Google Scholar]
- Kolattukudy, P.E. (2002) Cutin from Plants. Columbus, OH: Ohio State University. [Google Scholar]
- Kolattukudy, P.E. , Rogers, L. and Larson, J.D. (1981) Enzymatic reduction of fatty acids and alpha‐hydroxy fatty acids. Methods Enzymol. 71, 263–275. [DOI] [PubMed] [Google Scholar]
- Kolattukudy, P.E. , Rogers, L.M. , Li, D.X. , Hwang, C.S. and Flaishman, M.A. (1995) Surface signaling in pathogenesis. Proc. Natl Acad. Sci. USA. 92, 4080–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Kolattukudy, P.E. (1997) Cloning of cutinase transcription factor 1, a transactivating protein containing Cys6Zn2 binuclear cluster DNA‐binding motif. J. Biol. Chem. 272, 12462–12467. [DOI] [PubMed] [Google Scholar]
- Li, D. , Sirakova, T. , Rogers, L. , Ettinger, W. and Kolattukudy, P.E. (2002) Regulation of constitutively expressed and induced cutinase genes by different zinc finger transcription factors in Fusarium solani f. sp. pisi (Nectria haematococca ). J. Biol. Chem. 277, 7905–7912. [DOI] [PubMed] [Google Scholar]
- Liang, S.D. , Marmorstein, R. , Harrison, S.C. and Ptashne, M. (1996) DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2Cys6 binuclear cluster proteins recognises DNA. Mol. Cell. Biol. 16(7): 3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, I.B. and Kolattukudy, P.E. (1979) Prevention of fungal infection of plants by specific inhibition of cutinase. Science, 205, 507–508. [DOI] [PubMed] [Google Scholar]
- Martínez, C. , Geus, P. , Lauwereys, M. , Matthyssens, G. and Cambillau, C. (1992) Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature, 356, 615–618. [DOI] [PubMed] [Google Scholar]
- Podila, G.K. , Rosen, E. , San Francisco, M.J.D. and Kolattukudy, P.E. (1995) Targeted secretion of cutinase in Fusarium solani f. sp. pisi and Colletotrichum gloeosporioides . Phytopathology, 85, 238–242. [Google Scholar]
- Punt, P.J. and Van Den Hondel, C.A. (1992) Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216, 447–457. [DOI] [PubMed] [Google Scholar]
- Punt, P.J. , Oliver, R.P. , Dingemanse, M.A. , Pouwels, P.H. and Van Den Hondel, C.A.M.J.J. (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli . Gene, 56, 117–124. [DOI] [PubMed] [Google Scholar]
- Raeder, U. and Broda, P. (1985) Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1, 17–20. [Google Scholar]
- Rogers, L.M. , Flaishman, M.A. and Kolattukudy, P.E. (1994) Cutinase gene disruption in Fusarium solani f. sp. pisi decreases its virulence on pea. Plant Cell, 6, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán‐Arjona, T. , Pérez Espinosa, A. and Ruiz Rubio, M. (1999) Tomatinase from Fusarium Oxysporum f. sp. lycopersia defines a new class of saponinases. Mol. Plant Microbe. Interact. 12, 852–861. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Roldán, M.C. , Di Pietro, A. , Huertas‐González, M.D. and Roncero, M.I.G. (1999) Two xylanase genes of the vascular wilt pathogen Fusarium oxysporum are differentially expressed during infection of tomato plants. Mol. Gen. Genet. 261, 530–536. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D. (2001) Molecular Cloning: a Laboratory Manual, 3rd edn. New York: Cold Spring Harbor Laboratory. [Google Scholar]
- Schjerling, P. and Holmberg, S. (1996) Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucl. Acids Res. 24, 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag, J.D. , Li, Y.G. , Wu, S. and Cygler, M. (1991) Ser‐His‐Glu triad forms the catalytic site of the lipase from Geotrichum candidum . Nature, 351, 761–764. [DOI] [PubMed] [Google Scholar]
- Soliday, C.L. , Flurkey, W.H. , Okita, T.W. and Kolattukudy, P.E. (1984) Cloning and structure determination of cDNA for cutinase, an enzyme involved in fungal penetration of plants. Proc. Natl Acad. Sci. USA 81, 3939–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, D.J. and Schäfer, W. (1992) Cutinase is not required for fungal pathogenicity on pea. Plant Cell, 4, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard, J.A. , Chumley, F.G. and Valent, B. (1992) Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 232, 183–190. [PubMed] [Google Scholar]
- Todd, R.B. and Andrianopoulos, A. (1997) Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21, 388–405. [DOI] [PubMed] [Google Scholar]
- Turgeon, B.G. , Garber, R.C. and Yoder, O.C. (1987) Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7, 3297–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, C.A. , Schäfer, W. and Salomon, S. (2005) A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 42, 364–375. [DOI] [PubMed] [Google Scholar]
- Walton, T.J. and Kolattukudy, P.E. (1972) Determination of the structures of cutin monomers by a novel depolymerization procedure and combined gas chromatography and mass spectrometry. Biochemistry, 11, 1885–1896. [DOI] [PubMed] [Google Scholar]
- Winkler, U.K. and Stuckmann, M. (1979) Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 138, 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, C. and Köller, W. (1995) Diversity of cutinases from plant pathogenic fungi: different cutinases are expressed during saprophytic and pathogenic stages of Alternaria brassicicola . Mol Plant–Microbe Interact. 8, 122–130. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item


